Highlights
-
•
Up-regulated SMYD3 correlates with worse prognosis and controls DDP resistance of NSCLC.
-
•
ANKHD1 interacts with and is essential for SMYD3-induced DDP resistance.
-
•
CDK2 is identified to be a downstream effector of SMYD3-ANKHD1 in NSCLC.
-
•
SMYD3-ANKHD1 critically regulates the growth DDP-resistant NSCLC cells in vivo.
Keywords: NSCLC, Cisplatin resistance, SMYD3, ANKHD1, CDK2
Abstract
Background
Cisplatin (DDP) remains the backbone of chemotherapy for non-small cell lung cancer (NSCLC), yet its clinical efficacy is limited by DDP resistance. We aim to investigate the role of the SET and MYND domain-containing protein 3 (SMYD3) in DDP resistance of NSCLC.
Methods
Expression pattern of SMYD3 was determined in NSCLC tissues using qRT-PCR, which also validated its correlation with NSCLC clinicopathological stages. Impacts of SMYD3 on DDP resistance were evaluated by knocking down SMYD3 in DDP-resistant cells and overexpressing it in DDP-sensitive cells, and assessed for several phenotypes: IC50 by MTT, long-term proliferation by colony formation, apoptosis and cell-cycle distribution by flow cytometry. The interaction between Ankyrin Repeat and KH Domain Containing 1 (ANKHD1) and SMYD3 was examined by co-immunoprecipitation and immunofluorescence. The transcriptional regulation of SMYD3 on cyclin-dependent kinase 2 (CDK2) promoter regions was confirmed using chromatin-immunoprecipitation. The in vivo experiments using DDP-resistant cells with altered SMYD3 and ANKHD1 expression were further performed to verify the SMYD3/ANKHD1 axis.
Results
Highly expressed SMYD3 was observed in NSCLC tissues or cells, acted as a sensitive indicator for NSCLC, correlated with higher TNM stages or resistant to DDP treatment, and shorter overall survival. The promotion of SMYD3 on DDP resistance requires co-regulator, ANKHD1. CDK2 was identified as a downstream effector. In vivo, SMYD3 knockdown inhibited the growth of DDP-resistant NSCLC cells, which was abolished by ANKHD1 overexpression.
Conclusions
SMYD3 confers NSCLC cells chemoresistance to DDP in an ANKHD1-dependent manner, providing novel therapeutic targets to overcome DDP resistance in NSCLC .
Introduction
With an estimated 2.09 million new cases and 1.76 million deaths in 2018, lung cancer remains the leading cause of cancer-related deaths globally [1]. Of all lung cancer cases, non-small cell lung cancer (NSCLC) accounts for approximately 85% and more than 70% of NSCLC patients are diagnosed with locally advanced or metastatic disease that is associated with a high rate of relapse and a poor five-year survival [2]. Despite advances made in the treatment of NSCLC, such as targeted therapy for cases with actionable mutations, 85–90% NSCLC cases present no targetable mutations and for these cases, cisplatin (DDP)-based chemotherapy, remains the first-line treatment option [3]. Correspondingly, development of chemoresistance to DDP has become a clinical challenge that contributes to the recurrence and the poor prognosis of NSCLC patients. Therefore, understanding the molecular mechanisms responsible for DDP chemoresistance is urgently needed and will significantly benefit the prediction of treatment responses and the development of modification strategies to improve the efficacy of cisplatin-based therapy.
Besides genetic mutations, epigenetic modifications also significantly contribute to oncogenesis and malignant progression of cancers. The SET and MYND domain-containing protein 3 (SMYD3) is a member of the SMYD family of lysine methyltransferases that contains five members, SMYD1 to SMYD5. A common feature of all five SMYD members is their capability to methylate histone 3 lysine 4 (H3K4) [4]. In addition, the activities of SMYD3 in human physiology and pathology are regulated by interacting proteins, methylation targets (histone and non-histone proteins), as well as subcellular localizations [5]. The oncogenic activities of SMYD3 are well established. SMYD3 is minimally expressed in normal adult lung tissues [6], yet in lung cancer, not only the expression of SMYD3 is up-regulated, but higher SMYD3 level is significantly and negatively associated with disease-free survival of lung cancer patients [5]. The oncogenic phenotypes of SMYD3 include promoting proliferation, migration/invasion, epithelial-mesenchymal transition (EMT), and metastasis, while inhibiting apoptosis [5,7]. A recent study in breast cancer showed that SMYD3 contributed to the resistance of breast cancer cells to DDP. However, little is known whether SMYD3 also regulates DDP resistance of lung cancer.
In this study, we hypothesize that SMYD3 critically regulates DDP resistance in NSCLC. To test this hypothesis and to explore the underlying mechanisms, we used two distinct DDP-resistant NSCLC cell lines, A549/DDP and H1299/DDP, as well as their DDP-sensitive counterparts, A549 and H1299, to assess the significance of SMYD3 in regulating DDP resistance and explored the underlying mechanisms. We showed, for the first time, that SMYD3 is necessary and sufficient for promoting DDP resistance of NSCLC. The regulation of DDP resistance by SMYD3 is mediated at least partially through Ankyrin Repeat and KH Domain Containing 1 (ANKHD1) as a co-regulator, and cyclin-dependent kinase 2 (CDK2) as a target co-regulated by both SMYD3 and ANKHD1. This study provides novel mechanisms that may benefit the development of strategies to overcome DDP resistance.
Materials and methods
Human tissues
This study was reviewed and approved by the Ethics Committee of The Affiliated Cancer Hospital of Zhengzhou University (Zhengzhou, China; Approval No.2020086). A cohort of 38 matching pairs of cancer tissues and adjacent tumor-free lung tissues were obtained during surgery from NSCLC patients admitted into The Affiliated Cancer Hospital of Zhengzhou University. The isolated human tissues were immediately frozen in liquid nitrogen for expressional analysis. These patients included 18 cases of stage I/II and 20 cases of stage III/IV according to the 8th tumor-node-metastasis (TNM) classification of lung cancer [8]. All patients received standard DDP-based chemotherapy after the surgery. According to the revised Response Evaluation Criteria in Solid Tumors (RECIST) criteria [9], 16 patients developed complete or partial response after two rounds of DDP treatment, as shown by computed tomography (CT) scanning, and were classified as sensitive cases. The remaining 22 cases showed stable or progressive diseases and were classified as resistant cases. All patients were followed up for at least 60 months or till their death. All patients signed the informed consent to participate in this study.
Cell lines and cell culture
The normal human bronchial epithelial cell line 16HBE, human NSCLC cell lines A549, DDP-resistant A549 cells (A549/DDP), H1299, and H1299/DDP were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). All cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (all from Invitrogen, Carlsbad, CA, USA) at 37 °C with 5% CO2. For A549/DPP and H1299/DPP cells, DDP (Sigma, St. Louis, MO, USA) was added to the culture medium at 3.3 μM to maintain the DDP-resistant phenotype.
Plasmid construction and cell transfection
Control siRNA (si-NC), siRNA specifically targeting SMYD3 (si-SMYD3), si-ANKHD1, pcDNA3.1 vector (empty vector), pcDNA3.1-SMYD3 (SMYD3 vector), and pcDNA3.1-ANKHD1 (ANKHD1 vector) were purchased from GenePharma (Shanghai, China), and transfected into target cells using Lipofectamine 2000 (Invitrogen), according to the manufacturer's protocol. Briefly, for cells in 24-well plate, 500 ng of plasmid DNA and/or 15 pmol siRNA were diluted in 50 µL of Opti-MEM reduced serum medium (Invitrogen), while 1 µL of Lipofectamine 2000 was diluted in 50 µL of Opti-MEM reduced serum medium. Then, these two solutions were mixed and incubated at room temperature for 5 min before being applied to cells. At 48 h after the transfection, cells were used for further experiments.
