Abstract
Diffuse adult high‐grade gliomas (HGGs) with necrosis encompass anaplastic oligodendrogliomas (AOs) with necrosis (grade III), glioblastomas (GBM, grade IV) and glioblastomas with an oligodendroglial component (GBMO, grade IV). Here, we aimed to search for prognostic relevance of histological classification and molecular alterations of these tumors. About 210 patients were included (63 AO, 56 GBM and 91 GBMO). GBMO group was split into “anaplastic oligoastrocytoma (AOA) with necrosis grade IV/GBMO,” restricted to tumors showing intermingled astrocytic and oligodendroglial component, and “GBM/GBMO” based on tumors presenting oligodendroglial foci and features of GBM. Genomic arrays, IDH1 R132H expression analyses and IDH direct sequencing were performed. 1p/19q co‐deletion characterized AO, whereas no IDH1 R132H expression and intact 1p/19q characterized both GBM and GBM/GBMO. AOA with necrosis/GBMO mainly demonstrated IDH1 R132H expression and intact 1p/19q. Other IDH1 or IDH2 mutations were extremely rare. Both histological and molecular classifications were predictive of progression free survival (PFS) and overall survival (OS) (P < 10−4). Diffuse adult HGGs with necrosis can be split into three histomolecular groups of prognostic relevance: 1p/19q co‐deleted AO, IDH1 R132H‐GBM and 1p/19q intact IDH1 R132H+ gliomas that might be classified as IDH1 R132H+ GBM. Because of histomolecular heterogeneity, we suggest to remove the name GBMO.
Keywords: 1p/19q co‐deletion, adult high‐grade gliomas, GBMO, IDH1 R132H immunoexpression, necrosis
Introduction
Diffuse adult high‐grade gliomas (HGGs) with necrosis encompass anaplastic oligodendrogliomas (AOs) with necrosis (grade III), glioblastomas (GBM, grade IV) and glioblastomas with an oligodendroglial component (GBMO, grade IV) 16. The name of GBMO was first coined by two independent teams who were searching for genetic alterations in a subset of GBM containing “areas showing oligodendroglioma‐like tumor cell differentiation” 8, 14. Adding GBMO in the World Health Organization (WHO) 2007 classification was based on two independent studies, showing that these tumors have an intermediate prognosis between anaplastic oligoastrocytomas (AOAs) and GBM 18, 21. In the 2007 WHO classification, “GBMO” was used to describe an “anaplastic oligoastrocytoma with necrosis” in the chapter dedicated to anaplastic mixed gliomas, while it corresponds to “glioblastoma that contains foci that resemble oligodendroglioma” in the chapter devoted to GBM 16. Recent studies including a large number of patients have reported conflicting results regarding the clinical behavior, genetic alterations and outcome of GBMO 1, 5, 9, 23.
In France, the POLA network (Prise en charge des OLigodendrogliomes Anaplasiques) dedicated to de novo diffuse adult HGG with an oligodendroglial component was set up in 2008. Initially restricted to AO, it was subsequently extended to AOA and then to GBMO. One of the aims of this program was to provide a centralized histological review and molecular analysis of the cases, together with biobanking and recommendations for treatment. The main goal of the present study was to refine histomolecular characteristics of diffuse adult HGG with necrosis and to test whether these subgroups were sufficiently characterized histologically, molecularly and prognostically to be distinguished from each other. Our results suggest integrating histological and molecular data to classify diffuse adult HGG with necrosis into three groups of prognostic relevance according to 1p/19q co‐deletion and IDH1 R132H expression status. These results might contribute to the forthcoming update of the 4th Edition of the WHO classification.
Material and Methods
Material
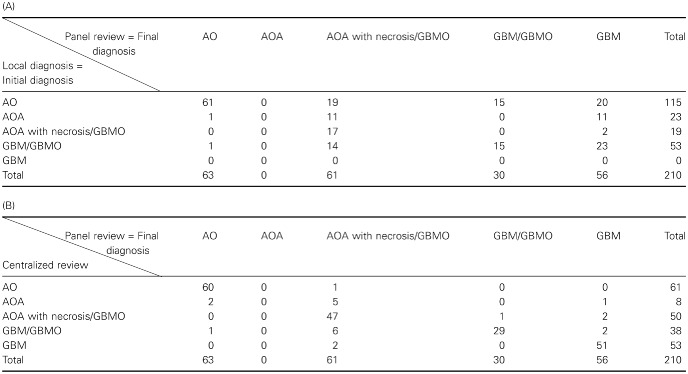
About 210 patients from 29 clinical centers were enrolled in the study. According to the POLA network recommendations, local pathologists from the clinical centers sent all cases demonstrating features of de novo malignant glioma with an oligodendroglial component for centralized review. Centralized review was made by Dr K. Mokhtari for Marseille cases, and by Professor D. Figarella‐Branger for all the other cases. Then, all cases benefited from a panel review by a board of four neuropathologists (Professor D. Figarella‐Branger, Professor E. Uro‐Coste, Dr K. Mokhtari and Dr A. Jouvet). The panel review was performed around a multi‐head microscope to provide consensus diagnosis. Importantly, this was performed without knowing the results of the centralized review, the immunohistochemistry (IHC) analysis or the genotyping. Only cases demonstrating features of diffuse adult HGG with necrosis after panel review were eligible. Table 1A shows the concordance between local diagnosis (initial diagnosis) and panel review (final diagnosis), and Table 1B shows the concordance between the centralized review and the panel review. For all cases, formalin‐fixed paraffin‐embedded (FFPE) tumor tissue was available for histological and immunohistochemical investigations. In addition, frozen material was available in 78% of cases. Patients included prospectively in the POLA network have provided their written consent for clinical data collection and genetic analysis according to the national and POLA network policy (see flowchart, Supporting Information Figure S1).
