Abstract
Thermal swamps are unique ecosystems where geothermally warmed waters mix with decomposing woody biomass, hosting novel biogeochemical-cycling and lignin-degrading microbial consortia. Assembly of shotgun metagenome libraries resolved 351 distinct genomes from hot-spring (30–45 °C) and mesophilic (17 °C) sediments. Annotation of 39 refined draft genomes revealed metabolism consistent with oligotrophy, including pathways for degradation of aromatic compounds, such as syringate, vanillate, p-hydroxybenzoate, and phenol. Thermotolerant Burkholderiales, including Rubrivivax ssp., were implicated in diverse biogeochemical and aromatic transformations, highlighting their broad metabolic capacity. Lignin catabolism was further investigated using metatranscriptomics of sediment incubated with milled or Kraft lignin at 45 °C. Aromatic compounds were depleted from lignin-amended sediment over 148 h. The metatranscriptomic data revealed upregulation of des/lig genes predicted to specify the catabolism of syringate, vanillate, and phenolic oligomers in the sphingomonads Altererythrobacter ssp. and Novosphingobium ssp., as well as in the Burkholderiales genus, Rubrivivax. This study demonstrates how temperature structures biogeochemical cycling populations in a unique ecosystem, and combines community-level metagenomics with targeted metatranscriptomics to identify pathways with potential for bio-refinement of lignin-derived aromatic compounds. In addition, the diverse aromatic catabolic pathways of Altererythrobacter ssp. may serve as a source of thermotolerant enzymes for lignin valorization.
Subject terms: Biogeochemistry, Water microbiology, Freshwater ecology, Biogeochemistry
Introduction
Few geothermally influenced wetlands exist on Earth—geothermal hot springs that feed marshlands can be found in Yellowstone National Park (USA) [1–3], Great Rift Valley (Kenya), Iceland, and New Zealand [4]—fewer still are forested. The Liard River Hot Springs in northern British Columbia, Canada feature geothermally warmed springs (30–55 °C) with organic sediments and biomats that receive inputs from the surrounding mixed-forest vegetation. Based on these characteristics, we hypothesized that this environment harbors thermotolerant microbiota capable of degrading lignin-derived aromatic compounds.
Geothermally warmed systems are hot spots of chemoautotrophy and other oligotrophic adaptations [5, 6]. Dilute chemical resources may select for high-affinity and high-yielding chemoautotrophy. Specific examples include complete ammonia oxidation (Comammox) [7, 8], the conversion of cyanate and urea to ammonium [7, 9] and carboxydotrophy [10, 11]. Pathways for aromatic catabolism are also associated with oligotrophic bacteria, as with the mesophilic sphingomonad, Sphingopyxis alaskensis RB2256 [12]. Thermophilic mineralization of lignin-derived aromatic compounds is of both ecological and biotechnological interest [13–15]. Lignin is a recalcitrant plant heteropolymer synthesized from three phenylpropanoids: p-coumaryl (H), coniferyl (G), and sinapyl (S) alcohols. The degradation of lignin yields a variety of aromatic compounds. Due to the convergent organization of bacterial catabolic pathways, they are well suited to the biocatalytic conversion of these biomass-derived aromatics to fuels and chemicals [16–18]. The bacterial catabolism of S-lignin compounds, such as syringate, is primarily limited to aerobic sphingomonads [17, 19, 20] and anaerobic Firmicutes [21–24]. Few S-lignin-degrading thermophiles have been reported [24], and no aerobic, thermotolerant S-lignin degradation pathways have been elucidated. There are two main routes for degradation of syringate in bacteria [25, 26]: one via gallate using vanillate/3-O-methylgallate O-demethylase (LigM), and gallate dioxygenase (DesB), and one via 3-O-methylgallate carried out by protocatechuate 4,5-dioxygenase (LigAB) and 3-O-methylgallate 3,4-dioxygenase (DesZ). The later route can result in two different products with unique oxidation pathways [26]. Identifying enzymes capable of metabolizing lignin-derived aromatic compounds at elevated temperature would further biorefining and the move away from fossil feedstocks.
Herein, we used genome-resolved shotgun metagenomics and metatranscriptomics to characterize the phylogenetic and metabolic diversity of a thermal swamp. We exploited the negative, unimodal relationship between temperature and microbial diversity [27–29] to accomplish high-coverage genome reconstruction from the hot-spring microbiome. We then used metatranscriptomics to test the hypothesis that thermal swamp communities contained active microbial populations involved in aromatic catabolism. Genomic reconstruction revealed diverse chemoautotrophic metabolism in the thermal microbiome but only partially elucidated aromatic pathways for lignin-derived compounds, while metatranscriptomics and analytical chromatography provided evidence for thermotolerant catabolism of aromatic compounds. In particular, this study characterized transcriptomes from oligotrophic sphingomonads associated with degradation of S- and G-lignin at elevated temperature.
Material and methods
Field site and sampling
Liard River Hot Springs (59.431N, 126.1W) is a complex of carbonate-hosted springs that feed geothermally warmed meteoric waters to extensive swampland underlaid by sandstone and shale [30, 31] (Supplementary Fig. 1). The pools are surrounded by unique thermal meadow vegetation, and mixed forest containing Populus tremuloides, Betula papyrifera, and Picea glauca. Four hot springs were sampled on September 27, 2018: the Alpha (50–55 °C), Beta (30–35 °C), Delta (30–40 °C), and Epsilon (40–45 °C) pools. Alpha pool contains a recreational bathing area, and was previously measured to have a pH of 6.5–6.8 and concentrations of 0.9–1.0 mg l−1 O2, 180–187 mg l−1 HCO3, 554–592 mg l−1 SO4, 0.4–33.8 mg l−1 H2S, < 1 mg l−1 NO3, and <0.005 mg l−1 Fe [31, 32]. Gas in the water column was primarily N2 (93.9%) with trace CO2 (4.2%) and methane (0.3%) [32]. Water cools as it traverses the marshland via the Epsilon and Alpha riverine outflows to about 17 °C at our final sampling location (Cool). In each pool, three replicate ~500 ml bulk samples of sediment were removed with a syringe pump into autoclaved 1 l Nalgene® bottles. Bottles were filled to the brim with spring water, and the pump was rinsed once with 70% ethanol and thrice with autoclaved distilled water between each sample. In addition, ~5 ml from each sampling location was placed in a Falcon® tube and immediately frozen on dry ice for DNA extraction.
DNA extraction, library preparation, and sequencing
DNA was extracted twice per sample, using 0.25 g sediment, soil, or biofilm, with NucleoSpin Soil kits (Machery-Nagel, Düren, Germany) then pooled. DNA quality was assessed with a 1% agarose gel and concentration was measured with Qubit dsDNA high-sensitivity assays (ThermoFisher, Waltham, USA). 1 ng of DNA from three replicates of selected sediment samples (Cool, Beta, Epsilon) was prepared for shotgun sequencing on one NextSeq550 (Illumina) run in High Output mode using NexteraXT library preparation (Illumina, San Diego, USA). Additional sampling and 16S rRNA gene sequencing are described in the Supplementary Methods.
Lignin incubations
2 g of sediment from Epsilon, the warmest undisturbed pool, was incubated in 5 ml of M9-Goodies minimal medium [33, 34] with 2 mg of vanillin (VAN), Eucalyptus milled-wood lignin (EMWL) (see Supplementary Methods), eucalyptus kraft lignin (EKL), or coniferyl alcohol dehydrogenase-polymer model lignin (DHP). Eucalyptus wood chips and EKL were provided by Suzano Canada. DHP was synthesized as in [35]. Control incubations were conducted with no exogenous carbon. All incubations took place in 50-ml sealed serum bottles at 45 °C and 150 RPM mixing in the dark. During incubations, substrate-induced respiration (SIR) was measured by adding 1 ml air to the vial headspace with a 1-ml glass syringe, mixing, and removing 1 ml headspace, which was manually injected into a 5890 Series II gas chromatograph (Agilent Technologies, Santa Clara, USA) equipped with a flame ionization detector and methanizer, and quantified against CO2 standards (Praxair, Danbury, USA). Three separate incubations for each substrate were destructively sampled at 0, 48, 96, and 148 h, except for VAN, which was sampled only at 24 h. At each time point, samples were centrifuged at 4 °C for 5 min at 18,000 × g, and the supernatant was stored at −80 °C prior to high-pressure liquid chromatography (HPLC). RNA was immediately extracted from the remaining sediment.
