Abstract
Hemerocallis citrina Borani (huang hua cai in Chinese) is an important horticultural crop whose flower buds are widely consumed as a delicious vegetable in Asia. Here we assembled a high-quality reference genome of H. citrina using single-molecule sequencing and Hi-C technologies. The genome assembly was 3.77 Gb and consisted of 3183 contigs with a contig N50 of 2.09 Mb, which were further clustered into 11 pseudochromosomes. A larger portion (3.25 Gb or 86.20%) was annotated as a repetitive content and 54,295 protein-coding genes were annotated in the genome. Genome evolution analysis showed that H. citrina experienced a recent whole-genome duplication (WGD) event at ~15.73 million years ago (Mya), which was the main factor leading to many multiple copies of orthologous genes. We used this reference genome to predict 20 genes involved in the rutin biosynthesis pathway. Moreover, our metabolomics data revealed neither colchicine nor its precursors in H. citrina, challenging the long-standing belief that this alkaloid causes poisoning by the plant. The results of our disruptive research are further substantiated by our genomic finding that H. citrina does not contain any genes involved in colchicine biosynthesis. The high-quality genome lays a solid foundation for genetic research and molecular breeding of H. citrina.
Subject terms: Comparative genomics, Metabolomics
Introduction
Hemerocallis citrina Borani is a perennial crop and its flower buds are one of the most commonly consumed vegetables in Asia. This plant has been widely grown in Asian countries, including China, Japan, and Korea, and has also been regarded as the traditional mother’s flower in Chinese culture for a thousand years1,2. H. citrina flower buds have been used to relieve depression and promote lactation, as documented in the medicinal book “Compendium of Materia Medica,” which is a famous Chinese encyclopedia of medicine3,4. Modern pharmaceutical studies have demonstrated that H. citrina extract has antidepressant, antioxidant, and anti-inflammatory effects5–7. The chemical components isolated from H. citrina mainly include flavonols, polyphenols, anthraquinones, and alkaloids8. Rutin is the main chemical constituent and plays an important role in the antidepressant activity of H. citrina5; however, the corresponding biosynthetic genes have rarely been reported in this plant. Here we predicted some candidate genes of the rutin biosynthesis pathway by the comparative genomic method. In addition, the relatively fast floral development of H. citrina severely restricts the harvest window and places a significant resource strain on post-harvest processing. Moreover, the edible value of H. citrina rapidly deteriorates after flowering due to a loss of flavor, leading to substantial food waste. Therefore, it is an urgent task to cultivate new varieties of H. citrina with staggered flowering periods or non-blooming buds via molecular breeding, which could generate tremendous economic value. However, the lack of genomic information restricts the cultivation of new varieties and a high-quality genome of H. citrina could provide the possibility of achieving this goal.
The market value of H. citrina has been ~1 billion US dollars for many years. One of the crucial reasons for the limited market value is that colchicine in the flower buds is widely recognized as a poisonous substance1. However, the existence of colchicine in H. citrina was questioned by our team several years ago9. This study aimed to further determine whether colchicine and its precursors exist in H. citrina or not, based on metabolic data, and to clarify why this alkaloid is not produced according to genomic data. The high-quality and chromosome-level genome of H. citrina will provide new insights into the rutin biosynthesis and the lack of colchicine.
Results
Sequencing and assembly
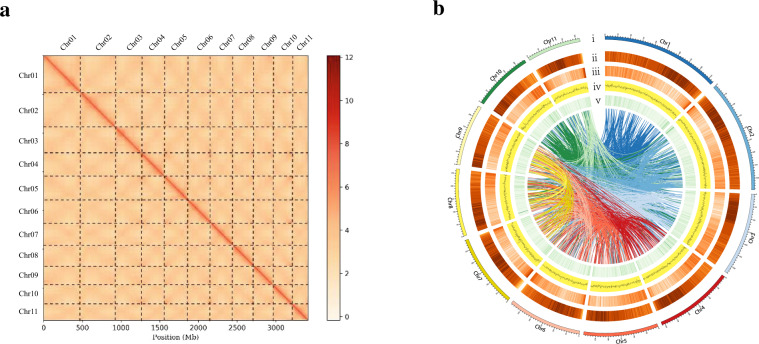
We generated 177.52 Gb of 150 bp paired-end reads and 157.53 Gb (coverage of ~41.46×) of short reads (Supplementary Table S1). Simultaneously, we generated 165-fold PacBio single-molecule long polymerase reads (625.85 Gb with an N50 length of 38.27 kb) and 172-fold Hi-C data (646.63 Gb) were used to construct the chromosome-level high-quality reference genome. The genome size was estimated to be ~3.80 Gb and the heterozygosity rate and repeat sequence contents were 1.28% and 78.85%, respectively (Supplementary Table S2), based on Illumina resequencing data. In the end, we obtained 3183 contigs with an N50 of 2.08 Mb and a size of 3.77 Gb, which was ~99% of the estimated size (Table 1). To construct chromosome-level genes, we used ~170× Hi-C data to anchor contigs to chromosomes. We successfully clustered 2919 contigs spanning 3.41 Gb (90.36% of the total length of all contigs) into 11 chromosome groups after further ordering and orienting the clustered contigs (Fig. 1a). Finally, we obtained the first chromosome-level and high-quality genome of H. citrina, with chromosome lengths ranging from 216.66 to 471.57 Mb, accounting for 90.42% of the whole sequence (Fig. 1b and Supplementary Table S3).
