Abstract
Diffuse gliomas progress by invading neighboring brain tissue to promote postoperative relapse. Transcription factor SOX2 is highly expressed in invasive gliomas and maps to chromosome region 3q26 together with the genes for PI3K/AKT signaling activator PIK3CA and effector molecules of mitochondria fusion and cell invasion, MFN1 and OPA1. Gene copy number analysis at 3q26 from 129 glioma patient biopsies revealed mutually exclusive SOX2 amplifications (26%) and OPA1 losses (19%). Both forced SOX2 expression and OPA1 inactivation increased LN319 glioma cell invasion in vitro and promoted cell dispersion in vivo in xenotransplanted D. rerio embryos. While PI3 kinase activity sustained SOX2 expression, pharmacological PI3K/AKT pathway inhibition decreased invasion and resulted in SOX2 nucleus‐to‐cytoplasm translocation in an mTORC1‐independent manner. Chromatin immunoprecipitation and luciferase reporter gene assays together demonstrated that SOX2 trans‐activates PIK3CA and OPA1. Thus, SOX2 activates PI3K/AKT signaling in a positive feedback loop, while OPA1 deletion is interpreted to counteract OPA1 trans‐activation. Remarkably, neuroimaging of human gliomas with high SOX2 or low OPA1 genomic imbalances revealed significantly larger necrotic tumor zone volumes, corresponding to higher invasive capacities of tumors, while autologous necrotic cells are capable of inducing higher invasion in SOX2 overexpressing or OPA1 knocked‐down relative to parental LN319. We thus propose necrosis volume as a surrogate marker for the assessment of glioma invasive potential. Whereas glioma invasion is activated by a PI3K/AKT‐SOX2 loop, it is reduced by a cryptic invasion suppressor SOX2‐OPA1 pathway. Thus, PI3K/AKT‐SOX2 and mitochondria fission represent connected signaling networks regulating glioma invasion.
Keywords: 3q26, cell invasion, Glioma, neuroimaging, tumor necrosis
Introduction
Diffuse gliomas are among the most aggressive human tumors. Glioblastoma (GB), the most malignant form of glioma either develops de novo, or from the progression of lower grade glioma (LGG). Currently, surgical resection and chemotherapy remain the standard therapies, although they are not curative, and have limited prognostic impact 22, 31. Unlike tumors of other tissue lineages, which are prone to metastasize into remote organs, diffuse gliomas progress by tumor cell invasion into adjacent healthy brain regions, which almost invariably gives rise to tumor recurrence. Thus, identification of molecular mechanisms underlying glioma cell invasion is an important goal toward improving glioma patient survival.
Glioma cells detach from the bulk of the tumor mass by losing adhesion, gaining motility and progress by interaction with extracellular matrix (ECM) and surrounding tissue. Migratory routes are either perivascular, or within the brain parenchyma 7, 12, 27. Among the known molecular mechanisms that regulate effectors of glioma invasion, TGF‐ß activates expression of matrix metalloproteinase‐2 (MMP‐2), as well as of the transcription factors Snail, Twist and ZEB family members. As a consequence, glioma cells undergo an epithelial‐to‐mesenchymal transition (EMT), a precursor state of invasive behavior 20, 36, 41. The PI3K/AKT pathway, activated by receptor tyrosine kinase signaling or PTEN loss, increases the activity of MMP‐2 and MMP‐9, while Notch signaling trans‐activates the ECM protein tenascin‐C 32. Further adding to these aspects, we find here SOX2 transcripts associated with higher invasion in LGG (data shown in Supplementary Figure S1). SOX2 is known to maintain glioma cell stemness, for driving EMT in many solid tumors 14, 21, 23, 26, 42 and for inducing cell invasion 2, 3, 13, 35. SOX2 is induced by TGF‐ß via SOX4 and activates the Notch pathway to mediate tenascin‐dependent glioma cell invasion 16, 32, 43.
SOX2 maps to chromosome band 3q26, which is known for frequent genetic imbalances in gliomas 25. Not surprisingly, this region contains several genes with potential or proven impact on tumor development (Figure 1A). Among them PIK3CA, which encodes p110α, the catalytic subunit of the predominant form of PI3 kinase, and thus is a key constituent of the canonical PI3K/AKT signaling axis. PIK3CA carries gain‐of‐function missense mutations in about 10% of GB, at the same hotspots as observed in other cancers 29. For example, in mammary carcinoma, where activating PIK3CA mutations are recurrent hits and the PI3K/AKT signaling pathway shown to sustain SOX2 nuclear expression, more specifically 25, 30, 34. Further relevant genes encoded on 3q26 comprise OPA1 and MFN1. Germline mutations in the OPA1 (optical atrophy 1) gene are responsible for autosomal dominant optic atrophy. OPA1, together with the mitofusin‐1 gene MFN1, are both involved in mitochondrial fusion 9, 33. Of interest, mitochondrial fission has been associated with cell polarization and an increased cell motility of lymphocytes as well as breast and thyroid cancer cells 4, 10, 11. Therefore, given the vicinity of these genes in the human genome, we hypothesized that they could interact with SOX2 to result in enhanced cell invasion.
Figure 1.
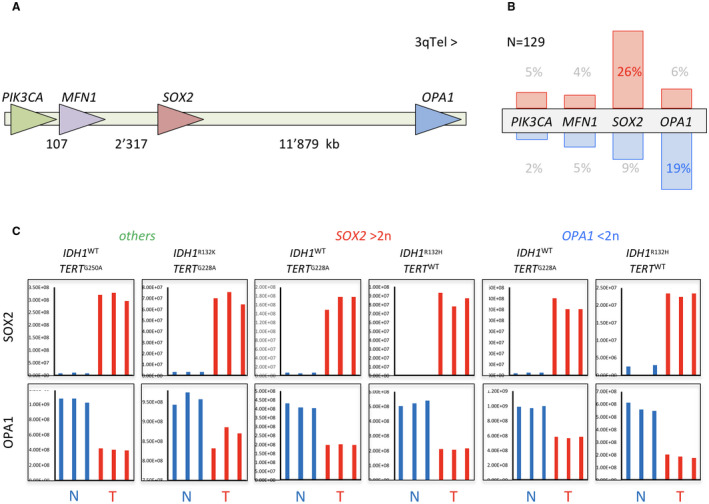
3q26 genes SOX2 and OPA1 are targeted in glioma. A. Schematic illustration of chromosome segment 3q26 with indication of relative position and orientation of PIK3CA, MFN1, SOX2 and OPA1 genes. B. Frequency of genomic amplification (red) and deletion (blue) of 3q26 genes as determined by quantitative PCR of DNA from 129 glioma biopsies. C. Protein levels of SOX2 and OPA1 determined by mass spectrometry in six representative glioma biopsies (T) compared with corresponding non‐tumorous white matter (N). The glioma biopsies were genotyped to be either IDH wild type or mutant, or carrying SOX2 amplification (SOX2 > 2n) or OPA1 deletion (OPA1 < 2n). “Others” stands for glioma with neither SOX2 amplification nor OPA1 deletion.
Neuroimaging parameters allow detection and measurement of distinct compartments of tumor anatomy of potential diagnostic or prognostic value 38. T2w‐hyperintense non‐enhancing domains represent the tumor parts of lower grade with intact blood‐brain barrier (BBB) 19, 44. Breakdown of BBB, an indicator for malignancy, allows contrast uptake and gives brightness to contrast‐enhanced T1‐weighted images. Measurement of contrast‐enhancing tumor by response assessment to neuro‐oncology (RANO) expresses—as one part of this assessment—the largest area of tumor extension in axial direction 39. The necrosis compartment, usually embedded in the enhancing volume, can be triggered by tumor necrosis factor secreted by glioma cells 28. Necrosis is a hallmark of GB, a survival marker and inducer of glioma cell invasion 1, 15, 44. As exemplified in Figure 6A, these three compartments represent altogether the neuroradiologically visible total volume of the tumor with invading glioma cells spreading far beyond the visible boundaries.
Figure 6.
