Abstract
Lactic acid bacterial exopolysaccharides (EPS) are used in the food industry to improve the stability and rheological properties of fermented dairy products. β-Lactoglobulin (BLG), the dominant whey protein in bovine milk, is well known to bind small molecules such as fatty acids, vitamins, and flavors, and to interact with neutral and anionic polysaccharides used in food and pharmaceuticals. While sparse data are available on the affinity of EPS–milk protein interactions, structural information on BLG–EPS complexes, including the EPS binding sites, is completely lacking. Here, binding sites on BLG variant A (BLGA), for oligosaccharides prepared by mild acid hydrolysis of two EPS produced by Streptococcus thermophilus LY03 and Lactobacillus delbrueckii ssp. bulgaricus CNRZ 1187, respectively, are identified by NMR spectroscopy and supplemented by isothermal titration calorimetry (ITC) and molecular docking of complexes. Evidence of two binding sites (site 1 and site 2) on the surface of BLGA is achieved for both oligosaccharides (LY03-OS and 1187-OS) through NMR chemical shift perturbations, revealing multivalency of BLGA for EPS. The affinities of LY03-OS and 1187-OS for BLGA gave KD values in the mM range obtained by both NMR (pH 2.65) and ITC (pH 4.0). Molecular docking suggested that the BLGA and EPS complexes depend on hydrogen bonds and hydrophobic interactions. The findings provide insights into how BLGA engages structurally different EPS-derived oligosaccharides, which may facilitate the design of BLG–EPS complexation, of relevance for formulation of dairy products and improve understanding of BLGA coacervation.
Introduction
β-Lactoglobulin (BLG) is the dominant and most well-studied whey protein in bovine milk.1 It belongs to the lipocalin family of small, extracellular proteins that bind and transport hydrophobic molecules.2,3 Of 10 isoforms, β-lactoglobulin variants A (BLGA) and B are the two most abundant in cow. They differ at two positions, D/G64 and V/A118,4 but have essentially identical three-dimensional structures.5 Both variants contain 162 residues and adopt an eight-stranded β-barrel fold, with an internal cavity referred to as the calyx, flanked by a three-turn α-helix (Figure 1).6 Two β-sheets of β-strands A–D and E–H, respectively, create the β-barrel with the calyx being the primary location for small guest molecules such as fatty acids,3,7−12 dodecyl sulfate,13 β-carotene,14 and irinotecan.15,16 A ninth β-strand I constitutes a large part of the interface in a BLG homodimer, formed in a pH sensitive monomer-to-dimer transition between pH 2.5 and 4.0 that depends on temperature and ionic strength.17−21 BLG is thus mostly a dimer under physiological conditions, that is BLG > 50 μM, NaCl ∼150 mM, 37 °C, and neutral pH 6–8.21−23 The tertiary structure of BLG is further stabilized by two disulfide bonds, C66–C160 and C106–C119, while C121 has a free thiol group. The AB, CD, EF, and GH loops, connecting the strands at the open end of the calyx, are longer and more flexible than the other barrel strand-connecting loops BC, DE, and FG (Figure 1). The EF loop forms a lid mediating the opening and closing of the calyx and exists in a closed conformation at pH < 7, as observed in crystal and NMR structures.6,17 In the closed conformation, the EF loop prevents ligand entry into the calyx, while at pH > 7, the open EF loop24,25 allows access to the calyx, typically hosting hydrophobic ligands.26,27 Dynamics measured by NMR revealed high flexibility of the D strand and of the EF and GH loops allowing malleability in BLG and accommodation of a wide range of ligands.28 Besides the calyx, several studies reported a secondary hydrophobic pocket on the outer surface of BLG that interacts with ligands such as the flavor compound β-ionone (involving Y102, L104, and D129), vitamin D3 affecting hydrophobic residues from the main α-helix (F136, A139, and L140), I147, and the buried R148 from β-strand I, and A142, L143, P144, and M145 from the loop connecting strand I and the main α-helix.27,29−33
Figure 1.
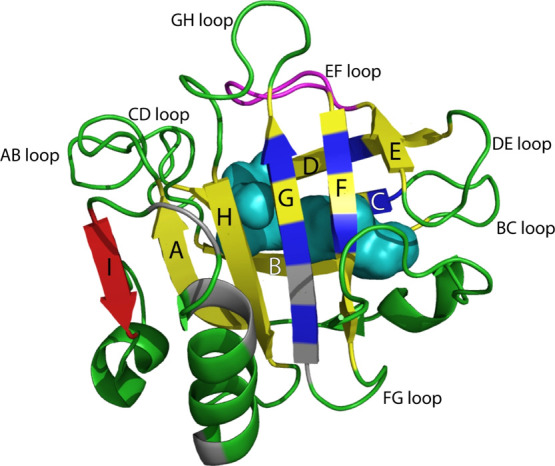
NMR solution structure of BLGA at pH 2.65 (PDB code 1DV9).6 The β-barrel formed by β-strands A–H (labelled) is shown in yellow, and the closed conformation of the EF loop is shown in magenta. The β-strand I in red contributes to the interface in the BLGA homodimer. Hydrophobic ligand binding residues are indicated in blue for the calyx-binding site, and the secondary hydrophobic surface-binding site is indicated in gray. The calyx is space filled (cyan) to show the spaciousness, and the remaining backbone is colored in green.
BLG has been found to interact with several types of polysaccharides, either occurring naturally or added as ingredients in foods and in pharmaceuticals,34,35 such as the anionic pectin,36 κ-carrageenan,37 and alginate,38 as well as the mostly neutral exopolysaccharides (EPS), produced by lactic acid bacteria common in fermented dairy foods.39−41 EPS comprise homo-EPS, composed of a single monosaccharide, such as in fructans and a variety of α-glucans,42−44 and hetero-EPS, composed of different monosaccharides and synthesized from lipid carrier-oligosaccharide repeats, typically of a degree of polymerization (DP) of 3–9, joined to form the corresponding polysaccharide.43−45 Hetero-EPS provide texturizing characteristics, for example, to yogurt,46−48 via formation of coacervates, which motivated the present choice of oligosaccharide repeat ligands, produced by mild acid hydrolysis of EPS from Streptococcus thermophilus LY03 and Lactobacillus delbrueckiissp. bulgaricus CNRZ 1187. Previously, binding of BLGA and other prominent milk proteins to seven EPS from lactic acid bacteria has been assessed by surface plasmon resonance analysis, and the two EPS selected here bound most strongly to native BLGA, as demonstrated in that study.41 Generally, the interactions with purified milk proteins vary with EPS monosaccharide composition, glycosidic linkage patterns, branching, and molecular weight.41−51 Recently, it was suggested that the affinity for EPS is governed by polar interactions with only a smaller contribution from hydrophobic interactions and involves avidity between sites.41,51 However, as the large molecular size of EPS hampers structural characterization of the protein complexes, the role of distinct EPS structures and the mechanisms of EPS–BLG interactions are poorly understood.
In the present study, oligosaccharides representing repeat units were prepared from EPS secreted by S. thermophilus LY0352 and L. delbrueckiissp. bulgaricus CNRZ 1187.53 Their binding sites on BLGA were determined and characterized by NMR revealing two distinct sites on the surface of BLGA, one of which has not been described for ligand binding before. The findings were complemented with affinity measurements by isothermal titration calorimetry (ITC) and molecular docking of the oligosaccharide repeats to the identified binding sites on BLGA. Overall, the results provide molecular details on where EPS-derived oligosaccharides bind to BLGA as well as their binding parameters, which together will facilitate molecular-based construction of optimized food texture and help understand BLGA-based coacervation.
Results
Preparation and Structure Elucidation of LY03-OS and 1187-OS
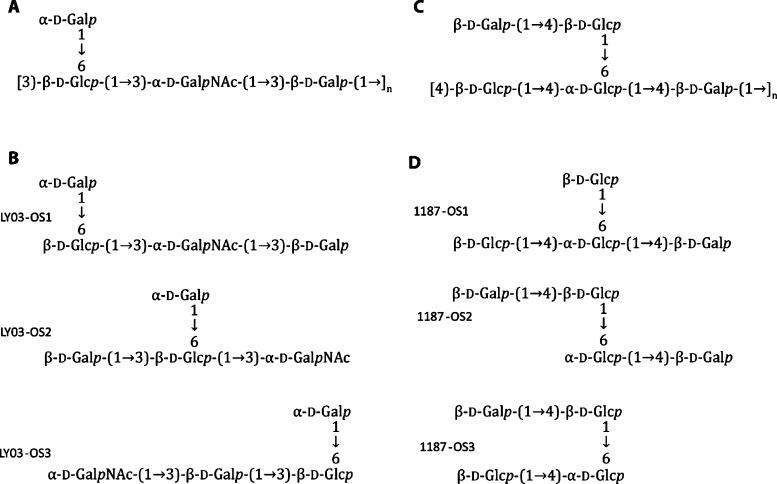
EPS were purified from two strains, S. thermophilus LY03 and L. delbrueckiissp. bulgaricus CNRZ 1187, respectively, grown in 10% skimmed milk medium. Different variations of the known oligosaccharide repeat unit of the EPS from S. thermophilus LY03 (Figure 2A,B), as well as oligosaccharides of smaller or larger DP than the known repeat structures of both EPS (onwards referred to as LY03-OS and 1187-OS, respectively) were produced by mild acid hydrolysis and purified by size-exclusion chromatography.54−56 After further purification by HPLC, LY03-OS and 1187-OS were essentially size homogeneous (Figure S1, inserts), as supported by MALDI-TOF MS (Figure S1). Due to the limited amount available, a purified larger oligosaccharide LY03-OS DP6 was used only for ITC experiments.
Figure 2.
