Abstract
This study aims (1) to evaluate ATRX expression in different grades and subtypes of gliomas and correlate with other hallmark genetic alterations, (2) to identify and characterize mosaic/heterogeneous staining in gliomas in terms of mutation status.
One hundred seventy six cases of glioma were assessed for ATRX immunohistochemistry and subdivided into positive, negative and mosaic/heterogeneous staining patterns. Five cases with heterogeneous staining were further subjected to next generation sequencing.
Higher frequency of ATRX immune‐negativity was detected in grade II/III astrocytic, oligoastrocytic tumors and secondary glioblastomas (GBMs), while infrequent in primary GBMs and rare in oligodendrogliomas. Loss of expression was significantly associated with IDH1 and/or TP53 mutation, while mutually exclusive with 1p/19q codeletion. Mosaic/heterogeneous staining was detected exclusively in GBMs (21.2%). Two different types of mosaic staining were identified (1) Admixture of positive and negative nuclei or intermixed mosaic and (2) Separate fragments with positive and negative/intermixed mosaic staining. ATRX mutation was identified in 2/5 (40%) cases with mosaic staining while one case showed DAXX mutation. All these cases were characterized by distinctly separate immune‐negative and positive/intermixed foci. Hence, it is suggested that cases with heterogeneous staining (especially those with distinctly negative fragments) should be subjected to mutation analysis.
Keywords: ATRX, Glioma, heterogeneous staining, mosaic staining
Introduction
Alpha Thalassemia/Mental Retardation Syndrome X‐linked (ATRX) is a recently described gene that plays a crucial role in normal telomere homeostasis. It is a core component of chromatin remodeling complex and regulates incorporation of histone H3.3 into telomeric chromatin 7, 11, 17. Loss of function of ATRX gene leads to alternative lengthening of telomeres (ALT), a telomerase independent mechanism of telomere lengthening which results in cellular immortality and tumorigenesis. Mutations in ATRX gene have recently been described in various tumors including pancreatic neuroendocrine carcinoma, neuroblastoma and gliomas 4, 10, 13, 14. This molecular alteration is reported to be most prevalent in grade II and grade III astrocytic tumors (range 45%–90.5% in grade II and 57.5%–89.4% in grade III) followed by oligoastrocytomas (25%–56% in grade II and 54%–77% in grade III), and secondary GBMs (57% in two separate reports). It is distinctly rare in pure oligodendrogliomas (0%–24% in grade II and 3%–7% in grade III) and primary Glioblastomas (GBMs) (4%–18.4%) 13, 19, 21, 24, 28. Further, ATRX mutation is significantly more frequent in IDH and/or TP53 mutated tumors and almost mutually exclusive with 1p/19q codeletion, the hallmark of oligodendroglial tumors 19, 24. Recently, Wiestler et al 28 and Reuss et al 24 separately proposed a molecular diagnostic algorithm for gliomas by combining ATRX with 1p/19q and IDH1 which appeared to be superior to classical neuropathological techniques. The ISN‐Haarlem guideline has also included ATRX in the integrated diagnoses for WHO grade II and grade III adult diffuse gliomas 20.
ATRX mutations are mostly inactivating type and almost exclusively associated with loss of protein expression. Hence, loss of ATRX staining on immunohistochemistry is often considered as a surrogate marker of gene mutation. Two recent studies on neuroblastoma and pancreatic neuroendocrine tumors also described a mosaic/heterogeneous staining pattern of ATRX staining on immunohistochemistry. On correlation with mutation status, it was observed that 66.7% cases of neuroblastoma with heterogeneous staining had ATRX mutation while the single case of pancreatic neuroendocrine tumor showed ALT 4, 10. However, to the best of our knowledge, no study till date has described mosaic/heterogeneous immunopositivity of ATRX in gliomas and characterized it in relation to mutation status. This may be important because diffuse gliomas especially GBMs are known for marked intratumoral heterogeneity.
Hence, the study was undertaken to analyze the frequency of ATRX mutation in different grades and subtypes of gliomas and to correlate ATRX status with other key molecular alterations in astrocytic, oligodendroglial and mixed oligoastrocytic tumors. Another specific aim of this study was to evaluate occurrence, if any, of heterogeneous/mosaic pattern of ATRX immunostaining in diffuse gliomas and if so, to correlate this with corresponding ATRX gene mutation status.
Material and Methods
A combined retrospective and prospective study was performed. One hundred and seventy six cases of gliomas diagnosed over a period of 7 years (2006–2012) in the Neuropathology Laboratory of the Department of Pathology, All India Institute of Medical Sciences, New Delhi were included, in which paraffin blocks with adequate tumor tissue and frozen tumor tissue were available. All experiments using patient samples were approved by the ethics committee of All India Institute of Medical Sciences, New Delhi, and informed consent was obtained. Hematoxylin and Eosin‐stained slides of the cases were reviewed and a concordant agreement was established for the confirmation of the diagnosis between three trained pathologists based on the WHO classification (2007). The cases included 19 diffuse astrocytomas (DA; grade II), 15 anaplastic astrocytomas (AA; grade III), 80 glioblastomas (GBM; grade IV), 15 oligodendrogliomas (OG; grade II), 12 anaplastic oligodendrogliomas (AO; grade III), 20 oligoastrocytoma (OA; grade II) and 15 anaplastic oligoastrocytoma (AOA; grade III). GBM cases were classified as primary if diagnosis was made at the first biopsy without clinical or histologic evidence of presence of a less malignant precursor lesion and with a clinical history of less than 3 months. On the other hand, tumors were considered as secondary GBMs, when there was a previous history of surgery with histopathologic evidence of a preceding lower‐grade glioma 12, 22.
