Abstract
We have identified a discrete, focal telomere DNA expansion phenotype in the photoreceptor cell layer of normal, non‐neoplastic human retinas. This phenotype is similar to that observed in a subset of human cancers, including a large fraction of tumors of the central nervous system, which maintain their telomeres via the non‐telomerase‐mediated alternative lengthening of telomeres (ALT) mechanism. We observed that these large, ultra‐bright telomere DNA foci are restricted to the rod photoreceptors and are not observed in other cell types. Additionally, focus‐positive rod cells are dispersed homogeneously throughout the posterior retinal photoreceptor cell layer and appear to be human‐specific. We examined 108 normal human retinas obtained at autopsy from a wide range of ages. These large, ultra‐bright telomere DNA foci were not observed in infants before 6 months of age; however, the prevalence of focus‐positive rod cells dramatically increased throughout life. To investigate associations between this phenotype and retinal pathology, we assessed adult glaucoma (N = 29) and diabetic retinopathy (N = 38) cases. Focus‐positive rod cells were prominent in these diseases. When compared to the normal group, after adjusting for age, logistic regression modeling revealed significantly increased odds of falling in the high category of focus‐positive rod cells for glaucoma and diabetic retinopathy. In summary, we have identified a dramatic telomere alteration associated with aging and diseases affecting the retina.
Keywords: diabetic retinopathy, glaucoma, retina, rod photoreceptors, telomeres
Introduction
Telomeres are composed of double‐stranded TTAGGG hexameric nucleotide repeats located at the ends of eukaryotic chromosomes and bound by telomere‐associated proteins, including the six‐member shelterin complex. This complex sequesters telomeric DNA, thereby masking double strand break DNA damage signals at the chromosomal termini 9. In addition, telomeres function to inhibit terminal exonucleolytic degradation and inhibit non‐homologous end joining as a mechanism to prevent end‐to‐end fusions 3, 5, 10, 25, 26. Irradiation induced damage experiments causing random double strand breaks in human and mouse embryonic fibroblasts have shown that telomeric regions are favored targets for a persistent DNA damage response 20. Furthermore, random double strand DNA breaks that are induced by oxidative stress are preferentially localized at telomeric regions, independent of telomerase activity and telomere length 20.
In cancer cells, telomeres can be maintained either by expressing the enzyme telomerase, a telomere‐specific reverse transcriptase 29, or through homology‐directed DNA repair 6, 12, 18 by a mechanism termed Alternative Lengthening of Telomeres (ALT). ALT‐positive tumors feature striking telomere length heterogeneity and the presence of ultra‐bright, intra‐nuclear, telomere DNA FISH signals representing large distinctive foci, termed ALT‐associated foci, which strongly correlate with other molecular hallmarks of ALT 8, 27. Previously, we comprehensively characterized the prevalence of ALT‐associated foci in thousands of cancer specimens using a highly sensitive telomere‐specific fluorescent in situ hybridization (FISH) assay 17. Importantly, ALT‐associated foci were not identified in diverse, non‐neoplastic human tissues or in any cancer‐adjacent benign cells 17.
Here, we analyze multiple regions of normal brain and healthy and diseased retinal tissue. Interestingly, we identify a large, ultra‐bright telomere DNA foci pattern, reminiscent of but distinct from the ALT‐associated foci, which is restricted to rod photoreceptors and not observed in other cell types within the retina or adult brain of varying ages. This dramatic telomere alteration is associated with aging and diseases affecting the retina.
Materials and Methods
Case selection
Formalin‐fixed, paraffin‐embedded autopsy eye specimens were retrieved from the autopsy pathology and ophthalmic pathology archives of The Johns Hopkins Hospital, Baltimore, MD. Eye specimens from 1994 to 2014 were obtained from 175 autopsied patients from new‐borns to 95 years of age. Among the subjects examined, autopsy diagnoses included two subjects with a primary systemic diagnosis of short telomere syndrome, 108 subjects had normal, non‐pathologic eye diagnoses, 29 with pathologic diagnosis of glaucoma, 38 with pathologic features of non‐proliferative or proliferative diabetic retinopathy and 16 with pathologic diagnosis of age‐related macular degeneration.
Animal collection
Formalin‐fixed, paraffin‐embedded archived animal eye specimens were provided by Comparative Medicine at Johns Hopkins, Veterinary School at Cornell and Comparative Ocular Pathology Laboratory of Wisconsin (COPLOW).
