Abstract
Obesity is currently an important health problem and is associated with an increased likelihood of various diseases. The efficacies of various natural treatments have been assessed for their utility in treating obesity. Alliin (S-allyl-L-cysteine sulfoxides) is considered the major component of garlic and has a wide range of natural antioxidant properties. However, the direct effects of alliin on obesity have not been well clarified. The present study investigated the effects and possible mechanisms of alliin on adipocyte differentiation. The 3T3-L1 cells were treated with alliin (0-40 µg/ml) during adipogenic differentiation. The effect of alliin on lipid accumulation was evaluated by Oil red O staining. Reverse transcription-quantitative PCR was performed to investigate the expression levels of adipogenic differentiation-related genes. The accumulation of lipid droplets was markedly inhibited following alliin treatment. The expression levels of multiple adipogenic transcription markers, such as CCAAT/enhancer-binding protein (C/EBP) β, C/EBP α and peroxisome proliferation-activity receptor γ, were markedly decreased following treatment with alliin during adipogenic differentiation. Expression levels of several adipocyte-related genes were subsequently suppressed. Additionally, alliin suppressed PKB/Akt and PI3K expression. These results suggested that alliin exhibits anti-adipogenic activity by downregulating major adipogenic differentiation-related genes and Akt/PI3K expression. Alliin may have a potential therapeutic effect on metabolic disease.
Keywords: alliin, adipogenesis, metabolic disease, peroxisome proliferation-activity receptor γ, CCAAT/enhancer-binding protein β, Akt
Introduction
Obesity is a significant risk factor for many metabolic diseases such as obstructive sleep apnea, cardiovascular diseases, diabetes, cancers, and osteoarthritis (1,2). Obesity can be considered a consequence of energy imbalance, which causes an increase in differentiated adipocytes and excessive fat accumulation (3,4). Adipogenesis, the process of maturation of preadipocytes, is an integrated process involving the activation of several signaling pathways (5). The transcription factors such as the peroxisome proliferation-activity receptor (PPAR) γ and CCAAT/enhancer-binding protein (C/EBP) family are crucial for adipogenesis (6-8). These major adipogenic-related factors control the expression of the lipid metabolizing enzymes to form mature adipocytes. C/EBP families are expressed at certain times in the process of adipogenesis. C/EBP β and C/EBP δ promoters are activated in the early stage of differentiation and act in the direct regulation of adipogenesis (9). Activated C/EBP β then regulate its neighboring promoter element, which subsequently encodes the PPAR γ and C/EBP α genes during the later stage of differentiation (10,11). The major transcription factors upregulate adipocyte differentiation-related genes such as adiponectin, leptin, and fatty acid binding protein (Fabp4) during adipocyte differentiation (12,13). Therefore, the control of adipogenesis involves suppressing this transcriptional regulation. Many signaling pathways influence adipocyte differentiation. The Akt pathway and the extracellular signal-regulated kinase (ERK) 1/2 pathway are shown to be responsible for adipogenesis (14,15). The Akt is in a key position to promote insulin-like growth factor-1 expression and adipocyte differentiation, while Akt pathway inhibition is shown to reduce adipogenesis (16).
Many studies have focused on developing anti-obesity drugs (17). Although several drugs are currently available to treat obesity, their long-term use may cause severe side effects (18). Thus, multiple research studies are focused on new natural products with potential for anti-obesity activity (19). The anti-obesity activity of various plant extracts is reported to be mediated by the regulation of adipogenesis (20). Garlic (Allium sativum), one of the oldest medicinal plants, is originally from Asia. Allium vegetables comprise one of the natural sources of organosulfur compounds and show advantages in therapeutic application, mostly owing to their cardiovascular protective effects, lowering of cholesterol, and anti-cancer properties (21). In addition, Allium has possibly beneficial effects in the treatment of obesity by adipogenesis inhibition (22), energy expenditure increase (23) and influence on expression of inflammatory mediators from serum (24). The synthesis of alliin (S-allyl-L-cysteine sulfoxide, SACSO), considered to be the major component of garlic, was first reported by Stoll and Seebeck in 1951(25). Alliin, which has strong antioxidant properties, has been used as a treatment remedy for some diseases (26,27). Previous research found that alliin can help in decreasing the serum levels of glucose and insulin (28). Alliin also can regulate the anti-inflammatory effects of preadipocytes by reducing cytokine levels, such as IL-6 and TNF (29). Alliin was shown to possess many biological effects. However, the direct effect of alliin in adipogenesis has not yet been explored. The aim of this study was to evaluate the effects of alliin on the adipogenic differentiation of 3T3-L1 cells and its potential association with the regulation of adipogenic transcription factors PPAR γ, C/EBP, adiponectin and Fabp4, as well as possible mechanism.
Materials and methods
Materials
Alliin, obtained from Abcam (ab141896), was dissolved in dimethylsulfoxide (DMSO), and stored at -20˚C.
Cell culture
3T3-L1 fibroblasts were obtained from Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan). Cells were cultured in DMEM (Sigma-Aldrich; Merck KGaA) supplemented with 10% fetal bovine serum (FBS; Gibco Life Technologies), 100 U/ml penicillin, 100 µg/ml streptomycin, and 0.25 µg/ml amphotericin B (Biological Industries) in a CO2 ncubator. The cells were then trypsinized after reaching 80-90% confluence. Cells at passages 2-15 were used in this study.
