Abstract
Adult glioblastomas, IDH‐wildtype represent a heterogeneous group of diseases. They are resistant to conventional treatment by concomitant radiochemotherapy and carry a dismal prognosis. The discovery of oncogenic gene fusions in these tumors has led to prospective targeted treatments, but identification of these rare alterations in practice is challenging. Here, we report a series of 30 adult diffuse gliomas with an in frame FGFR3‐TACC3 oncogenic fusion (n = 27 WHO grade IV and n = 3 WHO grade II) as well as their histological and molecular features. We observed recurrent morphological features (monomorphous ovoid nuclei, nuclear palisading and thin parallel cytoplasmic processes, endocrinoid network of thin capillaries) associated with frequent microcalcifications and desmoplasia. We report a constant immunoreactivity for FGFR3, which is a valuable method for screening for the FGFR3‐TACC3 fusion with 100% sensitivity and 92% specificity. We confirmed the associated molecular features (typical genetic alterations of glioblastoma, except the absence of EGFR amplification, and an increased frequency of CDK4 and MDM2 amplifications). FGFR3 immunopositivity is a valuable tool to identify gliomas that are likely to harbor the FGFR3‐TACC3 fusion for inclusion in targeted therapeutic trials.
Keywords: CD34, FGFR3, glioblastoma, fusion, IDH‐wildtype, TACC3
Introduction
Diffuse gliomas harbor a high degree of histologic, genetic and prognostic heterogeneity, making their diagnosis and clinical management difficult. Recently, the WHO 2016 edition improved their classification by an integrated diagnosis: this new method requires the combination of histological and genetic features to characterize tumor types with higher prognostic and predictive values 15. Integrated diagnosis of “glioblastoma, IDH‐wildtype” (GBM, IDHwt) corresponds to a resistant‐to‐treatment disease with a dismal prognosis and standard of care that includes concomitant radiochemotherapy. However, GBM, IDHwt is only a negative molecular definition, which requires the absence of mutations of the IDH1 and IDH2 genes as well as the absence of the K27M mutation in genes encoding Histone H3. Most frequent genetic alterations of this group are not constant (co‐occurrence of 7p gain and 10q loss, EGFR amplification or TERT promoter mutation), and GBM, IDHwt presents other highly variable genetic alterations, with a rare prevalence for each one 1, 24. More detailed molecular characterization has no impact on the standard of care, but opens the potential for clinical trials to assess targeted therapies at tumor relapse 2, 10, 21. Because of their rarity and high variability, detection of oncogenic drivers is challenging. Some histological variants of GBM, IDHwt are associated with a higher prevalence of specific drugable oncogenic drivers. Such variants are opportunities for pathologists to detect a potential therapeutic target. For example, the BRAF p.V600E mutation is rare in GBM, IDHwt but has a high prevalence in the variant epithelioid GBM 12.
Massive parallel sequencing allowed the identification of the FGFR3‐TACC3 gene fusion as a rare alteration in IDH‐wildtype diffuse gliomas of histological grade II, III and IV according to the WHO 2007 4, 23. The fusion point of FGFR3‐TACC3 varies from case to case and results in the production of a chimeric protein with the kinase domain of FGFR3 and coiled‐coil domain of TACC3. The fusion protein presents hyperphosphorylation and constitutive activation of the kinase domain 16, 23. The fusion protein is an oncogenic driver in vitro and in mouse models. Its oncogenic activity requires the kinase activity of FGFR3. Inhibitors of kinase activity can block tumor growth in a preclinical model of gliomas with the TACC3‐FGFR3 fusion. Two cases of glioblastoma with the FGFR3‐TACC3 fusion treated by FGFR3 inhibitors showed arrest of tumor growth 4. In spite of its rarity, the FGFR3‐TACC3 fusions open new therapeutic prospectives for selected patients 3.
Here, we describe the recurrent histological and immunohistochemical features of gliomas with the FGFR3‐TACC3 fusion, which can be considered to be a new histomolecular variant of GBM, IDHwt with diagnostic, and potential therapeutic impacts.
Patients and Methods
Patients
A multicentric series of 30 patients with diffuse gliomas containing the FGFR3‐TACC3 fusion was studied. All cases included an in‐frame FGFR3‐TACC3 fusion containing the tyrosine kinase domain of FGFR3. Eleven cases were previously reported 4. A control monocentric series of 30 consecutive GBM, IDHwt cases without the FGFR3‐TACC3 fusion was studied to assess the specificity of the histopathological features. A control group of 11 adult GBM, IDHwt patients without the FGFR3‐TACC3 fusion were matched according to the age and sex ratio for 11 GBM, IDHwt patients with the FGFR3‐TACC3 fusion to compare the vascular density. A monocentric prospective series of 256 IDH‐wildtype adults with diffuse gliomas was screened for the FGFR3‐TACC3 fusion by RT‐PCR and tested by FGFR3 immunohistochemistry. The study was approved by the appropriate institutional research ethics committee and was performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki.