RNA extraction and quantitative real-time PCR (qRT-PCR)
TRIzol reagent (Invitrogen) was used to extract total RNA from tissues and cells. The ImProm-II Reverse Transcription System (Promega, USA) was used to generate first-strand cDNA, and qPCR analysis was carried out using SYBR Green qPCR assay reagents (Takara, China) and the following specific primers for the corresponding human genes (Table 1). As a routine for qRT-PCR assays, melting curve was always checked to verify the specificity of the amplification reaction. The relative expression levels of RNAs were calculated using the comparative Ct method [10].
Table 1.
List of primers used for qRT-PCR analysis.
| Gene Name | Forward Primer (5′−3′) | Reverse Primer (5′−3′) |
|---|---|---|
| SMYD3 | TTACTGCGAGCAGTCCGAGACA | TTGTCCTGGGTTTGGCAACGGA |
| ANKHD1 | AGACCAATCGGAACACGGCTCT | CAGAAGCTGCTTCCATCAAGGG |
| CDK2 | ATGGATGCCTCTGCTCTCACTG | CCCGATGAGAATGGCAGAAAGC |
| CDK4 | CCATCAGCACAGTTCGTGAGGT | TCAGTTCGGGATGTGGCACAGA |
| CDK6 | GGATAAAGTTCCAGAGCCTGGAG | GCGATGCACTACTCGGTGTGAA |
| GAPDH | AGGTCGGTGTGAACGGATTTG | GGGGTCGTTGATGGCAACA |
Cell viability assay and determination of half maximal inhibitor concentration (IC50)
Cells were seeded in 96-well plates at a density of 2 × 104 cells/mL and incubated with fresh medium containing different concentrations of DDP (0, 20, 40, 60, or 80 µM). After 48 h, 20 μL of MTT solution (5 mg/mL in PBS; Sigma) was added to each well and the plates were incubated at 37 °C for a further 3 h. The medium was then discarded and 100 µL dimethyl sulfoxide (DMSO; Sigma) was added to each well and incubated for 2 h in the dark at room temperature. DMSO dissolved the formazan crystals and created a purple color. Finally, the optical density (proportional to the number of live cells) was assessed with a Microplate Reader Bio-Rad 550 at 570 nm. IC50 was calculated by as concentration of DDP that reduced cell viability by 50%.
FACS analysis for apoptosis and cell-cycle distribution
Apoptotic cells were detected using Annexin-V-FITC and PI kit (Thermo Fisher Scientific, Waltman, MA, USA) following the manufacturer's instructions, and presented as the percentage (%) of Annexin-V+PI+ cells.
To determine cell-cycle distribution, cells were collected, washed with PBS three times, and fixed in 70% ethanol overnight. Fixed cells were then stained using FxCycle™ PI/RNase Staining Solution (Thermo Fisher Scientific) according to the manufacturer's instructions. All FACS analysis was performed on FACSCalibur (BD Biosciences, San Jose, CA, USA).
Colony formation assay
The long-term proliferation of cells was measured using colony formation assay. Briefly, target cells were seeded into 24-well plate at 200 cells/well. After incubating the cells at 37 °C for 10 days, cells were fixed with 100% methanol (Sigma) at room temperature for 10 min and then stained with 0.5% crystal violet (Sigma) at room temperature for 5 min. The number of cell colonies was counted under a light microscope.
Co-immunoprecipitation (Co-IP)
Whole cell lysate was prepared in IP lysis buffer (Thermo Fisher Scientific) containing 1 × protease/phosphatase inhibitor cocktail (Cell Signaling, Danvers, MA, USA). Total protein (500 μg) was incubated with 1 μg anti-H3K4me3 antibody (ab8580, Abcam, Cambridge, MA, USA) or anti-ANKHD1 antibody (ab117788, Abcam), or rabbit IgG on a rotating shaker at 4 °C for 1 h. Then, protein A Sepharose beads (Abcam) were added to the mixture and shaken at 4 °C overnight. After washing the beads and associated protein complexes three times, they were boiled in 5 × sample loading buffer for 5 min and the supernatant was examined using western blot.
Subcellular fractionation and western blot
Cytoplasmic and nuclear fractions were prepared from cells using Minute™ Cytoplasmic & Nuclear Extraction Kits (Invent Biotechnologies, USA) according to the manufacturer's instructions.
Whole cell lysates were prepared in RIPA buffer (Thermo Fisher Scientific) containing protease/phosphatase inhibitor cocktail (Cell Signalling). Protein samples were separated by 10% SDS-PAGE gel electrophoresis and then transferred to PVDF membranes. The membrane was incubated with the following primary antibodies (all from Abcam): anti-SMYD3 (ab187149, 1:1000), anti-H3K4me3 (ab8580, 1:1000), anti-ANKHD1 (ab117788, 1:2000), anti-CDK2 (ab32147, 1:1000), anti-H3 (ab1791, 1:1000), and anti-GAPDH (ab181602, 1:5000). After washing the membrane with TBST three times, it was incubated with HRP-labelled goat anti-mouse/rabbit IgG (1:5000, Abcam) for 2 h at room temperature. Specific signals were detected using the enhanced chemiluminescence (ECL, Millipore, MA, USA) method and analysed with ImageJ software.
Immunofluorescence staining
Target cells were cultured on glass coverslips and treated as indicated. Upon fixation/permeabilization with cold 100% methanol for 5 min at room temperature, cells were washed with PBS twice and blocked in TBS buffer (50 mM Tris-Cl, 150 mM NaCl, pH 7.5) containing 0.1% Tween-20, 2% BSA and 0.1% NaN3 (Sigma) for one hour at room temperature. The primary rabbit anti-ANKDH1 antibody (ab117788) and mouse anti-H3K4me3 antibody (ab185637) diluted 1:200 in blocking solution were then added and incubated with cells at 4 °C overnight. After three washes in PBS, cells were incubated with Alexa Fluor 488 conjugated anti-rabbit and PE-conjugated anti-mouse secondary antibody for two hours at room temperature. Coverslips were washed four times with PBS and mounted on SuperFrost Plus slides with Vectashield anti-fade mounting medium with DAPI (Vector Labs, Burlingame, CA, USA). Cells were imaged under confocal microscopy.
Chromatin immunoprecipitation (ChIP)
ChIP assay was performed using SimpleChIP kit (Cell Signaling Technology) according to the protocols provided by the manufacturer. Briefly, cells were crosslinked with 1% formaldehyde and lysed to prepare nuclei. Then chromatin was partially digested in Micrococcal Nuclease followed by sonication to generate DNA/protein fragments of 150–900 base pairs (bps) in length. Upon incubating the digested chromatin with anti-SMYD3 (ab228015), anti-ANKHD1 (ab117788), anti-RNA polymerase II (anti-RNAP II; ab164350), or normal rabbit IgG (negative control; ab172730) at 4 °C overnight, the immune complexes were pulled down using ChIP-grade protein G magnetic beads. After eluting chromatin from the antibody/protein G magnetic beads, DNA was purified using the spin column provided with the kit and examined with qRT-PCR analysis.
Xenograft mouse model
All animal protocols were reviewed and approved by the Institutional Animal Care and Use Committee of Zhengzhou University. Athymic nude mice of four to six weeks old were purchased from Shanghai Laboratory Animal Center (Shanghai, China) and housed in a specific-pathogen-free facility with a 12-h light:dark cycle at a temperature of 21 °C ± 2 °C and a relative air humidity of 55% ±10%. To establish the xenograft model, A549/DDP cells transfected with si-NC, si-SMYD3, si-SMYD3 + empty vector, or si-SMYD3 + ANKHD1 vector were injected subcutaneously into the dorsal flank region of each mouse (N = 6 per group). The length (L) and width (W) of each xenograft tumor were measured using a vernier caliper every three days, and the tumor volume (V) was calculated as: V = 1/2 × L × W2. On day 18 after the inoculation of cancer cells, all mice were sacrificed, and the xenograft tumors were isolated, imaged, weighed, and snap frozen in liquid nitrogen for further analysis.