Table 1.
A. Concordance table between local diagnosis (initial diagnosis) and panel review (final diagnosis) for the five possible outcomes (AO, AOA, AOA with necrosis/GBMO, GBM/GBMO and GBM). B. Concordance table between the centralized review and the panel review for the same outcomes. Abbreviations: AO = anaplastic oligodendroglioma; AOA = anaplastic oligoastrocytoma; GBM = glioblastoma; GBMO = glioblastomas with an oligodendroglial component
Preoperative KPS (Karnofsky Performance Status) was known in 119/210 patients and the extent of surgical removal was assessed by the local neurosurgeon and was recorded in 156/210 patients. As postoperative contrast enhanced imaging was available for a minority of patients, two groups were established: biopsy (n = 30) vs. surgery (n = 126). According to the POLA network guidelines drawn up in 2008, it was strongly recommended that patients with 1p/19q co‐deletion received radiotherapy only or were included in the CODEL EORTC trial, whereas patients exhibiting non‐1p/19q co‐deleted AO were treated by concurrent and adjuvant temozolomide radiochemotherapy 20 or were included in the CATNON EORTC trial (NCT00626990). However, after the publication in 2013 of the long‐term results of the RTOG and EORTC trials in AO, the treatment recommendation for 1p/19q co‐deleted tumors was changed to radiotherapy followed by six cycles of adjuvant PCV (procarbazine, CCNU and vincristine) chemotherapy regimen 2, 21. Median follow‐up was 12.1 months (range: 0–63.8 months). During this period, 102 patients relapsed, 55 died and 21 were lost to follow‐up.
Methods
Histological review
During the centralized and panel reviews, the pathologists were specifically looking for the presence (or absence) of mitosis, marked atypia, areas of high cell density, microvascular proliferation, necrosis (palisading or not), branched vessels, oligodendroglial component, honeycomb‐like appearance and calcification. In addition, particular attention was paid to cell differentiation and the distribution of the oligodendroglial component. According to the WHO 2007 classification, tumors were classified as AO when typical features of oligodendrogliomas were recorded (cellular monomorphism, round regular nuclei giving a honeycomb‐like appearance and chicken wire vasculature often associated with microcalcifications). GBMO were split into two categories: the first one called “AOA with necrosis/GBMO” was restricted to tumors displaying intermingled astrocytic and oligodendroglial components associated with microvascular proliferation and necrosis. In these cases, cell differentiation was always preserved in all cells, and both astrocytic and oligodendroglial components were easily recognizable. The second GBMO subtype, named “GBM/GBMO,” was characterized by two distinctive patterns: one made of oligodendroglial foci (whatever their size) and the other displayed features of GBM with undifferentiated or poorly differentiated cells (Figure 1). Importantly, small cell GBM can be distinguished from oligodendroglioma at high magnification because it shows highly monomorphic nuclei with an elongated shape and hyperchromasia, a feature not encountered in oligodendroglial cells. Histological evaluations and classification were blinded to the molecular data.
Figure 1.
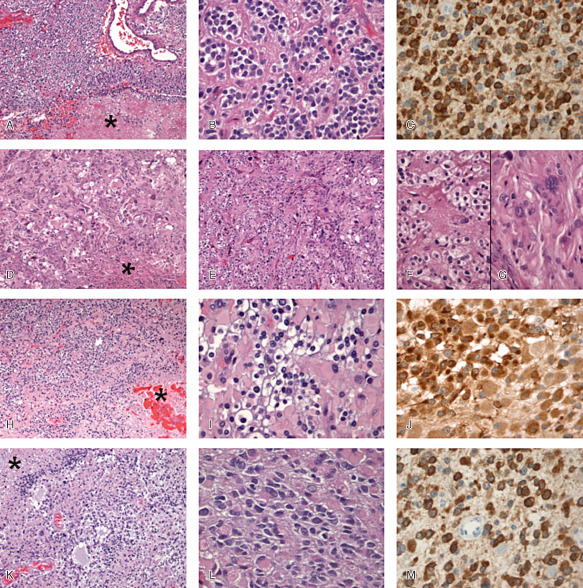
A–C. Anaplastic oligodendroglioma (AO) with focal necrosis. A. Oligodendroglial population with high cellularity, vascular proliferation and focal necrosis (asterisk) [hematoxylin and eosin (HE) ×100]. B. Perinuclear halo, moderate pleomorphism and hyperchromatism (HE ×400). C. Diffuse expression of IDH1 R132H (×400). D–G. Glioblastoma with oligodendroglial component (GBM/GBMO). D. Malignant glioma with high cellular pleomorphism and necrosis (asterisk) (HE ×100). E. On the left, tumor is composed of round cells with perinuclear halo and on the right by more pleomorphic cells (HE ×100). F. High magnification of oligodendroglial‐like component (HE ×400). G. High magnification of more classical GBM feature (HE ×400). H–J. Anaplastic oligoastrocytoma (AOA) with necrosis/GBMO. H. Mixture of oligodendroglial‐like and gemistocytic‐like cells with necrotic area (asterisk) (HE ×100). I. High magnification showing transitional appearances between oligodendroglial and astrocytic cells (HE ×400). J. Diffuse expression of IDH1 R132H (×400). K–M. AOA with necrosis/GBMO. K, L. Mixture of oligodendroglial cells and astrocytic cells usually pleomorphic with necrotic area (asterisk) (HE ×100). M. Diffuse expression of IDH1 R132H (×400).
