Abstract
Epithelioid glioblastoma (eGBM) and pleomorphic xanthoastrocytoma (PXA) with anaplastically transformed foci (ePXA) show overlapping features. Eleven eGBMs and 5 ePXAs were reviewed and studied immunohistochemically. Fluorescence in situ hybridization for EGFR amplification, PTEN deletion and ODZ3 deletion was also performed, with Ilumina 450 methylome analysis obtained in five cases. The average age for eGBM was 30.9 (range 2–79) years, including five pediatric cases and a M : F ratio of 4.5. The ePXA patients had a M : F ratio of 4 and averaged 21.2 (range 10–38) years in age, including two pediatric cases. Six eGBMs and two ePXAs recurred (median recurrence interval of 12 and 3.3 months, respectively). All tumors were composed of solid sheets of loosely cohesive, “melanoma‐like” cells with only limited infiltration. ePXAs showed lower grade foci with classic features of PXA. Both tumor types showed focal expression of epithelial and glial markers, retained INI1 and BRG1 expression, occasional CD34 positivity, and lack of mutant IDH1 (R132H) immunoreactivity. BRAF V600E mutation was present in four eGBMs and four ePXAs. ODZ3 deletion was detected in seven eGBMs and two ePXAs. EGFR amplification was absent. Methylome analysis showed that one ePXA and one eGBM clustered with PXAs, one eGBM clustered with low‐grade gliomas, and two eGBMs clustered with pediatric‐type glioblastomas. Common histologic, immunohistochemical, molecular and clinical features found in eGBM and ePXA suggest that they are closely related or the same entity. If the latter is true, the nomenclature and WHO grading remains to be resolved.
Keywords: BRAFV600E, epithelioid, glioblastoma, ODZ3, pediatric, pleomorphic, prognosis, xanthoastrocytoma
Introduction
Glioblastoma (GBM), WHO grade IV, is one of the most malignant cancer types in adult and pediatric populations. Part of its original name of “multiforme” reflects its remarkable histologic variability, with the World Health Organization (WHO) recognizing several subtypes as either clinically and molecularly distinct variants or merely morphologic patterns with no obvious differences from classic GBM 14.
Epithelioid glioblastoma (eGBM) is one of the rarest variants and is not yet recognized as a subtype in the 2007 WHO classification scheme 14. It is characterized by relatively solid aggregates of large “melanoma‐like” epithelioid cells with abundant eosinophilic cytoplasm, some eccentrically placed nuclei, prominent nucleoli and distinct cellular membranes 8, 22, 23. While one study makes a distinction between eGBM and rhabdoid GBM based on the focal loss of INI1 immunoreactivity in rhabdoid areas 9, there is considerable overlap and others use these terms interchangeably 11, 17, 27. The diagnosis of eGBM is often challenging and only a few small series have been reported in the adult and pediatric populations 2, 8, 19. Excluding other CNS tumors with epithelioid features is critical and it has been noted that a subset of anaplastic pleomorphic xanthoastrocytomas (PXAs) have similar pathologic features 7, 14 and even some overlapping molecular findings 5, 8. In our practice, we have occasionally encountered PXAs with anaplastic transformation showing cytologic characteristics that are indistinguishable from those of eGBM. In a similar fashion, a case report of eGBM arising in a PXA has recently been reported 26. As with eGBM, PXAs with anaplastic transformation most often afflict children and young adults, although cases in older patients have also been reported 3, 15.
A recent case report of an eGBM with BRAFV600E mutation used comparative genomic hybridization (CGH) to identify three copy number alterations located in different morphologic areas; there were three alterations restricted to the epithelioid areas: a homozygous deletion in intron 3 of LSAMP at 3q13, a hemizygous deletion of ODZ3 at 4q34.3–4q35.1 and a hemizygous deletion in LRP1B on chromosome 2 19. ODZ3 gene was reported as a potent tumor suppressor gene, which has also been implicated in the pathogenesis of lung adenocarcinoma, adrenal carcinoma and neuroblastoma 13, 16, 18. In this study, we analyzed the available clinical, histopathologic and molecular data of 16 eGBMs and PXAs with epithelioid forms of anaplastic transformation (ePXA), with the intent of establishing if these tumors share a common tumorigenic background. To explore this possibility further, we performed fluorescence in situ hybridization for ODZ3 deletion and methylome signature studies in subsets of both ePXA and eGBM.
Materials and Methods
Case selection
As defined previously by Kleinschmidt‐DeMasters et al, eGBM had to be composed in great part (more than 30%–40% of the tumor) of patternless sheets of loosely cohesive, epithelioid or melanoma‐like cells with distinct cellular borders, abundant eosinophilic cytoplasm, and central or slightly eccentric nuclei with distinct nucleoli 8. Anaplastic features such as high mitotic index, microvascular proliferation and necrosis are variably superimposed and some evidence of glial differentiation was required. ePXA was similarly defined as a glioma that is composed predominantly (more than 30%–40% of the tumor) of epithelioid cells with the same features as seen in eGBM, but also demonstrates at least a small area of classic PXA.