Adipocyte differentiation
The cells were differentiated following previously reported method with some modifications (30). Briefly, adipocyte differentiation begins after cell cultures reached 100% confluence. For adipogenic induction, the medium used was DMEM containing 10% FBS, 1 µM dexamethasone (Sigma-Aldrich; Merck KGaA), 0.5 mM 3-isobutyl-1-methylxanthine (Wako), and 10 µg/ml insulin (Funakoshi Co. Ltd.) for 2 days. The cells were then transferred to DMEM containing 10% FBS and 10 µg/ml insulin for 6 days. The medium was changed every 2-3 days. Mature adipocytes were obtained on day 7 or 8. To evaluate the effects of alliin on adipogenesis, various concentrations of alliin (10-40 µg/ml) were used during the induction process. Cells that were not treated with alliin were identified as the control.
Oil red O staining
To confirm the lipid accumulation in cultured cells, Oil red O staining (Sigma-Aldrich; Merck KGaA) was performed. Eight days after induction, mature adipocytes were fixed with 4% paraformaldehyde phosphate buffer solution for 30 min at room temperature and then stained with Oil red O working solution (6:4 of oil red stock solution: Distilled water) for 15 min. PBS was washed three times to remove the excess Oil red O dye. The images were observed under a parallel phase contrast microscope (Olympus IX70 inverted microscope, Olympus Optical CO, Ltd.). For quantitative analysis, the percentage of positively stained areas were calculated using imageJ. Results are expressed as percentage of Oil red O-stained area compared to control. For each sample, the experiments were performed in triplicates.
Measurement of cell viability
Cell viability was determined using a Cell Counting Kit-8 assay (CCK-8; DOJINDO Lab, Osaka, Japan). 3T3-L1 cells were seeded in 96-well plates (5x103 cells/well) for 2 days. Cells were then cultured with various concentrations of alliin (0-40 µg/ml) for 7 days. Cell Counting Kit-8 (CCK-8; Dojindo Laboratories) was used according the manufacturer's instruction. Cells were incubated with 10 µl of CCK-8 reagent for 2 h at 37˚C. The result was determined using a Synergy™ HTX Multi-Mode Microplate Reader (BioTek Instruments) at 450 nm.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells using TRIzol® reagent (Ambion®; Gibco Life Technologies) according to manufacturer's instructions. The quantity and quality of isolated RNA were detected using a Nano Drop spectrophotometer (Nano Drop® ND-1000, Thermo Fisher Scientific, Inc.). Complementary DNA (cDNA) was synthesized in a 20-µl reaction using 2 µg RNA, 20 pmol Oligo dT12-18 (Invitrogen; Thermo Fisher Scientific, Inc., 18418-012), 0.5 µl RNase inhibitor (Promega Corporation, N211A), 0.2 µl ReverTra Ace (Toyobo, TRT-101), 2 µl dNTP Mixture (Takara, 4030), 4 µl 5X RT buffer (Toyobo, TRT-101), and 10 mM DEPC water. RT-qPCR was performed in a 25-µl reaction, containing 1 µl cDNA, 10 pmol forward and reverse primers, and 12.5 µl SYBR Premix Ex Taq II (Takara, RR820A). PCR amplification was set as follows: 95˚C for 30 sec, 40 cycles of 95˚C for 5 sec, and 60˚C for 30 sec; then the final dissociation was set at 95˚C for 15 sec, 60˚C for 30 sec, and 95˚C for 15 sec. The data were quantified using the 2-∆∆Cq method and were normalized against the levels of β-actin (31). The primer sequences used for PCR are provided in Table I.
Table I.
Primer sequences for reverse transcription-quantitative PCR.
| Name of gene | Primers (5'-3') | Melting temperature (˚C) | Product length (bp) | Genbank code |
|---|---|---|---|---|
| β-actin | F: CATCCGTAAAGACCTCTATGCCAAC ATGGAGCCACCGATCCACA | 64.1 | 193 | NM_007393.5 |
| R: ATGGAGCCACCGATCCACA | 64.9 | |||
| PPAR γ | F: GTGCCAGTTTCGATCCGTAGA | 66.2 | 167 | NM_001113418.1 |
| R: GGCCAGCATCGTGTAGATGA | 66.2 | |||
| C/EBP α | F: GGACAAGAACAGCAACGAGTA | 61.8 | 237 | NM_001287514.1 |
| R: GCAGTTGCCATGGCCTTGA | 69.7 | |||
| C/EBP β | F: TGGACAAGCTGAGCGACGAG | 69.1 | 192 | NM_001287738.1 |
| R: TGTGCTGCGTCTCCAGGTTG | 70.0 | |||
| Adiponectin | F: GCACTGGCAAGTTCTACTGCAA | 66.4 | 156 | NM_009605.5 |
| R: GTAGGTGAAGAGAACGGCCTTGT | 66.4 | |||
| Leptin | F: CCACACACAGCTGGAAACTC | 63.4 | 216 | NM_008493.3 |
| R: GCCTTGCTTCAGATCCATCC | 65.9 | |||
| Fabp4 | F: CCAATGAGCAAGTGGCAAGA | 66.2 | 179 | NM_024406.3 |
| R: GATGCCAGGCTCCAGGATAG | 65.9 | |||
| PI3K | F: TCCTGCTTCATACCGAGCTT | 63.8 | 212 | NM_001024955.2 |
| R: CATGACATCCTCCCTCTCGT | 64.2 | |||
| AKT | F: CCCTTCTACAACCAGGACCA | 63.9 | 210 | NM_009652.3 |
| R: ATACACATCCTGCCACACGA | 64.1 | |||
| MAPK | F: TGCCAGGCTGAACTACAGTG | 64.1 | 169 | NM_008927.4 |
| R: CACAAGGCTCCCTCTCAGAC | 64.1 | |||
| ERK | F: TCAGAGGCAGGTGGATCTCT | 64.0 | 188 | NM_011949.3 |
| R: GGTGCCATCATCAACATCTG | 64.1 |
F, forward; R, reverse.
