Abstract
Whenever Pseudomonas aeruginosa is cultured from cystic fibrosis (CF) patient airways, the primary goal is eradication by antibiotic therapy. Success is defined by ≥6 months of negative bacterial airway cultures. However, we suspect that P. aeruginosa persists in airways without clinical detection for long periods.
Out of 298 P. aeruginosa-infected Copenhagen CF patients, we identified 80 with complete P. aeruginosa monitoring records and measured their maximum P. aeruginosa-free eradication periods (MEP). Isolates from 72 patients were whole-genome sequenced (n=567) and clone typed. Select isolate relatedness was examined through phylogenetic analysis and phenotypic multivariate modelling.
69 (86%) patients exhibited eradication in the monitoring period (2002–2018). Sequenced isolates bridged the MEP of 42 patients, and the same clone type persisted over the MEP in 18 (43%) patients. Patients with failed eradication were on average treated more intensively with antibiotics, but this may be linked to their more severe pre-MEP infection trajectories. Of the 42 patients, 26 also had sinus surgery; the majority (n=15) showed MEPs adjacent to surgery, and only five had persisting clone types. Importantly, combined phylogenetic–phenomic evaluation suggests that persisting clone types are a result of re-emergence of the same strain rather than re-infection from the environment, and similar relatedness is exhibited by paired lower and upper airway samples and in transmission cases.
In conclusion, nearly half of CF patients with supposed eradication may not truly be cleared of their original bacteria according to omics-based monitoring. This distinct cohort that is persistently infected would probably benefit from tailored antibiotic therapy.
Short abstract
For 80 cystic fibrosis patients, this study used omics and positive culture history of P. aeruginosa infections to show that strains routinely persist over lengthy Pseudomonas-free periods. The authors recommend using genomic data in “eradication” metrics. https://bit.ly/2H318Ca
Introduction
The positive experience of early and aggressive antibiotic treatment, which has prolonged the lives of cystic fibrosis (CF) patients, has promoted the clinical conclusion that populations of colonising bacteria early in the treatment timeline can be eradicated from CF airways, thus delaying the onset of chronic infection [1–3]. The progression of Pseudomonas aeruginosa lung infection in CF patients is currently divided into two components: 1) intermittent colonisation, where a bacterial species cannot be cultured from the next sputum sample after antibiotic therapy; and 2) chronic infection, denoted by continuous culturing of P. aeruginosa (the “Leeds criteria” [4]), and/or an increased antibody response or mucoidity of cultures (the “Copenhagen criteria”) [5, 6]. These guidelines have been structured around avoiding chronic infections of P. aeruginosa, the historically dominant pathogen in the lungs of CF patients [7]. At the Copenhagen CF Center, the frontline approach to avoiding chronic infections for more than two decades has been routine use of early eradication therapy (EET), a standardised treatment period predominantly consisting of inhaled colistin and oral ciprofloxacin, for every new positive culture of P. aeruginosa after a period of ≥6 months of P. aeruginosa-negative culture [1]. The EET approach has been recommended in best practice guidelines by the European Cystic Fibrosis Society, although no specific antibiotic regimen has been recommended due to lack of evidence [8, 9]. While several publications have stated that antibiotic therapy applied to “early P. aeruginosa infection” has in fact achieved eradication in 66–80% of patients [2, 10, 11] and postponed chronic infection in up to 80% [1], few of these incorporated strain genomics in their assessment of eradication. However, integration of pathogen genotyping and genomics in CF clinics is growing, offering increased resolution into strain dynamics within patient cohorts such as genotype prevalence within and between patients [10, 12–14], strain origin [15] and diversification [16–18] and transmission [7, 10]. The adoption of bacterial whole-genome sequencing (WGS) in particular provides a reliable, quantitative method of tracking the persistence and adaptation of specific strains.
Given P. aeruginosa's extensive toolset for evading treatment, we expect that strain persistence in CF patients is common; however, the incidence and underlying mechanisms of this persistence are poorly tracked in current clinical practice. The purpose of this study was to contextualise clinical bacteriology records with genomic and phenomics screening data to 1) further our understanding of infection dynamics and strain persistence; and 2) demonstrate the utility of WGS-based tools in routine care and in defining type of infection. We focus on the longest Pseudomonas-free interval in each patient's culture records (referred to as the maximum eradication period (MEP) when >0.5 years), as this represents the period most responsive to therapy and most likely to result in true strain eradication. We then compare strains via available genomic and phenomic data to assess strain presence, persistence over MEPs, response to treatment, and adaptation during these periods and between different airway environments. This work provides an important new assessment of true strain persistence and therefore treatment effectiveness in a large patient cohort using WGS-based tools, which illustrates the potential for treatment redesign from an omics-based perspective.
Methods
Patient cohort and sampling
Since 1971, all Danish CF patients have been followed approximately monthly in the outpatient clinic at the Copenhagen CF Center at Rigshospitalet (Copenhagen, Denmark) for clinical status, pulmonary function tests and bacteriological investigations of lower respiratory tract secretions [5, 19, 20]. Since late 2004, we have collected all first and most subsequent P. aeruginosa lung isolates from CF children and young adults in a frozen biobank. Since 2007, all P. aeruginosa isolates cultured from sinus surgery samples have also been stored [21]. In total, airway cultures of P. aeruginosa assessed in this study were derived from approximate monthly samples that included expectorate, endolaryngeal suction, bronchial and/or nasopharyngeal secretions, sinus surgery tissue, pus and fluid samples, bronchiolar lavage or saliva. Further details on standard culture methods and patient classification at the CF Center at Rigshospitalet are described in the supplementary methods. Through a database maintained by the department of clinical microbiology at Rigshospitalet with systematic entries of all airway bacteriological samples since 2002, we had access to records for 399 patients with CF treated at the Copenhagen CF Center between January 2002 and December 2018. During the study period, 298 (75%) out of 399 patients had one or more P. aeruginosa-positive culture from their airways, and we focus on 80 (27%) of these 298 patients, who were divided into the four cohorts described later. Further details on this selection based on factors such as sample frequency and exclusion parameters are discussed in detail in the supplementary methods (supplementary figures S1 and S2). The local ethics committee at the Capital Region of Denmark (Region Hovedstaden) approved the use of the stored P. aeruginosa isolates (registration numbers H-4-2015-FSP and H-19029688) and the sinus surgery procedure (registration number H-A-2008-141). Access to the microbiological and treatment records was approved by both the management of the department of clinical microbiology at Rigshospitalet and the Danish Agency for Patient Safety (registration number 31-1521-428), and all data analysis was performed on pseudonymised data.
We hereafter refer to four nested patient groups in our analysis (table 1):
1) Cohort 1 (C1): this group of 80 patients has a complete and continuous record of P. aeruginosa cultures with their first P. aeruginosa culture collected within the study period, i.e. after January 2002 and before July 2018. These patients were still under continuous care at the CF Center at Rigshospitalet at the end of 2018, and we thus had their complete P. aeruginosa infection history covered in the bacteriological database. For each patient, we calculated the length of every period where the clinic did not detect any P. aeruginosa in airway culture (Pseudomonas-free interval).