With Institutional Review Board approval, we searched the archives of the Department of Pathology at UCSF Medical Center and found 12 cases that fit the histologic criteria for eGBM or ePXA within the January 2013 to April 2015 interval. With the exception of one ePXA, the rest of the cases were consults submitted to one of the authors (AP). None of these 12 cases have been previously reported. In addition, four eGBMs were provided by the Institute of Pathology, Ruprecht‐Karls University, Heidelberg, Germany, and were also part of a separate previously reported study of pediatric GBMs 10. For the purpose of this study, patients 18 years old or younger were considered pediatric.
Clinical and radiographic information
All available clinical and neuroimaging studies (images and/or radiology reports) were reviewed from electronic patient records and submitted consult materials.
Histology
Histological review and immunohistochemical studies were performed on 4 μ sections of formalin‐fixed paraffin‐embedded tissue. Immunohistochemical assessment was performed using antibodies against: glial fibrillary acidic protein (GFAP) (DAKO 1:300, Carpinteria, CA), Olig2 (Immuno Bio Labs 1:200, Minneapolis, MN), neurofilament (NF) protein (Cell Marque predilute, Rocklin, CA), synaptophysin (Cell Marque 1:100, Rocklin, CA), CD34 (Leica Biosystems predilute, Buffalo Grove, IL), EMA (Leica Biosystems predilute, Buffalo Grove, IL), vimentin (Leica Biosystems predilute, Buffalo Grove, IL), Ki67 (Leica Biosystems predilute, Buffalo Grove, IL), NeuN (Chemicon International 1:4000, Doublin, OH), MAP2 (Sigma‐Aldrich 1:20 000, St. Louis, MO), p53 (Vector Laboratories, 1:100, Burlingame, CA), IDH1(R132H) (Dianova 1:500, Hamburg, Germany), BRAFV600E (Spring Bioscience 1:50, Pleasanton, CA) and INI1 (SMARCB1 gene product) (Becton Dickinson 1:100, San Jose, CA). An antibody against BRG1 (SMARCA4 gene product) (Abcam 1:100, Cambridge, MA) was applied to three ePXA and four eGBM cases in the histology laboratory at St. Jude's Children's Research Hospital. H3.3 K27M (Millipore 1:500, Billerica, CA) immunohistochemical stain was performed on eight of the eGBM cases. The presence or absence of tumoral staining, the general extents of expression (eg, diffuse, patchy, focal, rare) and staining localization patterns were noted. The p53 immunostain was reported as positive when greater than 10% p53 nuclear positivity was present 25. The immunohistochemical studies were limited in one case of eGBM and a case of ePXA because of insufficient material. Digital photomicrographs were taken using an Olympus DP72 camera.
Fluorescence in situ hybridization
To investigate the deletion status of the ODZ3 gene, we utilized a commercial ODZ3 gene bacterial artificial clone (BAC) (RP11‐713O21; 4q34.3–4q35.1) and confirmed a portion of its published sequence using polymerase chain reaction (PCR).
This probe was directly labeled with rhodamine and paired with a SpectrumGreen labeled CEP 4 commercial FISH probe (06J37‐014, Abbott Molecular) using previously reported methodology 20.
Green and red signals were enumerated under an Olympus BX 41 fluorescence microscope. Cases with more than 25% cells demonstrating a ratio of 1 red : 2 green signals were considered deleted. For control specimens, we performed FISH for ODZ3 gene deletion on slides from 10 epilepsy resections (five focal cortical dysplasia and five mesio‐temporal sclerosis).
The EGFR amplification and PTEN deletion FISH assays were designed and performed in the Molecular Laboratory in the Department of Pathology at UCSF Medical Center using the VysisTM dual‐color DNA probe set. EGFR amplification was diagnosed when the ratio of EGFR to CEP7 exceeded 2.2. PTEN (10q) deletion was identified when the ratio of PTEN to CEP10 is less than 0.8 and the percent of cells with <2 signals is greater than 20%. Monosomy 10 required that the percentage of cells with <2 CEP10 signals exceeded 30%.
Genome‐wide methylation studies
Genome‐wide methylation patterns were studied in four pediatric cases of eGBM and an adult case of ePXA in the Department of Pathology at Heidelberg University Hospital in Germany, utilizing the method previously reported by Korshunov et al 10. Briefly, the DNA was extracted from tumors and analyzed for genome‐wide methylation patterns using the Illumina Human Methylation450 BeadChip (450k) array. Custom approaches described by Sturm et al 24 were utilized to process the DNA methylation data and the detection of copy number aberrations. The most variably methylated probes across the dataset were used for unsupervised hierarchical clustering, and samples were clustered using 1‐Pearson correlation coefficient as the distance measure and average linkage. Methylation probes were reordered by hierarchical clustering using Euclidean distance and average linkage.
Results
Clinical data
A summary of clinical findings is found in Table 1. There were 13 male (81.3%) (nine eGBMs, four ePXAs) and 3 female (18.8%) (two eGBMs and one ePXA) patients. Seven of the cases (43.8%) (two ePXAs and five eGBMs) were pediatric and nine (56.2%) (three ePXAs and six eGBMs) were adult. The average ages for ePXA and eGBM patients were 21.2 (range 10–38) and 30.9 (range 2–79) years, respectively. All ePXAs arose in the cerebral hemispheres (one frontal, one fronto‐temporal, two occipital, one temporal). Two of the eGBMs arose in the thoracic spine and nine in the cerebral hemispheres (six temporal, one frontal, one parietal and one adjacent to the lateral ventricle).
