Abstract
Positive species interactions underlie the functioning of ecosystems. Given their importance, it is crucial to understand the stability of positive interactions over evolutionary timescales, in both constant and fluctuating environments; e.g., environments interrupted with periods of competition. We addressed this question using a two-species microbial system in which we modulated interactions according to the nutrient provided. We evolved in parallel four experimental replicates of species growing in isolation or together in consortia for 200 generations in both a constant and fluctuating environment with daily changes between commensalism and competition. We sequenced full genomes of single clones isolated at different time points during the experiment. We found that the two species coexisted over 200 generations in the constant commensal environment. In contrast, in the fluctuating environment, coexistence broke down when one of the species went extinct in two out of four cases. We showed that extinction was highly deterministic: when we replayed the evolution experiment from an intermediate time point we repeatably reproduced species extinction. We further show that these dynamics were driven by adaptive mutations in a small set of genes. In conclusion, in a fluctuating environment, rapid evolution destabilizes the long-term stability of positive pairwise interactions.
Subject terms: Microbial ecology, Bacterial genetics, Population genetics
Introduction
Positive interactions—mutualisms and commensalisms—underlie the functioning of virtually every ecosystem on Earth [1, 2]. One example of such positive interactions is the mutualism between microscopic algae and corals, which supports the diversity and stability of coral reefs [3]. More broadly, mutualistic and commensal interactions in microbial communities underlie the biogeochemical cycling of elements on Earth [4]. Thus, ecosystem functioning relies on the stability of positive interactions. Nevertheless, positive interactions may be destabilized by environmental change; e.g., changes associated to human activities [5–7]. Warmer ocean temperatures caused by climate change shifts the corral–microalgae symbiosis into parasitism [8]. Nitrogen pollution selects for less cooperative rhizobium disrupting the legume–rhizobium mutualism [6]. These two examples involve stresses affecting particular pair of species and their beneficial influence on each other; i.e., positive pairwise interactions. Given that we are losing such valuable partnerships in nature, it is pressing that we better understand the factors that destabilize positive pairwise interactions and lead to their breakdown.
Positive pairwise interactions may breakdown due to: (1) interactions changing from mutualism or commensalism into competition or parasitism, (2) interactions evolving toward neutralism, and (3) species going extinct [9]. Both ecological and evolutionary factors have been identified as major drivers destabilizing interactions [9]. Ecological changes such as alterations in nutrient availability can shift interactions from positive to negative [10, 11]. Interactions can also shift from positive to negative due to rapid evolution [12, 13]. Although we understand some aspects of how ecological and evolutionary factors break positive pairwise interactions, we lack a general understanding on how these factors influence the long-term stability of these interactions. For example, how resilient are positive interactions when species evolve and adapt to new environments? And, how resilient are positive interactions when, in addition to evolutionary change, they are interrupted with periods in which interactions become negative? More generally, how stable are interactions over evolutionary timescales in fluctuating environments?
Some of these questions have been addressed by studies of coexistence in variable environments. Environmental fluctuations have been long recognized as a mechanism that can promote coexistence [14], but only recently, evolution has been incorporated into coexistence theory [15–17]. Coexistence models in variable environment show that high rates of adaptive evolution may lead to coexistence breakdown [16]. In other words, rapid evolution destabilizes pairwise interactions that were otherwise stable in fluctuating environments. However, these theoretical predictions are based on negative interactions—e.g., competition, predation, and parasitism. It is unclear if we can expect the same outcomes for positive interactions. Moreover, these theoretical predictions are not well supported by experiments showing that adaptation—although slower in fluctuating environments—does not lead to coexistence breakdown [18, 19].
Another important aspect that has not been fully explored is the genetics of adaptation of interacting species. There is a rich body of theoretical and empirical work on the genetics of adaptation of single species in both constant and fluctuating environments. In a constant environment, the first step of adaptation in an asexual population is characterized by the fixation of the first beneficial mutation that succeeds at escaping clonal interference and genetic drift [20]. Instead, in a fluctuating environment, the fate of a mutation depends on (1) its averaged fitness effect across environments and (2) the rate of environmental fluctuations [21]. Even though these studies have provided us with a good understanding on the genetics of adaptation to multiple environments, they have focused on single species. Thus, we do not know the role that other species play in shaping these adaptive dynamics. In other words, what is the relative contribution of abiotic vs biotic factors shaping evolutionary trajectories? Do species mostly adapt to their physical environment or to their interactions with other species? Answering these questions is relevant because if biotic factors are important evolutionary forces, evolutionary responses might differ from those expected from single species. In such case, evolution should be studied in a community context [22, 23].
In this study we used a two-species system to start addressing some these questions. The two species, Acinetobacter johnsonii and Pseudomonas putida, interact through the use of resources in the external environment [12]. When grown in an environment limited by citrate, they compete for it, while when grown in an environment limited by benzyl alcohol, P. putida cross-feeds on the benzoate excreted by A. johnsonii. In a previous study, we showed that this cross-feeding interaction was stable over ecological timescales, i.e., over 6 days or ~40 generations [24]. In this study, we explored the stability of such commensal interaction over evolutionary timescales in a constant environment and in a fluctuating environment imposing ecological changes between positive and negative interactions (i.e., daily switches from benzyl alcohol to citrate). We observed that species coexisted over evolutionary timescales in a constant environment, but coexistence broke down in two out of four cases in the fluctuating environment. We further show that these dynamics were driven by evolutionary changes.
Materials and methods
Strains and growth conditions
We used a previously characterized consortium of A. johnsonii strain C6 and P. putida stain KT2440 [12, 24, 25].
Strains were grown in FAB minimal medium (1 mM MgCl2, 0.1 mM CaCl2, 0.003 mM FeCl3, 15 mM (NH4)2SO4, 33 mM Na2HPO4, 22 mM KH2PO4, and 50 mM NaCl) supplemented with either 0.6 mM benzyl alcohol or 1 mM citrate. All solutions were filter sterilized using filters that were washed with one liter of ddH2O to remove traces of carbon in the filters. We grew the cultures in 40 ml glass vials with screw caps containing TFE-lined silicone septa. Vials and screw caps were treated to eliminate any trace of assimilable organic carbon [24].