Immunohistochemistry (IHC)
Automated IHC was performed on 4‐μm‐thick FFPE sections with avidin biotin peroxidase complex on Benchmark XT (Ventana Medical System Inc., Tucson, AZ, USA) with Ventana kit including 3,3'‐diaminobenzidine (DAB) reagent. The cases were screened for Ki67 (clone Mib1, 1/100, Dako, Courtaboeuf, France), p53 (clone DO.7, 1/200, Dako) and the monoclonal antibody IDH1 R132H (clone H09, 1/75, Dianova GmbH, Hambourg, Germany), which is highly sensitive and specific for IDH1 R132H mutation 3 and allows the detection of up to 90% of IDH‐mutated cases 24. Expression of Ki67 and p53 was scored in percentage by counting the immunostained nuclei in 400 cells in the most positive area.
DNA extraction
Following the manufacturer's recommendations, tumor DNA was extracted from frozen tissue if available or from FFPE samples using the iPrep ChargeSwitch® Forensic Kit, Life Technologies, Saint Aubin, France. Qualification and quantification of tumor DNA was conducted using a NanoVue spectrophotometer and gel electrophoresis, respectively.
1p/19q co‐deletion, EGFR amplification and P16 deletion analysis
Chromosome 1p/19q, EGFR and P16 status were assessed for all tumors on SNP (single nucleotide polymorphism) array genomic profiles. By using this technique, tumors were considered as co‐deleted if they exhibited whole chromosome arm 1p deletion, whole chromosome arm 19q deletion, chromosome 1 centromeric breakpoint and chromosome 19 centromeric breakpoint. This pattern is highly suggestive of t(1;19)(q10;p10). Therefore, if these four criteria were not met concurrently, the tumor was classified as non–co‐deleted regardless of 1p and 19q status otherwise.
IDH1 and IDH2 mutation status
When IDH1 R132H IHC was negative, IDH1 and IDH2 mutational status was systematically addressed by direct sequencing using Sanger method and primers as previously described 10 in order to formally exclude IDH1 and IDH2 other mutations.
Statistical analysis
The combined kappa statistics for the two histological reviews (local diagnosis vs. panel review and centralized review vs. panel review) and four possible outcomes (AO, AOA with necrosis/GBMO, GBM/GBMO and GBM) were calculated.
The SNP array analysis was performed as previously described 11. For all arrays, genomic imbalances were classified as loss, gain, homozygous deletion or amplification. The association of chromosome arm imbalances with histological variables was estimated using either Fisher's exact test (for factors) or Student's t‐test (for quantitative variables). For other correlation analyses, the chi‐square test (or Fischer's exact test) was used to compare variables when they scored as positive or negative. In order to identify histological and/or molecular factors related to overall survival (OS) or progression free survival (PFS), survival curves were obtained according to the Kaplan–Meier method and compared using the log‐rank test. The following variables were searched for prognosis significance in the whole group of 210 cases: age at diagnosis (cut‐off = 50 years), sex, extent of surgical resection (biopsy vs. surgery), preoperative KPS (cut‐off = 70), histological groups, molecular subgroups according to IDH1 R132H expression and 1p/19q co‐deletion, EGFR amplification and P16 deletion. Age at diagnosis, extent of surgical removal, histological groups and molecular subgroups were used to build the multivariate Cox proportional hazards backward models. However, because of the short clinical follow‐up of our series, these analyses included a large number of censored patients at the endpoint. All statistical tests were two‐sided and the threshold for statistical significance was P = 0.05. Analyses were conducted using PASW Statistics version 17.02 (IBM SPSS Inc., Chicago, IL, USA).
Results
Histological review
After panel review, the 210 diffuse adult HGGs with necrosis (palisading or not) were classified according to the WHO 2007 classification as anaplastic oligodendroglioma grade III (AO, n = 63; Figure 1A–C), glioblastoma grade IV (GBM, n = 56), glioblastoma with an oligodendroglial component [n = 91, including 30 GBM/GBMO (Figure 1D–G) and 61 AOA with necrosis/GBMO (Figure 1H–M)]. In all cases, microvascular proliferation was recorded. Some of the AOs included in this study have been previously reported 6. It is worth noting that agreement was very poor between local pathologist and panel review in the assessment of glioma subtype (Table 1A, kappa = 0.27) but it was excellent between centralized review and panel review (Table 1B, kappa = 0.85). Although the panel review was performed in average of 3 months after central review without knowledge of the diagnosis performed by the central review, we cannot exclude that the two pathologists who had performed the central review might have influenced, to some extent, the other pathologists for final diagnosis contributing to the excellent agreement between centralized review and panel review.