Table 1.
Summary of the clinical information. Abbreviations: NA = not applicable; NK = not known; ePXA = epithelioid pleomorphic xanthoastrocytoma; eGBM = epithelioid glioblastoma; GTR = gross total resection; STR = subtotal resection
| Case No. | Age | Sex | Diagnosis | Location | Resection type | Recurrence/progression | Recurrence interval (months) | Follow‐up (months) | Alive at last follow‐up |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 38 | M | ePXA | Left occipital | GTR | Yes | 1.5 | 2 | Yes |
| 2 | 25 | M | ePXA | Right frontal | GTR | No | NA | 10 | Yes |
| 3 | 13 | M | ePXA | Right occipital | GTR | Yes | 5 | 7 | Yes |
| 4 | 20 | F | ePXA | Right temporal | GTR | No | NA | 13 | Yes |
| 5 | 10 | M | ePXA | Right fronto‐temporal | STR | No | NA | 3 | Yes |
| 6 | 38 | M | eGBM | T7–8 | STR | No | NA | 4 | No |
| 7 | 11 | M | eGBM | Right temporo‐parietal | GTR | Yes | 53 | 65 | Yes |
| 8 | 21 | F | eGBM | Right fronto‐temporal | NK | NK | NK | 0 | NK |
| 9 | 79 | M | eGBM | Right temporal | NK | Yes | 8 | 8 | Yes |
| 10 | 70 | F | eGBM | Right temporal | NK | No | NA | 5.2 | Yes |
| 11 | 12 | M | eGBM | Frontal | STR | Yes | 12 | 14 | No |
| 12 | 11 | M | eGBM | Temporal | STR | Yes | 24 | 38 | Yes |
| 13 | 2 | M | eGBM | Lateral ventricle | GTR | No | NA | 16 | Yes |
| 14 | 14 | M | eGBM | Parietal | GTR | Yes | 12 | 33 | Yes |
| 15 | 26 | M | eGBM | T9–12 | STR | No | NA | 2 | Yes |
| 16 | 56 | M | eGBM | Right temporal | NK | Yes | 2 | 2 | Yes |
The neuroradiology reports and/or images were reviewed in six eGBM and all ePXA cases. The tumors showed nonspecific features overall, but regardless of diagnosis, most commonly featured well demarcated, contrast enhancing, superficial cerebral masses. The associated edema was more pronounced in the eGBM cases (Figure 1). Three of the ePXAs (60%) and two of the eGBMs (18.2%) had leptomeningeal spread at diagnosis.
Figure 1.
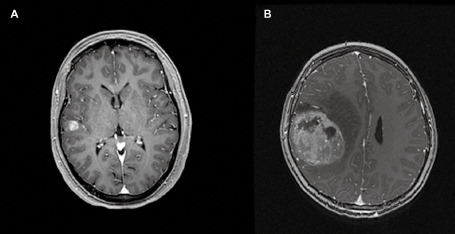
A. Post‐gadolinium t1 axial magnetic resonance image demonstrating a well‐defined enhancing superficial ePXA in the right temporal lobe, involving mostly the cortex; surrounding edema is not seen. B. Post‐gadolinium magnetic resonance t1 axial image demonstrating a well‐defined, heterogeneously enhancing superficial eGBM with marked surrounding edema.
The median follow‐up interval for all patients was 8 months (range 2–65 months). Two ePXAs (40%) and six eGBMs (54.5%) recurred; the median interval to recurrence for the former was 3.3 months and for the latter was 12 months. Two patients with eGBM, one adult and one pediatric, died of disease 4 and 14 months after diagnosis, respectively. However, most of the patients remain alive at last follow‐up with survival times already exceeding 2 years in three eGBM cases (33, 38 and 65 months at last follow‐up), suggesting that long survival times are seen in a subset.
