Abstract
Sunlight is the most important environmental control on diel fluctuations in phytoplankton activity, and understanding diel microbial processes is essential to the study of oceanic biogeochemical cycles. Yet, little is known about the in situ temporal dynamics of phytoplankton metabolic activities and their coordination across different populations. We investigated diel orchestration of phytoplankton activity in photosynthesis, photoacclimation, and photoprotection by analyzing pigment and quinone distributions in combination with metatranscriptomes in surface waters of the North Pacific Subtropical Gyre (NPSG). We found diel cycles in pigment abundances resulting from the balance of their synthesis and consumption. These dynamics suggest that night represents a metabolic recovery phase, refilling cellular pigment stores, while photosystems are remodeled towards photoprotection during daytime. Transcript levels of genes involved in photosynthesis and pigment metabolism had synchronized diel expression patterns among all taxa, reflecting the driving force light imparts upon photosynthetic organisms in the ocean, while other environmental factors drive niche differentiation. For instance, observed decoupling of diel oscillations in transcripts and related pigments indicates that pigment abundances are modulated by environmental factors extending beyond gene expression/regulation reinforcing the need to combine metatranscriptomics with proteomics and metabolomics to fully understand the timing of these critical processes in situ.
Subject terms: Biogeochemistry, Transcriptomics, Metabolomics
Introduction
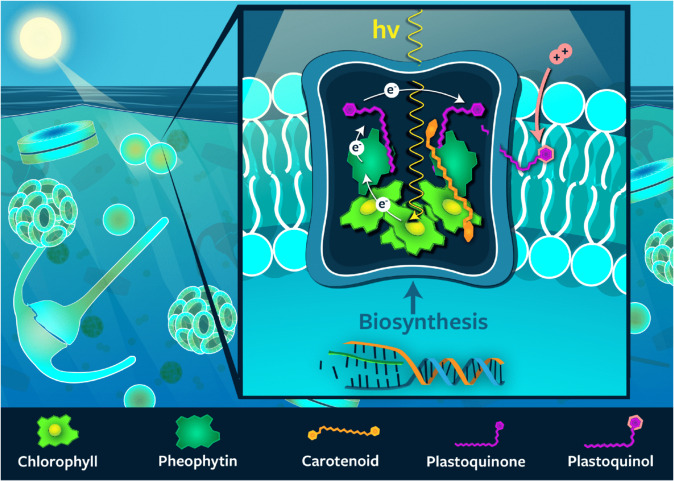
Sunlight regulates the growth and cellular processes of all photosynthetic organisms in the ocean. It plays a pivotal role for ocean ecosystem processes, many of which are driven by energy provided by photosynthesis. The conversion of light into chemical energy is achieved by the absorption of photons by pigments in light-harvesting complexes followed by the transfer of that energy to the reaction center, where it is used to initiate the electron transfer and charge separation processes. Plastoquinones mediate the transfer of electrons between photosystem II (PSII), the cytochrome b6f complex (Cytb6f), and photosystem I (PSI). The net result is the generation of adenosine triphosphate (ATP) and nicotinamide adenine dinucleotide phosphate (NADPH), which are then used for carbon fixation, biosynthesis, and aerobic respiration (see review by Eberhard et al. [1]). Pigments play a central role in energy transport within light-harvesting complexes and photosystems of all photosynthetic organisms (Fig. 1). In particular, chlorophyll (Chl) is vital in these processes, performing both light harvesting and electron transfer, and it is typically the most abundant pigment. Carotenoid pigments also participate in energy transport and have an additional role in photoacclimation.
Fig. 1. Schematic representation of the structure of photosystem II in photosynthetic membranes.
The relative locations of the major pigments and quinones as well as the pathway of electrons through these molecules is shown.
Phytoplankton have developed a wide variety of pigment-based mechanisms for adapting to the variable light field experienced in the upper ocean. These mechanisms include downsizing of the photosynthetic antenna size by reducing the pool size of Chl, using xanthophyll cycling to arrest photoinhibition, and quenching singlet oxygen by carotenes [2]. Pigment molecules thus play key roles in the acclimation (short-term) and adaptation (long-term) of phytoplankton to the variability in light. In addition to their physiological importance, evolutionary divergence has led to the formation of a variety of Chl and carotenoid pigments in different photosynthetic organisms, which allows us to delineate activity amongst phytoplankton groups [e.g., 3]. Also, the biosynthetic pathways of pigments are well known [e.g., 4]. This allows investigation of expression patterns of Chl- and carotenoid pigment-related genes to gain further insight into the activity of individual phytoplankton groups, enabling taxonomic resolution not necessarily provided by pigment analysis alone.
Diel oscillations in metabolic activities have been observed for both gene expression [e.g., 5–8] and pigment profiles [e.g., 9, 10] in natural phytoplankton populations as well as culture studies. However, there has been no study of the relationship between phytoplankton pigments and photosynthesis-related gene expression over the diel cycle in the ocean. Further, little is known about the synchronicity of photosynthetic metabolic pathways between different phytoplankton groups in nutrient deplete marine environments such as the subtropical gyres. Yet, the nutrient scarcity requires niche and/or resource partitioning by different phytoplankton to facilitate growth in the face of competition [11]. In addition, temporal anomalies of phytoplankton biomass and cellular pigment content impact the optical properties of the surface ocean influencing the interpretation of satellite-derived ocean color observations and global models using these data [12]. In these studies, diel oscillations are typically not considered because satellite observations take place only at a narrow interval of the diel cycle. Instead, most global ocean ecosystem models based on satellite data assume that the observed phytoplankton communities are under steady state growth conditions with respect to daily time scales [e.g., 13]. Thus, better understanding group-specific differences as well as bulk community acclimation over the diel cycle is important for informing models relying on photoacclimation parameters to determine growth and accumulation rates as well as primary production [12, 14].
In this study, we used a combined analysis of pigments, quinones, and metatranscriptomes to investigate the diel orchestration of phytoplankton activity involved in photosynthesis, photoacclimation, and photoprotection in the surface waters of the North Pacific Subtropical Gyre (NPSG). An intensive multidisciplinary field campaign near station ALOHA in summer 2015 made use of a Lagrangian sampling strategy that mitigates the effect of spatial variability and allowed us to investigate microbial populations within the same water parcel over time [15]. In light of the importance of the subtropical gyres of the oceans for global biogeochemical cycles [16, 17], it is crucial to know how diel cycles of photoacclimation in phytoplankton affect the pigment composition in these ecosystems.
Materials and methods
Sample collection and lipid analysis
Seawater samples were collected during R/V Kilo Moana cruise KM1513 (July/August 2015) near Station ALOHA (22° 45′ N, 158° 00′ W) in the oligotrophic NPSG using standard Niskin-type bottles attached to a CTD rosette. Triplicate samples were collected every 4 h from 2 a.m. (local time) July 27, 2015 to 6 a.m. (local time) on July 30, 2015 for a total of 20 time points. Samples (~2 L) were filtered using vacuum filtration (ca. −200 mm Hg) onto 47 mm diameter 0.2 µm hydrophilic Durapore filters (Millipore). Samples were immediately flash frozen and stored at −196 °C until processing.
Lipids, including Chls, carotenoids and quinones, were extracted from whole filters using a modified Bligh and Dyer protocol [18] with DNP-PE-C16:0/C16:0-DAG (2,4-dinitrophenyl phosphatidylethanolamine diacylglycerol; Avanti Polar Lipids, Inc., Alabaster, AL) used as an internal standard. Filter blanks were extracted and analyzed alongside environmental samples. The total lipid extract was analyzed by reverse phase high-performance liquid chromatography (HPLC) mass spectrometry (MS) on an Agilent 1200 HPLC coupled to a Thermo Fisher Exactive Plus Orbitrap high resolution mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). HPLC and MS conditions are described in detail in [19, 20]; modified after [21].