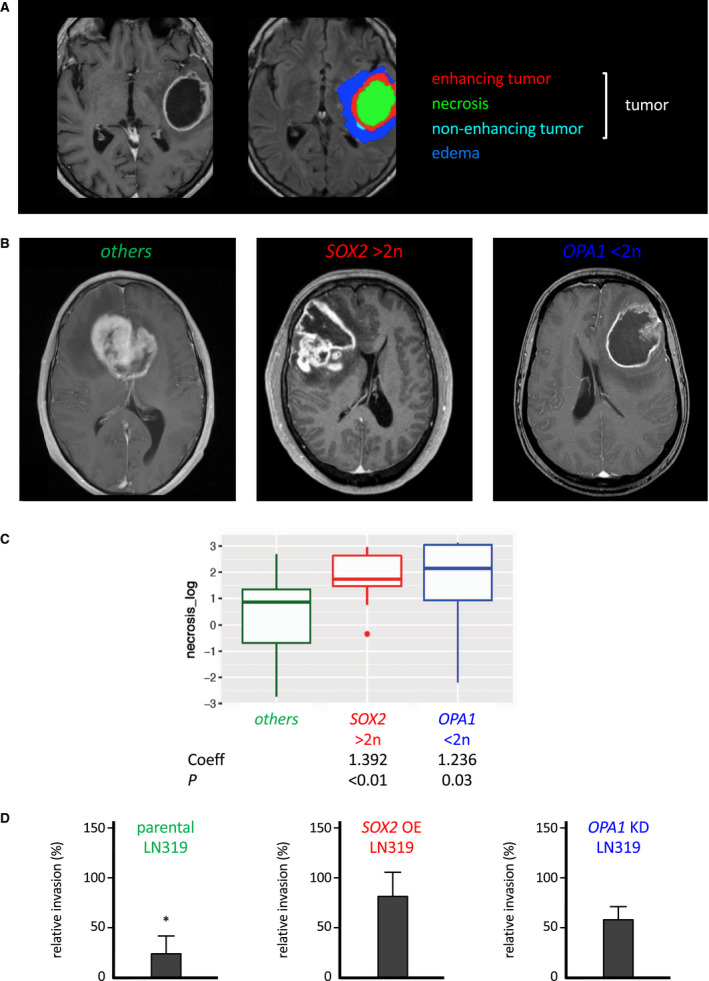
SOX2 amplification and OPA1 deletion are associated with larger necrotic volumes in the glioma mass. A. Neuroimaging (left) and color modelization (right) of the tumor compartments of the same glioma. B. Neuroimaging of representative glioma cases with neither SOX2 amplification nor OPA1 deletion (left) with SOX2 amplification (center) and OPA1 deletion (right). C. Boxplots visualizing the distribution of the log‐transformed volume of necrotic compartment of gliomas across different statuses of SOX2 and OPA1. The boxes display the median and the interquartile range. Coefficients estimates for a linear model with log‐transformed necrosis volume and adjusted for age and ratio of postoperative to preoperative volume are shown below the boxplots. D. Comparative invasion kinetics of LN319 cells in the presence of autologous necrotic LN319 cells. Parental LN319 (left), LN319 overexpressing cherry‐SOX2 under the control of 30 ng/ml doxycycline (middle) and L319 with OPA1 knocked down by shRNA (right). Invasiveness in absence of necrotic cells was set to 100% (reference score). Significance cutoff: *P < 0.05.
We here describe that molecular interactions between 3q26 gene products SOX2, PIK3CA and OPA1 combined with respective gene copy imbalances determine glioma invasion capacity in cell‐based in vitro assays and xenotransplant in vivo studies alike. These molecular findings meet a phenotypic manifestation in human gliomas, whose neuro radiologic examination revealed that tumors with SOX2 amplification or OPA1 deletion had larger volumes of tumor necrosis, an established inducer of invasion.
Materials and Methods
Cell culture, lentiviral transduction and plasmid transfection
Human glioma LN319 17 and human embryonic kidney HEK293 cells (ATCC #CRL‐1573) were grown in Dulbecco’s modified Eagle’s medium (DMEM, Sigma‐Aldrich, #D6429) supplemented with 5% heat‐inactivated fetal bovine serum (Sigma‐Aldrich, #F7524) and 1% Pen/Strep (Sigma‐Aldrich, #P4333). Short‐term ex vivo cell cultures derived from dissociated patient tumor tissue were grown in DMEM supplemented with 30mM Hepes, 10% fetal bovine serum and 1% Pen/Strep.
A mCherry color reporter was N‐terminally fused to human SOX2 cDNA, inserted into a pLVX‐puro expression vector (Clontech, Mountain View, CA, USA) and stably integrated into LN319 cells by lentiviral transduction. Expression of the corresponding fusion protein was driven from a doxycycline‐dependent (D9891, Sigma, St‐Louis, MO, USA) TetON gene induction system as described 30. Integrants were positively selected first by puromycin resistance and subsequently by FACS. PIK3CA WT , PIK3CA H1047R and PIK3CA E545K constructs encoding HA‐tagged variants of the catalytic subunit of PI3 kinase alpha p110α were PCR‐amplified from Addgene plasmids #12522, #12524, and #12525, BamHI‐inserted into pLVX‐puro, and likewise lentivirally integrated into LN319 and expressed in doxycycline‐inducible manner. Lentiviruses expressing shRNA against OPA1 and scrambled control (Santa Cruz Biotechnology #sc‐106808‐V, #sc‐108080‐V Dallas, TX, USA) were transduced into LN319 cells according to the manufacturer’s instructions and positively selected with puromycin (Sigma‐Aldrich, #P8833). HEK293 cells were transfected using reagent JET PEI™ (Polypus‐transfection #101‐01N; ArchiMed, Lyon, France).
Nucleic acid extraction and analysis
RNA from cell cultures was extracted using Trizol (Thermo Fischer Scientific #15596‐026) and RNeasy Mini Kit (Qiagen #74104; Hombrechtikon, Switzerland). Reverse transcription was performed with iScript cDNA Synthesis Kit (Bio‐Rad #170‐8890). Quantitative PCR was performed with Ssofast Evagreen supermix (Bio‐Rad #172‐5200) using the CFX90 thermocycler (Bio‐Rad, Hercules, CA, USA). Samples were normalized with GAPDH. Primers for cDNAs were purchased from Qiagen, and where PIK3CA: QT000114861, SOX2: QT00237601, MFN1: QT00077966, OPA1: QT00085519 and GAPDH: QT00079247. Genomic DNA from glioma biopsies was extracted using DNeasy Blood & Tissue Kit (Qiagen, Cat # 69504). Forward (F) and reverse (R) primers for copy number variation studies were: PIK3CA (F: gag aaa atg aaa gct cac tct gg and R: gtg cta tca aac cct gtt tgc, SOX2 (F: tga gag aga tcc tgg act tc and R: gca aac ttc ctg caa agc tc), MFN1 (F: gcc tcc tga aga aca aca gc and R: agg aac gct ttc cta gtg ca), OPA1 (F: cac tga ttt cct tga gtt ctc a and R: act gtt tgg ggt tga tat ttg g), IDH1 (F: acc aaa tgg cac cat acg a and R: ttc ata cct tgc tta atg ggt gt).
Western blotting and immunodetection
Cells were disrupted in 1x lysis buffer (#9803, Cell Signaling, Danvers, MA, USA) supplemented with protease and phosphatase inhibitors (#78442, Thermo Fisher Scientific, Waltham, MA, USA). Total cell lysates were cleared at 4°C for 10 minutes at 15 000 rpm and supplemented with Laemmli buffer. Alternatively (Figure 2B), cell lysates were extracted using the NE‐PER kit (Thermo Fischer Scientific #78833).
Figure 2.
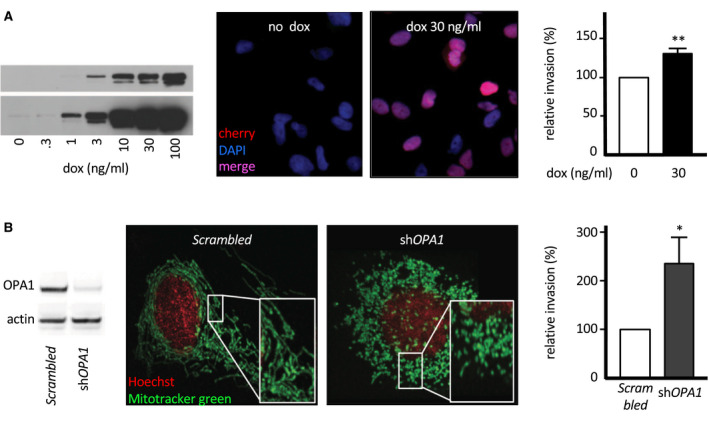
SOX2 and OPA1 modulate glioma cell invasion in vitro and in vivo. A. Inducible SOX2 overexpression in LN319 cells. LN319 stably transfected with an mCherry‐SOX2 fusion construct under control of a doxycycline‐inducible promoter. Western blot documenting dose‐dependent induction of the fusion protein (anti‐SOX2 staining, two exposure times, left). Fluorescence microscopy verifying effective nuclear localization of mCherry‐SOX2 24 h past induction (middle). SOX2‐dependent modulation of cell invasiveness as investigated by 24 h Boyden chamber assay (right). B. shRNA‐driven OPA1 knock‐down in LN319 cells. Western blot of transfected LN319 cells (left). Verification of knock‐down phenotype by mitochondria morphology in transfected LN319 (middle). Comparative cell invasion in response by 24 h Boyden chamber assay (right).