Human iPS cells‐derived retinal organoids
Three‐dimensional retinal organoids derived from human iPS cells were generated as previously described 32. Briefly, Human episomal iPS cells derived from CD34+ cord blood (A18945, ThermoFisher Scientific) 7 were maintained on Matrigel (growth‐factor‐reduced; BD Biosciences) coated plates with mTeSR1 medium (Stemcell Technologies) according to WiCell protocols. On day 0 (D0) of differentiation, iPS cell colonies were treated with Dispase (Stemcell Technologies), mechanically dissociated into small clumps and cultured in suspension to induce aggregate formation. After 24 h, aggregates were gradually transitioned into neural‐induction medium (NIM) to induce anterior neural differentiation. On D7, neural aggregates were seeded onto Matrigel (growth‐factor‐reduced; BD Biosciences) coated dishes and cultured in retinal differentiation media (RDM). On the 4th week of differentiation, horseshoe‐shaped neural retina (NR) domains were mechanically detached and cultured in suspension at 37°C in a humidified 5% CO2 incubator in RDM, where they gradually formed 3‐dimesional retinal organoids. For long‐term suspension culture, the medium was supplemented with 10% fetal bovine serum (FBS; Gibco), 100 µM Taurine (Sigma) and 2mM GlutaMAX (Invitrogen) beginning on D42, with the addition of 1 µM all‐trans retinoic acid (RA; Sigma) during week 10–14 and 0.5 µM RA thereafter.
ATRX and DAXX immunohistochemistry
Immunohistochemistry (IHC) was performed on formalin‐fixed, paraffin‐embedded tissue sections. Immunolabeling for ATRX was performed using an anti‐ATRX (HPA001906, Sigma‐Aldrich, 1:100) antibody. The labeling was performed in the Ventana Benchmark XT autostainer using CC1 antigen retrieval buffer for 30 min at 95°C, incubation with primary antibody for 32 min and detection with Ventana ultraView Universal DAB. Immunolabeling for DAXX was performed as previously described using an anti‐DAXX (HPA008736, Sigma‐Aldrich, 1:100) antibody 16. Briefly, tissue sections were deparaffinized in xylene and hydrated by serially diluting in decreasing concentrations of ethanol. For antigen retrieval, sections were steamed with citrate buffer for 30 min and then blocked against endogenous peroxidase activity with dual endogenous enzyme blocking agent (Dako) for 10 min. Sections were incubated with primary antibody (1:100 dilution) for 2 h at room temperature followed by secondary antibody (Leica Microsystems) for 30 min and detected with 3,30‐diaminobenzidine (Sigma‐Aldrich). Sections were counterstained with hematoxylin, rehydrated and coverslips were mounted. Only nuclear labeling was evaluated for ATRX and DAXX.
Telomere‐specific FISH and immunolabeling
Deparaffinized slides were hydrated, steamed for 25 min in citrate buffer (Vector Laboratories, Burlingame, CA), dehydrated and hybridized with a Cy3‐labeled peptide nucleic acid (PNA) probe complementary to the mammalian guanine‐rich telomere repeat sequence (N‐terminus to C‐terminus; Cy3‐CCCTAACCCTAACCCTAA) 17, 24. For some experiments, a complementary telomere PNA probe for the G‐rich strand is utilized instead (TelG‐FAM; Panagene). All normal and diseased human specimens were reviewed and visually scored (CMH and WRB) using a defined categorical scoring system. Utilizing a 20× objective, each case was visually scanned in both globes, if present, across the posterior or central retina and peripheral retina. Categorical scoring was based on the average percentage of focus‐positive rod cells within the outer nuclear layer in multiple high‐powered (20×) fields. Categories included “none” (0%), “low” (< 5%), “medium” (5%–30%) and “high” (> 30%). Categorical scoring of human retina specimens was done a blinded manner without knowledge of diagnosis. As a positive control for hybridization efficiency, an Alexafluor‐488 labeled PNA probe specific to human centromeric DNA repeats for the CENP‐B binding sequence (N‐terminus to C‐terminus; Alexafluor‐488‐ATTCGTTGGAAACGGGA) was included in the hybridization solution. If slides were also immunolabeled with primary antibodies, the following occurred. The slides were rinsed in PBST followed by application of primary antibody and incubated for 2 h. Antibodies include: Rhodopsin (1:200; GTX23267; GeneTex), Opsin Red/Green (1:750; AB5405; Millipore), Opsin Blue (1:750; AB5407; Millipore), TRF2 (1:1000; NB110–57130; Novus Biologicals) and PML (1:100; sc‐966; Santa Cruz Biotechnology). After the incubation, the slides were rinsed in PBST followed by application of fluorescent secondary antibody labeled with appropriate Alexa Fluor labeled secondary antibodies (Molecular Probes) diluted 1:100 in Dulbecco's PBS and incubated at room temperature for 30 min. The slides were then rinsed in PBST, thoroughly washed in deionized water, drained and counterstained with 4′‐6‐diamidino‐2‐phenylindole (DAPI) (500 ng/mL in deionized water, Sigma Chemical Co. Cat #D‐8417) for 5 min at room temperature. The slides were then mounted with Prolong anti‐fade mounting medium (catalogue No. P‐7481; Molecular Probes) and imaged.