For identification and quantification of pigments and quinones, we used LOBSTAHS, an open-source lipidomics software pipeline based on adduct ion abundances and several other orthogonal criteria [20]. Pigments and quinones identified using LOBSTAHS were quantified from MS data after pre-processing with XCMS [22] and CAMERA [23]. XCMS peak detection was validated by manual identification using retention time as well as accurate molecular mass and isotope pattern matching of proposed sum formulas in full-scan mode and tandem MS (MS2) fragment spectra of representative compounds [19]. For validation of accuracy and reliability of LOBSTAHS identification and quantification, quality control (QC) samples of known composition and spiked lipid standards were interspersed with samples as described previously [20]. Lipid abundances obtained from LOBSTAHS were corrected for relative response of commercially available standards. Abundances of quinones were corrected for the response of a ubiquinone (UQ10:10) standard, Chls and their associated compounds using a Chl a standard, and carotenoid pigments using a β-carotene standard. All standards were purchased from Sigma Aldrich (St. Louis, MO, USA). Individual response factors were obtained from external standard curves by triplicate injection of a series of standard mixtures in amounts ranging from 0.15 to 40 pmol on column per standard. Data were corrected for differences in extraction efficiency using the recovery of the DNP-PE internal standard (Avanti Polar Lipids, Inc., Alabaster, AL, USA).
Eukaryotic metatranscriptome analysis
Samples for the >5 μm microeukaryote metatranscriptomes were collected in triplicate at the same time as lipid samples following Harke et al. [24]. Briefly, 20 L of seawater was collected in acid-washed carboys every 4 h over the same study period at the same time points as the lipid samples plus an additional time point at 22 p.m. (local time) July 26, 2015. Seawater was prescreened through a 200 μm nylon mesh and then filtered onto two 5 μm polycarbonate filters (47 mm) via peristaltic pump, passing ~10 L across each filter. Samples were then flash frozen in liquid N until extraction. Total RNA was extracted from individual filter sets (n = 2 per time point) using Qiagen RNeasy Mini Kit (Qiagen, Hilden, Germany) as in Harke et al. [24] and then sequenced on an Illumina HiSeq 2000 at the JP Sulzberger Genome Center (CUGC) using center protocols. PolyA-selected samples were sequenced to a depth of 90 million 100 bp, paired-end reads. Raw sequence quality was visualized with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and cleaned and trimmed using Trimmomatic [25] version 0.27 (paired-end mode, 4 bp-wide sliding window for quality below 15, minimum length of 25 bp). Processed reads were mapped to a reference database after Alexander et al. [26], constructed from the Marine Microbial Eukaryotic Transcriptome Sequencing Project (MMETSP); [27]. Transcripts within the reference database were annotated with KEGG using UProC [28]. Mapping was conducted with the Burrows–Wheeler Aligner (BWA-MEM, parameters –k 10 –aM; [29]) and read counts generated with the HTSeq 0.6.1 package (options –a 0, --m intersection-strict, -s no; [30]). Read counts were filtered for contigs with average read counts ≥10 across the time series and then normalized with DESeq2 “varianceStabilizingTransformation” command [31]. These environmental sequence data are deposited in the Sequence Read Archive through the National Center for Biotechnology Information under accession no. SRP136571, BioProject no. PRJNA437978. To facilitate comparisons with pigment and quinone data, transcripts occurring in dinoflagellate, haptophyte, and diatom taxa were mined for the following KEGG pathways: Carotenoid biosynthesis [PATH:ko00906], Porphyrin, and Chl metabolism [PATH:ko00860], Photosynthesis [PATH:ko00195], Carbon fixation in photosynthetic organisms [PATH:ko00710], and Ubiquinone and other terpenoid-quinone biosynthesis [PATH:ko00130]. In addition, signals involved in plastoquinol biosynthesis were separated out from Ubiquinone and other terpenoid-quinone biosynthesis to mirror pigment and quinone data.
Prokaryotic metatranscriptome analysis
For this study, a previously published data set of transcriptomes was used [15, 32], which consisted of bacterioplankton samples collected every four hours over the same study period at the same time points as the eukaryotic metatranscriptome samples. Sampling was performed as follows: 2 L of seawater were filtered onto 25 mm, 0.2 μm Supor PES Membrane Disc filters (Pall) using a peristaltic pump. The filtration time was between 15 and 20 min and filters were placed in RNALater (Ambion) immediately afterwards, and preserved at −80 °C until processing. Molecular standard mixtures for quantitative transcriptomics were prepared as previously described [33], and, 50 μl of each standard group was added to sample lysate before RNA purification. Metatranscriptomic libraries were prepared for sequencing with the addition of 5–50 ng of RNA to the ScriptSeq cDNA V2 library preparation kit (Epicentre). Metatranscriptomic samples were sequenced with an Illumina NextSeq 500 system using V2 high output 300 cycle reagent kit with PHIX control added. Reads were mapped to the station ALOHA gfne catalog [34] using LAST [35]. Transcripts were quantified through normalization of raw hit counts using molecular standards [15]. Transcripts in Prochlorococcus and Crocosphaera were mined for the same KEGG pathways as the eukaryotes.
Statistical analysis
The statistical significance of diel oscillations of pigment, quinone, and transcript abundances was tested using the Rhythmicity Analysis Incorporating Non-parametric methods (RAIN) package in R [see 15, 32, 36]. Resulting p values from RAIN analysis were corrected for false discovery using the ‘padjust’ function in R with Benjamini–Hochberg method [37]. Corrected p values ≤ 0.05 were considered to have significant diel periodicity. Peak times were calculated with a harmonic regression, fitting the expression data to a sine curve.
To evaluate rank correlations between pigment and transcript time series, data were centered to a mean of zero and scaled to have a standard deviation of one to facilitate inter-comparability between data with different units. Then, pairwise Spearman rank correlations (Spearman’s rho) were calculated between all pairs of measured time series. To address autocorrelation, we evaluated whether observed correlations were stronger than independent identically distributed random walks, which are known to often be spuriously correlated [38]. To test this, the differences between consecutive observations were calculated for all measured time series. The frequency distributions of observed differences were then used to bootstrap sample 20 random differences and cumulatively summed to simulate a random walk of the same total duration as the data. This process was repeated and we bootstrap sampled 100,000 such random walks independently to create a null ensemble of time series with similar autocorrelation-1 structure to the data. Bootstrap sampling was then used to randomly select 100,000 pairs of random walks and rank correlations were calculated to generate a Monte Carlo simulation of the distribution of Spearman’s rho between independent identically distributed random walks with step sizes similar to those observed in the data (Fig. S1). This distribution was then used to calculate empirical one-tailed p values of observed rank correlations. The adaptive Benjamini–Hochberg method was applied to account for multiple testing [37]. All calculations were carried out using the R statistical computing language v 3.6.3 with functionalities from the ‘tidyverse’ set of packages version 1.2.1.
Results and discussion
Diel oscillations of chloropigments and carotenoids
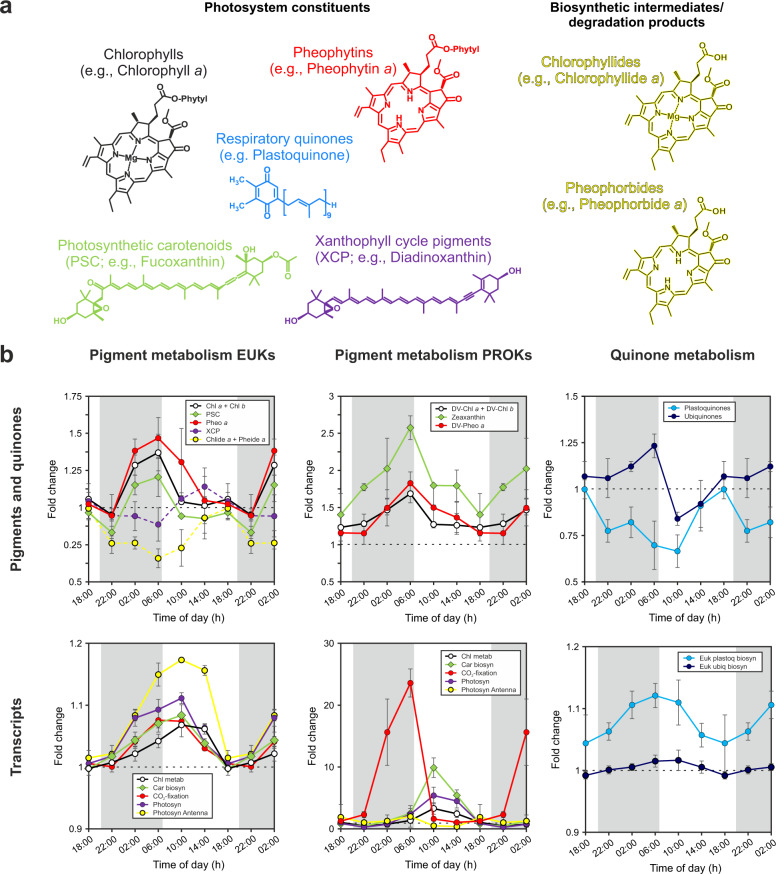
The majority of detected pigment and quinone molecules investigated here displayed clear, statistically significant diel oscillations in abundance based on the RAIN hypothesis-testing method ([15, 32, 36]; Supplementary Table 1). Diel oscillations of chloropigments, i.e., mono- and divinyl-Chls, showed maximum concentrations at dawn (6:00 h local time) and minimum concentrations during daylight hours (Figs. 2 and S2). This pattern is distinct from previous investigations of diel Chl, which showed a mid-day maximum and a night-time minimum especially in data integrated through the water column [10, 39, 40]. Based on these previous observations, it was thought that Chl synthesis directly reflects primary production by photoadaptation, with increasing cellular Chl during the day and Chl break down (or dilution by cell-division) at night [e.g., 41]. Alternatively, mid-day minima in surface water Chl that have frequently been observed in data from in vivo Chl fluorescence have been interpreted to result from non-photochemical quenching, not absolute decreases in Chl concentrations. The reasoning behind this interpretation is that variability in quantum yield for fluorescence is significant and complicates the interpretation of Chl estimates from in vivo fluorescence at high light intensities [e.g., 42–44]. However, here we used MS to determine Chl concentrations from lipid extracts, which is insensitive to quenching. Other studies, measuring extracted Chl, also showed clear and reproducible night-time maxima for both near surface waters [e.g., 9] and culture experiments at high light intensities [e.g., 45–48]. In addition, Fouilland et al. [49] found a depth-dependence of diel variations in Rubisco activity and Chl concentration per cell further supporting a light regime-dependence of Chl profiles.