Immunohistochemistry (IHC)
The mouse tissues were fixed in 10% neutral formalin and embedded in paraffin. Serial sections of 4 µm in thickness were prepared. For IHC detection of Ki-67 and CDK2, tissue sections were de-paraffinized in xylene and rehydrated in a series of diluted alcohol. Antigen retrieval was performed in boiling 10 mM citrate buffer (pH 6.0) for 10 min. After blocking endogenous peroxidase activity with 0.3% H2O2 in PBS for 10 min and non-specific binding with 5% normal goat serum for 1 h at room temperature, the tissue sections were incubated with anti-Ki67 (ab92742), anti-CDK2 (ab32147) and anti-SMYD3 (ab187149)antibodies. Upon three washes in 0.15 M NaCl containing 0.1% v/v Triton-X-100, pH 7.6 (TBST) buffer, the sections were incubated with biotinylated secondary antibody at room temperature for 30 min, washed three times in TBST, and then incubated in Vectastain ABC—HRP solution (Vector Labs) at room temperature for 30 min, according to the manufacturer's instructions. The signal of a target protein was developed using Diaminobenzidine (DAB) substrate (Vector Labs) and the slides were counterstained with hematoxylin.
Statistical analysis
GraphPad Prism 6 software (GraphPad Software; http://www.graphpad.com) was used to calculate and assess significant differences between experimental groups. The results are presented as the mean ± standard deviation (SD). Statistical significance between two groups was analysed using one-way ANOVA followed by Newman-Keuls post hoc test. Student's t-test was used for paired comparisons. P < 0.05 was considered statistically significant.
Results
Up-regulation of SMYD3 indicates the poor prognosis of NSCLC patients
To understand the molecular mechanisms by which SMYD3 functions in NSCLC, we first examined its expression in NSCLC. By comparing SMYD3 mRNA between 38 pairs of NSCLC tissues and matching normal lung tissues, we found that SMYD3 was significantly up-regulated in NSCLC tissues (Fig. 1A). Focusing on the NSCLC tissues, we found that SMYD3 level markedly higher in tumours of higher TNM stages (III/IV; N = 20) than in those of lower TNM stages (I/II; N = 18; Fig. 1B). Survival analysis using the Kaplan-Meier revealed that higher SMYD3 expression was significantly associated with shorter overall survival of NSCLC patients (Fig. 1C). In addition to the steady-state mRNA level, we also measured SMYD3 protein from randomly selected NSCLC and matching normal tissues (n = 6/group) and found that, SMYD3 protein was also significantly higher in tumours than in normal tissues (Fig. 1D). Collectively, these data demonstrated the clinical significance of SMYD3 as a biomarker for the diagnosis and an indicator of poor prognosis of NSCLC.
Fig. 1.
Up-regulation of SMYD3 indicated the poor prognosis of NSCLC patients. (A) The expression of SMYD3 was compared using qRT-PCR analysis between 38 pairs of NSCLC and normal tissues. (B) SMYD3 level was compared between NSCLC tumors of low TNM stage (I/II, N = 18) and those of high TNM stage (III/IV, N = 20) using qRT-PCR analysis. (C) The correlation of SMYD3 level and the overall survival of NSCLC patients was analyzed via the Kaplan-Meier method. (D) SMYD3 protein expression was examined by Western blot between six pairs of randomly selected NSCLC and normal tissues. GAPDH was detected as the internal control. Representative Western images were shown on the left and the quantification of relative SMYD3 protein (normalized to GAPDH) on the right. Data are presented as means ± SD of all samples in each group. **P < 0.01, ***P < 0.001.
Highly expressed SMYD3 is observed in DDP-resistant NSCLC tissues and cell lines
A previous study suggests that SMYD3 renders breast cancer more resistant to DDP [11]. To understand whether this is the case in NSCLC, we first compared SMYD3 expression between NSCLC tissues sensitive (N = 16) or resistant (N = 22) to DDP treatment. As shown in Fig. 2A, SMYD3 was significantly increased in resistant tissues, supporting its potential involvement in chemoresistance to DDP. To understand the underlying mechanisms, we used two DDP-resistant NSCLC cell lines, A549/DDP and H1299/DDP. Expressional analysis showed that SMYD3 expression on both mRNA (Fig. 2B) and protein (Fig. 2C) levels was significantly elevated in parental A549 and H1299 cells, when compared to normal bronchial epithelial cells 16HBE, but was much higher in A549/DDP and H1299/DDP cells (when compared to the corresponding parental cells). Concomitant with the up-regulation of SMYD3, A549/DDP and H1299/DDP cells were more resistant to DDP, as demonstrated by much higher IC50 for DDP on these cells (when compared to the corresponding parental cells; Fig. 2D), and also by the potently reduced apoptotic cells in response to DDP treatment (when compared to the corresponding parental cells; Fig. 2E). These data suggest the association between highly expressed SMYD3 and increased resistance of NSCLC tissues or cells to DDP.
Fig. 2.
SMYD3 was up-regulated in DDP-resistant NSCLC tissues and cell lines. (A) The expression of SMYD3 was compared using qRT-PCR analysis between 16 DDP-sensitive and 22 DDP-resistant NSCLC tissues. (B-C) The expression of SMYD3 was examined by qRT-PCR (B) and western blot (C) in 16HBE, A549, A549/DDP, H1299, and H1299/DDP cells. (D) The IC50 of DDP on A549, A549/DDP (upper panel), H1299, and H1299/DDP cells (lower panel) was determined by MTT assay. (E) A549, A549/DDP, H1299, and H1299/DDP cells were treated with DDP. Apoptosis was examined by dual staining of cells with Annexin V and PI followed by flow cytometry (left panels) and quantified as the percentage of AnnexinV+PI+ cells (right panels). Data are presented as mean ± SD of all samples in each group (A) or from three independent experiments (B to D). **P < 0.01, ***P < 0.001.