Statistical analysis
The results are presented as the mean ± standard error of the mean (SEM) of three experiments. SPSS 13.0 and GraphPad Prism 6 software were used for statistical analysis. The statistical significance between each group was computed using one-way analysis of variance followed by Dunnett's test. P<0.05 was considered statistically significant.
Results
Effect of alliin on intracellular lipid accumulation during adipogenic differentiation
3T3-L1 cells showed fibroblastic-like morphology before adipogenic induction (Fig. 1A-a and -b). Then, the suppression effect of alliin on adipogenic differentiation was examined. During adipogenic differentiation, the 3T3-L1 cells were treated with different concentrations of alliin (10, 20, 40 µg/ml) for 7 days, and the cells treated without alliin were used as control. 3T3-L1 cells showed a morphological change from fibroblastic-shape to round-shape, and then oil droplets were formed and maintained after 7 days of induction (Fig. 1A-c-f). Oil red O staining was performed to evaluate lipid accumulation levels. The Oil red O-stained area decreased significantly in the alliin-treated groups compared with the control group (Fig. 1A-g-j). Meanwhile, quantitative analysis showed that alliin-treated group displayed dramatically lower lipid accumulation in a dose-dependent manner compared to the control group. 40 µg/ml alliin treatment showed the greatest effects on the lipid accumulation which level dropped to 39.5% (Fig. 1B). The effect of alliin on cell viability was examined. 3T3-L1 cells were treated with different concentrations of alliin (10-40 µg/ml) for 7 days. Alliin has no effect on cell viability even at a concentration of 40 µg/ml (Fig. 1C). These data indicated that alliin may contain anti-adipogenic potential.
Figure 1.
Effect of alliin on lipid droplet accumulation in 3T3-L1 cells. (A) Phase-contrast images of 3T3-L1 cells. (A-a and -b) 3T3-L1 cells exhibited fibroblastic morphology before adipogenic induction, (c-f) and then the cells changed to a round-shape after 7 days of induction. (A-g-j) Oil-red O staining revealed that the intracellular lipids were decreased in alliin-treated groups compared with the control. (B) Quantitative analysis demonstrated that the positively stained areas were decreased in the alliin treatment group. (C) Cell viability was tested on day 7. Alliin (10-40 µg/ml) exhibited no effect on cell viability. Scale bar, 100 µm. Data are presented as the mean ± SEM. *P<0.05, **P<0.01, ***P<0.001.
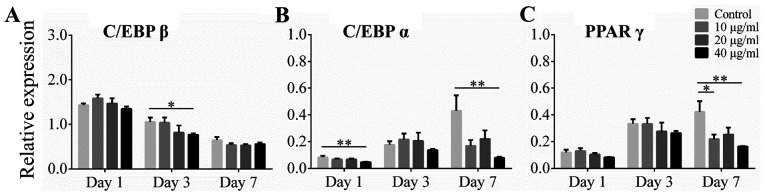
Effect of alliin on adipogenic transcription markers C/EBP β, C/EBP α, and PPAR γ
To examine the effect of alliin on adipogenic transcription markers during induction, cells were analyzed by RT-qPCR. The expression levels of C/EBP β, C/EBP α and PPAR γ were assessed on dayz 1, 3 and 7 of induction, respectively. The mRNA level of C/EBP β, induced by the differentiation medium, reached its peak on day 1, and then decreased gradually over time. Cells treated with 40 µg/ml alliin on day 3 showed a significant decrease in lipid accumulation compared to cells in the control group, but there was no significant difference on day 7 (Fig. 2A). The expression of C/EBP α and PPAR γ were gradually induce during adipogenic differentiation in control group. Expression level of C/EBP α was significantly reduced by treatment with 40 µg/ml alliin compared to without treatment on day 1 and day 7 (Fig. 2B) Treatment with 40 µg/ml alliin also significantly inhibited the expression of PPAR γ on day 7 (Fig. 2C). These results indicate that 40 µg/ml alliin suppress adipogenesis by downregulating the expression of C/EBP α and PPAR γ.
Figure 2.