Table 1.
H. citrina genome assembly results
| Mecat 2 assembly | Post Gcpp | Post Pilon | Haplotig purge | Hi-C assembly | |
|---|---|---|---|---|---|
| Size (Mb) | 5611.82 | 5611.82 | 5611.82 | 3774.13 | 3775.58 |
| No. contigs/Scaffold | 8877 | 8877 | 8877 | 3183 | 734 |
| No. contigs/Scaffold (>2 kb) | 8834 | 8834 | 8842 | 3174 | 725 |
| Max. contig/Scaffold length (bp) | 21,710,810 | 21,804,269 | 21,795,804 | 21,795,804 | 471,572,209 |
| Contig/Scaffold N50 size (bp) | 1,516,939 | 1,522,568 | 1,521,497 | 2,081,915 | 294,951,729 |
| Contig/Scaffold N90 size (bp) | 428,328 | 430,376 | 428,572 | 761,644 | 216,659,559 |
| BUSCO | 82.78% | — | 92.0% | 91.5% | 91.2% |
Fig. 1. H. citrina genome assembly.
a The Hi-C interaction heatmap for the H. citrina. The color in the figure from light to dark indicates the increase in the intensity of interaction. b Overview of H. citrina genomic features. (i) Circular representation of the pseudomolecule, (ii–v) repetitive sequence density (bin = 1 M), gene density (bin = 1 M), GC content (bin = 1 M), ncRNA densities (bin = 1 M), and genome collinearity
We first assessed the accuracy and completeness of our assembly results through Benchmarking Universal Single-Copy Orthologs (BUSCO) analysis, then identified 91.4% complete and 2.4% partial BUSCO genes (Supplementary Table S4). In addition, 99.49% of the filtered short reads (157.53 Gb, Supplementary Table S1) were mapped to the genome of H. citrina, which covered 99.86% of the assembly. Furthermore, a total of 22,310 homozygous single-nucleotide polymorphisms (SNPs) (0.0006% of the total H. citrina assembly) were identified. In summary, the above results demonstrate the high accuracy and completeness of the H. citrina genome.
Genome annotation
Repetitive sequence prediction of the H. citrina genome was mainly performed through two methods: homology annotation and ab initio prediction. A total of 3.25 Gb of repetitive elements was identified in our assembled genome, comprising 86.20% of the whole genome (Supplementary Table S5). Among these repetitive elements, long terminal repeats were the main type, accounting for 72.39% (2.73 Gb). The rest were short interspersed nuclear elements, DNA transposons, and long interspersed nuclear elements, which accounted for 0.15%, 14.24%, and 6.63%, respectively. Similarly, a total of 3540 transfer RNA (tRNA), 406 ribosomal RNA, 457 small nuclear RNA, and 127 microRNA genes were annotated in the H. citrina genome (Supplementary Table S6).
We predicted 54,295 protein-coding genes in the H. citrina genome, with an average length of 8339 bp and an average exon number of 4.53 for each gene (Supplementary Table S7). By comparing the genes annotated in the other six species, we found that the various indicators of the annotated genes (gene, CDS, exon, and intron lengths) were similar to those of other species (Supplementary Fig. S1). We functionally annotated ~44,398 (81.77%) protein-coding genes of H. citrina based on known genes, conserved domains, and Gene Ontology (GO) terms (Supplementary Table S8). Finally, 93.8% of the BUSCO genes were identified in the annotation of H. citrina (Supplementary Table S4), which showed that our annotations were complete and reliable by BUSCO analysis.