2) Cohort 2 (C2): this group of 72 patients from C1 has had at least one isolate whole-genome sequenced. Sequencing was performed either for genotypic monitoring in the clinic or, for 23 patients, during a 2015 study of 474 longitudinal WGS P. aeruginosa isolates [17].
3) Cohort 3 (C3): this group of 57 patients from C2 has at least one instance of a Pseudomonas-free interval lasting >6 months, qualifying according to the Copenhagen CF Center as a true “eradication period”.
4) Cohort 4 (C4): this group of 42 patients from C3 has a sufficient dataset of sequenced isolates (supplementary methods) to evaluate whether an early strain persists over the longest eradication period, or MEP.
TABLE 1.
Cohort overview: counts of males and females, median age in years at first Pseudomonas aeruginosa sample, the time covered by P. aeruginosa-positive cultures (timespan) in years and the total number of positive P. aeruginosa samples
| CF patient filtering | Patients | Patient age# years | Sample timespan¶ | Total samples+ | |||
| Total | Male | Female | |||||
| C1 | First P. aeruginosa-positive culture after 2002 | 80 | 36 | 44 | 4.3 (0.3–17.2) | 7.0 (0.0–16.7) | 2093 |
| C2 | ≥1 sequenced isolates | 72 | 30 | 42 | 5.1 (0.3–24.5) | 5.1 (0.0–15.3) | 567 |
| C3 | Eradication period >0.5 years | 57 | 24 | 33 | 5.8 (0.8–24.5) | 7.5 (0.0–15.3) | 525 |
| C4 | Sufficient data to assess persisting/switched strains | 42 | 20 | 22 | 4.6 (0.8–17.2) | 7.5 (0.7–15.3) | 438 |
Data are presented as n or median (range). #: C1: age at first P. aeruginosa-positive culture, C2–4: age at first sequenced P. aeruginosa isolate; ¶: C1 timespan from first P. aeruginosa-positive culture to last, C2–4: timespan of sequenced isolates; +: C1: total samples cultured, C2-4: total samples sequenced.
Definition of infection status
Since 1974, CF patients have been defined as intermittently colonised from the first positive culture of P. aeruginosa until chronic diagnosis according to either the Copenhagen definition or the Leeds criteria [4–6]. In this study, patients are defined as chronically infected when P. aeruginosa is cultured in six consecutive monthly sputum samples and/or there are elevated or increasing precipitating antibodies against P. aeruginosa and/or if the bacteria is observed to have a mucoid phenotype [5, 6]. We define eradication periods as Pseudomonas-culture-free intervals of ≥6 months in length, and the MEP as the longest lasting eradication period for all patients who have shown at least one eradication period.
Patient interventions
The Copenhagen CF Center follows a positive culture-based model of treatment supported by the approximately monthly patient clinic visits. CF patients treated at the Copenhagen CF Center undergo EET of 3 weeks (before 2008, this lasted 3 months) of predominantly oral ciprofloxacin and nebulised colistin for treatment of first P. aeruginosa isolation (a few patients are treated with ciprofloxacin and tobramycin based on patient-specific sensitivities) [1, 22]. If P. aeruginosa is subsequently cultured within the following 6 months, an intravenous course of 2 weeks (β-lactam+aminoglycoside based on antibiogram) is initiated followed by treatment with inhaled antibiotics, preferably colistin in combination with oral ciprofloxacin for 3 months. If P. aeruginosa is not detected within the 6 months after the early eradication, the next positive culture is treated as a new infection using EET with ciprofloxacin and colistin unless antibiotic susceptibility tests support use of alternative treatment. Sinus surgery is considered when patients have noncontinuous positive P. aeruginosa culture, they have declining lung function, specific antibody levels are increasing, lung transplantation occurred within the prior year, or symptoms of severe rhinosinusitis are exhibited [21].
Sequencing and genotyping
Both in-house and external sequencing was performed during the study period. For in-house sequencing, a DNEasy Blood and Tissue Kit (Qiagen Denmark, Copenhagen, Denmark) was used to purify DNA from overnight (in liquid or on blood agar plates) cultures of single colonies. Sequencing of libraries constructed using Nextera XT was performed on an Illumina MiSeq using a v2 250×2 kit (Illumina, San Diego, CA, USA). External sequencing was performed by BGI Genomics, Europe (Copenhagen, Denmark) on their DNBseq platform. De novo assemblies were aligned and clone types were demarcated on the basis of >10 000 differential single nucleotide polymorphisms (SNPs) post assembly alignment as described by Marvig et al. [17].
Statistics
Statistical comparisons between groups were performed using unpaired Wilcoxon–Mann–Whitney tests to compare group means in pairwise fashion. All data analysis and modelling was conducted in R (version 3.4.0; www.r-project.org).
Phylogenetic reconstruction
A maximum likelihood phylogeny was produced using MEGA (version 7.0.26) [23], based on concatenated SNPs from Marvig et al. [17] using 1000 bootstraps. This was performed for multipatient clone types as shown in supplementary figure S4.
Archetype analysis
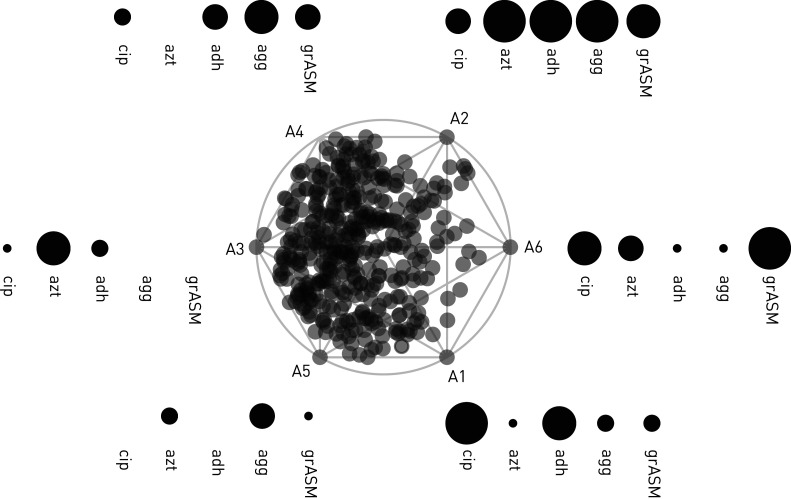
Isolates related to eradication periods in 19 patients of C4 were analysed with respect to their similarity to phenotypic “archetypes” defined by the archetypal model constructed in Bartell et al. [24] using 443 phenotyped P. aeruginosa isolates from our young Copenhagen CF cohort. In-depth analysis was performed for prominent and well-characterised clone types DK19 and DK06 [17]. Archetype analysis is a multidimensional statistical modelling approach where the extremal points of a multifeature dataset are defined as “archetypes” and all samples are defined with respect to their similarity to each archetype. In this analysis, we evaluate five phenotypic traits (features) that are known to adapt during CF infections and can be assessed by continuous values (growth rate, susceptibility to antibiotics ciprofloxacin and aztreonam, aggregation and adhesion). We refer to these measures as a phenomics dataset because they were collected at strain library scale for integration with genomics data via modelling. Archetype-based assessment combines and translates this multidimensional data into novel insights into systems-level pathogen evolution. This method has proven useful because the predicted archetypes match naïve and adapted evolutionary states. The projected simplex visualisations (figure 1) allow simultaneous assessment of multiple traits for each sample based on their localisation relative to the archetypes and illustrate the multitrait adaptive trajectory of an evolving clone type between archetypes.