SMYD3 knockdown inhibits the resistance to DDP of A549/DDP and H1299/DDP cells
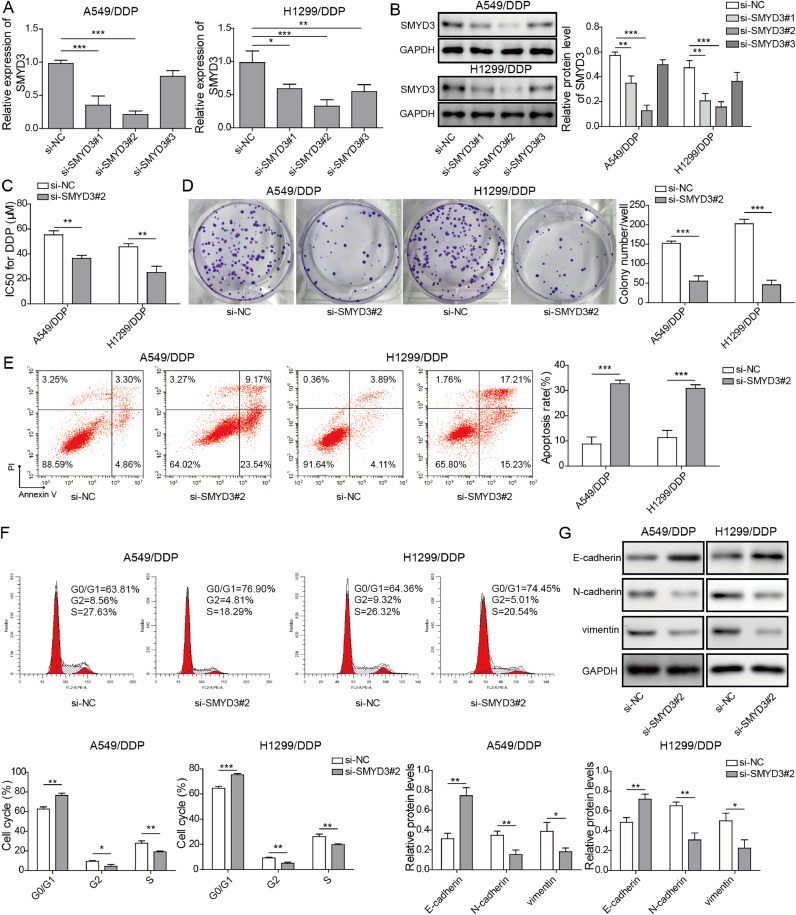
Considering the up-regulation of SMYD3 in A549/DDP and H1299/DDP cells, we applied the loss-of-function strategy in these cells and designed distinct three si-SMYD3 (#1 to #3). As shown in Fig. 3A and B, si-SMYD3#2 most potently reduced SMYD3 mRNA (Fig. 3A) and protein (Fig. 3B) levels in both A549/DDP and H1299/DDP cells, when compared to the control si-NC, and thus were chose for subsequent experiments. Next, we compared several phenotypes between si-NC and si-SMYD3#2 cells. We found that knockdown of SMYD3 markedly lowered IC50 of DDP (Fig. 3C), reduced long-term cell proliferation, as demonstrated by colony formation assay (Fig. 3D), increased apoptosis, as indicated by the percentage of both AnnexinV+PI− (early-stage) and AnnexinV+PI+ (late-stage) apoptotic cells (Fig. 3E), and induced cell-cycle arrest in G0/G1 phase (Fig. 3F) in DDP-challenged A549/DDP and H1299/DDP cells, suggesting that SMYD3 is essential for maintaining the resistance of NSCLC cells to DDP. Furthermore, we measured several EMT biomarkers, including E-cadherin, N-cadherin, and vimentin in between si-NC and si-SMYD3#2 cells. We found that E-cadherin was significantly up-regulated, while N-cadherin and vimentin down-regulated in si-SMYD3#2 cells, when compared to si-NC cells (Fig. 3G), implying that SMYD3 is critical for sustaining EMT of DDP-resistant NSCLC cells.
Fig. 3.
SMYD3 knockdown inhibited the chemoresistance of A549/DDP and H1299/DDP cells to cisplatin. (A-B) A549/DDP (left panel) and H1299/DDP (right panel) cells were transfected with si-NC, si-SMYD3#1, si-SMYD3#2, or si-SMYD3#3. The expression of SMYD3 was examined by qRT-PCR (A) and western blot (B). (C) IC50 of DDP on si-NC vs. si-SMYD3-transfected A549/DDP and H1299/DDP cells was examined by MTT assay. (D) The long-term proliferation of indicated cells was examined by colony formation assay. (E) Indicated cells were treated with DDP. Apoptosis was examined by dual staining of cells with Annexin V and PI followed by flow cytometry (left panels) and quantified as the percentage of AnnexinV+PI+ cells (right panels). (F) Cell cycle distribution of indicated cells was examined by PI staining followed by flow cytometry. Representative flow images shown on the left and percentage of cells in G0/G1, S, and G2/M phase quantified on the right. (G) Expressions of EMT biomarkers, E-cadherin, N-cadherin, and vimentin were examined by western blot. Data are presented as mean ± SD of all samples from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
SMYD3 overexpression promotes the chemoresistance of A549 and H1299 cells to DDP
Conversely, we applied the gain-of-function strategy in A549 and H1299 cells that presented relatively low expression of SMYD3, and overexpressed SMYD3 in these cells (SMYD3 vector). As the negative control, we used empty vector. As expected, overexpressing SMYD3 significantly boosted its mRNA (Fig. 4A) and protein (Fig. 4B) levels, when compared to empty vector. Opposite to si-SMYD3#2-induced phenotypes, we found that SMYD3 vector remarkably increased IC50 of DDP (Fig. 4C), boosted cell proliferation (Fig. 4D), inhibited apoptosis (as indicated by the percentage of both AnnexinV+PI− (early-stage) and AnnexinV+PI+ (late-stage) apoptotic cells; Fig. 4E), promoted cell-cycle progression from G0/G1 to S phase (Fig. 4F), and stimulated EMT (as represented by the reduction of E-cadherin and the increase of N-cadherin and vimentin; Fig. 4G) in DDP-challenged A549 and H1299 cells, implying that SMYD3 was sufficient to induce DDP-resistance and EMT of NSCLC cells.
Fig. 4.
SMYD3 overexpression promoted the chemoresistance of A549 and H1299 cells to cisplatin. A549 and H1299 cells were transfected with either empty vector or SMYD3-overexpressing vector. (A-B) The expression of SMYD3 was examined by qRT-PCR (A) and western blot (B). (C) IC50 of DDP on indicated cells was examined by MTT assay. (D) The long-term proliferation of indicated cells was examined by colony formation assay. (E) Indicated cells were treated with DDP. Apoptosis was examined by dual staining of cells with Annexin V and PI followed by flow cytometry (left panels) and quantified as the percentage of AnnexinV+PI+ cells (right panels). (F) Cell cycle distribution of indicated cells was examined by PI staining followed by flow cytometry. Representative flow images shown on the left and percentage of cells in G0/G1, S, and G2/M phase quantified on the right. (G) Expressions of EMT biomarkers, E-cadherin, N-cadherin, and vimentin were examined by western blot. Data are presented as mean ± SD of all samples from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
ANKHD1 is a co-regulator for SMYD3 in NSCLC cells
SMYD3 is a methyltransferase and trimethylation of histone H3K4 (H3K4me3) is critical for the transcriptional activation of SMYD3 on many oncogenes [12]. A previous study showed that ANKHD1 interacted with H3K4me3 and was required for SMYD3 to promote the metastasis of hepatocellular carcinoma [13]. To examine whether the same mechanism may be responsible for chemoresistance-promoting effect of SMYD3 in NSCLC, we examined the interaction between H3K4me3, ANKHD1, and SMYD3 in control or SMYD3-overexpressing A549 and H1299 cells. Co-IP analysis showed that SMYD3 overexpression elevated the expression of H3K4me3, yet not appreciably impacted ANKHD1 level (Fig. 5A). It also promoted the interaction between ANKHD1 and SMYD3, as in control cells minimal ANKHD1 was pulled down by anti-H3K4me3 and no H3K4me3 was detectable in precipitate using anti-ANKHD1 (Fig. 5A). Subcellular fractionation analysis showed that both SMYD3 and ANKHD1 were detectable in cytoplasm and nucleus, while H3K4me3 were only detected in nucleus and its level boosted in SMYD3-overexpressing cells (Fig. 5B). Immunofluorescence confirmed the co-localization of ANKHD1 and H3K4me3 in the nucleus of A549 and H1299 cells (Fig. 5C). Expressional analysis showed that, similar to the expression of SMYD3, ANKHD1 was also up-regulated in NSCLC tissues than in matching normal lung tissues (Fig. 5D). Furthermore, Pearson correlation analysis revealed a positive correlation between ANKHD1 and SMYD3 in NSCLC tissues (Fig. 5E). Collectively, these data suggest that ANKHD1 may function as a critical co-regulator for SMYD3 functions.
Fig. 5.