Evolution experiment
We performed our evolution experiment by daily transferring our strains: (1) into FAB medium supplemented with benzyl alcohol, referred as constant benzyl alcohol environment; (2) into FAB medium supplemented with citrate, referred as constant citrate environment, and (3) by switching daily between FAB medium supplemented with citrate and FAB medium supplemented with benzyl alcohol, referred as fluctuating environment. In total we evolved 36 experimental lines (Fig. S1): four replicated co-cultures in each of the three conditions, referred as consortia (i.e., a total of 12 consortia) and four replicated monocultures of each species in each of the three conditions, referred as populations (i.e., a total of 24 populations). We evolved these experimental lines in parallel for 30 days (∼200 generations).
To start the experimental lines, we streaked A. johnsonii and P. putida onto Lysogeny Broth (LB) agar plates supplemented with streptomycin (64 µg/ml) and gentamicin (10 µg/ml), respectively. We then picked four single colonies from each LB agar plate and grew them separately in 3 ml of LB broth supplemented with antibiotics. We incubated the cultures for ∼16 h at 30 °C with constant shaking (220 rpm). Part of the overnight culture was used to prepare the ancestors’ glycerol stocks (20% glycerol) and part was used to launch the experimental lines. To launch the experimental lines, we spun down 1 ml of the overnight culture and washed it three times with FAB medium without carbon source to remove LB and antibiotics. Washed cells were diluted 10,000-fold into 10 ml fresh FAB medium supplemented with the desired carbon source and incubated for 1 day at 26 °C and shaken at 150 rpm. These acclimated cultures were then diluted 100-fold into 10 ml fresh FAB medium supplemented with the desired carbon source to start the monocultures or were mixed at a 1:1 volumetric ratio and diluted 100-fold into 10 ml media to start the co-cultures. Initial population size was ~105 cells per species. Monocultures and co-cultures were propagated by daily transfers of 0.1 ml of culture into 9.9 ml of fresh medium—approximately 6.64 generations (log2 100) of binary fission per day—for 30 days. At the end of each day, population densities of A. johnsonii and P. putida from one of the replicates (replicate #2) were estimated by plating cultures on LB agar with streptomycin (64 µg/ml) and gentamicin (10 µg/ml), respectively.
Population samples of all 36 lines were frozen weekly and used to prepare the populations’ glycerol stocks (20%). In addition, we prepared glycerol stocks from single clones isolated at the end experiment. Briefly, we plated the populations in LB agar plates supplemented with antibiotics. We then picked one random colony from each plate and inoculated each colony in 3 ml of liquid LB for 16 h at 26 °C with continuous shaking (150 rpm) and resuspended this overnight culture in glycerol. We stored the glycerol stocks (20%) at −80 °C for further analyses; i.e., genome sequencing and growth curves.
Genome sequencing and variant calling
We sequenced the genomes of single clones isolated from: (1) 8 ancestors; (2) 36 experimental lines at the end of the experiment (i.e., at generation 200) and 3) consortia grown in the fluctuating environment at generation 92.
Single clones were revived from glycerol stocks in 3 ml of LB media and grown overnight at 30 °C with constant shaking (220 rpm). We used 1 ml of this overnight culture and isolated the genomic DNA using the Wizard Genomic DNA Purification Kit (Promega). Eurofins Genomics LLC prepared the libraries for sequencing (DNA fragmentation, adapter ligation, amplification, and size selection) and sequenced genomes using Illumina technology (paired end, read length: 2 × 150 bp, MiSeq; Eurofins Genomics Europe Shared Services GmbH). All of the sequences can be accessed at the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under Bioproject ID number PRJNA623337.
We used these data to do genome re-sequencing (Genetic Diversity Center, ETH Zürich). First, we controlled for the quality of the raw Illumina sequences with FastQC (version 0.11.4). We did a variant calling analysis using the breseq pipeline (version 0.30.1) and the utility program GDtools [26]. The sequence reads from each clone—from both the ancestors and the evolved lines—were aligned to the reference genomes of P. putida KT2440 (reference NC_002947; https://www.ncbi.nlm.nih.gov/nuccore/NC_002947) and A. johnsonii isolate C6 (reference FUUY01.1; https://www.ncbi.nlm.nih.gov/nuccore/FUUY00000000.1/). We used the following parameters for calling genetic changes: polymorphism-prediction; max-displayed-reads 300. We identified mutations that occurred de novo during the evolution experiment by pairwise comparison between an evolved clone and its respective ancestral clone. De novo mutations were those mutations that were uniquely observed in the evolved clone and not in the ancestral clone.
Selection and enrichment statistical tests
We identified genes under selection based on the observed genetic parallelism. We assumed that it would be highly unlikely that the same gene would be mutated in several independently evolved lines unless these mutations had been enriched by selection. Therefore, to identify genes under positive selection we estimated the strength of parallelism based on a Poisson distribution: ; with λ equal to the . The reported P values in Table 1 were estimated from the Poisson cumulative expectation, P(x ⩾ observed, 1), using the ppois function in R (version 3.2.3).
Table 1.
Parallel mutations are commonly observed across experimental conditions.
| Species | Target | Product/Function | Mut | P value | Frequency across replicates | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| cstBA mono | cstBA co | cstCi mono | cstCi co | Flu mono | Flu co | |||||
| A. johnsonii | oatA | O-acetyltransferase OatA | 15 | 1 × 10−40 | 1 | 1 | 0.75 | – | 1 | 0 (0/2) |
| P. putida | gacS | Sensor protein GacS | 9 | 5 × 10−24 | 0.25 | 0 | 0.75 | 0.5 | 0.25 | 0.5 |
| P. putida | flagelluma | Flagellar components | 7 | 3 × 10−17 | 0.25 | 0 | 0.25 | 0.25 | 0.75 | 0.25 |
| P. putida | fleQ | Transcriptional regulator FleQ/cyclic-di-GMP | 5 | 3 × 10−15 | 0.25 | 0.25 | 0 | 0.25 | 0.25 | 0.25 |
“Target” refers to the most commonly mutated gene or function; “Mut” refers to the number of mutations observed across lines. “P value” testing whether the number of mutations in a gene is random (Poisson cumulative expectation). “Frequency across replicates” in different conditions (cstBA constant Benzyl Alcohol, cstCi constant Citrate, Flu fluctuating environment, mono monoculture, co co-culture). Frequencies reflect how replicable are mutations across replicates: 1 = mutation in 4 out of 4 replicates; 0.75 = 3 out of 4; 0.5 = 2 out of 4; 0.25 = 1 out of 4; 0 = 0 out of 4.
aflagellum: genes related motility (GO: 2000147); mutations in flgH (5), fliQ (1), flhB (1). P value shown was calculated for flgH.
