Abstract
Background
Cucumber (Cucumis sativus L.) is cultivated worldwide, and it is essential to produce enough high-quality seeds to meet demand. Pre-harvest sprouting (PHS) in cucumber is a critical problem and causes serious damage to seed production and quality. Nevertheless, the genetic basis and molecular mechanisms underlying cucumber PHS remain unclear. QTL-seq is an efficient approach for rapid quantitative trait loci (QTL) identification that simultaneously takes advantage of bulked-segregant analysis (BSA) and whole-genome resequencing. In the present research, QTL-seq analysis was performed to identify QTLs associated with PHS in cucumber using an F2 segregating population.
Results
Two QTLs that spanned 7.3 Mb on Chromosome 4 and 0.15 Mb on Chromosome 5 were identified by QTL-seq and named qPHS4.1 and qPHS5.1, respectively. Subsequently, SNP and InDel markers selected from the candidate regions were used to refine the intervals using the extended F2 populations grown in the 2016 and 2017 seasons. Finally, qPHS4.1 was narrowed to 0.53 Mb on chromosome 4 flanked by the markers SNP-16 and SNP-24 and was found to explain 19–22% of the phenotypic variation in cucumber PHS. These results reveal that qPHS4.1 is a major-effect QTL associated with PHS in cucumber. Based on gene annotations and qRT-PCR expression analyses, Csa4G622760 and Csa4G622800 were proposed as the candidate genes.
Conclusions
These results provide novel insights into the genetic mechanism controlling PHS in cucumber and highlight the potential for marker-assisted selection of PHS resistance breeding.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-021-07548-8.
Keywords: Cucumber, Pre-harvest sprouting, QTL-seq, qPHS4.1
Introduction
Cucumber (Cucumis sativus L.) is an economically important vegetable globally. In 2018, cucumber was grown on 1,984,518 ha worldwide, and the cultivated area in China accounted for 52.72% of this area (www.fao.org/faostat/en). It is necessary to produce enough excellent-quality cucumber seeds to meet demand, especially in China. However, pre-harvest sprouting (PHS), also known as vivipary, a critical trait describing the untimely germination of seeds inside maternal fruits under certain conditions, severely decreases seed yields and quality [1]. Breeding for resistance to PHS would decrease the loss of usable seeds in cucumber.
In agriculture, it is widely accepted that PHS is a complex agronomic trait controlled by multiple genes or quantitative trait loci (QTLs) [2, 3]. PHS is tightly connected with seed dormancy which is characterized as the prevention of physiologically mature seeds from germinating under unfavorable environmental conditions [4, 5]. Low levels of seed dormancy lead to PHS [6], while excessive seed dormancy usually gives rise to PHS resistance but unfortunately causes undesirable results, such as nonuniform seedling establishment after sowing [7, 8]. Therefore, maintenance of the balance between seed dormancy and germination is critical.
Regarding the genetic and molecular basis of seed dormancy and PHS resistance, extensive QTLs or genes for this trait have been identified in cereal crops and other vegetables, such as rice (Oryza sativa), wheat (Triticum aestivum), maize (Zea mays), barley (Hordeum vulgare) and tomato (Solanum lycopersicum). To date, in rice, more than 165 QTLs associated with seed dormancy or PHS resistance and located on different chromosomes have been identified [9, 10]. Similar to rice, QTLs responsible for PHS identified in wheat, which has a much more complicated genome, were distributed on almost all of the chromosomes [11]. Among them, the major QTLs were detected mainly on chromosome 2B [12], 3AS [13], and 7B [14], while minor QTLs were detected on chromosomes 3B and 5A [13]. In barley, several QTLs associated with seed dormancy have been identified [15–17]. Among the QTLs, two QTLs, SD1 and SD2 on chromosome 5H, contributed the major effects on seed dormancy [18]. SD1 was a major regulator of dormancy [19], and SD2 was identified to prevent PHS [17]. However, to date, QTL genetic mapping for PHS in cucumber has not been reported.
Traditional QTL mapping requires a segregating population originating from two parents with extreme opposite traits and polymorphic markers linked to target genes. It is extremely time-consuming and labor-intensive to screen DNA markers and genotype individuals in the segregating populations [20]. Bulked-segregant analysis (BSA) is an effective method to rapidly identify polymorphic markers linked to traits of interest [21]. QTL-seq [22], a powerful new approach combining BSA and next-generation sequencing, is used for the rapid identification of QTLs. Recently, QTL-seq has been widely used in the detection of QTLs for many traits in various plants, including 100-seed weight trait in chickpea [23], branch angle in oilseed rape [24], fruit length in cucumber [25], stalk rot in maize [26], heat-tolerance and high-temperature stress response in tomato [27], and cooked grain elongation [28] and salt tolerance [29] in rice. Therefore, QTL-seq provides a convenient method for identifying key loci controlling PHS in cucumber.
Our previous studies have revealed that PHS was controlled by one major gene of additive-dominance effects plus additive-dominance polygene (D-1 model) via the method of mixed major-gene plus polygenes inheritance model [30]. However, the genetic mapping and QTL location have not been performed. In this paper, we performed QTL-Seq analysis using an F2 population derived from Q12 and P60, which are resistant and susceptible to PHS in cucumber, respectively. SNP and InDel markers generated from QTL-seq were developed to genotype all the individuals in the F2 population grown in two years. The major QTLs were refined, and annotated genes located in the associated regions were analyzed by quantitative RT-PCR. This study may have the potential for cucumber breeding of PHS resistance by marker-assisted selection (MAS) and gene cloning analysis.
Results
Phenotypic evaluation of PHS in cucumber
The seeds of the resistant parent Q12, susceptible parent P60, and their F1, F2 populations were sown directly into soil in the greenhouse on April 15 each year. For plant management, two female flowers were self-pollinated, and all the other female flowers and lateral branches were removed from each plant. The pollination date was recorded on labels hung on the peduncles of the fruits. The seeds in the cucumber fruits were harvested at 45 days after pollination (DAP), and the numbers of germinated seeds and total seeds were counted immediately. The PHS rate (%) was calculated as (germinated seeds/total seeds in fruit) × 100%. The average PHS rates of two cucumber fruits grown on the same plant were used for QTL analysis.