ANKHD1 was a co-regulator for SMYD3. (A) Co-IP analysis was performed in control or SMYD3-overexpressing A549 (upper panels) and H1299 (lower panels) cells using anti-H3K4me3 (left) or anti-ANKHD1 antibody (right). Normal IgG was used as the negative control. (B) Expression levels of SMYD3, ANKHD1, and H3K4me3 from whole cell lysate (total), cytoplasmic and nuclear fractions of control or SMYD3-overexpressing A549 (upper panels) and H1299 (lower panels) cells were examined by western blot. GAPDH and H3 were measured as markers for cytoplasm and nucleus, respectively. (C) The expressions of ANKHD1 (green signals) and H3K4me3 (red signals) in A549 (upper panels) and H1299 (lower panels) cells were examined by immunofluorescence followed by confocal microscopy. DAPI (blue) was used to stain the nucleus. (D) The expression of ANKHD1 was compared using qRT-PCR analysis between 38 pairs of NSCLC and normal tissues. (E) Correlation analysis on SMYD3 and ANKHD1 expressions was performed in NSCLC tissues (N = 38). Data are presented as mean ± SD of all samples from three independent experiments. ***P < 0.001.
ANKHD1 was essential for SMYD3-conferred chemoresistance to DDP
To understand the functional importance of ANKHD1 in SMYD3-induced chemoresistance to cisplatin, we knocked down ANKHD1 with si-ANKHD1 in SMYD1-overexpressing A549 and H1299 cells, and examined the impact on cell behaviours. As expected, SMYD3 overexpressing cells displayed the higher IC50 of DDP (Fig. 6A), stronger cell capacity proliferation (Fig. 6B), and decreased percentage of apoptotic cells (Fig. 6C), as well as reduced cell number in G0/G1 phase (Fig. 6D), whereas, these effects were dramatically abolished by ANKHD1 knockdown, with lowered IC50 of DDP (Fig. 6A), reduced cell proliferation (Fig. 6B), increased apoptosis (Fig. 6C), and induced cell-cycle arrest in G0/G1 phase (Fig. 6D) in response to DDP challenge, indicating that ANKHD1 is essential for SMYD3-induced chemoresistance to DDP.
Fig. 6.
ANKHD1 was required for SMYD3-mediated DDP resistance of NSCLC cells. A549 and H1299 cells were transfected with empty vector, SMYD3-overexpressing vector (SMYD3), SMYD3+si-NC, or SMYD3+si-ANKHD1. (A) IC50 of DDP on indicated cells was examined by MTT assay. (B) The long-term proliferation of indicated cells was examined by colony formation assay. (C) Indicated cells were treated with DDP. Apoptosis was examined by dual staining of cells with Annexin V and PI followed by flow cytometry (left panels) and quantified as the percentage of AnnexinV+PI+ cells (right panels). (D) Cell cycle distribution of indicated cells was examined by PI staining followed by flow cytometry. Representative flow images shown on the left and percentage of cells in G0/G1, S, and G2/M phase quantified on the right. Data are presented as mean ± SD of all samples from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
CDK2 is a downstream target of SMYD3-ANKHD1 in NSCLC cells
Cyclin-dependent kinases (CDKs) are central regulators for cell cycle progression and their aberrant regulations contribute to multiple malignant phenotypes including chemoresistance [14]. Previous studies reported SMYD3 targeted CDK2 in cancer cells [15,16]. To understand whether CDK2 might be a target regulated by SMYD3-ANKHD1, we first examined the expressions of CDK2, CDK4 and CDK6 in A549 and H1299 cells transfected empty vector, SMYD3 vector, SMYD3 vector+si-NC, and SMYD3 vector+si-ANKHD1. All three CDKs were potently up-regulated by SMYD3 overexpression on both mRNA (Fig. 7A) and protein (Fig. 7B) levels, while this effect was reversed by ANKHD1 knockdown, suggesting that ANKHD1 is required for SMYD3-induced expressions of all three CDKs. Among the three CDKs, CDK2 was most abundantly expressed and thus was further studied. ChIP assay showed that in A549 cells, endogenous SMYD3 specifically bound to the sequence close to the transcription start site (TSS) of human CDK2 gene (Fig. 7C and 7D). This binding was associated with and required for the binding of H3K4me3 and RNA polymerase II (RNAP II) to the promoter of human CDK2 gene, since knocking down SMYD3 with si-SMYD3 abolished the binding of both H3K4me3 and RNAP II (Fig. 7E). These data suggest that both ANKHD1 and SMYD3 are required for the active transcription of CDK2 in NSCLC cells.
Fig. 7.
CDK2 was a downstream target of SMYD3-ANKHD1 in NSCLC cells. A549 and H1299 cells were transfected with empty vector, SMYD3-expressing vector (SMYD3), SMYD3+si-NC, or SMYD3+si-ANKHD1. (A) The expression of CDK2, CDK4, and CDK6 in indicated cells was examined by qRT-PCR. (B) The expression of SMYD3, ANKHD1, and CDK2 from indicated cells was measured using western blot. (C) A schematic illustration showing relative positions of three PCR amplicons (P1, P2 and P3) in human CDK2 gene designed for ChIP assay. The transcription start site (TSS) was indicated as 0. (D) The binding of endogenous SMYD3 to different promoter regions (P1, P2, and P3) of CDK2 gene in A549 cells was determined by ChIP assay. (E) A549 cells were transfected with empty vector, SMYD3-overexpressing vector (SMYD3), si-NC, or si-SMYD3. The occupation of CDK2 promoter by H3K4me3 and RNAP II was examined by ChIP assay. Data are presented as mean ± SD of all samples from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
SMYD3-ANKHD1 critically regulated the xenograft growth DDP-resistant NSCLC cells in vivo
Lastly, we examined the in vivo significance of our in vitro findings using a xenograft model of A549/DDP cells. We separately and subcutaneously inoculated A549/DDP cells transfected with si-NC, si-SMYD3, si-SMYD3+empty vector, and si-SMYD3+ANKHD1 vector into nude mice (N = 6 per group). We found that si-NC and si-SMYD3+ANKHD1 vector both generated xenografts of decent sizes, while si-SMYD3 or si-SMYD3+empty vector cells resulted in significantly smaller xenografts (Fig. 8A), consistent with their corresponding weights (Fig. 8B) and growth curve in vivo (Fig. 8C). Corresponding to the tumor growth in different xenografts, cell proliferation, as represented by positive Ki67 signals, was noticeably more active in si-NC or si-SMYD3+ANKHD1 vector xenografts than in si-SMYD3 or si-SMYD3+empty vector xenografts (Fig. 8D). In addition, we measured the expressions of CDK2 and SMYD3 by IHC in different xenografts and found that both proteins were reduced in xenografts derived from si-SMYD3 or si-SMYD3+empty vector cells (Fig. 8D), when compared to those from si-NC or si-SMYD3+ANKHD1 vector, suggesting that the phenotypes we observed are due to the alteration of SMYD3 expression and also supporting our in vitro finding that CDK2 is a downstream target positively regulated by SMYD3-ANKHD1 interaction.
Fig. 8.
SMYD3 and ANKHD1 critically controlled the growth of DDP-resistant A549 cells in vivo. A549/DDP cells were transfected with si-NC, si-SMYD3, si-SMYD3+empty vector, or si-SMYD3+ANKHD1 vector, and subcutaneously injected into nude mice (N = 6/group). The picture (A), the growth curve (B), and the weight (C) of xenografts were compared between the indicated groups. (D) The expressions of Ki-67 (upper panels), CDK2 (middle panels), and SMYD3 (lower panels) were examined by immunohistochemistry (brown signals) from indicated xenografts. Data are presented as mean ± SD of all samples in each group. *P < 0.05, ***P < 0.001.