We used the Enrichment analysis tool from Panther (pantherdb.org) version 14.1 for the statistical overrepresentation test [27]. Genes involved in a particular cellular function were overrepresented based on a small and significant P value obtained from Fisher’s exact test indicating that is unlikely that the number of genes involved in that particular cellular function occurred randomly as determined by a reference gene list from P. aeruginosa.
Growth curves
We did growth curve experiments to characterize maximum growth rate in exponential phase, µmax, and yield, Y, defined as the amount of biomass produced per unit of resource of single clones grown in isolation. Bacteria were acclimated to their assay condition as previously described for the beginning of the serial dilution experiment. We transferred 2 µl of the saturated culture intro 198 µl of fresh media dispensed in each of the wells of the 96-wells plate. To avoid evaporation, we covered the plates with a lid, which we sealed with silicon grease. We incubated the plates at 30 °C with constant shaking inside a photospectrometer (EonTM, BioTekTM), whose lowest detection limit was 0.003. OD measurements were acquired every 5 min for 24 h. We replicated the experiments three times (technical replicates) and fitted µmax and Y from these three growth curves using Matlab (version R2017a, Mathworks). We then compared these parameters in a pairwise fashion between the ancestor and the evolved clone using a two-tailed t-test. Due to the large temporal variability between experiments, we compared the growth measurements for the ancestor and the evolve clone derived from the same day/experiment (i.e., temporal replicates).
Results
Coexistence is stable over evolutionary timescales in a constant but not in a fluctuating environment
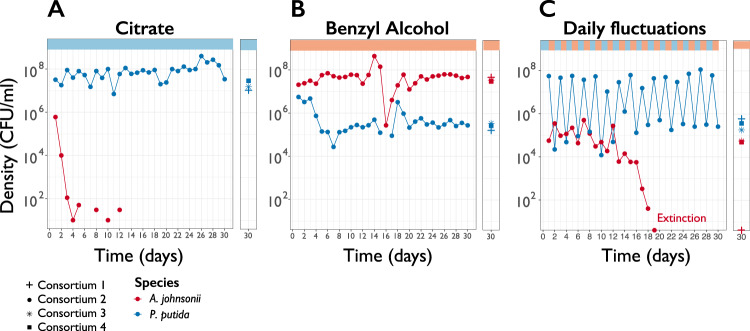
We explored if species coexisted over evolutionary timescales, both in constant and fluctuating environments (Fig. S1). In a constant citrate environment, in all four consortia, P. putida drove A. johnsonii to extinction after 1 week (Fig. 1a), which was expected from their ecological dynamics [24]. In contrast, in a constant benzyl alcohol environment, the two species coexisted for 200 generations in all four consortia (Fig. 1b). A. johnsonii maintained high population densities throughout the experiment, despite a steep drop in densities at day 16 followed by recovery (Fig. 1b). At day 30, the average population density across replicates of A. johnsonii was 4.0 × 107 CFU mL−1 ± 4.3 × 106 CFU mL−1 (mean ± standard error of the mean). P. putida coexisted with A. johnsonii and reached ~2.7 × 105 CFU mL−1 ± 3.8 × 104 CFU mL−1. P. putida densities in co-culture were one order of magnitude higher than in monoculture (Fig. S2) indicating that the positive effect that A. johnsonii has on P. putida’s was maintained for 200 generations. Thus, cross-feeding interactions were stable over evolutionary timescales in a constant environment. To summarize, in constant environments replicated communities converged into the same outcome: either competitive exclusion or coexistence.
Fig. 1. Two-species consortium displays different eco-evolutionary dynamics in different environments.
Population density (CFU ml−1) trajectories of co-cultures from consortium #2 are shown over 30 days of serial transfers (100-fold dilution) in three environments. Each circle represents the final density of each species after daily cycles of growth; lines connecting circles help visualization and do not have any biological meaning. Densities from the remaining three consortium at day 30 are also plotted (sums, stars and squares symbols). A. johnsonii’s extinction was confirmed by plating consortia for five consecutive days in LB supplemented with streptomycin and confirming zero CFU ml−1.
In contrast, in the fluctuating environment replicated communities diverged into two evolutionary outcomes. In two cases, coexistence broke down, while in the other two cases species coexisted until the end of the experiment (Fig. 1c). In consortia #1 and #2, A. johnsonii went extinct after 18 and 19 days, respectively. In contrast, in consortia #3 and #4, A. johnsonii coexisted with P. putida until the end of the experiment and reached a final population density of 5.6 × 104 CFU mL−1 and 5.0 × 104 CFU mL−1, respectively. To asses if fluctuations in carbon sources alone could lead to the extinction of A. johnsonii, we evolved A. johnsonii in isolation in a fluctuating environment. In all four monocultures, in the fluctuating condition, A. johnsonii stably persisted during the entire experiment (Fig. S3), suggesting there was something specific about the interaction between A. johnsonii and P. putida that caused the extinction of A. johnsonii in co-cultures.