Histopathology and molecular features
The histopathologic and immunohistochemical findings are summarized in Tables 2 and 3, while the molecular findings are summarized in Table 4. Epithelioid foci were defined by the presence of large “melanoma‐like” tumor cells with abundant eosinophilic cytoplasm and nuclei that are occasionally eccentrically placed and suggestive of rhabdoid cytology (Figure 2). This was typically accompanied by loose cellular cohesion, a high mitotic index and foci of tumor necrosis, with microvascular proliferation seen in a subset. All ePXAs showed at least focal areas of classic PXA by definition (Figure 3A), including spindled cells variably forming fascicles, large bizarre mono‐ and multinucleated cells with or without lipidized cytoplasm, perivascular lymphocytic cuffing and eosinophilic granular bodies; these features were absent from eGBMs and from the epithelioid portions of ePXAs (Figure 3B). The eGBMs in our series were composed almost exclusively of epithelioid cells with the cytologic features described above, and only three of the cases demonstrated a definite infiltrative component, resembling a classic diffuse astrocytoma. Frequent mitoses (more than 5 per 10 high‐power fields), areas of necrosis, including pseudopalisading, and microvascular proliferation were present in the epithelioid foci of all cases. The reticulin stain showed a rich intercellular meshwork in the classic PXA areas in three of the five cases, whereas the epithelioid areas were reticulin poor in all five. Classic PXA areas were extensively positive for GFAP (Figure 3C) and Olig2, whereas these glial markers highlighted only rare cells in epithelioid areas (Figure 3D). A NF immunostain demonstrated occasional entrapped axons at the periphery of the ePXAs, but otherwise highlighted the mostly solid growth patterns; similarly, eGBMs displayed a mostly solid growth pattern with infiltration only at the periphery in seven (30%) of the cases and a more diffusely infiltrative growth pattern in three (30%) cases. There was patchy EMA positivity in all ePXAs and in eight of the eGBMs (72.7%). Four ePXAs (80%) and four eGBMs (40%) showed patchy synaptophysin positivity in tumor cells. Three of the ePXAs demonstrated scattered cells with membranous CD34 immunopositivity involving both classic PXA and epithelioid foci (60%) (Figure 3E,F), whereas occasional CD34‐positive tumor cells were encountered in four eGBMs (40%). The vimentin immunostain was extensively positive in all cases tested. All cases had retained nuclear INI1 expression, with BRG1 expression similarly retained in all seven cases tested. All eGBMs were negative for IDH1 (R132H) mutant protein expression. A p53 immunostain was performed on 10 cases and it was positive in two ePXAs (40%) and four eGBMs (40%). The BRAFV600E immunostain was positive in both components of 4 ePXAs (80%) and in 4 of the 10 eGBMs in which it was performed (40%) (Figure 3G,H). The Ki‐67 labeling index was low in the PXA areas of ePXA (Figure 3I) and high in the epithelioid areas of all cases in which it was performed (Figure 3J). Case 15 was unique in the series in that it was the only case with H3 K27M mutant protein expression and loss of ATRX. It additionally had a small focus resembling ganglioglioma, which similarly expressed H3 K27M, but retained nuclear ATRX expression.
Table 2.
Histopathologic features of 5 EPXAs and 11 eGBMs. Abbreviations: ePXA = epithelioid pleomorphic xanthoastrocytoma; eGBM = epithelioid glioblastoma
| Case No. | Diagnosis | Mitotic rate (per 10 HPF) | Necrosis | Microvascular proliferation | Eosinophilic granular bodies |
|---|---|---|---|---|---|
| 1 | ePXA | 6 | Geographic, large foci | Present, one focus | Present |
| 2 | ePXA | 20 | Pseudopalisading and geographic | Present | Present; rare |
| 3 | ePXA | 5 | Geographic, small focus | Not present | Present; rare |
| 4 | ePXA | 5 | Pseudopalisading and geographic, small foci | Present | Present |
| 5 | ePXA | 7 | Pseudopalisading and geographic | Present | Present |
| 6 | eGBM | 30 | Geographic, extensive | Present | Absent |
| 7 | eGBM | 7 | Pseudopalisading and geographic | Present | Absent |
| 8 | eGBM | 15 | Pseudopalisading and geographic, extensive | Present | Absent |
| 9 | eGBM | 12 | Geographic and focally pseudopalisading | Present | Absent |
| 10 | eGBM | 5 | Geographic | Present | Absent |
| 11 | eGBM | 16 | Geographic; a small focus of pseudopalisading | Present | Absent |
| 12 | eGBM | 30 | Pseudopalisading; geographic occasionally | Present | Absent |
| 13 | eGBM | 7 | Geographic | Absent | Absent |
| 14 | eGBM | 15 | Pseudopalisading | Present | Absent |
| 15 | eGBM | 8 | Absent | Present, rare foci | Absent |
| 16 | eGBM | 4 | Pseudopalisading | Present | Absent |
Table 3.