Proteins were separated on Novex PAGE gels (Invitrogen, Carlsbad, CA, USA) and transferred onto PVDF membrane (#10600021, Amersham, GE Healthcare Life Sciences, Chalfont St. Giles, UK) in a semi‐dry blotting apparatus (Trans‐Blot Turbo, BioRad). Membranes were blocked with 10% w/v nonfat dry milk (#9999S, Cell Signaling) diluted in TBS containing 0.1% Tween‐20 (P1379, Sigma). Proteins were decorated with the following primary antibodies: anti PI3 kinase p110α (Cell Signaling Technology (CST), #4255), pAKT(Ser473) (CST, #4060), AKT (CST, #4691), SOX2 (CST, #3579), pRPS6(Ser235,Ser236) (CST, #4856), actin (CST, #3700) and detected by ECL reaction.
For immunofluorescence, cells were fixed with 4% formaldehyde for 10 minutes followed by permeabilization with 0.2% Tween in blocking buffer (PBS + 5% normal serum). Cells were incubated overnight with anti‐SOX2 (Millipore #MAB4343) then with Cy2‐conjugated secondary antibodies (Jackson ImmunoResearch, Cambridgeshire, UK) for 1 h. Cell nuclei were stained with 0.25 µg/ml DAPI (4’,6’‐diamidine‐2‐phenylindole, Sigma) added to the secondary antibodies. mCherry‐SOX2 expressing cells were only stained with DAPI. Slides were mounted with FluorSave Reagent (Calbiochem, Sigma‐Aldrich Chemie GmbH Buchs, Switzerland) and images taken with a DMRE microscope (Leica Microsystems, Wetzlar, Germany) combined with an F‐view camera (Soft Imaging System GmbH, Muenster, Germany). For mitochondria staining, cells were seeded on borosilicate 1.5‐mm glass chambers (Lab‐Tek II 8 well, Thermo Fischer Scientific) and co‐stained with 12 nM mito‐tracker green (Thermo Fischer Scientific #M7514) and 10 nM Hoechst dye (Thermo Fischer Scientific #H1398) for 30 minutes. Mitochondria were imaged with a spinning disk confocal microscope (Perkin Elmer, Waltham, MA, USA).
Invasion assay
Boyden chambers (Cell Biolabs Inc, San Diego, CA, USA, #CBA‐100‐C) were used according to manufacturer’s instructions, with 5 × 104 cells for 24 h, except for BTB251 short‐term cell culture (7 × 102 cells). Invading cells were stained with DAPI and counted over five microscopic fields per well at 10× magnification. Experiments were performed 3 times in triplicate. Generation of necrotic cells for invasion assay in the presence of necrotic cells is based on Ahn et al 1. Necrotic cells were added to the autologous live cells at a ratio of 3:1.
Zebrafish xenografts
Animal experiments and zebrafish husbandry were performed in accordance with the regulations of the veterinary office of the city of Basel (Kantonales Veterinäramt Basel Stadt, Basel, Switzerland). Cell Preparation: Preceding transplantation, LN319 cells were labeled with CellTrackerTM CM‐Dil (C7000, Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. In case of inducible SOX2 overexpression, LN319 TetON mCherry‐SOX2 and the corresponding LN319 TetON control cells were first incubated with Doxycycline in vitro (1 µg/ml for 24 h) and labeled with CellTrackerTM thereafter. Zebrafish Preparation: Wild‐type zebrafish (TU, Tuebingen, “zfin.org”) were maintained, collected and staged in E3 medium at 28.5°C according to standard procedures 40. For xenotransplantation experiments, zebrafish embryos were anesthetized in 0.4% tricaine (Sigma) at 48‐h postfertilization (hpf) and approximately 100 glioma cells microinjected into the vessel‐free area of the yolk sack essentially as described 18. Cell transfer was verified by fluorescence microscopy after a recovery time of 1–2 h and fish harboring red cells shifted to 35°C thereafter. On assay day 5 (72‐h post transplantation) animals were screened for persisting engraftment on a Zeiss Discovery V20 Stereo microscope supplied with an HXP120 fluorescent light source and further analyzed regarding tumor cell status on a Zeiss LSM 710 confocal microscope. Original image data were recorded in Zeiss Zen software and z‐stack projections composed in Fiji (http://imagej.nih.gov) for optimal illustration. Experiments were performed in three independent biological replicates (n = 3) involving zebrafish embryos derived from several independent clutches.
Pharmacology
PI3K/AKT/TOR signaling pathway inhibitors BYL‐719, MK‐2206, AKTi1/2 and rapamycin were all purchased from Selleckchem (Houston, TX, USA), dissolved and diluted according to manufacturer’s instructions, and applied at the indicated concentrations for 48 h. For induction of mCherry‐SOX2, p110α(WT), p110α(H1047R) and p110α(E545K) protein expression, cells were incubated with the indicated concentrations of doxycycline (D9891, Sigma, St‐Louis, MO, USA) or standardly with 1 µg/ml for a minimum of 24 h.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was according to Carey et al 5. Chromatin immunoprecipitation of LN319 (shSOX2 vs. shscrambled) was performed with either anti‐SOX2 (Abcam #59776; Cambridge, UK) or IgG control (antibody Millipore, #12‐170). Resulting DNA extract was subjected to quantitative real‐time PCR of fragments containing the predicted SOX2‐binding sites. Primers are listed in Table S2. Immunoprecipitation levels between LN319 shSOX2 (Santa Cruz Biotechnology sc‐38408‐V) and LN319 shscrambled samples were normalized with Ct values from the IgG control. In each sample, immunoprecipitation with anti‐SOX2 was calculated by subtraction of Ct value from control IgG immunoprecipitation.
Cloning and site‐directed mutagenesis
Genomic fragments of 3q26 gene promoters were amplified from HEK293 cell genomic DNA. PCR products were introduced into pGCL4.1 basic luciferase expression vector and sequenced. Site‐directed mutagenesis was performed on cloned wild‐type 3q26 promoters to destroy potential SOX2‐binding sites. MFN1 promoter was first produced as mutant and wild type was produced by reversing the mutation. Primers are listed in Table S2.
Luciferase assay
HEK cells were transfected with the 3q26 gene promoter‐luciferase reporter gene constructs using reagent JET PEI™ reagent (Polypus‐transfection #101‐01N). SOX2 was overexpressed by addition of an SOX2 expression vector (pMSCV‐Flag‐hSOX2, Addgene #2007). Luciferase activity was normalized with co‐transfection of Renilla expression vector (pRL‐null, Promega, Madison, WI, USA). Lysates were analyzed with the Dual Luciferase kit (Promega #E1980)
Neuro‐imaging
Volumetric imaging data were assessed for 65 GB patients from the most recent preoperative magnetic resonance imaging (MRI) from T2‐ and T1‐weighted (native and contrast enhanced) as well as from Fluid Attenuated Inversion Recovery (FLAIR) images. In every instance, volumes for enhancing tumor, non‐enhancing tumor, necrosis and total tumor were determined. These volumes were calculated using the ABC/2 method and expressed in mL. In addition, the tumor areas were calculated by multiplying the two largest axes, perpendicular to each other, determined from an axial contrast‐enhanced T1‐weighted image. This creates a two‐dimensional plane through the tumor that is expressed in mm2. Volume of recurrence was measured (mL) for tumors with documented recurrence.
Statistics
Summary statistics of baseline demographics characteristics (age and gender) of the patient population are presented overall and separately for patients with and without a suitable MRI, together with summary statistics of the outcome variables corresponding to the primary endpoint (OS events, summaries of time to and surgery type). For continuous variables the number (n) of observations, median and interquartile range (IQR) are reported (Table S5).
The status of the genetic alteration is coded through a three‐level categorical variable with the following genetic categories: (i) SOX2 not amplified and OPA1 not deleted, (ii) SOX2 amplified and OPA1 not deleted and (iii) SOX2 not amplified and OPA1 deleted. To assess the potential relationship between SOX2 amplification or OPA1 deletion and overall survival (OS) a Cox regression was fitted to the time of OS and the status of genomic aberration as factor of interest. The following variables are also included in the model to account for potential confounding: age and ratio of postoperative to preoperative tumor volume (log‐transformed) as a measure of the extent of intervention.