Immunohistochemical staining for ATRX
Immunohistochemical analysis was performed on serial 5 µm sections mounted on poly‐L‐lysine coated slides using monoclonal antibody against ATRX protein (M/S Sigma). The sections were deparaffinized in xylene and rehydrated through a decreasing concentration of alcohol. Antigen retrieval was performed by transferring the sections into 0.01 mol/L citrate buffer (pH 6.0) inside a 600‐watt microwave oven on full power for 30 min. Peroxidase activity was blocked with 3% H2O2 in methanol for 30 min at room temperature. The sections were then incubated with adequate amount and dilution (1:400) of primary antibodies at 4°C in a humidity chamber for 2 h. Sections were washed in Tris, treated with the biotin‐labeled secondary antibody for 60 min at RT, and then washed in Tris buffer. Peroxidase conjugated streptavidin was applied to cover the sections and incubated at room temperature for 30 min. Then the slides were rinsed with three changes of Tris‐HCl buffer for 5 min each. Sections were then stained with diaminobenzidine for 10 min, washed with distilled water, counterstained in hematoxylin for 1 min, and then mounted.
Two experienced pathologists independently examined the slides while blinded to diagnosis. Fragments of normal brain adjacent to tumor and endothelial cells of the intratumoral vessel which demonstrated crisp nuclear immunoreactivity for ATRX were taken as internal positive control.
ATRX staining was characterized as follows: (1) Positive (2) Negative and (3) Mosaic. Complete absence of nuclear staining with retained staining in internal controls such as endothelia or microglia was regarded as negative and this loss of ATRX nuclear expression was considered as an indirect evidence of ATRX mutation. Two distinct patterns of heterogeneous/mosaic staining were observed viz. (i) tumors with distinctly separate immunopositive and immunonegative/intermixed mosaic fragments and (ii) tumors with positively and negatively stained cells within the same fragment (intermixed). Concordance of staining pattern was established between three pathologists.
IDH1 and TP53 mutation analysis by Sanger sequencing
This was done in 170 and 143 cases of gliomas for IDH1and TP53, respectively, where frozen tissue was available. TP53 mutational status was not assessed in cases of OG and AO. For sequencing analysis, DNA was isolated from frozen tumor tissue using standard protocol. TP53 mutational analysis of coding regions from exons 2 to 11 were evaluated using the direct sequencing protocol as described in the International Agency for Research on Cancer (IARC) p53 database. Mutations in exon 4 of IDH1 were determined by direct sequencing in all the cases as described in the previous study 12.
Expression of NF‐1 using Real‐time PCR
This was done only in 74 cases of GBMs where frozen tissue was available. RNA was extracted from frozen tissue specimens by using commercially available mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) following manufacturer's protocol. One microgram of the total RNA from all samples were reverse‐transcribed to cDNA using SuperScript® VILO™ cDNA Synthesis Kits (Life technologies). To determine the expression profile of NF1 real‐time PCR was performed using primers designed via Primer3 software (v.0.4.0) (sequence available in Table 1). Primers sequence for NF‐1 was 5′‐GCATTTCTACCAGTAACCTTGATGATAC‐3′ (F) and 5′‐TCTGAACAAACAGTTAATTCCTGTAACC‐3′ (R). Quantification was performed on Light Cycler 480 (Roche Diagnostic, Basel, Switzerland) using SYBR‐Green chemistry. Expression levels of the mRNAs were calculated using comparative cycle threshold (Ct) method. Ct values of the target mRNAs were normalized in relation to reference genes (TBP). The fold change was calculated using the equation 2−ΔΔCt. Down‐regulation (≤0.5‐fold change) of NF‐1 expression was considered as a surrogate marker of gene mutation.
Table 1.
Frequency of ATRX loss in various morphological and molecular subgroups of gliomas.