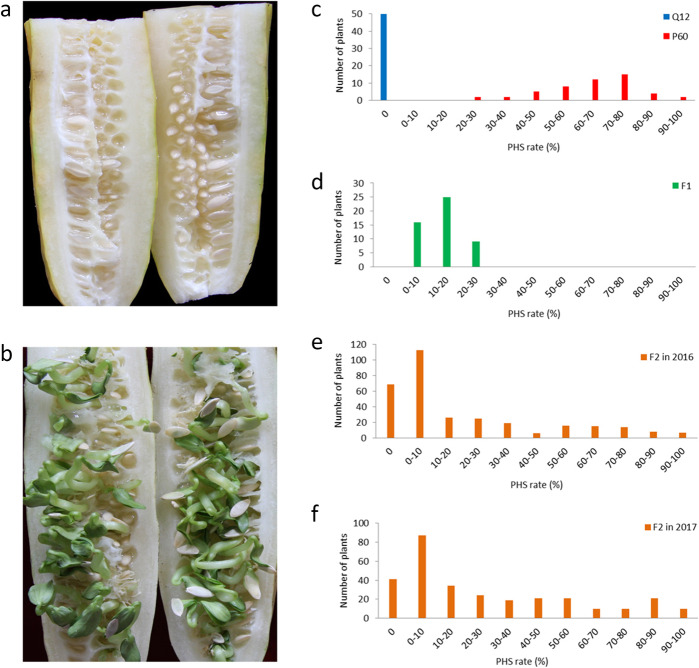
Phenotypic data of the PHS rate were collected from Q12, P60, and their F1, F2 populations (Additional file 1: Table S1). Q12 showed complete resistance to PHS; P60 displayed a wide range of variation for PHS (Fig. 1). The mean PHS rates of Q12, P60 and F1 progeny were 0, 64.97 and 13.88%, respectively. The PHS rates of the segregating mapping population of 328 F2 individuals grown in 2016 covered the full range from 0 to 100% (20.77% on average) and showed a skewed normal distribution (Fig. 1). The PHS rates of the 298 F2 individuals grown in 2017 showed a similar distribution to 2016. This phenotypic variation in the populations indicated that PHS is a quantitative trait controlled by a major-effect QTL.
Fig. 1.
Pre-harvest sprouting (PHS) and its frequency distribution in the parental lines, F1 and F2 populations. a: Phenotype of Q12, resistant to PHS; b: Phenotype of P60, susceptible to PHS; c: Frequency distribution of PHS in the parental lines, Q12 and P60; d: Frequency distribution of PHS in the F1 generation grown in 2016; e: Frequency distribution of F2 population grown in 2016; f: Frequency distribution of F2 population grown in 2017
Pool construction and QTL-seq
Based on the phenotypic data of F2 individuals (Additional file 1: Table S1), 30 extremely resistant and 30 extremely susceptible individuals were selected from the F2 population grown in 2017 for the construction of the R- and S-pool, respectively. The PHS rate of each extreme F2 individual in the R-pool was 0%, and the PHS rate of extreme individuals in the S-pool ranged from 80 to 100%. Each DNA pool, along with the R-parent (Q12) and S-parent (P60), were subjected to whole-genome resequencing (WGRS) using the Illumina HiSeq4000 platform, and 36.83 Gb raw data was generated. The clean data were mapped to the cucumber reference genome (http://www.cucurbitgenomics.org/organism/2; Chinese long; V2) [31] using the BWA 0.7.10 (Burrows-Wheeler Aligner) software [32], and 36.55 Gb remained after trimming and adapter removal. A total of 6.83 Gb clean data (18.59X coverage) for Q12, 8.43 Gb (22.30X coverage) for P60, 10.37 Gb for the R-pool (28.06X coverage) and 10.92 Gb (30.54X coverage) S-pool was generated. Detailed information is listed in Table 1.
Table 1.
Resequencing summary of the parental lines, R-pool and S-pool
| Sample | Clean bases (Gb) | Total reads | Mapped reads | Rate of mapped reads(%) | Sequencing depth (X) | Genome coverage |
|---|---|---|---|---|---|---|
| Q12 | 6.83 | 45,507,344 | 38,958,916 | 85.61 | 18.59 | 98.75 |
| P60 | 8.43 | 56,203,612 | 47,443,606 | 84.41 | 22.30 | 98.82 |
| R-pool | 10.37 | 69,130,804 | 61,513,704 | 88.98 | 28.06 | 98.99 |
| S-pool | 10.92 | 72,805,348 | 63,923,453 | 87.80 | 30.54 | 98.99 |
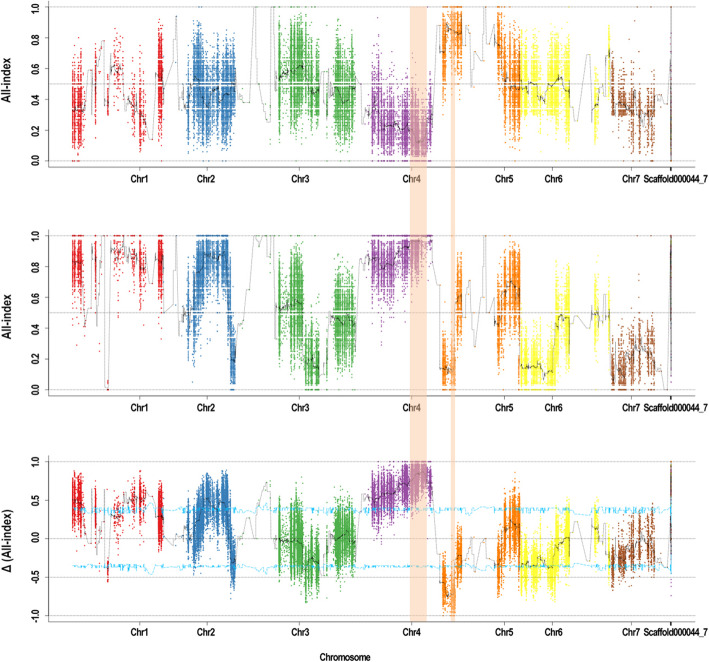
Using GATK 3.8 software [33], a total of 62,504 SNPs and 18,646 InDel variants were detected between the two parents. The Δ (SNP/InDel-index) of the polymorphic loci between R-pool and S-pool was calculated based on the SNP/InDel-index in R-pool and S-pool. The sliding window approach was used, and SNP/InDel-index plotted graphs against the genomic positions for R-pool (Fig. 2a) and S-pool (Fig. 2b) were generated. After calculating, Δ (SNP/InDel-index) plotted graph was constructed (Fig. 2c). Two regions harboring high Δ (SNP/InDel-index) values exceeding the confidence interval and containing variations with SNP/InDel-index = ‘0’ or ‘1’ were examined and defined as the predicted regions associated with PHS. As a result, the SNP-index of predicted regions for R-pool and S-pool appeared as mirror images [22]. One of the regions spanned 7.3 Mb on chromosome 4, and the other region spanned 0.15 Mb on chromosome 5. We named these two predicted regions that were putatively associated with PHS in cucumber qPHS4.1 and qPHS5.1, respectively. In qPHS4.1, several loci with the highest Δ (SNP/InDel-index) value equal to ‘1’ were detected. Conversely, qPHS5.1 region was harboring loci with the lowest Δ (SNP/InDel-index) value equal to ‘-1’. These results indicated that the QTLs were associated with PHS in cucumber. qPHS4.1 conferred a partial level of PHS resistance in the resistant donor Q12, while qPHS5.1 provided partial resistance for the parent P60.
Fig. 2.