To assess the relationship between alterations of SOX2 and OPA1 on parameters of tumor extension (including total volume, enhancing volume, necrosis volume, tumor area and recurrence volume) linear regression models were fitted to the genetic category as factor of interest. Age and the (log‐transformed) ratio of postoperative to preoperative tumor volume are considered to account for potential confounding.
Independence between genomic alterations in SOX2 and OPA1 was assessed by Fisher exact test. With regards to fish xenotransplant analyses, error bars represent the standard deviation (SD) of three independent biological replicates (n = 3), with a T‐test score of P ≤ 0.05 considered statistically significant (*).
Mass spectrometry
Proteomics analysis of glioma biopsies was according to Dazert et al 8.
Results
Regional imbalances at 3q26
PIK3CA, MFN1, SOX2 and OPA1 all map within 15 megabases at chromosome locus 3q26 (Figure 1A). In order to examine putative genetic imbalances at the gene level, we tested copy number variations of PIK3CA, MFN1, SOX2 and OPA1 in genomic DNAs from 129 glioma biopsies by quantitative PCR. Of the four genes, SOX2 had the highest relative amplification frequency (26%) ranging from two‐ to eightfold absolute amplification, and OPA1 the highest deletion frequency (19%) (Figure 1B). Similarly, data extraction of SOX2 and OPA1 CNV from the TCGA LGG and GB repositories revealed ca 10% amplification and deletion, respectively (Figure S3A). Quantitative comparison of OPA1 CNV to those of PTEN and CDKN2A (frequently subject to hemizygous and homozygous deletion, respectively) suggest OPA1 hemizygous rather than homozygous deletion. This was true for IDH‐wild type (IDH WT) glioma and IDH‐mutant (IDH mut) glioma (Figure S1B left), the prevailing marker used for glioma genetic classification 2. Moreover, we uncovered that SOX2 genomic amplification (SOX2amp) and OPA1 deletion (OPA1del) are mutually exclusive events (P = 0.0044, Figure S1B right). This raised the question whether both events lead to a common oncogenic phenotype. Consistent with an oncogenic relevance of SOX2amp and OPA1del, a comparison of glioma biopsies of various histology to non‐tumor brain tissue on the Rembrandt database (>400 glioma) showed higher SOX2 mRNA levels and lower OPA1 mRNA levels in GB and in LGG (astrocytoma and oligodendroglioma). Moreover, both variations were in addition significantly associated with shorter survival time (Figure S3B). We then tested the relative expression levels of SOX2 and OPA1 proteins in six representative biopsies by mass spectrometry. Regardless of IDH mutation, SOX2amp or OPA1del statuses, SOX2 protein levels were always considerably higher and OPA1 reduced in glioma tissue sample compared to non‐neoplastic white matter control tissues (Figure 1C). The discrepancy between SOX2 and OPA1 copy number variations and protein levels may be attributed to transcriptional and/or posttranslational regulation. Nevertheless, genomic, transcriptomic and protein data all point to SOX2 gain‐of‐function and OPA1 loss‐of‐function in glioma. Taken all results together, we accumulated arguments on the oncogenic relevance of SOX2 gain and OPA1 loss.
Mimicking 3q26 gene imbalances in glioma line LN319
To perform SOX2 gain and OPA1 loss functional investigations in vitro, we initially screened a panel of glioma‐derived cell lines for overt abnormalities in 3q26 gene expression. Out of eight tested cell lines, only two, LN319 and U373, showed robust SOX2 protein expression (Figure S1C, top). Of these, LN319 had nuclear localization of SOX2 (Figure S1C) predicting transcriptionally active SOX2, no aneuploidy at SOX2 and OPA1 genes and no PIK3CA coding sequence mutation, while U373 had mostly cytosolic SOX2 (Figure S1C), likely to be transcriptionally inactive, and triploidy at 3q26 37. For these reasons, we focused our investigations on LN319 as primary study tool. We mimicked SOX2 overexpression in LN319 cells with a stably integrated and inducible mCherry‐SOX2 fusion construct that showed a preferential nuclear localization upon induction (Figure 2A). Conversely, we inactivated OPA1 through stable knock‐down with shRNA, with mitochondria shortening as resulting phenotype (Figure 2B). Both SOX2 overexpression and OPA1 inactivation led to higher relative cell invasion rates (Figure 2A,B, right). In both cases, increased invasion rates were accompanied by reduced proliferation rates (Figure S4). Knock‐down of OPA1 in U373 cells led to a slight but nonsignificant invasion increase, consistent with the predicted low SOX2 transcriptional activity in parental U373 (Figure S1C). Among short‐term cell cultures grown ex vivo from resected human gliomas, only one, BTB251, showed no CNV at 3q26 genes, could be stably transfected for OPA1 knock‐down and propagated for assessment of invasion potential. Invasion rate of BTB251 was increased when OPA1 was knocked‐down, although not to a degree of significance (Figure S1D).
We then referred to xenotransplant experiments to examine the functional significance of our findings under in vivo conditions. Either LN319 OPA1 knock‐down or LN319 SOX2 overexpression cells were microinjected into the yolk sack of zebrafish embryos and the propagation and spreading of the injected specimen analyzed over time (Figure 3A,B, respectively). We found that, in comparison to respective controls, both OPA1 loss and SOX2 gain increased the propensity of cells to leave the site of injection, to spread and invade the yolk sack, and—in a fair number of cases—to even disseminate into distal zones of the fish body, that is, the caudal hematopoietic tissue (CHT) of the tail fin. It is noteworthy that these effects were more pronounced in OPA1 loss than in SOX2 gain cells, which may reflect the intrinsically high endogenous SOX2 level of the LN319 strain background. In any case though, we provide cumulative experimental evidence to suggest that OPA1 loss and SOX2 gain promote invasiveness of glioma cells in vitro and in vivo.
Figure 3.
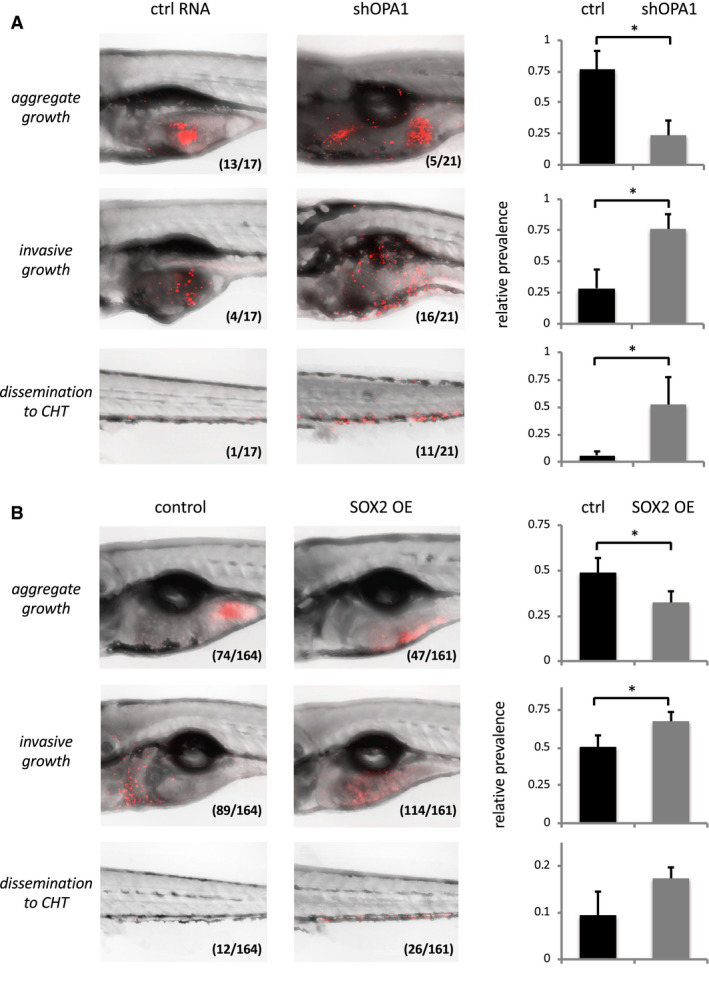
A. OPA1 knock‐down increases invasiveness of LN319 cells in vivo. Left: Confocal pictures of zebrafish embryos effectively xenotransplanted with LN319 control cells (shRNA non‐coding, left) or LN319 shOPA1 knock‐down cells (right). LN319 cells (Cell Tracker, red) either established stable cell masses near the site of injection (aggregate growth, top), dispersed throughout the yolk sack (invasive growth, center) or disseminated to the caudal hematopoietic tissue (CHT) of the tail fin (bottom). Inlays indicate absolute animal numbers per condition. Right: Relative quantification of experimental outcomes documenting aggravated invasiveness and dissemination of OPA1 knock‐down vs. respective control cells. B. SOX2 overexpression (OE) increases invasiveness of LN319 cells in vivo. Experimental setup as above, except that TetON (control, left) and TetON mCherry‐SOX2 (SOX2 OE, right) cells had been pretreated with doxycycline (1 µg/ml for 24 h) to selectively induce SOX2 protein formation prior to transplantation. Right: Assay quantification verifying increased invasiveness and dissemination to CHT (by trend) also for SOX2 OE cells when compared to equally treated controls. Significance cut‐off‐: *P < 0.05.