| Histomorphological diagnosis | Molecular profile (Number; %) | ATRX negative | ATRX mosaic |
|---|---|---|---|
|
DA (grade II) (n‐19) |
IDH1+, TP53+ (9/19; 47.3%) | 66.7% (6/9) | |
| IDH1+, TP53− (9/19; 47.3%) | 88.9% (8/9) | ||
| IDH1−, TP53+ (1/19; 5.3%) | 100% (1/1) | ||
| Total | 78.9% (15/19) | ||
|
AA (grade III) (n‐15) |
IDH1+, TP53+ (3/15; 20%) | 100% (3/3) | |
| IDH1+, TP53− (10/15; 66.7%) | 60% (6/10) | ||
| IDH1−, TP53+ (2/15; 13.3%) | 100% (2/2) | ||
| Total | 73.3% (11/15) | ||
|
OG (grade II) (n‐15) |
1p/19q codeletion+ IDH1+ (11/15; 73.3%) | 9.1% (1/11) | |
| 1p/19q codeletion+ IDH1− (4/15; 26.7%) | 0% (0/4) | ||
| Total | 6.7% (1/15) | ||
|
AO (grade III) (n‐12) |
1p/19q codeletion+ IDH1+ (11/12; 91.7%) | 9.1% (1/11) | |
| 1p/19q codeletion+ IDH1− (1/12; 8.3%) | 0% (0/1) | ||
| Total | 8.3% (1/12) | ||
|
OA (grade II) (n‐20) |
1p/19q codeletion+ IDH1± TP53− (8/20; 40%) | 0% (0/8) | |
| 1p/19q codeletion− IDH1± TP53± (12/20; 60%) | 83.3% (10/12) | ||
| Total | 50% (10/20) | ||
|
AOA (grade III) (n‐15) |
1p/19q codeletion+ IDH1± TP53− (8/15; 53.3%) | 12.5% (1/8) | |
| 1p/19q codeletion− IDH1± TP53± (7/15; 46.7%) | 71.4% (5/7) | ||
| Total | 40% (6/15) | ||
|
GBM (grade IV) (n‐80)* |
IDH1 mutation (18/74; 24.3%) | 44.4% (8/18) | 22.2% (4/18) |
| TP53 mutation (15/74; 20.3%) | 40% (6/15) | 20% (3/15) | |
| PDGFRA amplification (6/74; 8.1%) | 66.7% (4/6) | 33.3% (2/6) | |
| EGFR amplification (24/74; 32.4%) | 0% (0/24) | 20.8% (5/24) | |
| Decreased NF1 expression (15/74; 20.3%) | 0% (0/15) | 6.7% (1/15) | |
| None of the above (16/74; 21.6%) | 12.5% (2/16) | 31.25% (5/16) | |
| Total | 17.5% (14/80) | 21.2% (17/80) |
*Data of IDH1, TP53, PDGFRA, EGFR and NF1 was available in 74 out of 80 cases.
Assessment of EGFR, PDGFRA amplification and 1p/19q deletion by Fluorescence In Situ Hybridization (FISH)
FISH analysis was carried out in 74 cases of GBMs on formalin fixed paraffin embedded sections as previously described 12. Briefly a dual‐color FISH assay was performed on paraffin embedded sections with a locus‐specific probe for EGFR (spectrum orange) paired with a corresponding reference centromeric probe for chromosome 7 (spectrum green) (M/S Vysis, Inc., Abbott Laboratories SA, Downers Grove, IL, USA). For analysis of PDGFRA amplification Tricolor Rearrangement Probe was used (Spectrum aqua for PDGFRA, spectrum green and orange‐control). (M/S Vysis, Inc., Abbott Laboratories SA, Downers Grove, IL, USA). Assessment of 1p and 19q status was performed with locus‐specific probes for 1p36 and 19q13 paired, respectively, with the reference probes for 1q25 and 19p13 (M/S Vysis, Inc., Abbott Laboratories SA, Downers Grove, IL, USA). Signals were enumerated in 200 nonoverlapping nuclei.
Amplification (EGFR and PDGFRA) was considered when >10% of tumor cells showed either test: control signal ratio >2 or innumerable tight clusters of signals of the locus probe. An interpretation of deletion of 1p or 19q was made if greater than 50% of the nuclei showed test to reference ratio of 1/2 or 0/2.
Sequencing for ATRX gene mutations in the cases with mosaic pattern of immunostaining
Sequencing was performed in five cases of GBMs with mosaic staining (one intermixed mosaic and other four with positive and negative/mosaic stained areas) and in one case with negative staining. The positive and negatively stained areas of the tumors were macrodissected from the paraffin blocks and separate blocks were made for areas containing distinct staining patterns. ATRX staining of each of the fragments was reconfirmed and separately subjected to sequencing. Genomic DNA was extracted from the FFPE tissue using commercially available QIAamp DNA FFPE Tissue Kit.
Library construction, capture and sequencing
Small insert dual indexed Illumina paired end libraries were constructed on the SciClone instrument (Perkin Elmer, Waltham, MA, USA) utilizing the KAPA HTP sample prep kits according to the manufacturer's recommendations (KAPA Biosystems, Woburn, MA, USA) with a few exceptions: (i) 100–250 ng of genomic DNA was fragmented using a Covaris LE220 DNA Sonicator (Covaris, Woburn, MA, USA) targeting a size range between 100 and 400 bp using the following settings: volume = 50 µL, temperature = 4°C, duty factor = 30%, peak incident power = 450 W, cycles per burst = 200, and treatment time = 190 s. (ii) Libraries with a starting input of 100 ng were enriched for 10 PCR cycles and libraries with a starting input of 250 ng were enriched for eight PCR cycles. (iii) The final size selection of the library was achieved by AMPure XP paramagnetic beads (Agencourt, Beckman Coulter Genomics, Beverly, MA, USA) and reduced to a 1.2× bead to sample ratio. The libraries were assessed for quality and quantity using the HT DNA Hi Sens Dual Protocol Assay with the HT DNA 1K/12K chip on the LabChip GX instrument (Perkin Elmer, Waltham, MA, USA).