FIGURE 1.
Archetype analysis simplex projection. A simplex visualisation is presented centrally, with individual isolates projected as transparent grey dots. Archetypes are denoted on the edge of the simplex (A1 to A6) ordered with the archetypes showing the most naïve phenotypic characteristics on the left (A3 and A5) and the archetypes with the more adapted phenotypic profiles on the right (A1, A2 and A6). Generally, naïve isolates grow quickly and are antibiotic-susceptible, while adapted isolates grow slowly and are not antibiotic-susceptible. The degree of adaptation within a specific phenotype is indicated by size of the points and the phenotype evaluated stated beneath. cip: ciprofloxacin minimum inhibitory concentration (MIC); azt: aztreonam MIC; adh: adhesion; agg: aggregation; grASM: growth rate in artificial sputum medium (replicating cystic fibrosis sputum components). Absence of points indicate the archetypes with the most naïve version of the phenotype (e.g. A5 and cip or adh) and the largest circles indicate archetypes with the most evolved version of the phenotype (e.g. A1 and cip).
Results
Eradication assessed by culture
86% (n=69/80, C1) of patients exhibited an eradication period (≥0.5 years) at least once during the course of their care between 2002 and 2018, including eight patients with a single P. aeruginosa-positive culture. For each patient, we evaluated the longest period between P. aeruginosa cultures in the course of their monitoring period (figure 2a, supplementary figure S3), and define this as the MEP if >0.5 years in length. In those patients with at least two positive P. aeruginosa cultures, we observed a median±sd MEP of 2.4±2.8 years, with the longest period lasting 14.8 years. The length of colonisation time before an MEP ranges from 0 to 12.2 years (median±sd time to MEP 1.6±3.0 years; figure 2b), indicating that there is still a substantial therapeutic window for mitigation of P. aeruginosa for many patients in whom it has not been successfully eradicated after the first P. aeruginosa culture.
FIGURE 2.
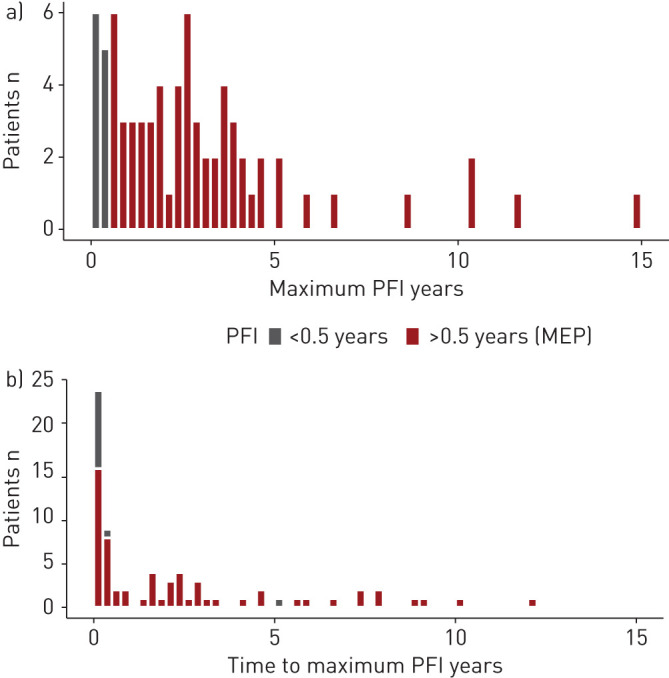
Pseudomonas-free interval (PFI) data by patient count. All PFIs >6 months qualify as eradication periods (and are marked in red). Histogram bins are 3 months. a) Maximum PFI for each patient over their complete monitoring period. b) Time between first Pseudomonas aeruginosa isolation in a patient and the occurrence of their maximum PFI. Many patients experience their max PFI directly after their first P. aeruginosa isolate (t=0). MEP: maximum eradication period.
Eradication assessed by clonal persistence
An eradication period theoretically represents the total clearance of an infecting strain, but despite lengthy periods without positive cultures, the strain may still be present in a patient's airways. In an effort to track strain presence, 72 patients had at least one isolate sequenced (C2), resulting in the identification of 96 distinct strain genotypes (or “clone types”). 20 of these clone types occurred in multiple patients, with the most abundant being DK06, DK19, DK26, DK36 and DK54 (range: six to nine patients). Supplementary figure S4 shows close genetic relationships between sequenced isolates from the same patient at the start and end point of eradication periods for clone types infecting multiple patients, supporting that recurrence of the same clone type is in fact undetected persistence of this strain specific to that patient's lungs. We thereafter identified 57 patients with at least one eradication period (C3) and then selected a group of patients with sufficient data (both sequencing frequency and the colonisation timespan over which isolates were sequenced) for further analysis (table 2, supplementary figures S1 and S2). The 42 patients in C4 had frequent isolate sequencing that established the genotypic profile of early colonisation strains and spanned their MEP.
TABLE 2.