Genome evolution and gene families expansion/contraction
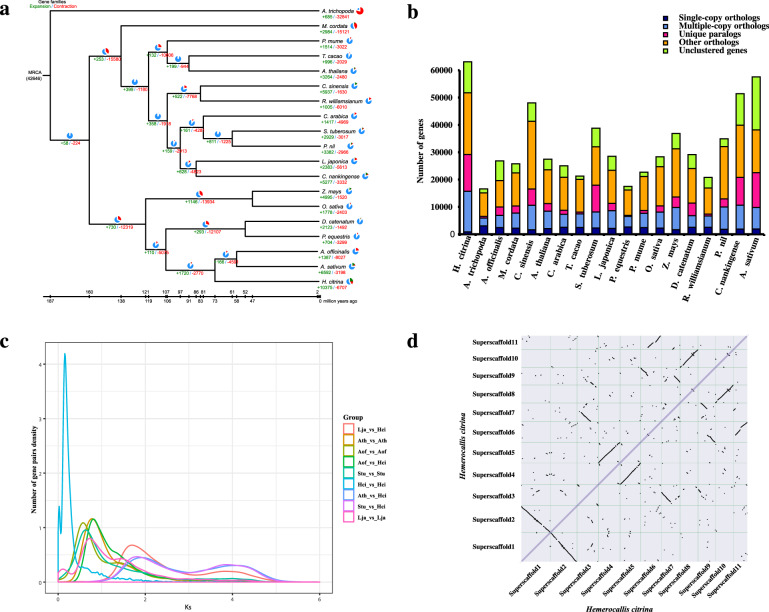
In this study, we first compared the protein sequences encoded by H. citrina with those encoded by 18 other species, namely, Amborella trichopoda, Macleaya cordata, Prunus mume, Arabidopsis thaliana, Theobroma cacao, Camellia sinensis, Rhododendron williamsianum, Solanum tuberosum, Pharbitis nil, Coffee arabica, Chrysanthemum nankingense, Lonicera japonica, Dendrobium catenatum, Phalaenopsis equestris, Asparagus officinalis, Allium sativum, Oryza sativa, and Zea mays. These species had 116 single-copy orthologous gene families according to gene family cluster analysis. In addition, we clustered 51,740 protein sequences (81.99%) encoded by H. citrina into 15,974 gene families. After length-based filtering of the shared single-copy orthologous gene families, 116 genes remained. The phylogenetic tree showed that the H. citrina, A. sativum, and A. officinalis were located on the same evolutionary branch, showed a closer relationship. In addition, our prediction results showed that H. citrina, A. sativum, and A. officinalis phylogenetically diverged from the common ancestor ~71.7 Mya, after the separation of Orchidaceae at 107.24 Mya (Fig. 2a), which is consistent with published research10.
Fig. 2. Evolution of the H. citrina genome and gene families.
a Phylogenetic tree of 19 plant species and the sizes of expanded and contracted gene families: A. trichopoda, M. cordata, P. mume, T. cacao, A. thaliana, C. sinensis, R. williamsianum, C. arabica, S. tuberosum, P. nil, L. japonica, C. nankingense, Z. mays, O. sativa, D. catenatum, P. equestris, A. officinalis, A. sativum, and H. citrina. b The distribution of single-copy, multiple-copy, unique, other orthologous and unclustered genes in the 19 plant species. c Distribution of Ks values between L. japonica, A. thaliana, A. officinalis, S. tuberosum, and H. citrina (Ks distribution chart showing that H. citrina experienced a relatively recent WGD event). d Dot plots of paralogs in the H. citrina genome
A total of 42,646 gene families in the most recent common ancestor of the 19 species were obtained by analyzing the gene family expansion and contraction. The number of expanded and contracted gene families in H. citrina were 10,375 and 6707, respectively (Fig. 2a). Compared with A. officinalis and A. sativum, it has 116 expanded and 4591 contracted gene families, which demonstrated that the number of expanded genes in H. citrina had increased significantly. This result indicated that H. citrina may have experienced more duplication events than A. officinalis and A. sativum. We found that these genes in H. citrina were also the most abundant based on the multicopy homologous genes number (Fig. 2b). In addition, we performed GO and KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analyses of these expanded and contracted genes in the H. citrina genome. We found lineage-specific expansions of genes related to the metabolic biosynthesis of flavonoids, which may affect the biosynthesis of rutin and enhance flavor and medicinal value (Supplementary Table S9).
Genome-wide duplication events
To identify the source of many genes (>50,000) in H. citrina, we performed whole-genome duplication (WGD) analysis using overlapping H. citrina genomes. The synonymous substitution rate (Ks) estimates were applied to detect WGD events. The distribution of Ks values results showed that H. citrina have one main peak at Ks values of ~0.18 (~15.73 Mya) (Fig. 2c), whereas A. officinalis have more ancient WGD event. Dot plots can be shown as paralogs (2–2 diagonal relationships) evolving from a recent WGD event in the H. citrina genome (Fig. 2d).
Prediction of rutin biosynthesis genes in H. citrina
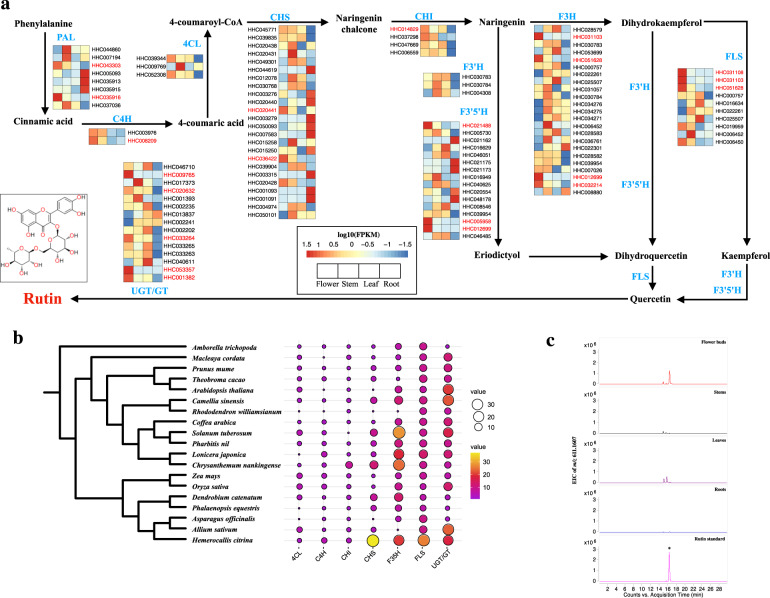
Rutin is the main ingredient and is the recognized one of the main antidepressant compounds in H. citrina. The biosynthetic precursor of rutin is derived from phenylalanine and then synthesized by ten enzymes11 (Supplementary Table S10 and Fig. 3a). Through analysis, the expanding and contracting of seven gene families involved in the biosynthesis of rutin. We found four homologous genes (CHS, F3’5’H, FLS, and UGT/GT) in H. citrina have been increased significantly compared with other species (Fig. 3b). We predicted 108 candidate genes in ten gene families involved in rutin biosynthesis by homologous alignment and a Pfam database search (Fig. 3a). Then, we found that rutin primarily accumulates in the flower buds, whereas the content of rutin in the stems, leaves, and roots is lower according to High-performance liquid chromatography/quadrupole time-of-flight (HPLC-Q-TOF) methods (Fig. 3c), which indicated that the candidate genes were mainly expressed in flower buds. Finally, 20 candidate genes were predicted in line with this coexpression pattern (Fig. 3a, red color).