Fig. 2. Diel oscillations of pigments and pigment-related transcripts.
a Molecular structure of investigated pigments and respiratory quinones associated with the photosystem. b Time-of-day average of fold change relative to 18:00 h (local time) of chloro- and carotenoid pigments, quinones, as well as transcripts associated with the metabolism of the pigment and quinone molecules. Error bars represent the standard deviation of averaged values for each time of day (n ≥ 6). Molecular structures and pigment and quinone data are colored according to compound group. If colors for transcripts match those of pigments or quinones, transcripts are related to the metabolism of the respective molecule.
The photosynthetic carotenoids (PSC) aligned with Chl over the diel cycle (Figs. 2 and S2) and no periodic variations were observed in ratios among PSC pigments (Fig. S3), which has previously been observed for both phytoplankton cultures [50] and surface ocean samples [51] and is consistent with PSC and Chl having similar functions [52]. The most abundant PSC in surface waters were fucoxanthin (diatom biomarker), 19′-butanoyloxyfucoxanthin (pelagophyte biomarker), and 19′-hexanoyloxyfucoxanthin (prymnesiophyte biomarker). All showed similar trends, with a nocturnal increase leading to a pre-dawn maximum and a minimum later, during daylight hours (Figs. 2 and S3). Similar to Chls, the nocturnal increase may seem counterintuitive as light-harvesting molecules are needed during the day for photosynthesis. However, at high photon flux, the rate of photon absorption by Chl antenna far exceeds the rate at which photons can be utilized for photosynthesis. To avoid overexcitation of the photosystem, high-light acclimation typically involves downsizing of the antenna of both photosystems [53, 54], in particular of PSII [55]. The observed profiles thus likely represent the balance of photoprotective and photorepair mechanisms.
While the observed depression in Chl and PSC during daylight might be explained by downsizing the photosystems due to high-light stress, the increase at night clearly indicates synthesis in the dark. Increased vertical mixing during the night could also explain this signal, but the surface mixed layer typically ranges from ~0–40 m [56], which is too shallow to transport large amounts of Chl, for example, from the deep Chl maximum (at ~100 m at station ALOHA our sampling site and times [15, see Fig. 1 therein]) to the sampling depth of 15 m. In addition, Prochlorococcus cell numbers remained constant over the diel cycle in our samples, which results in increasing divinyl-Chl/cell quotas during the night (Fig. S3) and supports the hypothesis of night-time synthesis. Further evidence can be found in dark Chl biosynthesis, which requires a distinct enzyme, light-independent protochlorophyllide oxidoreductase (see Fig. S4 for Chl metabolic pathways). This enzyme catalyzes the reduction of protochlorophyllide to chlorophyllide (see review by Armstrong [57]) and we detected expression of the three genes (chlB, chlL, and chlN) required for light-independent protochlorophyllide reduction in all major phytoplankton groups (Figs. S5–7). The energy and carbon for the de novo synthesis of Chls in the dark is likely provided by carbon stores, for example glycogen in cyanobacteria or triacylglycerols (TAGs) in eukaryotic phytoplankton. Consistent with this, we recently showed that nanoeukaryotic phytoplankton in the NPSG accumulate large amounts of TAGs during the day and subsequently consume them at night [19]. A similar mechanism has been proposed for dark synthesis of proteins in several phytoplankton [e.g., 58–60] and our findings extend the potential utilization of C stores by phytoplankton for dark synthesis of pigments.
Pheophytin and divinyl- pheophytin profiles were tightly coupled to those of Chl and divinyl-Chl, respectively, with peak times at night (Fig. 2). While the presence of pheophytin in marine samples has traditionally been interpreted to reflect dead or dying cells due to the high abundance of these pigments in zooplankton guts and sinking particles, which are unlikely to be consistently captured in our small volume samples [61–65], pheophytins are also associated with living phytoplankton. Most notably, pheophytin has been found to play an important role in electron transport in PSII by being the primary electron acceptor for excited Chl [66] and has been frequently detected in isolated PSII with a 3:1 stoichiometry of Chl and pheophytin [e.g., 67]. In addition, pheophytin and Chl share a biochemical pathway, where Chl is converted to pheophytin by a magnesium dechelatase [68] supporting the idea of simultaneous dark synthesis of Chls and pheophytins. Other Chl related structures, pheophorbide and chlorophyllide, were out of phase with Chl and pheophytin, both showing peak times at 14:00 h (Fig. 2). These Chl related structures are not known to participate in photosynthetic reactions, suggesting daytime peaks are likely associated with photodamage and photoprotective mechanisms. As an alternative to the simple Chl degradation during the day and de novo synthesis at night, the use of the Chl cycle as a mechanism for fine-tuning Chl ratios in response to changes in the light regime has been proposed [69, 70]. In this cycle, chlorophyllide is a key intermediate for biosynthesis and degradation of Chl (see Fig. S4), and it might accumulate at different times of day with the conversion to Chl happening at night. Daytime synthesis of chlorophyllide is indicated by the expression of light-dependent protochlorophyllide reductase in all phytoplankton groups (Fig. S8). However, transcript levels of genes involved in the final step of Chl synthesis, i.e., the esterification of chlorophyllide a with phytyl or geranylgeranyl pyrophosphate via a Chl synthase, did not reveal diel oscillations, except for a few genes in the haptophytes (Fig. S9). Similarly, pheophorbide a oxygenase, a key regulator of Chl catabolism [71], did not oscillate on a day/night cycle (Fig. S10) suggesting that light-dependent Chl cycling is at least not regulated at the transcriptional level.
High-light stress during daylight is also apparent from xanthophyll cycle pigments, essential molecules that channel excess energy away from Chls for protection against photooxidative damage [72]. Diadinoxanthin (Dd) and diatoxanthin (Dt), which are part of the Dd/Dt xanthophyll cycle predominantly possessed by haptophytes, diatoms, and dinoflagellates in the NPSG [73], were the most abundant xanthophyll cycle pigments detected (Fig. S2). The sum of Dt and Dd showed clear diel cycles with maximum abundances during the day (peak time at 14:00 h; Fig. 2). This pattern indicates that the cellular xanthophyll pigment pool increased during daylight hours likely due to photoadaptation triggered by a change of light intensity over the course of the day [51]. The de-epoxidation state (DES), which is defined as [Dt]/[Dt + Dd], showed a minimum during the day (Fig. S3), which contrasts observations from culture experiments [74] and the field [51]. This pattern is likely related to severe high-light stress at 15 m water depth where our samples were collected. Oxidative stress due to an increased formation of various reactive oxygen species (ROS) species consumes Dt by degrading it to low molecular weight compounds [75, 76], requiring re-synthesis for the pool to be refilled. Potentially, phytoplankton refill cellular Dt stores during the night. We therefore looked for Violaxanthin de-epoxidases, which convert Dd to Dt in the xanthophyll cycle [77], and find that transcript abundances for the diatoms and haptophytes were out of phase with XCPs (Fig. S11). In addition, Zeaxanthin epoxidases, which participate in non-photochemical quenching by regulating the level of epoxy-free xanthophylls in photosynthetic energy conversion, showed statistically significant diel oscillation for diatoms and haptophytes with peak times during the day at 10:00 h local time (Fig. S12). Our results from the pigment analysis thus show that high-light conditions in the upper photic zone results in severe photooxidative stress for phytoplankton, which is displayed in a distinct and different diel xanthophyll cycle pigment and Chl degradation product patterns compared to observations from the deeper photic zone [51]. Through this, phytoplankton are likely able to maintain the balance between dissipation and utilization of light energy to minimize the generation of ROS and resulting molecular damage. Since net primary production is typically invariable throughout the mixed layer at station ALOHA [78], the observed pigment signal appears to represent a light adaptation signal, as opposed to an increased photodegradation state of near surface phytoplankton cells.
