Abstract
Recent studies imply the importance of rapid and reliable diagnostic assessment of 1p/19q status in oligodendroglial tumors. To date, fluorescence in situ hybridization (FISH) is the most commonly applied technique. FISH, however, has several technical shortcomings that are suboptimal for diagnostic applications: results must be viewed in a fluorescence microscope, results are usually evaluated by a single investigator only, and signal fading excludes physical archiving. Also, in gliomas, the distinction of diffusely infiltrating tumor cells from reactively altered normal tissue may be challenging in fluorescence microscopy. Dual‐color chromogenic in situ hybridization (CISH) has started to replace FISH in some diagnostic tests performed in pathology. Here, we present the first single institute experience with a side‐by‐side analysis of 1p/19q FISH and CISH in a series of 42 consecutive gliomas. FISH and CISH produced identical results for 1p and 19q in 93% of cases (n = 39/42). Discrepant results were reevaluated by repeated FISH and a polymerase chain reaction (PCR)‐based microsatellite marker analysis for loss of heterozygosity. Reevaluation confirmed CISH data in all three cases. We conclude that CISH is a reliable alternative in 1p/19q testing in paraffin‐embedded tissues likely to be more sensitive to detect 1p/19q status than FISH analysis.
Keywords: chromogenic in situ hybridization, chromosome 19q, chromosome 1p, fluorescence in situ hybridization, oligodendroglioma
Introduction
There is abundant evidence that concurrent loss of both the complete 1p and 19q chromosome arms is a specific molecular marker for oligodendroglial tumors 27, 33, 36, 38, 45. It has been shown that a balanced whole‐arm translocation between chromosomes 1 and 19 leads to the formation of two derivative chromosomes, one composed of 1q and 19p, the other of 1p and 19q. Subsequent loss of the der 1, 19(p10;q10) then results in the simultaneous 1p and 19q loss observed in oligodendroglioma with retention of the der 1, 19(q10;p10) seen in these cases 16, 20, 39. Clinically, multiple studies provided evidence that oligodendrogliomas featuring combined loss of 1p/19q are associated with a more favorable prognosis due to less aggressive tumor behavior and better responsiveness to therapy than infiltrating astrocytomas of comparable grade 9, 13, 15, 30, 40, 41. Therefore, 1p/19q testing has become an essential diagnostic tool in clinical neuropathology. It aims at the identification of oligodendroglial tumors with 1p/19q loss prior to therapy and helps substantiate the diagnosis in gliomas lacking unequivocal features of oligodendroglioma. In the past, multiple different techniques have successfully been used for the detection of 1p/19q loss, that is, polymerase chain reaction (PCR)‐based microsatellite analysis 8, 18, 25, 26, 47, comparative genomic hybridization (CGH) 6, 7, 8, 19, 23, 24, 28, 32, 41, multiplex ligation‐ dependent probe amplification (MLPA) 11, 14, 21, 22, real‐time comparative quantitative PCR 10, 35, 47 and fluorescence in situ hybridization (FISH) 8, 25. From these techniques, FISH has become by far the most frequently used technique in clinical neuropathology as it allows for an on‐slide analysis of paraffin‐embedded tissue, partially preserves tumor morphology, and does not require leukocyte or normal tissue‐derived DNA for reference as does the PCR‐based microsatellite analysis. However, FISH analysis also has some major practical disadvantages, some of which make routine 1p/19q testing especially in small, less generously fitted neuropathology departments impossible. 1p/19q FISH is expensive. It necessitates a fluorescent microscope. This microscope has to be fitted with suitable filters to visualize the fluorescence signal for each locus specific fluorescent probe. For documentation and archival storage, the microscope has to be fitted with a digital camera that is incorporated into special software allowing multicolor‐overlay imaging. Because of its dependence on a darkened surrounding, FISH analysis is sometimes tedious and time‐consuming work. Therefore, hybridization results are for the most part evaluated by a single investigator only. Fluorescent signals tend to fade quickly when kept at room temperature or exposed to daylight. This excludes physical storage. Limited local digital storage capacities additionally hamper a hard‐copy archive of all hybridization results. Even more problematic is the fact that tissue preservation in FISH is low or it may be difficult and impossible to distinguish diffusely infiltrating tumor cells from reactively altered normal tissue. This is especially challenging in low‐grade lesions with only small diffusely infiltrating tumor areas. Chromogenic in situ hybridization (CISH) has become an attractive alternative to FISH due to its permanent stain, which is more familiar to pathologists, and because it can be viewed using light microscopy. Also, CISH allows simultaneous multi‐investigator evaluation of hybridization results on multiheaded microscopes that are available in most diagnostic institutions. Here, we evaluate the implementation of a novel CISH test for the analysis of 1p/19q status in paraffin‐embedded tissues. To this end, we investigated a total of 42 paraffin‐embedded gliomas with previously FISH‐established 1p/19q status implementing a novel CISH assay.