Immunohistochemistry
Mean Ki67 expression was 29.5% ± 17% (median 20%) and was not statistically correlated with histological diagnoses. p53 expression was scored as positive in 77/210 (37%) cases and was statistically correlated with histological diagnosis (P < 10−4): 11/63 AO (17%), 37/61 AOA with necrosis/GBMO (61%), 14/30 GBM/GBMO (47%) and 15/56 GBM (27%). IDH1 R132H positive expression was detected in 87/210 (41%). The 123 cases not expressing IDH1 R132H protein were successfully analyzed by direct sequencing: 4 cases of AO displayed IDH2 mutation and 1 case of GBM displayed IDH1 R132G mutation, both mutations being not detected by the IDH1 R132H immunolabeling. The other cases displayed no mutation.
Molecular data (Table 2)
Table 2.
A. Histological diagnosis distribution according to the molecular alteration studied and results of the correlation analyses between these variables using the chi‐square test. B. IDH sequencing results in IDH1 R132H‐ cases, according to histological diagnosis. Abbreviations: AO = anaplastic oligodendroglioma; AOA = anaplastic oligoastrocytoma; GBM = glioblastoma; GBMO = glioblastomas with an oligodendroglial component
| (A) | |||||
|---|---|---|---|---|---|
| AO | AOA with necrosis/GBMO | GBM/GBMO | GBM | P‐value | |
| 1p/19q co‐deletion | 45/63 | 7/61 | 1/30 | 0 | <0.0001 |
| IDH1 R132H expression | 50/63 | 39/61 | 1/30 | 1/56 | <0.0001 |
| EGFR amplification | 3/63 | 5/61 | 10/30 | 29/56 | <0.0001 |
| p16 deletion | 6/63 | 16/61 | 16/30 | 30/56 | <0.0001 |
| (B) | ||
|---|---|---|
| IDH1 R132H+ | IDH sequencing results in IDH1 R132H‐ cases | |
| AO | 50/63 | 4/13 IDH2 mutations (all 1p/19q co‐deleted) |
| AOA with necrosis/GBMO | 39/61 | 22/22 no mutation |
| GBM/GBMO | 1/30 | 29/29 no mutation |
| GBM | 1/56 | 1/55 IDH1 R132G mutation |
A 1p/19q co‐deletion was present in 53/210 cases (25%). All these cases were IDH‐mutated including 49 IDH1 R132H mutations and 4 IDH2 mutations. No GBM displayed 1p/19q co‐deletion. EGFR amplification was observed in 47/210 cases (22%) and P16 deletion occurred in 68/210 (32%) cases. All these molecular alterations were significantly related to histological diagnosis (P < 10−4). By combining IDH1 R132H expression and 1p/19q co‐deletion status, we defined three molecular subgroups: (i) 1p/19q co‐deletion; (ii) IDH1 R132H positive and 1p/19q intact; and (iii) IDH1 R132H negative and 1p/19q intact. These molecular subgroups were also significantly related to histological diagnosis (P < 10−4) (Figure 2A). Moreover, EGFR amplification, P16 deletion (Figure 2B) and p53 overexpression were differentially observed in these subgroups (P < 10−4). p53 expression was observed in 7/53 of 1p/19q co‐deleted cases and the percentage of immunostained nuclei was rather low (mean 31%, median 20%), whereas it was observed in 41/119 of the IDH1 R132H negative and 1p/19q intact cases (mean number of immunostained nuclei 46%, median 30%). Of interest, in the third group characterized by IDH1 R132H expression and 1p/19q intact, 29/38 cases strongly expressed p53 in a high number of nuclei (mean number of immunostained nuclei 67%, median 80%).
Figure 2.
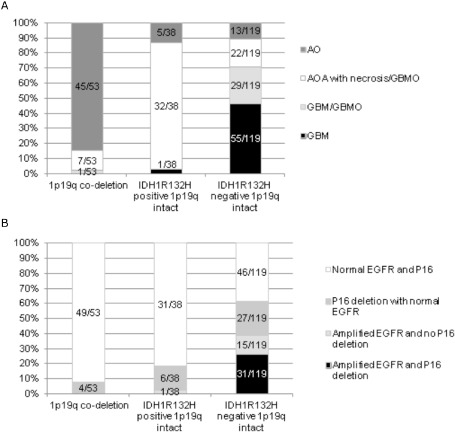
A. Histological diagnosis distribution according to the three molecular subgroups defined regarding 1p/19q co‐deletion and IDH1 R132H expression status. B. EGFR amplification and p16 deletion status in these three molecular subgroups.
Correlation between clinical, histological features, molecular data and follow‐up
Univariate analysis (Table 3A) showed that among the clinical variables analyzed, age <50 years at diagnosis was predictive of a longer PFS (P < 10−4) and a longer OS (P = 0.031). Surgical resection was also predictive of a longer PFS than biopsy (P = 0.001). Among the histopathological variables analyzed, histological groups were predictive of PFS (P < 10−4) (Figure 3A) and OS (P < 10−4) (Figure 3B), the best prognosis being observed for AO with a survival rate of 75% at the endpoint. Interestingly, when the subgroup of AO was split into two groups according to the 1p/19q status, it was obvious that AO 1p/19q co‐deleted had the longest PFS (37.3 months, 95% confidence interval (CI) [30.1–44.4]; Figure 3C) and OS (54.7 months, 95% CI [48.2–61.2]; Figure 3D), whereas AO with intact 1p/19q showed almost the same behavior as AOA with necrosis/GBMO (22.4 months, 95% CI [17–27.8] for PFS and 34.4 months, 95% CI [25.4–43.4] for OS). At the endpoint, AO 1p/19q co‐deleted had a survival rate of 87%, whereas AO with intact 1p/19q had a survival rate of 44%. Regarding the histomolecular variables analyzed, patients displaying 1p/19q co‐deletion (and IDH mutation) had the longest PFS (P < 10−4) (Figure 3E) and OS (P < 10−4) (Figure 3F).