Repeating units of the EPS from Streptococcus thermophiles LY03 and Lactobacillus delbrueckiibulgaricus CNRZ 1187. (A) LY03-OS repeat unit structure. (B) Prepared LY03 oligosaccharides LY03-OS1, LY03-OS2, and LY03-OS3 present at ratios of 8:1:1. (C) 1187-OS repeat unit structure. (D) Prepared 1187 oligosaccharides 1187-OS1, 1187-OS2, and 1187-OS3 present at ratios of 6:3:1. The structures in A and C have been determined previously.46,52,53
The reported structures of the repeating units of LY03-OS and 1187-OS of DP4 and 5, respectively (Figure 2A,C),52,53 were confirmed by NMR structural analysis, of oligosaccharide samples (Tables S1–S5, Figure S2). The LY03-OS sample contained one major and two minor tetrasaccharides, referred to as LY03-OS1-3, in approximate 8:1:1 molar ratios (Figure 2B). The 1187-OS sample contained three tetrasaccharides, 1187-OS1-3, in molar ratios of about 6:3:1 (Figure 2D). Notably, oligosaccharides corresponding to the full repeat motif of DP5 (Figure 2C), were not identified in the 1187-OS sample. The results of the detailed NMR structural analyses of LY03-OS1-3 and 1187-OS1-3 are given in Supporting Information (inclusive Tables S1–S5).
Chemical Shift Assignment of BLGA
The NMR interaction analyses were performed at pH 2.65, where BLGA is purely monomeric. At higher pH values, BLGA exists in a monomer–dimer equilibrium which complicates the NMR analysis with many peaks missing due to line broadening. To analyze the change in chemical shifts upon oligosaccharide binding, the BLGA was made up of the same amino acid sequence as the BLGA previously structure-determined by NMR.57 A similar heteronuclear single-quantum correlation (HSQC) spectrum to our recent study of BLGA–alginate interactions was obtained58 (Figure S3). BLGA has 162 residues including eight prolines; thus, disregarding the N-terminal amide, 153 backbone resonance peaks were expected in the 1H,15N-HSQC spectrum. The chemical shifts were taken from the previous assignments58 and matched perfectly to those published for BLGA,57,58 indicating correctly folded recombinant BLGA.
Interaction between BLGA and LY03-OS and 1187-OS
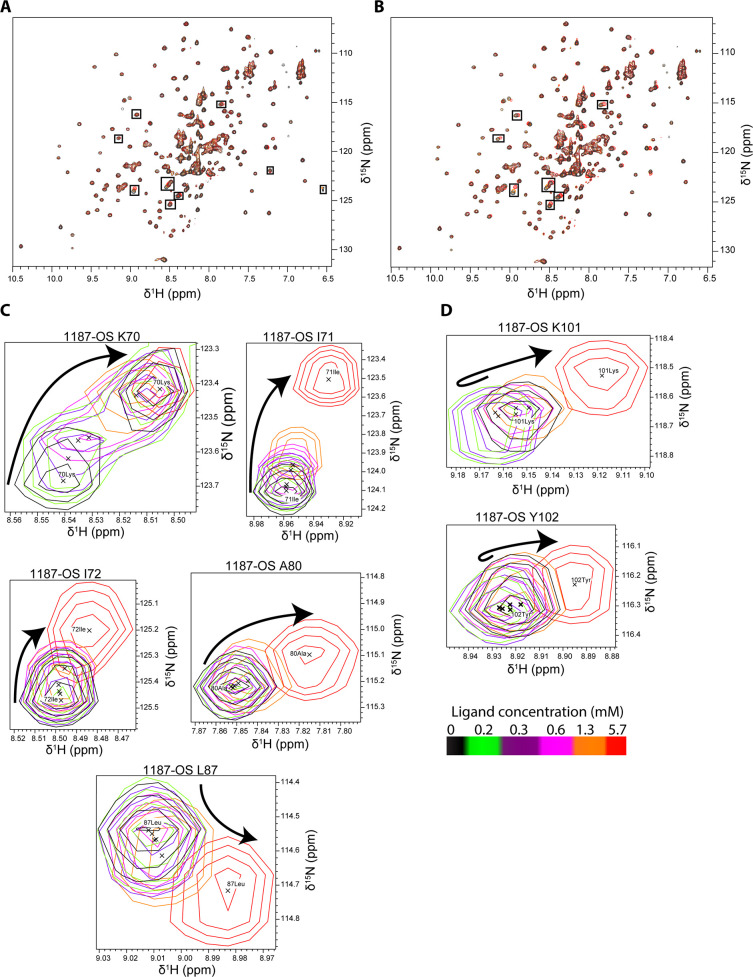
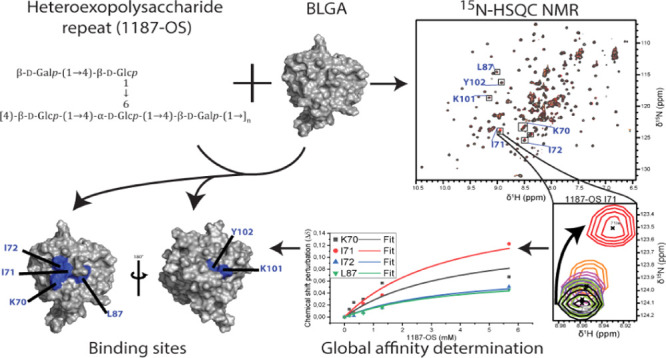
Titration of 15N-labeled BLGA, with increasing amounts of added oligosaccharides obtained from the two EPS, was monitored by changes in chemical shifts of backbone amides (Figure 3A,B) by chemical shift perturbation analysis (Figure 4).
Figure 3.
Overlay of 1H,15N-HSQC spectra of BLGA titrated with 1187-OS and LY03-OS at pH 2.65. (A) Addition of 0.0–5.7 mM 1187-OS and (B) addition of 0.0–6.3 mM LY03-OS. Residues with large chemical shift perturbations are labeled and marked with boxes. (C–D) Changes in peak positions upon titration with 1187-OS (0.0–5.7 mM), (C) chemical shift changes of K70, I71, I72, A80, and L87 corresponding to site 1. (D) Chemical shift changes of K101 and T102 corresponding to site 2. Arrows indicate the direction of the changes with (C) single directional movement and (D) double directional movement.
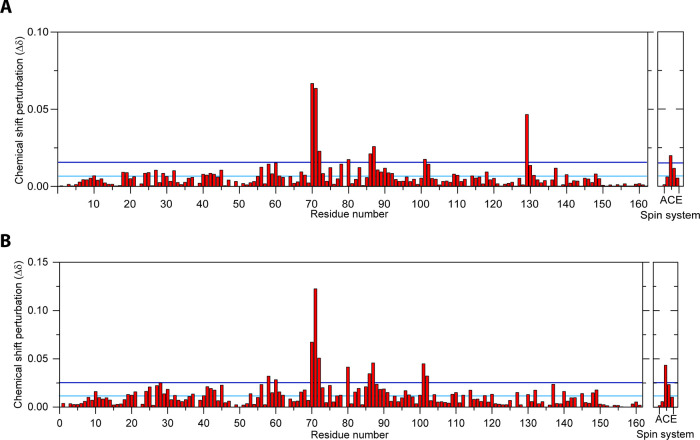
Figure 4.
Chemical shift perturbation analysis of backbone amides in BLGA (350 μM) titrated with (A) LY03-OS (6.3 mM) and (B) 1187-OS (5.7 mM). The horizontal cyan line represents the average chemical shift change, and the blue line represents the AVG + 1STD chemical shift perturbations. Unassigned peaks are labelled A–E.
The highest concentrations used of LY03-OS and 1187-OS were 6.3 and 5.7 mM, respectively, corresponding to BLGA/oligosaccharide molar ratios of 1:14.3 and 1:15.8, respectively. Although changes in chemical shifts were in general small and appeared linear with increasing oligosaccharide concentrations without reaching saturation (Figure 6), large chemical shift perturbations (>1 standard deviation from the mean) reaching saturation were in both cases observed for a subset of residues (Figure 4A,B). Both LY03-OS and 1187-OS bound with modest affinity to BLGA and gave comparable chemical shift perturbation patterns (Figure 4).
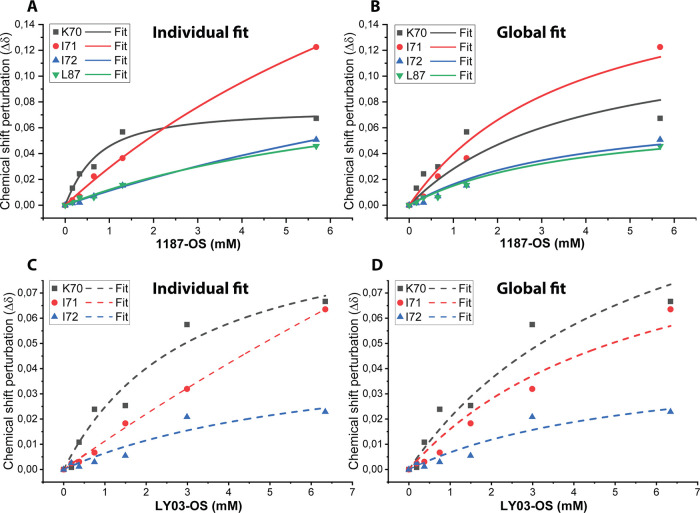
Figure 6.
Fit of the classical binding model to chemical shift perturbations as a function of HEPS oligosaccharide concentrations of binding site 1 (see the Materials and Methods section). (A) Chemical shift perturbations fitted individually for residues titrated with 1187-OS, where chemical shift perturbation is above AVG + 1STD in the 1H,15N-HSQC. (B) Chemical shift perturbations fitted globally for residues titrated with 1187-OS, where chemical shift perturbation is above AVG + 1STD in the 1H,15N-HSQC. (C) Chemical shift perturbations fitted individually for residues titrated with LY03-OS, where chemical shift perturbation is above AVG + 1STD in the 1H,15N-HSQC. (D) Chemical shift perturbations fitted globally for residues titrated with LY03-OS, where chemical shift perturbation is above AVG + 1STD in the 1H,15N-HSQC. See Figures 3C,D and S4 for zooms on peak shifts in the 1H,15N-HSQC for titration with 1187-OS and LY03-OS, respectively.