Materials and Methods
Tissue samples
Formalin fixed paraffin‐embedded (FFPE) tissue of 42 consecutive brain tumor biopsies with previously established 1p/19q status by FISH was available for a comparative analysis of CISH. All tumors were classified and graded according to the guidelines of the World Health Organization 31. In detail, we investigated 10 oligodendrogliomas WHO grade II (OII), 6 anaplastic oligodendrogliomas WHO grade III (OIII), 6 oligoastrocytomas WHO grade II (OAII), 1 anaplastic oligoastrocytoma WHO grade III (OAIII), 6 astrocytomas WHO grade II (AII), 10 anaplastic astrocytomas WHO grade III (AIII), 1 pilocytic astrocytoma WHO grade I (PAI), 1 ependymoma WHO grade II (EII) and 1 glioblastoma WHO grade IV (GBM). FISH analysis of 1p/19q was initiated in all cases during diagnostic work‐up and based on morphological features resembling oligodendroglioma.
FISH
FISH analysis was performed on FFPE tissue. Tissue was cut at 5 μm. First and last sections of each analysis were stained with hematoxylin and eosin (H&E) to delineate regions representing tumor and normal tissue. Tissue preparation for dual‐probe hybridization was facilitated by ZytoLight FISH‐tissue implementation kit according to manufacturers' instructions (ZytoVision, Bremerhaven, Germany). Dual‐color probes ZytoLight SPEC 1p36/1q25 and ZytoLight SPEC 19q13/19p13 were used for locus‐specific 1p and 19q analysis, respectively, following manufacturers' instructions (ZytoVision). Nuclei were counterstained with 4,6‐diamidino‐2 phenylindole (DAPI).
An Olympus BX50 microscope equipped with single‐pass filters for ZyOrange, ZyGreen, and DAPI (ZytoVision), and a dual‐pass filter for ZyOrange/ZyGreen (ZytoVision) were used to assess the number of FISH signals for each locus‐specific fluorescent probe. Individual FISH status for 1p/19q was established following the recommendation of the Research Committee of the European Confederation of Neuropathological Societies (Euro‐CNS) 46. In short, target‐to‐control signal ratios were evaluated for 100 individual, nonoverlapping nuclei. The cut‐off of nuclei that had to show deletion was set at 50% according to the examination guidelines of the International Society of Paediatric Oncology (SIOP) Europe Neuroblastoma Pathology and Biology and Bone Marrow Group 1 cited in 46.
CISH
Paraffin‐embedded tissue was cut as described for FISH analysis above. Tissue preparation for dual‐color in situ hybridization was done using the ZytoDot 2C CISH Implementation Kit following manufacturers' instructions. For 1p analysis, we implemented the dinitrophenyl (DNP)‐labeled 1p36‐target probe and the digoxigenin (DIG)‐labeled 1q25 reference probe. Accordingly, DNP‐labeled 19q13 target probe and DIG‐labeled 19p13‐reference probe were used for 19q testing. DNP‐labeled target probes result in light‐red signals visualized by AP‐Red Solution. DIG‐labeled reference probes result in dark‐green signals visualized by HRP‐Green Solution. Implementation kit, dual‐color probes for 1p and 19q, AP‐Red Solution and HRP‐Green Solution were provided by ZytoVision. An Olympus BX50 microscope was used to assess colored signals for each individual probe. The individual CISH status for 1p/19q was established as outlined above for the FISH analysis.