Pathology, immunohistochemistry, in situ hybridization
FB, DM and KM reviewed the histological features and integrated diagnosis according to WHO 2016. Formalin‐fixed paraffin‐embedded tissue sections were processed for deparaffinization and immunolabeling by a fully automatic immunohistochemistry system, the Ventana benchmark XT System (Roche, Basel, Switzerland), using a streptavidin‐peroxidase complex with diaminobenzidine as the chromogen. The references and dilutions of the primary antibodies are given in Table 1. FB and KM reviewed the FGFR3 immunolabeling and evaluated the maximal intensity (low, moderate or high) and percentage of positive tumor cells among the total tumor cells of the whole section. EGFR immunolabeling was evaluated by a score ranging from 0 to 400 as adapted from Hirsch et al 8. To quantify the vascular density, only cases with a sufficient surface of the histological section were selected (n = 11 gliomas with FGFR3‐TACC3 fusion and n = 11 gliomas without FGFR3‐TACC3 fusion, matched according to criteria described in Patients paragraph). The three tumor areas with the highest vascular density were selected in each tumor independently from the tumor cell density. Microphotography of CD34 immunolabeling was acquired at high power fields (X400) for the three areas. The total number of vascular sections in the three pictures was obtained by visual count. The whole process of quantification was performed blind to the presence or absence of the FGFR3‐TACC3 fusion. Formalin‐fixed paraffin‐embedded tissue sections were processed for deparaffinization and dual in situ hybridization by a fully automatic immunohistochemistry system, the Ventana benchmark XT System (Roche, Basel, Switzerland), using an MDM2 probe and centromeric probe of chromosome 12 (Roche, Basel, Switzerland).
Table 1.
List of primary antibodies used for immunohistochemistry.
| Antigen | Species | Dilution | Reference | Provider |
|---|---|---|---|---|
| ATRX | rabbit polyclonal | 1/200 | HPA‐001906 | Sigma |
| CD34 | monoclonal mouse QBEnd 10 | 1/50 | M7165 | Dako |
| EMA | mouse monoclonal E29 | prediluted | IS62930 | Dako |
| EGFR | mouse monoclonal | 1/200 | NCL‐L‐EGFR‐384 | Leica |
| FGFR3 | mouse monoclonal | 1/100 | sc‐13121 | Santa Cruz Biotech. |
| GFAP | mouse monoclonal 6F2 | 1/500 | M076101 | Dako |
| IDH1 R132H | mouse monoclonal H09 | 1/50 | DIA‐H09 | Dianova |
| INA | mouse monoclonal 2E3 | 1/100 | NB300‐140 | Novus Biologicals |
| Ki67 | mouse monoclonal MIB‐1 | 1/50 | M724001 | Dako |
| OLIG2 | rabbit monoclonal EP112 | 1/1000 | AC‐0106RUO | Epitomics |
| P53 | mouse monoclonal DO‐7 | 1/100 | M700101 | Dako |
Genetic analysis
In‐frame FGFR3‐TACC3 fusions were detected by reverse transcription and PCR of the fusion breakpoint followed by sequencing of the PCR products 4. Total RNA was extracted from frozen tissues using Trizol (Invitrogen) according to the manufacturer's instructions. Two‐ to three‐hundred nanograms of total RNA was retro‐transcribed with the Maxima First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, USA) or SuperScript II (Thermo Scientific, Waltham, USA). RT‐PCR was performed using AccuPrime Taq DNA Polymerase (Thermo Scientific, Waltham, USA). The primer pairs used for the FGFR3‐TACC3 fusions screening were: FGFR3ex12‐FW: 5′‐CGTGAAGATGCTGAAAGACGATG‐3′ and TACC3ex14‐RV: 5′‐AAACGCTTGAAGAGGTCGGAG; the amplification conditions were 94°C–3 min, (94°C–30 sec/61°C–30 sec/68°C–1 min 40 sec) for 35 cycles, 68°C–7 min. The PCR products were subjected to Sanger sequencing.
The mutational status of the IDH1 and IDH2 genes, and of the TERT promoter was analyzed, as well as copy number variations (CNV). IDH1 and IDH2 gene mutations were screened by Sanger sequencing 22. TERT promoter hotspot mutations were detected by Sanger sequencing 14.
Targeted gene capture followed by sequencing with parallel next‐generation sequencing for IDH1, IDH2 and TERT promoter mutations; EGFR, CDK4 and MDM2 amplifications; p16 deletions; and chromosomal gains and losses was performed in 7 samples. CNV analyses were performed by a SNP array, using Illumina Omni in 14 samples 6. Array processing was outsourced to Integragen. Raw copy numbers were estimated for each of the SNPs and copy‐number markers. The bio‐discovery property SNP‐FASST2 algorithm was then used to segment the copy number data. Segments were mapped to the hg18 genome assembly 17. The CNV magnitudes, called the log‐R ratio (LRR), were classified (deletion, loss, gain, or amplification) using simple thresholds according to the default Nexus 7.5 software.
Statistical analysis
The quantitative variables are shown as the mean ± standard deviation. The nominal variables were tested with the chi‐square test. The quantitative variables were tested by the Mann‐Whitney test if n < 30 and by the t test if n ≥ 30. The significance level was P < 0.05 according to the two‐sided test.
Results
Clinical features of diffuse gliomas with the FGFR3‐TACC3 fusion
The series gathered 30 gliomas with in‐frame FGFR3‐TACC3 fusions (Figure 1, Tables 2 and 3). The most frequent fusions were FGFR3 exon 17 fused to exon 10 of TACC3 (n = 9, 30%) or fused to exon 11 of TACC3 (n = 13, 43%). The sex ratio was 1:1. The mean age at diagnosis was 62 y ±12 (median age 62 y, range 42–87). All cases were treatment‐naive supratentorial tumors without a medical history of irradiation. The frontal (n = 11) and parietal (n = 11) lobes were the most frequently involved, followed by the temporal (n = 10), and occipital (n = 7) lobes. Seventy percent of surgical samples were resections, and 30% were biopsies.