Microscopy
Slides were imaged with a Zeiss AxioObserver microscope with LSM700 confocal module (NIH Award S10 OD016374) using the following objectives: 40×/1.30 PlanNeofluar oil with DIC, 63×/1.4 PlanApo oil with DIC and 100× PlanApo oil with DIC. All images were pseudo‐colored and merged. For quantified data, images were captured with a Nikon 50i epifluorescence microscope equipped with X‐Cite series 120 illuminator using a 40×/0.95 NA PlanApo lens with correction collar. The digitized fluorescent telomere FISH signals were quantified using ImageJ and a custom designed plugin (“Telometer”; http://bui2.win.ad.jhu.edu/telometer/). Short exposure times were used for image collection of the large, ultra‐bright telomere DNA foci to ensure that these FISH signals fell within the linear range of the camera. At a higher exposure time, telomere FISH signals were also quantified for all telomeres within individual adjacent focus‐negative rod nuclei or inner nuclear layer nuclei. For each nucleus, the telomere FISH signals were then summed, giving an intensity value representing the total telomere intensity per nucleus. Intensity ratios were calculated by dividing the normalized telomere FISH signals for individual large, ultra‐bright telomere DNA foci by the average per cell sum of individual telomere FISH signals for either the focus‐negative rod nuclei or inner nuclear layer nuclei within the same case, with appropriate linear adjustment for differences in exposure times during image collection.
Statistical analysis
Participant characteristics were summarized by frequencies and the Spearman's rank correlation was used to evaluate the correlation between age and the category of focus‐positive rod cells. A binary logistic regression model was performed to compute the odds of falling in the highest category (high) for each of the sampled tissue types. Adjusted Odds Ratios (ORs), 95% Confidence Intervals (CIs) and P‐values were presented adjusting for the participant's age. All analyses were conducted using SAS version 9.4 (Cary, NC), and a two‐sided. P < 0.05 was considered to be a statistically significant result.
Results
Alternative lengthening of telomeres (ALT) phenotype in cancer
As described previously, ALT‐positive tumors include striking telomere length heterogeneity and the presence of ultra‐bright, intra‐nuclear, telomere DNA FISH signals of variable size (Figure 1A) representing large distinctive foci. The presence of these ALT‐associated foci, indicative of ALT, have not been previously identified in cancer‐adjacent benign cells or non‐neoplastic human tissues.
Figure 1.
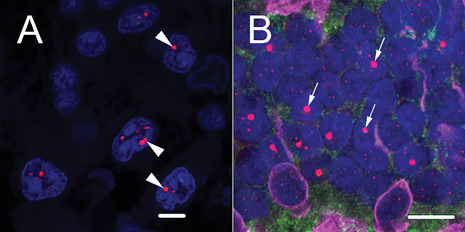
The large, ultra‐bright telomere DNA foci in retinal rod photoreceptors are reminiscent of ALT‐associated foci. A. For comparison, a representative image of ALT‐associated foci (arrowheads) from an ALT‐positive anaplastic astrocytoma; scale = 5 µm. B. Image of adult retinal photoreceptor layer demonstrates that the large, ultra‐bright telomere DNA foci (arrows) are seen in rod photoreceptors; scale = 10 µm. Telomeres are labeled with a Cy3‐labeled telomere‐specific FISH probe (red) and DNA is stained with DAPI (blue) in A and B. Shown in B, immunofluorescence for the combination of Red/Green and Blue opsins highlights the cone photoreceptor nuclei and cellular processes (magenta).