RNA extraction, library preparation, and sequencing
Approximately 0.5 g of sediment was separated into 2-ml screw-top microtubes containing 740 µl 0.1 M Na2PO4-NaH2PO4 pH 7.3, 60 µl 10% SDS and 800 µl 25:24:1 phenol:chloroform:isoamyl alcohol (Sigma Aldrich, St. Louis, USA), modified from [36]. RNases from thermophilic organisms are highly resistant to inhibition or denaturation. To recover high-quality RNA from thermophilic communities, 100 µl of 200 mM ribonucleoside vanadyl complex (RVC) (Sigma Aldrich) was added to the extraction buffer [37, 38]. RVC was essential for recovery of RNA from these samples (Supplementary Fig. 2A). Bead beating at 5.5 m/s for 30 s (twice) was followed by centrifugation for 5 min at 4 °C and 18,000 × g. The supernatant was added to 800 µl of chloroform:isoamyl alcohol (Sigma Aldrich), mixed by hand, and centrifuged again. The supernatant was purified using the RNeasy® Mini Kit (Qiagen), and DNA was removed using Turbo DNase (ThermoFisher). RNA concentration was measured using the Qubit™ RNA HS assay (ThermoFisher), and purified RNA stored at −80 °C for <1 week. RNA integrity was measured using an Agilent 2100 Bioanalyzer (Agilent Technologies) before and after rRNA removal with RiboMinus™ Transcriptome Isolation Kit, bacteria (ThermoFisher). Sequencing libraries were prepared using SuperScript™ Double-Stranded cDNA synthesis (ThermoFisher) and NexteraXT (Illumina). Twelve 100-bp paired-end libraries were sequenced per NextSeq550 (Illumina) High Output run.
Lignin analysis
Lignin substrates were analyzed using 13C nuclear magnetic resonance (NMR) and 2D heteronuclear single quantum coherence (HSQC) NMR analysis [39–41]. Full details are provided in Supplementary Methods.
Liquid chromatography
Monoaromatic compounds in incubation supernatants were analyzed by HPLC against 2,6-dimethoxy-1,4-benzoquinone, vanillic acid, syringic acid, VAN, and syringaldehyde standard curves obtained by injecting between 1 and 20 μM of authentic standards in 50 mM sodium phosphate. Full details are provided in Supplementary Methods.
Assembly of genomes from metagenomes
Shotgun metagenome libraries were quality filtered and adapter trimmed using Trimmomatic 0.38 with default settings [42] prior to MegaHit 1.1.3 co-assembly with “meta-large” presets [43]. Contig open reading frames (ORFs) were predicted using Prodigal 2.6.3 [44]. Taxonomy of each ORF was annotated using Kaiju 1.7.2 [45], which, in turn, was used to calculate contig consensus taxonomy if 50%+1 of contig ORFs shared taxonomic identity. Contig coverage was calculated with BBMap 38.22 (https://sourceforge.net/projects/bbmap/) prior to binning with Metabat2 [46]. CheckM 1.0.13 [47] was used to identify phylogenetic markers and calculate bin completeness, contamination, and heterogeneity. High-quality MAGs were manually refined (Supplementary Methods) and CheckM was re-run on the resulting refined genomes, followed by taxonomic classification and placement in whole-genome phylogenetic trees using GTDB-Tk [48] (github.com/Ecogenomics/GTDBTk). A metatranscriptome co-assembly was performed as above but using only 25-mers and binned without the support of coverage calculations.
Metabolic pathway annotation
Genes were annotated using a set of previously compiled Pfam and TIGRFAMs profile HMMs representing enzymes involved in biogeochemical cycling and energy metabolism [49–51], as well as Kofamscan profile HMMs [52]. Hits for ammonia oxidation (amoA) and nitrite oxidoreductase genes (nxrA) were validated by phylogenetic placement. Additional archaeal ammonia oxidation genes were identified using local alignment against Nitrososphaera viennensis EN76 amoABC [53]. Carbohydrate-active enzyme (CAZy) gene annotations were defined using dbCAN2 [54] with an E value of 1e−60. To facilitate the identification aromatic compound degradation and anoxygenic photosystem genes, custom thresholds were set for 25 KEGG HMMs (Supplementary Data 1, Supplementary Methods).
Calculating genomic abundance
Quality-filtered read files from metagenome and transcriptome libraries were aligned to the 39 refined genomes with BBMap. To capture potential biodiversity missed by the assembly approach, unmapped reads were subsequently aligned with 351 remaining contig bins and 24,668 reference genomes [48]. Genomes with >1% coverage were included in subsequent genomic analysis. To visualize the distribution of each genome across temperatures, feature scaling was used to normalize counts in each metagenome library between 0 and 1 for all genomes independently.
Statistical analysis
Comparison of OTU richness, assembly coverage, and genome richness across sequencing libraries was made using unpaired t-tests in R v3.6.2 [55] with a significance cutoff of p < 0.05. Headspace CO2 was also compared with unpaired t-tests. Genomic and transcriptomic bins were analyzed for transcription levels in incubations with DHP (48 h), EKL (96 h), EMWL (168 h), VAN (24 h), and NC (96 h), coinciding with sampling times when maximum RNA was recovered (Supplementary Fig. 2B). Transcript counts from metatranscriptome libraries were compared as transcripts per million with generalized linear models (GLM) and the multcomp Tukey-HSD implementation in R against NC controls. Differential gene expression was analyzed across metatranscriptomes using DESeq2 [56], using log2 fold change (L2FC) values, and a significance cutoff of padj < 0.05 following false-discovery rate correction.
Results
Influence of temperature on hot-spring microbial diversity
We first characterized community-level patterns across hot-spring samples using 16S rRNA amplicon sequences. Prokaryote community structure differed by sample type (sediment, biomat, soil) (p = 0.001) and temperature (p = 0.001) following PERMANOVA (Supplementary Fig. 3A). Shannon–Weiner diversity was negatively correlated with temperature (R2 = 0.73, p < 0.001) (Supplementary Fig. 3B). Generally, we saw lower 16S rRNA OTU richness (Fig. 1A) in warmer samples.
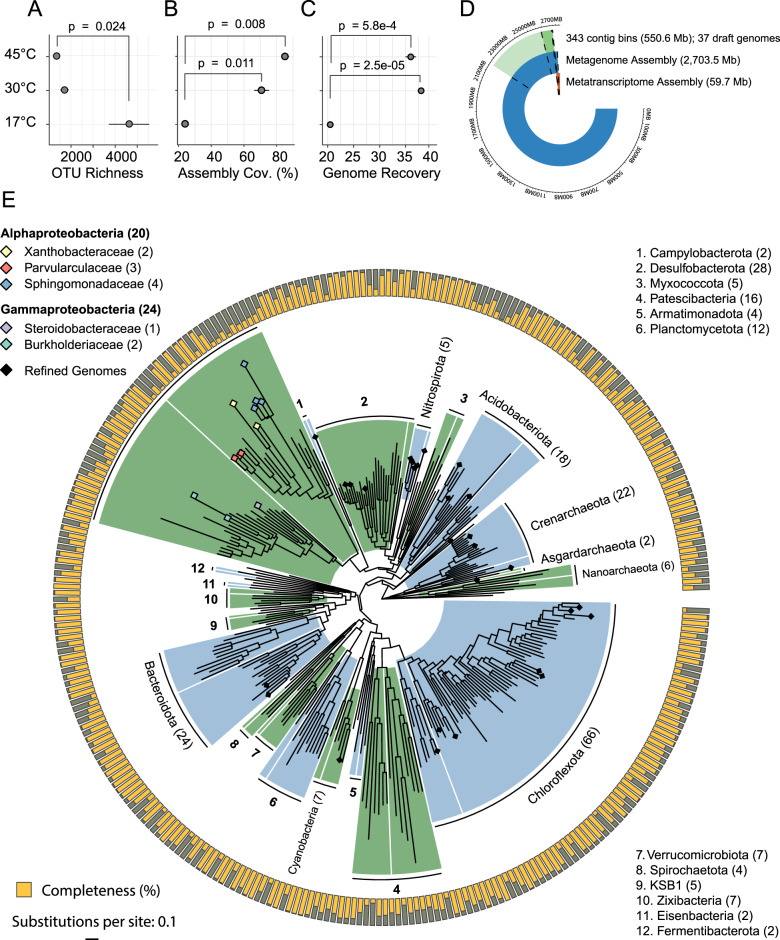
Fig. 1. Diversity, assembly, and phylogeny of a thermal swamp community.
A Richness of 16S rRNA gene OTUs in samples from Epsilon (EPS, 45 °C) and Beta (BETA, 30 °C) hot springs and from mesophilic sediment (COOL, 17 °C) (n = 3). B Assembly coverage across samples. C Number of recovered refined genomes across samples. D Length of the full metagenome assembly in Mb compared with metagenome-assembled genome (MAG) bins, refined (draft) genomes, and the metatranscriptome assembly. E Whole-genome phylogeny of the MAGs shaded by phylum. Number of bins in each phylum indicated in parentheses. The outer ring shows bin completeness assessed with CheckM 1.0.13. Tree scale represents substitutions per site. Bins belonging to proteobacterial taxa implicated in degradation of lignin-derived aromatic compounds are highlighted with shaded diamonds.