Fig. 3. Prediction of rutin biosynthesis genes in H. citrina.
a The biosynthesis pathway and the expression profile for rutin in four tissues: flowers, stems, leaves, and roots. Expression profiles of 108 candidate enzymes are illustrated using a gradient color. Gene expression values (FPKM) were scaled by log10 and the genes with an expected expression pattern were indicated in red. b The seven gene families are expanding and contracting, and the heat map mainly shows the number of genes in the 4CL, C4H, CHS, CHI, F3’5’H, FLS, and UGT/GT gene families. c EIC of m/z value of rutin (m/z 611.1607, [M + H]+) in the TICs of different tissues of H. citrina
Colchicine and its biosynthesis pathway is not existent in H. citrina
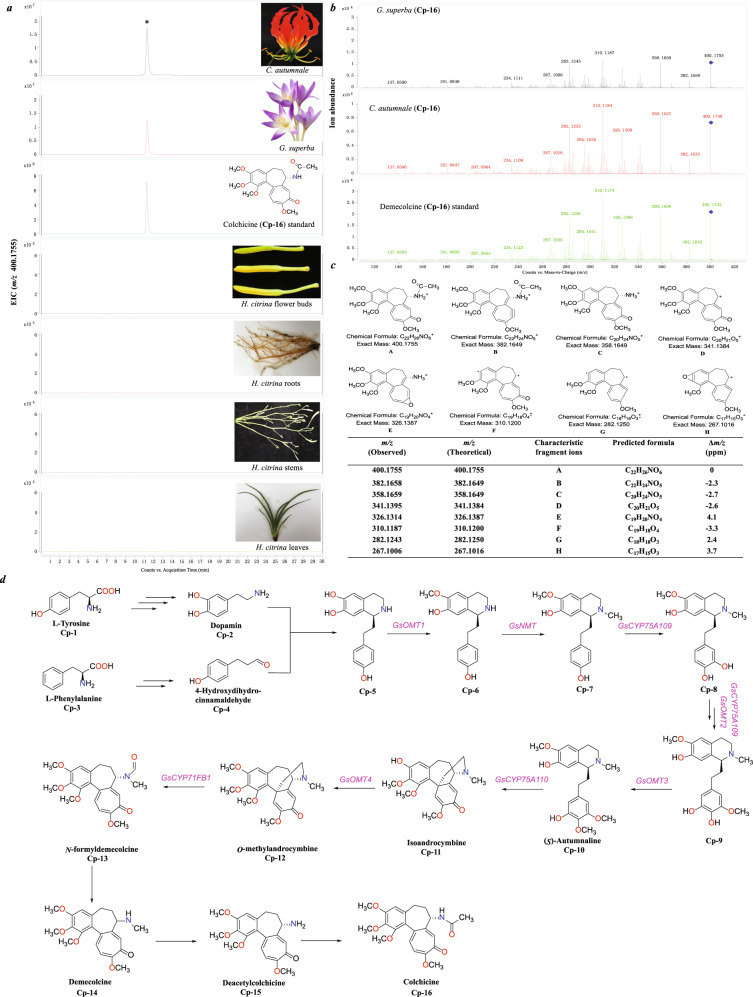
The extracted ion chromatogram (EIC) of the precise m/z value of colchicine standard (Cp 16, m/z 400.1074, [M + H]+) for the total ion chromatograms (TICs) of Gloriosa superba, Colchicum autumnale, and H. citrina were performed. Colchicine was found and identified unambiguously in G. superba and C. autumnale by comparing their retention time, precise m/z value, and characteristic fragment ions with those of the standard. However, the precise m/z value of this compound was not found in the TICs of different tissues of H. citrina (Fig. 4a–c), which proved that this plant did not contain colchicine.