Diel patterns of isoprenoid quinones
Analyses of quinones in the marine water column are still relatively rare [19, 79, 80], but due to their physiological importance in electron transport in both photosynthesis and cellular respiration, quinones provide valuable information on microbial activities. While plastoquinones (PQs) are primarily involved in electron transport of PSII, ubiquinones (UQs) are associated with aerobic respiration [81]. Recent studies suggested PQs and UQs might additionally be involved in photoprotective mechanisms by acting as ROS scavengers [82–85]. In our samples, both quinone groups showed distinct and statistically significant diel patterns (Fig. 2 and Table S1) with UQs peaking at the end of the night (6:00 h; cf. [19]) and PQs peaking at the end of the day (18:00 h, Fig. 2). These two groups of structurally similar molecules were thus out of phase by 12 h. The pre-dawn maximum of UQs may be associated with enhanced respiration of storage lipids, which have been shown to accumulate during the day and are consumed at night [19]. However, similar to Chl, the daytime minimum of UQ may also be related to oxidative damage. While ROS are produced throughout the day during mitochondrial respiration, additional ROS are produced during daylight hours due to photosynthesis and photorespiration [86], which may deplete UQ levels. During the night, the UQ stores are then refilled. As such, observed oscillations in UQ and PQ concentrations likely reflect a balance of production and consumption ensuring adequate intracellular concentrations are present of these important molecules to maintain homeostasis.
Similar to other photosystem constituents, photoacclimation mechanisms play a major role for intercellular PQ abundances. Increasing the rates of electron transport processes can protect PSII when absorption of quanta exceeds the requirements of photosynthetic carbon metabolism [87], which, in turn, requires an increase in the PQ pool size and may in part be responsible for observed daytime maxima of PQs. In addition, in plants, PQs have been shown to act as ROS scavengers under high-light stress [88, 89]. These processes deplete PQs due to photooxidation during daylight hours. Hence, PQs have to be re-synthesized to keep this important function. The daytime increase in the proportion of the PQ to Chl (Fig. S3) is likely due to the different organization of chloroplast structure. In plants, chloroplasts have fewer antenna Chl compared to other molecules in the electron transport chain during high-light versus low-light conditions [90].
Diel oscillations of transcripts involved in photosynthesis
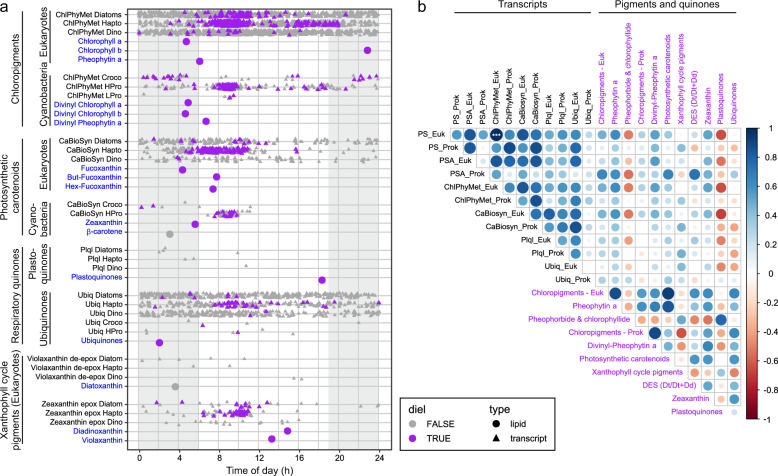
In addition to measuring diel oscillations in pigments and quinones, we further explored diel transcriptional rhythms of genes involved in photosynthesis and pigment metabolism of the dominant bacterial and eukaryotic groups at station ALOHA. Transcripts of all photosynthetic populations showed statistically significant diel oscillations for most investigated pathways (Supplementary Table 2). Almost all significant diel transcripts associated with photosynthesis and pigment metabolism peaked in the first part of the day (10:00 h; Fig. 3a), suggesting their transcriptional regulation in phototrophs is strongly synchronized across diverse taxa (within the temporal resolution of our measurements). These observations further imply that the temporal dynamics of pigment metabolism is conserved across domains in the oligotrophic open ocean (see Figs. S12–S19 for profiles of individual phytoplankton groups). This complements recent findings by Kolody et al. in coastal systems [8] who observed significantly periodic photosynthesis-related transcripts were conserved across diverse phototrophic lineages in a high-nutrient coastal environment. Thus, our data are consistent with multispecies synchronous diel cycles of genes associated with photosynthesis and pigment metabolism with increasing transcript abundances during dark hours and peak times in the mid-morning. Nightly expression of photosynthesis genes in natural photoheterotrophic bacteria, followed by a drawdown shortly after light onset, has been suggested to be related to preparing cells for efficient solar energy harvest in the early morning hours [6]. Our data from cyanobacteria and eukaryotic phytoplankton are consistent with this hypothesis. A notable exception of this pattern was Crocosphaera; transcripts for most pathways in this diazotroph only showed a weak relationship or no significant positive correlation with the pathways in the other phytoplankton populations. However, this is in agreement with other culture [91] and field-based observations [15, 92]. The process of N2 fixation in this unicellular cyanobacteria seems to require a tightly orchestrated diel cycle distinct from other diazotrophs and non-diazotrophic microorganisms [15, 92].
Fig. 3. Relationship between periodic pigment-associated gene expression in phytoplankton transcripts and actual pigment abundances.
a Peak times of expression of all transcripts assigned to KEGG pathways associated with pigment metabolism (triangles) and pigment molecule abundances (circles). Purple symbols denote transcripts or pigment molecules identified as significantly periodic (24-h period), whereas gray symbols denote peak times without a significant diel component in peak expression or pigment abundance, respectively. b Spearman’s rank correlation matrix of the different pigment and quinone classes (blue text) and aggregated genes within the KEGG pathways photosynthesis (PS), carotenoid biosynthesis (CaBiosyn), chlorophyll and porphyrin metabolism (ChlPhyMet), photosynthesis - antenna proteins (PSA), plastoquinol biosynthesis (Plql), ubiquinone and other terpenoid-quinone biosynthesis (Ubiq) separated into prokaryotes (Prok) and eukaryotes (Euk). Circle size and color is related to correlation value. Asterisks denote correlation significance (***p < 0.001).
Strong diel rhythmicity in transcripts of eukaryotic phytoplankton including haptophytes and diatoms is expected from earlier culture and field experiments [e.g., 5, 8, 24, 32, 93, 94]. However, peak times for pathways involved in photosynthesis and pigment metabolism seem to be variable in culture experiments. For example, light-harvesting antenna protein gene expression was enriched in late afternoon (2–6 p.m.) for iron-replete cultures of the diatom Phaeodactylum tricornutum, along with genes involved in porphyrin and Chl metabolism [94], while enrichment for the KEGG pathways photosynthesis and porphyrin and Chl metabolism was observed in the dawn for the diatom Thalassiosira pseudonana [93]. This variability in the timing of gene expression contrasts with our findings from natural phytoplankton populations in the NPSG, which showed synchronized expression patterns across diverse phytoplankton. This synchrony across taxa has recently also been shown for a coastal environment [8] and together the results indicate that responses to signals in the environment, including those resulting from organismal interactions, play an important role in shaping gene expression patterns of natural phytoplankton populations.