The largest chemical shift perturbations mapped to the regions K70–I72, L87 (Figures 3C and 6A,C), and K101–Y102 (Figures 3D, S4), with chemical shift perturbations of K70–I72 being the most prominent. Addition of the oligosaccharides also affected L58, K60, and A80, and some chemical shift perturbations occurred in residues C and D of the five unassigned spin systems A–E (Figure 4). In the case of 1187-OS, the signal from D129 broadened beyond detection during the titration, suggesting it to be involved in binding as well.
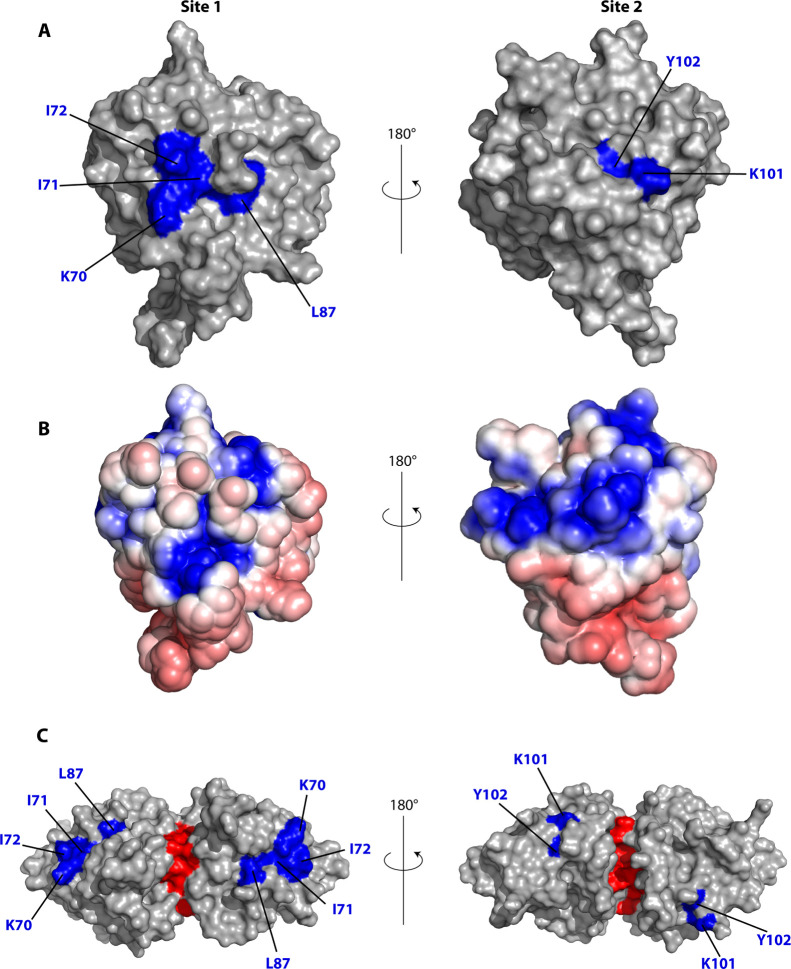
Generally, the largest chemical shift perturbations occurred for charged and polar residues, indicating that BLGA-oligosaccharide binding may involve polar interactions and electrostatics. Furthermore, the uniform chemical shift perturbation pattern in the presence of either of the neutral LY03-OS and 1187-OS, compared to a different chemical shift perturbation pattern induced by trisaccharides from the acidic polysaccharide alginate,58 indicated a certain site specificity of oligosaccharide binding to BLGA. To address such potential discrimination of oligosaccharide-binding sites, the chemical shift perturbations were therefore mapped onto the three-dimensional structure of BLGA (Figure 5). This revealed that K70–I72 plus L87 generate a well-defined, positively charged patch59 for LY03-OS and 1187-OS (site 1, Figure 5), while K101 and Y102 form a smaller and more neutral binding patch (site 2, Figure 5). The site 2 was previously identified to bind alginate trisaccharides.58 D129 has been reported to be involved in site 2.58 Indeed, the peak of D129 also disappeared upon titration with 1187-OS, indicating its involvement in binding, but this interaction could not be quantified. Notably, the direction of the chemical shift perturbations for site 2 residues K101 and Y102 was not unidirectional, suggesting these residues to be perturbed also by binding to site 1 and potentially being cooperatively matured by site 1 binding. This was most prominent for 1187-OS binding, indicating potential differences in orientation across the binding site or differences in binding affinity. Finally, as BLGA exists as a dimer at the milk-relevant pH, we mapped the two identified binding sites onto the dimer surface (Figure 5C). None of the two binding sites were obscured by dimer formation, and both were accessible also in the dimer.
Figure 5.
Chemical shift perturbations larger than AVG + 1STD mapped on the structure of BLGA.6 (A) LY03-OS, 1187-OS induced changes mapped onto the BLGA surface (PDB code 1DV9). (B) Electrostatic surface representation of BLGA (PDB code 1DV9) at pH 2.65 calculated using ProPka, APBS, and pymol.59 Red: negative potentials, blue: positive potentials, and white: uncharged regions. (C) LY03-OS, 1187-OS induced changes mapped onto the BLGA surface (PDB code 1BEB); the dimeric interface is marked in red.
Binding Affinities of BLGA for LY03-OS and 1187-OS
To quantify the interactions of the LY03-OS and 1187-OS with BLGA in more detail, their dissociation constants (KD) were determined from nonlinear global fits to the largest chemical shift perturbations observed in the 1H,15N-HSQCs, including all residues perturbed above average plus 1 standard deviation (AVG + 1STD) (Figure 6; Table S6). Due to peak overlap or line broadening, A86 and D129 were excluded from the fits. With the oligosaccharide concentrations used, saturation of site 2 was not achieved, and hence, reliable KD values for this site were not obtained. Residues which were located alone in the spectra and with large chemical shift perturbations (Figure 4) were selected, and only residues with reliable independent KD values (Table S6) were included in the global fitting procedure (see the Materials and Methods section), which gave very similar KD values for LY03-OS and 1187-OS of 5.4 ± 1.8 and 3.5 ± 1.2 mM, respectively (Figure 6 and Table S7).
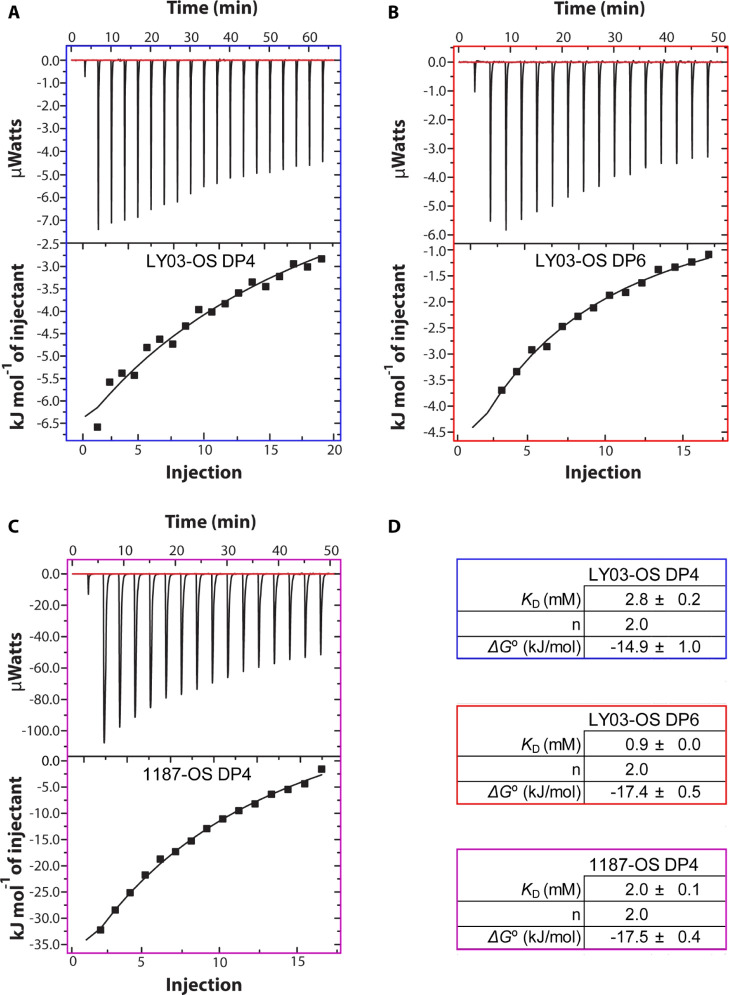
As the change in pH from pH 2.65 to pH 4.0 will affect the protonation states of acidic side chains, and especially that of D129 positioned in site 2,58 we addressed binding of the oligosaccharides at dimer conditions, at pH 4.0, that is also more relevant for milk by using ITC. The data of binding of oligosaccharides determined using ITC at pH 4.0 were fitted to a one site binding model using n = 2, as provided from the NMR analyses. The enthalpograms showed that the interactions between BLGA and LY03-OS DP4 and 1187-OS DP4 were both exothermic (Figure 7) and yielded KD values of 2.8 ± 0.2 and 2.0 ± 0.1 mM, respectively, close to the values obtained by NMR at monomer conditions. Fitting using n = 1 or n = 3 provided poorer fits to the data (with χ2 values of 255, 240, and 260 for n = 1, 2, or 3, respectively); still, KDs varied only ±0.5, 0.2, and 0.4 mM for LY03-OS DP4, LY03-OS DP6, and 1187-OS DP4, respectively. Thus, the change in pH did not seem to affect the affinity considerably, underscoring the dominance of lysine side chains in the binding sites. However, D129 may get protonated at pH 2.65. We therefore calculated its pKa value using the program PropKa59,60 and the three-dimensional structures of the monomer and the dimer. D129 is fully accessible in both states, and a pKa value of 2.87 (PDB 1BEB) and 2.51 (PDB 1DV9) suggests that even at pH 2.65, a considerable fraction of D129 will still be negatively charged.
Figure 7.
ITC analysis of 37 μM BLGA binding to LY03-OS and 1187-OS (DP4 and DP6). ITC data of (A) LY03-OS of DP4 (5 mM), (B) LY03-OS of DP6 (4 mM), (C) 1187-OS of DP4 (5.5 mM), and (D) binding parameters from fitting the data to a one site model with n fixed to 2.