Normal retinal rod photoreceptors display large, ultra‐bright telomere DNA foci not observed in other cell types within the retina or adult brain
Here, we expand our analysis to include multiple regions of normal brain and healthy and diseased ocular tissues. Unexpectedly, we identified a unique, focal telomere DNA expansion pattern, reminiscent of ALT‐associated foci, in the photoreceptor cell layer of normal retinas (Figure 1B). Intriguingly, using multiplex staining, coupling telomere‐specific FISH staining with immunolabeling with rod and cone‐specific antibodies, we determined that these large, ultra‐bright telomere DNA foci, which manifest as single foci in focus‐positive cells, are restricted to the rod photoreceptors and were not observed in other cell types within the retina (Figure 2A,B). When quantified, accounting for different exposure times to ensure FISH signals fell within the linear range of the camera, these large, ultra‐bright telomere DNA foci represent an order of magnitude more total telomere content compared to the total telomere content in any adjacent focus‐negative photoreceptor nuclei or inner nuclear layer nuclei. Additionally, normal‐appearing distributions of telomeres and centromeres are similar between focus‐positive and focus‐negative rod cells (Figure 2C); thus, these foci are not the result of clustering of multiple telomeres. These distinctive foci were observed using DNA FISH probes specific for either telomere strand (G‐rich or C‐rich) and were also observed via antibody staining for telomere‐binding proteins (eg, TRF2), thereby strongly suggesting that these foci are associated with telomeric DNA (Figure 3), rather than telomeric repeat‐containing RNA (TERRA) 1, 2. Finally, these large, ultra‐bright foci were not observed in the neocortex, basal ganglia, cerebellum or brainstem of adult brains of varying ages.
Figure 2.
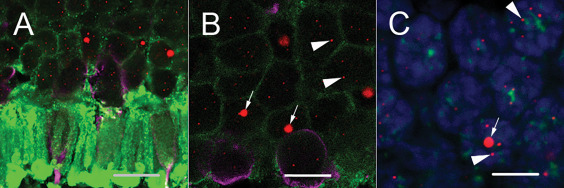
Representative retinal images demonstrate the presence of large, ultra‐bright telomere DNA foci restricted to rod photoreceptors. A and B. Retinal photoreceptor layer at the edge of the outer nuclear layer in the central posterior retina of an adult demonstrates that the large, ultra‐bright foci (arrows) are seen in rod photoreceptors and not in cone photoreceptors. C. Centromeres are marked with a FITC‐labeled centromere‐specific FISH probe (green) demonstrating the large, ultra‐bright telomere DNA foci do not co‐localize with centromere DNA. Telomeres are labeled with a Cy3‐labeled telomere‐specific FISH probe (red), DNA is stained with DAPI (blue) and immunofluorescence for rhodopsin, labeling the rod photoreceptors (green), and a combination of Red/Green and Blue opsins, labeling the cone photoreceptor nuclei and cellular processes (magenta). The smaller telomere FISH signals are typical of labeled telomere ends (arrowheads) present in all cell types, including rod cells with large, ultra‐bright foci. Scale: 10 µm.
Figure 3.
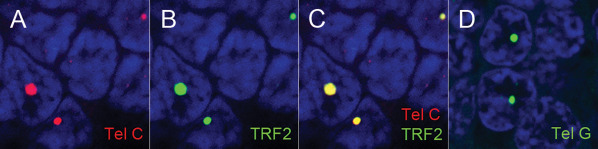
The large, ultra‐bight foci are specific for telomeric DNA. A. Telomere DNA is labeled with a Cy3‐labeled telomere sequence‐specific probe complementary to the G‐rich telomere repeat strand sequence (Tel‐C, red); B. Same area as in A, showing anti‐TRF‐2 antibody staining (green) (B); and C. co‐localization of A&B (yellow). D. FAM labeled telomere DNA probe (Tel‐G, green; guanine‐rich probe complementary to the C‐rich strand telomere repeat sequence) demonstrates a similar staining pattern, thus indicating the large, ultra‐bright foci consist of telomere DNA, which is G‐rich and not telomeric RNA, which is transcribed from only one strand of telomeric DNA, producing RNA consisting of the C‐rich telomere repeat sequence. For all images, DNA is stained with DAPI (blue).
Differences between ALT‐associated foci and rod‐specific large, ultra‐bright telomere DNA foci
In contrast to the ALT‐associated foci observed in a subset of cancers, these large, ultra‐bright telomere DNA foci are (i) present in normal, non‐neoplastic cells; (ii) uniform in size, shape, number and intensity; and (iii) localized to the perinucleolar region. Focus‐positive rod cells are dispersed throughout the posterior retinal photoreceptor cell layer in a roughly homogenous distribution. Additionally, in contrast to ALT‐associated foci, these large, ultra‐bright foci do not co‐localize with promyelocytic leukemia protein (PML), for form so‐called ALT‐associated PML bodies 19 (Figure 4).
Figure 4.