Fig. 4. Detection and identification of colchicine.
a EIC of the theoretical m/z value (400.1755, [M + H]+) of colchicine in the TICs of G. superba, C. autumnale, H. citrina, and the standard. b MS/MS spectra of the parent ions at m/z 400.1755 and 400.1748 from the TICs of G. superba and C. autumnale, respectively, compared to the colchicine standard (Cp 16, m/z 400.1742). c The putative structures and tabulated list for the fragment ions from the MS/MS analysis of the mother ion at m/z 400.1755. d Near-complete biosynthesis pathway of colchicine and related functional genes
The near-complete biosynthesis pathway and related functional genes of colchicine have been identified in G. superba12 (Fig. 4d). The EIC of the theoretical m/z values of 15 compounds (Cp 1 to Cp 15), which were the precursors of colchicine, were performed for the TICs of G. superba, C. autumnale, and H. citrina. These precursors were detected and determined from G. superba and C. autumnale by their precise m/z values and characteristic fragment ions. However, only two original amino acids, l-tyrosine (Cp 1) and l-phenylalanine (Cp 3), were observed and identified in H. citrina (Supplementary Figs. S2 and S4), and the theoretical m/z values of the remaining 13 precursors were not found (Supplementary Figs. S3 and S5–S16). In addition, the candidate genes involved in the colchicine biosynthesis pathways were detected by BLASTP searching with eight known genes from G. superba12 (with an E-value ≤ 1e – 5, a coverage ≥ 0.5, and an identity ≥ 0.5) (Fig. 4d and Supplementary Table S11). Unsurprisingly, none of the orthologous genes were obtained from H. citrina. Therefore, the genomic analysis demonstrated that H. citrina does not contain any genes involved in colchicine biosynthesis.
Discussion
We construct a high-quality and chromosome-level reference genome by combining PacBio SMRT and Hi-C technology. We found that the genome data of H. citrina have high heterozygosity and repetitive content features. More importantly, we can use the genome to research the phylogenetic and evolutionary characteristics at a deeper level, and to cultivate new varieties of H. citrina with staggered flowering periods or non-blooming buds via molecular breeding. Based on the multi-omics analysis, we deduced a gene coexpression rule and predicted that 20 candidate genes match this rule. These results lay the groundwork for further research on the functional genes involved in the biosynthesis pathway of rutin.
Numerous journals, magazines, newspapers, and other news outlets have reported that H. citrina contains colchicine, which was first identified in Hemerocallis by microchemical methods in 192913; however, the identification method and result were doubted by other scientists in 194914. In 1977, colchicine was first reported from H. citrina in China15. In the next few decades, more than 30 poisoning incidents were recorded in China due to the consumption of the fresh flower buds of H. citrina, which resulted to more than 830 people with symptoms of poisoning. All reports stated that the poisoning was caused by colchicine in H. citrina (Supplementary Table S12). Moreover, H. citrina containing colchicine was even recorded in college textbooks and popular science books in China16–19. However, this compound was not found in different tissues of H. citrina using HPLC-Q-TOF-mass spectrometry (MS) technologies in this study. In addition, none of the orthologous genes involved in the colchicine biosynthesis pathway were identified in the H. citrina genome, which further clarified that this alkaloid was absent at the genomic level. Both results unambiguously demonstrate that H. citrina does not contain colchicine. In past studies, colchicine was never isolated and identified from H. citrina by phytochemical methods. In addition, this alkaloid was only determined by thin-layer chromatography or HPLC by comparing the Rf value or the retention time (Rt) with that of the standard20,21, so another compound (m/z 455.1455 in positive mode) had Rf and Rt values close to those of colchicine (m/z 400.1755 in positive mode). Therefore, this compound was incorrectly identified as colchicine9. This study challenges the long-standing belief that colchicine present in H. citrina leads to poisoning.
Conclusion
Here, a high-quality and chromosome-scale H. citrina genome was reported. The genome was ~3.8 Gb in size, with a heterozygosity rate of ~1.28% and contig N50 of 2.09 Mb. Subsequently, Hi-C technology was applied and we anchored 90.42% of the assembled contigs to 11 pseudochromosomes. We identified a total of 54,295 protein-coding genes and 63,105 transcripts. Based on comparative genomics, we found that H. citrina experienced a recent WGD event at ~15.73 Mya that increased the number of genes by more than 50 thousand and expanded gene families by more than 10 thousand. A total of 4 gene families involved in the rutin biosynthesis pathway were expanded and 20 candidate genes were predicted by multi-omic data. Finally, we proved for the first time that the biosynthesis pathway of colchicine does not exist in the genome of H. citrina. Our research provided the first chromosome-level genome of the Hemerocallis genus, which laid the foundation for genetic research and molecular breeding of H. citrina.
Materials and methods
Sample collection and high-throughput sequencing
The H. citrina was cultivated at Hunan Agricultural University. We collected the healthy leaves from the best-growing H. citrina. A modified cetyltrimethyl ammonium bromide (CTAB) method22 was used for DNA extraction. RNA contaminants were removed by RNase A and the integrity of DNA was obtained. The DNA molecules were used to construct a library after being cut into ~30 kb fragments and then sequenced on the PacBio Sequel II platform (Frasergen, China). Simultaneously, a library with an insert size of 350 bp was constructed for the Illumina HiSeq X Ten platform (Illumina, Inc., San Diego, CA, USA). These short reads for whole-genome sequencing were mainly used for genome survey, error correction, and polishing after initial assembly. A Hi-C library was established using the young leaves of H. citrina and the BGI MGISEQ-2000 platform (BGI, China) was used for sequencing. In addition, the size of H. citrina genome was evaluated by k-mer analysis with GCE23 (Supplementary Fig. S17).