Due to the scarcity of the oligosaccharides, and the overall correspondence of the results with the NMR data, only one repetition was carried out for each ligand. Because of this, we did not attempt to extract the enthalpies and entropies for binding. Comparing the ITC and NMR results, 1187-OS appears to have a slightly higher affinity for BLGA than LY03-OS. An oligosaccharide of DP6 was also purified from the acid hydrolysate of LY03 and showed KD of 0.9 ± 0.0 mM for BLGA (Figure 7D).
Overall, the analyses of chemical shift perturbations from NMR and the ITC titrations jointly indicated that BLGA uses two sites for binding EPS oligosaccharides, which all bound with low mM KD, and hence modest affinity, to BLGA. With two sites accessible there is potential avidity effects driving coacervation of BLGA and EPS.
Docking of Modeled LY03-OS and 1187-OS Repeats on BLGA
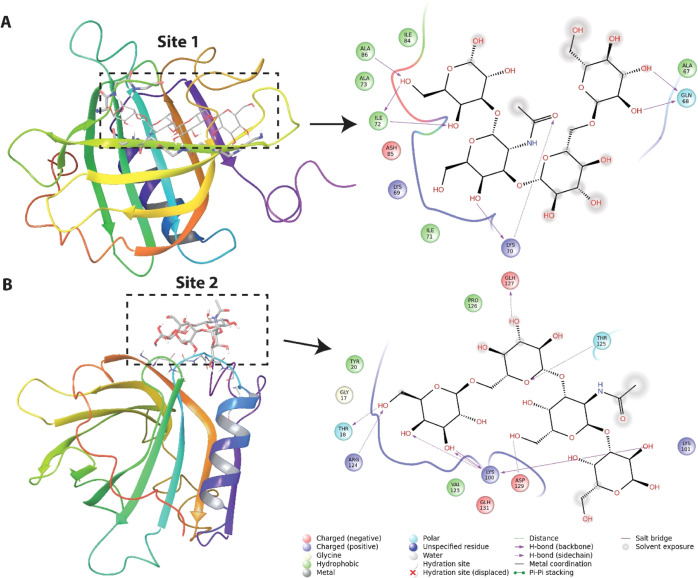
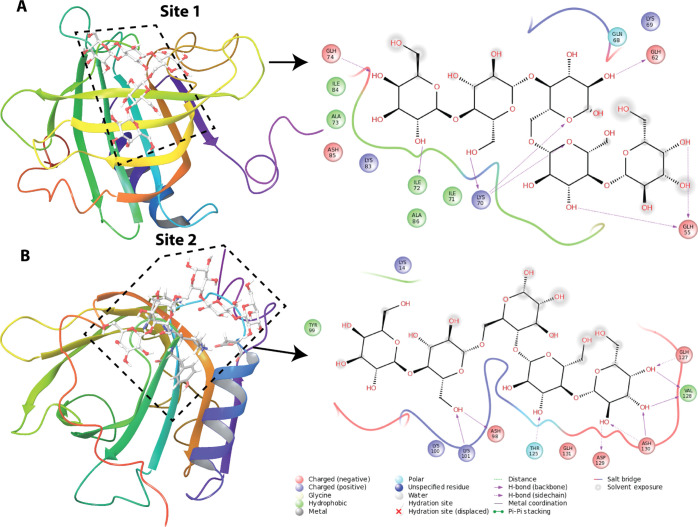
The chemical shift perturbations from the oligosaccharide titrations were used to model complexes of BLGA with LY03-OS and 1187-OS (Figure 2A,C), respectively. The docking experiments were independently performed at site 1 (K70, I71, and I72) and site 2 (K100, K101, Y102, and D129) by initially using the Glide SP docking mode. Four structures of the complex were selected using the criterion that the poses (a candidate-binding mode) are different (>1 Å rmsd between the pose with the best Glide SP score and the selected pose). These structures were subsequently subjected to induced-fit docking (IFD) with final rescoring using Glide XP. For sites 1 and 2, the best ligand conformation for the LY03-OS tetrasaccharide repeat (Figure 2A) resulted in IFD scores (Glide XP score) of −306.5 kcal/mol (−11.8 kcal/mol) and −305.2 kcal/mol (−10.7 kcal/mol), respectively. For the 1187-OS pentasaccharide repeat (Figure 2C), the scores for sites 1 and 2 were −304.5 kcal/mol (−13.4 kcal/mol) and −303.0 kcal/mol (−11.4 kcal/mol), respectively. Poses with the best IFD scores for the LY03-OS and 1187-OS repeat units at the two sites are presented in Figures 8 and 9. Although the energy differences are relatively small for a particular oligosaccharide binding at the two sites, the poses for LY03-OS and 1187-OS are clearly distinct. Both oligosaccharides bind to β-strand D in site 1 and form several hydrogen bonds with backbone atoms, especially of K70 and I72. The docking poses of the oligosaccharides at site 1 show that LY03-OS forms fewer hydrogen bonds with protein residues than 1187-OS (see Figures 8A and 9A). Furthermore, 1187-OS has several interactions with protonated acidic residues (E55, D62, and E74), which are not observed for LY03-OS. Site 2 is located partly in the FG loop and in the loop between strand H and the main α-helix, and the binding of the oligosaccharides is favored by hydrogen bonds with backbones and side chains. For 1187-OS, backbone hydrogen bonds are observed with T18, K101, and R124, whereas for LY03-OS, backbone hydrogen bonds are only observed with K100 (Figures 8B and 9B). We also investigated the binding of 1187 tetrasaccharides (Figure 2D) to site 1/site 2. The IFD score (Glide XP score) for 11987-OS1, 1187-OS2, and 1187-OS3 are, respectively, −303.2/–301.2 (−11.7/–10.1), −305.6/–302.5 (−13.4/–12.0), and −302.7/–304.3 (−11.9/–11.7). Numbers are given in kcal/mol. Although the IFD scores are close to each other, the interaction pattern for the tetrasaccharides with BGLA is different in utilizing hydrogen bonds to residue side chains and backbones (see Figures S9–S10). Considering the latter: 1187-OS1 forms backbone hydrogen bonds with I72, D85, and A86 (site 1) and T125, K100, E125, and D129 (site 2); 1187-OS2 forms backbone hydrogen with K70, I72, D85, and A86 (site 1) and K100, R124, and V128 (site 2); and 1187-OS3 forms backbone hydrogen bonds with W61, K70, I72, and D85 (site 1) and T18, K101, and R124 (site 2). Furthermore, the energies are close to the docking score for the 1187-OS pentasaccharide repeat, indicating that an extension of the saccharide from 4 to 5 units does not significantly change the binding score.
Figure 8.
Structure of BLGA with bound LY03-OS repeat model (Figure 2A) at: (A) site 1 (K70, I71, and I72) and (B) site 2 (K100, K101, Y102, and D129). The LY03-OS repeat model is displayed as sticks. The backbone of BLGA residues within 3 Å of the oligosaccharide is shown as sticks. The color coding is as follows: carbon—gray, oxygen—red, and nitrogen—blue. The tertiary structure of BLGA is displayed in rainbow color on the left. To the right, the oligosaccharide interaction diagrams are shown. The type of interactions are listed in the inset of the interaction diagram.
Figure 9.
Structure of BLGA with the bound 1187-OS repeat model (Figure 2C) at: (A) site 1 (K70, I71, and I72) and (B) site 2 (K100, K101, Y102, and D129). The 1187-OS repeat model is displayed as stick (light gray). The backbone of residues that are within 3 Å of the oligosaccharide is shown as sticks, and the color coding is as follows: carbon-gray, oxygen-red, and nitrogen-blue. The tertiary structure of BLGA is displayed in rainbow color on the left. To the right, the oligosaccharide interaction diagrams are shown. The type of interactions are listed in the inset of the interaction diagram.
Discussion
Due to its ability to bind a wide range of hydrophobic compounds, BLG is an extensively studied milk protein relevant for a large number of industrial and biotechnological applications. The primary binding site for these ligands is in the calyx,3,8,27,29 whereas a secondary binding site is located in the hydrophobic pocket at the homodimer interface formed by the α-helix and the β-barrel.8,10,11,27 We recently reported two surface-exposed binding sites for anionic alginate trisaccharides on BLGA, different to both the primary and secondary sites.58 The present study also identified two surface-exposed binding sites for the neutral oligosaccharides of DP4 LY03-OS and 1187-OS, prepared from two EPS from lactic acid bacteria. One of these sites, site 1, is distinct from all previously reported binding sites, while site 2 is shared with one of the two recently identified binding sites for alginate trisaccharides.58 Jointly, this surmounts to a total of five different ligand-binding sites identified on BLGA.
Site 1, consisting of K70, I71, I72, and L87, is situated at the mouth of the calyx where access to the cavity of the hydrophobic β-barrel is prevented by the EF loop forming a lid at pH 2.65. The mouth of the calyx is often involved in hydrogen bonding with hydroxyl groups of amphipathic molecules having their hydrophobic part buried inside the calyx.26,27 Notably, under the conditions used, site 1 was not involved in binding alginate trisaccharides.58 In contrast, site 2 which is composed of K101, Y102, and for LY03-OS including D129, is distinct from both site 1 and the previously reported surface-residing hydrophobic pocket accessible in the dimer. However, some of the residues perturbed by β-ionone, that is Y102 and D129, but not K60 and L104,33 are shared with site 2. The present BLGA EPS-oligosaccharide titrations were carried out at pH 2.65, where side chains of aspartic and glutamic acids are protonated. However, from the chemical shift perturbations (Figure 4), the only acidic amino acid affected upon binding of EPS-oligosaccharides is D129 and its pKa value suggests that this is fractionally deprotonated at pH 2.65. As there is also a possibility of an interaction between the deprotonated carboxylate form of D129 with the OH groups of the EPS-oligosaccharides, this suggests that site 2 is available also at higher pH, as investigated by ITC at pH 4.0. EPS-oligosaccharide-binding site 2 has also been reported for the negatively charged alginate trisaccharides; however, binding of alginate trisaccharide also caused selective perturbation of E127, D130, and D137,58 which were assumed to be protonated at pH 2.65. These perturbations likely stem from longer ranged negative electrostatic effect of the alginate trisaccharide, an effect we do not observe for the neutral EPS-oligosaccharides LY03-OS and 1187-OS.