Assembly of genomes from thermal sediment metagenomes
We then sequenced nine metagenomic libraries, three each from two hot springs receiving woody carbon inputs (Epsilon (45 °C), Beta (30 °C)) and three from mesophilic riverine samples (Cool (17 °C)). In total, 168 million paired-end reads (41.65 gigSuzano abases (Gb)) passing quality filtering were assembled into 2.7 Gb of contigs (Supplementary Data 2). About 20% of the combined assembly was binned into 343 MAGs based on GC content, tetranucleotide frequency, sequencing coverage across samples, and presence of SCGs [50–52] (Fig. 1D). Metagenome assembly coverage was significantly higher for 30 °C (p = 0.011) and 45 °C samples (p = 0.008) than for the 17 °C samples following unpaired t-tests (Fig. 1B). All pools clustered distinctly following PCoA ordination (Supplementary Fig. 3C). Likewise, refined genome recovery was significantly greater for the 30 and 45 °C samples (p < 0.001) than for the 17 °C samples (Fig. 1C).
Phylogenetic distribution of assembled genomes
Sixty-six (19%) of MAGs were placed into phylum Chloroflexota (Fig. 1E and Supplementary Data 3), including families Roseiflexaceae (L.E.CH.5) and Chloroflexaceae (L.E.CH.39) (Fig. 2). Forty-four MAGs (13%) were placed into the Proteobacteria. In particular, two Burkholderiaceae and four Sphingomonadaceae MAGs were identified. Notably, two Asgardarchaeota MAGs were recovered, representing potentially novel genera in the class, Lokiarchaeia (Supplementary Data 3).
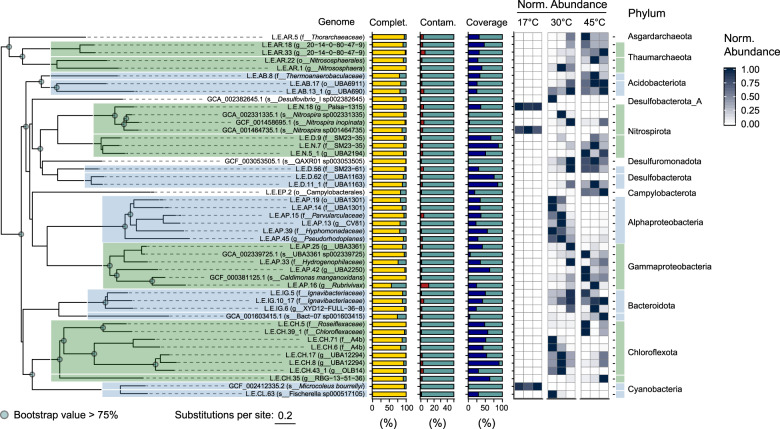
Fig. 2. Refined genome phylogeny, assembly statistics, and abundance in Epsilon (45 °C) and Beta (30 °C) hot springs and mesophilic sediment (17 °C) metagenomic libraries.
Whole-genome phylogeny assessed as in Fig. 1. Tree scale represents estimated site divergence and green dots indicate branching calculated with bootstrapped confidence > 75%. Completeness (yellow) and contamination (red) were estimated with CheckM 1.0.13. Genome abundance calculated by alignment of quality-filtered reads against refined genomes and GTDB reference genomes using BBMap 38.22 and retaining targets > 0.5% coverage (blue). Abundance counts normalized using feature scaling between 0 and 1 for each genome.
MAGs can contain erroneously binned sequences that can lead to faulty ecological interpretation. While MetaBat2 performs favorably compared to other binning algorithms [46], we nonetheless manually refined 39 MAGs with contamination < 5% and completeness > 80% (unless otherwise noted) into our final “refined genome” data set (Supplementary Fig. 4). Subsequent analyses focused on the refined genomes because: (1) they are taxonomically representative of the larger MAG data set; (2) many MAGs have incomplete or erroneous assembly; and (3) the refined genomes are the most abundant in the metagenome libraries based on read coverage. Only one Burkholderiaceae genome (Rubrivivax sp. L.E.AP.16) was included in the refined genome pool, despite a completion estimate of 56%, due to its unique metabolic capacity and possible thermotolerance. Sphingomonadaceae MAGs had a mean completeness of only 28%, and none were included in the analysis of refined genome.
Environmental distribution of refined genomes
The temperature at which each refined genome was found to have the highest relative abundance was used to infer their potential thermotolerance. We quantified the relative abundance of the refined genomes across a thermal gradient by aligning quality-filtered reads from shotgun metagenome libraries to refined genomes. We additionally assessed genomes potentially missed by our approach by aligning remaining reads against a collection of species-level reference genomes. Both refined and reference genomes were used to evaluate relative abundance and diversity across thermal samples. Comammox Nitrospira genomes were abundant at 17 °C (see Supplementary Figs. 5 and 6 for Amo and Nxr phylogenies), highlighting the oligotrophy of this environment. Twelve of 41 genomes used in this analysis were most abundant at 30 °C (Fig. 2). These included many of the Proteobacteria, excluding L.E.AP.16, and the manganese-oxidizing thermophile, Caldimonas manganoxidans [57]. Two of the most abundant genomes at 45 °C were the Chloroflexota L.E.CH.5 and L.E.CH.39, at 9.8% ± 10.9% and 7.1% ± 7.6%, respectively.
Inferring genome metabolic potential across temperature ranges
The metabolic capacity of each genome was inferred using profile HMMs from select Pfam [50], TIGRFAMs [49, 51], CAZy [58], and KEGG [52] protein families (Supplementary Data 1), as well as local alignment against the UniRef90 database [59]. Metabolic pathways were positively annotated when they contained essential enzymes such as monooxygenases or dioxygenases, and also contained at least 50%+1 of all genes identified using the above pHMMs (Fig. 3). Gene-level annotation is provided in Supplementary Fig. 7 and Supplementary Data 4.
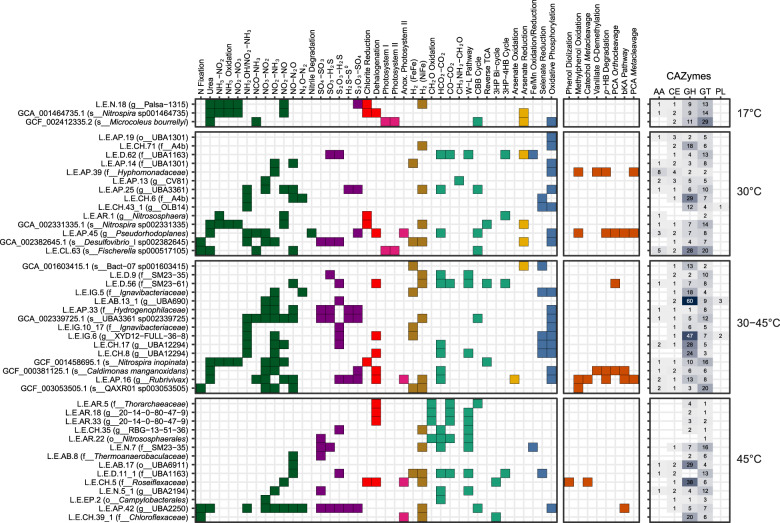
Fig. 3. Genome functional annotation.
Annotation of key genes and pathways involved in energy and carbon metabolism used kofamscan 1.1.0 profile hidden Markov models (HMMs) and select PFAM and TIGRFAM HMMs. Nitrogen cycling genes are shown in green, sulfur cycling in purple, dehalogenation in red, photosynthesis in pink, H2-oxidation in gold, carbon fixation in teal, arsenate metabolism in yellow, oxidative phosphorylation in blue, and aromatic degradation in orange. Number of CAZymes in each class were determined using dbCAN2. W–L Wood–Ljungdahl, 3HP 3-hydroxypropionate, 4-HB 4-hydroxybutyrate, p-HB p-hydroxybenzoate, PCA protocatechuate, bKa beta-ketoadipate, GH glycoside hydrolases, GT glycosyltransferases, PL polysaccharide lyases, CE carbohydrate esterases, AA auxiliary activities.
Multiple refined genomes were found only at 30 or 45 °C, while 14 were similarly abundant in both pools (Fig. 2). The genomes from three temperature categories (30, 45, and 30–45 °C) encoded similar biogeochemical cycling capacity, with differences in the taxonomy of microorganisms involved in nitrogen, sulfur, hydrogen, and halogen metabolism (Figs. 3 and 4). At 30 °C, a phylogenetically diverse group of genomes encoded transformation of nitrogen, including a cyanobacterium, Fischerella sp. (L.E.CL.63), which encoded capacity for both N-fixation and urea mineralization. A Nitrososphaera genome (L.E.AR.1), containing a full complement of archaeal ammonia-monooxygenase genes (Fig. 3 and Supplementary Figs. 5 and 6), was maximally abundant at 30 °C. The abundance of Comammox Nitrospira sp. and group I.1b Thaumarchaeota ammonia-oxidizing archaea indicates highly oligotrophic conditions, i.e., [NH3 + NH4+] < 5 µM [7]. The capacity for dissimilatory nitrogen respiration was encoded by two alphaproteobacterial genomes, L.E.AP.45 and L.E.AP.39, which also contained pathways for aerobic aromatic catabolism via meta- and ortho-cleavage of protocatechuate (Fig. 4), indicating the potential for facultative anaerobic respiration and aerobic biomass decomposition.