Number of isolates sequenced per patient by cohort
| Min | 1st quartile | Median | 3rd quartile | Maximum | |
| C2 | 1 | 2 | 3 | 12.3 | 34 |
| C3 | 1 | 2 | 6 | 15 | 34 |
| C4 | 2 | 3 | 8 | 16.8 | 34 |
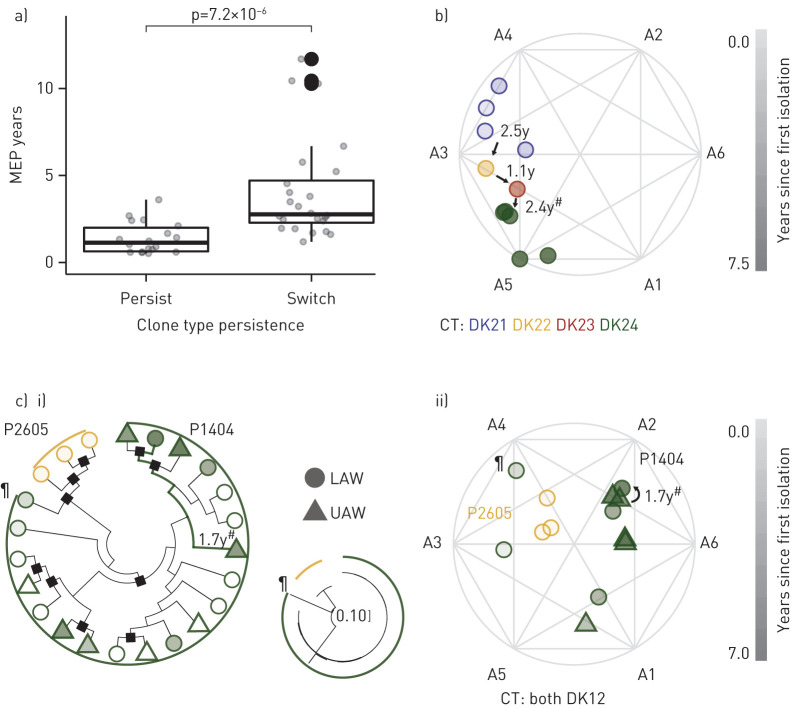
To evaluate how often patients retain a clone type over the MEP, we assessed the post-MEP recurrence of any clone types appearing before the MEP in the 42 patients of C4. Ultimately, ≥43% (n=18) of patients showed the same clone type persisting over their MEP (ranging from 0.5 to 3.6 years, median 1.1 years). In the 22 patients where the clone type switched after the MEP, indicating truly successful eradication, the period ranged from 1.2 to 11.7 years (median 2.8 years) (supplementary figure S3). Length of colonisation before the MEP occurred did not differ significantly between patient groups, but there was a significant difference between mean length of MEP of patients whose clone types persisted or switched (1.4 versus 4.1 years, respectively; p=2.2×10−5, adjusted Wilcoxon) (figure 3a).
FIGURE 3.
Eradication of Pseudomonas aeruginosa. a) The maximum eradication period (MEP) is significantly different (Wilcoxon, p=7.2×10−6) in patients with clone types that “persist” versus “switch” over the MEP in C4 (n=42). b) Example of phenotypic clustering of four clone types (CT) (DK21–DK24) sequentially colonising and then being eradicated in a “switch” patient (illustrated by an archetypal analysis simplex plot [24]). The last eradication was supported by a functional endoscopic sinus surgery (FESS) (marked by #). Years since first isolation are indicated by symbol opacity, and clone type is indicated by hue. c) Phenotypic and phylogenetic clustering of clone type DK12 in “persist” patient P1404, where genetic and trait similarity is maintained over a 1.7-year eradication period, supported by a FESS, marked by #, and between the upper (UAW) and lower airway (LAW). Years since first isolation are indicated by symbol opacity and patient of origin is indicated by colour. Phylogenetic relationships are indicated by i) the cladogram with accompanying phylogenetic tree (showing the distance between isolates to scale) of concatenated single nucleotide polymorphisms (SNPs) accumulated within the clone type [17] with black squares denoting bootstrap values ≥50; ii) isolates from P1404 with trait data (filled symbols) are shown in the archetypal analysis simplex plot. For context, samples from patient P2605 infected with the same clone type are included to show the phenotypic and phylogenetic difference between patients that are likely infected from different environmental sources. A single isolate from P1404 (marked by ¶) suggests a heterogeneous initial infection population, as it shows higher similarity to isolates from P2605 than others from P1404. Note the true scale of genotypic differences as indicated by the small phylogeny in the middle.
Persistence versus patient outcomes
During our monitoring period, seven patients of C4 were diagnosed as chronically infected. Four of these patients had clone types persisting over their MEP, and the most extreme case was initially colonised by clone type DK41 for 6.98 years (including 2.41 years of eradication) prior to diagnosis of chronic infection. Interestingly, in our C4 analysis, we observe a potential link between the MEP and functional endoscopic sinus surgery (FESS) combined with i.v. antibiotics and a colistin rinse, a common intervention at the Copenhagen CF Center for patients with intermittent yet progressing infection [25]. The MEP was adjacent to an FESS in the majority (15 out of 26 patients) who received this intervention, which aligns with past studies indicating that FESS reduces positive culture [26]. We extend these findings by determining that only five out of these 15 patients had a clone type which persisted over both the FESS and MEP.
Phenotypes for persistent versus switched strains
Leveraging our genetic and phenotypic evolution study in a subset of the patients [24], we compared patients with switched versus persisting clone types within the eradication cohort to assess other potential differences between these groups. Figure 3b illustrates an extreme case of clone type switching, where four different clone types sequentially colonise patient P2204 and then are eradicated over a period of 7.5 years. All isolates show naïve phenotypic profiles (normal growth rate, low antibiotic resistance and low biofilm-linked traits) based on their localisation near the naïve archetypes (A3 and A5). In contrast to this “switch” patient, the “persist” patient of figure 3c shows both genetic and phylogenetic adaptation of the same clone type over 7 years in patient P1404. Trait profiles of isolates shift from the “naïve” archetype A3 towards prototypical “adapted” traits, adapting towards slow growth rate and increased ciprofloxacin resistance at A1 and then slow growth and increased adherence at A2/A6. The last set of traits are maintained in isolates spanning an FESS and consequent MEP of 1.7 years. The isolates spanning this MEP are also more similar to each other than to the deviating early isolate (denoted with ¶) similar in trait profile to A4 as well as other DK12 isolates colonising a different patient (P2605). This also holds true with respect to genetic adaptation in the included phylogenetic tree (figure 3c, ii). In examining the other “persist” patient for whom we have trait data for isolates at the start and end of their MEP, we saw the same similarity in genetic and phenotypic traits between the isolates bridging the MEP. As a final summary, 17 additional patients had phenotypic data available for isolates sampled at the start or end of an eradication period, which we highlight in the archetype model in supplementary figure S5 (showing either isolates linked to a patient's MEP or longest eradication period for which we have trait data). This figure emphasises the diversity of adaptive trajectories of strains colonising different patients in alignment with our previous studies of adaptive trajectories [24]; different adaptive paths can lead to equal strain persistence. While “starting” isolates for “switch” patients tend to have more naïve traits than the more diverse traits observed in “persist” patients (as illustrated in the guide in figure 1), starting isolates do not cluster dramatically with a specific, shared archetypal profile.