SNP/InDel-index Manhattan graphs of R-pool, S-pool and Δ (SNP/InDel-index) from QTL-seq approach for mapping the genomic regions controlling pre-harvest sprouting in cucumber. a: SNP/InDel-index plot of R-pool; b: SNP/InDel-index plot of S-pool; c: the Δ (SNP/InDel-index) plot of all chromosomes with the statistical confidence interval under the null hypothesis of no QTLs (blue line P = 0.05). The significant genomic regions on Chromosome 4 and 5 are highlighted in shaded color
These two regions contained 443 SNPs and 124 InDels, of which 272 SNPs and 82 InDels were found to be intergenic, 70 SNPs and 19 InDels intronic, 4 SNPs synonymous, 6 SNPs nonsynonymous, 39 SNPs and 11 InDels in upstream and 47 SNPs and 10 InDels in downstream (Table 2). In qPHS5.1, there were only two InDels detected in upstream. The other variations were identified in qPHS4.1interval.
Table 2.
Categorization of Detected Variations in qPHS4.1 and qPHS5.1
| Category | qPHS4.1 | qPHS5.1 | |||
|---|---|---|---|---|---|
| SNPs | InDels | SNPs | InDels | ||
| Exonic | Synonymous | 4 | 0 | ||
| Non-Synonymous | 6 | 0 | |||
| Non-Frameshift Insertion | – | 1 | |||
| Intronic | 70 | 19 | |||
| Upstream | 39 | 9 | 2 | ||
| Downstream | 47 | 10 | |||
| Upstream/Downstream | 5 | 1 | |||
| Intergenic | 272 | 82 | |||
| transition | 277 | – | |||
| transversion | 166 | – | |||
| Insertion | – | 62 | |||
| Deletion | – | 62 | |||
| Total | 443 | 124 | |||
Based on the gene annotation via ANNOVAR (Version 2013Aug23) software [34], genes containing stop loss, stop gain or nonsynonymous mutations were preferentially selected as candidate genes (Additional file 2: Table S2) from the associated regions.
Validation and narrowing down the associated region
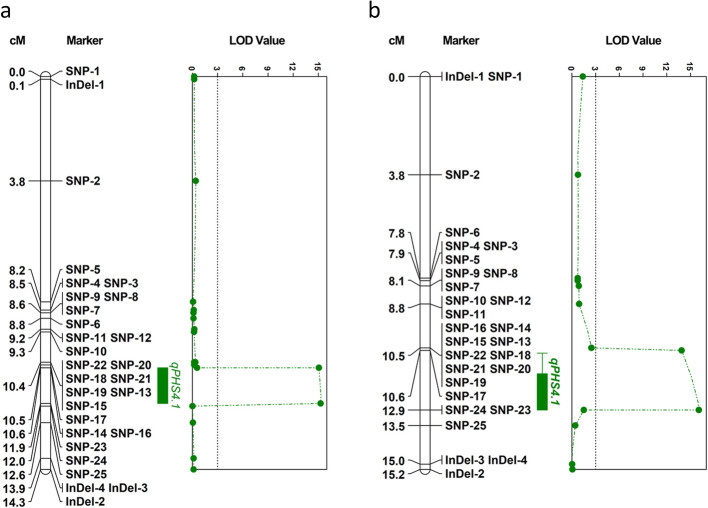
To verify the results detected by QTL-seq and narrow down the candidate intervals, a traditional QTL mapping method was used. We genotyped all F2 individuals grown in 2016 and 2017 for 62 SNP and/or InDel markers selected from the qPHS4.1 and qPHS5.1 intervals, respectively. Finally, twenty-nine markers in qPHS4.1 were accurately genotyped and applied to construct the local genetic linkage maps by JoinMap 4.0 software [35]. Two InDel markers on Chromosome 5 were unmapped. After calculation by MapQTL version 6 software [36], two loci with LOD scores over the threshold, SNP-16 and SNP-23, were found by using the 2016 F2 population. As shown in Table 3, the peak LOD scores of SNP-16 and SNP-23 were 15.07 and 15.28, respectively. This interval explained 19.6–19.8% of the phenotypic variation in PHS. In the 2017 F2 population, two peak SNP loci, SNP-17 (LOD = 13.89) and SNP-24 (LOD = 16.06), were detected (Table 3, Additional file 3: Table S3). The interval explained 19.3–22.0% of the phenotypic variation in PHS. By taking the overlapping regions into account, these results reduced the candidate genomic interval associated with qPHS4.1 from 7.3 Mb to the 0.53 Mb flanked by the markers SNP-17 to SNP-23 on chromosome 4 in cucumber (Fig. 3).
Table 3.
LOD Values, Additive Effects, and Variance Explained for the Significant Loci Associated with pre-harvest sprouting in Cucumber
| Year | The SNP markers | Physical position on Chromosome 4 (bp) | Interval(Mb) | LODa | Additive effectb | Dominance | Variance explained(%)c |
|---|---|---|---|---|---|---|---|
| 2016 | SNP-16 | 19,973,741 | 0.53 | 15.07 | −0.136391 | −0.0594552 | 19.6 |
| SNP-23 | 20,505,510 | 15.28 | −0.141843 | −0.0345112 | 19.8 | ||
| 2017 | SNP-17 | 19,973,782 | 0.55 | 13.89 | −0.159806 | −0.0408643 | 19.3 |
| SNP-24 | 20,521,004 | 16.06 | −0.182258 | 0.0193450 | 22.0 |
aPeak LOD score of the QTL. bAdditive or dominant effect of the SNPs. cPercentage of variance explained by the QTL peak
Fig. 3.