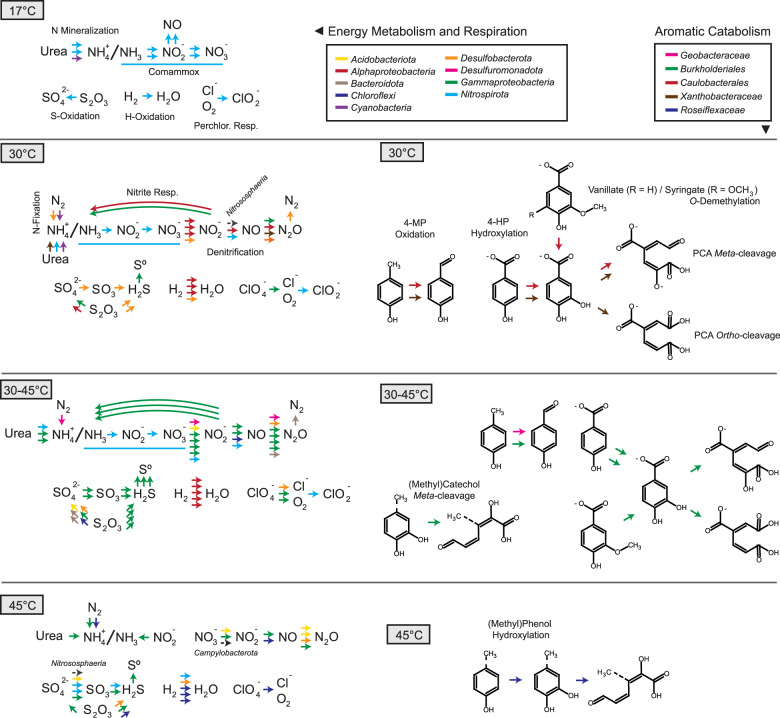
Fig. 4.
Predicted energy metabolism, respiration, and aromatic degradation pathways encoded in thermal swamp genomes shown in Fig. 3.
The genomes that showed equivalent abundance at 30 and 45 °C (30–45 °C) may represent thermotolerant organisms. Burkholderiales genomes of L.E.AP.16 (Rubrivivax sp.) and C. manganoxidans encoded aromatic degradation capacity though the catechol meta-cleavage and protocatechuate meta- and ortho-cleavage pathways (Figs. 3 and 4). The capacity for vanillate degradation was encoded in two-component vanillate O-demethylase (vanAB) in C. manganoxidans, rather than in the tetrahydrofolate-dependent vanillate/syringate O-demethylase (ligM/desA) found in a Caulobacterium genome from 30 °C. The vanAB genes were not detected in the L.E.AP.16 genome, despite expression of their homologs in Rubrivivax transcriptomes (see below). The Burkholderiales genomes abundant in the 30–45 °C range also showed substantial carbohydrate degradation capacity based on the presence of high numbers of glycoside hydrolases (GHs) (Fig. 4 and Supplementary Fig. 8).
Genomes found only 45 °C may represent moderate thermophiles. The Chloroflexota genome L.E.CH.5 encoded the ability to hydroxylate alkylated phenols with an actinobacterial-like two-component monooxygenase [16], in addition to catechol meta-cleavage. In total, thermotolerant and thermophilic Burkholderiales genomes encoded a large portion of the biogeochemical cycling and aromatic degradation at elevated temperatures, with a range of possible electron donors and acceptors allowing substantial metabolic flexibility.
Lignin incubations
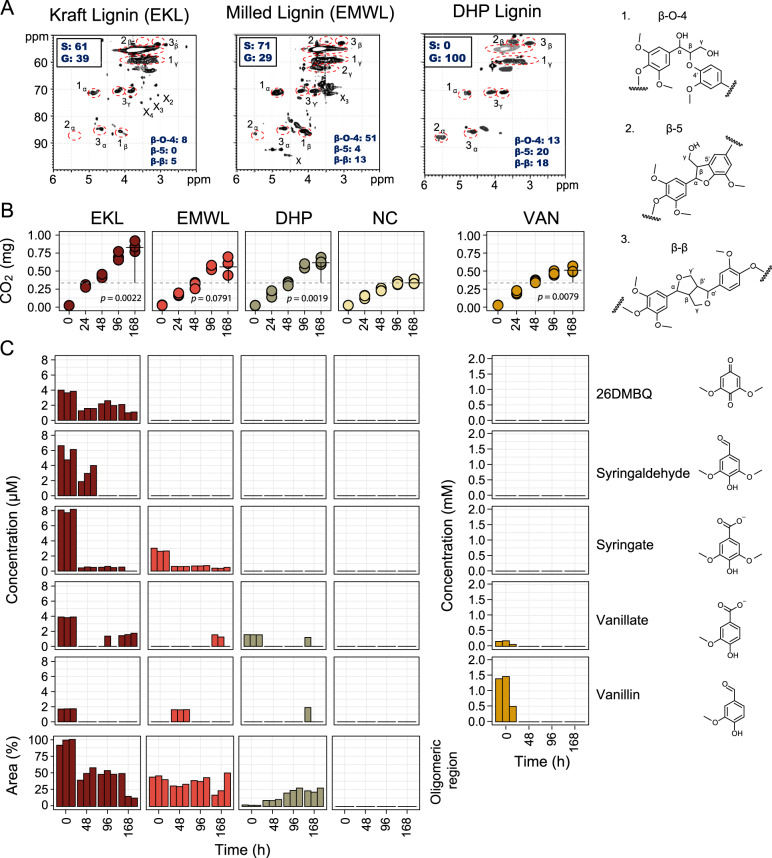
To characterize thermotolerant modification of lignin and the catabolism of lignin-derived aromatic compounds, we incubated Epsilon pool (45 °C) sediment with one of three preparations of lignin (EMWL, EKL, and DHP lignin), VAN, or a no-carbon control. Epsilon sediment was chosen as it was the warmest undisturbed sampling location. 2D HSQC NMR analysis revealed that EMWL, generated by extracting lignin from enzymatically treated, milled Eucalyptus wood chips, was much less modified than EKL, generated by the Kraft process (Fig. 5A). Specifically, EMWL contained 51 β-aryl ether bonds per 100 aromatic subunits, while EKL contained only eight. Further, β-5 bonds were detected in EMWL but not EKL, and EMWL contained a higher S:G ratio (7:3) than EKL (6:4). EMWL, and to a lesser extent EKL, contained residual carbohydrates (annotated with an “X” in Fig. 5A). The synthetic DHP lignin contained a higher β-5 ratio than EMWL and, consistent with its synthesis, no carbohydrate and only G-lignin units. Finally, EMWL and EKL contained small amounts of monoaromatic compounds, such as vanillate and syringate, as well as aromatic oligomers, while DHP lignin was relatively free of these compounds. EKL specifically contained about three times as much S-lignin-derived monoaromatics than G-lignin-derived compounds.
Fig. 5. Lignin structure and aromatic compound concentrations in Epsilon pool sediment incubations.
A 2D HSQC NMR spectra of initial lignin substrates showing bond types and proportions. Values for bond types and syringyl (S) and guaiacyl (G) subunits (13C NMR) are normalized per 100 aromatic units. Examples of each bond type are provided (1–3). B CO2 evolution in incubation vial headspace. Total CO2 compared with no exogenous carbon (NC) controls at 168 h using Student’s t test. C Concentrations of five monoaromatic compounds in triplicate incubations over 168 h during incubation experiments. Bars representing lignin-derived aromatic oligomers scaled to percent of max peak area. 26DMBQ 2,6-dimethoxy-1,4-benzoquinone, VAN vanillin.
SIR during lignin incubations was monitored by quantifying CO2 in the headspace. CO2 production significantly increased by 168 h in the presence of EKL (240%, p = 0.0022), EMWL (170%, p = 0.079), DHP (180%, p = 0.0019), and VAN (150%, p = 0.0079) relative to NC control incubations (Fig. 5B). The aromatic constituents of the incubation supernatant were analyzed with HPLC. At time zero, we detected monoaromatic compounds in the lignin-amended samples, and a large peak provisionally assigned to aromatic oligomers based on compound size. The area of this large peak decreased ~50% by 48 h. In EKL, syringate concentrations were reduced by 94% after 48 h, syringaldehyde disappeared from the supernatant after 96 h, and VAN was completely removed after 48 h.