RNA extraction and Iso-Seq sequencing
H. citrina was grown in Qidong County (Hunan, China, coordinates: 111°52′22.44″E, 26°53′23.75″N) for RNA extraction. We sampled fresh, healthy roots, stems, leaves, and flowers from five different periods with three biological duplication. We used TRIzol reagent (Invitrogen, USA) to extract total RNA based on the recommended protocol. DNA was removed via RQ1 DNase (Promega, USA). Finally, RNA from all samples was mixed to construct the library.
The cDNA synthesis kit (ClontechSMARTer®) was used to establish the cDNA libraries. AMPure PB beads were employed for the cDNA product purification. A total of 376.06 Mb was sequenced with 30 h movies by PacBio Sequel II platform (Supplementary Table S1). Simultaneously, these RNAs were used to construct short-fragment libraries and then processed on the BGI platform, which yielded 30.74 Gb of raw RNA sequence data with a read quality Q30 of 91.0%.
Genome assembly
All subread data from SMRT sequencing were used for H. citrina genome assembly. The draft genome assembly was obtained using mecat 2 (20,190,226) with the default parameters. The gcpp in the SMRT link 4 toolkit was performed to correct errors after the initial assembly of the genome. Then, we used 157.53 Gb of short reads to correct any remaining errors with Pilon24 (v1.22). Due to the heterozygosity of the genome, Haplotigs purge was used to filter redundant sequences25.
Pseudochromosomes were determined using Hi-C analysis, as described previously26. Briefly, 646.63 Gb of clean read pairs were produced from the Hi-C library and mapped to the polished H. citrina contig assembly using BWA (bwa-0.7.17) with the default parameters27. LACHESIS28 tool was used to cluster contigs into chromosome-level scaffolds by the genomic proximity signal of Hi-C data.
Evaluation of genome quality
Genome assembly accuracy and completeness were first assessed using the continuous long reads subreads. A total of 96.60% of subreads were mapped to 99.97% of the genome, with an average depth of 129.89×. Then, a window of size 10 kb was used to continuously slide along the genome without overlapping (when the sequence length was <10 kb, the actual length prevailed), calculate the average sequencing depth of the sequence in the window and the percentage of GC content. Finally, draw the contig GC content distribution-sequencing depth distribution density map based on the statistical data (Supplementary Fig. S18). Second, the single-base level genome assembly was evaluated using Illumina short-read by BWA 0.7.17 software27. Furthermore, homozygous SNPs were filtered by the GATK 4.0.8.129 package. The assembled genome was also subjected to BUSCO v3.0.230 analysis with embryophyta_odb10 to evaluate the completeness of the genome and annotation.
Annotation of repetitive sequences and genes
De novo and homology-based prediction methods were employed to annotate the repeat sequences in the genome of H. citrina. The known transposable elements within the H. citrina genome were identified by combining RepeatMasker31, RepeatProteinMask, and RepeatModeler. In addition, the tRNA-related genes were mainly identified by tRNAscan-SE (v1.3.1)32 and Infernal (v1.1.2)33 software with default parameters.
The assembled genome of H. citrina was hard and soft masked by RepeatMasker prior to gene prediction. First, we used homologous proteins to train the gene models of Augustus (v3.3.1)34 and SNAP35, and then performed ab initio gene prediction based on these models. Second, the protein sequences were predicted genes using Exonerate (v2.2.0)36 with the default parameters. Third, the clean RNA-Sequencing reads were assembled into transcripts via Trinity37 to perform RNA-based gene prediction and the gene structure was further predicted using PASA38. Finally, Maker (v3.00)39 was employed to integrate the prediction results of the three strategies.
Gene functions were inferred by aligning our annotated gene models with known databases. BLAST+ (v2.6.0+)40 was performed against the National Center for Biotechnology Information (NCBI), Non-Redundant, TrEMBL, and Swiss-Prot41. The protein domains were annotated using PfamScan42 and InterProScan (v5.35–74.0)43 based on InterPro protein databases. The motifs and domains were identified by Pfam44. GO45 IDs for each gene were obtained from Blast2GO46. KEGG Automatic Annotation Server was used to annotate the KEGG pathways47.
Gene family identification
To cluster families of protein-coding genes, proteins from the longest transcripts of each gene from H. citrina and other closely related species, including A. trichopoda, M. cordata, P. mume, A. thaliana, T. cacao, C. sinensis, R. williamsianum, S. tuberosum, P. nil, C. arabica, C. nankingense, L. japonica, D. catenatum, P. equestris, A. officinalis, A. sativum, O. sativa, and Z. mays, were used. All proteins were extracted and aligned with each other using BLASTP40 programs (NCBI blast v2.6.0) with a maximal E-value of 1e − 5. We filtered out and excluded putative fragmented genes with an identity <30%, a coverage <50%, and protein-encoding sequences shorter than 50 bp. Then, we used OrthoMCL (v14–137)48 to cluster genes from different species into gene families.