Summary of the immunohistochemical stains. Abbreviations: ND = not done; ePXA = epithelioid pleomorphic xanthoastrocytoma; eGBM = epithelioid glioblastoma
| Case No. | Diagnosis | Reticulin | GFAP | Olig2 | NF | Synaptophysin | CD34 | EMA | Ki67 | Vimentin |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ePXA | Rich in PXA/poor in epithelioid | Focal in PXA/negative in epithelioid | Positive in PXA/negative in epithelioid | Mostly solid; occasional tumor cells | Weak; occasional epithelioid cells positive | Patchy; arborescent pattern | Patchy | 3% in PXA; 90% in epithelioid | Extensive |
| 2 | ePXA | Poor | Focal | Focal in PXA | Mostly solid; rare tumor cells | Focal in tumor cells | Negative | Patchy | 8% in PXA; 30% in epithelioid | Extensive |
| 3 | ePXA | Rich in PXA areas/poor in epithelioid | Occasional | Negative | Mostly solid; negative in tumor cells | Negative | Negative | Patchy | 10% in epithelioid; not done in PXA | Extensive |
| 4 | ePXA | Rich in PXA/poor in epithelioid | Extensive | ND | Mostly solid; negative in tumor cells | Occasional cells | Positive; arborescent pattern | Patchy | 2.5% in PXA; 11.8% in epithelioid | Extensive |
| 5 | ePXA | Rich in PXA/poor in eGBM | Positive in PXA/negative in eGBM | Patchy; more in PXA | Mostly solid; rare tumor cells | Occasional tumor cells | Focal; arborescent pattern | Patchy | 12% in PXA; 50% in epithelioid | Extensive |
| 6 | eGBM | ND | Patchy | Focal | ND | Negative | Negative | Patchy | 50% | ND |
| 7 | eGBM | ND | Negative | Negative | Mostly solid; negative in tumor cells | ND | Negative | Patchy | ND | Extensive |
| 8 | eGBM | ND | Focal | Negative | Mostly solid; occasional tumor cells | Focal in tumor cells | Rare; membrane staining | Patchy | ND | Extensive |
| 9 | eGBM | ND | Occasional | Negative | Mostly solid; occasional tumor cells | Rare tumor cells | Negative | Negative | ND | Extensive |
| 10 | eGBM | ND | Rare cells | Negative | Mostly solid; negative in tumor cells | Positive in a subset of tumor cells | Negative | Patchy | ND | Extensive |
| 11 | eGBM | ND | Focal | ND | Infiltrative; negative in tumor cells | Negative | Focal; membrane staining | Rare tumor cells | ND | ND |
| 12 | eGBM | ND | Extensive | ND | Mostly solid; occasional tumor cells | Negative | Negative | Patchy | ND | ND |
| 13 | eGBM | ND | Focal | ND | Mostly solid; negative in tumor cells | Negative | Negative | Negative | ND | ND |
| 14 | eGBM | ND | Extensive | ND | Mostly solid; negative in tumor cells | Negative | Rare; membrane staining | Rare tumor cells | ND | ND |
| 15 | eGBM | ND | Focal; less in epithelial areas | Extensive | Infiltrative; occasional tumor cells | Occasional tumor cells | Extensive; membrane staining | Negative | 17% | Extensive |
| 16 | eGBM | ND | Negative | Rare cells | Infiltrative; negative in tumor cells | Occasional entrapped neurons | ND | Patchy | ND | ND |
Table 4.
Molecular characteristics of the cases of ePXA and eGBM. Abbreviations: ND = not done; NI = not interpretable
| Case No. | p53 (IHC) | IDH1 (R132H) (IHC) | ATRX (IHC) | INI1(BAF47) (IHC) | BRG1 (IHC) | BRAF V600E (IHC) | EGFR amplification (FISH) | PTEN deletion (FISH) | ODZ3 deletion (FISH) | H3 K27M (IHC) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Negative | ND | ND | Retained | Retained | Positive | ND | ND | Negative | ND |
| 2 | Positive | ND | ND | Retained | Retained | Negative | Negative | Negative | Negative | ND |
| 3 | Negative | ND | ND | Retained | Retained | Positive | ND | ND | Deletion | ND |
| 4 | Negative | ND | ND | ND | ND | Positive | ND | ND | Negative | ND |
| 5 | Positive | ND | ND | Retained | ND | Positive | Negative | Negative | Deletion | ND |
| 6 | Positive | Negative | Retained | Retained | Retained | Negative | Negative | ND | Deletion | Negative |
| 7 | Positive | Negative | Retained | Retained | Retained | Positive | Negative | Monosomy 10 | Negative | Negative |
| 8 | Negative | Negative | Retained | Retained | Retained | Positive | Negative | ND | Deletion | Negative |
| 9 | Negative | Negative | Retained | Retained | Retained | Negative | Negative | Deletion | Deletion | ND |
| 10 | Positive | Negative | Retained | Retained | ND | Negative | Negative | ND | Deletion | ND |
| 11 | Negative | Negative | ND | Retained | ND | Negative | Negative | ND | Deletion | Negative |
| 12 | Negative | Negative | ND | Retained | ND | Positive | Negative | Negative | Deletion | Negative |
| 13 | Negative | Negative | ND | Retained | ND | Positive | Negative | Negative | ND | Negative |
| 14 | Negative | Negative | ND | Retained | ND | Negative | Negative | Negative | Deletion | Negative |
| 15 | Positive | Negative | Lost | Retained | ND | Negative | Negative | Negative | Negative | Positive |
| 16 | ND | Negative | ND | ND | ND | ND | Negative | ND | NI | ND |
Figure 2.
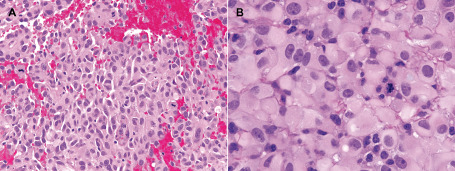
A. Hematoxylin–eosin‐stained (H&E) section of an epithelioid area in a case of ePXA (case 5), demonstrating sheets of loosely cohesive cells with abundant eosinophilic cytoplasm, large, occasionally eccentric nuclei, occasional nucleoli and distinct cellular borders (melanoma‐like features). A high mitotic rate is observed. B. A high‐power microscopic field of an eGBM (case 6), demonstrating epithelioid cells with the same features as described above. Occasional cells with paranuclear globular inclusions are seen (rhabdoid features); furthermore, there are occasional cells with enlarged red macronucleoli (melanoma‐like features).