Microsatellite analysis for loss of heterozygosity (LOH) on 1p and 19q
In cases of data discrepancy between FISH and CISH, microsatellite analysis was implemented to verify 1p/19q status by an independent molecular method. DNA extracted from leukocytes was available for reference in all cases that needed microsatellite analysis. To identify LOH on 1p we used the following tetranucleotide microsatellite markers: D1S1608 (1p36.31), D1S548 (1p36.23), D1S1597 (1p36.21), D1S1592 (1p36.13) and D1S1161 (1p35.1). For determining LOH on 19q, the tetranucleotide markers D19S431 (19q12), D19S433 (19q12), D19S559 (19q13.2) and D19S601 (19q13.33) were implemented. Primer sequences and PCR conditions were described elsewhere 17. Amplified DNA was separated on 8% denaturizing urea gels and visualized by silver staining. LOH was scored as previously described 44.
Results
FISH
In our series, oligodendrogliomas revealed combined 1p/19q loss in 81% (n = 13/16) with 80% (n = 8/10) in OII and 83% (n = 5/6) in OIII, respectively. For one of the six OIII, the 1p/19q status was not safely determinable due to unbalanced signal counts. Oligoastrocytomas featured combined 1p/19q loss in 86% (n = 6/7) with 83% (n = 5/6) in OAII and 100% (n = 1/1) in OAIII. None of the investigated AII (n = 6) or AIII (n = 10) harbored a combined 1p/19q loss (n = 0/16). One AIII featured an isolated 1p loss (10%, 1/10). One case each of PAI, EII and GBM IV was included in the series and all demonstrated retained 1p/19q status. Table 1 provides detailed information regarding individual FISH data of the investigated gliomas.
Table 1.
Clinical and molecular data of all investigated gliomas. Abbreviations: CISH = chromogenic in situ hybridization; FISH = fluorescence in situ hybridization
Table 2.
Reevaluation of gliomas with discrepant FISH/CISH data. Abbreviations: CISH = chromogenic in situ hybridization; FISH = fluorescence in situ hybridization; LOH = loss of heterozygosity; RET = retained copy numbers; DEL = deleted, loss of copy number; ND = data were not suitable to unequivocally decide on 1p/19q status in this glioma as data suggested an unbalanced tetraploidy
| ID | Age | Sex | Tissue | 1p | 19q | Urea gel | FISH R | Figure | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FISH | CISH | FISH | CISH | 1p | 19q | 1p | 19q | |||||
| 56140 | 21 | m | O II | RET | DEL | RET | DEL | LOH | LOH | DEL | DEL | Figure 2 |
| 56604 | 66 | f | O III | ND | DEL | ND | DEL | – | – | DEL | DEL | Figure 1 |
| 56134 | 38 | f | A III | RET | DEL | RET | DEL | – | – | DEL | DEL | – |
Reevaluation by repeated FISH analysis confirmed CISH data in all three gliomas with initially discrepant FISH/CISH results. Of note, CISH and repeated FISH results could additionally be confirmed for case ID56140 by independent polymerase chain reaction (PCR)‐based microsatellite loss of heterozygosity analysis. Representative CISH and validation data are illustrated in Figures 1 and 2 as indicated in the table.
CISH
In a second step, all 42 glioma specimens were investigated by CISH. To ensure an unbiased analysis the investigators were blinded to the FISH results.
In the group of oligodendrogliomas, CISH revealed combined 1p/19q loss in 94% (n = 15/16) with 90% (n = 9/10) in OII and 100% (n = 6/6) in OIII, respectively. As by FISH, CISH demonstrated combined 1p/19q loss in 86% of all oligoastrocytomas (n = 6/7), with 83% (n = 5/6) in OAII and 100% (n = 1/1) in OAIII. One of the investigated astrocytomas (n = 1/16, 6%) revealed combined 1p/19q loss by CISH. CISH confirmed the isolated 1p loss detected by FISH in one AIII (n = 1/10, 10%). The single PAI, EII and GBM IV all revealed retained copies of 1p and 19q. For details, see Table 1.
FISH/CISH data side‐by‐side comparison
FISH and CISH produced identical results for 1p and 19q in 93% of cases (n = 39/42). Discrepant results affected tumor ID56140 (OII), ID56604 (OIII) and ID56134 (AIII). While FISH analysis was interpreted as retained 1p/19q for ID56140 (OII) and ID56134 (AIII), CISH demonstrated combined 1p/19q loss in both cases. In ID56604 (OIII), 1p/19q status was not determinable by FISH but revealed combined 1p/19q loss by CISH. For details, see Table 1.