For this project 16 libraries were multiplexed with 70 samples from another project of diffuse gliomas using the same capture probes. The indexed libraries were pooled prior to hybridization. Hybridization was performed using Integrated DNA Technologies (IDT) manufacturer's protocol. Capture targets included 184 120 bp biotinylated oligonucleotide probes specific for the coding regions of the genes ATRX, TP53, TERT, EGFR and DAXX. The 86 indexed samples were combined and captured as a pool and run on an Illumina HiSeq 2500, which produces approximately 60 Gb/lane of sequence. The average total sequence per sample was ∼700 Mb per sample. Sequence was aligned to the GRCh37‐lite reference sequence using BWA‐mem v0.7.10 (params ‐t 4 –M) 18 and deduplicated using picard v1.113 (https://broadinstitute.github.io/picard/). Single nucleotide variants and small insertions and deletions were detected using VarScan v2.3.2 15 (params—min‐coverage 3—min‐var‐freq 0.08—P‐value 0.10—strand‐filter 1—map‐quality 10) and filtered by false‐positive v1 (params: bam‐readcount‐min‐base‐quality: 15, bam‐readcount‐version: 0.4, max‐mm‐qualsum‐diff:50).
Results
ATRX immunohistochemistry
Loss of ATRX immunostaining
Grade II and grade III astrocytomas showed highest frequency of ATRX immunonegativity, a surrogate marker of gene mutation (79% and 73%, respectively). There were no significant differences between the frequencies of ATRX loss in cases with both IDH‐1 and TP53 mutations or either of these alterations (Table 1).
Overall, 17.5% (14/80) of the GBMs showed loss of ATRX expression (Figure 1). The frequency of ATRX immunonegativity was significantly lower in primary GBMs (15%) as compared to their lower‐grade counterpart (P‐value <0.001). However, 43% cases of secondary GBM showed this alteration. Interestingly, ATRX negative cases showed significantly higher frequency of IDH1 mutation (8/14; 57.1%), TP53 mutation (6/14; 42.9%) and PDGFRA amplification (4/14; 28.6%) as compared to the positive cases. As a corollary, approximately 86% of the ATRX negative cases were associated with IDH1 and/or TP53 mutation and/or PDGFRA amplification and none of the cases showed EGFR amplification or NF1 alteration. Thus, only 14% of negative cases did not show any of the above genetic alterations (Table 1 and Figure 2).
Figure 1.
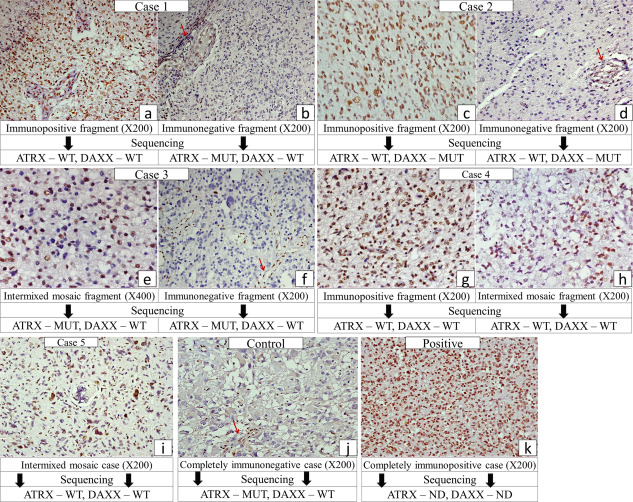
Representative photomicrographs of ATRX immunostaining in cases subjected to sequencing [arrow—endothelial positivity in negatively stained areas/cases (internal control)] with the corresponding ATRX sequencing results.
Figure 2.

Correlation of different ATRX staining pattern with key molecular genetic alterations (EGFR amplification, PDGFRA amplification, TP53 mutation, IDH‐1 mutation and NF1 alteration) in GBMs.
ATRX loss was almost mutually exclusive with 1p/19q codeletion. Only single case each of OG and AO (6.7% and 8.3%, respectively) with 1p/19q codeletion showed concomitant ATRX loss.
Loss of ATRX expression was identified in 45.7% (16/35) of cases showing histomorphological features of OA (50% grade II, 40% grade III), frequency intermediate between astrocytic and oligodendroglial tumors of same grade. However, cases with a signature of molecular astrocytoma (TP53 and/or IDH‐1 mutated without 1p/19q codeletion) showed a significantly higher frequency of ATRX loss [83.3% (10/12) and 71.4% (5/7) for grade II and grade III, respectively], similar to grade II/III astrocytomas. In contrast, cases with molecular signature of OG (1p/19q codeleted ± IDH1 mutation), demonstrated ATRX loss only in a single case (6.2%; 1/16), similar to oligodendrogliomas.
No significant difference in the frequency of ATRX immunonegativity was observed in relation to the tumor location (supratentorial vs. infratentorial) or demographic parameters.