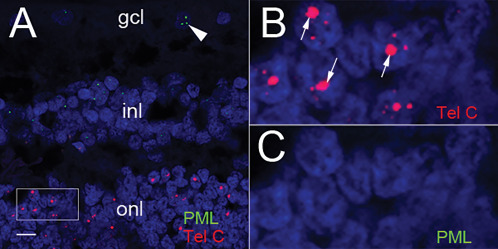
PML does not co‐localize with the large, ultra‐bright telomere DNA foci. A. Telomere DNA is labeled with a Cy3‐labeled telomere sequence‐specific probe complementary to the G‐rich telomere repeat sequence (Tel‐C, red) and PML nuclear bodies are labeled with an anti‐PML antibody and detected with a Cy5‐conjugated secondary antibody (green). PML nuclear bodies (green) are observed only in the retinal ganglion cell layer (arrowhead) and neurons of in the inner nuclear layer; however, not present in the rod photoreceptors containing large telomeric foci (red), scale: 10 µm. B. Higher magnification of box selected region in panel A highlighting the Cy3‐labeled telomeres (red) and large telomeric foci (arrows). C. Higher magnification of box selected region panel A, omitting telomere signals, showing no PML nuclear bodies in focus‐positive rod photoreceptor cells (panel B) and no PML nuclear bodies in any photoreceptor cells. For all images, DNA is stained with DAPI (blue).
The prevalence of focus‐positive rod photoreceptor cells is significantly associated with increasing age
In 108 normal retinas obtained at autopsy, covering a wide range of donor ages (new‐borns to 95 years), we did not observe the large, ultra‐bright telomere DNA foci in infants before 6 months of age (n = 21). However, the prevalence of focus‐positive rod cells dramatically increased in children, adolescents and young adults, with an additional gradual increase throughout life (Figure 5). Thus, the prevalence of focus‐positive rod cells was significantly associated with increasing age (Spearman R, 0.73, P < 0.001). In the oldest decades, most people have either medium or high levels of focus‐positive rods cells. Accounting for relative differences in rod cell density, the distribution of the focus‐positive rod cells is relatively homogenous in a given region, and most common in the posterior pole caudal to the anatomic equator of the globe, with diminished numbers of focus‐positive rod cells in the retinal extreme periphery. Additionally, the frequency of focus‐positive rod cells was consistent between eyes from the same individual. Finally, we did not observe obvious evidence of diminished cell numbers, pyknotic nuclear debris or apoptotic bodies in the cases with high levels of focus‐positive rod cells.
Figure 5.
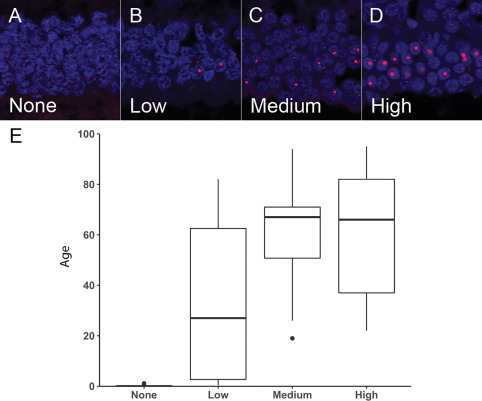
Prevalence of focus‐positive rod cells significantly increases with age. Specimens were scored as having (A) none, (B) low, (C) medium or (D) high levels of focus‐positive rod cells in the outer photoreceptor nuclear layer. E. Box plot showing the distribution of age for the different categories (none, low, medium or high) of focus‐positive rod cells in the normal retinas. The median values are represented as dark horizontal lines. Telomeres are labeled with a Cy3‐labeled telomere‐specific FISH probe (red) and DNA is stained with DAPI (blue).
Focus‐positive rod cells are still present in patients with inherited short telomere syndrome and are not associated with protein loss of ATRX or DAXX
Retinas of two adult patients over the age of 60, with an inherited short telomere syndrome caused by reduced telomerase function, were examined and both displayed widespread focus‐positive rod cells, suggesting that, as in cancer‐associated ALT, this retinal phenotype is independent of telomerase activity. To further address the molecular basis of the large, ultra‐bright telomere DNA foci, we examined expression of ATRX and DAXX, chromatin remodelling proteins that are frequently mutated or functionally lost in cancers associated with ALT 16, 22. Immunostaining for ATRX and DAXX revealed uniform expression in the posterior, peripheral and extreme peripheral regions of the retina in rod photoreceptors, as well as other retina cell neurons (ie, ganglion cells, amacrine cells, bipolar cell neurons or cone photoreceptors).
The focal telomere DNA expansion phenotype may be specific to humans
To determine if this rod‐specific focal telomere DNA expansion phenotype was a general phenomenon, we analyzed retinal neurons from numerous other mammals of varying ages, including rodents, cat, dog, sheep, rabbit, pig and macaque (Figure 6). However, we did not observe the presence of large, ultra‐bright telomere DNA foci in any of the photoreceptor cells, suggesting this phenotype may be specific to humans. To determine if these large, ultra‐bright telomere DNA foci could be visualized in 3D retinal organoids with functional photoreceptors derived from human induced pluripotent stem cells (hiPSC) in vitro 32, we assessed retinal organoids at different time intervals. However, even after 65 weeks in culture, the retinal organoids did not display the focal telomere DNA expansion phenotype (Figure 7), suggesting other factors such as light exposure or longer periods of aging may contribute to their formation.