Effect of alliin on adipogenic transcriptional markers during induction. The 3T3-L1 cells were treated with or without alliin during adipogenic induction. Reverse transcription-quantitative PCR revealed the expression levels of (A) C/EBP β, (B) C/EBP α and (C) PPAR γ in 3T3-L1 cells during induction. Data are presented as the mean ± SEM. *P<0.05, **P<0.01. C/EBP, CCAAT/enhancer-binding protein; PPAR γ, peroxisome proliferation-activity receptor γ.
Effect of alliin on adipocyte-related genes adiponectin, Fabp4, and leptin
Expression of adipocyte-related genes was also analyzed using RT-qPCR. During adipogenic differentiation, expression of adiponectin mRNA increased over time. The level of adiponectin mRNA was significantly lower with treatment of 40 µg/ml alliin than without treatment at all time points examined (Fig. 3A). The level of Fabp4 mRNA was significantly decreased in cells cultured with 40 µg/ml alliin than in those without treatment on day 1 and day 7 (Fig. 3B). Whereas, the level of leptin mRNA was significantly lower in the 40 µg/ml alliin treatment group than in the control group on day 1 (Fig. 3C). These results indicate that 40 µg/ml alliin inhibits adipogenesis and leads to reduce expression of adiponectin and Fabp4.
Figure 3.
Effect of alliin on adipocyte-related genes during induction. Alliin was administered at various concentrations during adipogenic differentiation. Reverse transcription-quantitative PCR analysis was used to evaluate the gene expression levels of (A) adiponectin, (B) Fabp4 and (C) leptin. Data are presented as the mean ± SEM. *P<0.05, **P<0.01. Fabp4, fatty acid binding protein 4.
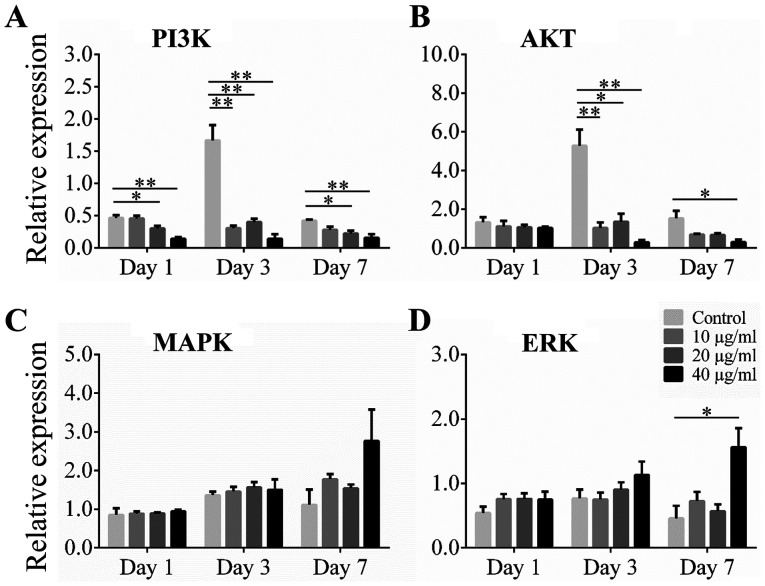
Effect of Alliin on the Akt and ERK 1/2 pathways related genes during adipogenic differentiation
Akt and ERK 1/2 are upstream regulators in the adipocyte differentiation pathway, including C/EBP α and PPAR γ pathways. To investigate the possible mechanism of alliin on adipogenic differentiation, the Akt and ERK 1/2 pathways related genes were examined. The 3T3-L1 cells were cultured with various concentrations of alliin during the differentiation of adipocyte. Results showed that expression of PI3K and Akt was significantly increased at the early stage of differentiation. Expression of PI3K was significantly inhibited in the 20 or 40 µg/ml alliin treatment group compared to that in the control group at all time points examined (Fig. 4A). The level of AKT mRNA was significantly lower with treatment of 40 µg/ml alliin than without treatment on days 3 and 7 (Fig. 4B). However, increased expression of MAPK and ERK was observed in the treatment groups compared to that in the control groups on day 7 (Fig. 4C). The level of ERK mRNA increased significantly with 40 µg/ml alliin treatment compared to that without treatment after 7 days (Fig. 4D). These results suggest that 40 µg/ml alliin inhibits adipocyte differentiation by reducing the expression of PI3K and Akt.
Figure 4.
Effect of alliin on the Akt and ERK 1/2 pathway-related genes during induction. The 3T3-L1 cells were treated with different concentration of alliin during adipogenesis. The expression levels of (A) PI3K, (B) Akt, (C) MAPK and (D) ERK were analyzed by reverse transcription-quantitative PCR. Data are presented as the mean ± SEM. *P<0.05, **P<0.01.
Discussion
In the current study, we indicated that alliin treatment may contribute to decreased adipogenesis of 3T3-L1 adipocyte. This study revealed, for the first time, the direct effect of alliin on adipogenesis. Moreover, we found an effect of alliin on adipogenesis may be achieved by downregulating the Akt and PI3K expression. Several recent reports indicate the benefit of plant extracts as drugs (19,32). Compared to conventional drugs, herbal medicines show less potentially dangerous side effects. Many plant compounds, such as ginsenoside, caffeic acid, berberine, anthocyanin, and capsaicin, have been shown to inhibit adipogenesis (33-35).