Table 3.
Clinical, histological and molecular markers predictive of prognosis in diffuse adult high‐grade gliomas with necrosis on (A) univariate analysis and (B) multivariate analysis. Abbreviations: AO = anaplastic oligodendroglioma; AOA = anaplastic oligoastrocytoma; CI = confidence interval; GBM = glioblastoma; GBMO = glioblastomas with an oligodendroglial component; HR = hazard ratio; KPS = Karnofsky Performance Status; OS = overall survival; PFS = progression free survival
| (A) | ||||
|---|---|---|---|---|
| Univariate analysis | ||||
| PFS (mean [95% CI]) (months) | P‐value | OS (mean [95% CI]) (months) | P‐value | |
| Age at diagnosis | <0.0001 | 0.031 | ||
| <50 years | 29.8 [24.5–35.2] | 43 [36.6–49.6] | ||
| >50 years | 15.5 [13–18] | 31.2 [26–36.3] | ||
| Sex | 0.531 | 0.710 | ||
| Male | 22.9 [18.5–27.3] | 40.4 [33.8–46.9] | ||
| Female | 24.1 [18.9–29.3] | 36.7 [30.4–43] | ||
| Preoperative KPS | 0.399 | 0.152 | ||
| <70 | 17.5 [10.7–24.2] | 23.3 [15.5–31.1] | ||
| >70 | 28.2 [23–33.4] | 42.1 [36.3–48] | ||
| Extent of surgical removal | 0.001 | 0.092 | ||
| Biopsy | 14.5 [7.7–21.3] | 36.3 [25.2–47.4] | ||
| Surgery | 25 [21.1–28.9] | 39.8 [34.4–45.1] | ||
| Histological diagnosis | <0.0001 | <0.0001 | ||
| AO | 32.7 [27–38.4] | 47.6 [41.5–53.6] | ||
| AOA with necrosis/GBMO | 19.9 [16.1–23.8] | 33.2 [25.6–40.9] | ||
| GBM/GBMO | 14.2 [9.8–18.6] | 19 [13–25] | ||
| GBM | 10.7 [7.8–13.8] | 22.7 [16.6–28.7] | ||
| Molecular subgroups | <0.0001 | <0.0001 | ||
| 1p/19q co‐deletion | 37.1 [30.5–43.9] | 55.4 [49.4–61.5] | ||
| IDH1R 132H positive/ 1p/19q intact | 21.8 [16.6–27.1] | 31.6 [23.7–39.5] | ||
| IDH1R 132H negative/ 1p/19q intact | 14.7 [12.1–17.3] | 26.5 [21.6–31.5] | ||
| EGFR gene status | 0.003 | 0.004 | ||
| Normal | 26.1 [21.9–30.4] | 41.8 [36.5–47] | ||
| Amplified | 13.7 [10.4–16.9] | 22.9 [17.5–28.3] | ||
| P16 gene status | 0.004 | 0.111 | ||
| Normal | 27.1 [22.4–31.8] | 41.5 [35.8–47.2] | ||
| Deleted | 17.6 [13.1–22] | 31.7 [25.2–38.2] | ||
| (B) | ||||||
|---|---|---|---|---|---|---|
| Multivariate analysis | ||||||
| PFS | OS | |||||
| HR | 95% CI | P‐value | HR | 95% CI | P‐value | |
| Histological diagnosis | 0.003 | 0.020 | ||||
| AO | ||||||
| AOA with necrosis/GBMO | ||||||
| GBM/GBMO | 1.350 | 1.107–1.647 | 1.397 | 1.053–1.852 | ||
| GBM | ||||||
| Molecular subgroups | <0.0001 | <0.0001 | ||||
| 1p/19q co‐deletion | 0.491 | 0.369–0.653 | 0.297 | 0.181–0.486 | ||
| IDH1R 132H positive/ 1p/19q intact | ||||||
| IDH1R 132H negative/ 1p/19q intact | ||||||
Figure 3.
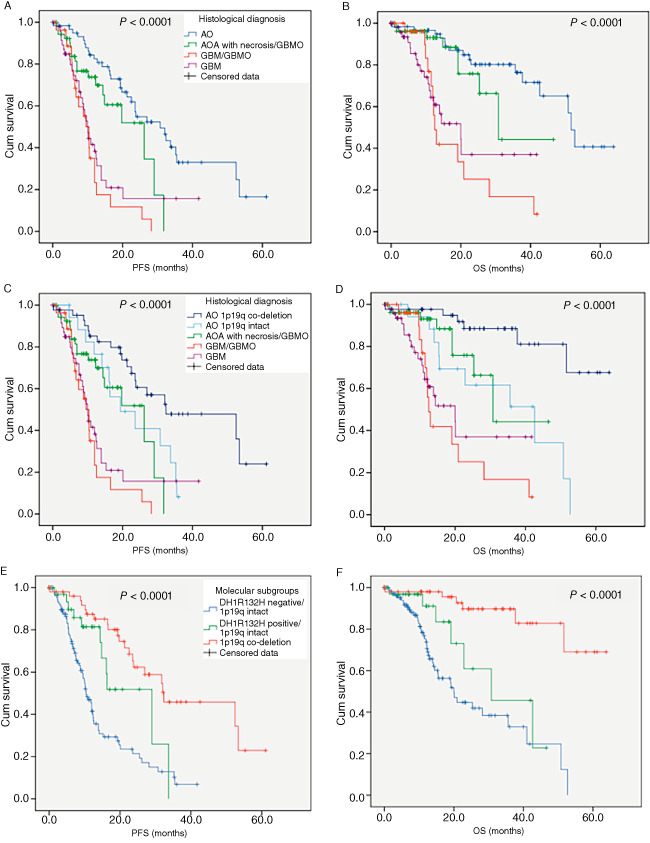
A–D. Histological subgroups display significant different progression free survival (PFS) and overall survival (OS): AO with 1p/19q co‐deletion (C, D) are associated with best prognosis. E–F. Three prognostically relevant molecular subgroups of patients can be defined on the basis of IDH1 R132H expression and 1p/19q co‐deletion status: IDH1 R132H positive and 1p/19q co‐deleted cases are associated with best prognosis.