Figure 3.
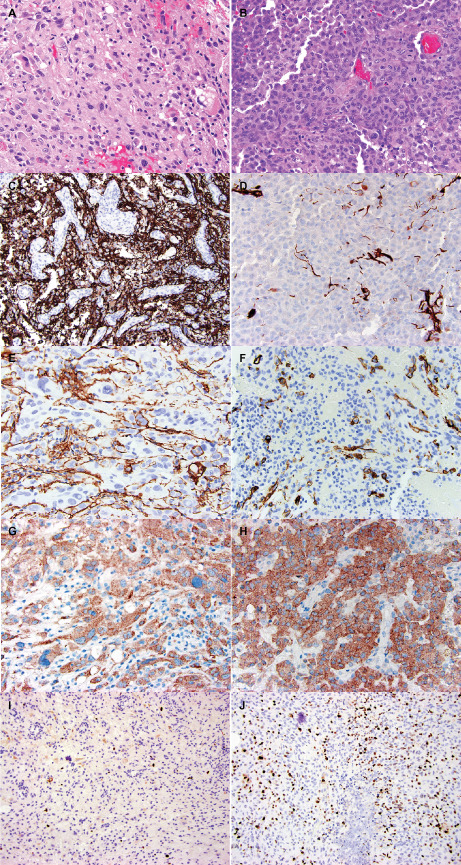
Comparison of morphologic and immunohistochemical features of PXA (left) and epithelioid (right) areas in a single case of ePXA (case 5). A. Illustrates an area composed of spindled and oval cells with irregular nuclear contours and variably vacuolated (lipidized) cytoplasm. Eosinophilic granular bodies and occasional bizarre, large cells are also seen. B. Shows the epithelioid regions of anaplastic transformation. The PXA areas were extensively positive for GFAP (cytoplasmic and fibrillary pattern) (C), whereas the epithelioid areas showed mostly loss of GFAP expression (D). Occasional tumor cells in the PXA areas demonstrated CD34 expression in an arborescent pattern (E). The epithelioid areas also contained occasional CD34‐positive tumor cells (F). Both areas demonstrated BRAF V600E mutant protein expression (G,H). The Ki67 immunostain revealed a low proliferation index in the PXA areas (I), and a considerably higher labeling index in the epithelioid areas (J).
Fluorescence in situ hybridization
FISH for ODZ3 gene could not be performed on one consult case due to lack of additional unstained slides and paraffin blocks. Furthermore, in one case, the signals could not be accurately interpreted. Seven of the remaining nine eGBM cases (77.8%) and two of the ePXA cases (40%) demonstrated hemizygous deletion of ODZ3 gene at 4q34.3–4q35.1 (Figure 4). Of these eGBMs, three were pediatric and four were adult. The ePXAs with deletion were both pediatric. Both eGBMs and ePXAs in this study were composed in great part of areas with epithelioid morphologic features and the majority of cells in which the FISH signals were counted were therefore epithelioid.
Figure 4.
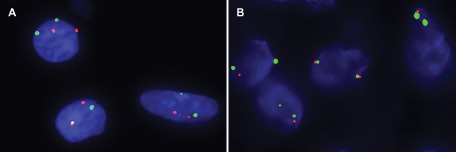
A. Illustrates a non‐neoplastic control case with two CEP4 signals (green) and two ODZ3 signals (red) in most nuclei. B. Illustrates a case of eGBM in which the majority of nuclei contain one ODZ3 (red) and two CEP4 (green) signals.
Five eGBMs (45.5%) showed polysomy 7 (3–5 centromeres and EGFR signals per cell), but none had evidence of EGFR gene amplification. Monosomy of chromosome 10 was present in a pediatric case of eGBM and more localized PTEN (10q) deletion was found in one adult case of eGBM.
Methylome signatures
The adult ePXA (case 2) had a methylation pattern that clustered with that of other histologically classic PXAs studied in Heidelberg, Germany (not part of current study) 10. One pediatric eGBM with ODZ3 gene deletion (case 12) similarly had a methylation pattern that clustered with the PXAs and another pediatric GBM (case 13) had a methylation pattern that clustered with the low‐grade astrocytomas. The remaining two cases of eGBM clustered with histologically classic examples of pediatric GBM studied in Heidelberg, Germany (not part of current study). The results of Illumina Human Methylation450 BeadChip (450k) array for the five cases are summarized in Table 5.
Table 5.