Phylogenetic analysis
We construct a phylogenetic tree of H. citrina and other closely related species by the protein sequences of 186 single-copy orthologous genes, which were aligned with the MUSCLE (v3.8.31)49 program, and we further employed RAxML (v8.2.11)50 to build the phylogenetic tree.
Gene families expansion/contraction
According to the identified gene families and the constructed phylogenetic tree with the predicted divergence times of those species, we used CAFÉ51 to analyze gene families expansion and contraction. Families with a p-value < 0.05 were considered to have an accelerated rate of gene gain or loss. These gene families in H. citrina (p-value ≤ 0.05) were mapped to KEGG pathways for functional enrichment analysis, which was conducted using enrichment methods. For this process, hypergeometric test algorithms were implemented and the Q-value (false discovery rate) was calculated to adjust p-values utilizing the R environment (https://github.com/StoreyLab/qvalue).
Whole-genome duplication analysis
We used the synonymous substitution rate (Ks) to detect WGD events. First, syntenic paralogous blocks were identified with MCSCAN between L. japonica, A. thaliana, A. officinalis, S. tuberosum, and H. citrina. Then, the protein sequences of these plants in the syntenic paralogous blocks were aligned against each other with Blastp (E-value ≤ 1e − 5) to identify the conserved paralogs of each plant. Third, the Ks values of these gene pairs were calculated. Finally, the Ks distribution was used to evaluate the WGD events.
Sample collection and preparation for metabolomic analysis
G. superba, C. autumnale, and H. citrina plants were collected from Kunming University of Science and Technology, China Pharmaceutical University, and Hunan Agriculture University, respectively. All samples (whole G. superba and C. autumnale, and flower buds, roots, stems, and leaves of H. citrina) were freeze-dried and crushed by a disintegrator. Approximately 0.4 g of powdered sample was extracted using ultrasonic bath for 120 min with 10 mL of 70% methanol-water (v/v). The extract solution was filtered by a 0.22 μm microporous membrane and stored in a bottle.
HPLC-Q-TOF-MS conditions
HPLC-Q-TOF-MS conditions were optimized based on the previous method1. The gradient of elution was modified as follows: 0–3 min, 10–15% (B); 3–8 min, 15–30% (B); 8–16 min, 30–65% (B); and 16–30 min, 65–95% (B). The injection volume was reduced to 2 μL and the MS/MS data of each compound were obtained using different collision energy (10–35 eV).
Supplementary information
Acknowledgements
This work was supported by the “National Key R&D Program of China (2017YFD0501500),” “Hunan Provincial Key Research and Development Project (2020NK2031),” and “The Special Funds for Development of Local Science and Technology from Central Government (2019XF5067).”
Author contributions
Z.Q., P.H., and J.Z. conceived and designed the study. Z.Q., J. Liu, and P.H. collected the sample. X.Y., G.H., and J. Lao estimated the genome size and assembled the genome. X.Z. and P.H. performed DNA, RNA-sequencing, and Hi-C experiments. M.S. and P.H. performed the genome annotation and functional genomic analysis. Z.Q., X.L., and Z.Y. performed the data analysis of metabolome. Z.Q., X.Y., and P.H. wrote the manuscript.
Data availability
All sequencing data were deposited in the NCBI Sequence Read Archive (SRA) database with BioProject accession number PRJNA647253. The assembled genome was submitted to DDBJ/ENA/GenBank with accession number JACEHZ000000000. The version is JACEHZ010000000.
Conflict of interest
The authors declare no competing interests.
Footnotes
These authors contributed equally: Zhixing Qing, Jinghong Liu, Xinxin Yi, Xiubin Liu
Contributor Information
Peng Huang, Email: huangpeng@hunau.edu.cn.
Jianguo Zeng, Email: zengjianguo@hunau.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41438-021-00539-6.