Fine mapping of the major-effect QTLqPHS4.1in cucumber using F2 populations grown in 2016 (a) and 2017 (b). SNP and InDel markers in candidate regions generated by QTL-seq were selected and genotyped in the 318 F2 individuals grown in 2016 and 298 F2 individuals grown in 2017. One major-effect QTL in the overlapping region was identified. The interval of qPHS4.1 was narrowed down to 0.53 Mb on Chromosome 4
Gene annotation and expression analysis of candidate genes
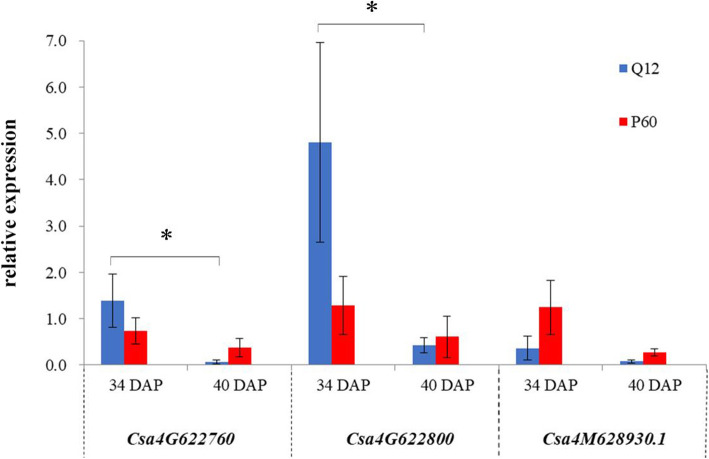
On the basis of the gene annotations, within the qPHS4.1 region, Csa4G622760, Csa4G622800 and Csa4M628930.1 (Table 4), in which nonsynonymous or upstream mutations occurred, were selected as candidate genes for further analysis. The relative expression levels of the candidate genes in seed cavity flesh tissues between Q12 and P60 were examined by Real-time Quantitative PCR (qRT-PCR) at 34 DAP (PHS not occurred) and 40 DAP (PHS occurred) stages, as shown in Fig. 4. The expression level of Csa4G622760, which is predicted to encode a chalcone isomerase-like protein, was 1.9-fold higher in Q12 than in P60 at the 34 DAP stage. However, its expression level was 5.4-fold lower in Q12 than in P60 at 40 DAP. This indicated that the expression level of the Csa4G622760 gene significantly decreased, by approximately 20-fold, from 34 DAP to 40 DAP in Q12 but was only 2-fold down-regulated in P60. The Csa4G622800 gene is annotated as a peptide methionine sulfoxide reductase msrB. Its expression level was 3.7-fold higher in Q12 than in P60 at the 34 DAP stage. At the 40 DAP stage, the expression level was down-regulated 11.2-fold in Q12 and 2.1-fold in P60. Gene expression of Csa4G622800 also decreased significantly. Csa4M628930.1 is a putative ERI1 exoribonuclease 3 protein. At 34 DAP, the expression level in P60 was 3.43-fold higher than that in Q12. From 34 DAP to 40 DAP, gene expression decreased approximately 4.6-fold in both parental lines. At 40 DAP, the expression level in P60 was 3.41-fold higher than that in Q12. The expression pattern did not show significant differences.
Table 4.
Candidate Genes Underlying qPHS4.1 Control of Preharvest Sprouting in Cucumber
| Gene ID | SNP location | SNP locus | Physical position (bp) | Mutation | Functional prediction | |
|---|---|---|---|---|---|---|
| Q12 | P60 | |||||
| Csa4G622760 | upstream | SNP-14 | 19,973,692 | G | T | Chalcone isomerase-like protein |
| upstream | SNP-15 | 19,973,724 | C | A | ||
| upstream | SNP-16 | 19,973,741 | T | C | ||
| upstream | SNP-17 | 19,973,782 | A | T | ||
| Csa4G622800 | upstream | SNP-18 | 19,995,077 | A | G | Peptide methionine sulfoxide reductase msrB |
| upstream | SNP-19 | 19,995,107 | G | A | ||
| upstream | SNP-20 | 19,995,109 | C | G | ||
| upstream | SNP-21 | 19,995,123 | A | C | ||
| upstream | SNP-22 | 19,995,137 | C | A | ||
| Csa4M628930.1 | nonsynonymous | SNP-23 | 20,505,510 | T | C | ERI1 exoribonuclease 3 |
Fig. 4.
The relative quantitative expression analysis of the predicted genes in cucumber cavity flesh tissue of Q12 and P60. The blue bars represent Q12, the red bars represent P60. 34 DAP indicates the relative gene-expression levels in the cucumber cavity flesh tissue sampled from cucumber fruits at 34 days after pollination (DAP), at which point the seeds had not germinated in the cucumber cavities. 40 DAP indicates the relative gene-expression levels in the cucumber fruits at 40 days after pollination, at which point the seeds had germinated in those cucumbers that were susceptible to pre-harvest sprouting. Data are the means of three biological and technical replicates ± the standard error. * P < 0.05 in the t-test
Taken together, these data show that the expression levels of the three genes were both down-regulated in Q12 and P60 with increasing ripeness of cucumber fruits. The Csa4M628930.1 gene showed a different expression pattern from that of Csa4G622760 and Csa4G622800. Csa4G622760 and Csa4G622800 gene expression levels significantly decreased (p < 0.05) in Q12 but decreased slightly in P60 from the 34 DAP stage to 40 DAP stage. These results suggested that Csa4G622760 and Csa4G622800 gene expression levels were higher in resistant cucumbers than in susceptible cucumbers before PHS occurred. Subsequently, accompanying the occurrence of PHS, its gene expression levels decreased significantly in resistant cucumbers compared to susceptible cucumbers. Therefore, we hypothesized that Csa4G622760 and Csa4G622800 are possible candidate genes involved in PHS in cucumber, but further functional analysis of these genes needs to be conducted.
Discussion
In cucumber and other seed-bearing crops, pre-harvest sprouting (PHS) is a critical problem that causes devastating losses to seed yields and quality [1] and widely limits seed dispersal. To promote the process of cucumber PHS resistance breeding, it is greatly important to identify key loci controlling PHS resistance and develop molecular markers for marker-assisted selection (MAS). In cereal crops, including wheat, rice, maize and barley, PHS is a very popular research topic, and the investigation of genetic mapping and molecular mechanisms underlying PHS is extensive and intensive. However, unfortunately, few published studies have focused on the PHS trait in cucumber [32]. In this study, we identified two QTLs associated with PHS by a QTL-seq approach in the F2 population derived from the two parents Q12 and P60, which showed opposite extremes of PHS phenotypes. Q12 is a typical resistant line in which PHS never occurs in favorable environments, while PHS occurs in the P60 line (Fig. 1). The frequency distribution of PHS in P60 was normal. Subsequently, in the F2 population, the frequency distribution was skewed normal rather than normal (Fig. 1), suggesting that PHS was a quantitatively inherited trait in cucumber and controlled by a major-effect QTL. This is consistent with our previous research on the inheritance of PHS.
The application of high-throughput next-generation sequencing technology promotes the development of rapid molecular marker discovery and physical map construction. QTL-seq is a new method that combines next-generation sequencing with BSA for the rapid detection of QTLs and links molecular markers associated with traits of interest. It was first developed by Takagi et al. and applied in rice [22]. Since that time, QTL-seq has been successfully used in many species [23–29]. However, the candidate regions generated from QTL-Seq are often too rough or too broad, and additional QTL analysis performed by traditional methods is necessary to refine gene locations and narrow chromosomal intervals. In the present study, a QTL-seq approach was performed in the F2 population grown in 2017. Two QTLs associated with PHS, qPHS4.1 and qPHS5.1, were initially identified, which spanned 7.3 Mb on chromosome 4 and 0.15 Mb on chromosome 5, respectively. The predicted regions in R-pool and S-pool appeared as mirror images [22] in Fig. 2. These results confirmed that qPHS4.1 was derived from the resistant donor Q12 and qPHS5.1 provided PHS resistance for P60. However, P60 was identified to be a susceptible genotype to PHS. Therefore, qPHS5.1 could be a putative minor-effect QTL for PHS.