Persistence versus strain origin
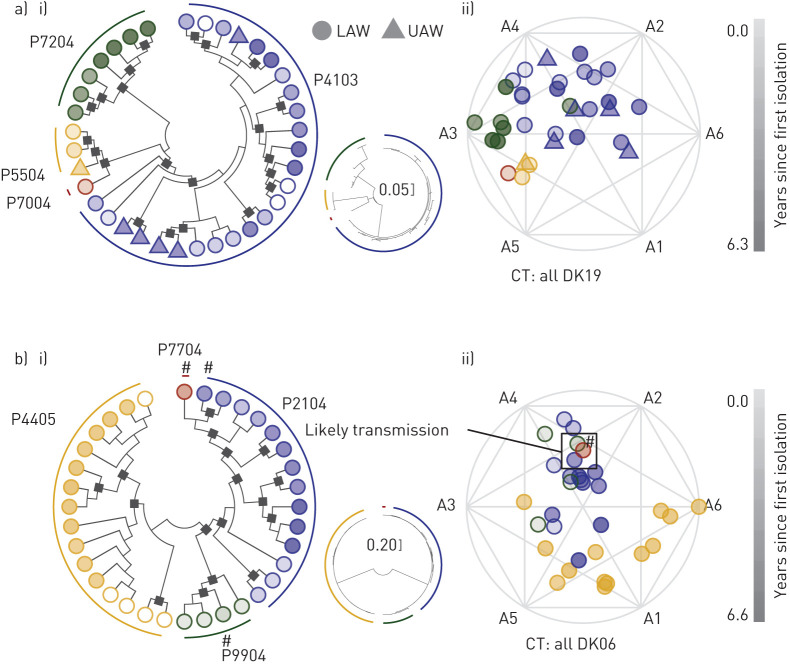
Strain persistence and adaptation is theorised to be influenced by patient-specific environmental effects that include inherent differences such as immune function, external differences such as individualised antibiotic therapy, and spatial differences as strains may localise in different regions of the airway. For example, it has been argued that the sinuses may serve as an infectious focus, and consequently more adapted P. aeruginosa may repeatedly spread from the upper to the lower airways [3, 21, 27]. This assertion has been investigated primarily in older chronically infected patients [16, 28]. In this young cohort, we do not see any evidence of the upper airway (UAW) and lower airway (LAW) being segregated (figure 4a), supportive of the hypothesis of united airways. For each of the clone types that were cultured in both the UAW and the LAW (15 clone types in 18 patients, three of which were shared clone types DK03, DK19 and DK36), we again looked for clustering of isolates both through archetypal and phylogenetic analyses. However, we were not able to identify specific clades or clustering of UAW isolates as compared to LAW isolates (figures 3c and 4a, and supplementary figure S4).
FIGURE 4.
Maximum likelihood phylogenetic analysis and archetypal analysis (multidimensional clustering) of two clone types a) DK19 and b) DK06. Isolates are indicated by circles (lower airway (LAW)) or triangles (upper airway (UAW)) and the colour indicates patient origin as indicated by the phylogenies in a) and b), with the hue indicating years since the clone type was first isolated from any patient. Isolates with a white centre are not included in the cluster analysis. i) Cladograms with accompanying phylogenetic trees (smaller tree showing the distance between isolates to scale) of concatenated single nucleotide polymorphisms (SNPs) accumulated within the clone type [17] with black squares denoting bootstrap values ≥50. Likelihoods: −1136.76 (DK19) and −1466.05 (DK06). ii) Cluster analysis (archetype analysis) of five phenotypes (see main text and prior study [24] for further explanation). Isolates marked by (#) indicate the phylogenetically closest isolates of three different patients all infected by DK06, and this similarity is reflected in ii) where the same isolates are also identified in the (#) box.
A more extreme example of environmental differences is presented by cases of patient-to-patient transmission of P. aeruginosa clone types, mainly a concern before introduction of appropriate infection control [10, 29, 30]. We evaluated three probable direct transmission cases (0–29 SNPs different, mean of 10.2) and 10 clone types present in multiple patients by likely indirect means (54–504 SNPs different, mean of 211.5). We find remarkable similarities in traits between transmitted strains showing close genetic relatedness, as shown for DK06 in figure 4b. In patients without obvious evidence of a direct transmission, strains from the same clone type are often less phenotypically similar (environment) (figures 3c and 4).
Persistence versus patient treatment
The lung environment of CF patients is probably dramatically affected by the intense regimen of individualised antibiotic therapy that each patient receives in an effort to eradicate bacteria. Using clinical treatment records, we examined the effect of treatment on patients with persisting versus switching clone types over their MEPs (n=42, C4). We evaluated the months in which 12 antibiotics which target P. aeruginosa (amikacin, azithromycin, aztreonam, ciprofloxacin, ceftazidime, clarithromycin, colistin, imipenem, meropenem, ofloxacin, piperacillin/tazobactam and tobramycin) were in use among the 42 patients within 6 months of the start of their MEP; the antibiotics were grouped by antibiotic class (figure 5a). In some patients, a given antibiotic was in use every month of the 12-month period (6 months before and after the start of the MEP to account for the influence of prolonged treatment on the continuation of the eradication period). The four classes most often in use were aminoglycosides, fluoroquinolones, macrolides and polymyxins (relative number of patient cohort treated with a given antibiotic class is indicated by boxplot width in figure 5a), and supplementary figure S6 shows that most patients received combination therapy using at least three of these classes within their MEP timeframe. Interestingly, figure 5 shows that the distribution of each antibiotic class's usage in the “persist” patients is generally similar or elevated compared to the “switch” patients, with significantly different means between months in use for aminoglycosides and macrolides (by Wilcoxon test). To add context to this finding, we estimated age of each patient's P. aeruginosa infection (figure 5b) and estimated severity of infection by counting the positive P. aeruginosa cultures from their approximately monthly clinic visits in the year prior their MEP (figure 5c). Patients in the “persist” cohort tend to have MEPs that occur later in their infection period, though the means of each cohort are not significantly different. However, there is a significant difference in the average positive P. aeruginosa cultures which occur the year prior to the MEP for “persist” versus “switch” patients, supporting that “persist” patients have more serious infections than “switch” patients.
FIGURE 5.