Discussion
Cisplatin (DDP) is a most widely used anticancer drug and remains the backbone of chemotherapy for NSCLC [3]. The therapeutic efficacy of DDP is mediated preferentially by forming DNA adducts that lead to DNA lesions, inhibited DNA transcription and replication, and apoptosis [17], but is severely limited in clinic due to the development of intrinsic or acquired chemoresistance. Several mechanisms have been suggested to account for DDP resistance: changes in efflux/uptake reducing accumulation of cisplatin, glutathione- or metallothionein-induced detoxification of cisplatin, improved DNA repair, elevated toleration for DNA damages, and alterations in apoptosis [18]. Therefore, understanding and targeting these mechanisms will help to resume the therapeutic benefits of cisplatin-based chemotherapy. In this study, we revealed the significance of SMYD3 in controlling DDP resistance of NSCLC cells and evidenced the SMYD3-ANKHD1 interaction as a novel critical mechanism for up-regulating CDK expressions and promoting DDP resistance.
The oncogenic activities of SMYD3 have been demonstrated in a panel of human cancers, including breast cancer, ovarian cancer, bladder cancer, colorectal cancer, gastric cancer, liver cancer, and pancreatic cancer [12,16,[19], [20], [21], [22], [23], [24], [25], [26]. Few studies have looked into the status or clinical significance of SMYD3 in lung cancer. In this study, we showed that SMYD3 expression was significantly up-regulated in NSCLC tissues and cells (A549 and H1299), when compared to matching normal lung tissues and normal cells, respectively. More importantly, the increased expression of SMYD3 was a sensitive marker for NSCLC diagnosis, and significantly associated with NSCLC tumors of higher TNM stages as well as those leading to shorter overall survival, supporting the clinical value of SMYD3 as a diagnostic biomarker and an indicator for malignant NSCLC with worse prognosis. Clearly, the clinical significance of SMYD3 should be further verified in studies with larger numbers of NSCLC cases.
The more robust up-regulation of SMYD3 in DDP-resistant NSCLC tumors or cells (A549/DDP and H1299/DDP) implied that SMYD3 may be involved in cisplatin chemoresistance. Our subsequent experiments applying the loss-of-function strategy in DDP-resistant and the gain-of-function strategy in DDP-sensitive A549 and H1299 cells corroborated the necessary and sufficient roles of SMYD3 in promoting DDP resistance, as measured by IC50, colony formation, apoptosis, and cell-cycle distribution. We noticed that few studies have examined the impacts of SMYD3 in chemoresistance, except that Wang et al. reported that SMYD3 inhibited the sensitivity of breast cancer cells to cisplatin, and cisplatin suppressed SMYD3 expression via miR-124 [11]. However, Wang's study fails to provide mechanistic insights on how SMYD3 regulates chemoresistance of cancer cells. Besides chemoresistance, other oncogenic phenotypes induced by SMYD3 include the regulations on cell cycle progression, proliferation, apoptosis, migration, invasion, EMT, and cancer stemness [15,16,19,[26], [27], [28]], many closely related to chemoresistance. The mechanisms employed by SMYD3 to achieve its oncogenic phenotypes depend on its methyltransferase activity but vary by the methylation targets, the subcellular localization, and interacting partners. Although SMYD3 is well characterized as a lysine methyltransferase for H3K4 [22,29], it is also reported to methylate H4K20 [19,30], H2A.Z.1K101 [31], H4K5 [32] in the nucleus, as well as non-histone proteins such as MAP3K2 [33,34], VEGFR1 [35], HER2 [36], and AKT1 [37] in the cytoplasm. For interacting partners, RNAPII, RNA helicase, heat shock protein 90 (HSP90), and estrogen receptor have been shown to interact with SMYD3 in different cancer types [38], [39], [40], [41]. In this study, we detected that SMYD3, ANKHD1, and H3K4me3 were in the same complex and higher SMYD3 level elevated the level of H3K4me3 (although not ANKHD1 level) and promoted the interaction between ANKHD1 and H3K4me3. In addition, both SMYD3 and ANKHD1 were detectable in both nucleus and cytoplasm and nuclear ANKHD1 co-localized with H3K4me3. Functionally, ANKHD1 was required for the SMYD3-induced DDP chemoresistance. Furthermore, ANKHD1 level was markedly higher in NSCLC than in normal tissues and positively correlated with SMYD3 level in NSCLC tissues. These findings suggest clinical importance and biological significance of SMYD3-ANKHD1 interaction as well as the potential involvement of H3K4me3 in mediating DDP resistance.
As indicated by its name, ANKHD1 contains an ankyrin repeat and a KH domain, the former enabling interactions with other proteins and methylated histone lysines [42,43] that allow ANKHD1 to become a scaffold protein, while the latter engaging ANKHD1 into interactions with RNA molecules [44]. The knowledge of ANKHD1 in cancer is mostly obtained during the past decade. Initially reported to be up-regulated in leukemia [45], the increased expression of ANKHD1 is later confirmed in multiple myeloma [46], breast cancer [47], prostate cancer [48], renal cell carcinoma [49], colorectal cancer [50], and hepatocellular carcinoma [13], and frequently associated with worse prognosis of cancer patients, suggesting its nature as an oncogene. The only study on ANKHD1 in NSCLC was recently available, which showed that, similar to our finding here, not only ANKHD1 was up-regulated in NSCLC tissues and cells, and correlated with advanced TNM stage, lymph node metastasis, and poor prognosis, but it presented pro-proliferation and pro-invasion activities that were mediated via up-regulating YAP expression [51]. In hepatocellular carcinoma, Zhou et al. recently reported that ANKHD1 was a co-regulator and required for SMYD3-activated transcription of Slug as well as SMYD3-promoted metastasis of cancer cells, and higher SMYD3 expression led to elevated H3K4me3, H3K9Ac and H3K14Ac levels [13]. Although Zhou's study as well as ours provide the oncogenic importance of interactions among SMYD3, ANKHD1, and H3K4me3 within the nucleus, little is known on the cytoplasmic SMYD3 and ANKHD1 in NSCLC, whether they also interact in the cytoplasm and if so, what is the biological significance of this interaction?
The oncogenic roles of SMYD3 are accomplished mainly through H3K4me3-mediated transcriptional up-regulation of oncogenes and/or activation of oncogenic signaling pathways [7]. In renal cell carcinoma, SMYD3 cooperated with SP1 and transactivated EGFR gene [52]. In bladder cancer, SMYD3 induced H3K4me3, up-regulated IGF-1R and activated the downstream AKT/E2F1 signaling [24]. In ovarian cancer, SMYD3 induced H3K4me3-enhanced integrin expression [22]. In hepatocellular carcinoma, SMYD3 up-regulated CDK2 and MMP2 to promote tumorigenicity and metastasis [16]. In this study, we showed that both SMYD3 and ANKHD1 essentially controlled the up-regulation of multiple CDKs, which are critical regulators of cell-cycle progression and chemoresistance. While focusing on CDK2, we showed that the up-regulation was induced at the transcription level by SMYD3 elevating H3K4me3 binding to the promoter and bringing RNAP II to the transcription start site.
Although this study was performed mostly in vitro between DDP-resistant and their corresponding parental NSCLC cells, and suggested the critical role of SMYD3 in controlling the malignant phenotypes (growth, apoptosis, proliferation, and EMT) and DDP-resistant NSCLC cells. The finding may also apply to the malignant progression of NSCLC in general, considering the significant up-regulation of SMYD3 in NSCLC than in matching control tissues, as well as in A549 and H1299 cells than in normal 16HEB cells, and the positive correlation between SMYD3 and ANKHD1 in NSCLC tissues. In addition, malignant progression and drug resistance are not two mutually exclusive but closely interrelated processes as they both share similar mechanisms such as growth inhibition and EMT and thus could positively feedback on each other [53]. It would be interesting for future studies to examine effects of SMYD3 on the malignant progression of NSCLC in general, including drug-sensitive NSCLC.