Figure 6.
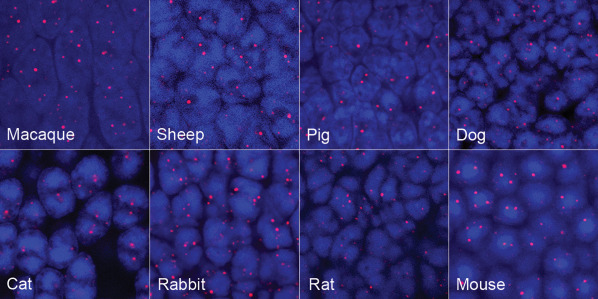
Telomere‐specific FISH across a variety of different animal retinal photoreceptor layer. While telomere‐FISH labeling demonstrates variation in intensity, size and shape across several animal species, no evidence of single, large, ultra‐bright telomere foci are identified. Of note, although only the photoreceptor nuclei are presented here, the telomere FISH pattern for each animal was similar in the other retina cell neurons (ie, ganglion cells, amacrine cells, bipolar cell neurons or cone photoreceptors).
Figure 7.
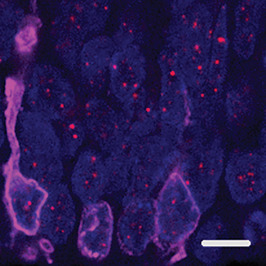
Telomere‐specific FISH in functional retinal organoids do not show large, ultra‐bright telomere foci. Induced pluripotent stem cells (iPS) grown in culture for 65 weeks and differentiated into full thickness functional retinal organoids. Combination of Red/Green and Blue opsin labeling of the cone photoreceptors (magenta) is shown at the edge of the outer nuclear layer of photoreceptor cells. DNA is stained with DAPI (blue); scale: 5 µm.
Prevalence of focus‐positive rod cells significantly increases in adults with glaucoma or diabetic retinopathy
To test for associations between the prevalence of focus‐positive rod cells and diseases that impact the retina, we assessed adult (N = 79; age range: 18–92) cases with pathological conditions, including 29 glaucoma, 38 diabetic retinopathy and 16 age‐related macular degeneration cases (Table 1). Diseased cases were limited to adults, and for comparison, the distribution of normal retinas was also limited to adults (18 years and older). Focus‐positive rod cells were prominent in these conditions, particularly in the glaucoma cases, which were more frequently categorized as having a high level of focus‐positive rod cells (83%; 24/29 cases) as compared to the normal cases (44%; 21/79 cases). Similarly, focus‐positive rod cells more prevalent in the diabetic retinopathy cases (55%; 21/38 cases) compared to the normal group. As shown in Table 1, compared to the normal group and after adjusting for age, logistic regression modeling revealed increased odds of falling in the high category of focus‐positive rod cells for the glaucoma (OR = 11.90, P < 0.0001) and diabetic retinopathy (OR = 2.87, P < 0.01) groups. Similarly, age‐related macular degeneration cases also demonstrated high levels of focus‐positive rod cells (63%; 10/16 cases); however, the increase in focus‐positive rod cells over the high baseline in these older patients (74–92 years) was not significant (OR = 2.65, P = 0.1). Taken together, these associations suggest that the presence of focus‐positive rod cells either contribute to, or result from, aging and disease in the retina.
Table 1.
Association * of tissue type with the high category of focus‐positive rod cells among adults.
| N | High (%) | OR | (95% CI) | P‐value | |
|---|---|---|---|---|---|
| Tissue type | |||||
| Normal | 79 | 21 (44%) | 1.00 | (Ref) | |
| AMD | 16 | 10 (63%) | 2.65 | (0.78–8.92) | 0.1 |
| Diabetic retinopathy | 38 | 21 (55%) | 2.87 | (1.25–6.58) | 0.01 |
| Glaucoma | 29 | 24 (83%) | 11.90 | (3.92–36.12) | <0.0001 |
*Logistic regression modeling odds of falling into the high category of focus‐positive rod cells adjusting for age.