Obesity is commonly caused by an excessive increase of adipocytes. Adipogenesis is a complex regulated cellular differentiation process involving many signaling pathways and related molecules (36,37). The inhibition of adipogenesis process can provide a target for the control and treatment of obesity. During 3T3-L1 adipocyte differentiation, C/EBP β is rapidly induced and is responsible for activating the adipogenic regulators C/EBP α and PPAR γ (38). PPAR γ plays a crucial role in adipogenesis of embryonic stem cells and fibroblasts. PPAR γ levels are significantly induced when preadipocytes are converted into adipocytes (39,40). The research suggested that white adipose tissue under obesity condition a significant increment in oxidative stress, pro-inflammatory status and depletion of n-3 long chain polyunsaturated fatty acid (n-3 LCPUFA) (41). Whereas PPAR γ transcription factor is regulated by n-3 LCPUFA that participate in the metabolism of adipose tissue (42). So, the regulation of adipogenesis occurs by controlling PPAR γ levels (20). Moreover, PPAR γ has been shown to induce and activate the transcriptional factor C/EBP α promoter (43). C/EBP α is a transcriptional factor of the C/EBP family and also plays an important role in regulating adipogenesis (44). Thus, PPAR γ and C/EBP α genes are upregulated and together activate the adipogenesis pathways (45). In this study, a direct effect of alliin on adipocyte differentiation was performed. Alliin significantly decreased the lipid droplet accumulation. Expression level of C/EBP β was significantly decreased in presence of 40 µg/ml alliin on day 3. Expression of C/EBP α and PPAR γ was lower with 40 µg/ml alliin treatment on day 7, while the level of C/EBP β was not affected. These result may correspond to C/EBP β induced in the early stage of differentiation that subsequently activates the adipogenesis markers, PPAR γ and C/EBP α, at the later stage of differentiation. The results suggest that alliin may inhibit adipogenesis by reducing C/EBP β, C/EBP α, and PPAR γ levels during adipocyte differentiation.
In addition, PPAR γ and C/EBP α can be part of a feedback loop and promote adipogenesis by leading the expression of downstream genes, such as adiponectin, Fabp4, and leptin. These downstream genes play major roles in inducing and maintaining mature adipocytes (5). A recent study indicated that the expression of adipocyte-related genes during induction is PPAR γ dependent (46). In the alliin treatment group, expression of leptin was lower during the early stage of differentiation, while the level was not affected at day 7. This may be due to the existence of different regulatory mechanisms of leptin expression. Recent study noted that leptin can directly induce the adipocyte differentiation in the early stage of adipogenesis, while showing anti-adiposity effects after maturation of adipocytes (47). Expression of adiponection and Fabp4 was significantly inhibited with alliin treatment after adipocyte induction. These results clearly imply that alliin inhibits adipogenic differentiation by reducing the expression of adipocyte-related genes, which may be through the repression of PPAR γ and C/EBP α related pathways.
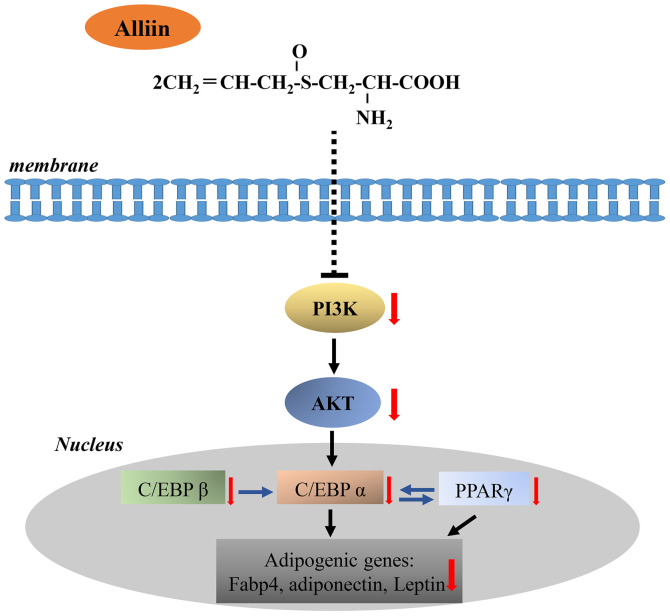
Insulin signaling pathway plays a critical role in modulating adipogenesis. In the presence of insulin, the insulin receptor is autophosphorylated, and subsequently proteins in the insulin receptor substrate family are also phosphorylated, thereby activating the two main signaling pathways, PI3K/Akt and MAPK/ERK pathways (48). The function of PI3K/Akt signaling is to activate adipogenic transcription marker such as C/EBP α and promote adipocyte maturation during adipogenesis (49). A study also showed that expression of PPAR γ is significantly impeded in Akt-deficient mice (50). Thus, inhibition of PI3K/Akt signaling pathway may provide a therapeutic target for obesity (51). The MAPK/ERK signaling pathway plays a complicated role in the regulation of adipogenesis. Activation of the MAPK/ERK pathway can both inhibit and promote adipocyte differentiation. Tanabe et al (52) reported that the MAPK/ERK pathway works in a suppressive manner on adipocyte differentiation when receiving a long-term or sustained stimulation. In this study, the higher expression levels of ERK with alliin treatment may correspond to the inhibition effects of MAPK pathway on adipogenesis. In contrast, the expression level of P13K and Akt mRNA expression were markedly decreased with alliin treatment. Collectively, our study indicated that alliin resulted in PI3K/Akt inhibition, thereby suppressing the expression of C/EBP β, C/EBP α, and PPAR γ and adipocyte-related genes (Fig. 5). Although the results from this study indicated that the Akt signaling pathway play an important role in alliin inhibit adipocyte differentiation, there are still limitations. First, the phosphorylation levels of Akt signaling related proteins should be test. Second, whether the validity of this theory still needs further experimental investigation.