And then, traditional QTL mapping methods were conducted to validate and narrow down the candidate regions. The phenotype identification and QTL mapping using the extended F2 population grown in 2016 was consistent with the findings from the 2017 season, which indicated that the experimental results were reliable and accurate. Subsequently, the regions from the two seasons were found to overlap. Therefore, qPHS4.1 was refined and narrowed down to 0.53 Mb on Chromosome 4. Unfortunately, qPHS5.1 was unmapped by JoinMap 4.0 and MapQTL version 6 software. This result demonstrated that qPHS4.1 was a major-effect QTL controlling PHS in cucumber and qPHS5.1 was a merely minor-effect QTL. We supposed that only two InDel markers detected from QTL-seq were used in the validation. We need to develop more molecular markers to further analyze the minor-effect involved in qPHS5.1 controlling PHS. As a major-effect QTL, qPHS4.1 was identified to explain about 20% of the phenotypic variation. The available tightly linked markers in qPHS4.1 can be used in MAS to promote the breeding process. We propose the introgression of qPHS4.1 could provide a partial level of PHS resistance for a susceptible background genotype and decrease the PHS rate of susceptible cucumber lines in a certain extent.
Functional annotation of the qPHS4.1 region, a total of 39 candidate genes was identified by ANNOVAR software. Based on gene expression analysis by qRT-PCR, two genes, Csa4G622760 and Csa4G622800, containing upstream polymorphic SNPs, were considered candidate PHS regulating genes in cucumber (Fig. 4). The Csa4G622760 gene is predicted to encode a chalcone isomerase-like protein that catalyzes the biosynthesis of flavonoids and secondary metabolism in plants [37]. Flavonoids are important secondary metabolites found in various plant tissues, such as leaves, flowers, fruits and seeds. In Arabidopsis, overexpression of the chalcone isomerase-like gene increased the accumulation of proanthocyanidin and flavonol, which are flavonoids, while loss of function of the chalcone isomerase-like gene led to a strong reduction in proanthocyanidin and flavonol levels and influenced the seed phenotype [38]. However, the correlation between flavonoids and PHS in cucumber is unclear. The Csa4G622800 gene is predicted to encode the peptide methionine sulfoxide reductase msrB. In the promoters of methionine sulfoxide reductase genes, cis-regulatory elements were found from Arabidopsis, poplar and rice [39, 40]. Methionine sulfoxide reductase can play protective roles in redox homeostasis in plant growth, including seed development [41, 42]. In plant seeds, methionine sulfoxide reductase plays a decisive role in the establishment and preservation of seed longevity [43]. Higher activity of this enzyme leads to better preservation of the seeds and higher germination capacity [43]. In our research, the SNPs were identified in the promoter of Csa4G622800 gene. We consider that the mutations in promoter of Csa4G622800 gene would putatively alter the gene-expression levels and then affect the development and germination of seeds in cucumber fruits. However, further experiments need to be performed to test the functionality of the candidate genes in the genetic mechanisms of cucumber PHS.
In some cereal crops, e.g., wheat, rice, barley, etc., extensive QTLs and numerous genes associated with seed dormancy and PHS have been reported. However, only one major-effect QTL in cucumber was identified in this study. In contrast to cereal crops, cucumber seeds are surrounded by flesh tissues in seed cavities, in which the water content is higher than 95%. Therefore, PHS in cucumber is less likely to be influenced by the humidity of the environment. Cucumber PHS is a very specific and interesting trait, and the molecular mechanisms underlying PHS need further study.
Conclusion
In this study, two QTLs associated with PHS in cucumber were detected using QTL-seq approach. The major-effect QTL qPHS4.1 was refined to 0.53 Mb on chromosome 4. Based on the gene annotation and qRT-PCR analysis, two genes located in qPHS4.1 were proposed to be the candidate genes associated with cucumber PHS. To our knowledge, this is the first report on the identification of QTLs associated with PHS trait in cucumber. This study provides novel insights into the genetic mechanism controlling PHS in cucumber and highlights the potential for PHS resistance MAS breeding.
Materials and methods
Plant materials and phenotypic evaluation
The high-generation inbred cucumber lines Q12 (North China fresh market cucumber, derived from Chinese commercial variety ‘Jinyan No.4’ crossed with ‘Sipingcigua’, PHS resistant, P1) and P60 (North China fresh market cucumber, generated from a Chinese commercial variety ‘YuanFengYuan No.6’, PHS susceptible, P2) were crossed to obtain F1. F1 plants were self-pollinated to generate an F2 segregating population. P1 and P2 populations were evaluated for PHS in the experimental farm of the Tianjin Kernel Cucumber Research Institute (Tianjin, China) in 2016. Significance test was conducted between P1 and P2 populations. The F2 population was evaluated in 2016 (328 plants) and 2017 (299 plants) seasons. All the plants were grown in greenhouse conditions under whole-day light exposure. The day/night temperature in the greenhouse was controlled at 28–35 °C/15–26 °C.
Pool construction and whole-genome re-sequencing
The genomic DNA of Q12, P60 and F2 individuals was extracted from seedling leaves using a Quick Prep Plant Genome DNA Kit (HUALIKEXI, Tianjin, China). Q12 and P60 genomic DNA were used to construct the P1 pool and P2 pool. Based on the phenotype data of F2 individuals grown in the 2017 season (Additional file 1: Table S1), 30 extreme resistant plants and 30 extreme susceptible plants were selected to construct a resistant pool (R-pool) and susceptible pool (S-pool), respectively. Equal amounts of DNA from the selected individuals were mixed and subsequently processed to generate sequencing libraries using the TruSeq Nano DNA HT Sample preparation Kit (Illumina Inc., United States) by Novogene Co., Ltd. (http://www.novogene.com/). According to the protocol, briefly, DNA samples were randomly fragmented by sonication to a size of 350 bp, then DNA fragments were end polished, A-tailed, and ligated with the full-length adapter for Illumina sequencing with further PCR amplification. PCR products were purified (AMPure XP system) and libraries were analyzed for size distribution by Agilent2100 Bioanalyzer and quantified using real-time PCR [44]. These libraries were resequenced and 150 bp paired-end reads were generated with insert size around 350 bp using the Illumina HiSeq4000 platform (Illumina Inc., United States) by Novogene Co., Ltd. (http://www.novogene.com/).
QTL-seq
The raw sequencing data were filtered to get high-quality clean reads by removing the reads with ≥10% unidentified nucleotides, removing the reads with > 50% bases having phred quality < 5 and the reads with > 10 nucleotides aligned to the adapter. The clean reads obtained from four pools were aligned to the cucumber reference genome (Chinese long; V2) [31] using the BWA 0.7.10 (Burrows-Wheeler Aligner) software [32]. Variant calling was performed for the samples by using the Unified Genotyper function in GATK 3.8 software [33]. To determine the genomic regions associated with PHS, we calculated the SNP/InDel-index and Δ (SNP/InDel-index) to locate the QTLs. The SNP/InDel-index refers to the proportion of reads carrying a SNP/InDel different from the reference reads of either parent. The Δ (SNP/InDel-index) of each locus was determined based on the difference in the SNP/InDel-index between the R-pool and S-pool. To eliminate background interference, we filtered out all loci with an SNP/InDel-index of less than 0.3 [22]. Using the slicing window method with a 1 Mb window size and 1 kb increment, the average SNP/InDel-index of loci in a given genomic interval was calculated. The SNP/InDel-index of the R-pool and S-pool and the corresponding Δ (SNP/InDel-index) in the slicing window were plotted in a graph to generate SNP/InDel-index plots. We calculated statistical confidence intervals of Δ (SNP/InDel-index) for all SNP and InDel loci with a given read depth under the null hypothesis of no QTL, following the detail procedures of Takagi et al. [22]. The confidence intervals of Δ (SNP/InDel -index) were defined to be 95% (p = 0.05). By examining the Δ (SNP/InDel-index), the candidate genomic regions harboring high average Δ (SNP/InDel-index) values exceeding the confidence intervals and containing variations with SNP/InDel-index = ‘0’ or ‘1’ were defined as predicted regions for association with PHS. SNP/InDel-index was equal to ‘0’ or ‘1’ when the candidate variations in pools were entirely from P1 or P2, respectively, and the corresponding Δ (SNP/InDel-index) was equal to ‘-1’ or ‘1’.