Results of the genome‐wide methylation studies performed on five of the cases 10. Abbreviations: LGA = low‐grade glioma; Mut. = mutated; WT = wild type; PXA = pleomorphic xanthoastrocytoma; GBM = glioblastoma
| Case No. | 450k array clustering pattern | H3F3A | IDH1 (R132H) | BRAF V600E | Amplifications | Homozygous deletions | Gains | Losses |
|---|---|---|---|---|---|---|---|---|
| 2 | PXA | WT | WT | WT | None | CDKN2A | 5, 7, 9q, 12p, 14q, 16q, 22q | 1, 6, 13q, 14q, 21q |
| 11 | GBM | WT | WT | WT | None | None | 7 | 1p, 3p, 6, 8, 9p, 10p, 13q, 14q, 17p, 22q |
| 12 | PXA | WT | WT | Mut. | None | None | None | None |
| 13 | LGA | WT | WT | Mut. | None | None | None | None |
| 14 | GBM | WT | WT | WT | None | None | None | None |
Discussion
eGBM is a very rare variant of GBM that is not included in the 2007 WHO classification scheme 14. Only case reports and a few small series have been previously reported 2, 8. It is usually seen within the first three decades of life and the majority of cases are primary (ie, no previously established lower grade precursor). The course of eGBM is an aggressive one and is often complicated by early recurrence, intratumoral hemorrhage and leptomeningeal spread 2. Nevertheless, surprisingly long survival times have been reported in a subset 1, 8 as was found also in a subset of our own patients. Additionally, dramatic responses to BRAF inhibitors have been reported anecdotally in V600E mutant examples, emphasizing that this variant may have several important differences from that of conventional GBM 4, 12, 21. Most of the time, this variant also differs in that it does not show EGFR amplification, IDH1 gene mutation or PTEN deletion, but instead, about half of these cases harbor BRAF V600E mutation 8, a finding that remained true in our study as well.
eGBM is difficult to distinguish from PXA with anaplastic transformation with epithelioid features (ePXA), another subtype that is not described in the 2007 WHO scheme, but has been reported rarely 6. In our series, ePXA behaved clinically similar to the eGBMs, with recurrences within months of initial resection, and leptomeningeal dissemination, although in both tumor types, some long survival times were also seen. In the current study, we found no reliable demographic, radiologic, histologic, immunohistochemical or molecular differences between the two glioma subtypes, with the only distinction being definitional, in that a classic area of low‐grade PXA is found in ePXA but not in eGBM. This raises the possibility that at least in some eGBM, a precursor PXA component may be overrun by the more malignant epithelioid component or was not sampled in the resection specimen. Supporting this hypothesis is the fact that in one of our cases, the PXA component accounted for no more than 5% of the entire tumor and was relatively inconspicuous in comparison to the adjacent, more abundant epithelioid elements. Furthermore, methylome analysis showed that one of our four tested eGBMs had a methylation pattern that clustered with conventional PXA. Lastly, a case recently reported by Tanaka et al 26 showed an eGBM developing within the tumor bed of a PXA 13 years after initial resection. All these data suggest that eGBM and ePXA are either the same entity or highly related.
Nevertheless, one of our eGBM cases was associated with the H3 K27M molecular subtype of high‐grade glioma and one of the eGBM cases previously reported by Kleinschmidt‐DeMasters et al was IDH mutant 8. As such, it is possible that the epithelioid phenotype can also represent a secondary pattern in other more classically described molecular subtypes of GBM. Therefore, there remain a number of unanswered questions regarding WHO nomenclature (PXA vs. GBM), grade (III vs. IV), and whether such cases should be considered part of a unique entity, a distinct variant, or merely a morphologic pattern that can be encountered in more than one molecularly defined GBM subtype.
Acknowledgments
We thank Roxanne Marshall for her help and expertise in setting up and performing fluorescence in situ hybridization. The authors have no conflict of interest to declare.
References
- 1. Babu R, Hatef J, McLendon RE, Cummings TJ, Sampson JH, Friedman AH, Adamson C (2013) Clinicopathological characteristics and treatment of rhabdoid glioblastoma. J Neurosurg 119:412–419. [DOI] [PubMed] [Google Scholar]
- 2. Broniscer A, Tatevossian RG, Sabin ND, Klimo P Jr, Dalton J, Lee R et al (2014) Clinical, radiological, histological and molecular characteristics of paediatric epithelioid glioblastoma. Neuropathol Appl Neurobiol 40:327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chakrabarty A, Mitchell P, Bridges LR, Franks AJ (1999) Malignant transformation in pleomorphic xanthoastrocytoma—a report of two cases. Br J Neurosurg 13:516–519. [PubMed] [Google Scholar]
- 4. Chamberlain MC (2013) Salvage therapy with BRAF inhibitors for recurrent pleomorphic xanthoastrocytoma: a retrospective case series. J Neurooncol 114:237–240. [DOI] [PubMed] [Google Scholar]