The initial concentrations of monoaromatic compounds in lignin-amended samples (t = 0) were in the low µM range. As this is much lower than the concentration of VAN (~1.4 mM), their presence alone does not explain the measured respiration, as the lignins induced more CO2 production than VAN. As SIR is a very broad measure of metabolic activity, we also cannot rule out the impact of the potential toxicity of VAN [60] on the CO2 production, or the possible effect of added lignins on the degradation of endogenous carbon. Vanillate and VAN were only detected after several days of incubation with EMWL and DHP, consistent with depolymerization. However, further analysis of lignin substrates would be required to confirm the possibility of depolymerization. Such analysis was not possible in the current study as the samples were sacrificed to obtain RNA yields suitable for metatranscriptomics analysis described in the next section.
Metatranscriptomics of lignin-incubated thermal sediment
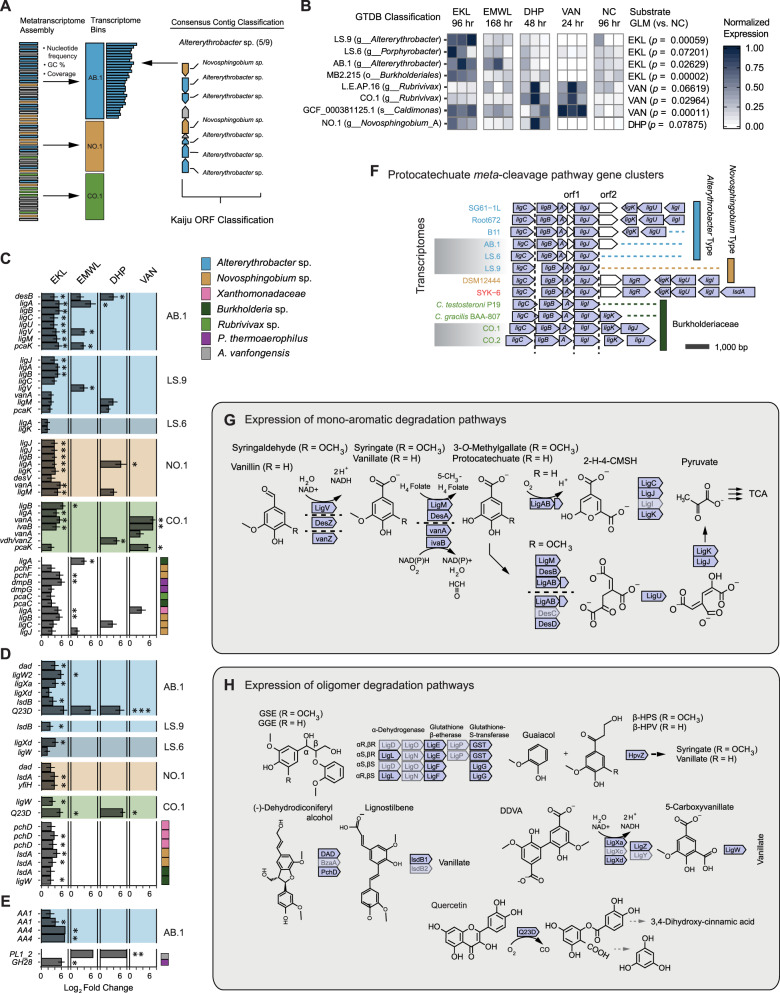
To understand how thermotolerant communities can modify lignin, and to identify mineralization pathways for LMW aromatic compounds, we sequenced samples from all incubation time points with >100 ng total RNA. Transcriptome contig taxonomy was assessed with Kaiju (“Material and methods”), as well as analysis of SGCs in MetaBat2 “transcriptomes” (Fig. 6A). Taxonomically classified transcriptomes were added to the refined and reference genomes for read alignment and quantification as above. Transcriptomic reads mapped to 37 genomes and 6 transcriptomes. GLM analysis with Tukey post hoc testing indicated that four bins had higher normalized read-mapping rates in EKL relative to controls (Fig. 6B). These included two Altererythrobacter transcriptomes, LS.9 and AB.1, and one Burkholderiales genome, MB2.215. Two Burkholderiales showed higher overall gene expression in VAN incubations: the Rubrivivax transcriptome CO.1, and the C. manganoxidans genome.
Fig. 6. Metatranscriptomics of hot-spring sediment incubated with 0.02% (m/v) DHP, eucalyptus kraft lignin (EKL), Eucalyptus milled-wood lignin (EMWL), vanillin (VAN), and control incubations with no exogenous carbon (NC).
A Strategies for taxonomic identification and binning of metatranscriptome contigs. B BBMap read mapping to MAGS (MB2), refined genomes (L.E.), GTDB reference genomes (GC), and binned transcriptome assemblies (AB, CO, LS). Mapped read counts per bin were normalized using length of the contig assembly and depth of sampling. p values generated from the multcomp implementation of Tukey-HSD pairwise-GLM analysis of normalized counts for lignin substrate shotgun metatranscriptome libraries against NC libraries (p < 0.05 shown). C Transcripts putatively involved in catabolic pathways for methoxylated monoaromatics (vanillate/syringate). Taxonomy determined using Kaiju and GTDB-Tk classification of assembled transcriptomes as shown in (A). Results of DESeq2 differential expression (DE) analysis against NC libraries provided as log2 fold change values if (padj < 0.1). Transcripts that are differentially expressed with padj < 0.05 are denoted with an asterisk (*). D Transcripts putatively involved in aromatic oligomer degradation. E CAZyme transcripts. F Proteobacterial protocatechuate meta-cleavage (lig) gene clusters identified in genome and transcriptome bins. Clusters are aligned to the start position of the ligJ/I reading frame. G Vanillate and syringate meta-cleavage by des, van, and lig gene products in the eucalyptus kraft lignin (EKL) transcriptomes. H Degradation of guaiacylglycerol-β-guaiacyl ether (GGE), 5,5′-dehydrodivanillate (DDVA), and other oligomeric compounds. Lightened genes were not detected in transcriptome.
Differential expression analysis was performed with DESeq2 to determine transcript-level expression patterns during lignin incubations (Fig. 6C–E). Twenty-six transcripts in aromatic degradation pathways were found to be significantly upregulated on EKL using a cutoff of padj < 0.05, compared to only nine in EMWL and three in VAN incubations (Fig. 6C). Several upregulated transcripts were identified as homologs of syringate or vanillate degradation pathway genes (Fig. 6G). Phylogenetic analysis (Supplementary Figs. 9–14) and syntenic conservation (Fig. 6F) of the protocatechuate meta-cleavage pathway genes were assessed to validate these assignments. Transcripts assigned to this pathway were found in AB.1, LS.6, LS.9, NO.1, CO.1, and CO.2 transcriptomes (Supplementary Data 5).
The protein phylogenies derived from metagenomic and metatranscriptomic sequences for vanillate O-demethylase (LigM), syringate O-demethylase (DesA), protocatechuate 4,5-dioxygenase α (LigA) and β (LigB) subunits were assessed by sequence alignment and phylogenetic tree estimation (Supplementary Figs. 9–14). The AB.1 and LS.9 bins encoded all of these enzymes, except LigM, which was only encoded in AB.1. Differences existed between these two bins, despite both being classified as Altererythrobacter on the basis of SCG phylogeny. The two bins contained identical desA genes; however, LS.9 encoded protocatechuate 4,5-dioxygenase subunits that clustered with those of SYK-6, while AB.1 encoded homologs that clustered with those of Altererythrobacter. Furthermore, the presence of an uncharacterized ORF directly upstream of the ligA was detected in only Altererythrobacter reference genomes and AB.1, while the corresponding genes in LS.9 were syntenous to those of SYK-6 and N. aromaticivorans (Fig. 6F).
The sphingomonads SYK-6 and N. aromaticivorans have been widely investigated for their ability to depolymerize model lignin compounds [61, 62]. We identified potential homologous depolymerization systems in moderately thermophilic Altererythrobacter and Novosphingobium transcriptomes (Fig. 6D). Two putative Altererythrobacter ligX homologs (L2FC = 4.1, padj = 0.03; L2FC = 2.2, padj = 0.06) and one putative ligW (L2FC = 5.8, padj = 0.02) implicated in the catabolism of the methoxylated biphenyl compound 5,5′-dehydrodivanillate (DDVA) were upregulated on EKL after 96 h. The NO.1 and LS.9 transcripts encoding ligW sequences clustered with SYK-6, Novosphingobium, and Altererythrobacter homologs following phylogenetic placement (Supplementary Fig. 13). However, ligXc and ligY homologs were not identified (Fig. 6H). Several glutathione S-transferases (GSTs) were also upregulated in the EKL metatranscriptome. While GSTs can catalyze diverse reactions, when added to an existing phylogeny [63], these sequences were identified as encoding stereospecific β-etherases (ligE, ligF1), Omega-class (ligG), or Nu-class (GST3) GST homologs (Supplementary Fig. 15). Two hpvZ sequences implicated in the carboxylation of hydroxypropiovanillone or hydroxypropiosyringone were expressed, and several transcripts possibly involved in dehydrodiconiferyl alcohol degradation (encoding DAD, phcD, lsdB) were identified in EKL and EMWL metatranscriptomes. A putative quercetin 2,3-dioxygenase (Q23D) was also upregulated on EKL (L2FC = 6.6, padj = 0.0002).