The ANNOVAR (Version 2013Aug23) software was used to annotate the candidate genes in the regions [34].
Genotyping, regional linkage mapping and QTL analysis
To verify the candidate SNP and InDel markers and narrow down the regions identified by QTL-seq, significant SNPs and InDels in the candidate regions were first selected and validated in the two parents and their F1 plants. Then, polymorphic SNP and InDel markers were used to genotype the extended F2 individuals sown in the 2016 season and 2017 season. This validation was performed using Hi-SNP high-throughput genotyping method (Shanghai Biowing Applied Biotechnology CO. LTD, Shanghai, China). The specific multiplex PCR primers of the markers were designed by Primer 3 online software (http://frodo.wi.mit.edu/, Version 0.4.0) based on the cucumber reference genome (Chinese long; V2) [31] (listed in Additional file 4: Table S4). Multiplex PCR and high-throughput sequencing genotyping were performed as previously described [45, 46]. Based on the genotypes of significant SNPs and InDels in candidate regions of F2 individuals sown in 2016 (328 plants) and 2017 (299 plants), regional linkage maps were constructed using JoinMap 4.0 software [35] with the maximum likelihood mapping algorithm and Kosambi mapping function [47], respectively. According to the phenotyping datasets of the F2 individuals, QTL analysis was performed by the software MapQTL version 6 [36]. The “MQM mapping” algorithm with an LOD threshold score of > 3.0 was used to perform the calculation. The output logarithm of odds (LOD) scores were plotted along the genetic distances of the markers analyzed.
Candidate gene annotation
According to the further narrowed region of the QTLs, effective SNPs or InDels associated with PHS were identified. Based on the Cucurbit Genomics Database (http://www.icugi.org/cgi-bin/ICuGI/index.cgi), the functions of candidate PHS-associated genes that contained non-synonymous or upstream/downstream variations were predicted.
Gene expression analysis by qRT-PCR
We used qRT-PCR to investigate the relative expression levels of the candidate genes between the two parents. The cucumber cavity flesh tissues surrounding the seeds at 34 DAP and 40 DAP were sampled, and RNA was extracted using TRNzol Universal Reagent following the manufacturer’s protocol (TIANGEN, Beijing, China). The RNA quality was evaluated by agarose gel electrophoresis. cDNA was synthesized using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc., MA, USA). Primers for candidate genes were designed by Primer 3 and synthesized by Sangon Biotech (Shanghai) Co., Ltd. (Sangon, Shanghai, China). Details of the primer sequences are presented in Table 5. qRT-PCR was conducted by using TB Green Premix Ex Taq II (Takara Bio Inc., Dalian, China) on a real-time PCR system (Step One Plus; Applied Biosystems). The qRT-PCR conditions were set as follows: 95 °C for 30 s; followed by 40 cycles of 95 °C for 5 s and then 60 °C for 30 s; and then denaturation at 95 °C for 15 s, 60 °C for 60 s, a temperature increase of 0.3 °C per 15 s, and finally 95 °C for 15 s. The tubulin gene (GenBank ID: AF044573.1) was used as the reference gene for normalization of the relative expression of the candidate genes. The relative expression levels of the target genes were calculated using the 2−ΔΔCt method [48]. The experiments were conducted with three biological and technical replicates. A Student’s t-test was used to check the significant differences in expression levels among the samples.
Table 5.
qRT-PCR primers for candidate genes and reference gene (Tubulin)
| Gene ID | F | R | Size (bp) |
|---|---|---|---|
| Csa4G622760 | TAACTCTGCCAGGCTGCTCAAC | GTCTCGGACAAGAACAATCTGTAAAG | 242 |
| Csa4G622800 | GCATCAAAGAGGCTGGCACC | TATCCCTGCCGTTTTGGTGTTC | 152 |
| Csa4M628930.1 | GGAGTTGACCGTGTCTGGC | TGATGTGGGACCTGAGTTT | 173 |
| Tubulin | GCAAGGAAGATGCTGCCAATA | TCCATAGTCAACAGACAAACGCTC | 203 |
Supplementary Information
Additional file 1: Table S1. Phenotypic evaluation of pre-harvest sprouting trait for parents and their F1 and F2 populations.
Additional file 2: Table S2. Candidate genes by annotation and loci generated from QTL-seq.
Additional file 3: Table S3. Physical position, genetic distance, LOD values and variations explained generated from software MapQTL version 6.
Additional file 4: Table S4. Detailed multiplex PCR Primers of the Markers.
Acknowledgements
We are grateful to Yu Ning (Institute of Vegetable Crops, Jiangsu Academy of Agricultural Sciences) and Qingzhen Wei (Institute of Vegetable Research, Zhejiang Academy of Agricultural Sciences) for their excellent suggestions and revision on our manuscript.
Authors’ contributions
CM performed the linkage analysis, genotyping work, mapping the QTLs and wrote the manuscript. LS aided research design, assisted data analysis and revised the manuscript. DQ ang WH did the PCR and electrophoresis detection. YR did the target trait testing. All authors read and approved the final manuscript.
Funding
This research was financially supported by Natural Science Foundation of Tianjin City (17JCYBJC29400) and Innovation Team of Tianjin Vegetables Research System (ITTVRS) 202102.
Availability of data and materials
The raw datasets are stored in NCBI. The accessions of the datasets are SRR13637896, SRR13637895, SRR13637894 and SRR13637893. The analyzed data during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
All of the materials are owned by the authors and no permissions are required.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mingming Cao, Email: caoming2013@126.com.
Shuju Li, Email: lishuju1964@126.com.
Qiang Deng, Email: dengqiang022@126.com.
Huizhe Wang, Email: wanghuizhe@126.com.
Ruihuan Yang, Email: yruihuan@126.com.