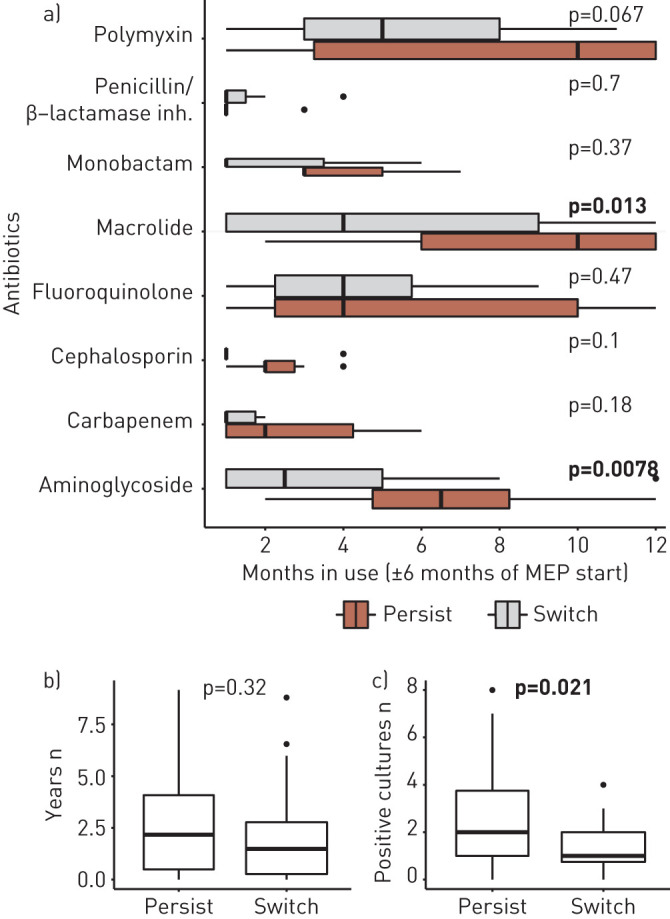
Use of Pseudomonas-targeting drugs in treatment period surrounding maximum eradication period (MEP). a) In C4 patients, eight drug classes (covering 13 drugs that target P. aeruginosa) were in use in ≥1 month in one patient within 6 months of their MEP (both before and after MEP start). The months an antibiotic class was in use by each patient treated with that antibiotic class are shown by boxplots, where the width of each box (in the y-axis direction) correlates with the number of patients treated with that drug (patients with 0 months of treatment with a given drug are not included in the analysis). Patients with clone types that persisted versus switched over their MEP are separated for comparison, and significant differences between these groups are shown by bolded p-values (Wilcoxon test of means). b) We show the years from first ever positive culture of P. aeruginosa to start of the MEP for “persist” and “switch” cohorts (difference between cohorts is nonsignificant, Wilcoxon test of means). c) We show the number of positive P. aeruginosa cultures in the year prior to the start of the MEP for “persist” and “switch” cohorts (difference between cohorts is significant, Wilcoxon test of means).
Discussion
We leverage the implementation of bacterial WGS in the clinical routine at the Copenhagen CF Center to demonstrate that nearly half of patients in a longitudinal infection cohort are infected with the same clone type before and after their maximum eradication period, which should represent the most likely instance of true eradication. Through omics methods and statistical modelling, we observe remarkable phylogenetic and phenotypic similarity in isolates separated by long eradication periods, infection niche and during transmission between patients. We see a significant difference in months of treatment with three classes of drugs in “persist” patients versus “switch” patients; the elevated treatment in “persist” patients accompanied by a higher number of P. aeruginosa-positive cultures in the “persist” patients before their MEP suggests a worse infection that requires more antibiotics to achieve the MEP, yet still does not result in true eradication. Our experience with WGS implementation in the clinic has major implications for effective monitoring and eradication of persisting infections. We support the pairing of positive P. aeruginosa culture records with omics data for an improved understanding of patient infection trajectory, and recommend the reassessment of studies of treatment efficacy that might benefit from improved, omics-based eradication metrics.
First, our observations of continuous persistence independent of a chronic diagnosis highlight a shortcoming of the current view of P. aeruginosa airway infections in CF [4, 31]. Treatment regiments are generally based on the infection status as either “intermittently colonised” or “chronically infected” [4, 10, 31]. In reality, the infecting clone type in many “intermittently colonised” patients is not being eradicated. This has been suspected at other CF centres which track the genotype of infecting bacterial isolates via multilocus sequence typing [14], but WGS offers new resolution in identifying re-emergent strains via genetic relatedness to other adapted isolates previously found in the patient (supplementary figure S4). We estimate that 43% (n=18) of patients in our initially nonchronic, young cohort (C4) experience this clonal persistence over at times quite lengthy eradication periods, and improved diagnostic approaches should highlight these patients who could benefit from customised therapy in the clinic. Our work suggests, for example, that sinus surgery may be an effective way of initiating a lengthy eradication period and reducing clone type recurrence rates, and an omics-based assessment of eradication versus treatment could highlight particularly beneficial antibiotic regimens which evaded prior positive culture only studies showing no difference between regimens.
Currently, we observe that many patients with a persistent clone type have been treated with more antibiotics in the timeframe of their MEP (figure 5a). This unexpected trend could be interpreted to mean that intensive antibiotic therapy is actually counterproductive, but alternative explanations are available. We speculate that this treatment difference can be partly explained by the timing of MEPs with persisting versus switching clone types. The mean time between first ever P. aeruginosa and MEP is approximately a year shorter for “switch” patients versus “persist” patients (2.07 versus 2.92 years, C4), although the difference is not statistically significant (figure 5b). These “persist” patients with longer infection histories may be undergoing heavier treatment as their strains have adapted and persisted, while a true eradication is likely easier in the “switch” patients with less adapted strains. There is also a significant difference in the average number of positive P. aeruginosa cultures between “persist” and “switch” patients in the year prior to their MEP, with “persist” patients showing more frequent positive cultures, which lead to additional antibiotic courses per CF Center treatment guidelines (figure 5c). Ultimately, our findings support the idea of tracking age of infection by both P. aeruginosa culture incidence and clone type recurrence as a predictor of strain persistence and mode of assessing treatment. Effectiveness will be dependent on the frequency of clinical sampling and ability to perform comprehensive sequencing of patient bacterial isolates to identify clonal incidence at the earliest possible date. We acknowledge that while this is becoming feasible to implement in Danish clinics, it may require advances in at-home sampling and analytics in other treatment environments. Larger studies of the outcome of eradication periods of similar duration and timing paired with our omics-centered approach will hopefully provide answers as to 1) the utility of this approach in customising treatment strategies supporting eradication and 2) the scope of data necessary to implement this approach to infection monitoring.
Our analysis of genetic and phenotypic similarity of isolates spanning eradication periods and spatial separation is notable as we also have observed clear adaptive trajectories in other persisting infections. Here, we show that the UAW and LAW are not separate entities with different subpopulations. We instead highlight the shared persistence in UAW and LAW by similar populations of bacteria in similar timeframes, which may ultimately render a diagnosed infection focus irrelevant for most patients. However, infecting strains are able to persist and adapt within patients’ airways to a significant degree in these early stages of colonisation, and the likelihood of persistence of a specific clone type from its first appearance in lab culture should be incorporated in treatment decisions. We previously found that bacterial growth rate and resistance to ciprofloxacin adapt rapidly in an initial 2–3-year period after first colonisation [24]. The re-emergence of phylogenetically and phenotypically similar isolates after long eradication periods in this study adds a new perspective to our prior work, as the clone type persists, but may undergo limited adaptation while surviving in an inactive “persister” state [32]. However, acknowledging that the same adapted bacterial strain can re-emerge after eradication is important for accurately estimating its exposure to antibiotics. This study highlights the likely underestimation of risk of treatment tolerance, resistance and persistence for these patients.