Patterns between transcription and pigment production
The combination of pigment analysis and metatranscriptomics further shed light on the complex interplay between metabolic pathways and actual metabolite concentrations. In a previous study of energy storage mechanisms of natural phytoplankton populations at station ALOHA, we showed that gene expression and lipid abundances were decoupled—despite distinct diel patterns in storage lipid abundance, transcript abundance for the genes involved in the final step of their biosynthesis remained relatively constant over the diel cycle [19; Fig. 4]. In addition, the correlations we observe between transcripts and pigments (shown in Fig. 3b) are not significantly stronger than the correlations expected from comparing independent time series (see Fig. S1). In culture experiments with the marine diatom P. tricornutum, during high-light acclimation, Chl metabolism at the transcriptional level was downregulated at an early stage, while Chl a concentrations showed a lag time to this initial transcriptional response [95], which is what would be expected for an ideal system. However, the change of concentrations of pigments or any metabolite is governed by the difference between production and consumption. For example, in our data set, the observed offset in peak time between transcripts (mid-morning) and associated pigments (dawn, Fig. 3a) suggests the influence of additional regulatory processes for Chl and carotenoid pigment abundances. Irradiance at 15 m at station ALOHA (700 µmol m−2 s−1) was higher than in the “high-light” experiments conducted by Nymark et al. [95] (500 µmol m−2 s−1) and the different patterns between the culture experiments and our field-based observations may be related to photoacclimation mechanisms among other factors. Our observations that transcripts peak ~20 h earlier than their corresponding pigments suggests that phytoplankton actively synthesize Chls and PSC at high rates, but that degradation processes (i.e., a shift towards photoprotection) consume more molecules than are produced. Alternatively, post-transcriptional processes affecting metabolite concentrations may be responsible for the observed decoupling of transcriptional and pigment concentration rhythms. Additional measurements of enzyme activity and/or proteins will be needed to further constrain the metabolic network. Time delays between changes in transcript levels and changes in pigment concentrations over the diel cycle have been observed in plant cultures, such as Arabidopsis [96]. In addition, Waldbauer et al. [97] found large (~12 h) offsets between mRNA and protein abundances, including the Chl biosynthesis gene chlP encoding geranylgeranyl diphosphate reductase, in culture experiments with the marine cyanobacterium Prochlorococcus. In the latter study, chlP showed peak abundances at the end of the dark phase, matching our Chl measurements from the NPSG where Prochlorococcus dominates. This could indicate that Chl biosynthesis is regulated on the protein level at least in these organisms. Examples from culture experiments indicate that caution is warranted when extrapolating from temporal patterns of transcript abundance to changes in metabolic fluxes. Our environmental data support the growing evidence that metatranscriptome data should not be interpreted as real-time readouts of biochemical or metabolic processes, unless verified by independent measurements of endpoint activities. A complete understanding of metabolic networks requires quantitative data about transcript and protein abundances, enzymatic activities, and ultimately metabolite fluxes. While additional measurements of proteins and enzyme activities would help resolve the regulation of the in situ pigment dynamics observed here, the diel patterns of both transcripts and their downstream products highlight how closely tuned these microbes are to the day–night cycle.
The response of cells toward ambient light conditions involves optimization of their photosystems in such a way that energy generation and utilization are in balance. Kana et al. [98] proposed that pigment abundance is modulated by environmental factors extending beyond gene expression/regulation, i.e., the initial response to high light intensities is self-adjusting. The light sensor that in a first step regulates photoacclimation in phytoplankton has been suggested to be the redox state of the quinone pool [98]. Although our methods do not allow direct determination of the redox state of the PQ pool because plastoquinols become spontaneously oxidized to PQs when exposed to oxygen during sample analysis, we found significant diel oscillations in the expression of Chl a/b-binding protein (cab) genes (Fig. S20). The cab genes are important for both light-harvesting and photoprotection in eukaryotic phytoplankton and are under transcriptional control by the redox poise of the PQ pool [99]. In cultures, under high light, it has been shown that the PQ pool becomes more reduced resulting in a suppression of cab mRNA synthesis and light-harvesting complex production, and an ultimate decrease in cellular photosynthetic pigments [99]. We observed a decline in cab transcript abundances during daylight hours (Fig. S20) suggesting that this light intensity-dependent photon-sensing system is active in the NPSG. This process then likely triggers the downregulation of photosynthesis and pigment metabolism-related genes to keep pigments levels consistently low to prevent overexcitation of the photosystems at the high irradiance levels phytoplankton experience in the surface waters of the NPSG. Besides the insights our data provided on the (de)coupling between transcriptional and metabolite rhythms, our data suggest that the night reflects a metabolic recovery phase that is used by all photosynthetic organisms across domains to be prepared for photosynthetic reaction as soon as the sun rises and allowing cells to make optimal use of sunlight energy.
Conclusions
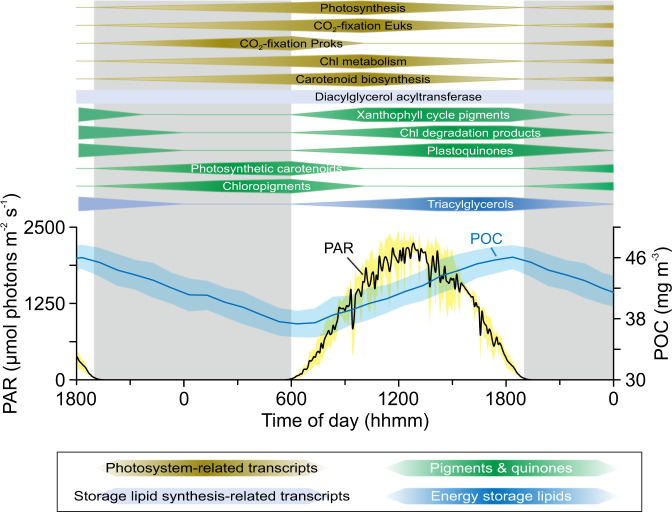
We are just beginning to understand in detail the mechanisms by which marine phytoplankton maintain homeostasis and coordinate growth under metabolic and energetic shifts driven by the perpetual rising and setting sun. Our work demonstrates that combined lipidomics (here pigment and quinone profiles) and transcriptomics can provide mechanistic insights into rapid (i.e., sub-daily) microbial processes (Fig. 4). While pigments involved in light harvesting and energy transfer peaked at dawn, photoprotective pigments and Chl degradation products peaked later in the day. This succession of pigment metabolism according to their function suggests a recovery of cellular Chl and PSC stores at night and a dominance of photoprotective mechanisms during daylight. The diel cycles of combined pigment, quinone and transcriptomics data from the NPSG thus highlight the fundamental connection between sunlight and phytoplankton photosynthetic metabolism [98]. Furthermore, diel metabolic cycles were similar across all major phytoplankton taxa with few exceptions. Thus, harvesting light energy is not the basis for temporal niche differentiation among these taxa, which stands to reason because photons are in excess at 15 m depth and therefore are not a resource that phytoplankton compete with one another to obtain in this environment. Niche differentiation among phytoplankton occurs for processes involving competitive and growth-limiting substrates, such as nitrogen, which is scarce in the surface of the NPSG [100–102]. Yet the daily synthesis of pigments—and the machinery required for this synthesis—place considerable burden on phytoplankton for growth-limiting substrates. Thus, any temporal partitioning in accessing growth-limiting substrates must also be accompanied by different modes for their storage, which are rarely considered [103, 104]. In summary, the data presented herein advances our knowledge of this fundamental process within photosynthetic microbes from the surface ocean, providing valuable detailed observations of transcriptional and pigment dynamics across kingdoms. These data will thus prove useful for future models linking remotely sensed ocean color to temporal dynamics of pigment concentrations or photosynthetic activity.
Fig. 4. Conceptualized temporal separation of phytoplankton pigment, plastoquinone and transcript abundances during the dark–light cycle in surface waters (15 m) at station ALOHA.
The timing was set based on Fig. 2 with the center of the polygon indicating the approximate peak time. In addition, the particulate organic carbon (POC) and photosynthetically available radiation (PAR) profiles averaged for one day/night cycle are shown. Surface PAR data were obtained from the HOT program database (http://hahana.soest.hawaii.edu/hoelegacy/hoelegacy.html), POC data from White et al. [105] and TAG (triacylglycerols & diacylglycerol acyltransferase) data from Becker et al. [19].