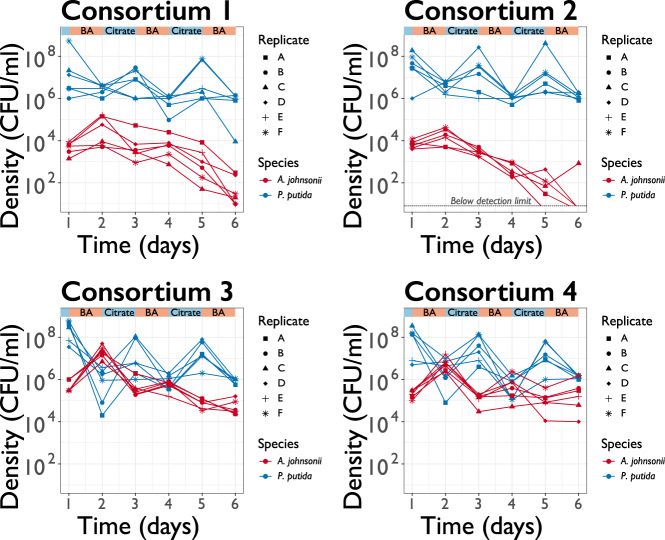
A key question in evolutionary biology is the extent to which extinctions are stochastic or deterministic events [28, 29]. On the one hand, A. johnsonii’s extinction may have been caused by random demographic fluctuations, occasionally resulting in populations that are so small that they are unable to overcome the serial dilution regime. On the other hand, deterministic changes in organismal features—such as adaptive mutations in either A. johnsonii or P. putida—may have determined whether or not A. johnsonii went to extinction. For example, P. putida could have mutated to attach to A. johnsonii to better extract benzoate (i.e., evolution of parasitism; 12). Another possibility is that A. johnsonii accumulated deleterious mutations leading to its collapse [29]. In this case, once such mutations have risen to large enough frequency, population dynamics become deterministic and extinction becomes a predictable event. We assessed if extinction was a random or deterministic outcome. To do so, we replayed the evolution experiment from an intermediate time point and quantified the frequency in which A. johnsonii goes extinct. We revived frozen population samples from each consortium at day 14 (~92 generations) and transferred the populations for 6 days in a fluctuating environment, with six replicates per consortium. In all six replicates derived from consortium #1, A. johnsonii’s densities dropped close to extinction after 6 days (Fig. 2). For consortium #2, A. johnsonii went extinct after 5 days in five out of six cases. In contrast, in consortia #3 and #4, A. johnsonii sustained an intermediate population density and reached a final population density of ~6.1 × 104 CFU mL−1 ± 2.2 × 104 CFU mL−1 and 4.1 × 105 CFU mL−1 ± 2.3 × 105 CFU mL−1 for all six replicates, respectively. In conclusion, when we replayed the evolution experiment from an intermediate time point we saw the extinction of A. johnsonii in consortia #1 and 2 but not in consortia #3 and 4, which is consistent with the original evolution experiment. Therefore, in our experimental setting, extinction was deterministic.
Fig. 2. Coexistence and extinction were reproducible when we replayed the evolution experiment from an intermediate time point.
Population density (CFU ml−1) trajectories of six replicates launched from populations samples frozen at day 14 during the evolution experiment. Each dot represents the final density of each species after daily cycles of growth; lines connecting dots help visualizing replicates dynamics (represented by different symbols) and do not have any biological meaning.
Species show signs of adaptive evolution after 200 generations
Based on this reproducible outcome we hypothesized that genetic changes occurring as early as 92 generations underlay these dynamics. Before exploring if these early genetic changes in fact occurred, we first characterized the genetic changes accumulated during the entire length of the experiment, i.e., 200 generations. In particular we looked for signs of adaptation. To do so, we sequenced the genome of a single clone isolated from each of the 36 experimental lines at generation 200 and identify all de novo mutations relative to their respective ancestral genome (see “Material and methods”).
On average each clone accumulated two mutations after 200 generations (Table S1). Each clone had on average 1.3 point mutations, 0.5 short insertions and deletions, and 0.2 large deletions (>30 bp). Importantly, the point mutations displayed a strong signal of adaptive evolution: over all 42 genomes, the ratio of nonsynonymous to synonymous mutations was 3.9, and most of the mutations occurred in open reading frames (Table S1). Moreover, many mutations were shared among replicate lines, e.g., in some cases, the exact same nonsynonymous point mutation was shared among two or more lines (Table S1). More generally, mutations targeting the same gene were found in parallel across lines and conditions for each species (Table 1). This strong convergence observed at the gene level is consistent with previous studies showing that the gene level is the appropriate level to assess convergence [30]. For P. putida, the most commonly mutated gene was the gacS gene transcribing the sensor protein GacS (mutations in 9 out of 24 lines). Mutations affecting regulation of motility were also commonly found across lines of P. putida (GO: 2000147; test for statistical overrepresentation, P = 1.31 × 10−4; see “Materials and methods”). In the case of A. johnsonii, 15 of the 20 lines had mutations in the oatA gene. The targeting of 15 different mutations to one gene suggests the oatA gene was under strong selection given that the probability of observing 15 random mutations in a target gene of 1926 bp is extremely low (P = 9 × 10−38). The oatA gene encodes the O-acetyltransferase, OatA. OatA from other bacterial species has been reported to modify the bacterial cell wall, for example, to prevent the antibacterial activity of lysozymes [31]. It is unclear what the function of OatA is in our experimental conditions but given the observed parallelism (Table 1), we hypothesized these mutations were adaptive.
To better understand the nature of this putative adaptive benefit, we analyzed whether strains with these mutations would show increased growth when grown in isolation, without their partner. We characterized the growth of single clones in benzyl alcohol or benzoate, as well as in citrate, and compared it to the growth of the ancestor. We did growth curves in triplicates for each clone in each carbon source and estimated the maximum growth rate in exponential phase, µmax, and the amount of biomass produced per unit of resource, i.e., the yield, Y (Table S2).
Overall, evolved clones increased both their growth rates and their yields on both benzyl alcohol and citrate compared to the ancestor (Figs. 3, S4). That being said, A. johnsonii’s clones evolved in the fluctuating environment did not improved as much as clones evolved in constant environments, and, in some cases, grew significantly worse than the ancestor (Table S2, Fig. S4). In contrast, P. putida clones evolved in constant citrate and in the fluctuating environment grew significantly better than the ancestor. Importantly, some of the evolved clones had only one mutation (e.g., on the oatA or gacS gene; Table S1), therefore, the grow improvement observed can be directly attributed to the effect of the mutation.
Fig. 3. Evolved clones tended to grow better than the ancestor in all carbon sources.