Chemical shift perturbation analysis showed that LY03-OS and 1187-OS bind weakly albeit similarly to BLGA, yielding KD values in the low mM range (Figure 6, Tables S6 and S7). However, due to lack of saturation for some residues in the binding site, as well as the observed line broadening effect at the applied oligosaccharide concentrations, a thorough differentiation between sites 1 and 2 was not possible. One reason may be that several different oligosaccharide orientations are possible at the binding site, illustrated by the docking experiments, or that the oligosaccharide binding to one site may affect binding at the other site, as indicated by the non-linear changes in the chemical shift perturbations for site 2 residues (Figure 3D). Thus, residues showing saturation as K70 and I71 (site 1), as well as line broadening of D129 (site 2), are likely core residues in the binding sites, whereas residues not saturated by oligosaccharide might still be involved in binding if the oligosaccharide adopts a certain orientation. We thus note that in the 1H,15N-HSQC, D129 showed line broadening effects and that K101 and Y102 indicated a complex binding model (Figure 3D), suggesting that site 2 in fact may have a different binding mechanism and affinity from site 1, for which we were able to extract affinity by NMR. The docking procedure resulted in docking scores for the best ligand conformation for site 1 and site 2 of −11.8 kcal mol–1 and −10.7 kcal mol–1, respectively, supporting a slightly higher affinity of site 1 for the LY03-OS repeat (DP4, Figures 2A and 8). Similarly, for the 1187 repeat (DP5, Figures 2C and 9), the docking score was −13.4 kcal mol–1 for site 1 and −11.7 kcal mol–1 for site 2, again indicating higher affinity for site 1. Both sites showed higher affinity for the 1187-OS repeat than that for the LY03-OS repeat, fully in line with affinities determined by NMR and ITC. However, to further rank the conformations in the different poses, it may require allowing for the effect of water molecules and protein flexibility by using molecular dynamics simulations.
The ITC analysis at pH 4.0 quantifying BLGA binding to the LY03-OS and 1187-OS of DP4 (Figure 2B,D) yielded KD values of 2.8 ± 0.2 mM and 2.0 ± 0.1 mM, respectively (Figure 7), while KD was 0.9 mM ± 0.0 mM for binding of DP6 LY03-OS (Figure 7D). These KD values are in the same order of magnitude and only slightly lower than the values obtained by chemical shift perturbations at pH 2.65, indicating that the dimer, which is present at pH 4.0, maintains both interaction sites. Notably, the full-length EPS binding analysis carried out using SPR at pH 4.0 gave KD values normalized to the repeats of LY03 and 1187 of 1.5 and 4.7 μM, respectively,41 indicating that a distinct avidity effect was also reflected in the difference in KD between LY03 DP4 and DP6 determined by ITC in the present work. This multivalency of BLGA may enable formation of coacervates linking more chains to one BLGA monomer, and it is therefore possible that the presence of two different sites is an underlying feature of BLGA that promotes higher order structures of relevance to its use in the food industry. Studying larger oligosaccharides will be crucial for further addressing the mechanism of coacervation.
Conclusions
Two EPS binding sites, sites 1 and 2, are identified on the surface of BLGA at pH 2.65. Site 1 is distinctly different from all previously reported binding sites on BLG, while site 2 shares residues involved in binding of alginate trisaccharides and hydrophobic compounds. The chemical shift perturbations showed site-specific binding of LY03-OS and 1187-OS to BLGA with the modest affinity driven by polar interactions. As shown by ITC, the binding sites are preserved in the BLGA dimer. These results provide new insights into polysaccharide binding and may form a basis for designing interactions in BLGA–EPS particles to optimize food texture. The data furthermore offer a platform for exploring the binding ability of BLG with various other hydrophilic compounds and open for understanding BLGA coacervation.
Materials and Methods
EPS Production and Purification
EPS LY03 and 1187 were produced in 10% skimmed milk medium by S. thermophilus LY03 (kind gift of Prof. Luc De Vuyst, Brussels, Belgium) and by L. delbrueckiissp. bulgaricus CNRZ 1187, respectively, as described previously.41 Briefly, proteins and cells were precipitated after fermentation by addition of 80% (v/v) trichloroacetic acid to a final concentration of 20% (v/v) and removed by centrifugation (15,000g, 30 min, 4 °C). Subsequently, EPS LY03 and 1187 were precipitated from the supernatants overnight at 5 °C by ethanol (two volumes) and acetone (one volume), respectively, collected by centrifugation (15,000g, 30 min, 4 °C), dissolved in Milli-Q (MQ)-water (1 h, 50 °C), and purified by size-exclusion chromatography (XK26/100 Sephacryl S-400 column; GE Healthcare, Uppsala, Sweden) with MQ-water as the mobile phase at RT and a constant flow rate (1 mL min–1). EPS were quantified by the phenol–sulfuric acid method using monosaccharide mixtures mimicking the repeating unit compositions as standard.61
Preparation of LY03-OS and 1187-OS
EPS (10 mg mL–1) was hydrolyzed by 0.1 M TFA (20 min, 90 °C) as slightly modified from a method described previously.62 The hydrolysate was adjusted on ice to pH 5–7 by 5 M NaOH and diluted with MQ-water to 0.1 M NaOH. Oligosaccharides (<3 kDa) were separated from larger EPS fragments by ultrafiltration (3 kDa Amicon filter; Millipore, Billerica, MA) and collected in the run-through. EPS fragments (>3 kDa, ∼1 mL in the filter) were diluted with MQ-water (∼15 mL) and a second run-through containing oligosaccharides (<3 kDa) was collected. This step was repeated thrice. EPS >3 kDa was collected and resubjected to hydrolysis by adding 99% TFA to a final concentration of 0.1 M and heating for 5 min (90 °C). Such further partial hydrolysis was continued until negligible amounts of high molecular mass EPS remained, as evaluated by thin-layer chromatography. The oligosaccharide mixtures were separated (XK16/100 Bio-Gel P-2; Bio-Rad, California, USA) in MQ-water at a constant flow rate (0.1 mL min–1) and RT. Oligosaccharides were collected, and acetonitrile was added to 50% (v/v) and further purified by HPLC (UltiMate 3000; Dionex, Sunnyvale, CA) equipped with a refractive index detector (RI-101; Showa Denko, Kanagawa, Japan) using a TSKgel Amide-80 column (5 μm particle size; 4.6 by 250 mm with 4.6 by 10 mm guard column at 1 mL min–1; Tosoh, Tokyo, Japan]) in acetonitrile/water (70:30, v/v) at 70 °C. Oligosaccharides were lyophilized and stored at −20 °C. They were dissolved in appropriate buffers depending on the experiment. Oligosaccharides were quantified using the phenol–sulfuric acid method61 and purified after HPLC was assessed by thin-layer chromatography (silica gel 60 F254, Merck), as described58 (Figure S1).
Structure Elucidation of LY03-OS and 1187-OS
LY03-OS and 1187-OS were dissolved in 99.9% D2O, and NMR spectra were recorded at 25 °C on a Bruker Avance III (799.90 MHz for 1H and 201.15 MHz for 13C) equipped with a 5 mm TCI 1H/(13C, 15N) cryoprobe using 3-(trimethylsilyl)-1-propanesulfonic (DSS) as an internal standard (0.0002 ppm and −2.032 ppm for 1H and 13C, respectively).62 The mixing times for rotating frame nuclear Overhauser effect spectroscopy (ROESY) and heteronuclear single quantum coherence–total correlation spectroscopy (HSQC–TOCSY) were 200 and 80 ms, respectively. Heteronuclear multiple bond spectroscopy was optimized for 10 Hz long range coupling constants. All two-dimensional spectra were recorded using standard Bruker pulse sequences and were acquired and processed using TopSpin 3.5.
Isothermal Titration Calorimetry
BLGA (in house preparation isolated from cow milk)63 was dissolved in 10 mM sodium acetate (pH 4.0) at 5.0 mg mL–1 (272 μM), centrifuged (12,000g, 20 min, 4 °C), and dialyzed 3 × 5 h at 4 °C against 10 mM sodium acetate (pH 4.0) (3 kDa cutoff, SpectraPor membrane; Spectrum). LY03-OS (DP4 and DP6) and 1187-OS (DP4) were dissolved to 4–5.5 mM in 10 mM sodium acetate (pH 4.0) from the dialysis of BLGA and centrifuged (12,000g, 20 min, 4 °C). ITC titrations were conducted using ITC200 (Thermo Scientific, USA) at 25 °C with 4.0 mM LY03-OS DP4 and 5.0 mM DP6 and 5.5 mM 1187-OS DP4 (all diluted in dialysate) in the syringe (sample volume 40 μL) and 37 μM BLGA (diluted in dialysis buffer) in the cell (sample volume 200 μL), in a series of 16 injections, each of 2.5 μL including an initial 0.4 μL injection (deleted from the data set). A blank titration was subtracted from the data to correct for heat of dilution. Integrated and normalized binding data were fitted to a one-site binding model and n = 1, 2, or 3 using the accompanying origin-based software (Microcal analysis). For analysis of LY03-OS DP 6, the second titration step was removed as it was an outlier which impaired the fitting.