On multivariate analysis (Table 3B), occurrence of 1p/19q co‐deletion was predictive of longer PFS (P < 10−4, hazard ratio (HR) 0.491; 95% CI [0.369–0.653]) and OS (P < 10−4, HR 0.297; 95% CI [0.181–0.486]) and patients being diagnosed with GBM/GBMO displayed a shorter PFS (P = 0.003, HR 1.350; 95% CI [1.107–1.647]) and a shorter OS (P = 0.020, HR 1.397; 95% CI [1.053–1.852]).
Discussion
In this article, we have first shown that according to histological features, we can divide diffuse adult HGGs with necrosis into four subgroups (AO, GBM, AOA with necrosis/GBMO and GBM/GBMO) and that each subgroup is associated with its own molecular markers, although some overlapping exists. We were aware that because of inclusion criteria of the POLA program, standard GBM was underrepresented in this series. Nevertheless, their number was sufficient to draw conclusions. We have also shown that the histological subgroups that we have defined were of prognostic relevance. Moreover, IDH1 R132H expression and 1p/19q co‐deletion were sufficient to stratify diffuse adult HGG with necrosis into three prognostically relevant molecular subgroups of patients. These results might impact the future WHO classification of diffuse adult HGG with necrosis.
Histological classification of diffuse adult HCG with necrosis and molecular characterization
In this study, we have shown that careful analysis (with trained pathologists) of the histological features of diffuse adult HGG with necrosis is of utmost importance because it has enabled us to consider four subgroups that are associated with distinct molecular features. Of particular interest was the distinction of GBMO into two categories: AOA with necrosis/GBMO and GBM/GBMO. All the cases that we have classified as AO demonstrated histological features classic for oligodendrogliomas 7 and were associated with the molecular signature of oligodendrogliomas (characterized by 1p/19q co‐deletion) recorded in 45/63 (65%) cases, which is slightly lower than other AO series not restricted to the necrotic cases 6, 7. In this group of 45 patients, 41 showed IDH R132H expression, whereas the remaining 4 cases were IDH2‐mutated. Therefore, as previously reported 15, all 1p/19q co‐deleted tumors were IDH‐mutated. The remaining AO demonstrated IDH1 R132H expression and intact 1p/19q (5/63 cases) or IDH1 R132H negative (and no IDH mutation) and intact 1p/19q (13/63 cases). As previously reported by our team, AO with histological features classic for oligodendroglioma remains a molecularly heterogeneous entity 6. The group of AOA with necrosis/GBMO was characterized by IDH1 R132 H expression and intact 1p/19q in 39/61 (63.9%) cases, whereas the remaining cases were 1p/19q co‐deleted in 7 cases and IDH1 R132H negative in 15 cases. Importantly, other IDH mutation was not observed in this group. In contrast, the GBM/GBMO subgroup shared with GBM absence of IDH1R132H expression and intact 1p/19q. Only 1/30 GBM/GBMO was 1p/19q co‐deleted and only one IDH1 mutation (IDH1 R132G) was detected in one tumor that we classified as GBM. Therefore, according to our histological criteria, it was obvious that GBMO represents a highly heterogeneous entity, GBM/GBMO being GBM‐like, that is, IDH1 R132H negative (IDH wild‐type) and intact 1p/19q, and AOA with necrosis/GBMO being mainly characterized by IDH1 R132H expression and intact 1p/19q. Although a search for ATRX expression was not performed in this series, we have recorded that cases characterized by IDH1 R132H expression and intact 1p/19q strongly expressed p53, suggesting an astrocytic‐like signature of a subset of AOA with necrosis/GBMO 12. In the literature, we can find contrasting results regarding molecular characterization of GBMO and this is likely to reflect the loose pathological definition of this subgroup, which encompasses, in fact, two distinct subclasses as defined in our study. As an example, IDH mutation was recorded in 19% of the GBMO reported by Hegi and co‐workers 9, whereas it reached 31% 23 and even 55% in other series 13. Therefore, it is not surprising that the prognostic relevance of this subgroup should vary from one study to another. In the same way, the number of GBMO retrieved from large GBM series account for 15% 9, 18.3% 23 and 25% 5.
EGFR amplification, a common feature of GBM 22, was detected in 29/56 GBM, 10/30 GBM/GBMO, 5/61 AOA with necrosis/GBMO and 3/63 AO. However, according to previous results, none of the 53 diffuse adult HGG with necrosis that display 1p/19q co‐deletion demonstrated EGFR amplification 4, 6.