Discussion
We have identified and characterized a dramatic telomere alteration associated with aging and several diseases affecting the retina. These unique telomere DNA alterations may be linked to the long‐lived post‐mitotic nature of human photoreceptor neurons, as well as their high metabolic demands and/or continuous exposure to light‐induced DNA damage or oxidative stress 11. The fact that focus‐positive rod cells are not observed early in life and increase progressively over many years would be consistent with an environmental role in their formation. This concept is supported by the observation that focus‐positive rod cells are most common in the posterior portions of the retina, and thereby exposed to the greatest amount of light. A recent study measuring relative telomere length in several ocular tissues found retinal pigment epithelial cell and corneal cell telomere lengths were significantly shorter than neural retinal cells, thereby suggesting that neural retinal cells possess antioxidant resistance to telomere erosion 13. Our observations suggest that, specifically the rod photoreceptors, are the cell type containing increased telomere content compared to other cell types examined and the presence of the large, ultra‐bright telomere foci may be related to light induced DNA damage or oxidative stress. In addition, recent studies have shown irradiated cells that induce increased oxidative damage, also display DNA breaks that are preferentially localized at telomeric regions, regardless of telomerase activity 20. While these photoreceptor cells are undergoing excessive exposure to light‐induced DNA damage or oxidative stress, they likely have persistent DNA damage products localized to telomeric regions; although it remains unclear why these large, ultra‐bright telomere DNA foci are seen only in post‐mitotic rod photoreceptors. Thus, future experiments will need to be conducted using the retinal organoids as a model system to further evaluate the potential role of DNA damage and/or oxidative stress in the formation of these ultra‐bright telomere DNA foci.
Glaucoma and diabetes are known to stress retinal photoreceptors 4, 15, 21, 28, and this could relate to the accelerated accumulation of focus‐positive rod cells in these conditions as compared to age‐adjusted controls. Interestingly, this focal telomere DNA expansion phenotype does not correlate with the previous finding that rod cells from nocturnal animals have a unique inverted pattern of heterochromatin, in contrast to diurnal animals, which display a conventional heterochromatic pattern where the nuclear periphery is enriched for heterochromatin and euchromatin resides toward interior of the nucleus 30, 31. Thus, it remains unclear why only rod cells display these large, ultra‐bright telomere DNA foci and it does not appear to exist in other vertebrate species.
A recent study in Saccharomyces cerevisiae demonstrated a metabolically induced spatial reorganization of genomic chromatin in quiescent cells 14. These cells, able to sustain viability and respiration over an extended duration, were shown to form telomeric hyperclusters on induced carbon source starvation. More recently, studies on telomere maintenance and stability in non‐dividing, quiescent fission yeast cells have observed that eroded telomeres have high levels of rearrangements. Interestingly, these rearrangements are characterized by increased homologous recombination that correspond to duplications of subtelomeric regions 23. Furthermore, TERRA (telomeric repeat‐containing RNA) is increased in post‐mitotic fission yeast with short telomeres and correlates with these subtelomere rearrangements 23. While telomere clustering is unlikely causing the formation of the large, ultra‐bright telomere DNA foci we observe in human rod photoreceptors, these foci may represent rearrangements in these post‐mitotic cells undergoing DNA damage that are in many ways similar to observations in fission yeast; thus, further study is required to elucidate their function(s) and mechanism of formation.
In summary, we identify and characterize a large, ultra‐bright telomere DNA foci pattern that is restricted to human rod photoreceptors and not observed in other cell types within the retina or adult brain of varying ages. This unique focal telomere DNA expansion phenotype is associated with aging and diseases affecting the retina.
Acknowledgment
We thank Joseph Mankowski and Georg Furtmueller (Johns Hopkins); Leandro Teixeira and Gillian Shaw (Comparative Ocular Pathology Laboratory of Wisconsin); and Elizabeth Buckles and Nancy Lorr (Cornell) for providing archived animal specimens. Unrestricted grant to the Wilmer Eye Institute from Research to Prevent Blindness.
Contributor Information
Charles G. Eberhart, Email: ceberha@jhmi.edu.
Christopher M. Heaphy, Email: cheaphy@jhmi.edu.