Mosaic/heterogeneous pattern of ATRX immunostaining
Mosaic or heterogeneous staining was identified exclusively in GBMs [Primary GBMs—20.5% (15/73); Secondary GBMs—28.6% (2/7); Total—21.2% (17/80)]. Two distinct patterns of mosaic staining were identified viz. (i) Admixture of immunopositive and immunonegative nuclei within the same tumor fragment—referred to henceforth as intermixed mosaic (Figure 1E,H,I) and (ii) Separate areas/tumor fragments with positive and negative staining (Figure 1A–D). The genetic landscape of mosaic cases was entirely different from the cases with complete ATRX loss. Thus, mosaic cases demonstrated a significantly higher frequency of EGFR amplification (5/17; 29.4%) while only 23.5% (4/17) cases showed IDH1 mutation. Interestingly, 29.4% (5/17) of the mosaic cases did not show any genetic alterations as mentioned above (Figure 2).
Sequencing of the Mosaic cases
Sequencing for ATRX was carried out in five of the mosaic cases. While two of these cases (Cases 1 and 2) had distinct immunopositive and immunonegative areas, three cases (cases 3, 4 and 5) harbored an intermixed staining pattern either through‐out (case 5) or admixed with distinctly immunonegative (case 3) or distinctly immunopositive areas/tumor fragments (Case 4). In addition, a completely immunonegative case was used as control. The positive and negative areas were separately subjected to sequencing. Interestingly, case 1 showed presence of a missense mutation in the immunonegative area only; while cases 2 and 3 showed DAXX and ATRX (Stop gain) mutation, respectively, in both immunonegative and positive/intermixed mosaic stained areas (Table 2 and Figures 1, 3). On the other hand, the control case with complete negative staining showed frame‐shift type of ATRX mutation (Figure 3). Hence, there was heterogeneity in terms of ATRX mutation in between the cases albeit with homogeneous pattern in a given case. However, cases 4 (with positive and intermixed mosaic fragments) and 5 (completely intermixed mosaic) did not reveal ATRX or DAXX mutation. Thus, interestingly all cases with completely negatively stained fragments had an ATRX or DAXX mutation. However, intermixed mosaic areas were heterogeneous in terms of mutation status, in that only one of the three cases showed ATRX mutation.
Table 2.
Sequencing of the cases with mosaic immunostaining
| Age/Sex | ATRX staining | ATRX mutation | Type of ATRX mutation | Alterations in other genes | Photomicrographs | |
|---|---|---|---|---|---|---|
| Case 1 (Pr GBM) | 55/M | Positive area (65%–70%) | Absent | – | EGFR amplification | Figure 1A,B |
| Negative area (30%–35%) | Present | Missense mutation ENSP00000362441.4:p.Gly548 Val | ||||
| Case 2 (Pr GBM) | 48/F | Positive area (75%–80%) | Absent | – | EGFR amplification DAXX mutation (in both areas) | Figure 1C,D |
| Negative area (20%–25%) | Absent | – | ||||
| Case 3 (Sec. GBM) | 25/M | Intermixed Mosaic area (20%–25%) | Present | Stop gain and splice region variant ENSP00000362441.4:p.Ser1245Ter | TP53 mutation | Figure 1E,F |
| Negative area (75%–80%) | Present | Stop gain and splice region variant ENSP00000362441.4:p.Ser1245Ter | IDH1 mutation PDGFRA amplification | |||
| Case 4 (Pr GBM) | 59/F | Positive area (35%–40%) | Absent | – | EGFR amplification | Figure 1G,H |
| Intermixed Mosaic area (60%–65%) | Absent | – | ||||
| Case 5 (Pr GBM) | 63/M | Intermixed Mosaic | Absent | – | TP53 mutation | Figure 1I |
| Control (Sec. GBM) | 40/M | Complete loss of staining | Present | Frameshift variant ENSP00000362441.4:p.Lys1045Ter | TP53 mutation | Figure 1J |
Pr.—Primary, Sec.—Secondary.
Figure 3.
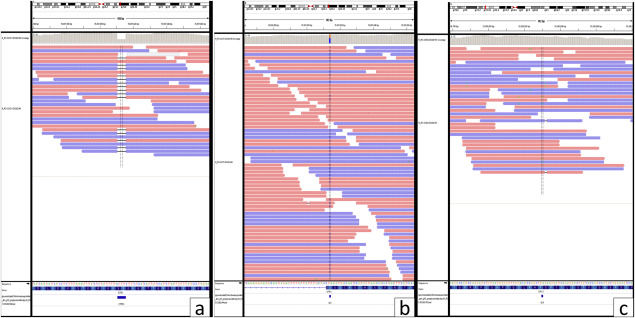
Representative photographs showing ATRX ( A,B ) and DAXX ( C ) mutations Red reads mapped to forward strand and Blue mapped to reverse strand. Bases that differ from the reference are displayed with their letters in color. A. ATRX mutation—Frameshift variant ENSP00000362441.4:p.Lys1045Ter. B. ATRX mutation‐ Stop gain‐ and splice region variant ENSP00000362441.4:p.Ser1245Ter. C. DAXX mutation—Missense variant NSP00000363668.5:p.Asp331Asn.