Finally, few CAZymes showed significant upregulation on lignins (Fig. 6E). Of these, two Altererythrobacter AA1 transcripts were classified by the laccase engineering database [64] as 3-domain CopA-type laccase-like multicopper oxidases. Two AA4 vanillyl-alcohol oxidases in the Altererythrobacter transcriptome were upregulated on EKL (L2FC = 7.0, padj = 0.03; L2FC = 6.9, padj = 0.00003) and EMWL (L2FC = 9.3, padj = 0.01). Two putative pectinases were upregulated on EKL (GH28: L2FC = 6.0, padj = 3.23e−08), EMWL (PL1_2: L2FC = 6.6, padj = 0.02), and DHP (PL1_2: L2FC = 7.9, padj = 0.003).
Discussion
Thermotolerant Alphaproteobacteria and Gammaproteobacteria from a geothermal environment expressed aromatic degradation pathways for lignin-derived monoaromatic compounds and aromatic oligomers in incubation with lignin. This finding is highly relevant to understanding the ecology of carbon cycling in geothermal environments, and has substantial biotechnology implications. In this study, two Altererythrobacter ssp. transcriptomes expressed complete aromatic monomer and oligomer degradation pathways during incubation with lignin (Fig. 6), despite not being fully reconstructed by genomic analysis. The syringate meta-cleavage pathway was primary expressed on EKL, which contained about three times higher concentration of dimethoxylated S-lignin-derived substrates than G-lignin monomers. Significantly elevated expression of ligAB in the Altererythrobacter ssp. transcriptomes suggests that syringate meta-cleavage in these strains proceeds via 3-O-methylgallate. Expression of desB in EKL, EMWL, and DHP also suggests that some syringate degradation occurs via gallate oxidation. Strain-level characterization of Altererythrobacter ssp. should resolve the relative contributions of these pathways to syringate catabolism, and characterization of the thermotolerance of these enzymes may provide new targets for biological upgrading of lignin-derived compounds.
No clear extracellular lignin-depolymerization systems were expressed in the metatranscriptomes, consistent with the apparent low mineralization of intact lignins. However, transcripts of two CopA-type LCMOs and Q23D were present in the Altererythrobacter transcripts. Although CopA-type LCMOs are predicted to play a role in copper detoxification, several of these enzymes have shown lignin oxidation activity [65, 66]. Similarly, a Q23D was associated with lignin depolymerization in Pseudomonas putida KT2440 [64], but the physiological role and substrate range of these enzymes remain uncharacterized. Finally, Altererythrobacter expressed genes associated with the intracellular degradation of the lignin-derived oligomers, e.g., guaiacylglycerol β-guaiacyl ether and DDVA. To date, aromatic oligomer depolymerization pathways have been identified only in the widely studied sphingomonads, SYK-6, and N. aromaticivorans DSM 12446 [19, 20, 61, 62, 67–69]. Altererythrobacter ssp. may also use Nu-class GSTs to break β-O-4 bonds [62]. As in other sphingomonads, aryl O-demethylation may supply essential C1 metabolites in Altererythrobacter strains, based on the prevalence of expressed tetrahydrofolate-dependent O-demethylases during lignin degradation [70]. Altererythrobacter spp. are typically associated with oligotrophic environments such as deep-sea sediments [71] and 45 °C hot springs [72]. Our metatranscriptomic experiment provides evidence that Altererythrobacter spp. catabolizes lignin-derived monoaromatic compounds via syringate and protocatechuate meta-cleavage pathways, and lignin-derived aromatic oligomers through several pathways including GST β-etherases.
Rubrivivax ssp. have diverse mechanisms to generate energy and utilize carbon including vanillate O-demethylation and protocatechuate meta-cleavage. Based on genome analyses, these thermotolerant organisms have the potential for facultative photoheterotophic growth, denitrification, CO-oxidation, H2-metabolism, and degradation of a variety of aromatic compounds [73–76]. Vanillate O-demethylase (vanA-ivaB) and protocatechuate 4,5-dioxygenase (ligAB) transcripts associated with Rubrivivax ssp. significantly increased in samples incubated with EKL or VAN. The Rubrivivax VanA had an amino acid sequence identity of 74.7% and 89.5%, respectively, to the VanA Rieske-type monooxygenases of Comamonas testosteroni and Curvibacter delicatus. A putative oxidoreductase encoded in the Rubrivivax transcriptome had 82.9% identity to IvaB of C. testosteroni, C. testosteroni VanA O-demethylates vanillate and veratrate to protocatechuate and isovanillate, respectively [77].
Genes encoding aromatic catabolic pathways, including ring-cleaving dioxygenases and GSTs, are associated with genomic adaptation to oligotrophic environments [12]. These enzymes may serve to supply carbon and energy, or carry out detoxification reactions. While we assembled protocatechuate dioxygenases in both 30 and 45 °C pools, the genes encoding ortho- and meta-cleavage enzymes were, respectively, encoded in Alphaproteobacteria (caulobacteria, Xanthobacteraceae) and Burkholderiales at each temperature. Additionally, the putative catabolism of alkylated phenols was encoded in thermophilic Chloroflexota. This is the first report of homologs for alkylated phenol hydroxylation encoded in Chloroflexota genomes, although catechol meta-cleavage potential was previously reported for pelagic Chloroflexota genomes [78]. These data demonstrate that biochemical transformations may be encoded by phylogenetically distinct organisms adapted to specific temperatures. The abundant Burkholderiales described herein encode multiple protocatechuate cleavage pathways and diverse energy generation mechanisms, and appear to be adapted to these highly oligotrophic geothermal environments. However, our metatranscriptomics data indicate that syringate and protocatechuate meta-cleavage pathways are the primary mechanisms for aromatic catabolism in these thermal swamp communities.
In this study, we demonstrate that genomic reconstruction could only partially distinguish aromatic degradation pathways, which co-occurred with pathways for facultative-photoautotrophy, halogen respiration, and arsenate detoxification in refined Burkholderiales (Rubrivivax sp.) and Chloroflexota (Roseiflexaceae) genomes. However, the targeted combination of metatranscriptomics and metabolite analysis indicated that significantly elevated expression of Rubrivivax sp. and sphingomonad (Altererythrobacter sp., Novosphingobium sp.) aromatic degradation pathways were associated with removal of lignin-derived substrates at 45 °C. These results supported our hypothesis that thermal swamp communities contain untapped sources of thermotolerant enzymes for lignin valorization, including copper oxidases, GSTs, and Q23Ds. The apparent preference for G- and S-lignin substrates between strains in families Sphingomonadaceae and Comamonadaceae, respectively, has implications for the use of these organisms in upgrading lignin-derived compounds. Additional approaches to characterizing hot-spring microorganisms, including isolation of identified strains, and characterization of putative lignin-depolymerization enzymes, will be critical to improve our understanding of the contribution of thermotolerant bacteria to degradation of lignin-derived aromatic compounds.
Supplementary information
Acknowledgements
This study was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (STPGP 506595–17) and a research contract from Genome BC (SIP004). LDE is the recipient of Canada Research Chair. Sequencing was performed by Microbiome Insights (Vancouver, CAN) and The University of British Columbia Sequencing + Bioinformatics Consortium (Vancouver, CAN). We thank Raymond Ma and Suzano Canada Inc. for supplying eucalyptus kraft lignin and eucalyptus wood chips. We thank Dr. Jason Grigg (The University of British Columbia) for assistance with preliminary LC-MS/MS analysis, Dr. Morgan Fetherolf (The University of British Columbia) for DHP synthesis, Dr. Peter Dunfield (University of Calgary), Dr. Stephen Grasby (Natural Resources Canada) for assistance with sampling, and Al Hansen (British Columbia Parks) for access to Liard River Hot Springs and assistance with sampling.
Author contributions
DJLB designed and implemented field sampling and experiments, performed genomic and transcriptomics analysis, and wrote the paper. AH performed metabolic pathway annotation and edited the paper. RR performed HPLC metabolite analysis and edited the paper. LYL and SR performed lignin chemistry analysis. LDE designed experiments and edited the paper. WWM designed experiments and co-wrote the paper.
Data availability
Sequence accessions are provided in Supplementary Data 2 and as part of NCBI BioProject PRJNA564648 (SRR10095339–SRR10095374). Refined genome information and NCBI accessions are provided in Supplementary Data 3.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-020-00820-x) contains supplementary material, which is available to authorized users.