Prognostic relevance of histological and molecular classifications
In addition to being predictive of molecular characteristics, the histological classification of diffuse adult HCG with necrosis into four groups was also of prognostic relevance. The current WHO classification classifies AO as grade III even if necrosis is observed, in contrast to mixed oligoastrocytomas that have to be classified as GBMO grade IV in case necrosis is present. The survival curves that demonstrated the best survival for AO in comparison to AOA with necrosis/GBMO and GBM/GBMO are in accordance with this recommendation. However, what our study clearly shows is that, in addition to sharing with GBM the same molecular signature, the group of GBM/GBMO shares with GBM the same dismal prognosis. Therefore, it is of utmost importance to distinguish AOA with necrosis/GBMO from GBM/GBMO. However, it is worth noting that when the initial diagnosis performed by the local pathologist was taken into account, the prognostic relevance of histological classification disappeared (data not shown). This emphasizes the difficulties in establishing accurate diagnosis for this group of gliomas, and to achieve this goal, we show here that some molecular markers are of utmost diagnosis relevance.
Actually, 1p/19q and IDH R132H expression are sufficient to stratify diffuse adult HGG with necrosis into three subgroups of prognostic relevance, the best prognosis with a 5‐year OS that reached 87% being observed for 1p/19q co‐deleted tumors. This molecular signature was also highly significant on multivariate analysis and was associated with longer PFS (P < 10−4, HR 0.491; 95% CI [0.369–0.653]) and OS (P < 10−4, HR 0.297; 95% CI [0.181–0.486]).
Interestingly, in this subgroup of diffuse adult HGG with necrosis, very few cases demonstrated IDH mutations that were not detected by IDH1 R132H immunostaining (5/210). Moreover, the prognostic relevance of the IDH mutation status in this group of patients relies on IDH1 R132H expression recorded by IHC, which is easy to perform in routine practice. These results are in agreement with the current recommendations 17, suggesting that additional IDH1/2 sequence analysis are required in the setting of a low‐grade or anaplastic tumor in a young adult patient in which the likelihood of IDH mutation is high, but not in the setting of an elderly patient with a clinically and histologically classic primary GBM. Moreover, when a diffuse glioma has whole‐arm 1p/19q co‐deletion, it seems unnecessary to assess the IDH mutation status.
At the meeting that was recently held in Haarlem (The Netherlands) to guide next steps in brain tumor classification and grading 17, it was also suggested that, in the future WHO classification, “integrated diagnosis” including histological classification, WHO grade and molecular information, might be proposed as the final diagnosis for brain tumor classification. Following this line, according to our results and in order to maintain consistency between histological features, molecular data, grade and final diagnosis, we suggest removing the name GBMO and integrating histological and molecular data to classify diffuse adult HGG with necrosis into “canonical oligodendroglioma” grade III when 1p/19q co‐deletion is present, “IDH1 R132H+ GBM” grade IV when IDH1 R132H is expressed and intact 1p/19q is recorded and as “GBM” grade IV when IDH1 R132H expression is lacking. This classification, which suggests removing the GBMO subgroup from the diffuse adult HGG with necrosis, is in the line of the recent published paper 19 suggesting to exclude the mixed gliomas from the WHO classification. Obviously, the histomolecular classification of diffuse adult HGG with necrosis that we have suggested should be confirmed by independent teams.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Figure S1. Flowchart of the histological review process, genotyping and biobanking in the POLA network.
Acknowledgments
This work was funded by the French Institut National du Cancer (INCa) and part of the national program Cartes d'Identité des Tumeurs® (CIT) (http://cit.ligue‐cancer.net/), funded and developed by the Ligue Nationale Contre le Cancer.
Patients from the AP‐HM institution are included in the SIRIC‐Marseille Glioma program (Grant INCa‐DGOS‐INSERM 6038).
Frozen specimens from the AP‐HM institution were stored then provided by the AP‐HM tumor bank (authorization number AC‐2013‐1786). Frozen specimens from Bordeaux were stored at Hôpital Haut Levèque CRB, 33604, Pessac, France. Frozen specimens from Montpellier were stored at CHU Montpellier, CCBH‐M, 34825, Montpellier, France. Frozen specimens from Nantes were stored in the IRCNA tumor bank, at CHU Nantes, Institut de Cancérologie de l'ouest, 44800 Saint‐Herblain, France. Frozen specimens from Saint‐Etienne were stored at CHU Saint‐Etienne, CRB 42, 42055 Saint‐Etienne, France. Frozen specimen from Lyon were stored at NeuroBioTec, Groupement Hospitalier Est, 69677 Bron Cedex, France. Frozen specimens from Amiens were stored at Tumorothèque de Picardie, CHU Amiens, 80054 Amiens, France.