Validation of discrepant CISH data by repeated FISH analysis and PCR‐based microsatellite analysis
In an effort to clarify the 1p/19q status in the three discrepant tumors, FISH was repeated. For one of the three tumors (ID56140), leukocyte DNA was available and an additional PCR‐ based microsatellite analysis for LOH was performed. The initial FISH analysis of tumor ID56604 (OIII) was interpreted as unbalanced tetraploidy for 1p and 19q chromosomes. In detail, for 1p36 the majority of tumor cells revealed three signals, three to four signals were detected for the reference probe 1q25. Two signals were seen for the 19q13 target probe in the majority of tumor cells, while again three to four signals were seen for the reference probe 19q13. As the tumor featured a clear oligodendroglial morphology, the investigator discussed the possibility of a reduplicated hemizygous deletion and suggested a MLPA analysis to substantiate this theory. While CISH analysis confirmed the FISH results in a significant part of the tumor, it also identified a small tumor area without signs of tetraploidy and clear 1p/19q loss (Figure 1). In an effort to confirm the CISH results, we repeated the FISH analysis focused on both the tumor in general and the tumor area identified by CISH with the clear 1p/19q loss. FISH of the CISH‐pre‐identified tumor area confirmed combined 1p/19q loss, whereas in other parts of the tumor it showed signal numbers reported for the initial FISH analysis (Figure 1). Repeated FISH analysis revealed combined loss of 1p and 19q in the other two discrepant tumors (ID56140 and ID56134). In line with this, microsatellite analysis also indicated a LOH 1p19q. Figure 2 summarizes CISH, repeated FISH and microsatellite analysis for tumor ID56140 (OII). Following thorough reevaluation of the tumors with discrepant FISH results the rate of identical FISH/CISH data changed from 93% (n = 39/42) prior to validation to 100% (n = 42/42) following data validation.
Figure 1.
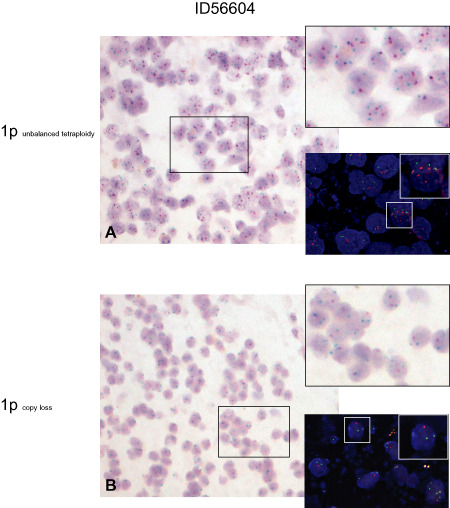
Chromogenic in situ hybridization (CISH) and repeated fluorescence in situ hybridization (FISH) analysis in tumor ID56604 (OIII). Upper panel: overview of a tumor area suggestive for an unbalanced tetraploidy by CISH analysis (A), magnification ×200. Insets to the right: CISH analysis (upper panel) and FISH analysis (lower panel) identify a high number of tumor cells harboring three signals for the target probe 1p36 (red in both FISH and CISH) and three to four signals for the control probe 1q25 (green in both FISH and CISH). Magnification ×400, inset in FISH analysis ×1000. Lower panel: overview of a CISH‐identified tumor area with clear evidence of 1p loss (B), magnification ×200. Insets to the right: CISH analysis (upper panel) and FISH analysis (lower panel) in the CISH‐preselected tumor area confirm 1p loss in the majority of tumor cells. Tumor cells show two signals for reference probe 1q25 and one signal for the target probe 1p36 in both the FISH and CISH analysis. Magnification ×400, inset in FISH analysis ×1000.
Figure 2.