PI3K/AKT activates SOX2
Overexpression of PIK3CA (alias p110α, the catalytic subunit of PI3 kinase) in LN319 increases phosphorylation of AKT, which leads to an activation of the mammalian target of Rapamycin complex 1 (mTORC1) and further downstream effectors such as ribosomal protein RPS6. More importantly, overexpression of PIK3CA coincided with elevated steady‐state levels of also SOX2, thus underscoring a functional relevance of the PI3K/AKT signaling axis for SOX2 expression (Figure 4A, left). The latter effect was even more pronounced when constitutively active alleles of PIK3CA (H1047R or E545K) were expressed (Figure 4A, right). Based on these findings and the previously described role of PIK3CA on posttranslational SOX2 activation in breast cancer 30, we went on to selectively inactivate PI3K, AKT or mTORC1 with respective inhibitory reagents in LN319 glioma cells (see Figure 4E for schematic illustration of compounds and targets). Indeed, suppression of PI3 kinase activity by BYL‐719 or of its downstream factor AKT by either MK‐2206 or AKTi1/2, as all indicated by drug‐induced loss of pSer437 AKT signal, led to decreased SOX2 levels. In contrast, inhibition of even further downstream mTORC1 by rapamycin, resulted in strong reduction of pRPS6, but did not affect SOX2 protein levels (Figure 4B). Accordingly, PI3K/AKT signaling influenced SOX2 protein turnover independently of mTORC1 (Figure 4B,E). What is more, AKT inhibitor MK‐2206 effectively displaced SOX2 from the nucleus and largely depleted cells of SOX2 (Figure 4C), which correlates once again with largely decreased relative invasion rates (Figure 4D). From these observations, we concluded that PI3K influences the localization and stability of SOX2 protein, thus fostering its activity in glioma cells. These findings further underscore a functional connectivity between products of the 3q26 segment and emphasize PI3K/AKT pathway inhibitors as potential drug candidates in glioma.
Figure 4.
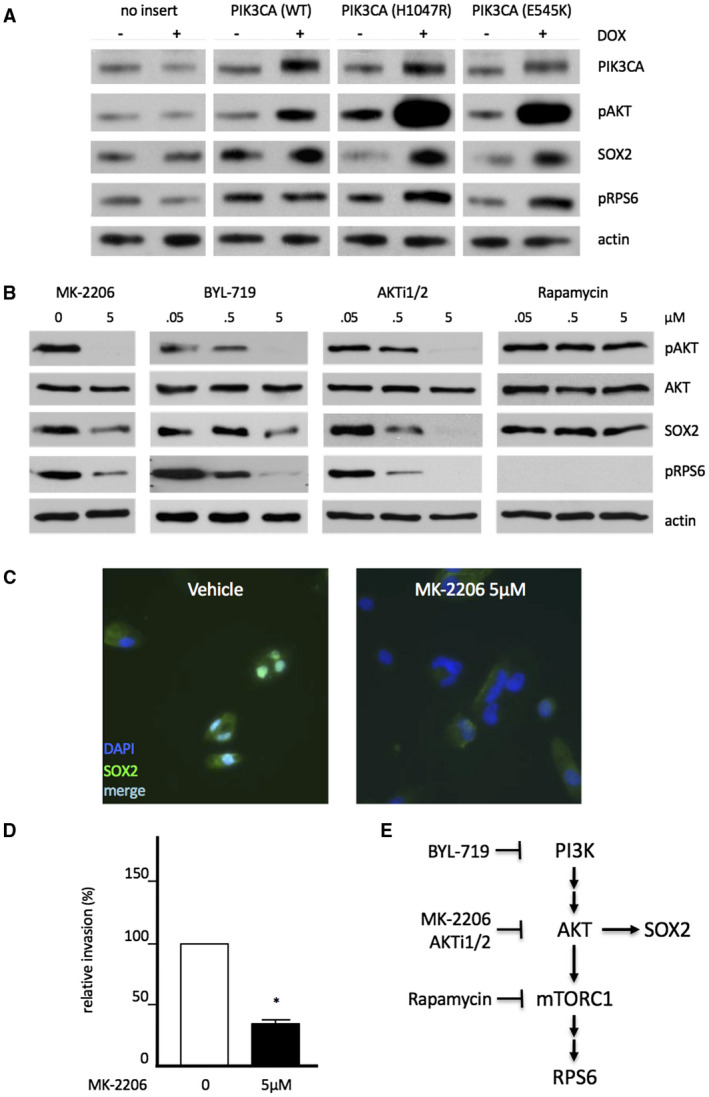
Modulation of SOX2 protein stability by canonical PI3K/AKT signaling. A. Increased PI3K activity fosters SOX2 protein expression in LN319 cells. Cell were stably transfected with Doxycycline‐inducible variants of PIK3CA encoding the catalytic subunit of PI3 kinase p110. From left to right: no insert (control), PIK3CA wild type, and the constitutively active mutant alleles H1047R or E545K. Note elevated SOX2 protein expression in response to aggravated PI3K activity, as indicated by increased pAKT (Ser473) and pS6 (Ser235,236) signatures. Actin staining is shown for loading control. B. Effects of 48 h‐exposure to PI3K/AKT/TOR pathway inhibitors on SOX2 protein levels as assessed by Western blot. SOX2 expression is strongly impaired by pan PI3K‐inhibitor (BYL‐719) and AKT‐inhibitors (AKTi1/2, MK‐2206), but not by further downstream inhibitor rapamycin (anti mTORC1). C. Fluorescence microcopy to document nucleo‐cytoplasmatic displacement and reduction of SOX2 protein expression by PI3K inhibitor MK‐2206. D. Impaired invasiveness of MK‐2206‐treated, SOX2‐depleted LN319 cells as assayed by Boyden chamber experiments. E. Schematic illustration of SOX2 protein expression and turnover in dependence of PI3K/AKT signaling.
SOX2 trans‐activates PIK3CA, OPA1, but not MFN1
Given that SOX2 is a transcriptional modulator, we tested whether 3q26 genes could be regulated by SOX2, which was supported by the presence of potential SOX2‐binding sites upstream of PIK3CA, MFN1 and OPA1 transcription initiation sites (Figure 5A). Physical binding of SOX2 to genomic fragments containing the predicted SOX2‐binding sites was tested by chromatin immunoprecipitation (ChIP) first. We observed significantly higher immunoprecipitation in the presence of anti‐SOX2 compared to isotype control antibodies for the PIK3CA site and for both OPA1 sites (Figure 5B). However, no binding was seen for the MFN1 fragment. To test the potential activation of transcription by SOX2 on 3q26 gene promoters, we constructed plasmids in which the luciferase reporter gene was under the control of PIK3CA, MFN1 and OPA1 promoter fragments containing the potential SOX2‐binding sites. In parallel, we disrupted the predicted potential SOX2‐binding sites by site‐directed mutagenesis. Constructs were transfected into human embryonic kidney HEK293 cells, in the presence or absence of a plasmid expressing additional SOX2. Figure 5C shows an active PIK3CA promoter that responded to additional SOX2, while introduction of mutations into the potential SOX2‐binding site abrogated promoter activity and response to SOX2 (left). Reflecting the presence of even two binding sites, the OPA1 promoter showed an even higher activity and response to SOX2 than the aforementioned PIK3CA. Consistently, introduction of mutations in either site partly diminished promoter activity and responsiveness to SOX2 (right). Instead, we found no experimental evidence for a SOX2 dependent promoter regulation of MFN1 (middle). Altogether, ChIP and luciferase assays consistently showed that PIK3CA and especially OPA1 are target genes of SOX2, whereas MFN1 is not.
Figure 5.