Overall, we provide the first evidence that an elevated expression of SMYD3 may serve as a sensitive diagnostic marker, an indicator for advanced disease with worse prognosis, or predictor for DDP-resistant NSCLC. The interaction between SMYD3 and ANKHD1 is critical for activating the transcription of multiple CDKs that mediates the chemoresistance-boosting activities of SMYD3. Therefore, targeting SMYD3 or ANKHD1 may improve the sensitivity of NSCLC to cisplatin-based chemotherapy.
Author contributions statement
Hong-Wei Lv: Conceptualization, Methodology, Data curation, Visualization, Investigation, Validation, Writing- Original draft preparation, Supervision; Wen-Qun Xing: Visualization, Investigation; Yu-Feng Ba: Validation, Writing- Reviewing and Editing; Hao-Miao Li: Visualization, Investigation; Hao-Ran Wang: Software, Validation; Yin Li: Conceptualization, Methodology, Supervision, Writing- Reviewing and Editing.
Funding acknowledgement
None.
Ethics approval
This study was reviewed and approved by the Ethics Committee of The Affiliated Cancer Hospital of Zhengzhou University (Zhengzhou, China). All patients signed the informed consent to participate in this study.
All animal protocols were reviewed and approved by the Institutional Animal Care and Use Committee of Zhengzhou University.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Bade B.C., Dela Cruz C.S. Lung cancer 2020: epidemiology, etiology, and prevention. Clin. Chest Med. 2020;41:1–24. doi: 10.1016/j.ccm.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Miller K.D., Nogueira L., Mariotto A.B., Rowland J.H., Yabroff K.R., Alfano C.M. Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 2019;69:363–385. doi: 10.3322/caac.21565. [DOI] [PubMed] [Google Scholar]
- 3.Fennell D.A., Summers Y., Cadranel J., Benepal T., Christoph D.C., Lal R. Cisplatin in the modern era: the backbone of first-line chemotherapy for non-small cell lung cancer. Cancer Treat. Rev. 2016;44:42–50. doi: 10.1016/j.ctrv.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Tracy C., Warren J.S., Szulik M., Wang L., Garcia J., Makaju A. The SMYD family of methyltransferases: role in cardiac and skeletal muscle physiology and pathology. Curr. Opin Physiol. 2018;1:140–152. doi: 10.1016/j.cophys.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giakountis A., Moulos P., Sarris M.E., Hatzis P., Talianidis I. SMYD3-associated regulatory pathways in cancer. Semin Cancer Biol. 2017;42:70–80. doi: 10.1016/j.semcancer.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Kim M.S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bottino C., Peserico A., Simone C., Caretti G. SMYD3: an oncogenic driver targeting epigenetic regulation and signaling pathways. Cancers (Basel) 2020;12 doi: 10.3390/cancers12010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstraw P., Chansky K., Crowley J., Rami-Porta R., Asamura H., Eberhardt W.E. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J. Thorac Oncol. 2016;11:39–51. doi: 10.1016/j.jtho.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Eisenhauer E.A., Therasse P., Bogaerts J., Schwartz L.H., Sargent D., Ford R. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur. J. Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 10.Rao X., Huang X., Zhou Z., Lin X. An improvement of the 2^(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013;3:71–85. [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L., Xu M.L., Wang C., Dong Q.Q., Miao Z., Chen X.Y. SET and MYND domain-containing protein 3 inhibits tumor cell sensitivity to cisplatin. Oncol. Lett. 2020;19:3469–3476. doi: 10.3892/ol.2020.11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarris M.E., Moulos P., Haroniti A., Giakountis A., Talianidis I. SMYD3 is a transcriptional potentiator of multiple cancer-promoting genes and required for liver and colon cancer development. Cancer Cell. 2016;29:354–366. doi: 10.1016/j.ccell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Z., Jiang H., Tu K., Yu W., Zhang J., Hu Z. ANKHD1 is required for SMYD3 to promote tumor metastasis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019;38:18. doi: 10.1186/s13046-018-1011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonelli P., Tuccillo F.M., Borrelli A., Schiattarella A., Buonaguro F.M. CDK/CCN and CDKI alterations for cancer prognosis and therapeutic predictivity. Biomed. Res. Int. 2014;2014 doi: 10.1155/2014/361020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren T.N., Wang J.S., He Y.M., Xu C.L., Wang S.Z., Xi T. Effects of SMYD3 over-expression on cell cycle acceleration and cell proliferation in MDA-MB-231 human breast cancer cells. Med. Oncol. 2011;28 Suppl 1:S91–S98. doi: 10.1007/s12032-010-9718-6. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y., Xie B.H., Lin W.H., Huang Y.H., Ni J.Y., Hu J. Amplification of SMYD3 promotes tumorigenicity and intrahepatic metastasis of hepatocellular carcinoma via upregulation of CDK2 and MMP2. Oncogene. 2019;38:4948–4961. doi: 10.1038/s41388-019-0766-x. [DOI] [PubMed] [Google Scholar]
- 17.Jamieson E.R., Lippard S.J. Structure, recognition, and processing of cisplatin-DNA adducts. Chem. Rev. 1999;99:2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 18.Koberle B., Tomicic M.T., Usanova S., Kaina B. Cisplatin resistance: preclinical findings and clinical implications. Biochim. Biophys. Acta. 2010;1806:172–182. doi: 10.1016/j.bbcan.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Jiang Y., Lyu T., Che X., Jia N., Li Q., Feng W. Overexpression of SMYD3 in ovarian cancer is associated with ovarian cancer proliferation and apoptosis via methylating H3K4 and H4K20. J. Cancer. 2019;10:4072–4084. doi: 10.7150/jca.29861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B., Pan R., Zhou C., Dai J., Mao Y., Chen M. SMYD3 promoter hypomethylation is associated with the risk of colorectal cancer. Future Oncol. 2018;14:1825–1834. doi: 10.2217/fon-2017-0682. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y., Liu H., Luo X., Deng J., Pan Y., Liang H. Overexpression of SMYD3 and matrix metalloproteinase-9 are associated with poor prognosis of patients with gastric cancer. Tumour Biol. 2015;36:4377–4386. doi: 10.1007/s13277-015-3077-z. [DOI] [PubMed] [Google Scholar]
- 22.Lyu T., Jiang Y., Jia N., Che X., Li Q., Yu Y. SMYD3 promotes implant metastasis of ovarian cancer via H3K4 trimethylation of integrin promoters. Int. J. Cancer. 2020;146:1553–1567. doi: 10.1002/ijc.32673. [DOI] [PubMed] [Google Scholar]
- 23.Sponziello M., Durante C., Boichard A., Dima M., Puppin C., Verrienti A. Epigenetic-related gene expression profile in medullary thyroid cancer revealed the overexpression of the histone methyltransferases EZH2 and SMYD3 in aggressive tumours. Mol Cell Endocrinol. 2014;392:8–13. doi: 10.1016/j.mce.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 24.Wang G., Huang Y., Yang F., Tian X., Wang K., Liu L. High expression of SMYD3 indicates poor survival outcome and promotes tumour progression through an IGF-1R/AKT/E2F-1 positive feedback loop in bladder cancer. Aging (Albany NY) 2020;12:2030–2048. doi: 10.18632/aging.102718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L., Wang Q.T., Liu Y.P., Dong Q.Q., Hu H.J., Miao Z. ATM signaling pathway is implicated in the SMYD3-mediated proliferation and migration of gastric cancer cells. J. Gastric Cancer. 2017;17:295–305. doi: 10.5230/jgc.2017.17.e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu C.L., Huang Q. Overexpression of the SMYD3 promotes proliferation, migration, and invasion of pancreatic cancer. Dig. Dis. Sci. 2020;65:489–499. doi: 10.1007/s10620-019-05797-y. [DOI] [PubMed] [Google Scholar]