References
- 1.Gonsior M, Hertkorn N, Hinman N, Dvorski SE-M, Harir M, Cooper WJ, et al. Yellowstone Hot Springs are organic chemodiversity hot spots. Sci Rep. 2018;8:1–13. doi: 10.1038/s41598-018-32593-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Channing A, Edwards D, Sturtevant S. A geothermally influenced wetland containing unconsolidated geochemical sediments. Can J Earth Sci. 2004;41:809–27. [Google Scholar]
- 3.Channing A. A review of active hot-spring analogues of Rhynie: environments, habitats and ecosystems. Philos Trans R Soc Lond B Biol Sci. 2018;5:373. doi: 10.1098/rstb.2016.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Owen RB, Renaut RW, Jones B. Geothermal diatoms: a comparative study of floras in hot spring systems of Iceland, New Zealand, and Kenya. Hydrobiologia. 2008;610:175–92. [Google Scholar]
- 5.Wächtershäuser G. From volcanic origins of chemoautotrophic life to Bacteria, Archaea and Eukarya. Philos Trans R Soc Lond B Biol Sci. 2006;361:1787–808. doi: 10.1098/rstb.2006.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eloe-Fadrosh EA, Paez-Espino D, Jarett J, Dunfield PF, Hedlund BP, Dekas AE, et al. Global metagenomic survey reveals a new bacterial candidate phylum in geothermal springs. Nat Commun. 2016;7:1–10. doi: 10.1038/ncomms10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kits KD, Sedlacek CJ, Lebedeva EV, Han P, Bulaev A, Pjevac P, et al. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature. 2017;549:269–72. doi: 10.1038/nature23679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa E, Pérez J, Kreft J-U. Why is metabolic labour divided in nitrification? Trends Microbiol. 2006;14:213–9. doi: 10.1016/j.tim.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Palatinszky M, Herbold C, Jehmlich N, Pogoda M, Han P, Bergen MV, et al. Cyanate as an energy source for nitrifiers. Nature. 2015;524:105–8. doi: 10.1038/nature14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parshina SN, Kijlstra S, Henstra AM, Sipma J, Plugge CM, Stams AJM. Carbon monoxide conversion by thermophilic sulfate-reducing bacteria in pure culture and in co-culture with Carboxydothermus hydrogenoformans. Appl Microbiol Biotechnol. 2005;68:390–6. doi: 10.1007/s00253-004-1878-x. [DOI] [PubMed] [Google Scholar]
- 11.Brady AL, Sharp CE, Grasby SE, Dunfield PF. Anaerobic carboxydotrophic bacteria in geothermal springs identified using stable isotope probing. Front Microbiol. 2015;6:897. doi: 10.3389/fmicb.2015.00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S, et al. The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA. 2009;106:15527–33. doi: 10.1073/pnas.0903507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceballos SJ, Yu C, Claypool JT, Singer SW, Simmons BA, Thelen MP, et al. Development and characterization of a thermophilic, lignin degrading microbiota. Process Biochem. 2017;63:193–203. [Google Scholar]
- 14.Fernandes TAR, da Silveira WB, Passos FML, Zucchi TD. Characterization of a thermotolerant laccase produced by Streptomyces sp. SB086. Ann Microbiol. 2014;64:1363–99. [Google Scholar]
- 15.Taylor CR, Hardiman EM, Ahmad M, Sainsbury PD, Norris PR, Bugg TDH. Isolation of bacterial strains able to metabolize lignin from screening of environmental samples. J Appl Microbiol. 2012;113:521–30. doi: 10.1111/j.1365-2672.2012.05352.x. [DOI] [PubMed] [Google Scholar]
- 16.Levy-Booth DJ, Fetherolf MM, Stewart GR, Liu J, Eltis LD, Mohn WW. Catabolism of alkylphenols in Rhodococcus via a meta-cleavage pathway associated with genomic islands. Front Microbiol. 2019;10:1862. doi: 10.3389/fmicb.2019.01862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez JM, Kontur WS, Alherech M, Coplien J, Karlen SD, Stahl SS, et al. Funneling aromatic products of chemically depolymerized lignin into 2-pyrone-4-6-dicarboxylic acid with Novosphingobium aromaticivorans. Green Chem. 2019;21:1340–50. [Google Scholar]
- 18.Beckham GT, Johnson CW, Karp EM, Salvachúa D, Vardon DR. Opportunities and challenges in biological lignin valorization. Curr Opin Biotechnol. 2016;42:40–53. doi: 10.1016/j.copbio.2016.02.030. [DOI] [PubMed] [Google Scholar]
- 19.Masai E, Kamimura N, Kasai D, Oguchi A, Ankai A, Fukui S, et al. Complete genome sequence of Sphingobium sp. strain SYK-6, a degrader of lignin-derived biaryls and monoaryls. J Bacteriol. 2012;194:534–5. doi: 10.1128/JB.06254-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cecil JH, Garcia DC, Giannone RJ, Michener JK. Rapid, parallel identification of catabolism pathways of lignin-derived aromatic compounds in Novosphingobium aromaticivorans. Appl Environ Microbiol. 2018;84:e01185–18.. doi: 10.1128/AEM.01185-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tasaki M, Kamagata Y, Nakamura K, Mikami E. Utilization of methoxylated benzoates and formation of intermediates by Desulfotomaculum thermobenzoicum in the presence or absence of sulfate. Arch Microbiol. 1992;157:209–12. doi: 10.1007/BF00245151. [DOI] [PubMed] [Google Scholar]
- 22.Kato S, Chino K, Kamimura N, Masai E, Yumoto I, Kamagata Y. Methanogenic degradation of lignin-derived monoaromatic compounds by microbial enrichments from rice paddy field soil. Sci Rep. 2015;5:1–11. doi: 10.1038/srep14295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daniel SL, Keith ES, Yang H, Lin Y-S, Drake HL. Utilization of methoxylated aromatic compounds by the acetogen Clostridium thermoaceticum: Expression and specificity of the co-dependent O-demethylating activity. Biochem Biophys Res Commun. 1991;180:416–22. doi: 10.1016/s0006-291x(05)81309-9. [DOI] [PubMed] [Google Scholar]
- 24.Daniel SL, Wu Z, Drake HL. Growth of thermophilic acetogenic bacteria on methoxylated aromatic acids. FEMS Microbiol Lett. 1988;52:25–8. [Google Scholar]
- 25.Kasai D, Masai E, Miyauchi K, Katayama Y, Fukuda M. Characterization of the gallate dioxygenase gene: three distinct ring cleavage dioxygenases are involved in syringate degradation by Sphingomonas paucimobilis SYK-6. J Bacteriol. 2005;187:5067–74. doi: 10.1128/JB.187.15.5067-5074.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasai D, Masai E, Katayama Y, Fukuda M. Degradation of 3-O-methylgallate in Sphingomonas paucimobilis SYK-6 by pathways involving protocatechuate 4,5-dioxygenase. FEMS Microbiol Lett. 2007;274:323–8. doi: 10.1111/j.1574-6968.2007.00855.x. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Yang Q, Li J, Gao H, Li P, Zhou H. The impact of temperature on microbial diversity and AOA activity in the Tengchong Geothermal Field, China. Sci Rep. 2015;5:17056. doi: 10.1038/srep17056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nottingham AT, Fierer N, Turner BL, Whitaker J, Ostle NJ, McNamara NP, et al. Microbes follow Humboldt: temperature drives plant and soil microbial diversity patterns from the Amazon to the Andes. Ecology. 2018;99:2455–66. doi: 10.1002/ecy.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharp CE, Brady AL, Sharp GH, Grasby SE, Stott MB, Dunfield PF. Humboldt’s spa: microbial diversity is controlled by temperature in geothermal environments. ISME J. 2014;8:1166–74. doi: 10.1038/ismej.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heron J, Sheffield C. First Canadian record of the water mite Thermacarus nevadensis Marshall, 1928 (Arachnida: Acariformes: Hydrachnidiae: Thermacaridae) from hot springs in British Columbia. Biodivers Data J. 2016;4:e9550. doi: 10.3897/BDJ.4.e9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grasby SE, Hutcheon I, Krouse HR. The influence of water–rock interaction on the chemistry of thermal springs in western Canada. Appl Geochem. 2000;15:439–54. [Google Scholar]
- 32.Grasby SE, Ferguson G, Brady A, Sharp C, Dunfield P, McMechan M. Deep groundwater circulation and associated methane leakage in the northern Canadian Rocky Mountains. Appl Geochem. 2016;68:10–18. [Google Scholar]
- 33.Bauchop T, Elsden SR. The growth of micro-organisms in relation to their energy supply. Microbiol. 1960;23:457–69. doi: 10.1099/00221287-23-3-457. [DOI] [PubMed] [Google Scholar]
- 34.Elder R. Cloning Techniques. BioScience. 1983;33:721–2. https://www.jstor.org/stable/1309366.