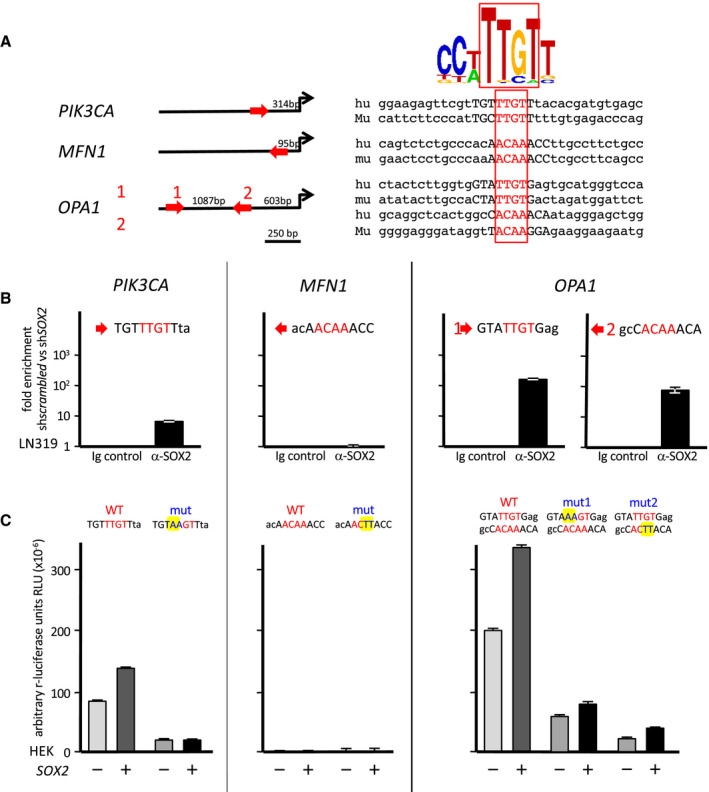
SOX2 trans‐activates 3q26 genes PIK3CA and OPA1 . A. Theoretical SOX2‐binding sequences found upstream of PIK3CA, MFN1 and OPA1 transcription initiation sites. B. ChIP of PIK3CA, MFN1 and OPA1 promoter regions containing the potential SOX2‐binding sites (LN319 cells). C. Luciferase assays of PIK3CA, MFN1 and OPA1 promoter regions containing the potential SOX2‐binding sites (HEK293 cells). Co‐transfection of SOX2 expressing vector and comparison between wild‐type and mutated sites.
Alterations of SOX2 and OPA1 impact neuroimaging parameters of tumor extension
We wanted to see if there is an association between tumor size (measured by neuroimaging) and SOX2amp or OPA1del on glioma invasion (Figure S5 and Table S6). Towards this goal, we implemented linear models linking the genomic alteration to the following parameters: non‐enhancing volume (lower grade tumor compartment), enhancing volume (malignant tumor component), necrosis (within tumor core, hallmark of GB), total volume (enhancing + non‐enhancing + necrosis) and preoperative maximal tumor area on the axial plane as parameters. An illustrated example is given in Figure 6A. The postoperative parameter was recurrence volume. To account for confounding, we also included age at surgery, and the log‐transformed ratio of postoperative to preoperative volume in the model. As SOX2amp and OPA1del are mutually exclusive events, we considered them as distinct glioma genetic entities to be analyzed for neuroimaging. From the linear model linking the genetic alteration with the log‐transformed necrosis volume, where we also included age at surgery and the log‐transformed ratio of postoperative to preoperative volume to account for confounding, we saw that both SOX2amp and OPA1del are associated with the necrosis tumor volume (Table S7, Figure 6A,B). In particular, we observed that the presence of SOX2amp was associated with a fourfold increase, while OPA1del, with a 3.4‐fold increase in the tumor necrosis volume with respect to the case with neither SOX2amp nor OPA1del (Figure 6B,C). In addition, higher tumor area extension tended to be associated with both genetic markers (Figure S8 and Table S9). To mimic a possible role of necrotic tissue on tumor cell invasion, we performed invasion assays of parental LN319 and LN319 cells engineered to overexpress SOX2 or inactivate OPA1 in the presence of necrotic autologous cells. Addition of necrotic cells impaired parental LN319 invasiveness (Figure 6D, left). However, relative to parental LN319, both SOX2‐overexpressing and OPA1‐inactivating LN319 strains had higher invasion rates (3.4‐ and 2.3‐fold, respectively) in the presence of necrotic cells (Figure 6D, middle and right), suggesting that that an overall invasion‐inhibitory effect of necrotic cells is largely overruled by SOX2 amplification or OPA1 loss in LN319 cells.
Based on our observations that SOX2 gain or OPA1 loss both induce glioma cell invasion in vitro, in vivo in xenotransplant studies, and are both associated with larger necrotic area revealed by neuroimaging of human glioma, we postulate a functional connectivity between necrosis and invasion in glioblastoma.
Discussion
SOX2 and OPA1 status in glioma
Approximately 10% of LGG and GB patient samples deposited in the TCGA database show SOX2 amplification or OPA1 deletion (Figure S3A), with no further reported investigation. Preferential SOX2amp and OPA1del occur independently of the IDH status, the major genetic determinant in diffuse gliomas. This indicates that SOX2amp and OPA1del promote cell invasiveness regardless of the glioma developmental pathway. Two hypotheses may account for the mutual exclusion between SOX2amp and OPA1del. First, mutual exclusivity can be an indicator for a commonly targeted phenotype, like invasion in this case. Alternatively, as SOX2 and OPA1 cluster within only 15 megabases at 3q26, amplification or deletion of one is likely to involve the neighboring gene too, which reduces the likelihood of concomitant SOX2amp and OPA1del events. This hypothesis is supported by the analysis of the TCGA database, where individual samples with OPA1 amplification show common amplification of SOX2, PIK3CA and MFN1, suggesting that SOX2 amplification events may extend to OPA1 gene (Figure S3A, red top bands). While we observed a mutual exclusion of SOX2 gene amplification and OPA1 deletion in our patient collection, we noticed that at the protein level, SOX2 gain and OPA1 loss are concomitant in distinct glioma genotypes, considering IDH mutation, SOX2amp or OPA1del statuses. This concomitance suggests that optimization of invasion requires further mechanisms downstream of gene copy variation, possible epigenetic and posttranslational regulation events.
Interplay between 3q26 genes
Consistent with previous data on breast cancer cells 30, we found that SOX2 was displaced from nucleus to cytosol in response to inhibition of the PI3K/AKT pathway. Maintaining SOX2 activation by PI3K/AKT pathway activity provided a first molecular link between the 3q26 gene products PIK3CA and SOX2. Of note, mTOR, a downstream target of PI3K/AKT signaling, was previously identified as a regulator of glioma cell invasion and proposed as target for controlling cell invasion 6. However, we found that PI3K/AKT directly regulates SOX2 activity in ways that do not (evidently) involve mTORC1. Furthermore, SOX2 trans‐activates PIK3CA as part of a positive feedback loop to further maintain cell invasion. Interestingly, SOX2 has been previously shown to activate the Notch pathway to promote glioma cell invasion 32, 43. Proteins involved in mitochondria fusion (OPA1 and MFN1) or fission (DRP1) have been shown to positively (DRP1) and negatively (OPA1 and MFN1) regulate cell motility 10, 11. mTOR, an activator of glioma invasion, has also been shown to induce mitochondria fission through DRP1 recruitment 6, 24. Thus, AKT seems to drive two distinct mechanisms of glioma cell invasion activation, an mTOR‐dependent, presumably through DRP1 activation and an mTOR‐independent mechanism through SOX2. However, the concomitant trans‐activation of OPA1 and PIK3CA results in a conflicting situation between the pro‐invasive PI3K/AKT‐mTOR‐DRP1 and PI3K/AKT‐SOX2 pathways, and invasion suppression by the PI3K/AKT‐SOX2‐OPA1 axis. Loss of OPA1 function—otherwise activated by SOX2 as shown here by Chip and Luciferase reporter assay—would therefore be a response to optimize invasion capacity from PI3K/AKT and SOX2 pathways (Figure 7).
Figure 7.