It is important to note that the findings of this study are affected by several factors. First, the young patients in our cohort cannot expectorate easily so we probably underestimate the true presence of P. aeruginosa via positive culture. In addition, we do not address the potential impact of other CF pathogens on P. aeruginosa clearance. It can also be possible that a post-eradication isolate genetically related to prior adapted isolates from a patient has been re-acquired from the patient's living environment after ≥6 months of true eradication, but this argument against clone type persistence seems very unlikely outside of young cohabiting siblings infected with the same strain. Furthermore, the ability to monitor the reappearance of a previously adapted strain by omics-based approaches is still relevant for these patients and their treatment design. With respect to our isolate similarity studies via archetypal analysis, we constructed these models using in vitro screening of five phenotypes (growth rate, susceptibility to antibiotics ciprofloxacin and aztreonam, aggregation and adhesion), but further phenotypes may also be undergoing adaptation in the lungs. We currently offer detailed examples of the potential of this approach (figures 3 and 4) rather than an expansive analysis of the spectrum of archetypes associated with MEPs; preliminary evidence suggests they are probably quite diverse (supplementary figure S5). We view the present work as a foundational surveillance study from which we can expand our understanding of bacterial adaptation in the complex environment of the CF lungs as well as improve our means of precision medicine in the CF clinic via integrated WGS and omics assays. We envision a treatment approach where persistence and adaptation of a colonising strain is tracked both by functional genetic and phenotypic markers individualised to each patient, and a strain of the same clone type re-emerging after an eradication period will be quickly compared to previous isolates to check degree of adaptation and any novel traits. This data can then inform a new antibiotic regimen design rather than repeating a prior regimen that has not prevented the adapted strain's persistence. We ultimately aim to leverage omics data that is feasible for a clinic to obtain and support a more structured and evidence-based approach to treatment for specific infection cases such as intermittently cultured persistent strains.
Our findings support the centring of bacterial genomics data in evaluating infection and achieving strain eradication in CF airways. Clinicians presented with a positive P. aeruginosa culture following a substantial period of P. aeruginosa-negative samples (including multiple years in length) should consider it equally likely that a previously treated, possibly adapted strain has re-emerged versus assuming that a new, susceptible strain has colonised the airways. Our findings are important for updating definitions of P. aeruginosa airway infections, diagnosis and treatment protocols in CF patients.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary methods and figures ERJ-00512-2020.Supplement (1.5MB, pdf)
Shareable PDF
Acknowledgements
We thank Katja Bloksted, Ulla Rydahl Johansen, Helle Nordbjerg Andersen, Sarah Buhr Bendixen, Camilla Thranow, Pia Poss, Bonnie Horsted Erichsen, Mette Pedersen and Rakel Schiøtt of the Department of Clinical Microbiology at Rigshospitalet, Copenhagen, Denmark, for excellent technical assistance.
Footnotes
This article has supplementary material available from erj.ersjournals.com
Author contributions: H.K. Johansen collected all the bacterial isolates and contributed to the conception and study design together with J.A. Bartell, L.M. Sommer and S. Molin. M. Skov and T. Pressler provided the clinical data. J.A. Bartell, L.M. Sommer, R.L. Marvig, S. Molin and H.K. Johansen contributed to the analysis and interpretation of data. J.A. Bartell, L.M. Sommer and H.K. Johansen drafted the manuscript and all authors commented on the manuscript prior to submission and approved the final version.
Conflict of interest: J.A. Bartell reports grants from Cystic Fibrosis Foundation (grant number BARTEL18F0), during the conduct of the study.
Conflict of interest: L.M. Sommer has nothing to disclose.
Conflict of interest: R.L. Marvig reports grants from Danish National Research Foundation (grant number 126), during the conduct of the study.
Conflict of interest: M. Skov has nothing to disclose.
Conflict of interest: T. Pressler has nothing to disclose.
Conflict of interest: S. Molin reports grants from Cystic Fibrosis Foundation (grant MOLIN18G0), during the conduct of the study.
Conflict of interest: H.K. Johansen has nothing to disclose.
Support statement: The Novo Nordisk Foundation supported H.K. Johansen as a clinical research stipend. Rigshospitalet Rammebevilling 2015-17 (R88-A3537), Lundbeckfonden (R167-2013-15229), Det Frie Forskningsråd FSS (DFF-4183-00051), RegionH rammebevilling (R144-A5287), The Novo Nordisk Foundation (NNF15OC0017444 and NNF18OC0052776) and Savværksejer Jeppe Juhl og Hustru Ovita Juhls Memorial Fund supported H.K. Johansen. The Cystic Fibrosis Foundation supported J.A. Bartell with a postdoctoral fellowship (BARTEL18F0). The Novo Nordisk Foundation supported S. Molin and J.A. Bartell. R.L. Marvig is supported by the Danish National Research Foundation (grant number 126). Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Hansen CR, Pressler T, Høiby N. Early aggressive eradication therapy for intermittent Pseudomonas aeruginosa airway colonization in cystic fibrosis patients: 15 years experience. J Cyst Fibros 2008; 7: 523–530. doi: 10.1016/j.jcf.2008.06.009 [DOI] [PubMed] [Google Scholar]
- 2.Mayer-Hamblett N, Kloster M, Rosenfeld M, et al. Impact of sustained eradication of new Pseudomonas aeruginosa infection on long-term outcomes in cystic fibrosis. Clin Infect Dis 2015; 61: 707–715. doi: 10.1093/cid/civ377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen SK, Rau MH, Johansen HK, et al. Evolution and diversification of Pseudomonas aeruginosa in the paranasal sinuses of cystic fibrosis children have implications for chronic lung infection. ISME J 2012; 6: 31–45. doi: 10.1038/ismej.2011.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee TWR, Brownlee KG, Conway SP, et al. Evaluation of a new definition for chronic Pseudomonas aeruginosa infection in cystic fibrosis patients. J Cyst Fibros 2003; 2: 29–34. doi: 10.1016/S1569-1993(02)00141-8 [DOI] [PubMed] [Google Scholar]
- 5.Johansen HK, Nørregaard L, Gøtzsche PC, et al. Antibody response to Pseudomonas aeruginosa in cystic fibrosis patients: a marker of therapeutic success? – a 30-year cohort study of survival in Danish CF patients after onset of chronic P. aeruginosa lung infection. Pediatr Pulmonol 2004; 37: 427–432. doi: 10.1002/ppul.10457 [DOI] [PubMed] [Google Scholar]