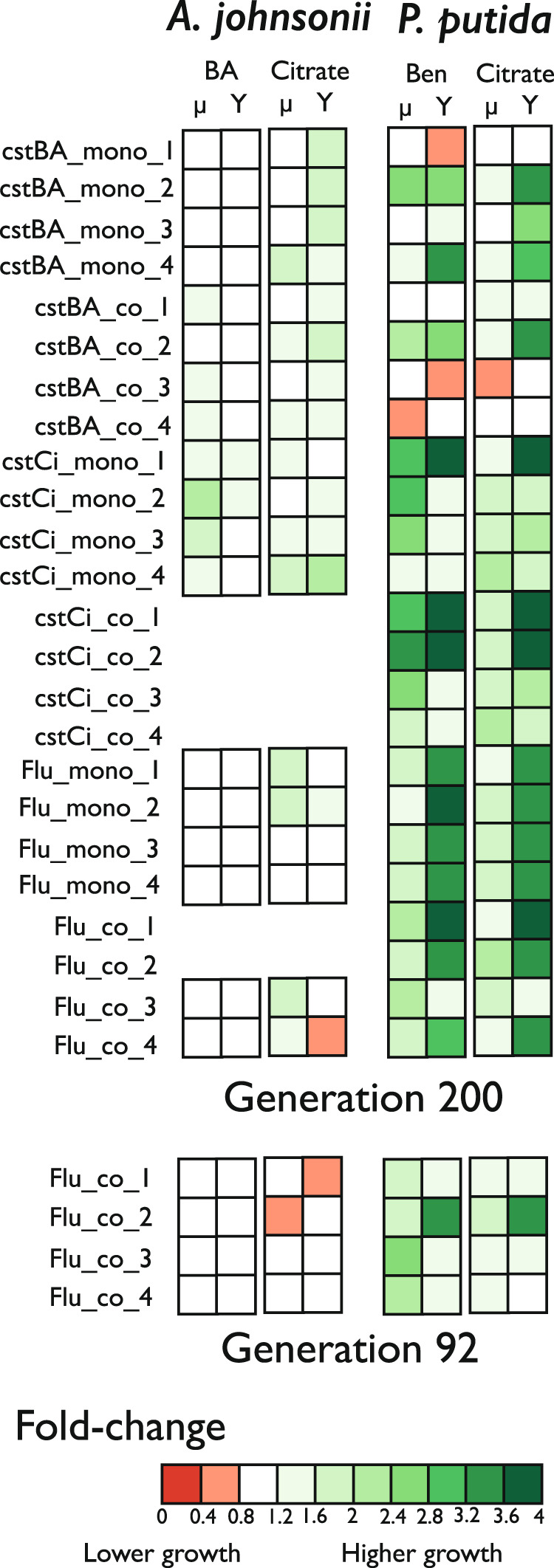
Summary of the growth fold changes (evolved clones vs ancestor) from two parameters, the maximum growth rate (µ) and yield (Y) estimated from growth curves in benzyl alcohol (BA), benzoate (Ben), and citrate. The column in the left lists all the experimental lines from which single clones were isolated; i.e., three environments, constant Citrate (cstCi), constant Benzyl Alcohol (cstBA), fluctuating (Flu), in monoculture (mono) and co-culture (co). Red colors indicate lower average growth and green colors higher average growth. Data can be found in Table S2.
In summary, based on the type of mutations observed, their parallelism and their link to growth improvements, it is likely that half of the mutations observed at generation 200 are adaptive. To confirm if these and other mutations are indeed adaptive future experiments would require inserting single mutations into the ancestral background and measure their fitness effects.
Early adaptation of P. putida influences A. johnsonii’s evolutionary trajectory
We established that clones acquired adaptive mutations after 200 generations of evolution in constant and fluctuating environments. Based on this rapid evolution we tested if mutations appeared as early as 92 generations in the fluctuating environment and if this adaptation could have been associated with the extinction or persistence of A. johnsonii. We characterized the first steps of adaptation by sequencing the genome of single clones isolated from the consortia evolved in the fluctuating environment at generation 92; i.e., before the extinction of A. johnsonii.
In all four consortia, P. putida accumulated one or more mutations at generation 92 (Table S2). Interestingly, these mutations targeted the same genes and functions mutated in clones isolated from generation 200 and evolved under different conditions. For example, clones isolated from generation 92 had mutations on either the uvrY or the gacS gene; the exception being the clone from consortium#3, which had a unique mutation not observed in any other line or condition. In addition, P. putida’s clones from generation 92 grew better than the ancestor: both the maximum growth rate and the yield were on average 1.9-fold that of the ancestor in benzoate and 1.8-fold that of the ancestor in citrate.
In the case of A. johnsonii, clones isolated at generation 92 from consortia #1 and #2—in which A. johnsonii went later on extinct—did not have mutations and were identical to the ancestor (Table S1). Accordingly, clones grew as well as the ancestor (Fig. 3, Table S2). In contrast, clones from consortia #3 and #4—in which A. johnsonii persisted until the end of the evolution experiment—had one or two mutations. These mutations targeted the epsL gene coding the putative sugar transferase EpsL and the hyaD gene transcribing the hyaluronan synthase and were only observed in the co-cultures evolving in the fluctuating environment (Table 1). One caveat of our experimental design is that we only sequenced one clone per time point; thus, we do not know if these mutations were representative of the entire population. That said, we observed the exact same mutation at generation 92 and at generation 200 (Table S1). If these mutations were in low frequency in the population, e.g., at 1%, the probability of randomly sampling the same mutation twice is very low (P = 0.01). Thus, it is likely these mutations were already at high frequency in the population or that they reached high frequency by generation 200. In either case, the fact that we observed these mutations both at generations 92 and 200 suggests that the mutations were numerically well-established in the population.
Intriguingly, clones carrying these mutations did not have significant growth improvements when A. johnsonii was growing in isolation on benzyl alcohol or on citrate (Figs. 3, S4, Table S2). Despite not knowing their specific function or phenotypic effects, we posit that these mutations prevented the extinction of A. johnsonii, potentially by modulating the interaction that A. johnsonii has with P. putida.
Taken together, we hypothesize that P. putida’s adaptation to a fluctuating environment could have exerted an additional selective pressure on A. johnsonii and this indirect effect could have led to the extinction of A. johnsonii. To test this hypothesis, we used a mathematical model that we previously developed and that accurately predicted community dynamics from single-species behavior (i.e., growth essays) [24]. This mathematical model explicitly modeled the changes in resources and the bacterial growth. The model was informed by growth and microbial activities measured in single-species experiments; i.e., growth curves and metabolite analysis of monocultures. The five parameters used to parametrize our model were: (1) the maximum growth rate, (2) the maximum resource uptake rate, (3) the half-saturation constant, (4) the duration of the lag phase, and (5) the benzoate excretion rate. In our current study, we implemented this model to test eco-evolutionary dynamics (e.g., dynamics observed in Fig. 1C). We parametrized the model using the growth curves of the ancestor and of the clone of P. putida at generation 92 and 200 from consortium #2 (Table S3). We contrasted two scenarios: (1) a co-culture of both ancestors in a fluctuating environment and (2) a co-culture of A. johnsonii’s ancestor with the evolved clone of P. putida. Our model was able to capture the extinction of A. johnsonii (Fig. S5), although the speed at which extinction occurred was not the same as in the evolution experiment (Fig. 1c). A. johnsonii’s extinction occurred after 28 and 22 days in competition with a clone from generations 92 and 200, respectively (Fig. S5). These results show that, in principle, the growth improvement of one species can indirectly affect a second species and drive it to extinction. In conclusion, early adaptation of P. putida could have played a decisive role in causing extinction of A. johnsonii.