- 35.Wilhelm RC, Singh R, Eltis LD, Mohn WW. Bacterial contributions to delignification and lignocellulose degradation in forest soils with metagenomic and quantitative stable isotope probing. ISME J. 2018;13:413–29. doi: 10.1038/s41396-018-0279-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffiths RI, Whiteley AS, O’Donnell AG, Bailey MJ. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol. 2000;66:5488–91. doi: 10.1128/aem.66.12.5488-5491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo XZ, Stevens SE. Isolation of full-length RNA from a thermophilic cyanobacterium. BioTechniques. 1997;23:904–10. doi: 10.2144/97235st08. [DOI] [PubMed] [Google Scholar]
- 38.Shieh T-M, Chen C-Y, Hsueh C, Yu C-C, Chen C-C, Wang T-H. Application of ribonucleoside vanadyl complex (RVC) for developing a multifunctional tissue preservative solution. PLoS ONE. 2018;13:e0194393. doi: 10.1371/journal.pone.0194393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das L, Li M, Stevens J, Li W, Pu Y, Ragauskas AJ, et al. Characterization and catalytic transfer hydrogenolysis of deep eutectic solvent extracted sorghum lignin to phenolic compounds. ACS Sustain Chem Eng. 2018;6:10408–20. [Google Scholar]
- 40.Balakshin MY, Capanema EA, Santos RB, Chang H, Jameel H. Structural analysis of hardwood native lignins by quantitative 13C NMR spectroscopy. Holzforschung. 2016;70:95–108. [Google Scholar]
- 41.Holtman KM, Chang H, Jameel H, Kadla JF. Quantitative 13C NMR characterization of milled wood lignins isolated by different milling techniques. J Wood Chem Technol. 2006;26:21–34. [Google Scholar]
- 42.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–6. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 44.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menzel P, Ng KL, Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun. 2016;7:1–9. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang D, Li F, Kirton ES, Thomas A, Egan RS, An H, et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7:e7359. doi: 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 49.Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen IT, et al. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res. 2001;29:41–3. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–32. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun. 2016;7:1–11. doi: 10.1038/ncomms13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, et al. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 2019;36:2251–2. doi: 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T, et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. PNAS. 2011;108:8420–5. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.R Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2018. https://www.R-project.org.
- 56.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takeda M, Kamagata Y, Ghiorse WC, Hanada S, Koizumi J. Caldimonas manganoxidans gen. nov., sp. nov., a poly(3-hydroxybutyrate)-degrading, manganese-oxidizing thermophile. Int J Syst Evol Microbiol. 2002;52:895–900. doi: 10.1099/00207713-52-3-895. [DOI] [PubMed] [Google Scholar]
- 58.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–5. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015;31:926–32. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pattrick CA, Webb JP, Green J, Chaudhuri RR, Collins MO, Kelly DJ. Proteomic profiling, transcription factor modeling, and genomics of evolved tolerant strains elucidate mechanisms of vanillin toxicity in Escherichia coli. mSystems. 2019;4:e00163–19.. doi: 10.1128/mSystems.00163-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoshikata T, Suzuki K, Kamimura N, Namiki M, Hishiyama S, Araki T, et al. Three-component O-demethylase system essential for catabolism of a lignin-derived biphenyl compound in Sphingobium sp. strain SYK-6. Appl Environ Microbiol. 2014;80:7142–53. doi: 10.1128/AEM.02236-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kontur WS, Bingman CA, Olmsted CN, Wassarman DR, Ulbrich A, Gall DL, et al. Novosphingobium aromaticivorans uses a Nu-class glutathione S-transferase as a glutathione lyase in breaking the β-aryl ether bond of lignin. J Biol Chem. 2018;293:4955–68. doi: 10.1074/jbc.RA117.001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kontur WS, Olmsted CN, Yusko LM, Niles AV, Walters KA, Beebe ET, et al. A heterodimeric glutathione S-transferase that stereospecifically breaks lignin’s β(R)-aryl ether bond reveals the diversity of bacterial β-etherases. J Biol Chem. 2018;294:1877–90. doi: 10.1074/jbc.RA118.006548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salvachúa D, Werner AZ, Pardo I, Michalska M, Black BA, Donohoe BS, et al. Outer membrane vesicles catabolize lignin-derived aromatic compounds in Pseudomonas putida KT2440. PNAS. 2020;17:9302–10. doi: 10.1073/pnas.1921073117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Granja-Travez RS, Bugg TDH. Characterization of multicopper oxidase CopA from Pseudomonas putida KT2440 and Pseudomonas fluorescens Pf-5: Involvement in bacterial lignin oxidation. Arch Biochem Biophys. 2018;660:97–107. doi: 10.1016/j.abb.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 66.Strachan CR, Singh R, VanInsberghe D, Ievdokymenko K, Budwill K, Mohn WW, et al. Metagenomic scaffolds enable combinatorial lignin transformation. PNAS. 2014;111:10143–8. doi: 10.1073/pnas.1401631111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Masai E, Sasaki M, Minakawa Y, Abe T, Sonoki T, Miyauchi K, et al. A novel tetrahydrofolate-dependent O-demethylase gene is essential for growth of Sphingomonas paucimobilis SYK-6 with syringate. J Bacteriol. 2004;186:2757–65. doi: 10.1128/JB.186.9.2757-2765.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abe T, Masai E, Miyauchi K, Katayama Y, Fukuda M. A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6. J Bacteriol. 2005;187:2030–7. doi: 10.1128/JB.187.6.2030-2037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sonoki T, Obi T, Kubota S, Higashi M, Masai E, Katayama Y. Coexistence of two different O demethylation systems in lignin metabolism by Sphingomonas paucimobilis SYK-6: cloning and sequencing of the lignin biphenyl-specific O-demethylase (LigX) gene. Appl Environ Microbiol. 2000;66:2125–32. doi: 10.1128/aem.66.5.2125-2132.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sonoki T, Masai E, Sato K, Kajita S, Katayama Y. Methoxyl groups of lignin are essential carbon donors in C1 metabolism of Sphingobium sp. SYK-6. J Basic Microbiol. 2009;49:S98–102. doi: 10.1002/jobm.200800367. [DOI] [PubMed] [Google Scholar]
- 71.Maeda AH, Nishi S, Ishii S, Shimane Y, Kobayashi H, Ichikawa J, et al. Complete genome sequence of Altererythrobacter sp. strain B11, an aromatic monomer-degrading bacterium, isolated from deep-sea sediment under the seabed off Kashima, Japan. Genome Announc. 2018;6:e00200–18. doi: 10.1128/genomeA.00200-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuan C-G, Chen X, Jiang Z, Chen W, Liu L, Xian W-D, et al. Altererythrobacter lauratis sp. nov. and Altererythrobacter palmitatis sp. nov., isolated from a Tibetan hot spring. Antonie Van Leeuwenhoek. 2017;110:1077–86. doi: 10.1007/s10482-017-0882-y. [DOI] [PubMed] [Google Scholar]
- 73.Nagashima S, Kamimura A, Shimizu T, Nakamura-Isaki S, Aono E, Sakamoto K, et al. Complete genome sequence of phototrophic betaproteobacterium Rubrivivax gelatinosus IL144. J Bacteriol. 2012;194:3541–2. doi: 10.1128/JB.00511-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sheu S-Y, Li Z-H, Young C-C, Chen W-M. Rubrivivax albus sp. nov., isolated from a freshwater pond. Int J Syst Evol Microbiol. 2020;70:805–13. doi: 10.1099/ijsem.0.003829. [DOI] [PubMed] [Google Scholar]
- 75.Ramana ChV, Sasikala CH, Arunasri K, Anil Kumar P, Srinivas TNR, Shivaji S, et al. Rubrivivax benzoatilyticus sp. nov., an aromatic, hydrocarbon-degrading purple betaproteobacterium. Int J Syst Evol Microbiol. 2006;56:2157–64. doi: 10.1099/ijs.0.64209-0. [DOI] [PubMed] [Google Scholar]
- 76.Wawrousek K, Noble S, Korlach J, Chen J, Eckert C, Yu J, et al. Genome annotation provides insight into carbon monoxide and hydrogen metabolism in Rubrivivax gelatinosus. PLoS ONE. 2014;9:e114551. doi: 10.1371/journal.pone.0114551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Providenti MA, O’Brien JM, Ruff J, Cook AM, Lambert IB. Metabolism of isovanillate, vanillate, and veratrate by Comamonas testosteroni strain BR6020. J Bacteriol. 2006;188:3862–9. doi: 10.1128/JB.01675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Colatriano D, Tran PQ, Guéguen C, Williams WJ, Lovejoy C, Walsh DA. Genomic evidence for the degradation of terrestrial organic matter by pelagic Arctic Ocean Chloroflexi bacteria. Commun Biol. 2018;1:90. doi: 10.1038/s42003-018-0086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence accessions are provided in Supplementary Data 2 and as part of NCBI BioProject PRJNA564648 (SRR10095339–SRR10095374). Refined genome information and NCBI accessions are provided in Supplementary Data 3.