Supplementary information
Acknowledgements
We are grateful to the officers and crew of the R/V Kilo Moana cruise KM1513 (HOE Legacy II). We thank Helen F. Fredricks for help with lipidomics analysis, Daniel J. Repeta for help with onboard sample collection, and two anonymous reviewers for constructive criticism. This work was funded by a grant from the Simons Foundation (SCOPE, Award # 329108, BASVM, STD, EFD, JSW), and Gordon and Betty Moore Foundation (grant #3777, EFD). KWB was further supported by the Postdoctoral Scholarship Program at Woods Hole Oceanographic Institution & U.S. Geological Survey.
Author contributions
BASVM, STD, EFD. conceived the study. KWB, MJH, and DRM conducted and analyzed lipidomic, eukaryotic metatranscriptomic and prokaryote transcriptomic studies, respectively. JSW and DM supported data analysis. KWB and MJH wrote the paper with contributions from all authors.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kevin W. Becker, Matthew J. Harke
Supplementary information
The online version of this article (10.1038/s41396-020-00793-x) contains supplementary material, which is available to authorized users.
References
- 1.Eberhard S, Finazzi G, Wollman F-A. The dynamics of photosynthesis. Annu Rev Genet. 2008;42:463–515. doi: 10.1146/annurev.genet.42.110807.091452. [DOI] [PubMed] [Google Scholar]
- 2.Dubinsky Z, Stambler N. Photoacclimation processes in phytoplankton: mechanisms, consequences, and applications. Aquat Micro Ecol. 2009;56:163–76. [Google Scholar]
- 3.Wright SW, Jeffrey SW. Pigment markers for phytoplankton production. In: Volkman JK (ed). Marine organic matter: biomarkers, isotopes and DNA. Berlin, Heidelberg: Springer Berlin Heidelberg; 2006. p. 71–104.
- 4.Armstrong GA. Eubacteria show their true colors: genetics of carotenoid pigment biosynthesis from microbes to plants. J Bacteriol. 1994;176:4795–802. doi: 10.1128/jb.176.16.4795-4802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ottesen EA, Young CR, Eppley JM, Ryan JP, Chavez FP, Scholin CA, et al. Pattern and synchrony of gene expression among sympatric marine microbial populations. Proc Natl Acad Sci USA. 2013;110:E488–97. doi: 10.1073/pnas.1222099110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ottesen EA, Young CR, Gifford SM, Eppley JM, Marin R, Schuster SC, et al. Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages. Science. 2014;345:207–12. doi: 10.1126/science.1252476. [DOI] [PubMed] [Google Scholar]
- 7.Aylward FO, Eppley JM, Smith JM, Chavez FP, Scholin CA, DeLong EF. Microbial community transcriptional networks are conserved in three domains at ocean basin scales. Proc Natl Acad Sci USA. 2015;112:5443–8. doi: 10.1073/pnas.1502883112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolody BC, McCrow JP, Allen LZ, Aylward FO, Fontanez KM, Moustafa A, et al. Diel transcriptional response of a California Current plankton microbiome to light, low iron, and enduring viral infection. ISME J. 2019;13:2817–33. doi: 10.1038/s41396-019-0472-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neveux J, Dupouy C, Blanchot J, Le Bouteiller A, Landry MR, Brown SL. Diel dynamics of chlorophylls in high-nutrient, low-chlorophyll waters of the equatorial Pacific (180°): interactions of growth, grazing, physiological responses, and mixing. J Geophys Res Ocean. 2003;108:8140. 10.1029/2000JC000747.
- 10.Le Bouteiller A, Herbland A. Diel variation of chlorophyll a as evidence from a 13-day station in the equatorial Atlantic ocean. Oceano Acta. 1982;5:433–41. [Google Scholar]
- 11.Litchman E. Resource Competition and the ecological success of phytoplankton. In: Falkowski PG, Knoll AH (eds). Evolution of primary producers in the sea. Burlington: Academic Press; 2007. p. 351–75.
- 12.Graff JR, Behrenfeld MJ. Photoacclimation responses in subarctic atlantic phytoplankton following a natural mixing-restratification event. Front Mar Sci. 2018;5:209. [Google Scholar]
- 13.Behrenfeld MJ, Boss E, Siegel DA, Shea DM. Carbon-based ocean productivity and phytoplankton physiology from space. Global Biogeochem Cycles. 2005;19:GB1006. 10.1029/2004GB002299.
- 14.Tomkins M, Martin AP, Nurser AJG, Anderson TR. Phytoplankton acclimation to changing light intensity in a turbulent mixed layer: a Lagrangian modelling study. Ecol Model. 2020;417:108917. [Google Scholar]
- 15.Wilson ST, Aylward FO, Ribalet F, Barone B, Casey JR, Connell PE, et al. Coordinated regulation of growth, activity and transcription in natural populations of the unicellular nitrogen-fixing cyanobacterium Crocosphaera. Nat Microbiol. 2017;2:17118. doi: 10.1038/nmicrobiol.2017.118. [DOI] [PubMed] [Google Scholar]
- 16.Emerson S, Quay P, Karl D, Winn C, Tupas L, Landry M. Experimental determination of the organic carbon flux from open-ocean surface waters. Nature. 1997;389:951–4. [Google Scholar]
- 17.Sarmiento JL, Slater R, Barber R, Bopp L, Doney SC, Hirst AC, et al. Response of ocean ecosystems to climate warming. Glob Biogeochem Cycles. 2004;18:GB3003. [Google Scholar]
- 18.Popendorf KJ, Fredricks HF, Van, Mooy BAS. Molecular ion-independent quantification of polar glycerolipid classes in marine plankton using triple quadrupole MS. Lipids. 2013;48:185–95. doi: 10.1007/s11745-012-3748-0. [DOI] [PubMed] [Google Scholar]
- 19.Becker KW, Collins JR, Durham BP, Groussman RD, White AE, Fredricks HF, et al. Daily changes in phytoplankton lipidomes reveal mechanisms of energy storage in the open ocean. Nat Commun. 2018;9:5179. doi: 10.1038/s41467-018-07346-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins JR, Edwards BR, Fredricks HF, Van Mooy BAS. LOBSTAHS: an adduct-based lipidomics strategy for discovery and identification of oxidative stress biomarkers. Anal Chem. 2016;88:7154–62. doi: 10.1021/acs.analchem.6b01260. [DOI] [PubMed] [Google Scholar]
- 21.Hummel J, Segu S, Li Y, Irgang S, Jueppner J, Giavalisco P. Ultra performance liquid chromatography and high resolution mass spectrometry for the analysis of plant lipids. Front Plant Sci. 2011;2:54. doi: 10.3389/fpls.2011.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–87. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 23.Kuhl C, Tautenhahn R, Böttcher C, Larson TR, Neumann S. CAMERA: an integrated strategy for compound spectra extraction and annotation of LC/MS data sets. Anal Chem. 2012;84:283–9. doi: 10.1021/ac202450g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harke MJ, Frischkorn KR, Haley ST, Aylward FO, Zehr JP, Dyhrman ST. Periodic and coordinated gene expression between a diazotroph and its diatom host. ISME J. 2019;13:118–31. doi: 10.1038/s41396-018-0262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexander H, Rouco M, Haley ST, Wilson ST, Karl DM, Dyhrman ST. Functional group-specific traits drive phytoplankton dynamics in the oligotrophic ocean. Proc Natl Acad Sci USA. 2015;112:E5972–9. doi: 10.1073/pnas.1518165112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA, et al. The marine microbial eukaryote transcriptome sequencing project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12:e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meinicke P. UProC: tools for ultra-fast protein domain classification. Bioinformatics. 