Figure 1.

Histological, immunohistochemical and molecular features of gliomas with FGFR3‐TACC3 fusion Abbreviations: AII IDHwt = diffuse astrocytoma, IDH‐wildtype grade II; endoc. vasc. network, endocrinoid vascular network; GBM IDHwt = glioblastoma IDH‐wildtype; microvascular prolif. = microvascular proliferation; RMF = recurrent morphological features (monomorphous ovoid nuclei, endocrinoid network of thin capillaries, nuclear palisading, attachment of tumor cells to vessels by equidistant thin parallel cytoplasmic processes producing vague pseudorosettes).
Table 2.
Clinical and radiological features of diffuse gliomas with FGFR3‐TACC3 fusion.
| # | Sex | age | Biopsy or Resection | Tumor side | Tumor site | Type of FGFR3 – TACC3 fusion | Series |
|---|---|---|---|---|---|---|---|
| 1 | M | 50 | Resection | R | Parieto‐occipital | EXON17 – EXON8 | Di Stefano et al. 4 |
| 2 | M | 53 | Resection | R | Parieto‐occipital | EXON17 – EXON6 | Di Stefano et al. 4 |
| 3 | M | 58 | Resection | R | Fronto‐temporal | EXON17 – EXON10 | Present study |
| 4 | M | 58 | Resection | L | Parieto‐occipital | EXON17 – EXON8 | Di Stefano et al 4 |
| 5 | M | 75 | Resection | R | Parieto‐occipital | EXON17 – EXON11 | Di Stefano et al 4 |
| 6 | F | 66 | Resection | L | Frontal | EXON17 – EXON11 | Di Stefano et al 4 |
| 7 | F | 79 | Resection | R | Frontal | EXON17 – EXON13 | Present study |
| 8 | M | 60 | Biopsy | R | Frontal | EXON17 – EXON10 | Di Stefano et al 4 |
| 9 | F | 78 | Biopsy | L | Temporal | EXON17 – EXON7 | Di Stefano et al 4 |
| 10 | F | 60 | Resection | L | Frontal | EXON17 – EXON11 | Present study |
| 11 | F | 60 | Biopsy | L | Parietal | EXON17 – EXON8 | Di Stefano et al 4 |
| 12 | M | 76 | Resection | R | Parietal | EXON17 – EXON11 | Present study |
| 13 | M | 64 | Biopsy | R | Frontal | EXON17 – EXON11 | Present study |
| 14 | M | 61 | Resection | L | Frontal | EXON17 – EXON11 | Present study |
| 15 | F | 87 | Resection | R | Parietal | EXON17 – EXON10 | Present study |
| 16 | M | 64 | Resection | R | Temporal | EXON17 – EXON11 | Present study |
| 17 | M | 46 | Resection | L | Frontal | EXON17 – EXON11 | Present study |
| 18 | M | 46 | Resection | L | Parieto‐occipital | EXON17 – EXON10 | Present study |
| 19 | F | 64 | Biopsy | R | Temporal | EXON17 – EXON11 | Present study |
| 20 | M | 35 | Resection | R | Temporal | EXON17 – EXON10 | Present study |
| 21 | F | 44 | Resection | L | Frontal | EXON17 – EXON11 | Present study |
| 22 | M | 42 | Resection | R | Frontal | EXON17 – EXON11 | Present study |
| 23 | F | 43 | Biopsy | R | Temporal | EXON18 – EXON13 | Present study |
| 24 | F | 68 | Resection | L | Temporo‐parietal | EXON17 – EXON11 | Present study |
| 25 | F | 77 | Biopsie | L | Parieto‐occipital | EXON17 – EXON10 | Present study |
| 26 | F | 63 | Resection | R | Temporal | EXON17 – EXON10 | Present study |
| 27 | M | 75 | Biopsy | R | Parieto‐occipital | EXON17 – EXON11 | Present study |
| 28 | F | 74 | Biopsy | L | Frontal | EXON17 – EXON10 | Di Stefano et al 4 |
| 29 | F | 59 | Resection | L | Temporal | EXON18 – EXON5 | Di Stefano et al 4 |
| 30 | F | 72 | Resection | R | Temporal | EXON17 – EXON10 | Di Stefano et al 4 |
Table 3.