- 6.Pressler T, Bohmova C, Conway S, et al. Chronic Pseudomonas aeruginosa infection definition: EuroCareCF Working Group report. J Cyst Fibros 2011; 10: Suppl. 2, S75–S78. doi: 10.1016/S1569-1993(11)60011-8 [DOI] [PubMed] [Google Scholar]
- 7.Parkins MD, Somayaji R, Waters VJ. Epidemiology, biology, and impact of clonal Pseudomonas aeruginosa infections in cystic fibrosis. Clin Microbiol Rev 2018; 31: e00019-18. doi: 10.1128/CMR.00019-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smyth AR, Bell SC, Bojcin S, et al. European Cystic Fibrosis Society standards of care: best practice guidelines. J Cyst Fibros 2014; 13: Suppl. 1, S23–S42. doi: 10.1016/j.jcf.2014.03.010 [DOI] [PubMed] [Google Scholar]
- 9.Proesmans M, Vermeulen F, Boulanger L, et al. Comparison of two treatment regimens for eradication of Pseudomonas aeruginosa infection in children with cystic fibrosis. J Cyst Fibros 2013; 12: 29–34. doi: 10.1016/j.jcf.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 10.Wiehlmann L, Cramer N, Ulrich J, et al. Effective prevention of Pseudomonas aeruginosa cross-infection at a cystic fibrosis centre – results of a 10-year prospective study. Int J Med Microbiol 2012; 302: 69–77. doi: 10.1016/j.ijmm.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 11.Schelstraete P, Deschaght P, Van Simaey L, et al. Genotype based evaluation of Pseudomonas aeruginosa eradication treatment success in cystic fibrosis patients. J Cyst Fibros 2010; 9: 99–103. doi: 10.1016/j.jcf.2009.11.006 [DOI] [PubMed] [Google Scholar]
- 12.Jeukens J, Boyle B, Kukavica-Ibrulj I, et al. Comparative genomics of isolates of a Pseudomonas aeruginosa epidemic strain associated with chronic lung infections of cystic fibrosis patients. PLoS One 2014; 9: e87611. doi: 10.1371/journal.pone.0087611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marvig RL, Dolce D, Sommer LM, et al. Within-host microevolution of Pseudomonas aeruginosa in Italian cystic fibrosis patients. BMC Microbiol 2015; 15: 218. doi: 10.1186/s12866-015-0563-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kidd TJ, Ramsay KA, Vidmar S, et al. Pseudomonas aeruginosa genotypes acquired by children with cystic fibrosis by age 5-years. J Cyst Fibros 2015; 14: 361–369. doi: 10.1016/j.jcf.2014.12.007 [DOI] [PubMed] [Google Scholar]
- 15.Schelstraete P, Van Daele S, De Boeck K, et al. Pseudomonas aeruginosa in the home environment of newly infected cystic fibrosis patients. Eur Respir J 2008; 31: 822–829. doi: 10.1183/09031936.00088907 [DOI] [PubMed] [Google Scholar]
- 16.Jorth P, Staudinger BJ, Wu X, et al. Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe 2015; 18: 307–319. doi: 10.1016/j.chom.2015.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marvig RL, Sommer LM, Molin S, et al. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet 2015; 47: 57–64. doi: 10.1038/ng.3148 [DOI] [PubMed] [Google Scholar]
- 18.Klockgether J, Cramer N, Fischer S, et al. Long-term microevolution of Pseudomonas aeruginosa differs between mildly and severely affected cystic fibrosis lungs. Am J Respir Cell Mol Biol 2018; 59: 246–256. doi: 10.1165/rcmb.2017-0356OC [DOI] [PubMed] [Google Scholar]
- 19.Mauch RM, Levy CE. Serum antibodies to Pseudomonas aeruginosa in cystic fibrosis as a diagnostic tool: a systematic review. J Cyst Fibros 2014; 13: 499–507. doi: 10.1016/J.JCF.2014.01.005 [DOI] [PubMed] [Google Scholar]
- 20.Høiby N, Koch C. Cystic fibrosis. 1. Pseudomonas aeruginosa infection in cystic fibrosis and its management. Thorax 1990; 45: 881–884. doi: 10.1136/thx.45.11.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansen HK, Aanaes K, Pressler T, et al. Colonisation and infection of the paranasal sinuses in cystic fibrosis patients is accompanied by a reduced PMN response. J Cyst Fibros 2012; 11: 525–531. doi: 10.1016/j.jcf.2012.04.011 [DOI] [PubMed] [Google Scholar]
- 22.Skov M, Hansen CR, Pressler T. Cystic fibrosis – an example of personalized and precision medicine. APMIS 2019; 127: 352–360. doi: 10.1111/apm.12915 [DOI] [PubMed] [Google Scholar]
- 23.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 2016; 33: 1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartell JA, Sommer LM, Haagensen JAJ, et al. Evolutionary highways to persistent bacterial infection. Nat Commun 2019; 10: 629. doi: 10.1038/s41467-019-08504-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aanaes K, Johansen HK, Skov M, et al. Clinical effects of sinus surgery and adjuvant therapy in cystic fibrosis patients – can chronic lung infections be postponed? Rhinology 2013; 51: 222–230. doi: 10.4193/Rhino12.207 [DOI] [PubMed] [Google Scholar]
- 26.Aanaes K, von Buchwald C, Hjuler T, et al. The effect of sinus surgery with intensive follow-up on pathogenic sinus bacteria in patients with cystic fibrosis. Am J Rhinol Allergy 2013; 27: e1–e4. doi: 10.2500/ajra.2013.27.3829 [DOI] [PubMed] [Google Scholar]
- 27.Bonestroo HJC, de Winter-de Groot KM, van der Ent CK, et al. Upper and lower airway cultures in children with cystic fibrosis: do not neglect the upper airways. J Cyst Fibros 2010; 9: 130–134. doi: 10.1016/J.JCF.2010.01.001 [DOI] [PubMed] [Google Scholar]
- 28.Markussen T, Marvig RL, Gómez-Lozano M, et al. Environmental heterogeneity drives within-host diversification and evolution of Pseudomonas aeruginosa. mBio 2014; 5: e01592-14. doi: 10.1128/mBio.01592-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marvig RL, Johansen HK, Molin S, et al. Genome analysis of a transmissible lineage of Pseudomonas aeruginosa reveals pathoadaptive mutations and distinct evolutionary paths of hypermutators. PLoS Genet 2013; 9: e1003741. doi: 10.1371/journal.pgen.1003741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fothergill JL, Walshaw MJ, Winstanley C. Transmissible strains of Pseudomonas aeruginosa in cystic fibrosis lung infections. Eur Respir J 2012; 40: 227–238. doi: 10.1183/09031936.00204411 [DOI] [PubMed] [Google Scholar]
- 31.Høiby N, Flensborg E, Beck B, et al. Pseudomonas aeruginosa infection in cystic fibrosis. Diagnostic and prognostic significance of Pseudomonas aeruginosa precipitins determined by means of crossed immunoelectrophoresis. Scand J Respir Dis 1977; 58: 65–79. [PubMed] [Google Scholar]
- 32.Fisher RA, Gollan B, Helaine S. Persistent bacterial infections and persister cells. Nat Rev Microbiol 2017; 15: 453–464. doi: 10.1038/nrmicro.2017.42 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary methods and figures ERJ-00512-2020.Supplement (1.5MB, pdf)
This one-page PDF can be shared freely online.
Shareable PDF ERJ-00512-2020.Shareable (283.3KB, pdf)