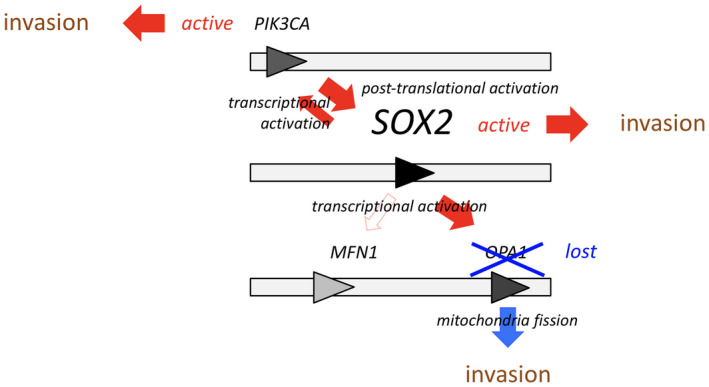
3q26 gene products modulate glioma cell invasion in various entangled ways. SOX2 balances glioma cell invasion through intrinsically antagonistic mechanisms: While PI3K/AKT/mTOR 41 and PI3K/AKT‐SOX2 loops promote glioma invasion 9, 20, SOX2 trans‐activates OPA1, which results in an effective inhibition of invasion. OPA1 knock‐down mutations resolve this functional antagonism and lead to an effective deregulation of glioma cell invasion.
Necrosis and invasion
The mutual exclusion between SOX2amp and OPA1del suggested that both glioma genetic entities could result in a similar phenotype. We used this assumption as a rationale for stratifying the alterations in SOX2 and OPA1 as a categorical variable with three levels in order to assess the association between SOX2/OPA1 status and tumor size parameters. Indeed, considering SOX2 gain vs. no SOX2 gain, no SOX2 gain would have also included OPA1 loss and conversely, OPA1 loss vs. no OPA1 loss, no OPA1 loss would have included SOX2 gain. Thus, in both cases, SOX2 gain and OPA1 loss would mask each other, potentially reducing the difference seen in the phenotype associated to each category. In a homogenous population of GB, tumor area tended to be associated with SOX2amp or OPA1del (P < 0.15). For the necrotic compartment volume, we observed a stronger association and significance in both cases (P = 0.01 and P = 0.03, respectively). This is consistent with recent results showing that the vicinity of necrotic tissue induces chemokine IL‐8 production by GB cells resulting in enhanced invasion in CRT‐MG cells 1. In LN319 cells, relative to parental strain, we observed that the presence of autologous necrotic cells increased invasion under conditions of either SOX2 gain or OPA1 loss. This suggests that SOX2 gain or OPA1 loss may play a role in this process, either in the necrotic cells, or in the invading cells, the underlying mechanism remaining to be investigated.
This could imply that the volume of the necrotic compartment could indeed represent an indirect biomarker to assess invasion. Nevertheless, due to the limited sample size, no strong relationship could be established between SOX2amp or OPA1del with overall survival (Table S9). The non‐enhancing compartment visualized by MRI may also include vasogenic edema of inflammatory origin which may lead to an overestimation of the volume (illustrated in Figure 6A). The infiltration zone, representing a gradient of progressive dilution of sparse invading tumor cells, cannot be accurately determined. Based on these observations, the necrotic volume compartment of the tumor turned out to be the most accurate value and is therefore a suitable surrogate marker for assessing invasion.
Pathways to be targeted to control invasion
Several pathways have been identified to induce cell motility and have been postulated as potential targets for controlling glioma cell invasion, such as, for example, mTOR 6. The PI3K/AKT/SOX2 axis described here imposes a natural limitation to any such efforts, as PI3K and SOX2 interact essentially independent of mTOR (see Figure 4B/E). Further upstream PI3KCA or AKT, both at the crossroads of mTOR‐dependent and SOX2‐driven branches of invasion modulation, may thus provide more suitable targets to restrict glioma cell invasion for clinical purposes. Also, mitochondria fusion protein OPA1 inactivation could potentially be counteracted by inactivation of, for example, mitochondria fission protein DRP1. Regarding cause‐and‐effect relationship, whether SOX2 gain or OPA1 loss either trigger tumor cell necrosis itself or sensitize tumor cells for paracrine responses to induce necrosis remains to be resolved.
Taken together, we unraveled a functional interplay of 3q26 gene products in the regulation of glioma cell invasion. OPA1 loss and SOX2 gain mutations in particular stimulate glioma cell invasiveness in vitro and in vivo, and coincide with elevated necrotic volume scoring in neuroimages of patients. Our molecular findings thus meet a phenotypic disease manifestation that may influence disease scoring in the future. This strengthens the need for combinatorial therapies that would target multiple pathways and pathway branches for controlling invasive tumor expansion.
Author’s Contributions
Conception and design: JL Boulay, L Mariani, S Frank, T Schaefer
Development of methodology: MF Ritz, C Tostado, A Ramadoss
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): JL Boulay, L Mariani, MF Ritz, C Lengerke, H Neddersen, S Falbo, C Stippich, A Bink, P Demougin, P Jenö, S Moes, C Tostado, L Tintignac, Christoph Schürch, Joëlle Müller, Jonas Schärer, S Leu, A Ramadoss, T Schaefer
Analysis and interpretation of data (eg, statistical analysis, biostatistics, computational analysis): JL Boulay, L Mariani, MF Ritz, H Bucher, A Bink, P Demougin, P Jenö, S Moes, G Moffa, L Tintignac, S Leu, A Ramadoss, T Schaefer
Writing, review, and/or revision of the manuscript: JL Boulay, L Mariani, S Frank, C Lengerke, A Bink, P Jenö, G Moffa, S Leu , T Schaefer
Administrative, technical, or material support (ie, reporting or organizing data, constructing databases): C Tostado, T Schaefer, S Leu,
Conflict of Interest
No potential conflicts of interest were disclosed
Supporting information
Figure S1. 3q26 genes SOX2 and OPA1 are targeted in glioma.
Figure S2. List of the oligonucleotides used in this report.
Figure S3. Data extracted from public databases.
Figure S4. SOX2 overexpression and OPA1 knock‐down in LN319 alter cell proliferation.
Figure S5. Patient cohort.
Table S6. Characteristics of patients for statistical analysis.
Table S7. Coefficient estimates from tumor size parameters used to evaluate the oncogenic relevance of SOX2amp and OPA1del.
Figure S8. Tumor size parameters other than tumor necrosis volume used to evaluate the oncogenic relevance of SOX2amp and OPA1del.
Table S9. Impact of SOX2amp and OPA1del statuses on overall survival.
Acknowledgments
The study was approved by the local Ethics Committee of Basel. This work was supported through fund from the Swiss Cancer League (Grant KLS 2765‐02‐2011 awarded to JLB LM and SF), from the Department of Surgery, University Hospital Basel and University of Basel, and from the University of Basel Forschungsfonds (Grant DMS‐2351 awarded to TS). We acknowledge Dr. Martina Konantz for supervision of fish husbandry and for technical advice concerning fish xenotransplants, Dr. Hui Wang for cloning and provision of the pLVX‐mCherry ‐SOX2 expression plasmid, and we are grateful to Monika Hegi for providing us LN319 cells.
Contributor Information
Luigi Mariani, Email: luigi.mariani@usb.ch.
Jean‐Louis Boulay, Email: jean-louis.boulay@unibas.ch.