Discussion
ATRX is a member of the SNF2 family of ATP dependent chromatin‐remodeling proteins 8, 23, 26. It forms stable complexes with DAXX and are colocated with pericentric heterochromatin which has crucial role in chromatin remodeling, nucleosome assembly, telomere maintenance. It also facilitates the incorporation of histone variant H3.3, into pericentric, telomeric and ribosomal repeat sequences 2, 5, 6, 9, 17, 26. Loss of function of ATRX usually leads to chromosomal instability such as aneuploidy along with numerous cellular aberrations including abnormal methylation and gene expression patterns as well as chromosome misaggregation 3, 25.
Essentially all mutations in ATRX gene are inactivating type, and almost exclusively associated with loss of protein expression 10. Loss of ATRX leads to alternative lengthening of telomeres (ALT), a nontelomerase dependent mechanism of telomere lengthening, along with widespread genomic instability. Mutation of ATRX or ALT has recently emerged as a critical factor involved in the pathogenesis of few human malignancies. Liu et al 19 in their study on 140 adult gliomas of different grades, identified ATRX alteration in 45% of grade II and 57.6% of grade III atrocytomas by immunohistochemistry and sequencing methods. Another study by Jiao et al 13 on 298 cases of gliomas showed a significantly higher frequency of this alteration (grade II—67% and grade III—73%). Another recent study on 395 gliomas reported a significantly higher frequency of loss of ATRX expression in approximately 90% of grade II and grade III astrocytic tumors 24. Notably all studies revealed that loss of ATRX was relatively infrequent in adult primary GBMs (4%–24%). However, the frequency in secondary GBMs was significantly higher 13, 19, 24. Thus, Jio et al 13 reported mutation in ATRX in 57% (8/14) cases of secondary GBMs. These studies also showed that ATRX mutations characterize IDH and TP53 mutant gliomas. IDH mutation was observed in 92%–99% cases of adult gliomas with ATRX alteration in three different studies 13, 19, 28. In contrast to astrocytic tumors, ATRX loss was rare in oligodendrogliomas. Liu et al 19 in their study of 19 cases of oligodendroglioma (grade II—9, grade III—10) did not find ATRX loss/mutation in any of the cases by immunohistochemistry or sequencing. Another study by Jiao et al 13 on a larger sample size, showed ATRX mutation in 24% cases of grade II and 7% cases of grade III oligodendroglioma. Further, loss of ATRX expression was reported to be mutually exclusive with 1p/19q codeletion, the molecular hallmark of oligodendroglial tumors. Recently, a comprehensive and integrative Genomic analysis of Diffuse Lower‐Grade Gliomas has been published by TCGA. The authors subdivided gliomas based on the 1p/19q codeletion and IDH status and found ATRX mutation frequently (86%) in cases with IDH mutations and no 1p/19q codeletion which morphologically corresponded mostly to astrocytic and oligoastrocytic tumors 27. The present study on various grades and histologic subtypes of diffuse gliomas revealed that ATRX mutation/loss of expression is more prevalent in grade II/III astrocytic tumors and secondary GBMs while rare in pure oligodendrogliomas and primary GBMs. Further, loss of ATRX expression was found to be more frequent in IDH1 mutated and/or TP53 mutated tumors and almost mutually exclusive with 1p/19q codeletion, the hallmark of oligodendroglial tumors. Our result is thus in concordance with the previous published literature. Interestingly, amongst the oligoastrocytic tumors the cases with a molecular signature of astrocytomas (1p/19q codeletion−, IDH1±, TP53±) had a significantly higher frequency of ATRX loss, similar to the grade II/III astrocytic tumors. In contrast, majority of the cases with a molecular signature of oligodendroglioma (1p/19q codeletion+, IDH1±, TP53−) were immunopositive for ATRX. Hence, loss of ATRX expression possibly is a surrogate marker of astrocytic tumor, irrespective of the histomorphology of the tumor which is subject to interobserver variation. Wiestler et al 28 also reported similar findings in their study. Further, in a recent study Reuss et al 24 proposed an “integrated” approach for the diagnosis of gliomas based on the concepts of the “ISN‐Haarlem” and reclassified all the cases of mixed oligoastrocytomas into either astrocytoma or oligodendroglioma groups 20.