Figure 5.
Schematic representation of the possible mechanism of the anti-adipogenic effect of alliin. Alliin (C6H11NO3S) inhibits adipogenesis by downregulating PI3K and Akt activity, thereby attenuating the expression levels of C/EBP β, C/EBP α, PPAR γ and lipid metabolizing enzymes. C/EBP, CCAAT/enhancer-binding protein; PPAR γ, peroxisome proliferation-activity receptor γ.
In conclusion, the results demonstrated that alliin in garlic can inhibit adipogenesis by reducing expression of major transcriptional activators and their downstream genes, which may be mediated by regulation of Akt signaling pathway. Alliin may provide a possible naturally occurring therapeutic method for the prevention and treatment of metabolic disease.
Acknowledgements
Not applicable.
Funding Statement
Funding: The present study was supported in part by Grants-in-Aid for Scientific Research (C) from Japan Society for the Promotion of Science (JSPS; KAKENHI grant no. JP19K10192).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
NL and KC performed all experiments, acquired data, analyzed the data and drafted the manuscript. . KC and HD contributed to the acquisition of data. JY, MY, HK and XL designed the experiment. XL revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wilson PW, Bozeman SR, Burton TM, Hoaglin DC, Ben-Joseph R, Pashos CL. Prediction of first events of coronary heart disease and stroke with consideration of adiposity. Circulation. 2008;118:124–130. doi: 10.1161/CIRCULATIONAHA.108.772962. [DOI] [PubMed] [Google Scholar]
- 2.Jehan S, Myers AK, Zizi F, Pandi-Perumal SR, Jean-Louis G, McFarlane SI. Obesity, obstructive sleep apnea and type 2 diabetes mellitus: Epidemiology and pathophysiologic insights. Sleep Med Disord. 2018;2:52–58. [PMC free article] [PubMed] [Google Scholar]
- 3.Faria SL, Faria OP, Menezes CS, de Gouvêa HR, de Almeida Cardeal M. Metabolic profile of clinically severe obese patients. Obes Surg. 2012;22:1257–1262. doi: 10.1007/s11695-012-0651-y. [DOI] [PubMed] [Google Scholar]
- 4.Jéquier E, Tappy L. Regulation of body weight in humans. Physiol Rev. 1999;79:451–480. doi: 10.1152/physrev.1999.79.2.451. [DOI] [PubMed] [Google Scholar]
- 5.Moseti D, Regassa A, Kim WK. Molecular regulation of adipogenesis and potential anti-adipogenic bioactive molecules. Int J Mol Sci. 2016;17(124) doi: 10.3390/ijms17010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosen ED, Hsu CH, Wang X, Sakai S, Freeman MW, Gonzalez FJ, Spiegelman BM. C/EBPalpha induces adipogenesis through PPARgamma: A unified pathway. Genes Dev. 2002;16:22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Jack AK, Hamm JK, Pilch PF, Farmer SR. Reconstitution of insulin-sensitive glucose transport in fibroblasts requires expression of both PPARgamma and C/EBPalpha. J Biol Chem. 1999;274:7946–7951. doi: 10.1074/jbc.274.12.7946. [DOI] [PubMed] [Google Scholar]
- 8.Lefterova MI, Lazar MA. New developments in adipogenesis. Trends Endocrinol Metab. 2009;20:107–114. doi: 10.1016/j.tem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka T, Yoshida N, Kishimoto T, Akira S. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J. 1997;16:7432–7443. doi: 10.1093/emboj/16.24.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke SL, Robinson CE, Gimble JM. CAAT/enhancer binding proteins directly modulate transcription from the peroxisome proliferator-activated receptor gamma2 promoter. Biochem Biophys Res Commun. 1997;240:99–103. doi: 10.1006/bbrc.1997.7627. [DOI] [PubMed] [Google Scholar]
- 11.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei S, Zan LS, Wang HB, Cheng G, Du M, Jiang Z, Hausman GJ, Mcfarland DC, Dondson MV. Adenovirus-mediated interference of FABP4 regulates mRNA expression of ADIPOQ, LEP and LEPR in bovine adipocytes. Genet Mol Res. 2019;12:494–505. doi: 10.4238/2013.January.4.21. [DOI] [PubMed] [Google Scholar]
- 13.Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- 14.Tseng YH, Butte AJ, Kokkotou E, Yechoor VK, Taniguchi CM, Kriauciunas KM, Cypess AM, Niinobe M, Yoshikawa K, Patti ME, Kahn CR. Prediction of preadipocyte differentiation by gene expression reveals role of insulin receptor substrates and necdin. Nat Cell Biol. 2005;7:601–611. doi: 10.1038/ncb1259. [DOI] [PubMed] [Google Scholar]