- 5. Dias‐Santagata D, Lam Q, Vernovsky K, Vena N, Lennerz JK, Borger DR et al (2011) BRAF V600E mutations are common in pleomorphic xanthoastrocytoma: diagnostic and therapeutic implications. PLoS ONE 6:e17948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iwaki T, Fukui M, Kondo A, Matsushima T, Takeshita I (1987) Epithelial properties of pleomorphic xanthoastrocytomas determined in ultrastructural and immunohistochemical studies. Acta Neuropathol 74:142–150. [DOI] [PubMed] [Google Scholar]
- 7. Kepes JJ (1993) Pleomorphic xanthoastrocytoma: the birth of a diagnosis and a concept. Brain Pathol 3:269–274. [DOI] [PubMed] [Google Scholar]
- 8. Kleinschmidt‐Demasters BK, Aisner DL, Birks DK, Foreman NK (2013) Epithelioid GBMs show a high percentage of BRAF V600E mutation. Am J Surg Pathol 37:685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kleinschmidt‐DeMasters BK, Alassiri AH, Birks DK, Newell KL, Moore W, Lillehei KO (2010) Epithelioid versus rhabdoid glioblastomas are distinguished by monosomy 22 and immunohistochemical expression of INI‐1 but not claudin 6. Am J Surg Pathol 34:341–354. [DOI] [PubMed] [Google Scholar]
- 10. Korshunov A, Ryzhova M, Hovestadt V, Bender S, Sturm D, Capper D et al (2015) Integrated analysis of pediatric glioblastoma reveals a subset of biologically favorable tumors with associated molecular prognostic markers. Acta Neuropathol 129:669–678. [DOI] [PubMed] [Google Scholar]
- 11. Lath R, Unosson D, Blumbergs P, Stahl J, Brophy BP (2003) Rhabdoid glioblastoma: a case report. J Clin Neurosci 10:325–328. [DOI] [PubMed] [Google Scholar]
- 12. Lee EQ, Ruland S, LeBoeuf NR, Wen PY, Santagata S (2014) Successful treatment of a progressive BRAF V600E‐mutated anaplastic pleomorphic xanthoastrocytoma with vemurafenib monotherapy. J Clin Oncol 32. doi: 10.1200/JCO.2013.51.1766. [DOI] [PubMed] [Google Scholar]
- 13. Letouze E, Rosati R, Komechen H, Doghman M, Marisa L, Fluck C et al (2012) SNP array profiling of childhood adrenocortical tumors reveals distinct pathways of tumorigenesis and highlights candidate driver genes. J Clin Endocrinol Metab 97:E1284–E1293. [DOI] [PubMed] [Google Scholar]
- 14. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK (2007) WHO Classification of Tumours of the Central Nervous System, 4th edn. IARC: Lyon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marton E, Feletti A, Orvieto E, Longatti P (2007) Malignant progression in pleomorphic xanthoastrocytoma: personal experience and review of the literature. J Neurol Sci 252:144–153. [DOI] [PubMed] [Google Scholar]
- 16. Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I et al (2012) Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 483:589–593. [DOI] [PubMed] [Google Scholar]
- 17. Momota H, Iwami K, Fujii M, Motomura K, Natsume A, Ogino J et al (2011) Rhabdoid glioblastoma in a child: case report and literature review. Brain Tumor Pathol 28:65–70. [DOI] [PubMed] [Google Scholar]
- 18. Nagayama K, Kohno T, Sato M, Arai Y, Minna JD, Yokota J (2007) Homozygous deletion scanning of the lung cancer genome at a 100‐kb resolution. Genes Chromosomes Cancer 46:1000–1010. [DOI] [PubMed] [Google Scholar]
- 19. Nobusawa S, Hirato J, Kurihara H, Ogawa A, Okura N, Nagaishi M et al (2014) Intratumoral heterogeneity of genomic imbalance in a case of epithelioid glioblastoma with BRAF V600E mutation. Brain Pathol 24:239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reis GF, Pekmezci M, Hansen HM, Rice T, Marshall RE, Molinaro AM et al (2015) CDKN2A loss is associated with shortened overall survival in lower‐grade (World Health Organization Grades II–III) astrocytomas. J Neuropathol Exp Neurol 74:442–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robinson GW, Orr BA, Gajjar A (2014) Complete clinical regression of a BRAF V600E‐mutant pediatric glioblastoma multiforme after BRAF inhibitor therapy. BMC Cancer 14:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rodriguez FJ, Scheithauer BW, Giannini C, Bryant SC, Jenkins RB (2008) Epithelial and pseudoepithelial differentiation in glioblastoma and gliosarcoma: a comparative morphologic and molecular genetic study. Cancer 113:2779–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rosenblum MK, Erlandson RA, Budzilovich GN (1991) The lipid‐rich epithelioid glioblastoma. Am J Surg Pathol 15:925–934. [DOI] [PubMed] [Google Scholar]
- 24. Sturm D, Witt H, Hovestadt V, Khuong‐Quang DA, Jones DT, Konermann C et al (2012) Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22:425–437. [DOI] [PubMed] [Google Scholar]
- 25. Takami H, Yoshida A, Fukushima S, Arita H, Matsushita Y, Nakamura T et al (2015) Revisiting TP53 mutations and immunohistochemistry—a comparative study in 157 diffuse gliomas. Brain Pathol 25:256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tanaka S, Nakada M, Nobusawa S, Suzuki SO, Sabit H, Miyashita K, Hayashi Y (2014) Epithelioid glioblastoma arising from pleomorphic xanthoastrocytoma with the BRAF V600E mutation. Brain Tumor Pathol 31:172–176. [DOI] [PubMed] [Google Scholar]
- 27. Wyatt‐Ashmead J, Kleinschmidt‐DeMasters BK, Hill DA, Mierau GW, McGavran L, Thompson SJ, Foreman NK (2001) Rhabdoid glioblastoma. Clin Neuropathol 20:248–255. [PubMed] [Google Scholar]