Discussion
We have shown that in a constant environment, a cross-feeding interaction between two species is stable over evolutionary timescales despite species changing through adaptation. In contrast, in a fluctuating environment, in which cross-feeding periodically switches to competition, coexistence sometimes breaks down. We have shown that extinction is highly deterministic: we repeatably reproduce species extinction when we replay the evolution experiment from an intermediate time point. We hypothesize that early adaptation of one of the species affects the other species and drives it to extinction, unless this second species evolves to counteract this effect.
The nature of the environment is important to predict how stable are species interactions over evolutionary timescales
Although we understand some aspects of the factors that control the long-term stability of positive interactions, most evolutionary studies on positive pairwise interactions have focused on well-established mutualistic interactions between species that have been co-evolving for tens of thousands to many millions of years [32, 33]. Instead, here we have focused on the long-term stability of a newly established positive pairwise interaction, which is more dynamic in respect of its environments; i.e., it changes from positive to negative according to the culture conditions. A previous experimental evolution study showed that a newly established mutualistic interaction between a bacterium and an archaeon was stable for thousands of generations in a constant environment despite species evolving [34]. Interestingly, interactions were less stable during the first 300 generations (two out 24 co-cultures went extinctic) suggesting the first step of adaptation might be relatively unstable [35]. Similarly, in our study we have shown that despite a period of erratic growth (Fig. 1b), evolution does not breakdown the cross-feeding interaction between A. johnsonii and P. putida in a constant benzyl alcohol environment. This was not the case in another study, in which P. putida evolved to exploit A. johnsonii, leading to commensalism breakdown [12]. The main difference between this previous study and our study is a lack of spatial structure in our environment. Spatial structure is necessary for the selection of mutants that physically attach to the surface of A. johnsonii’s microcolonies [12, 36]. Thus, spatial structure is an important driver for the evolution of species interactions [37, 38].
The evolution of species interactions is not only shaped by spatial structure but also by temporal structure. In a fluctuating environment, starting from an ecologically stable consortium, evolution can destabilize species interactions leading to the extinction of one of the community members (Fig. 1c). Interestingly, extinction is only one of the two evolutionary outcomes—two out of four co-cultures show stable coexistence. This observation agrees with previous theoretical predictions anticipating the collapse of coexistence in variable environments when evolution is rapid [16]. That said, more experimental studies on the evolution of positive species interactions are needed to confirm if this is a general trend [39]. Future directions to confirm if this is a general trend may require studies in variable environments that are not so predictable as in our study, such as environments that change stochastically or in environments with several stresses acting together, which may reflect better nature [40, 41].
In summary, rapid evolution may lead to coexistence breakdown in a fluctuating environment. This is not the case in the constant environment in which all four replicates converge into a single community composition. Thus, the environmental context—e.g., spatial and temporal structure—is important to predict the evolution of species interactions.
Adaptation to abiotic and not biotic conditions prevails in our experimental system
In natural environments it is difficult to disentangle the relative contribution of abiotic and biotic factors to adaptation. This limitation can be overcome by studying adaptation in the laboratory. One advantage of the laboratory setting, besides the well-controlled environment, is that it is possible to do evolution experiments in monocultures and co-cultures, controlling for the effect of biotic interactions [42].
In our study, we evolved our strains in monoculture and co-cultures and concluded that A. johnsonii and P. putida mostly adapted to the abiotic conditions; i.e., adaptation to the medium. This conclusion is based on two lines of evidence. First, the mutations showing high level of parallelism – i.e., putative adaptive mutations—are observed both in monoculture and co-cultures (Table 1). Second, these same mutations are observed in all three conditions: constant benzyl alcohol, constant citrate and fluctuating environment. It is likely that some these mutations are adaptation to features of the culture conditions other than just the carbon source. For example, mutations inactivating the flagellar machinery—observed in all conditions—have been reported to be an adaptation to shaking culture conditions through the energy saved by reducing flagella motility [43]. Interestingly, P. putida grows better in both benzoate and citrate, regardless of the carbon source in which it was evolved (Fig. 3, Fig. S4). This is surprising given that specialization to one carbon source has often been observed to be associated with fitness trade-offs in other carbon sources [44–46] but see [47], which is not observed in our study. In conclusion, both species predominantly adapted to abiotic conditions of the culture conditions. Assuming that the relative importance of abiotic vs biotic factors shaping evolutionary trajectories depends on the strength of species interactions [48], we can infer that the strength of the commensal interaction is weak. Consistent with this conjecture, we observe that P. putida is not strictly dependent on A. johnsonii for growing in benzyl alcohol.
In contrast, in our system, the strength of the negative interaction is strong and when the commensal interaction is interrupted with periods of competition, the two surviving lines of A. johnsonii accumulate mutations not observed in any other conditions (Table 1). Given their specificity, we posit that these mutations are associated with responses to the presence of P. putida; i.e., biotic adaptation. However, one limitation of our study is that we do know the mechanistic effects of these mutations. It is somewhat puzzling that we do not detect a beneficial effect of these mutations given that they persisted for 200 generations. Previous theoretical work has shown that in fluctuating environments, selection is less effective at discriminating between beneficial and neutral or deleterious mutations [21]. Therefore, in theory, it is possible for a neutral or deleterious mutation to achieve fixation. Alternatively, the beneficial effect of A. johnsonii’s mutations might be expressed exclusively in the presence of P. putida, which would support our conjecture that these mutations are adaptations to the biotic conditions.
Another limitation of our study is that we do not know how representative are these mutations in the entire population given that we only sequenced one clone per time point. We hypothesize that these mutations are well-established in the population given that we observe them both a generation 92 and 200. To confirm this hypothesis whole-population sequencing or whole genome sequencing of several clones per time point may be required. In conclusion, our predictions about A. johnsonii showing signs of adaptation to biotic conditions need to be followed by functional studies as well as by population genetic studies.
Extinction was a deterministic outcome of rapid evolution
A current debate in ecology and evolutionary biology is whether extinctions are caused by stochastic or deterministic factors [28, 29]. In our experiment, deterministic factors played a major role in the extinction of A. johnsonii. We have reached this conclusion based on our replay experiment in which we could repeatably recreate extinction events when starting from an intermediate time point. To our knowledge, highly deterministic extinctions during evolution experiments are rather uncommon. In a previous experiment in which an E. coli strain evolved the ability to consume citrate and outcompeted the strain unable to utilize citrate, extinction could not be replayed and was attributed to stochastic factors [49].