References
- 1. Azzalin CM, Lingner J (2015) Telomere functions grounding on TERRA firma. Trends Cell Biol 25:29–36. [DOI] [PubMed] [Google Scholar]
- 2. Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318:798–801. [DOI] [PubMed] [Google Scholar]
- 3. Bae NS, Baumann P (2007) A RAP1/TRF2 complex inhibits nonhomologous end‐joining at human telomeric DNA ends. Mol Cell 26:323–334. [DOI] [PubMed] [Google Scholar]
- 4. Benoist d'Azy C, Pereira B, Chiambaretta F, Dutheil F (2016) Oxidative and anti‐oxidative stress markers in chronic glaucoma: a systematic review and meta‐analysis. PLoS One 11:e0166915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blackburn EH (1991) Structure and function of telomeres. Nature 350:569–573. [DOI] [PubMed] [Google Scholar]
- 6. Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR (1995) Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J 14:4240–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burridge PW, Thompson S, Millrod MA, Weinberg S, Yuan X, Peters A et al (2011) A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One 6:e18293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cesare AJ, Reddel RR (2010) Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet 11:319–330. [DOI] [PubMed] [Google Scholar]
- 9. d'Adda di Fagagna F, Teo SH, Jackson SP (2004) Functional links between telomeres and proteins of the DNA‐damage response. Genes Dev 18:1781–1799. [DOI] [PubMed] [Google Scholar]
- 10. de Lange T (2009) How telomeres solve the end‐protection problem. Science 326:948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deneris ES, Hobert O (2014) Maintenance of postmitotic neuronal cell identity. Nat Neurosci 17:899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dilley RL, Verma P, Cho NW, Winters HD, Wondisford AR, Greenberg RA (2016) Break‐induced telomere synthesis underlies alternative telomere maintenance. Nature 539:54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Drigeard Desgarnier MC, Zinflou C, Mallet JD, Gendron SP, Methot SJ, Rochette PJ (2016) Telomere length measurement in different ocular structures: a potential implication in corneal endothelium pathogenesis. Invest Ophthalmol Vis Sci 57:5547. [DOI] [PubMed] [Google Scholar]
- 14. Guidi M, Ruault M, Marbouty M, Loiodice I, Cournac A, Billaudeau C et al (2015) Spatial reorganization of telomeres in long‐lived quiescent cells. Genome Biol 16:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guzman DC, Olguin HJ, Garcia EH, Peraza AV, de la Cruz DZ, Soto MP (2017) Mechanisms involved in the development of diabetic retinopathy induced by oxidative stress. Redox Rep 22:10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C et al (2011) Altered telomeres in tumors with ATRX and DAXX mutations. Science 333:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heaphy CM, Subhawong AP, Hong SM, Goggins MG, Montgomery EA, Gabrielson E et al (2011) Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol 179:1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Henson JD, Neumann AA, Yeager TR, Reddel RR (2002) Alternative lengthening of telomeres in mammalian cells. Oncogene 21:598–610. [DOI] [PubMed] [Google Scholar]
- 19. Henson JD, Hannay JA, McCarthy SW, Royds JA, Yeager TR, Robinson RA et al (2005) A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin Cancer Res 11:217–225. [PubMed] [Google Scholar]
- 20. Hewitt G, Jurk D, Marques FD, Correia‐Melo C, Hardy T, Gackowska A et al (2012) Telomeres are favoured targets of a persistent DNA damage response in ageing and stress‐induced senescence. Nat Commun 3:708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Izzotti A, Bagnis A, Sacca SC (2006) The role of oxidative stress in glaucoma. Mutat Res 612:105–114. [DOI] [PubMed] [Google Scholar]
- 22. Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T et al (2012) Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet 8:e1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maestroni L, Audry J, Matmati S, Arcangioli B, Geli V, Coulon S (2017) Eroded telomeres are rearranged in quiescent fission yeast cells through duplications of subtelomeric sequences. Nat Commun 8:1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meeker AK, Gage WR, Hicks JL, Simon I, Coffman JR, Platz EA et al (2002) Telomere length assessment in human archival tissues: combined telomere fluorescence in situ hybridization and immunostaining. Am J Pathol 160:1259–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD et al (1988) A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA 85:6622–6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O'Sullivan RJ, Karlseder J (2010) Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol 11:171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pickett HA, Reddel RR (2015) Molecular mechanisms of activity and derepression of alternative lengthening of telomeres. Nat Struct Mol Biol 22:875–880. [DOI] [PubMed] [Google Scholar]
- 28. Roy S, Kern TS, Song B, Stuebe C (2017) Mechanistic insights into pathological changes in the diabetic retina: implications for targeting diabetic retinopathy. Am J Pathol 187:9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shay JW, Bacchetti S (1997) A survey of telomerase activity in human cancer. Eur J Cancer 33:787–791. [DOI] [PubMed] [Google Scholar]
- 30. Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T et al (2009) Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137:356–368. [DOI] [PubMed] [Google Scholar]
- 31. Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M et al (2013) LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152:584–598. [DOI] [PubMed] [Google Scholar]
- 32. Zhong X, Gutierrez C, Xue T, Hampton C, Vergara MN, Cao LH et al (2014) Generation of three‐dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun 5:4047. [DOI] [PMC free article] [PubMed] [Google Scholar]