References
- 1. Ahn SH, Park H, Ahn YH, Kim S, Cho MS, Kang JL, Choi YH (2016) Necrotic cells influence migration and invasion of glioblastoma via NF‐κB/AP‐1‐mediated IL‐8 regulation. Sci Rep 6:24552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alonso MM, Diez‐Valle R, Manterola L, Rubio A, Liu D, Cortes‐Santiago N, et al (2011) Genetic and epigenetic modifications of Sox2 contribute to the invasive phenotype of malignant gliomas. PLoS One 6:e26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ben‐Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA (2008) An embryonic stem cell‐like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40:499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Campello S, Lacalle RA, Bettella M, Mañes S, Scorrano L, Viola A (2006) Orchestration of lymphocyte chemotaxis by mitochondrial dynamics. J Exp Med 203:2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carey KT, Tan KH, Ng J, Liddicoat DR, Godfrey DI, Cole TJ (2013) Nfil3 is a glucocorticoid‐regulated gene required for glucocorticoid‐induced apoptosis in male murine T cells. Endocrinology 154:1540–1552. [DOI] [PubMed] [Google Scholar]
- 6. Chandrika G, Natesh K, Ranade D, Chugh A, Shastry P (2016) Suppression of the invasive potential of Glioblastoma cells by mTOR inhibitors involves modulation of NFκB and PKC‐α signaling. Sci Rep 6:22455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cuddapah VA, Robel S, Watkins S, Sontheimer H (2014) A neurocentric perspective on glioma invasion. Nat Rev Neurosci 15:455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dazert E, Colombi M, Boldanova T, Moes S, Adametz D, Quagliata L, et al (2016) Quantitative proteomics and phosphoproteomics on serial tumor biopsies from a sorafenib‐treated HCC patient. Proc Natl Acad Sci U S A 113:1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, et al (2000) Nuclear gene OPA1, encoding a mitochondrial dynamin‐related protein, is mutated in dominant optic atrophy. Nat Genet 26:207–210. [DOI] [PubMed] [Google Scholar]
- 10. Desai SP, Bhatia SN, Toner M, Irimia D (2013) Mitochondrial localization and the persistent migration of epithelial cancer cells. Biophys J 104:2077–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ferreira‐da‐Silva A, Valacca C, Rios E, Pópulo H, Soares P, Sobrinho‐Simões M, et al (2015) Mitochondrial dynamics protein Drp1 is overexpressed in oncocytic thyroid tumors and regulates cancer cell migration. PLoS One 10:e0122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Friedl P, Alexander S (2011) Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 147:992–1009. [DOI] [PubMed] [Google Scholar]
- 13. Gangemi RM, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, et al (2009) SOX2 silencing in glioblastoma tumor‐initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells 27:40–48. [DOI] [PubMed] [Google Scholar]
- 14. Gao H, Teng C, Huang W, Peng J, Wang C (2015) SOX2 promotes the epithelial to mesenchymal transition of esophageal squamous cells by modulating slug expression through the activation of STAT3/HIF‐α signaling. Int J Mol Sci 16:21643–21657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hammoud MA, Sawaya R, Shi W, Thall PF, Leeds NE (1996) Prognostic significance of preoperative MRI scans in glioblastoma multiforme. J Neurooncol 27:65–73. [DOI] [PubMed] [Google Scholar]
- 16. Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K (2009) Autocrine TGF‐beta signaling maintains tumorigenicity of glioma‐initiating cells through Sry‐related HMG‐box factors. Cell Stem Cell 5:504–514. [DOI] [PubMed] [Google Scholar]
- 17. Ishii N, Maier D, Merlo A, Tada M, Sawamura Y, Diserens AC, Van Meir EG (1999) Frequent co‐alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol 9:469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Konantz M, Balci TB, Hartwig UF, Dellaire G, André MC, Berman JN, Lengerke C (2012) Zebrafish xenografts as a tool for in vivo studies on human cancer. Ann N Y Acad Sci 1266:124–137. [DOI] [PubMed] [Google Scholar]
- 19. Kondziolka D, Lunsford LD, Martinez AJ (1993) Unreliability of contemporary neurodiagnostic imaging in evaluating suspected adult supratentorial (low‐grade) astrocytoma. J Neurosurg 79:533–536. [DOI] [PubMed] [Google Scholar]
- 20. Kubiatowski T, Jang T, Lachyankar MB, Salmonsen R, Nabi RR, Quesenberry PJ, et al (2001) Association of increased phosphatidylinositol 3‐kinase signaling with increased invasiveness and gelatinase activity in malignant gliomas. J Neurosurg 95:480–488. [DOI] [PubMed] [Google Scholar]
- 21. Li X, Xu Y, Chen Y, Chen S, Jia X, Sun T, et al (2013) SOX2 promotes tumor metastasis by stimulating epithelial‐to‐mesenchymal transition via regulation of WNT/β‐catenin signal network. Cancer Lett 336:379–389. [DOI] [PubMed] [Google Scholar]
- 22. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella‐Branger D, Cavenee WK, et al (2016) The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 131:803–820. [DOI] [PubMed] [Google Scholar]
- 23. Luo W, Li S, Peng B, Ye Y, Deng X, Yao K (2013) Embryonic stem cells markers SOX2, OCT4 and Nanog expression and their correlations with epithelial‐mesenchymal transition in nasopharyngeal carcinoma. PLoS One 8:e56324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morita M, Prudent J, Basu K, Goyon V, Katsumura S, Hulea L, et al (2017) mTOR Controls Mitochondrial Dynamics and Cell Survival via MTFP1. Mol Cell 67:922–35.e5. [DOI] [PubMed] [Google Scholar]
- 25. Network CGAR (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455:1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park GB, Kim D (2017) TLR5/7‐mediated PI3K activation triggers epithelial‐mesenchymal transition of ovarian cancer cells through WAVE3‐dependent mesothelin or OCT4/SOX2 expression. Oncol Rep 38:3167–3176. [DOI] [PubMed] [Google Scholar]
- 27. Paw I, Carpenter RC, Watabe K, Debinski W, Lo HW (2015) Mechanisms regulating glioma invasion. Cancer Lett 362:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Raza SM, Lang FF, Aggarwal BB, Fuller GN, Wildrick DM, Sawaya R (2002) Necrosis and glioblastoma: a friend or a foe? A review and a hypothesis. Neurosurgery 51:2–12; discussion‐3. [DOI] [PubMed] [Google Scholar]
- 29. Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al (2004) High frequency of mutations of the PIK3CA gene in human cancers. Science 304:554. [DOI] [PubMed] [Google Scholar]
- 30. Schaefer T, Wang H, Mir P, Konantz M, Pereboom TC, Paczulla AM, et al (2015) Molecular and functional interactions between AKT and SOX2 in breast carcinoma. Oncotarget 6:43540–43556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Scherer HJ (1940) A critical review: the pathology of cerebral gliomas. J Neurol Psychiatry 3:147–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sivasankaran B, Degen M, Ghaffari A, Hegi ME, Hamou MF, Ionescu MC, et al (2009) Tenascin‐C is a novel RBPJkappa‐induced target gene for Notch signaling in gliomas. Cancer Res 69:458–465. [DOI] [PubMed] [Google Scholar]
- 33. Song Z, Ghochani M, McCaffery JM, Frey TG, Chan DC (2009) Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell 20:3525–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stemke‐Hale K, Gonzalez‐Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, et al (2008) An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res 68:6084–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suvà ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, et al (2014) Reconstructing and reprogramming the tumor‐propagating potential of glioblastoma stem‐like cells. Cell 157:580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tan EJ, Olsson AK, Moustakas A (2015) Reprogramming during epithelial to mesenchymal transition under the control of TGFβ. Cell Adh Migr 9:233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Torsvik A, Stieber D, Enger P, Golebiewska A, Molven A, Svendsen A, et al (2014) U‐251 revisited: genetic drift and phenotypic consequences of long‐term cultures of glioblastoma cells. Cancer Med 3:812–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wangaryattawanich P, Hatami M, Wang J, Thomas G, Flanders A, Kirby J, et al (2015) Multicenter imaging outcomes study of The Cancer Genome Atlas glioblastoma patient cohort: imaging predictors of overall and progression‐free survival. Neuro Oncol 17:1525–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, et al (2010) Updated response assessment criteria for high‐grade gliomas: response assessment in neuro‐oncology working group. J Clin Oncol 28:1963–1972. [DOI] [PubMed] [Google Scholar]
- 40. Westfield M (2000) The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (danio rerio), 4th edn. University of Oregon Press: Eugene. [Google Scholar]
- 41. Wick W, Platten M, Weller M (2001) Glioma cell invasion: regulation of metalloproteinase activity by TGF‐beta. J Neurooncol 53:177–185. [DOI] [PubMed] [Google Scholar]
- 42. Yang N, Hui L, Wang Y, Yang H, Jiang X (2014) Overexpression of SOX2 promotes migration, invasion, and epithelial‐mesenchymal transition through the Wnt/β‐catenin pathway in laryngeal cancer Hep‐2 cells. Tumour Biol 35:7965–7973. [DOI] [PubMed] [Google Scholar]
- 43. Ying M, Wang S, Sang Y, Sun P, Lal B, Goodwin CR, et al (2011) Regulation of glioblastoma stem cells by retinoic acid: role for Notch pathway inhibition. Oncogene 30:3454–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zee C, P C, S D, MLJ A (1995) Imaging Features of Benign Gliomas. Benign Cerebral Glioma, pp. 247–274. American Association of Neurological Surgeons: Park Ridge, IL. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. 3q26 genes SOX2 and OPA1 are targeted in glioma.
Figure S2. List of the oligonucleotides used in this report.
Figure S3. Data extracted from public databases.
Figure S4. SOX2 overexpression and OPA1 knock‐down in LN319 alter cell proliferation.
Figure S5. Patient cohort.
Table S6. Characteristics of patients for statistical analysis.
Table S7. Coefficient estimates from tumor size parameters used to evaluate the oncogenic relevance of SOX2amp and OPA1del.
Figure S8. Tumor size parameters other than tumor necrosis volume used to evaluate the oncogenic relevance of SOX2amp and OPA1del.
Table S9. Impact of SOX2amp and OPA1del statuses on overall survival.