We postulate that extinction was caused by P. putida’s rapid evolution. In particular, we hypothesize that P. putida’s adaptation to abiotic conditions indirectly influenced the evolutionary trajectories of A. johnsonii. This hypothesis is supported with our mathematical simulations which show that stable coexistence breaks down when inputting growth parameters relevant to adaptation. That said, extinction occurred faster in our evolution experiment than in our simulations. Therefore, it is possible that, in addition to indirect effects, direct effects played a role in extinction. For example, P. putida could have exerted an additional selective pressure on A. johnsonii by evolving feature that antagonized A. johnsonii. Future directions to confirm that P. putida’s rapid adaptation caused the extinction of A. johnsonii may require head-to-head competition between clones isolated in the middle of the experiment; e.g., P. putida isolates from generation 92 could be co-cultured with ancestors of A. johnsonii to see if P. putida mutants lead to coexistence breakdown.
In conclusion, our study shows that rapid evolution can lead to highly deterministic outcomes in simple communities. Furthermore, it highlights the need for more experimental studies to further our understanding on the evolution of positive pairwise interactions in fluctuating environments.
Supplementary information
Acknowledgements
We thank Jean-Claude Walser from ETH Zürich Genetic Diversity Center for assistance analyzing the sequences. Thanks to the Microbial Systems Ecology group for providing useful feedback during this research. We thank three anonymous reviewers for helpful comments that improved the paper. ARV was supported by the ETH Zürich Adaptation to a Changing Environment (ACE) postdoctoral fellowship, the European Molecular Biological Organization (EMBO) long-term postdoctoral fellowship (ALTF 241-2015) and a grant from ACE. MA was supported by the Swiss National Science Foundation grant 31003A_169978. ARV and MA were supported by ETH Zürich and Eawag.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-020-00787-9) contains supplementary material, which is available to authorized users.
References
- 1.Kearns CA, Inouye DW, Waser NM. Endangered mutualisms: the conservation of plant-pollinator interactions. Annu Rev Ecol Syst. 1998;29:83–112. [Google Scholar]
- 2.Wenny DG, Sekercioglu CH, Cordeiro NJ, Rogers HS, Kelly D. Seed dispersal by fruit-eating birds. In: Wenny DG, Whelan CJ, editors. Why birds matter. Chicago, IL: University of Chicago Press; 2016. p. 107–45.
- 3.Fisher R, O’Leary RA, Low-Choy S, Mengersen K, Knowlton N, Brainard RE, et al. Species richness on coral reefs and the pursuit of convergent global estimates. Curr Biol. 2015;25:500–5. doi: 10.1016/j.cub.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 4.Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth’s biogeochemical cycles. Science. 2008;320:1034–9. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 5.Hoegh-Guldberg O, Poloczanska ES, Skirving W, Dove S. Coral reef ecosystems under climate change and ocean acidification. Front Mar Sci. 2017;4:1–20. [Google Scholar]
- 6.Weese DJ, Heath KD, Dentinger BT, Lau JA. Long-term nitrogen addition causes the evolution of less-cooperative mutualists. Evolution. 2015;69:631–42. doi: 10.1111/evo.12594. [DOI] [PubMed] [Google Scholar]
- 7.de Fouw J, Govers LL, van de Koppel J, Belzen J, Dorigo W, Cheikh MAS, et al. Drought, mutualism breakdown, and landscape-scale degradation of seagrass beds. Curr Biol. 2016;26:1051–6. doi: 10.1016/j.cub.2016.02.023. [DOI] [PubMed] [Google Scholar]
- 8.Baker DM, Freeman CJ, Wong JCY, Fogel ML, Knowlton N. Climate change promotes parasitism in a coral symbiosis. ISME J. 2018;12:921–30. doi: 10.1038/s41396-018-0046-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiers ET, Palmer TM, Ives AR, Bruno JF, Bronstein JL. Mutualisms in a changing world: an evolutionary perspective. Ecol Lett. 2010;13:1459–74. doi: 10.1111/j.1461-0248.2010.01538.x. [DOI] [PubMed] [Google Scholar]
- 10.Hoek TA, Axelrod K, Biancalani T, Yurtsev EA, Liu J, Gore J. Resource availability modulates the cooperative and competitive nature of a microbial cross-feeding mutualism. PLoS Biol. 2016;14:e1002540. doi: 10.1371/journal.pbio.1002540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piccardi P, Vessman B, Mitri S. Toxicity drives facilitation between 4 bacterial species. Proc Natl Acad Sci USA. 2019;116:15979–84. doi: 10.1073/pnas.1906172116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen SK, Rainey PB, Haagensen JAJ, Molin S. Evolution of species interactions in a biofilm community. Nature. 2007;445:533–6. doi: 10.1038/nature05514. [DOI] [PubMed] [Google Scholar]
- 13.Andrade-Domínguez A, Salazar E, Vargas-Lagunas MC, Kolter R, Encarnación S. Eco-evolutionary feedbacks drive species interactions. ISME J. 2014;8:1041–54. doi: 10.1038/ismej.2013.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chesson P. Mechanisms of maintenance of species diversity. Annu Rev Ecol Syst. 2000;31:343–66. [Google Scholar]
- 15.Lankau RA. Rapid evolutionary change and the coexistence of species. Annu Rev Ecol Syst. 2011;42:335–54. [Google Scholar]
- 16.Kremer CT, Klausmeier CA. Coexistence in a variable environment: eco-evolutionary perspectives. J Theor Biol. 2013;339:14–25. doi: 10.1016/j.jtbi.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Kremer CT, Klausmeier CA. Species packing in eco-evolutionary models of seasonally fluctuating environments. Ecol Lett. 2017;20:1158–68. doi: 10.1111/ele.12813. [DOI] [PubMed] [Google Scholar]
- 18.Harrison E, Laine AL, Hietala M, Brockhurst MA. Rapidly fluctuating environments constrain coevolutionary arms races by impeding selective sweeps. Proc Biol Sci. 2013;280:20130937. doi: 10.1098/rspb.2013.0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiltunen T, Ayan GB, Becks L. Environmental fluctuations restrict eco-evolutionary dynamics in predator-prey system. Proc Biol Sci. 2015;282:20150013. doi: 10.1098/rspb.2015.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sniegowski PD, Gerrish PJ. Beneficial mutations and the dynamics of adaptation in asexual populations. Philos Trans R Soc Lond B Biol Sci. 2010;365:1255–63. doi: 10.1098/rstb.2009.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cvijović I, Good BH, Jerison ER, Desai MM. Fate of a mutation in a fluctuating environment. Proc Natl Acad Sci USA. 2015;112:E5021–8. doi: 10.1073/pnas.1505406112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.terHorst CP, Zee PC, Heath KD, Miller TE, Pastore AI, Patel S, et al. Evolution in a community context: trait responses to multiple species interactions. Am Nat. 2018;191:368–80. [Google Scholar]