Main histological, immunohistochemical and molecular features of diffuse gliomas with FGFR3‐TACC3 fusion.
| Features | Diffuse gliomas with FGFR3‐TACC3 fusion |
|---|---|
| N | 30 |
| M:F | 1:1 |
| Mean age at diagnosis | 62y ±12 (range 42–87 y) |
| Resection/Biopsy | 21/9 (70%)/(30%) |
| Localization | |
| Right/left side | 18 (60%)/12 (40%) |
| Frontal | 10 (33%) |
| Temporal | 8 (27%) |
| Parieto‐occipital | 7 (23%) |
| Parietal | 3 (10%) |
| Temporo‐parietal | 1 (3%) |
| Fronto‐temporal | 1 (3%) |
| Tumor type | |
| Glioblastoma, IDH‐wildtype grade IV | 25 (83%) |
| Gliosarcoma, IDH‐wildtype grade IV | 1 (3%) |
| Glioblastoma, NOS grade IV | 1 (3%) |
| Diffuse astrocytoma, IDH‐wildtype grade II | 3 (10%) |
| Histological features | |
| Recurrent morphological features* | 22 (73%) |
| Monomorphous ovoid nuclei | 26 (87%) |
| Calcifications | 17 (57%) |
| Desmoplastic changes | 15 (50%) |
| Endocrinoid vascular network | 26 (87%) |
| Immunohistochemical features | |
| IDH1 R132H positivity | 0/28 (0%) |
| ATRX loss of expression | 0/28 (0%) |
| P53 | 4/28 (14%) |
| Mean Ki67 index grade II | 3% ±2 |
| Mean Ki67 index grade IV | 16% ±11 |
| CD34 | 16/29 (55%) |
| EGFR score (from 0 to 400) | 131 ± 93 (n = 29) |
| EMA | 16/29 (55%) |
| Molecular features | |
| IDH1 or IDH2 mutation | 0/28 (0%) |
| TERT promoter mutation | 17/23 (74%) |
| 1p/19q codeletion | 0/26 (0%) |
| 7p gain and 10q loss | 16/25 (64%) |
| 10q loss, 13q loss, 14q loss | 2/22 (9%) |
| EGFR amplification | 0/29 (0%) |
| MDM2 amplification | 5/26 (19%) |
| CDK4 amplification | 5/26 (10%) |
| P16 deletion | 11/26 (42%) |
* Recurrent morphological features are monomorphous ovoid nuclei, endocrinoid network of thin capillaries, nuclear palisading, attachment of tumor cells to vessels by equidistant thin parallel cytoplasmic processes producing vague pseudorosettes.
Histopathological features of diffuse gliomas with the FGFR3‐TACC3 fusion
The 30 tumors were classified according to WHO 2016 (Figure 1, Table 3). Twenty five cases were GBM, IDHwt, and one case was a gliosarcoma, IDH‐wildtype. One case was GBM, not otherwise specified. Two cases were diffuse astrocytoma, IDH‐wildtype with molecular features of GBM, IDH‐wildtype (7p gain, 10q loss, mutation of TERT promoter). One case was a diffuse astrocytoma, IDH‐wildtype grade II without analysis of copy number variations (CNV). All of the 27 high‐grade cases showed microvascular proliferation, 74% showed necrosis (n = 24/27) and 67% showed thrombosis (n = 18/27).
The histological review revealed recurrent morphological features (RMF). RMF were diffusely present in grade II cases. RMF were restricted to some tumor areas devoid of microvascular proliferation and necrosis in glioblastoma (Figure 2, Supporting Information Figure S1). RMF associated: monomorphous ovoid nuclei, endocrinoid network of thin capillaries, nuclear palisading, and attachment of tumor cells to vessels by equidistant thin parallel cytoplasmic processes producing vague pseudorosettes. Tumor cells sometimes organized into pseudo‐ependymal pseudorosettes. Isolated tumor cells infiltrated the neuropile and harbored an ovoid nucleus that was sometimes surrounded by a pale ovoid cytoplasm. They also spread along vessels and organized into perivascular pseudorosettes (Supporting Information Figure S1). In glioblastoma, tumor areas with RMF were associated with different areas that presented with a high cellular density, anisocaryosis, microvascular proliferation and necrosis. The former areas corresponded to grade II/III, and the latter areas presented criteria for grade IV.
Figure 2.
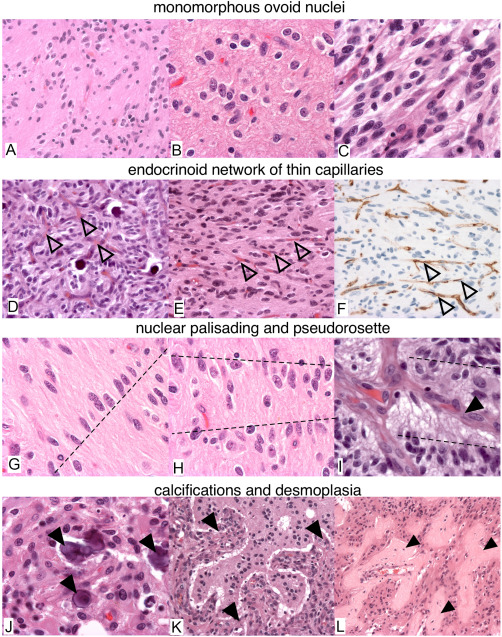
Histological features of gliomas with FGFR3‐TACC3 fusion. (A–E,G–L) H&E. F. CD34 immunostaining. (A,D,E,F,K,L) 100X. (B,C,G–J) 400X. Recurrent morphological features are: monomorphous ovoid nuclei (A–C), endocrinoid network of thin capillaries (open arrowhead in D–F), nuclear palisading (dash lines in G,H). Tumor cells form pseudorosettes with aligned nuclei (dash line in I) and presence of thin cytoplasmic cell processes between tumor nuclei and vessels (arrowhead in I). J. Microcalcifications (arrowheads). (K,L) Desmoplasia (arrowheads).