BLGA Cloning and Pichia Pastoris Transformation
The gene encoding bovine BLGA (CAA32835) was modified to encode the amino acid sequence of BLGA, previously analyzed by NMR spectroscopy, which has three extra residues (Glu-Ala-Glu) at the N-terminus and the first two residues of BLGA WT (1L-2I) replaced by 1A-2Y and a point mutation (V108F).57,58 Cloning and mutagenesis were carried out, as described previously.58
Production of Isotope-Labeled Recombinant BLGA
Production of BLGA was carried out, as described.58 Briefly, the best expressing transformant was precultured in 25 mL of buffered glycerol-complex medium [BMGY: 1% (w/v) yeast extract, 2% (w/v) peptone, 0.1 M KH2PO4 (pH 6.0), 0.34% (w/v) yeast nitrogen base, 4 × 10–5% (w/v) biotin, 1% (v/v) glycerol] and further propagated in 1 L BMGY medium until OD600 = 2–6 (150 rpm, 30 °C). The cells were harvested (1500g, 5 min, 22 °C), resuspended in 1 L buffered minimal methanol [0.1 M KH2PO4 (pH 6.0), 0.34% (w/v) yeast nitrogen base without amino acids and ammonium chloride, 4 × 105% (w/v) biotin, 1% 15N-ammonium sulfate (Cambridge Isotope Laboratories Inc, Andover, USA), 0.5% (v/v) glycerol] medium for expression, as described previously.58
Purification of Recombinant BLGA
BLGA was purified using a desalting Hiprep Desalt 26/10 column (GE Healthcare) and size exclusion Hiload Superdex 75 16/60, (GE Healthcare), as described previously.58 The protein was dialyzed (Spectra/Por, Spectrumlabs, 3 kDa, 4 °C) against MQ-water thrice with 100-fold dilution for at least 4 h and lyophilized. For NMR experiments, BLGA WT was dissolved in 55 mM KH2PO3 (pH 2.65) and dialyzed as above against the buffer. All protein concentrations were determined spectrophotometrically at 280 nm using ε = 18,450 M–1 cm–1 calculated by ProtParam64 from the amino acid sequence.
NMR Experiments of BLGA
15N-BLGA was diluted in 50 mM potassium phosphate (pH 2.65), H2O/D2O (90/10 v/v), 0.1 mM 2,2-dimethyl-2-silapentanesulfonic acid (DSS) to a concentration of 100 μM and analyzed by NMR spectroscopy. These conditions were chosen to stabilize the BLGA monomer, which is stable for many days at 37 °C and for months at 4 °C.57 Immediately prior to NMR analysis in 5 mm Shigemi microtubes, the protein samples were centrifuged (20,000g, 10 min, 25 °C) to remove the precipitate. Titrations were performed under the same conditions but with the addition of 0–6.3 mM LY03-OS or 0–5.7 mM 1187-OS. The 1H,15N-HSQC spectra were recorded on either an 800 MHz Varian INOVA spectrometer equipped with a 5 mm triple resonance room temperature probe with a Z-field gradient at 37 °C and mixing time 150 ms using Varian/Agilent BioPack sequence or a 600 MHz Bruker AVANCE system equipped with a cryoprobe. Recorded free induction decays were processed using the nmrPipe.65 Proton chemical shifts were referenced to internal 2,2-dimethyl-2-silapentanesulfonic acid at 0.00 ppm and the assignments of BLGA transferred from our previous study.58 For determination of chemical shift perturbations and KDs, the change in chemical shift (in ppm) in the proton and nitrogen dimension was calculated as:66
where a scaling factor of 5 was chosen to match the proton and nitrogen chemical shift change difference. The dissociation constant, KD, was then determined using the following equation
where [P]0 is the total concentration of protein and is set constant at 0.1 mM, whereas [L]0 is the total ligand concentration at each step (the variable). For global fits, the same equation was used, with KD being the global variable and Δδmax being kept as an individual variable.
Docking of LY03-OS and 1187-OS Repeat Models on BLGA
The NMR structure of BLGA was obtained from the protein data bank (PDB entry code: 1DV9)6 choosing arbitrarily the first model. The LY03-OS and 1187-OS repeat models (Figure 2A,C) were built in the GLYCAM carbohydrate builder.67 BLGA was prepared using Protein Preparation Wizard and LigPrep programs, respectively, within the Schrödinger suite 2016-168 applying besides for pH, which was set to 2.65, the default settings. The repeat models were prepared using LigPrep with default settings at target pH 2.65 using Epik.69 Docking calculations were performed using Glide within the Schrödinger suite 2016-168 and applying standard precision (Glide SP),70,71 extra precision (Glide XP),72 and IFD73 with default settings. According to the NMR results, two binding areas were of interest; site 1 (K70, I71, and I72) and site 2 (K100, K101, Y102, and D129). Both sites were independently investigated, and for each docking study, these residues were used to define the center of the docking region (grid) with an extension of ≤22 Å (larger than the sizes of the repeat models). The IFD docking protocol consists of several sequential steps; (i) Glide SP docking using a softened potential, where the scaling factor for van der Waals radii was set to 0.5 for the BLGA and oligosaccharides, (ii) Prime refinement was applied to optimize side-chain conformation of residues within 5 Å of the oligosaccharides using the OPLS3 force field,74 and (iii) Glide XP74 redocking of the oligosaccharides into all induced fit protein structures. Each pose was ranked by using the composite induced fit score calculated as a linear combination of 0.05 × Prime energy plus the Glide2 XP score (IFD score).
Acknowledgments
Karina Jansen is thanked for general laboratory technical assistance. All the participants of the HEXPIN project are thanked for discussion and guidance.
Glossary
Abbreviations
- BLG
β-lactoglobulin
- BLGA
β-lactoglobulin variant A
- EPS
exopolysaccharide
- HPLC
high-performance liquid chromatography
- HSQC
heteronuclear single quantum correlation
- HSQC–TOCSY
heteronuclear single quantum correlation–total correlation spectroscopy
- ITC
isothermal titration calorimetry
- NMR
nuclear magnetic resonance
- ROESY
rotating-frame nuclear Overhauser effect correlation spectroscopy
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c00060.
Structure elucidation of LY03-OS and 1187-OS, as well as NMR spectra of 15N-BLG (PDF)
Author Contributions
J.B., B.B.K., J.Ø.D., and B.S. conceived the study. J.B., S.K., M.M., C.K., M.S.M., and E.G.P.S. performed experiments and analyzed data. G.H.J.P. performed molecular docking. All authors contributed to the writing of the manuscript and reviewed the final version as completed by M.M., B.B.K., and B.S.
The project Associative interactions between EPS from lactic acid bacteria and milk proteins gaining insights deployable in the design of optimized food texture (HEXPIN) is supported by the Independent Research Fund Denmark Technology and Production Sciences (grant no. DFF–1335-00221). The oligosaccharides spectra were recorded at the NMR Center DTU, and the protein spectra were recorded at the Structural Biology and NMR Laboratory at BIO, UCPH, both supported by Villum Fonden. Some of the spectra were recorded at cOpenNMR, Department of Biology, UCPH, supported by the Novo Nordisk Foundation (#NNF18OC0032996). Novo Nordisk Foundation is thanked for funding by J.Ø.D and C.K. for grant no. 5371 and by B.S. for grant no. NNFOC0027616. J.B., M.M., and E.G.P.S. thank the Technical University of Denmark (DTU) for 1/3 PhD scholarships.
The authors declare no competing financial interest.
Supplementary Material
References
- Bordin G.; Cordeiro Raposo F.; de la Calle B.; Rodriguez A. R. Identification and Quantification of Major Bovine Milk Proteins by Liquid Chromatography. J. Chromatogr. A 2001, 928, 63–76. 10.1016/s0021-9673(01)01097-4. [DOI] [PubMed] [Google Scholar]
- Flower D. R. The Lipocalin Protein Family: Structure and Function. Biochem. J. 1996, 318, 1–14. 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.-Y.; Pérez M. D.; Puyol P.; Sawyer L. β-Lactoglobulin Binds Palmitate within Its Central Cavity. J. Biol. Chem. 1999, 274, 170–174. 10.1074/jbc.274.1.170. [DOI] [PubMed] [Google Scholar]
- Farrell H. M.; Jimenez-Flores R.; Bleck G. T.; Brown E. M.; Butler J. E.; Creamer L. K.; Hicks C. L.; Hollar C. M.; Ng-Kwai-Hang K. F.; Swaisgood H. E. Nomenclature of the Proteins of Cows’ Milk-Sixth Revision. J. Dairy Sci. 2004, 87, 1641–1674. 10.3168/jds.s0022-0302(04)73319-6. [DOI] [PubMed] [Google Scholar]
- Qin B. Y.; Jameson G. B.; Bewley M. C.; Baker E. N.; Creamer L. K. Functional implications of structural differences between variants A and B of bovine β-lactoglobulin. Protein Sci. 2008, 8, 75–83. 10.1110/ps.8.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrínová S.; Smith M. H.; Jameson G. B.; Uhrín D.; Sawyer L.; Barlow P. N. Structural Changes Accompanying pH-Induced Dissociation of the β-Lactoglobulin Dimer. Biochemistry 2000, 39, 3565–3574. 10.1021/bi992629o. [DOI] [PubMed] [Google Scholar]