- 27.Fenizia C., Bottino C., Corbetta S., Fittipaldi R., Floris P., Gaudenzi G. SMYD3 promotes the epithelial-mesenchymal transition in breast cancer. Nucleic Acids Res. 2019;47:1278–1293. doi: 10.1093/nar/gky1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang T., Wu H., Liu S., Lei Z., Qin Z., Wen L. SMYD3 controls a Wnt-responsive epigenetic switch for ASCL2 activation and cancer stem cell maintenance. Cancer Lett. 2018;430:11–24. doi: 10.1016/j.canlet.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Wu X., Xu Q., Chen P., Yu C., Ye L., Huang C. Effect of SMYD3 on biological behavior and H3K4 methylation in bladder cancer. Cancer Manag. Res. 2019;11:8125–8133. doi: 10.2147/CMAR.S213885. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Vieira F.Q., Costa-Pinheiro P., Almeida-Rios D., Graca I., Monteiro-Reis S., Simoes-Sousa S. SMYD3 contributes to a more aggressive phenotype of prostate cancer and targets Cyclin D2 through H4K20me3. Oncotarget. 2015;6:13644–13657. doi: 10.18632/oncotarget.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai C.H., Chen Y.J., Yu C.J., Tzeng S.R., Wu I.C., Kuo W.H. SMYD3-mediated H2A.Z.1 methylation promotes cell cycle and cancer proliferation. Cancer Res. 2016;76:6043–6053. doi: 10.1158/0008-5472.CAN-16-0500. [DOI] [PubMed] [Google Scholar]
- 32.Van Aller G.S., Reynoird N., Barbash O., Huddleston M., Liu S., Zmoos A.F. Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics. 2012;7:340–343. doi: 10.4161/epi.19506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colon-Bolea P., Crespo P. Lysine methylation in cancer: SMYD3-MAP3K2 teaches us new lessons in the Ras-ERK pathway. Bioessays. 2014;36:1162–1169. doi: 10.1002/bies.201400120. [DOI] [PubMed] [Google Scholar]
- 34.Mazur P.K., Reynoird N., Khatri P., Jansen P.W., Wilkinson A.W., Liu S. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature. 2014;510:283–287. doi: 10.1038/nature13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunizaki M., Hamamoto R., Silva F.P., Yamaguchi K., Nagayasu T., Shibuya M. The lysine 831 of vascular endothelial growth factor receptor 1 is a novel target of methylation by SMYD3. Cancer Res. 2007;67:10759–10765. doi: 10.1158/0008-5472.CAN-07-1132. [DOI] [PubMed] [Google Scholar]
- 36.Yoshioka Y., Suzuki T., Matsuo Y., Tsurita G., Watanabe T., Dohmae N. Protein lysine methyltransferase SMYD3 is involved in tumorigenesis through regulation of HER2 homodimerization. Cancer Med. 2017;6:1665–1672. doi: 10.1002/cam4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshioka Y., Suzuki T., Matsuo Y., Nakakido M., Tsurita G., Simone C. SMYD3-mediated lysine methylation in the PH domain is critical for activation of AKT1. Oncotarget. 2016;7:75023–75037. doi: 10.18632/oncotarget.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamamoto R., Furukawa Y., Morita M., Iimura Y., Silva F.P., Li M. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 39.Kim H., Heo K., Kim J.H., Kim K., Choi J., An W. Requirement of histone methyltransferase SMYD3 for estrogen receptor-mediated transcription. J. Biol. Chem. 2009;284:19867–19877. doi: 10.1074/jbc.M109.021485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Proserpio V., Fittipaldi R., Ryall J.G., Sartorelli V., Caretti G. The methyltransferase SMYD3 mediates the recruitment of transcriptional cofactors at the myostatin and c-Met genes and regulates skeletal muscle atrophy. Genes Dev. 2013;27:1299–1312. doi: 10.1101/gad.217240.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown M.A., Foreman K., Harriss J., Das C., Zhu L., Edwards M. C-terminal domain of SMYD3 serves as a unique HSP90-regulated motif in oncogenesis. Oncotarget. 2015;6:4005–4019. doi: 10.18632/oncotarget.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J., Mahajan A., Tsai M.D. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006;45:15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 43.Collins R.E., Northrop J.P., Horton J.R., Lee D.Y., Zhang X., Stallcup M.R. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat. Struct. Mol. Biol. 2008;15:245–250. doi: 10.1038/nsmb.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valverde R., Edwards L., Regan L. Structure and function of KH domains. FEBS J. 2008;275:2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 45.Traina F., Favaro P.M., Medina Sde S., Duarte Ada S., Winnischofer S.M., Costa F.F. ANKHD1, ankyrin repeat and KH domain containing 1, is overexpressed in acute leukemias and is associated with SHP2 in K562 cells. Biochim. Biophys. Acta. 2006;1762:828–834. doi: 10.1016/j.bbadis.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 46.Dhyani A., Duarte A.S., Machado-Neto J.A., Favaro P., Ortega M.M., Olalla Saad S.T. ANKHD1 regulates cell cycle progression and proliferation in multiple myeloma cells. FEBS Lett. 2012;586:4311–4318. doi: 10.1016/j.febslet.2012.10.037. [DOI] [PubMed] [Google Scholar]
- 47.Sansores-Garcia L., Atkins M., Moya I.M., Shahmoradgoli M., Tao C., Mills G.B. Mask is required for the activity of the Hippo pathway effector Yki/YAP. Curr. Biol. 2013;23:229–235. doi: 10.1016/j.cub.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Machado-Neto J.A., Lazarini M., Favaro P., Franchi G.C., Jr., Nowill A.E., Saad S.T. ANKHD1, a novel component of the Hippo signaling pathway, promotes YAP1 activation and cell cycle progression in prostate cancer cells. Exp. Cell Res. 2014;324:137–145. doi: 10.1016/j.yexcr.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 49.Fragiadaki M., Zeidler M.P. Ankyrin repeat and single KH domain 1 (ANKHD1) drives renal cancer cell proliferation via binding to and altering a subset of miRNAs. J. Biol. Chem. 2018;293:9570–9579. doi: 10.1074/jbc.RA117.000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao P., Li Y., Shen W., Xu X., Zhu W., Yang X. ANKHD1 silencing suppresses the proliferation, migration and invasion of CRC cells by inhibiting YAP1-induced activation of EMT. Am. J. Cancer Res. 2018;8:2311–2324. [PMC free article] [PubMed] [Google Scholar]
- 51.Liu X.F., Han Q., Rong X.Z., Yang M., Han Y.C., Yu J.H. ANKHD1 promotes proliferation and invasion of nonsmallcell lung cancer cells via regulating YAP oncoprotein expression and inactivating the Hippo pathway. Int. J. Oncol. 2020;56:1175–1185. doi: 10.3892/ijo.2020.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu C., Liu L., Wang K., Li X.F., Ge L.Y., Ma R.Z. VHL-HIF-2alpha axis-induced SMYD3 upregulation drives renal cell carcinoma progression via direct trans-activation of EGFR. Oncogene. 2020;39:4286–4298. doi: 10.1038/s41388-020-1291-7. [DOI] [PubMed] [Google Scholar]
- 53.Housman G., Byler S., Heerboth S., Lapinska K., Longacre M., Snyder N. Drug resistance in cancer: an overview. Cancers (Basel) 2014;6:1769–1792. doi: 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]