References
- 1. Appin CL, Gao J, Chisolm C, Torian M, Alexis D, Vincentelli C et al (2013) Glioblastoma with oligodendroglioma component (GBM‐O): molecular genetic and clinical characteristics. Brain Pathol 23:454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cairncross G, Wang M, Shaw E, Jenkins R, Brachman D, Buckner J et al (2013) Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long‐term results of RTOG 9402. J Clin Oncol 31:337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Capper D, Weissert S, Balss J, Habel A, Meyer J, Jager D et al (2010) Characterization of R132H mutation‐specific IDH1 antibody binding in brain tumors. Brain Pathol 20:245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dehais C, Laigle‐Donadey F, Marie Y, Kujas M, Lejeune J, Benouaich‐Amiel A et al (2006) Prognostic stratification of patients with anaplastic gliomas according to genetic profile. Cancer 107:1891–1897. [DOI] [PubMed] [Google Scholar]
- 5. Elmahdi A, Frary AJ, Scotton WJ, O'Donovan DG, Price SJ (2013) Glioblastomas with oligodendroglial component have the same clinical phenotype as classical glioblastomas. Br J Neurosurg 27:419–424. [DOI] [PubMed] [Google Scholar]
- 6. Figarella‐Branger D, Mokhtari K, Dehais C, Jouvet A, Uro‐Coste E, Colin C et al (2014) Mitotic index, microvascular proliferation, and necrosis define 3 groups of 1p/19q codeleted anaplastic oligodendrogliomas associated with different genomic alterations. Neuro‐Oncol 16:1244–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Giannini C, Burger PC, Berkey BA, Cairncross JG, Jenkins RB, Mehta M et al (2008) Anaplastic oligodendroglial tumors: refining the correlation among histopathology, 1p 19q deletion and clinical outcome in Intergroup Radiation Therapy Oncology Group Trial 9402. Brain Pathol 18:360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He J, Mokhtari K, Sanson M, Marie Y, Kujas M, Huguet S et al (2001) Glioblastomas with an oligodendroglial component: a pathological and molecular study. J Neuropathol Exp Neurol 60:863–871. [DOI] [PubMed] [Google Scholar]
- 9. Hegi ME, Janzer RC, Lambiv WL, Gorlia T, Kouwenhoven MC, Hartmann C et al (2012) Presence of an oligodendroglioma‐like component in newly diagnosed glioblastoma identifies a pathogenetically heterogeneous subgroup and lacks prognostic value: central pathology review of the EORTC_26981/NCIC_CE.3 trial. Acta Neuropathol 123:841–852. [DOI] [PubMed] [Google Scholar]
- 10. Houillier C, Wang X, Kaloshi G, Mokhtari K, Guillevin R, Laffaire J et al (2010) IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low‐grade gliomas. Neurology 75:1560–1566. [DOI] [PubMed] [Google Scholar]
- 11. Idbaih A, Ducray F, Dehais C, Courdy C, Carpentier C, de Bernard S et al (2012) SNP array analysis reveals novel genomic abnormalities including copy neutral loss of heterozygosity in anaplastic oligodendrogliomas. PLoS ONE 7:e45950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF et al (2012) Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget 3:709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joseph NM, Phillips J, Dahiya S, M Felicella M, Tihan T, Brat DJ, Perry A (2012) Diagnostic implications of IDH1‐R132H and OLIG2 expression patterns in rare and challenging glioblastoma variants. Mod Pathol 26:315–326. [DOI] [PubMed] [Google Scholar]
- 14. Kraus JA, Lamszus K, Glesmann N, Beck M, Wolter M, Sabel M et al (2001) Molecular genetic alterations in glioblastomas with oligodendroglial component. Acta Neuropathol 101:311–320. [DOI] [PubMed] [Google Scholar]
- 15. Labussiere M, Idbaih A, Wang XW, Marie Y, Boisselier B, Falet C et al (2010) All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology 74:1886–1890. [DOI] [PubMed] [Google Scholar]
- 16. Louis D, Ohgaki H, Wiestler O, Cavenee W (2007) World Health Organization Classification of Tumours of the Central Nervous System. IARC Press: Lyon. [DOI] [PubMed] [Google Scholar]
- 17. Louis DN, Perry A, Burger P, Ellison DW, Reifenberger G, von Deimling A et al (2014) International Society of Neuropathology‐Haarlem Consensus Guidelines for Nervous System Tumor Classification and Grading. Brain Pathol 24:429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miller CR, Dunham CP, Scheithauer BW, Perry A (2006) Significance of necrosis in grading of oligodendroglial neoplasms: a clinicopathologic and genetic study of newly diagnosed high‐grade gliomas. J Clin Oncol 24:5419–5426. [DOI] [PubMed] [Google Scholar]
- 19. Sahm F, Reuss D, Koelsche C, Capper D, Schittenhelm J, Heim S et al (2014) Farewell to oligoastrocytoma: in situ molecular genetics favor classification as either oligodendroglioma or astrocytoma. Acta Neuropathol 128:551–559. [DOI] [PubMed] [Google Scholar]
- 20. Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ et al (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352:987–996. [DOI] [PubMed] [Google Scholar]
- 21. van den Bent MJ, Carpentier AF, Brandes AA, Sanson M, Taphoorn MJ, Bernsen HJ et al (2006) Adjuvant procarbazine, lomustine, and vincristine improves progression‐free survival but not overall survival in newly diagnosed anaplastic oligodendrogliomas and oligoastrocytomas: a randomized European Organisation for Research and Treatment of Cancer phase III trial. J Clin Oncol 24:2715–2722. [DOI] [PubMed] [Google Scholar]
- 22. Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD et al (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y, Li S, Chen L, You G, Bao Z, Yan W et al (2012) Glioblastoma with an oligodendroglioma component: distinct clinical behavior, genetic alterations, and outcome. Neuro‐Oncol 14:518–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W et al (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Flowchart of the histological review process, genotyping and biobanking in the POLA network.