References
- 1.Liu J, et al. Systematic identification metabolites of Hemerocallis citrina Borani by high-performance liquid chromatography/quadrupole-time-of-flight mass spectrometry combined with a screening method. J. Pharm. Biomed. Anal. 2020;186:113314. doi: 10.1016/j.jpba.2020.113314. [DOI] [PubMed] [Google Scholar]
- 2.Ma G, et al. iTRAQ-based quantitative proteomic analysis reveals dynamic changes during daylily flower senescence. Planta. 2018;248:859–873. doi: 10.1007/s00425-018-2943-5. [DOI] [PubMed] [Google Scholar]
- 3.Li CF, et al. Evaluation of the toxicological properties and anti-inflammatory mechanism of Hemerocallis citrina in LPS-induced depressive-like mice. Biomed. Pharmacother. 2017;91:167–173. doi: 10.1016/j.biopha.2017.04.089. [DOI] [PubMed] [Google Scholar]
- 4.Yang RF, Geng LL, Lu HQ, Fan XD. Ultrasound-synergized electrostatic field extraction of total flavonoids from Hemerocallis citrina baroni. Ultrason. Sonochem. 2017;34:571–579. doi: 10.1016/j.ultsonch.2016.06.037. [DOI] [PubMed] [Google Scholar]
- 5.Lin SH, et al. The antidepressant-like effect of ethanol extract of daylily flowers (Jīn Zhēn Huā) in rats. J. Tradit. Complement. Med. 2013;3:53–61. doi: 10.4103/2225-4110.106548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, et al. Ethyl acetate fraction of Hemerocallis citrina Baroni decreases tert-butyl hydroperoxide-induced oxidative stress damage in BRL-3A cells. Oxid. Med. Cell Longev. 2018;2018:1–13. doi: 10.1155/2018/1526125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian H, et al. Effects of phenolic constituents of daylily flowers on corticosterone-and glutamate-treated PC12 cells. BMC Complement. Alter. Med. 2017;17:69. doi: 10.1186/s12906-017-1582-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu P, et al. Antidepressant-like effects and cognitive enhancement of the total phenols extract of Hemerocallis citrina Baroni in chronic unpredictable mild stress rats and its related mechanism. J. Ethnopharmacol. 2016;194:819–826. doi: 10.1016/j.jep.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 9.Tang MN, Liu XB, Huang JL, Deng FM, Zeng JG. Questioning and arguable research on edible Hemerocallis citrina containing colchicine. Chin. Tradit. Herb. Drugs. 2016;047:3293–3300. [Google Scholar]
- 10.Li SF, et al. Chromosome-level genome assembly, annotation and evolutionary analysis of the ornamental plant Asparagus setaceus. Hortic. Res. 2020;7:48. doi: 10.1038/s41438-020-0271-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, et al. The tartary buckwheat genome provides insights into rutin biosynthesis and abiotic stress tolerance. Mol. Plant. 2017;10:1224–1237. doi: 10.1016/j.molp.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Nett RS, Lau W, Sattely ES. Discovery and engineering of colchicine alkaloid biosynthesis. Nature. 2020;584:148–153. doi: 10.1038/s41586-020-2546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein G, Soos G. Der mikrochemische Nachweis der Alkaloide in der Pflanze. Oesterr Bot. Z. 1929;78:157–163. doi: 10.1007/BF02716541. [DOI] [Google Scholar]
- 14.Traub HP. Colchicine poisoning in relation to Hemerocallis and some other plants. Science. 1949;110:686–687. doi: 10.1126/science.110.2869.686. [DOI] [PubMed] [Google Scholar]
- 15.Li ZH. Plant poisoning. Barefoot Dr. Mag. 1977;8:44–45. [Google Scholar]
- 16.Zong, W., Zhang, L. & Wang, M. Z. (eds) Food Safety (Chemical Industry, 2016).
- 17.Zhang, Z. J. et al. (eds) Introduction of Food Safety (Chemical Industry, 2015).
- 18.Peng, W. X., Pan, T., Yuan, Y. Y. & Wang, L. (eds) Food Safety and Food Poisoning-First Aid Knowledge (GuiZhou, 2012).
- 19.Zhou, C. Q. et al. (eds) Food Nutrition (China Metrology, 2006).
- 20.Hong YF, Cheng ZW, Li JH, Hu C. On different methods to treat the fresh Hemerocallis citrina and lead to the change of colchicine. J. Hunan Agric. Univ. 2003;29:500–502. [Google Scholar]
- 21.Zhang N, et al. Optimization of HPLC detection system for colchicine content in flower buds of Hemerocallis. J. Agric. Univ. Hebei. 2017;9:48–54. [Google Scholar]
- 22.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- 23.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roach, M. J., Schmidt, S. & Borneman, A. R. Purge Haplotigs: synteny reduction for third-gen diploid genome assemblies. BMC Bioinformatics19, 460 (2018). [DOI] [PMC free article] [PubMed]
- 26.Yin D, et al. Genome of an allotetraploid wild peanut Arachis monticola: a de novo assembly. Gigascience. 2018;7:giy066. doi: 10.1093/gigascience/giy066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at arXiv:1303.3997 (2013).
- 28.Burton JN, et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013;31:1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simão FA, et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 31.Tarailo‐Graovac M, Chen N. Using repeatMasker to identify repetitive elements in genomic gequences. Curr. Protoc. Bioinformatics. 2004;25:4.10.1–4.10.14. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- 32.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:435–439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slater GSC, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haas BJ, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cantarel BL, et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camacho C, et al. BLAST plus: architecture and applications. BMC Bioinformatics. BioMed. Cent. 2009;10:1. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boeckmann B, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mistry J, Bateman A, Finn RD. Predicting active site residue annotations in the Pfam database. BMC Bioinformatics. 2007;8:298. doi: 10.1186/1471-2105-8-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hotz GC, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:S281–S288. doi: 10.1093/nar/gkn226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conesa, A. & Götz, S. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics. 2008, 619832 (2008). [DOI] [PMC free article] [PubMed]
- 47.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han MV, Thomas GW, Lugo-Martinez J, Hahn MW. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 2013;30:1987–1997. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data were deposited in the NCBI Sequence Read Archive (SRA) database with BioProject accession number PRJNA647253. The assembled genome was submitted to DDBJ/ENA/GenBank with accession number JACEHZ000000000. The version is JACEHZ010000000.