RMF were present at least focally in 73% (n = 22/30) of tumors. Diffuse gliomas with the FGFR3‐TACC3 fusion frequently presented microcalcifications (57%, n = 17/30), desmoplastic changes (50% n = 15/30), and an endocrinoid vascular network of thin capillaries (which is one of the RMF) 87% (n = 26/30). In resection cases, these features were observed at higher percentages of 57%, 66% and 90%. These features were significantly more present in diffuse gliomas with the FGFR3‐TACC3 fusion than in a control series of GBM, IDHwt without the FGFR3‐TACC3 fusion: RMF (n without fusion = 3/30, P < 10−3), microcalcifications (nwithout fusion=2/30 P < 10−3), desmoplastic changes (n without fusion = 3/30, P = 0.002), and an endocrinoid vascular network of thin capillaries (n without fusion = 11/30 P < 10−3). The mitotic activity was significantly lower (15 per 10 high power field) in GBM, IDHwt with the FGFR3‐TACC3 fusion compared to those without the fusion (19 per 10 HPF, P = 0.028). In one case, desmoplasia was marked and tumor cells were isolated in small islets separated by larger and predominant collagenous bands. Eight samples did not present RMF, but presented various histological aspects of GBM. Quantification confirmed the statistically significant higher vascular density in diffuse gliomas with the FGFR3‐TACC3 fusion compared to the control group of GBM, IDHwt (meanFGFR3‐TACC3 = 817 sections/mm2, meanwithout fusion = 423 sections/mm2, nFGFR3‐TACC3 = 11, n without_fusion = 11, P = 0.045, Mann‐Whitney test, Figure 3). This higher vascular density was observed in diffuse gliomas with the FGFR3‐TACC3 fusion, whereas the mean tumor cell density in quantified areas was lower than in the control group (Supporting Information Figure S2). The higher vascular density was thus not explained by a higher tumor cell density or biased selection of the tumor core versus the tumor infiltrative edge.
Figure 3.
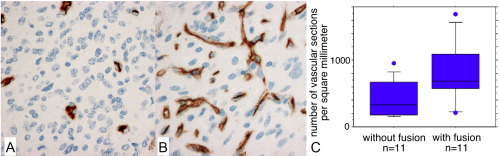
Quantification of vascular density. (A,B). CD34 immunolabeling of a glioblastoma, IDH‐wildtype without FGFR3‐TACC3 fusion (A) or with FGFR3‐TACC3 fusion (B). C. Quantification of the number of CD34 immunopositive vascular sections per square millimeter. Glioblastomas, IDH‐wildtype with FGFR3‐TACC3 fusion had significantly higher vascular density than glioblastomas, IDH‐wildtype without FGFR3‐TACC3 fusion (meanwith fusion=817 sections/mm2, meanwithout fusion=423 sections/mm2, P = 0.045, Mann‐Whitney test).
Immunohistochemical features of diffuse gliomas with the FGFR3‐TACC3 fusion
The constant immunohistochemical features of the series were: negative IDH1 R132H (n = 28), retained ATRX expression (n = 28), broad expression of Olig2 (n = 29) and GFAP (n = 28) (Figure 4, Supporting Information Table S1). The ratio of intense P53 immunolabeling was often low (meanP53=3% ±7), exceeding 10% of tumor cells in only 14% of cases (n = 4/28).
Figure 4.

Immunohistochemical features of gliomas with FGFR3‐TACC3 fusion. (A–F,J–L). 200X. (G,H) 100x. (I) 1000X. (M–O) 20X. A. positive Olig2 immunolabeling. B. positive GFAP immunolabeling. C. P53 intense immunolabeling inferior to the threshold of ten percent of tumor cells. D. stellate extravascular CD34 immunolabeling. E. extravascular CD34 immunolabeling as membrane frames. F. positive ramified EMA immunolabeling. G. Ki67 in a grade II tumor. H. Ki67 in a grade IV tumor. I. chromogenic in situ hybridization for centromere of chromosome 12 (red) and MDM2 (black) showed high copy number amplification in case #8. J. intense FGFR3 immunolabeling. K. moderate FGFR3 immunolabeling. L. weak FGFR3 immunolabeling. M. Diffuse intense FGFR3 immunolabeling. N. Diffuse FGFR3 immunolabeling with heterogeneous intensity from weak to intense. O. Focal FGFR3 immunolabeling and negative areas.
CD34 immunolabeling highlighted the dense branched vascular network vessels. CD34 was expressed by a minority of tumor cells in 55% of cases (n = 16/29) according to two patterns: either ramified cell processes or a frame corresponding to the cytoplasmic membrane. EMA was focally positive as an ill‐defined fibrillary staining in 55% of cases (n = 16/29, Figure 4), but no case presented EMA immunopositive dots, which are commonly observed in ependymomas. The mean score of EGFR expression was 132/400 ±95. The mean Ki67 proliferation index was 3% ±2 in grade II tumors (n = 3) and was 16% ±11 in grade IV tumors (n = 27). Internexin alpha immunolabeling identified a solid tumor area without residual immunopositive axons in 54% of cases (n = 15/28).
FGFR3 immunolabeling was constantly positive, but often heterogeneous. The maximal intensity of FGFR3 labeling was high in 23 cases, moderate in 5 cases and low in 1 case. Intense labeling was associated with areas with RMF. The mean percentage of the FGFR3‐immunoreactive tumor surface was 72% ±29.