2014;31:1382–8. doi: 10.1093/bioinformatics/btu843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aylward FO, Boeuf D, Mende DR, Wood-Charlson EM, Vislova A, Eppley JM, et al. Diel cycling and long-term persistence of viruses in the ocean’s euphotic zone. Proc Natl Acad Sci USA. 2017;114:11446 LP–11451. doi: 10.1073/pnas.1714821114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gifford SM, Becker JW, Sosa OA, Repeta DJ, DeLong EF. Quantitative transcriptomics reveals the growth- and nutrient-dependent response of a streamlined marine methylotroph to methanol and naturally occurring dissolved organic matter. MBio. 2016;7:e01279–16. doi: 10.1128/mBio.01279-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mende DR, Bryant JA, Aylward FO, Eppley JM, Nielsen T, Karl DM, et al. Environmental drivers of a microbial genomic transition zone in the ocean’s interior. Nat Microbiol. 2017;2:1367–73. doi: 10.1038/s41564-017-0008-3. [DOI] [PubMed] [Google Scholar]
- 35.Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–93. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thaben PF, Westermark PO. Detecting rhythms in time series with RAIN. J Biol Rhythms. 2014;29:391–400. doi: 10.1177/0748730414553029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 38.Coenen AR, Hu SK, Luo E, Muratore D, Weitz JS. A primer for microbiome time-series analysis. Front Genet. 2020;11:310. doi: 10.3389/fgene.2020.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuhrman JA, Eppley RW, Hagström Å, Azam F. Diel variations in bacterioplankton, phytoplankton, and related parameters in the Southern California Bight. Mar Ecol Prog Ser. 1985;27:9–20. [Google Scholar]
- 40.Behrenfeld MJ, Falkowski PG. A consumer’s guide to phytoplankton primary productivity models. Limnol Oceanogr. 1997;42:1479–91. [Google Scholar]
- 41.Post AF, Dubinsky Z, Wyman K, Falkowski PG. Kinetics of light-intensity adaptation in a marine planktonic diatom. Mar Biol. 1984;83:231–8. [Google Scholar]
- 42.Falkowski PG, Kolber Z. Variations in chlorophyll fluorescence yields in phytoplankton in the world oceans. Funct Plant Biol. 1995;22:341–55. [Google Scholar]
- 43.Vaulot D, Marie D. Diel variability of photosynthetic picoplankton in the equatorial Pacific. J Geophys Res Ocean. 1999;104:3297–310. [Google Scholar]
- 44.Nicholson DP, Wilson ST, Doney SC, Karl DM. Quantifying subtropical North Pacific gyre mixed layer primary productivity from Seaglider observations of diel oxygen cycles. Geophys Res Lett. 2015;42:4032–9. [Google Scholar]
- 45.Yentsch CS. Distribution of chlorophyll and phaeophytin in the open ocean. Deep Sea Res Oceanogr Abstr. 1965;12:653–66. [Google Scholar]
- 46.Yentsch CS, Reichert CA. The effects of prolonged darkness on photosynthesis, respiration, and chlorophyll in the marine flagellate Dunaliella euchlora. Limnol Oceanogr. 1963;8:338–42. [Google Scholar]
- 47.Glooschenko WA, Curl H, Jr., Small LF. Diel periodicity of chlorophyll a concentration in Oregon coastal waters. J Fish Res Board Can. 1972;29:1253–9. [Google Scholar]
- 48.Cosper E. Influence of light intensity on diel variations in rates of growth, respiration and organic release of a marine diatom: comparison of diurnally constant and fluctuating light. J Plankton Res. 1982;4:705–24. [Google Scholar]
- 49.Fouilland E, Courties C, Descolas-Gros C. Size-fractionated phytoplankton carboxylase activities in the Indian sector of the Southern Ocean. J Plankton Res. 2000;22:1185–201. [Google Scholar]
- 50.Ragni M, d’Alcalà MR. Circadian variability in the photobiology of Phaeodactylum tricornutum: pigment content. J Plankton Res. 2007;29:141–56. [Google Scholar]
- 51.Bidigare RR, Buttler FR, Christensen SJ, Barone B, Karl DM, Wilson ST. Evaluation of the utility of xanthophyll cycle pigment dynamics for assessing upper ocean mixing processes at Station ALOHA. J Plankton Res. 2014;36:1423–33. [Google Scholar]
- 52.Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. In Methods in Enzymology (vol 148). Academic Press; 1987. p. 350–82.
- 53.Salomon E, Bar-Eyal L, Sharon S, Keren N. Balancing photosynthetic electron flow is critical for cyanobacterial acclimation to nitrogen limitation. Biochim Biophys Acta. 2013;1827:340–7. doi: 10.1016/j.bbabio.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 54.Ünlü C, Drop B, Croce R, van Amerongen H. State transitions in Chlamydomonas reinhardtii strongly modulate the functional size of photosystem II but not of photosystem I. Proc Natl Acad Sci USA. 2014;111:3460–5. doi: 10.1073/pnas.1319164111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.MacIntyre HL, Kana TM, Anning T, Geider RJ. Photoacclimation of photosynthesis irradiance response curves and photosynthetic pigments in microalgae and cyanobacteria. J Phycol. 2002;38:17–38. [Google Scholar]
- 56.Karl DM, Church MJ. Microbial oceanography and the Hawaii Ocean Time-series programme. Nat Rev Microbiol. 2014;12:699–713. doi: 10.1038/nrmicro3333. [DOI] [PubMed] [Google Scholar]
- 57.Armstrong GA. Greening in the dark: light-independent chlorophyll biosynthesis from anoxygenic photosynthetic bacteria to gymnosperms. J Photochem Photobio B Biol. 1998;43:87–100. [Google Scholar]
- 58.Foy RH, Smith RV. The role of carbohydrate accumulation in the growth of planktonic Oscillatoria species. Br Phycol J. 1980;15:139–50. [Google Scholar]
- 59.Cuhel RL, Ortner PB, Lean DRS. Night synthesis of protein by algae. Limnol Oceanogr. 1984;29:731–44. [Google Scholar]
- 60.Lacour T, Sciandra A, Talec A, Mayzaud P, Bernard O. Diel variations of carbohydrates and neutral lipids in nitrogen‐sufficient and nitrogen‐starved cyclostat cultures of Isochrysis sp. 1. J Phycol. 2012;48:966–75. doi: 10.1111/j.1529-8817.2012.01177.x. [DOI] [PubMed] [Google Scholar]
- 61.Lorenzen CJ. A note on the chlorophyll and phaeophytin content of the chlorophyll maximum. Limnol Oceanogr. 1965;10:482–3. [Google Scholar]
- 62.Jeffrey SW. Profiles of photosynthetic pigments in the ocean using thin-layer chromatography. Mar Biol. 1974;26:101–10. [Google Scholar]
- 63.Head EJH, Horne EPW. Pigment transformation and vertical flux in an area of convergence in the North Atlantic. Deep Sea Res Part II Top Stud Oceanogr. 1993;40:329–46. [Google Scholar]
- 64.Sun M-Y, Lee C, Aller RC. Anoxic and oxic degradation of 14C-labeled chloropigments and a 14C-labeled diatom in Long Island Sound sediments. Limnol Oceanogr. 1993;38:1438–51. [Google Scholar]
- 65.Champalbert G, Neveux J, Gaudy R, Le Borgne R. Diel variations of copepod feeding and grazing impact in the high-nutrient, low-chlorophyll zone of the equatorial Pacific Ocean (0°; 3° S, 180°). J Geophys Res Ocean. 2003;108:8145. 10.1029/2001JC000810.