- 15.Prusty D, Park BH, Davis KE, Farmer SR. Activation of MEK/ERK signaling promotes adipogenesis by enhancing peroxisome proliferator-activated receptor gamma (PPARgamma) and C/EBPalpha gene expression during the differentiation of 3T3-L1 preadipocytes. J Biol Chem. 2002;277:46226–46232. doi: 10.1074/jbc.M207776200. [DOI] [PubMed] [Google Scholar]
- 16.Guo LX, Chen G, Yin ZY, Zhang YH, Zheng XX. P-Synephrine exhibits anti-adipogenic activity by activating the Akt/GSK3β signaling pathway in 3T3-L1 adipocytes. J Food Biochem. 2019;43(e13033) doi: 10.1111/jfbc.13033. [DOI] [PubMed] [Google Scholar]
- 17.Powell AG, Apovian CM, Aronne LJ. New drug targets for the treatment of obesity. Clin Pharmacol Ther. 2011;90:40–51. doi: 10.1038/clpt.2011.82. [DOI] [PubMed] [Google Scholar]
- 18.Yanovski SZ, Yanovski JA. Long-term drug treatment for obesity: A systematic and clinical review. JAMA. 2014;311:74–86. doi: 10.1001/jama.2013.281361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar P, Bhandari U. Common medicinal plants with antiobesity potential: A special emphasis on fenugreek. Anc Sci Life. 2015;35:58–63. doi: 10.4103/0257-7941.165629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatano T, Sameshima Y, Kawabata M, Yamada S, Shinozuka K, Nakabayashi T, Mizuno H. St. John's wort promotes adipocyte differentiation and modulates NF-κB activation in 3T3-L1 cells. Biol Pharm Bull. 2014;37:1132–1138. doi: 10.1248/bpb.b13-00989. [DOI] [PubMed] [Google Scholar]
- 21.Mikaili P, Maadirad S, Moloudizargari M, Aghajanshakeri S, Sarahroodi S. Therapeutic uses and pharmacological properties of garlic, shallot, and their biologically active compounds. Iran J Basic Med Sci. 2013;16:1031–1048. [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HJ, Lee MJ, Jang JY, Lee SH. Allium hookeri root extract inhibits adipogenesis by promoting lipolysis in high fat diet-induced obese mice. Nutrients. 2019;11(2262) doi: 10.3390/nu11102262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kagawa Y, Ozaki-Masuzawa Y, Hosono T, Seki T. Garlic oil suppresses high-fat diet induced obesity in rats through the upregulation of UCP-1 and the enhancement of energy expenditure. Exp Ther Med. 2020;19:1536–1540. doi: 10.3892/etm.2019.8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu C, Mathews AE, Rodrigues C, Eudy BJ, Rowe CA, O'Donoughue A, Percival SS. Aged garlic extract supplementation modifies inflammation and immunity of adults with obesity: A randomized, double-blind, placebo-controlled clinical trial. Clin Nutr ESPEN. 2018;24:148–155. doi: 10.1016/j.clnesp.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Stoll A, Seeback E. Chemical investigations on alliin, the specific principle of garlic. Adv Enzymol Relat Subj Biochem. 1951;11:377–400. doi: 10.1002/9780470122563.ch8. [DOI] [PubMed] [Google Scholar]
- 26.Slusarenko AJ, Patel A, Portz D. Control of plant diseases by natural products: Allicin from garlic as a case study. Eur J Plant Pathol. 2008;121:313–322. [Google Scholar]
- 27.Borlinghaus J, Albrecht F, Gruhlke MC, Nwachukwu ID, Slusarenko AJ. Allicin: Chemistry and biological properties. Molecules. 2014;19:12591–12618. doi: 10.3390/molecules190812591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Upadhyay RK. Garlic: A potential source of pharmaceuticals and pesticides: A review. Int J Green Phar. 2016;10:1–28. [Google Scholar]
- 29.Quintero-Fabián S, Ortuño-Sahagún D, Vázquez-Carrera M, López-Roa RI. Alliin, a Garlic (Allium sativum) compound, prevents LPS-induced inflammation in 3T3-L1 adipocytes. Mediators Inflamm. 2013;2013(381815) doi: 10.1155/2013/381815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang M, Sheng S, Zhang W, Zhang J, Zhang Z, Zhang M, Hatch GM, Chen L. MiR27a promotes the development of macrophage-like characteristics in 3T3-L1 preadipocytes. Int J Biol Sci. 2018;14:1599–1609. doi: 10.7150/ijbs.26274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Ekor M. The growing use of herbal medicines: Issues relating to adverse reactions and challenges in monitoring safety. Front Pharmacol. 2014;4(177) doi: 10.3389/fphar.2013.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aoyagi R, Funakoshi-Tago M, Fujiwara Y, Tamura H. Coffee inhibits adipocyte differentiation via inactivation of PPARγ. Biol Pharm Bull. 2014;37:1820–1825. doi: 10.1248/bpb.b14-00378. [DOI] [PubMed] [Google Scholar]
- 34.Liao CC, Ou TT, Huang HP, Wang CJ. The inhibition of oleic acid induced hepatic lipogenesis and the promotion of lipolysis by caffeic acid via up-regulation of AMP-activated kinase. J Sci Food Agric. 2014;94:1154–1162. doi: 10.1002/jsfa.6386. [DOI] [PubMed] [Google Scholar]