Performance of FGFR3 immunolabeling for screening of diffuse gliomas with the FGFR3‐TACC3 fusion
We analyzed 256 IDH‐wildtype diffuse gliomas by FGFR3 immunolabeling and RT‐PCR detection of the FGFR3‐TACC3 fusion. The sensitivity and specificity of FGFR3 immunopositivity for the detection of the FGFR3‐TACC3 fusion was 100% and 92%, respectively. The positive predictive value was 56% and negative predictive value was 100% (n true positive = 24, n false positive = 19, n true negative = 213, n false negative = 0).
Genetic features
We confirmed previously described molecular features of diffuse gliomas with the FGFR3‐TACC3 fusion; 18/21 cases showed CNVs typical of the integrated diagnosis of GBM, IDHwt: 16 cases with 7p+/10q– and 2 cases with 10q–/13q–/14q– (Table 3, Supporting Information Table S2). No EGFR amplification was detected in the 29 tested cases. The TERT promoter mutation was present in 74% of cases (n = 17/23). MDM2 (Figure 3) and CDK4 amplification was found in 19% (5/26) and 19% (5/26), respectively, which is higher than the mean frequency in GBM, IDHwt 24.
Discussion
We present the histopathological features of adult diffuse gliomas with the FGFR3‐TACC3 fusion. Our findings facilitate the diagnostic approach of this peculiar subgroup of diffuse gliomas, IDH‐wildtype, and improve the understanding of their oncogenesis.
Key genetic alterations of diffuse gliomas, co‐occurring or excluding each other, are the basis of the current histomolecular integrated classification. Nevertheless, some groups, such as GBM, IDHwt, remain heterogeneous. On the other hand, entities such as grade II and III astrocytoma, IDH‐wildtype should be better characterized since some of these tumors share molecular features and poor prognosis with GBM, IDH‐wildtype 20. The FGFR3‐TACC3 fusion is an oncogenic driver mutually exclusive with IDH mutation and EGFR amplification 4 and mainly occurs in GBM. Interestingly, three cases of our series had a low grade diffuse astrocytoma histology but presented a CNV similar to those of GBM, IDHwt. These findings support the similarities between some diffuse astrocytomas, IDH‐wildtype and GBM, IDHwt. The median age of GBM, IDH‐wildtype with the FGFR3‐TACC3 fusion (62 y) is the same as that of other forms of GBM, IDH‐wildtype 15. However, the age distribution was wide, from 35 to 87 years. We observed a balanced sex ratio, in contrast to the female predominance previously reported in a smaller cohort 7. The occurrence of the FGFR3‐TACC3 fusion in the pediatric population is unknown because only tumors of adult patients were investigated.
We identified recurrent morphological features (RMF) associated with frequent microcalcifications and desmoplasia. These histological findings in a diffuse glioma IDH‐wildtype should prompt pathologists to consider the FGFR3‐TACC3 fusion and look for additional genetic alterations that are required for the diagnosis of GBM, IDHwt. Some features of the RMF are shared with other tumors, representing a challenging differential diagnosis: (i) nuclear monomorphism, endocrinoid vascular network, and microcalcifications are also present in oligodendrogliomas, IDH‐mutant and 1p/19q co‐deleted, (ii) perivascular pseudorosettes are shared with ependymomas, astroblastomas, angiocentric gliomas and (iii) solid tumor areas, desmoplasia and CD34 ramified labeling are shared with glioneuronal tumors (eg gangliogliomas) 15. One FGFR3‐TACC3 fusion was recently reported in a low‐grade epilepsy associated tumor, called a polymorphous low‐grade neuroepithelial tumor of the young (PLNTY) 9. PLNTY shares some histological features and CD34 immunopositivity with those that we observed in GBM, IDHwt with FGFR3‐TACC3. However, the tumors of our series had an evolution of high grade diffuse gliomas and did not correspond to PLNTY.
FGFR3‐TACC3 fusions are typically IDH1 R132H negative, ATRX retained, OLIG2 positive, and GFAP positive, with low to moderate EGFR positivity, mostly negative P53, and are frequently CD34 positive. FGFR3 immunolabeling can be used to predict the presence of the FGFR3‐TACC3 fusion, with high specificity and sensitivity. No case with the FGFR3‐TACC3 fusion was FGFR3 negative, making immunohistochemistry a simple and reliable method for systematic screening of IDH‐wildtype diffuse gliomas. We thus confirmed in a larger cohort the recently reported high sensitivity of FGFR3 immunolabeling 7. The most intense FGFR3 immunostaining often highlighted tumor areas with the RMF, and such tumors have a high probability to harbor a FGFR3‐TACC3 fusion. Noteworthy, histological diagnosis of diffuse gliomas is a preliminary condition to analyze FGFR3 labeling. FGFR3 immunopositivity should not be considered to be a marker that is specific to diffuse gliomas. Moreover, the recurrent morphological features of diffuse gliomas with the FGFR3‐TACC3 fusion are not specific and sensitive enough to identify these tumors, and molecular confirmation is mandatory. FGFR3 immunostaining can be used clinically as a first step of screening to prioritize the tumors that are analyzed in a second molecular step. Confirmation of the FGFR3‐TACC3 fusion requires RT‐PCR followed by sequencing of the in‐frame fusion breakpoint. Direct detection of FGFR3‐TACC3 by RNA‐FISH could be useful 13. Indeed it could become an important theranostic marker if the benefit of FGFR3 inhibitors in these tumors is confirmed by ongoing clinical trials.