- Qin B. Y.; Creamer L. K.; Baker E. N.; Jameson G. B. 12-Bromododecanoic Acid Binds inside the Calyx of Bovine Beta-lactoglobulin. FEBS Lett. 1998, 438, 272. 10.2210/pdb1bso/pdb. [DOI] [PubMed] [Google Scholar]
- Kontopidis G.; Holt C.; Sawyer L. The Ligand-binding Site of Bovine β-Lactoglobulin: Evidence for a Function?. J. Mol. Biol. 2002, 318, 1043–1055. 10.1016/s0022-2836(02)00017-7. [DOI] [PubMed] [Google Scholar]
- Yi J.; Fan Y.; Yokoyama W.; Zhang Y.; Zhao L. Characterization of Milk Proteins-Lutein Complexes and the Impact on Lutein Chemical Stability. Food Chem. 2016, 200, 91–97. 10.1016/j.foodchem.2016.01.035. [DOI] [PubMed] [Google Scholar]
- Loch J.; Polit A.; Górecki A.; Bonarek P.; Kurpiewska K.; Dziedzicka-Wasylewska M.; Lewiński K. Two modes of fatty acid binding to bovine β-lactoglobulin-crystallographic and spectroscopic studies. J. Mol. Recognit. 2010, 24, 341–349. 10.1002/jmr.1084. [DOI] [PubMed] [Google Scholar]
- Loch J. I.; Polit A.; Bonarek P.; Olszewska D.; Kurpiewska K.; Dziedzicka-Wasylewska M.; Lewiński K. Structural and thermodynamic studies of binding saturated fatty acids to bovine β-lactoglobulin. Int. J. Biol. Macromol. 2012, 50, 1095–1102. 10.1016/j.ijbiomac.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Loch J. I.; Bonarek P.; Polit A.; Riès D.; Dziedzicka-Wasylewska M.; Lewiński K. Binding of 18-carbon unsaturated fatty acids to bovine β-lactoglobulin-Structural and thermodynamic studies. Int. J. Biol. Macromol. 2013, 57, 226–231. 10.1016/j.ijbiomac.2013.03.021. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Magdaleno G.; Bello M.; Portillo-Téllez M. C.; Rodríguez-Romero A.; García-Hernández E.; Rodríguez-Romero A.; García-Hernández E. Ligand binding and self-association cooperativity of β-lactoglobulin. J. Mol. Recognit. 2013, 26, 67–75. 10.1002/jmr.2249. [DOI] [PubMed] [Google Scholar]
- Mehrad B.; Ravanfar R.; Licker J.; Regenstein J. M.; Abbaspourrad A. Enhancing the physicochemical stability of β-carotene solid lipid nanoparticle (SLNP) using whey protein isolate. Food Res. Int. 2018, 105, 962–969. 10.1016/j.foodres.2017.12.036. [DOI] [PubMed] [Google Scholar]
- Bijari N.; Ghobadi S.; Derakhshandeh K. β-lactoglobulin-irinotecan inclusion complex as a new targeted nanocarrier for colorectal cancer cells. Res. Pharm. Sci. 2019, 14, 216. 10.4103/1735-5362.258488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijari N.; Ghobadi S.; Derakhshandeh K. Irinotecan binds to the internal cavity of beta-lactoglobulin: A multi-spectroscopic and computational investigation. J. Pharm. Biomed. Anal. 2017, 139, 109–115. 10.1016/j.jpba.2017.02.050. [DOI] [PubMed] [Google Scholar]
- Khan S.; Ipsen R.; Almdal K.; Svensson B.; Harris P. Revealing the Dimeric Crystal and Solution Structure of β-Lactoglobulin at pH 4 and Its pH and Salt Dependent Monomer-Dimer Equilibrium. Biomacromolecules 2018, 19, 2905–2912. 10.1021/acs.biomac.8b00471. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee C.; Saha S.; Biswas A.; Kundu M.; Ghosh L.; Das K. P. Structural Changes of ?-Lactoglobulin during Thermal Unfolding and Refolding ? An FT-IR and Circular Dichroism Study. Protein J. 2005, 24, 27–35. 10.1007/s10930-004-0603-z. [DOI] [PubMed] [Google Scholar]
- Abascal D.; Lencki R. W. Effect of pH on the Ternary Solution Behavior of β-Lactoglobulin. Biotechnol. Prog. 2004, 20, 1741–1748. 10.1021/bp0499350. [DOI] [PubMed] [Google Scholar]
- Brownlow S.; Cabral J. H. M.; Cooper R.; Flower D. R.; Yewdall S. J.; Polikarpov I.; North A. C.; Sawyer L. Bovine β-lactoglobulin at 1.8 Å resolution - still an enigmatic lipocalin. Structure 1997, 5, 481–495. 10.1016/s0969-2126(97)00205-0. [DOI] [PubMed] [Google Scholar]
- Gottschalk M.; Nilsson H.; Roos H.; Halle B. Protein self-association in solution: The bovine β -lactoglobulin dimer and octamer. Protein Sci. 2009, 12, 2404–2411. 10.1110/ps.0305903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercadante D.; Melton L. D.; Norris G. E.; Loo T. S.; Williams M. A. K.; Dobson R. C. J.; Jameson G. B. Bovine β-Lactoglobulin Is Dimeric Under Imitative Physiological Conditions: Dissociation Equilibrium and Rate Constants over the pH Range of 2.5-7.5. Biophys. J. 2012, 103, 303–312. 10.1016/j.bpj.2012.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson G. B.; Adams J. J.; Creamer L. K. Flexibility, functionality and hydrophobicity of bovine β-lactoglobulin. Int. Dairy J. 2002, 12, 319–329. 10.1016/s0958-6946(02)00028-6. [DOI] [Google Scholar]
- Qin B. Y.; Bewley M. C.; Creamer L. K.; Baker H. M.; Baker E. N.; Jameson G. B. Structural Basis of the Tanford Transition of Bovine β-Lactoglobulin. Biochemistry 1998, 37, 14014–14023. 10.1021/bi981016t. [DOI] [PubMed] [Google Scholar]
- Tanford C.; Bunville L. G.; Nozaki Y. The Reversible Transformation of β-Lactoglobulin at pH 7.5. J. Am. Chem. Soc. 1959, 81, 4032–4036. 10.1021/ja01524a054. [DOI] [Google Scholar]
- Kontopidis G.; Holt C.; Sawyer L. Invited Review: β-Lactoglobulin: Binding Properties, Structure, and Function. J. Dairy Sci. 2004, 87, 785–796. 10.3168/jds.s0022-0302(04)73222-1. [DOI] [PubMed] [Google Scholar]
- Yang M.-C.; Guan H.-H.; Liu M.-Y.; Lin Y.-H.; Yang J.-M.; Chen W.-L.; Chen C.-J.; Mao S. J. T. Crystal Structure of a Secondary Vitamin D3 Binding Site of Milk β-Lactoglobulin. Proteins Struct. Funct. Genet. 2008, 71, 1197–1210. 10.1002/prot.21811. [DOI] [PubMed] [Google Scholar]
- Konuma T.; Sakurai K.; Goto Y. Promiscuous Binding of Ligands by β-Lactoglobulin Involves Hydrophobic Interactions and Plasticity. J. Mol. Biol. 2007, 368, 209–218. 10.1016/j.jmb.2007.01.077. [DOI] [PubMed] [Google Scholar]
- Monaco H. L.; Zanotti G.; Spadon P.; Bolognesi M.; Sawyer L.; Eliopoulos E. E. Crystal Structure of the Trigonal Form of Bovine β-Lactoglobulin and of Its Complex with Retinol at 2.5 Å Resolution. J. Mol. Biol. 1987, 197, 695–706. 10.1016/0022-2836(87)90476-1. [DOI] [PubMed] [Google Scholar]
- Dufour E.; Genot C.; Haertlé T. β-Lactoglobulin Binding Properties during Its Folding Changes Studied by Fluorescence Spectroscopy. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1994, 1205, 105–112. 10.1016/0167-4838(94)90098-1. [DOI] [PubMed] [Google Scholar]
- Tromelin A.; Guichard E. Interaction between Flavour Compounds and β-Lactoglobulin: Approach by NMR and 2D/3D-QSAR Studies of Ligands. Flavour Fragrance J. 2005, 21, 13–24. 10.1002/ffj.1696. [DOI] [Google Scholar]
- Guichard E. Interactions between β-Lactoglobulin and Flavour Compounds. Food Chem. 2000, 71, 301–308. 10.1016/s0308-8146(00)00181-3. [DOI] [Google Scholar]
- Lübke M.; Guichard E.; Tromelin A.; Le Quéré J. L. Nuclear Magnetic Resonance Spectroscopic Study of β-Lactoglobulin Interactions with Two Flavor Compounds, γ-Decalactone and β-Ionone. J. Agric. Food Chem. 2002, 50, 7094–7099. 10.1021/jf020513k. [DOI] [PubMed] [Google Scholar]
- Ci S. X.; Huynh T. H.; Louie L. W.; Yang A.; Beals B. J.; Ron N.; Tsang W.-G.; Soon-Shiong P.; Desai N. P. Molecular Mass Distribution of Sodium Alginate by High-Performance Size-Exclusion Chromatography. J. Chromatogr. A 1999, 864, 199–210. 10.1016/s0021-9673(99)01029-8. [DOI] [PubMed] [Google Scholar]
- Johnson F. A.; Craig D. Q. M.; Mercer A. D.; Chauhan S. The Effects of Alginate Molecular Structure and Formulation Variables on the Physical Characteristics of Alginate Raft Systems. Int. J. Pharm. 1997, 159, 35–42. 10.1016/s0378-5173(97)00266-4. [DOI] [Google Scholar]
- Girard M.; Turgeon S. L.; Gauthier S. F. Quantification of the Interactions between β-Lactoglobulin and Pectin through Capillary Electrophoresis Analysis. J. Agric. Food Chem. 2003, 51, 6043–6049. 10.1021/jf034266b. [DOI] [PubMed] [Google Scholar]
- Jones O. G.; Adamcik J.; Handschin S.; Bolisetty S.; Mezzenga R. Fibrillation of β-Lactoglobulin at Low pH in the Presence of a Complexing Anionic Polysaccharide. Langmuir 2010, 26, 17449–17458. 10.1021/la1026619. [DOI] [PubMed] [Google Scholar]
- Hosseini S. M. H.; Emam-Djomeh Z.; Razavi S. H.; Moosavi-Movahedi A. A.; Saboury A. A.; Atri M. S.; Van der Meeren P. β-Lactoglobulin-sodium alginate interaction as affected by polysaccharide depolymerization using high intensity ultrasound. Food Hydrocolloids 2013, 32, 235–244. 10.1016/j.foodhyd.2013.01.002. [DOI] [Google Scholar]
- Girard M.; Schaffer-Lequart C. Attractive Interactions between Selected Anionic Exopolysaccharides and Milk Proteins. Food Hydrocolloids 2008, 22, 1425–1434. 10.1016/j.foodhyd.2007.09.001. [DOI] [Google Scholar]
- Gentès M.-C.; St-Gelais D.; Turgeon S. L. Exopolysaccharide-Milk Protein Interactions in a Dairy Model System Simulating Yoghurt Conditions. Dairy Sci. Technol. 2013, 93, 255–271. 10.1007/s13594-013-0121-x. [DOI] [Google Scholar]