- 23.Gorter FA, Manhart M, Ackermann M. Understanding the evolution of interspecies interactions in microbial communities. Philos Trans R Soc Lond B Biol Sci. 2020;375:20190256. doi: 10.1098/rstb.2019.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodríguez-Verdugo A, Vulin C, Ackermann M. The rate of environmental fluctuations shapes ecological dynamics in a two-species microbial system. Ecol Lett. 2019;22:838–46. doi: 10.1111/ele.13241. [DOI] [PubMed] [Google Scholar]
- 25.Kaas RS, Mordhorst H, Leekitcharoenphon P, Jensen JD, Haagensen JA, Molin S, et al. Draft genome sequence of Acinetobacter johnsonii C6, an environmental isolate engaging in interspecific metabolic interactions. Genome Announc. 2017;5:e00155–17. doi: 10.1128/genomeA.00155-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 2014;1151:165–88. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X, et al. Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0) Nat Protoc. 2019;14:703–21. doi: 10.1038/s41596-019-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lande R. Risks of population extinction from demographic and environmental stochasticity and random catastrophes. Am Nat. 1993;142:911–27. doi: 10.1086/285580. [DOI] [PubMed] [Google Scholar]
- 29.Wootton JT, Pfister CA. Experimental separation of genetic and demographic factors on extinction risk in wild populations. Ecology. 2013;94:2117–23. doi: 10.1890/12-1828.1. [DOI] [PubMed] [Google Scholar]
- 30.Tenaillon O, Rodríguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, et al. The molecular diversity of adaptive convergence. Science. 2012;335:457–61. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- 31.Ragland SA, Criss AK. From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog. 2017;13:e1006512. doi: 10.1371/journal.ppat.1006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Werner GDA, Cornelissen JHC, Cornwell WK, Soudzilovskaia NA, Kattge J, West SA, et al. Symbiont switching and alternative resource acquisition strategies drive mutualism breakdown. Proc Natl Acad Sci USA. 2018;115:5229–34. doi: 10.1073/pnas.1721629115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sachs JL, Skophammer RG, Regus JU. Evolutionary transitions in bacterial symbiosis. Proc Natl Acad Sci USA. 2011;108:10800–7. doi: 10.1073/pnas.1100304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hillesland KL, Lim S, Flowers JJ, Turkarslan S, Pinel N, Zane GM, et al. Erosion of functional independence early in the evolution of a microbial mutualism. Proc Natl Acad Sci USA. 2014;111:14822–7. doi: 10.1073/pnas.1407986111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hillesland KL, Stahl DA. Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc Natl Acad Sci USA. 2010;107:2124–9. doi: 10.1073/pnas.0908456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haagensen JA, Hansen SK, Christensen BB, Pamp SJ, Molin S. Development of spatial distribution patterns by biofilm cells. Appl Environ Microbiol. 2015;81:6120–8. doi: 10.1128/AEM.01614-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harcombe W. Novel cooperation experimentally evolved between species. Evolution. 2010;64:2166–72.. doi: 10.1111/j.1558-5646.2010.00959.x. [DOI] [PubMed] [Google Scholar]
- 38.Marchal M, Goldschmidt F, Derksen-Müller SN, Panke S, Ackermann M, Johnson DR. A passive mutualistic interaction promotes the evolution of spatial structure within microbial populations. BMC Evol Biol. 2017;17:106. doi: 10.1186/s12862-017-0950-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brockhurst MA, Koskella B. Experimental coevolution of species interactions. Trends Ecol Evol. 2013;28:367–75. doi: 10.1016/j.tree.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 40.Botero CA, Weissing FJ, Wright J, Rubenstein DR. Evolutionary tipping points in the capacity to adapt to environmental change. Proc Natl Acad Sci USA. 2015;112:184–9. doi: 10.1073/pnas.1408589111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Theodosiou L, Hiltunen T, Becks L. The role of stressors in altering eco-evolutionary dynamics. Funct Ecol. 2019;33:78–83.. [Google Scholar]
- 42.Hall JPJ, Harrison E, Brockhurst MA. Competitive species interactions constrain abiotic adaptation in a bacterial soil community. Evol Lett. 2018;2:580–9. doi: 10.1002/evl3.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martínez-Garcí aE, Nikel PI, Chavarrí aM, de Lorenzo V. The metabolic cost of flagellar motion in Pseudomonas putida KT2440. Environ Microbiol. 2014;16:291–303. doi: 10.1111/1462-2920.12309. [DOI] [PubMed] [Google Scholar]
- 44.Cooper VS, Lenski RE. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature. 2000;407:736–9. doi: 10.1038/35037572. [DOI] [PubMed] [Google Scholar]
- 45.Travisano M, Lenski RE. Long-term experimental evolution in Escherichia coli. IV. Targets of selection and the specificity of adaptation. Genetics. 1996;143:15–26. doi: 10.1093/genetics/143.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Travisano M, Vasi F, Lenski RE. Long-term experimental evolution in Escherichia coli. III. Variation among replicate populations in correlated responses to novel environments. Evolution. 1995;49:189–200. doi: 10.1111/j.1558-5646.1995.tb05970.x. [DOI] [PubMed] [Google Scholar]
- 47.Wenger JW, Piotrowski J, Nagarajan S, Chiotti K, Sherlock G, Rosenzweig F. Hunger artists: yeast adapted to carbon limitation show trade-offs under carbon sufficiency. PLoS Genet. 2011;7:e1002202. doi: 10.1371/journal.pgen.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawrence D, Fiegna F, Behrends V, Bundy JG, Phillimore AB, Bell T, et al. Species interactions alter evolutionary responses to a novel environment. PLoS Biol. 2012;10:e1001330. doi: 10.1371/journal.pbio.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turner CB, Blount ZD, Lenski RE. Replaying evolution to test the cause of extinction of one ecotype in an experimentally evolved population. PLoS One. 2015;10:e0142050. doi: 10.1371/journal.pone.0142050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.