Tumor areas with RMF showed the highest immunoreactivity of FGFR3 and could correspond to the highest activation of FGFR3 signaling. FGFR3 is involved in the development of astrocytes through the maintenance of progenitors and specification 5, 18, 19. It is also involved in the activation of astrocytes in reactive gliosis through migration, extension of cytoplasmic processes and induction of GFAP expression 11. The FGFR3‐TACC3 fusion could deregulate the normal function of FGFR3 signaling in astrocytic differentiation and could induce tumor cells to present RMF. Alternatively, RMF are characterized by a rich vascular network and could result from the requirement of the perivascular niche or of oxidative metabolism in FGFR3‐TACC3 fusion‐driven oncogenesis. The RMF was present in all cases with low‐grade histology. In glioblastoma, we observed areas with RMF corresponding to grade II/III (without microvascular proliferation and/or necrosis) as well as areas without RMF and presenting microvascular proliferation and/or necrosis. These findings suggest that RMF reflects the initial step of tumorigenesis in this oncogenic pathway and that tumor progression results in anaplastic areas with microvascular proliferation, necrosis and variable morphological aspects.
Our findings facilitate the diagnostic approach of this peculiar subgroup of diffuse gliomas, IDH‐wildtype. They may become particularly important to ongoing clinical FGFR targeted trial, and open avenues to understand their oncogenesis.
Conflict of Interest
The authors declare that they have no conflict of interest.
Supporting information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
Figure S1. Histological spectrum of diffuse gliomas with FGFR3‐TACC3 fusion. (A,B) Pseudo‐ependymal features with nuclei localized at distance of vessels (A), and a perivascular pseudorosette with thin perivascular cytoplasmic processes (B). (C) Pseudo‐astroblastic perivascular pseudorosette in tumor infiltrative edge. (d) Pseudo‐oligodendroglial tumor cells with clear cytoplasm (arrowheads) form perineuronal satellitosis. (E) Tumor cells with pale eosinophilic cytoplasm (arrowheads). (F) Tumor cells with eosinophilic cytoplasm (arrowheads).
Figure S2. Quantification of vascular density by CD34 immunolabeling. CD34 immunolabeling is showed for eleven gliomas without FGFR3‐TACC3 fusion and eleven gliomas with FGFR3‐TACC3 fusion.
Table S1. Detailed histological and immunohistochemical features of diffuse gliomas with FGFR3‐TACC3 fusion. Abbreviations: CD34 = positive or negative immunolabeling of tumor cells; FGFR3% = percentage of tumor cells with FGFR3 immunolabeling; FGFR3 int. = FGFR3 immunolabeling maximal intensity scored as intense (3), intermediate (2) or weak (1); INA solid tum. = presence of a solid tumor area without INA positive residual axons; INA tum. = negative or positive INA immunolabeling of tumor cells; Ki67 = maximal Ki67 proliferation index; m = maintained expression; Mitos. = Mitoses; MVP = microvascular proliferation; NA = not available; neg = negative; pos = positive.
Acknowledgments
This work was supported by funding from Fondation ARC pour la recherche sur le cancer (PJA 20151203562) and by grants from Institut National du Cancer (grant INCa‐DGOS‐Inserm 6038). ALDS was supported by Premio Riquier. Tissue specimens from the AP‐HM institution were stored and then provided by the AP‐HM tumor bank (authorization number AC‐2013‐1786; CRB number: BB‐0033‐00097).
References
- 1. Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A (2015) Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol 129:829–848. [DOI] [PubMed] [Google Scholar]
- 2. Cloughesy TF, Cavenee WK, Mischel PS (2014) Glioblastoma: from molecular pathology to targeted treatment. Annu Rev Pathol 9:1–25. [DOI] [PubMed] [Google Scholar]
- 3. Costa R, Carneiro BA, Taxter T, Tavora FA, Kalyan A, Pai SA et al (2016) FGFR3‐TACC3 fusion in solid tumors: mini review. Oncotarget 7:55924–55938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Di Stefano AL, Fucci A, Frattini V, Labussiere M, Mokhtari K, Zoppoli P et al (2015) Detection, characterization, and inhibition of FGFR‐TACC fusions in IDH wild‐type glioma. Clin Cancer Res 21:3307–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gaber ZB, Butler SJ, Novitch BG, Polleux F (2013) PLZF regulates fibroblast growth factor responsiveness and maintenance of neural progenitors. PLoS Biol 11:e1001676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gonzalez‐A0067uilar A, Idbaih A, Boisselier B, Habbita N, Rossetto M, Laurenge A et al (2012) Recurrent mutations of MYD88 and TBL1XR1 in primary central nervous system lymphomas. Clin Cancer Res 18:5203–5211. [DOI] [PubMed] [Google Scholar]
- 7. Granberg KJ, Annala M, Lehtinen B, Kesseli J, Haapasalo J, Ruusuvuori P et al (2017) Strong FGFR3 staining is a marker for FGFR3 fusions in diffuse gliomas. Neuro Oncol 19:1206–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hirsch FR, Varella‐Garcia M, Bunn PA, Jr. , Di Maria MV, Veve R, Bremmes RM et al (2003) Epidermal growth factor receptor in non‐small‐cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol 21:3798–3807. [DOI] [PubMed] [Google Scholar]