References
- 1.Mu Y, Liu Y, Bai L, Li S, He C, Yan Y, Yu X, Li Y. Cucumber CsBPCs regulate the expression of CsABI3 during seed germination. Front Plant Sci. 2017;8:459. doi: 10.3389/fpls.2017.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogbonnaya FC, Imtiaz M, Ye G, Hearnden PR, Hernandez E, Eastwood RF, van Ginkel M, Shorter SC, Winchester JM. Genetic and QTL analyses of seed dormancy and preharvest sprouting resistance in the wheat germplasm CN10955. Theor Appl Genet. 2008;116(7):891–902. doi: 10.1007/s00122-008-0712-8. [DOI] [PubMed] [Google Scholar]
- 3.Vetch JM, Stougaard RN, Martin JM, Giroux MJ. Review: revealing the genetic mechanisms of pre-harvest sprouting in hexaploid wheat (Triticum aestivum L.) Plant Sci. 2019;281:180–185. doi: 10.1016/j.plantsci.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. New Phytol. 2006;171(3):501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- 5.Bentsink L, Koornneef M. Seed dormancy and germination. Arabidopsis Book. 2008;6:e0119. doi: 10.1199/tab.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodríguez M, Barrero J, Corbineau F, Gubler F, Benech-Arnold R. Dormancy in cereals (not too much, not so little): about the mechanisms behind this trait. Seed Sci Res. 2015;25(2):99–119. doi: 10.1017/S0960258515000021. [DOI] [Google Scholar]
- 7.Takahashi N. Effect of environmental factors during seed formation on pre-harvest sprouting. Cereal Res Commun. 1980;8(1):175–183. [Google Scholar]
- 8.Nakamura S, Pourkheirandish M, Morishige H, Sameri M, Sato K, Komatsuda T. Quantitative trait loci and maternal effects affecting the strong grain dormancy of wild barley (Hordeum vulgare ssp. spontaneum). Front. Plant Sci. 2017;8:1840. doi: 10.3389/fpls.2017.01840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magwa RA, Zhao H, Xing Y. Genome-wide association mapping revealed a diverse genetic basis of seed dormancy across subpopulations in rice (Oryza sativa L.) BMC Genet. 2016;17:28. doi: 10.1186/s12863-016-0340-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizuno Y, Yamanouchi U, Hoshino T, Nonoue Y, Nagata K, Fukuoka S, Ando T, Yano M, Sugimoto K. Genetic dissection of pre-harvest sprouting resistance in an upland rice cultivar. Breed Sci. 2018;68(2):200–209. doi: 10.1270/jsbbs.17062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu K, Meng YJ, Shuai HW, Liu WG, Du JB, Liu J, Yang WY. Dormancy and germination: how does the crop seed decide? Plant Biol (Stuttg) 2015;17(6):1104–1112. doi: 10.1111/plb.12356. [DOI] [PubMed] [Google Scholar]
- 12.Somyong S, Ishikawa G, Munkvold JD, Tanaka J, Benscher D, Cho YG, Sorrells ME. Fine mapping of a preharvest sprouting QTL interval on chromosome 2B in white wheat. Theor Appl Genet. 2014;127(8):1843–1855. doi: 10.1007/s00122-014-2345-4. [DOI] [PubMed] [Google Scholar]
- 13.Shao M, Bai G, Rife TW, Poland J, Lin M, Liu S, Chen H, Kumssa T, Fritz A, Trick H, Li Y, Zhang G. QTL mapping of pre-harvest sprouting resistance in a white wheat cultivar Danby. Theor Appl Genet. 2018;131(8):1683–1697. doi: 10.1007/s00122-018-3107-5. [DOI] [PubMed] [Google Scholar]
- 14.Cao L, Hayashi K, Tokui M, Mori M, Miura H, Onishi K. Detection of QTLs for traits associated with pre-harvest sprouting resistance in bread wheat (Triticum aestivum L.) Breed Sci. 2016;66(2):260–270. doi: 10.1270/jsbbs.66.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hickey LT, Lawson W, Arief VN, Fox G, Franckowiak J, Dieters MJ. Grain dormancy QTL identified in a doubled haploid barley population derived from two non-dormant parents. Euphytica. 2012;188(1):113–122. doi: 10.1007/s10681-011-0577-9. [DOI] [Google Scholar]
- 16.Vanhala TK, Stam P. Quantitative trait loci for seed dormancy in wild barley (Hordeum spontaneum C. Koch) Genet Resour Crop Evol. 2006;53(5):1013–1019. doi: 10.1007/s10722-004-7368-2. [DOI] [Google Scholar]
- 17.Nakamura S, Pourkheirandish M, Morishige H, Kubo Y, Nakamura M, Ichimura K, Seo S, Kanamori H, Wu J, Ando T, Hensel G, Sameri M, Stein N, Sato K, Matsumoto T, Yano M, Komatsuda T. Mitogen-activated protein kinase kinase 3 regulates seed dormancy in barley. Curr Biol. 2016;26(6):775–781. doi: 10.1016/j.cub.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 18.Hori K, Sato K, Takeda K. Detection of seed dormancy QTL in multiple mapping populations derived from crosses involving novel barley germplasm. Theor Appl Genet. 2007;115(6):869–876. doi: 10.1007/s00122-007-0620-3. [DOI] [PubMed] [Google Scholar]
- 19.Sato K, Yamane M, Yamaji N, Kanamori H, Tagiri A, Schwerdt JG, Fincher GB, Matsumoto T, Takeda K, Komatsuda T. Alanine aminotransferase controls seed dormancy in barley. Nat Commun. 2016;7(1):11625. doi: 10.1038/ncomms11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salvi S, Tuberosa R. To clone or not to clone plant QTLs: present and future challenges. Trends Plant Sci. 2005;10(6):297–304. doi: 10.1016/j.tplants.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci U S A. 1991;88(21):9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S, Innan H, Cano LM, Kamoun S, Terauchi R. QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013;74(1):174–183. doi: 10.1111/tpj.12105. [DOI] [PubMed] [Google Scholar]
- 23.Singh VK, Khan AW, Jaganathan D, Thudi M, Roorkiwal M, Takagi H, Garg V, Kumar V, Chitikineni A, Gaur PM, Sutton T, Terauchi R, Varshney RK. QTL-seq for rapid identification of candidate genes for 100-seed weight and root/total plant dry weight ratio under rainfed conditions in chickpea. Plant Biotechnol J. 2016;14(11):2110–2119. doi: 10.1111/pbi.