- Birch J.; Hardarson H. K.; Khan S.; Van Calsteren M.-R.; Ipsen R.; Garrigues C.; Almdal K.; Hachem M. A.; Svensson B. Effect of Repeat Unit Structure and Molecular Mass of Lactic Acid Bacteria Hetero-Exopolysaccharides on Binding to Milk Proteins. Carbohydr. Polym. 2017, 177, 406–414. 10.1016/j.carbpol.2017.08.055. [DOI] [PubMed] [Google Scholar]
- Mozzi F.; Gerbino E.; Font De Valdez G.; Torino M. I. Functionality of exopolysaccharides produced by lactic acid bacteria in anin vitrogastric system. J. Appl. Microbiol. 2009, 107, 56–64. 10.1111/j.1365-2672.2009.04182.x. [DOI] [PubMed] [Google Scholar]
- Torino M. I.; Font de Valdez G.; Mozzi F.. Biopolymers from Lactic Acid Bacteria. Novel Applications in Foods and Beverages. Front. Microbiol. 2015, 6. DOI: 10.3389/fmicb.2015.00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwig G. J.Structural Analysis of Exopolysaccharides from Lactic Acid Bacteria. In Lactic Acid Bacteria: Methods and Protocols; Kanauchi M., Ed.; Springer New York: New York, NY, 2019; Vol. 1887; pp 67–84. [DOI] [PubMed] [Google Scholar]
- Górska-Frączek S.; Sandström C.; Kenne L.; Rybka J.; Strus M.; Heczko P.; Gamian A. Structural Studies of the Exopolysaccharide Consisting of a Nonasaccharide Repeating Unit Isolated from Lactobacillus Rhamnosus KL37B. Carbohydr. Res. 2011, 346, 2926–2932. 10.1016/j.carres.2011.10.024. [DOI] [PubMed] [Google Scholar]
- Birch J.; Van Calsteren M.-R.; Pérez S.; Svensson B. The Exopolysaccharide Properties and Structures Database: EPS-DB. Application to Bacterial Exopolysaccharides. Carbohydr. Polym. 2019, 205, 565–570. 10.1016/j.carbpol.2018.10.063. [DOI] [PubMed] [Google Scholar]
- Marshall V. M.; Rawson H. L. Effects of exopolysaccharide-producing strains of thermophilic lactic acid bacteria on the texture of stirred yoghurt. Int. J. Food Sci. Technol. 1999, 34, 137–143. 10.1046/j.1365-2621.1999.00245.x. [DOI] [Google Scholar]
- Han X.; Yang Z.; Jing X.; Yu P.; Zhang Y.; Yi H.; Zhang L. Improvement of the Texture of Yogurt by Use of Exopolysaccharide Producing Lactic Acid Bacteria. BioMed Res. Int. 2016, 2016, 7945675. 10.1155/2016/7945675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babol L. N.; Svensson B.; Ipsen R. Using Surface Plasmon Resonance Technology to Screen Interactions Between Exopolysaccharides and Milk Proteins. Food Biophys. 2011, 6, 468–473. 10.1007/s11483-011-9227-y. [DOI] [Google Scholar]
- Diemer S. K.; Svensson B.; Babol L. N.; Cockburn D.; Grijpstra P.; Dijkhuizen L.; Folkenberg D. M.; Garrigues C.; Ipsen R. H. Binding Interactions Between α-glucans from Lactobacillus reuteri and Milk Proteins Characterised by Surface Plasmon Resonance. Food Biophys. 2012, 7, 220–226. 10.1007/s11483-012-9260-5. [DOI] [Google Scholar]
- Khan S.; Birch J.; Van Calsteren M.-R.; Ipsen R.; Peters G. H. J.; Svensson B.; Harris P.; Almdal K. Interaction between structurally different heteroexopolysaccharides and β-lactoglobulin studied by solution scattering and analytical ultracentrifugation. Int. J. Biol. Macromol. 2018, 111, 746–754. 10.1016/j.ijbiomac.2018.01.050. [DOI] [PubMed] [Google Scholar]
- Degeest B.; Vaningelgem F.; Laws A. P.; De Vuyst L. UDP-N-Acetylglucosamine 4-Epimerase Activity Indicates the Presence of N-Acetylgalactosamine in Exopolysaccharides of Streptococcus Thermophilus Strains. Appl. Environ. Microbiol. 2001, 67, 3976–3984. 10.1128/aem.67.9.3976-3984.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber E. J.; Kamerling J. P.; Vliegenthart J. F. G. Structure of the Extracellular Polysaccharide Produced by Lactobacillus Delbrueckii Subsp. Bulgaricus 291. Carbohydr. Res. 2001, 331, 183–194. 10.1016/s0008-6215(01)00012-x. [DOI] [PubMed] [Google Scholar]
- Van Calsteren M.-R.; Gagnon F.; Calzas C.; Goyette-Desjardins G.; Okura M.; Takamatsu D.; Gottschalk M.; Segura M. Structure determination ofStreptococcus suisserotype 14 capsular polysaccharide. Biochem. Cell Biol. 2013, 91, 49–58. 10.1139/bcb-2012-0036. [DOI] [PubMed] [Google Scholar]
- Van Calsteren M.-R.; Gagnon F.; Lacouture S.; Fittipaldi N.; Gottschalk M. Structure determination ofStreptococcus suisserotype 2 capsular polysaccharide. Biochem. Cell Biol. 2010, 88, 513–525. 10.1139/O09-170. [DOI] [PubMed] [Google Scholar]
- Gerwig G. J.; Dobruchowska J. M.; Shi T.; Urashima T.; Fukuda K.; Kamerling J. P. Structure Determination of the Exopolysaccharide of Lactobacillus Fermentum TDS030603-A Revision. Carbohydr. Res. 2013, 378, 84–90. 10.1016/j.carres.2013.04.026. [DOI] [PubMed] [Google Scholar]
- Taulier N.; Chalikian T. V. Characterization of pH-induced transitions of β-lactoglobulin: ultrasonic, densimetric, and spectroscopic studies 1 1Edited by C. R. Matthews. J. Mol. Biol. 2001, 314, 873–889. 10.1006/jmbi.2001.5188. [DOI] [PubMed] [Google Scholar]
- Stender E. G. P.; Birch J.; Kjeldsen C.; Nielsen L. D.; Duus J. Ø.; Kragelund B. B.; Svensson B. Alginate Trisaccharide Binding Sites on the Surface of β-Lactoglobulin Identified by NMR Spectroscopy: Implications for Molecular Network Formation. ACS Omega 2019, 4, 6165–6174. 10.1021/acsomega.8b03532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M. H. M.; Søndergaard C. R.; Rostkowski M.; Jensen J. H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical PKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- Søndergaard C. R.; Olsson M. H. M.; Rostkowski M.; Jensen J. H. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values. J. Chem. Theory Comput. 2011, 7, 2284–2295. 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- Dubois M.; Gilles K. A.; Hamilton J. K.; Rebers P. A.; Smith F. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956, 28, 350–356. 10.1021/ac60111a017. [DOI] [Google Scholar]
- Lemoine J.; Chirat F.; Wieruszeski J. M.; Strecker G.; Favre N.; Neeser J. R. Structural Characterization of the Exocellular Polysaccharides Produced by Streptococcus Thermophilus SFi39 and SFi12. Appl. Environ. Microbiol. 1997, 63, 3512–3518. 10.1128/aem.63.9.3512-3518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen K. R.; Otte J.; Ipsen R.; Qvist K. B. Large-scale Preparation of β-Lactoglobulin A and B by Ultrafiltration and Ion-exchange Chromatography. Int. Dairy J. 1998, 8, 113–118. 10.1016/s0958-6946(98)00028-4. [DOI] [Google Scholar]
- Gasteiger E.; Hoogland C.; Gattiker A.; Duvaud S.; Wilkins M. R.; Appel R. D.; Bairoch A.. Protein Identification and Analysis Tools on the ExPASy Server. The Proteomics Protocols Handbook; Humana Press, 2005; pp 571–607. [Google Scholar]
- Delaglio F.; Grzesiek S.; Vuister G.; Zhu G.; Pfeifer J.; Bax A. NMRPipe: A Multidimensional Spectral Processing System Based on UNIX Pipes. J. Biomol. NMR 1995, 6, 277. 10.1007/bf00197809. [DOI] [PubMed] [Google Scholar]
- Teilum K.; Kunze M. B. A.; Erlendsson S.; Kragelund B. B. (S)Pinning down protein interactions by NMR. Protein Sci. 2017, 26, 436–451. 10.1002/pro.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods G.Carbohydrate builder. http://glycam.org.
- Madhavi Sastry G.; Adzhigirey M.; Day T.; Annabhimoju R.; Sherman W. Protein and Ligand Preparation: Parameters, Protocols, and Influence on Virtual Screening Enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- Greenwood J. R.; Calkins D.; Sullivan A. P.; Shelley J. C. Towards the Comprehensive, Rapid, and Accurate Prediction of the Favorable Tautomeric States of Drug-like Molecules in Aqueous Solution. J. Comput. Aided Mol. Des. 2010, 24, 591–604. 10.1007/s10822-010-9349-1. [DOI] [PubMed] [Google Scholar]
- Halgren T. A.; Murphy R. B.; Friesner R. A.; Beard H. S.; Frye L. L.; Pollard W. T.; Banks J. L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Friesner R. A.; Banks J. L.; Murphy R. B.; Halgren T. A.; Klicic J. J.; Mainz D. T.; Repasky M. P.; Knoll E. H.; Shelley M.; Perry J. K.; Shaw D. E.; Francis P.; Shenkin P. S. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Friesner R. A.; Murphy R. B.; Repasky M. P.; Frye L. L.; Greenwood J. R.; Halgren T. A.; Sanschagrin P. C.; Mainz D. T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein–Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Sherman W.; Day T.; Jacobson M. P.; Friesner R. A.; Farid R. Novel Procedure for Modeling Ligand/Receptor Induced Fit Effects. J. Med. Chem. 2006, 49, 534–553. 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- Harder E.; Damm W.; Maple J.; Wu C.; Reboul M.; Xiang J. Y.; Wang L.; Lupyan D.; Dahlgren M. K.; Knight J. L.; Kaus J. W.; Cerutti D. S.; Krilov G.; Jorgensen W. L.; Abel R.; Friesner R. A. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.