- 66.Holzwarth AR, Müller MG, Reus M, Nowaczyk M, Sander J, Rögner M. Kinetics and mechanism of electron transfer in intact photosystem II and in the isolated reaction center: Pheophytin is the primary electron acceptor. Proc Natl Acad Sci USA. 2006;103:6895–6900. doi: 10.1073/pnas.0505371103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Grondelle R, Dekker JP, Gillbro T, Sundstrom V. Energy transfer and trapping in photosynthesis. Biochim Biophys Acta. 1994;1187:1–65. [Google Scholar]
- 68.Shimoda Y, Ito H, Tanaka A. Arabidopsis STAY-GREEN, Mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell. 2016;28:2147–60. doi: 10.1105/tpc.16.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oster U, Tanaka R, Tanaka A, Rüdiger W. Cloning and functional expression of the gene encoding the key enzyme for chlorophyll b biosynthesis (CAO) from Arabidopsis thaliana. Plant J. 2000;21:305–10. doi: 10.1046/j.1365-313x.2000.00672.x. [DOI] [PubMed] [Google Scholar]
- 70.Vavilin D, Vermaas W. Continuous chlorophyll degradation accompanied by chlorophyllide and phytol reutilization for chlorophyll synthesis in Synechocystis sp. PCC 6803. Biochim Biophys Acta. 2007;1767:920–9. doi: 10.1016/j.bbabio.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 71.Pružinská A, Tanner G, Anders I, Roca M, Hörtensteiner S. Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron–sulfur protein, encoded by the accelerated cell death 1 gene. Proc Natl Acad Sci USA. 2003;100:15259–64. doi: 10.1073/pnas.2036571100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baroli I, Niyogi KK. Molecular genetics of xanthophyll–dependent photoprotection in green algae and plants. Philos Trans R Soc Lond Ser B Biol Sci. 2000;355:1385–94. doi: 10.1098/rstb.2000.0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andersen RA, Bidigare RR, Keller MD, Latasa M. A comparison of HPLC pigment signatures and electron microscopic observations for oligotrophic waters of the North Atlantic and Pacific Oceans. Deep Sea Res Part II Top Stud Oceanogr. 1996;43:517–37. [Google Scholar]
- 74.Obata M, Taguchi S. The xanthophyll-cycling pigment dynamics of Isochrysis galbana (Prymnesiophyceae) during light-dark transition. Plankt Benthos Res. 2012;7:101–10. [Google Scholar]
- 75.Sajilata MG, Singhal RS, Kamat MY. The carotenoid pigment zeaxanthin—a review. Compr Rev Food Sci Food Saf. 2008;7:29–49. [Google Scholar]
- 76.Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylidès C, Havaux M. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA. 2012;109:5535–40. doi: 10.1073/pnas.1115982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goss R. Substrate specificity of the violaxanthin de-epoxidase of the primitive green alga Mantoniella squamata (Prasinophyceae) Planta. 2003;217:801–12. doi: 10.1007/s00425-003-1044-1. [DOI] [PubMed] [Google Scholar]
- 78.Viviani DA, Karl DM, Church MJ. Variability in photosynthetic production of dissolved and particulate organic carbon in the North Pacific Subtropical Gyre. Front Mar Sci. 2015;2:73. [Google Scholar]
- 79.Elling FJ, Becker KW, Könneke M, Schröder JM, Kellermann MY, Thomm M, et al. Respiratory quinones in Archaea: phylogenetic distribution and application as biomarkers in the marine environment. Environ Microbiol. 2016;18:692–707. doi: 10.1111/1462-2920.13086. [DOI] [PubMed] [Google Scholar]
- 80.Becker KW, Elling FJ, Schröder JM, Lipp JS, Goldhammer T, Zabel M, et al. Isoprenoid quinones resolve the stratification of redox processes in a biogeochemical continuum from the photic zone to deep anoxic sediments of the Black Sea. Appl Environ Microbiol. 2018;84:e2736–17. [DOI] [PMC free article] [PubMed]
- 81.Nowicka B, Kruk J. Occurrence, biosynthesis and function of isoprenoid quinones. Biochim Biophys Acta. 2010;1797:1587–605. doi: 10.1016/j.bbabio.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 82.Kruk J, Trebst A. Plastoquinol as a singlet oxygen scavenger in photosystem II. Biochim Biophys Acta. 2008;1777:154–62. doi: 10.1016/j.bbabio.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 83.Szymańska R, Kruk J. Plastoquinol is the main prenyllipid synthesized during acclimation to high light conditions in arabidopsis and is converted to plastochromanol by tocopherol cyclase. Plant Cell Physiol. 2010;51:537–45. doi: 10.1093/pcp/pcq017. [DOI] [PubMed] [Google Scholar]
- 84.Agrawal S, Jaswal K, Shiver AL, Balecha H, Patra T, Chaba R. A genome-wide screen in Escherichia coli reveals that ubiquinone is a key antioxidant for metabolism of long chain fatty acids. J Biol Chem. 2017;292:20086–99. doi: 10.1074/jbc.M117.806240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ksas B, Légeret B, Ferretti U, Chevalier A, Pospíšil P, Alric J, et al. The plastoquinone pool outside the thylakoid membrane serves in plant photoprotection as a reservoir of singlet oxygen scavengers. Plant Cell Environ. 2018;41:2277–87. doi: 10.1111/pce.13202. [DOI] [PubMed] [Google Scholar]
- 86.Foyer CH, Noctor G. Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol Plant. 2003;119:355–64. [Google Scholar]
- 87.Long SP, Humphries S, Falkowski PG. Photoinhibition of photosynthesis in nature. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:633–62. [Google Scholar]
- 88.Triantaphylidès C, Havaux M. Singlet oxygen in plants: production, detoxification and signaling. Trends Plant Sci. 2009;14:219–28. doi: 10.1016/j.tplants.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 89.Pospíšil P. Molecular mechanisms of production and scavenging of reactive oxygen species by photosystem II. Biochim Biophys Acta. 2012;1817:218–31. doi: 10.1016/j.bbabio.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 90.Lichtenthaler HK. Biosynthesis, accumulation and emission of carotenoids, α-tocopherol, plastoquinone, and isoprene in leaves under high photosynthetic irradiance. Photosynth Res. 2007;92:163–79. doi: 10.1007/s11120-007-9204-y. [DOI] [PubMed] [Google Scholar]
- 91.Shi T, Ilikchyan I, Rabouille S, Zehr JP. Genome-wide analysis of diel gene expression in the unicellular N2-fixing cyanobacterium Crocosphaera watsonii WH 8501. ISME J. 2010;4:621. doi: 10.1038/ismej.2009.148. [DOI] [PubMed] [Google Scholar]
- 92.Muñoz-Marín M, del C, Shilova IN, Shi T, Farnelid H, Cabello AM, et al. The transcriptional cycle is suited to daytime N2 fixation in the unicellular cyanobacterium “Candidatus Atelocyanobacterium thalassa” (UCYN-A) MBio. 2019;10:e02495–18. doi: 10.1128/mBio.02495-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ashworth J, Coesel S, Lee A, Armbrust EV, Orellana MV, Baliga NS. Genome-wide diel growth state transitions in the diatom Thalassiosira pseudonana. Proc Natl Acad Sci USA. 2013;110:7518–23. doi: 10.1073/pnas.1300962110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smith SR, Gillard JTF, Kustka AB, McCrow JP, Badger JH, Zheng H, et al. Transcriptional orchestration of the global cellular response of a model pennate diatom to diel light cycling under iron limitation. PLOS Genet. 2016;12:e1006490. doi: 10.1371/journal.pgen.1006490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nymark M, Valle KC, Brembu T, Hancke K, Winge P, Andresen K, et al. An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum. PLoS ONE. 2009;4:e7743. doi: 10.1371/journal.pone.0007743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, Trethewey R, et al. Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol. 2006;7:R76. doi: 10.1186/gb-2006-7-8-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Waldbauer JR, Rodrigue S, Coleman ML, Chisholm SW. Transcriptome and proteome dynamics of a light-dark synchronized bacterial cell cycle. PLoS ONE. 2012;7:e43432. doi: 10.1371/journal.pone.0043432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kana TM, Geider RJ, Critchley C. Regulation of photosynthetic pigments in micro-algae by multiple environmental factors: a dynamic balance hypothesis. N Phytol. 1997;137:629–38. [Google Scholar]
- 99.Escoubas J-M, Lomas M, LaRoche J, Falkowski PG. Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA. 1995;92:10237–41. doi: 10.1073/pnas.92.22.10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Van Mooy BAS, Devol AH. Assessing nutrient limitation of Prochlorococcus in the North Pacific Subtropical Gyre by using an RNA capture method. Limnol Oceanogr. 2008;53:78–88. [Google Scholar]
- 101.Moore CM, Mills MM, Arrigo KR, Berman-Frank I, Bopp L, Boyd PW, et al. Processes and patterns of oceanic nutrient limitation. Nat Geosci. 2013;6:701–10. [Google Scholar]
- 102.Muratore D, Boysen AK, Harke MJ, Becker KW, Casey JR, Coesel SN, et al. Community-scale synchronization and temporal partitioning of gene expression, metabolism, and lipid biosynthesis in oligotrophic ocean surface waters. 2020. https://www.biorxiv.org/content/10.1101/2020.05.15.098020v1.
- 103.Saito MA, Bertrand EM, Dutkiewicz S, Bulygin VV, Moran DM, Monteiro FM, et al. Iron conservation by reduction of metalloenzyme inventories in the marine diazotroph Crocosphaera watsonii. Proc Natl Acad Sci USA. 2011;108:2184–9. doi: 10.1073/pnas.1006943108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marchetti A, Parker MS, Moccia LP, Lin EO, Arrieta AL, Ribalet F, et al. Ferritin is used for iron storage in bloom-forming marine pennate diatoms. Nature. 2008;457:467. doi: 10.1038/nature07539. [DOI] [PubMed] [Google Scholar]
- 105.White AE, Barone B, Letelier RM, Karl DM. Productivity diagnosed from the diel cycle of particulate carbon in the North Pacific Subtropical Gyre. Geophys Res Lett. 2017;44:3752–60. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.