Similar to grade II and grade III diffuse gliomas, ATRX mutation is reported to be significantly more prevalent in IDH‐1 and TP53 mutated GBMs. Liu et al 19 identified loss of ATRX expression/ATRX mutation in 80% and 60% cases of GBM with IDH1 and TP53 mutation, respectively. Further, analysing TCGA dataset, the authors identified a subset of GBMs with proneural signature having decreased ATRX expression. Interestingly, analysis of all proneural GBMs (n = 106) in this dataset using gene expression data revealed that the cluster with low ATRX expression contained all cases with IDH1 mutations (ATRX‐low 12/39 vs. ATRX‐high 0/67; P‐ 0.0001). In a recent study, Reuss et al 24 detected loss of ATRX expression in approximately 48% cases of GBMs with IDH1 mutation. In contrast, Abedalthagafi et al 1 did not find any significant correlations between IDH status and either ALT positivity or loss of ATRX protein expression in pediatric and adult high‐grade astrocytomas. In the present study, loss of ATRX expression showed significant association with IDH‐1 mutation, TP53 mutation and PDGFRA amplification which altogether are the characteristic genetic signature of Proneural GBMs. Overall, all but one case with ATRX loss showed Proneural signature. However, cases with heterogeneous/mosaic staining displayed a distinctly different molecular profile as compared to the negative cases. These cases demonstrated a significantly higher frequency of EGFR amplification and relatively lower prevalence of IDH1 mutation. Taken together 44.4% (8/18) of the IDH1 mutated cases showed complete loss of ATRX expression and 22.2% (4/18) showed heterogeneous/mosaic staining. Similarly, 40% (6/15) and 20% (3/15) of the TP53 mutated cases demonstrated negative and mosaic ATRX staining, respectively. Interestingly, all the cases with PDGFRA amplification revealed either ATRX immunonegativity (4/6, 66.7%) or mosaic (2/6, 33.3%) staining pattern. In contrast, 20.8% cases with EGFR amplification and 6.7% cases with NF1 down‐regulation revealed mosaic ATRX staining while none of the cases with NF1 or EGFR alteration showed complete loss of ATRX expression.
Loss of ATRX nuclear expression by immunohistochemistry has been shown to be significantly associated with gene mutation and telomere lengthening. But the significance of mosaic or heterogeneous staining is not well elucidated. Cheung et al 4 identified ATRX mosaic staining in 11% cases (3/27) of neuroblastoma. Out of the three cases, two had ATRX mutation (Exon 6–12 deletion and A1 690D). Heaphy et al 10 reported heterogeneous immunostaining only in 1 out of 41 cases of pancreatic neuroendocrine tumors which showed ALT on FISH but did not reveal any gene mutation. Recently, Koelsche et al 16 analyzed ATRX expression in 573 cases soft tissue sarcomas and found heterogeneous ATRX expression in 3.5% (20/573) cases. Interestingly, 25% (2/8) of these cases also showed ALT. Reuss et al 24 proposed that ATRX immunohistochemistry is significantly attributable to the quality of tumor material, ie, fixation, thermal alteration, etc. Hence, tumor tissue which is not optimally fixed or thermally altered does not provide satisfactory staining. The authors described focal loss of ATRX staining in a single case of gliosarcoma which could not be attributed to the overall quality of the material and assumed that focal loss of ATRX expression may be diagnostic if internal controls in those areas demonstrate characteristic nuclear staining. However, the significance of heterogenous/mosaic staining of ATRX in terms of gene mutation or ALT has not been described till date in gliomas.
The present study is the first to evaluate the significance of mosaic staining pattern in diffuse gliomas. Two out of five cases (40%) with mosaic staining showed ATRX mutation. Interestingly, focal but complete loss of ATRX staining appeared to be more important in the context of ATRX gene mutation as both these cases showed mutation in immunonegative area. The other case with focal loss of ATRX staining revealed DAXX mutation. As ATRX and DAXX together form a functional complex and colocalize in the cell, it is likely that DAXX mutation may affect the expression of ATRX protein. Two of the three cases with intermixed mosaic areas lacked ATRX mutation. However, the only case of intermixed mosaic with ATRX mutation also had a separate negatively stained area and showed similar mutation in both these areas. Hence, identification of negatively stained fragment of tumor is possibly more important than the diffuse intermixed mosaic staining. Further, mosaic ATRX staining is possibly not completely attributable to quality of the tumor tissue and therefore these cases (especially with separate ATRX immunonegative areas) require further screening for ALT or gene mutation for confirmation. These findings also raise challenges in missing some “true” mutant cases in studies using tissue microarrays, with single tiny tissue cores. However, since ATRX/DAXX sequencing was performed only in limited number of cases firm conclusions regarding exact frequency/percentage of ATRX/DAXX mutation in ATRX mosaic cases cannot be ascertained from the present study.
We therefore recommend a two‐step assessment of ATRX status. Cases which are diffusely immunopositive and completely immunonegative can be regarded as mutant and wild‐type, respectively. However, cases with heterogeneous/mosaic ATRX staining, which is seen exclusively in GBMs, should be assessed further for ALT by FISH or mutation status by sequencing. Though our sample size is relatively small, the results suggest that it is important to subject cases with mosaic/heterogeneous staining for ATRX/DAXX sequencing studies.
Acknowledgments
The authors would like to sincerely thank: Washington University School of Medicine and AIIMS (Research Section) for signing the Memorandum of understanding (MOU), which facilitated Sequencing related work in Washington University School of Medicine, USA. Division of Neuropathology, Department of Pathology and Immunology, Washington University School of Medicine, USA, for kindly providing funds towards the sequencing of these cases. Department Of Biotechnology, India for providing funds for AIIMS study. David Carrell at Washington University School of Medicine for his technical assistance in DNA extraction. All technical staff from the Neuropathology laboratory, AIIMS.