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Cheng H, Wang W, Liu J, Hao M, Mei D, Zhou R, Fu L, Hu Q. Identification of BnaYUCCA6 as a candidate gene for branch angle in Brassica napus by QTL-seq. Sci Rep. 2016;6(1):38493. doi: 10.1038/srep38493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei QZ, Fu WY, Wang YZ, Qin XD, Wang J, Li J, Lou QF, Chen JF. Rapid identification of fruit length loci in cucumber (Cucumis sativus L.) using next-generation sequencing (NGS)-based QTL analysis. Sci Rep. 2016;6:27496. doi: 10.1038/srep27496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Q, Song J, Du WP, Xu LY, Jiang Y, Zhang J, Xiang XL, Yu GR. Identification, mapping, and molecular marker development for Rgsr8.1: a new quantitative trait locus conferring resistance to Gibberella stalk rot in maize (Zea mays L.) Front Plant Sci. 2017;8:1355. doi: 10.3389/fpls.2017.01355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen J, Jiang F, Weng Y, Sun M, Shi X, Zhou Y, Yu L, Wu Z. Identification of heat-tolerance QTLs and high-temperature stress-responsive genes through conventional QTL mapping, QTL-seq and RNA-seq in tomato. BMC Plant Biol. 2019;19(1):398. doi: 10.1186/s12870-019-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arikit S, Wanchana S, Khanthong S, Saensuk C, Thianthavon T, Vanavichit A, Toojinda T. QTL-seq identifies cooked grain elongation QTLs near soluble starch synthase and starch branching enzymes in rice (Oryza sativa L.) Sci Rep. 2019;9(1):8328. doi: 10.1038/s41598-019-44856-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei L, Zheng H, Bi Y, Yang L, Liu H, Wang J, Sun J, Zhao H, Li X, Li J, Lai Y, Zou D. Identification of a Major QTL and Candidate Gene Analysis of Salt Tolerance at the Bud Burst Stage in Rice (Oryza sativa L.) Using QTL-Seq and RNA-Seq. Rice (N Y) 2020;13(1):55. doi: 10.1186/s12284-020-00416-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gai J, Wang J. Identification and estimation of a QTL model and its effects. Theor Appl Genet. 1998;97(7):1162–1168. doi: 10.1007/s001220051005. [DOI] [Google Scholar]
- 31.Huang S, Li R, Zhang Z, Li L, Gu X, Fan W, Lucas WJ, Wang X, Xie B, Ni P, Ren Y, Zhu H, Li J, Lin K, Jin W, Fei Z, Li G, Staub J, Kilian A, van der Vossen EA, Wu Y, Guo J, He J, Jia Z, Ren Y, Tian G, Lu Y, Ruan J, Qian W, Wang M, Huang Q, Li B, Xuan Z, Cao J, Asan WZ, Zhang J, Cai Q, Bai Y, Zhao B, Han Y, Li Y, Li X, Wang S, Shi Q, Liu S, Cho WK, Kim JY, Xu Y, Heller-Uszynska K, Miao H, Cheng Z, Zhang S, Wu J, Yang Y, Kang H, Li M, Liang H, Ren X, Shi Z, Wen M, Jian M, Yang H, Zhang G, Yang Z, Chen R, Liu S, Li J, Ma L, Liu H, Zhou Y, Zhao J, Fang X, Li G, Fang L, Li Y, Liu D, Zheng H, Zhang Y, Qin N, Li Z, Yang G, Yang S, Bolund L, Kristiansen K, Zheng H, Li S, Zhang X, Yang H, Wang J, Sun R, Zhang B, Jiang S, Wang J, Du Y, Li S. The genome of the cucumber, Cucumis sativus L. Nat Genet. 2009;41(12):1275–1281. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Ooijen JW. JointMap 4. Software for the calculation of genetic linkage maps in experimental populations. Wageningen: Kyazma BV; 2006. [Google Scholar]
- 36.Van Ooijen JW. MapQTL 6.0, software for the mapping of quantitative trait loci in experimental populations of dihaploid species. The Netherlands: Kyazma BV, Wageningen; 2009. [Google Scholar]
- 37.Lin LM, Guo HY, Song X, Zhang DD, Long YH, Xing ZB. Adaptive evolution of Chalcone Isomerase superfamily in Fagaceae. Biochem Genet. 2020;11:1–15. doi: 10.1007/s10528-020-10012-z. [DOI] [PubMed] [Google Scholar]
- 38.Jiang W, Yin Q, Wu R, Zheng G, Liu J, Dixon RA, Pang Y. Role of a chalcone isomerase-like protein in flavonoid biosynthesis in Arabidopsis thaliana. J Exp Bot. 2015;66(22):7165–7179. doi: 10.1093/jxb/erv413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarrago L, Laugier E, Rey P. Protein-repairing methionine sulfoxide reductases in photosynthetic organisms: gene organization, reduction mechanisms, and physiological roles. Mol Plant. 2009;2(2):202–217. doi: 10.1093/mp/ssn067. [DOI] [PubMed] [Google Scholar]
- 40.Stolarska E, Bilska K, Wojciechowska N, Bagniewska-Zadworna A, Rey P, Kalemba EM. Integration of MsrB1 and MsrB2 in the redox network during the development of orthodox and recalcitrant Acer seeds. Antioxidants (Basel). 2020;9(12):1250. doi: 10.3390/antiox9121250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rey P, Tarrago L. Physiological roles of plant methionine Sulfoxide Reductases in redox homeostasis and signaling. Antioxidants (Basel) 2018;7(9):114. doi: 10.3390/antiox7090114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalemba EM, Stolarska E. Regulation of gene expression of methionine Sulfoxide Reductases and their new putative roles in plants. Int J Mol Sci. 2019;20(6):1309. doi: 10.3390/ijms20061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Châtelain E, Satour P, Laugier E, Ly Vu B, Payet N, Rey P, Montrichard F. Evidence for participation of the methionine sulfoxide reductase repair system in plant seed longevity. Proc Natl Acad Sci U S A. 2013;110(9):3633–3638. doi: 10.1073/pnas.1220589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bronner IF, Quail MA. Best practices for Illumina library preparation. Curr Protoc Hum Genet. 2019;102(1):e86. doi: 10.1002/cphg.86. [DOI] [PubMed] [Google Scholar]
- 45.Chen K, Zhou YX, Li K, Qi LX, Zhang QF, Wang MC, Xiao JH. A novel three-round multiplex PCR for SNP genotyping with next generation sequencing. Anal Bioanal Chem. 2016;408(16):4371–4377. doi: 10.1007/s00216-016-9536-6. [DOI] [PubMed] [Google Scholar]
- 46.Ruff TM, Marston EJ, Eagle JD, Sthapit SR, Hooker MA, Skinner DZ, See DR. Genotyping by multiplexed sequencing (GMS): a customizable platform for genomic selection. PLoS One. 2020;15(5):e0229207. doi: 10.1371/journal.pone.0229207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kosambi DD. The estimation of map distances from recombination values. In: Ramaswamy R, editor. DD Kosambi: selected works in mathematics and statistics. New Delhi: Springer India; 2016. pp. 125–130. [Google Scholar]
- 48.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Phenotypic evaluation of pre-harvest sprouting trait for parents and their F1 and F2 populations.
Additional file 2: Table S2. Candidate genes by annotation and loci generated from QTL-seq.
Additional file 3: Table S3. Physical position, genetic distance, LOD values and variations explained generated from software MapQTL version 6.
Additional file 4: Table S4. Detailed multiplex PCR Primers of the Markers.
Data Availability Statement
The raw datasets are stored in NCBI. The accessions of the datasets are SRR13637896, SRR13637895, SRR13637894 and SRR13637893. The analyzed data during this study are included in this published article and its supplementary information files.