- 35.Wu T, Jiang Z, Yin J, Long H, Zheng X. Anti-obesity effects of artificial planting blueberry (Vaccinium ashei) anthocyanin in high-fat diet-treated mice. Int J Food Sci Nutr. 2016;67:257–264. doi: 10.3109/09637486.2016.1146235. [DOI] [PubMed] [Google Scholar]
- 36.Prins JB, O'Rahilly S. Regulation of adipose cell number in man. Clin Sci (Lond) 1997;92:3–11. doi: 10.1042/cs0920003. [DOI] [PubMed] [Google Scholar]
- 37.Guo L, Li X, Tang QQ. Transcriptional regulation of adipocyte differentiation: A central role for CCAAT/enhancer-binding protein (C/EBP) β. J Biol Chem. 2015;290:755–761. doi: 10.1074/jbc.R114.619957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salma N, Xiao H, Imbalzano AN. Temporal recruitment of CCAAT/enhancer-binding proteins to early and late adipogenic promoters in vivo. J Mol Endocrinol. 2006;36:139–151. doi: 10.1677/jme.1.01918. [DOI] [PubMed] [Google Scholar]
- 39.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPARgamma2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 40.Wang F, Mullican SE, DiSpirito JR, Peed LC, Lazar MA. Lipoatrophy and severe metabolic disturbance in mice with fat-specific deletion of PPARγ. Proc Natl Acad Sci USA. 2013;110:18656–18661. doi: 10.1073/pnas.1314863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Illesca P, Valenzuela R, Espinosa A, Echeverría F, Soto-Alarcon S, Ortiz M, Videla LA. Hydroxytyrosol supplementation ameliorates the metabolic disturbances in white adipose tissue from mice fed a high-fat diet through recovery of transcription factors Nrf2, SREBP-1c, PPAR-γ and NF-κB. Biomed Pharmacother. 2019;109:2472–2481. doi: 10.1016/j.biopha.2018.11.120. [DOI] [PubMed] [Google Scholar]
- 42.Echeverría F, Ortiz M, Valenzuela R, Videla LA. Long-chain polyunsaturated fatty acids regulation of PPARs, signaling: Relationship to tissue development and aging. Prostaglandins Leukot Essent Fatty Acids. 2016;114:28–34. doi: 10.1016/j.plefa.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 43.Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, McKeon C, Darlington GJ, Spiegelman BM. Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell. 1999;3:151–158. doi: 10.1016/s1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- 44.Tang QQ, Otto TC, Lane MD. CCAAT/enhancer-binding protein beta is required for mitotic clonal expansion during adipogenesis. Proc Natl Acad Sci USA. 2003;100:850–855. doi: 10.1073/pnas.0337434100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghaben AL, Scherer PE. Adipogenesis and metabolic health. Nat Rev Mol Cell Biol. 2019;20:242–258. doi: 10.1038/s41580-018-0093-z. [DOI] [PubMed] [Google Scholar]
- 46.Gerhold DL, Liu F, Jiang G, Li Z, Xu J, Lu M, Sachs JR, Bagchi A, Fridman A, Holder DJ, et al. Gene expression profile of adipocyte differentiation and its regulation by peroxisome proliferator-activated receptor-gamma agonists. Endocrinology. 2002;143:2106–2118. doi: 10.1210/endo.143.6.8842. [DOI] [PubMed] [Google Scholar]
- 47.Lukaszewski MA, Eberlé D, Vieau D, Breton C. Nutritional manipulations in the perinatal period program adipose tissue in offspring. Am J Physiol Endocrinol Metab. 2013;305:E1195–E1207. doi: 10.1152/ajpendo.00231.2013. [DOI] [PubMed] [Google Scholar]
- 48.Son YH, Ka S, Kim AY, Kim JB. Regulation of adipocyte differentiation via microRNAs. Endocrinol Metab (Seoul) 2014;29:122–135. doi: 10.3803/EnM.2014.29.2.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 50.Peng XD, Xu PZ, Chen ML, Hahn-Windgassen A, Skeen J, Jacobs J, Sundararajan D, Chen WS, Crawford SE, Coleman KG, Hay N. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 2003;17:1352–1365. doi: 10.1101/gad.1089403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang QQ, Gronborg M, Huang H, Kim JW, Otto TC, Pandey A, Lane MD. Sequential phosphorylation of CCAAT enhancer-binding protein beta by MAPK and glycogen synthase kinase 3beta is required for adipogenesis. Proc Natl Acad Sci USA. 2005;102:9766–9771. doi: 10.1073/pnas.0503891102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanabe Y, Koga M, Saito M, Matsunaga Y, Nakayama K. Inhibition of adipocyte differentiation by mechanical stretching through ERK-mediated downregulation of PPARgamma2. J Cell Sci. 2004;117:3605–3614. doi: 10.1242/jcs.01207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.