Conflict of Interest: None
References
- 1. Abedalthagafi M, Phillips JJ, Kim GE, Mueller S, Haas‐Kogen DA, Marshall RE, Croul SE et al. (2013) The alternative lengthening of telomere phenotype is significantly associated with loss of ATRX expression in high‐grade pediatric and adult astrocytomas: a multi‐institutional study of 214 astrocytomas. Mod Pathol 26:1425–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bassett AR, Cooper SE, Ragab A, Travers AA (2008) The chromatin remodelling factor dATRX is involved in heterochromatin formation. PLoS One 3:e2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baumann C, Viveiros MM, De La Fuente R (2010) Loss of maternal ATRX results in centromere instability and aneuploidy in the mammalian oocyte and pre‐implantation embryo. PLoS Genet 6:e1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheung NK, Zhang J, Lu C, Parker M, Bahrami A, Tickoo SK, Heguy A et al (2012) Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA 307:1062–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Elsässer SJ, Huang H, Lewis PW, Chin JW, Allis CD, Patel DJ (2012) DAXX envelops a histone H3.3‐H4 dimer for H3.3‐specific recognition. Nature 491:560–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Emelyanov AV, Konev AY, Vershilova E, Fyodorov DV (2010) Protein complex of Drosophila ATRX/XNP and HP1a is required for the formation of pericentric beta‐heterochromatin in vivo. J Biol Chem 285:15027–15037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eustermann S, Yang JC, Law MJ, Amos R, Chapman LM, Jelinska C, Garrick D et al (2011) Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat Struct Mol Biol 18:777–782. [DOI] [PubMed] [Google Scholar]
- 8. Gibbons RJ, Picketts DJ, Villard L, Higgs DR (1995) Mutations in a putative global transcriptional regulator cause X‐linked mental retardation with alpha‐thalassemia (ATR‐X syndrome). Cell 80:837–845. [DOI] [PubMed] [Google Scholar]
- 9. Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S et al (2010) Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140:678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C et al (2011) Altered telomeres in tumors with ATRX and DAXX mutations. Science 333:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Iwase S, Xiang B, Ghosh S, Ren T, Lewis PW, Cochrane JC, Allis CD et al (2011) ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental‐retardation syndrome. Nat Struct Mol Biol 18:769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jha P, Suri V, Singh G, Jha P, Purkait S, Pathak P, Sharma V et al (2011) Characterization of molecular genetic alterations in GBMs highlights a distinctive molecular profile in young adults. Diagn Mol Pathol 20:225–232. [DOI] [PubMed] [Google Scholar]
- 13. Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF, Rodriguez FJ et al (2012) Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget 3:709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD et al (2011) DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331:1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA et al (2012) VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22:568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koelsche C, Renner M, Johann P, Leiss I, Sahm F, Schimmack S, Wardelmann E et al (2015) Differential nuclear ATRX expression in sarcomas. Histopathology [Epub ahead of print; doi: 10.1111/his.12812]. [DOI] [PubMed] [Google Scholar]
- 17. Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD (2010) Daxx is an H3.3‐specific histone chaperone and cooperates with ATRX in replication‐independent chromatin assembly at telomeres. Proc Natl Acad Sci USA 107:14075–14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu XY, Gerges N, Korshunov A, Sabha N, Khuong‐Quang DA, Fontebasso AM, Fleming A et al. (2012) Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol 124:615–625. [DOI] [PubMed] [Google Scholar]
- 20. Louis DN, Perry A, Burger P, Ellison DW, Reifenberger G, von Deimling A, Aldape K et al (2014) International Society Of Neuropathology—Haarlem. International Society of Neuropathology—Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol 24:429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nguyen DN, Heaphy CM, de Wilde RF, Orr BA, Odia Y, Eberhart CG, Meeker AK et al (2013) Molecular and morphologic correlates of the alternative lengthening of telomeres phenotype in high‐grade astrocytomas. Brain Pathol 23: 237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ohgaki H, Kleihues P (2007) Genetic pathways to primary and secondary glioblastoma. Am J Pathol 170:1445–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OW, Gibbons RJ (1996) ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR‐X syndrome. Hum Mol Genet 5:1899–1907. [DOI] [PubMed] [Google Scholar]
- 24. Reuss DE, Sahm F, Schrimpf D, Wiestler B, Capper D, Koelsche C, Schweizer L et al (2015) ATRX and IDH1‐R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an “integrated” diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol 129:133–146. [DOI] [PubMed] [Google Scholar]
- 25. Ritchie K, Seah C, Moulin J, Isaac C, Dick F, Bérubé NG (2008) Loss of ATRX leads to chromosome cohesion and congression defects. J Cell Biol 180:315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tang J, Wu S, Liu H, Stratt R, Barak OG, Shiekhattar R, Picketts DJ et al (2004) A novel transcription regulatory complex containing death domain‐associated protein and the ATR‐X syndrome protein. J Biol Chem 279:20369–20377. [DOI] [PubMed] [Google Scholar]
- 27. Cancer Genome Atlas Research Network The (2015) Comprehensive, integrative genomic analysis of diffuse lower‐grade gliomas. N Engl J Med 372:2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wiestler B, Capper D, Holland‐Letz T, Korshunov A, von Deimling A, Pfister SM, Platten M et al. (2013) ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol 126:443–451. [DOI] [PubMed] [Google Scholar]