- 9. Huse JT, Snuderl M, Jones DT, Brathwaite CD, Altman N, Lavi E et al (2017) Polymorphous low‐grade neuroepithelial tumor of the young (PLNTY): an epileptogenic neoplasm with oligodendroglioma‐like components, aberrant CD34 expression, and genetic alterations involving the MAP kinase pathway. Acta Neuropathol 133:417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jue TR, McDonald KL (2016) The challenges associated with molecular targeted therapies for glioblastoma. J Neurooncol 127:427–434. [DOI] [PubMed] [Google Scholar]
- 11. Kang K, Lee SW, Han JE, Choi JW, Song MR (2014) The complex morphology of reactive astrocytes controlled by fibroblast growth factor signaling. Glia 62:1328–1344. [DOI] [PubMed] [Google Scholar]
- 12. Kleinschmidt‐DeMasters BK, Aisner DL, Birks DK, Foreman NK (2013) Epithelioid GBMs show a high percentage of BRAF V600E mutation. Am J Surg Pathol 37:685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kurobe M, Kojima T, Nishimura K, Kandori S, Kawahara T, Yoshino T et al (2016) Development of RNA‐FISH assay for detection of oncogenic FGFR3‐TACC3 fusion genes in FFPE samples. PLoS One 11:e0165109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Labussiere M, Boisselier B, Mokhtari K, Di Stefano AL, Rahimian A, Rossetto M et al (2014) Combined analysis of TERT, EGFR, and IDH status defines distinct prognostic glioblastoma classes. Neurology 83:1200–1206. [DOI] [PubMed] [Google Scholar]
- 15. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK (2016) WHO Classification of Tumours of the Central Nervous System, Revised. 4th update Edition, International Agency for Research on Cancer: Lyon. [Google Scholar]
- 16. Nelson KN, Meyer AN, Siari A, Campos AR, Motamedchaboki K, Donoghue DJ (2016) Oncogenic gene fusion FGFR3‐TACC3 is regulated by tyrosine phosphorylation. Mol Cancer Res 14:458–469. [DOI] [PubMed] [Google Scholar]
- 17. Olshen AB, Venkatraman ES, Lucito R, Wigler M (2004) Circular binary segmentation for the analysis of array‐based DNA copy number data. Biostatistics 5:557–572. [DOI] [PubMed] [Google Scholar]
- 18. Peters K, Ornitz D, Werner S, Williams L (1993) Unique expression pattern of the FGF receptor 3 gene during mouse organogenesis. Dev Biol 155:423–430. [DOI] [PubMed] [Google Scholar]
- 19. Pringle NP, Yu WP, Howell M, Colvin JS, Ornitz DM, Richardson WD (2003) Fgfr3 expression by astrocytes and their precursors: evidence that astrocytes and oligodendrocytes originate in distinct neuroepithelial domains. Development 130:93–102. [DOI] [PubMed] [Google Scholar]
- 20. Reuss DE, Kratz A, Sahm F, Capper D, Schrimpf D, Koelsche C et al (2015) Adult IDH wild type astrocytomas biologically and clinically resolve into other tumor entities. Acta Neuropathol 130:407–417. [DOI] [PubMed] [Google Scholar]
- 21. Sanson M, Idbaih A (2013) Neuro‐oncology: novel molecular targets in treatment of glioblastoma. Nat Rev Neurol 9:612–613. [DOI] [PubMed] [Google Scholar]
- 22. Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F et al (2009) Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol 27:4150–4154. [DOI] [PubMed] [Google Scholar]
- 23. Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A et al (2012) Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 337:1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. TCGA (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455:1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found online in the Supporting Information section at the end of the article.
Figure S1. Histological spectrum of diffuse gliomas with FGFR3‐TACC3 fusion. (A,B) Pseudo‐ependymal features with nuclei localized at distance of vessels (A), and a perivascular pseudorosette with thin perivascular cytoplasmic processes (B). (C) Pseudo‐astroblastic perivascular pseudorosette in tumor infiltrative edge. (d) Pseudo‐oligodendroglial tumor cells with clear cytoplasm (arrowheads) form perineuronal satellitosis. (E) Tumor cells with pale eosinophilic cytoplasm (arrowheads). (F) Tumor cells with eosinophilic cytoplasm (arrowheads).
Figure S2. Quantification of vascular density by CD34 immunolabeling. CD34 immunolabeling is showed for eleven gliomas without FGFR3‐TACC3 fusion and eleven gliomas with FGFR3‐TACC3 fusion.
Table S1. Detailed histological and immunohistochemical features of diffuse gliomas with FGFR3‐TACC3 fusion. Abbreviations: CD34 = positive or negative immunolabeling of tumor cells; FGFR3% = percentage of tumor cells with FGFR3 immunolabeling; FGFR3 int. = FGFR3 immunolabeling maximal intensity scored as intense (3), intermediate (2) or weak (1); INA solid tum. = presence of a solid tumor area without INA positive residual axons; INA tum. = negative or positive INA immunolabeling of tumor cells; Ki67 = maximal Ki67 proliferation index; m = maintained expression; Mitos. = Mitoses; MVP = microvascular proliferation; NA = not available; neg = negative; pos = positive.