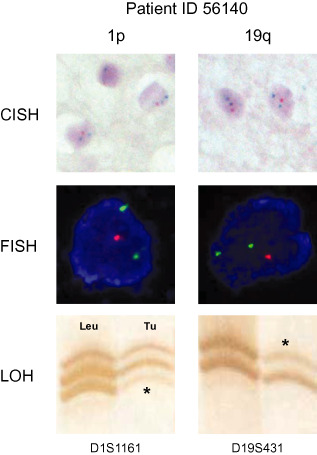
Validation of chromogenic in situ hybridization (CISH) results by repeated fluorescence in situ hybridization (FISH) and loss of heterozygosity (LOH) analysis in tumor ID56140 (OII). Upper panel: representative high‐magnification illustrations of CISH analysis for 1p (left) and 19q (right). Majority of tumor cells harbor one signal of the target probes on 1p and 19q (red signals) and two signals for the reference probes 1q and 19p (green signals). Magnification ×1000. Middle panel: high‐magnification illustrations of repeated FISH analysis in single tumor cells in CISH‐preselected tumor area. Of note, as in CISH analysis, tumor cells harbor one signal for target probes on 1p and 19q (red) and two signals for reference probes 1q and 19p (green) confirming CISH analysis data. Magnification ×1000. Lower panel: polymerase chain reaction (PCR)‐based microsatellite loss of heterozygosity analysis for 1p (left) and 19q (right) confirms CISH and FISH data of loss of chromosomal material on 1p and 19q. Microsatellite markers D1S1161 and D19S431 amplify a polymorphic chromosomal DNA fragment within the commonly deleted target zone on 1p and 19q. Leu: patient's leukocyte DNA serves as indicator of heterozygosity for the investigated microsatellite marker. Tu: patient's tumor DNA with evidence for loss of one allele on 1p and 19q (*) illustrated by a significant decrease in allele signal intensity. The second allele remains detectable at lower intensity and is not completely lost due to the presence of non‐tumorous cells within the biopsy (leukocytes, astrocytes, microglia, neurons) that are not affected by 1p/19q loss.
Discussion
The present study investigated the feasibility of a novel CISH‐based 1p/19q assay in a large series of gliomas that had been previously tested for 1p/19q by FISH in a diagnostic setting. For the vast majority of the gliomas tested, FISH and CISH generated identical results (39/42, 93%). This high concordance rate does not surprise as the locus‐specific CISH probes for 1p and 19q were deliberately designed to hybridize to nearly identical chromosomal sites as the commercially available FISH probes (Figure 3). Identical locus‐specific hybridization probes exclude the possibility of discrepant test results due to different hybridization sites. This should facilitate exchange of both assays. The fact that validation of discrepant findings by repeated FISH analysis and independent molecular testing for 1p/19q loss in our series substantiated the CISH results in all three cases suggests a superiority of CISH‐based testing in almost 10% of cases. This superiority likely roots in the main practical advantage of CISH—data analysis implementing the light microscope. The investigator obtains all visual information in one picture and can better appreciate the tumor morphology and easily and quickly identify areas of special interest, that is, areas with clear oligodendroglial morphology in a mixed glioma or solid tumor areas in low‐grade gliomas. Alternatively, one could argue that a sampling error, that is, using a different tissue block for analysis or using slides cut deeper into the tissue block, may account for the varying results. However, to avoid this issue it was made sure that the tissue slides used for FISH and CISH were cut from the same tissue block for all cases. Also, none of the tissue blocks had been recut for other purposes between the FISH and CISH analyses. Thus, FISH and CISH tissue slides were separated only by a few micrometers necessary to recut the tissue block. Also, H&E‐stained slides prior to FISH and CISH analysis did not show significant differences in tumor morphology or content. The second major advantage of CISH is the permanent color stain. Dual colors visualized under the light microscope are more familiar to histopathologists and a permanent stain allows archiving of test results without the need for time‐ and computational space‐occupying digital fixation of test results in clinical practice. Also, as CISH results can be viewed under the light microscope, simultaneous multi‐investigator evaluation of hybridization results is possible. As CISH does not require access to a fluorescence microscope equipped with hardware and software for FISH analysis, it is far more cost‐effective and will make 1p/19q testing available also to less generously equipped neuropathological institutions in the near future. It is these advantages coupled with high concordance rates that have led to the introduction of CISH assays in a continuously increasing number of tests in pathology that had formally been done by FISH. The most prominent among these is the evaluation of the HER2 status in breast cancer 2, 3, 4, 5, 34, 37. But to investigate chromosomal aberrations in pancreatic carcinoma, CISH also yielded similar results as the concurrently performed FISH analyses 12, 42 and proofed a useful tool even in small samples including tissue microarrays 43. Also, the detection of gene rearrangements in Ewing sarcoma by dual‐color break‐apart CISH is possible and a common practice in pathology 29.
Figure 3.
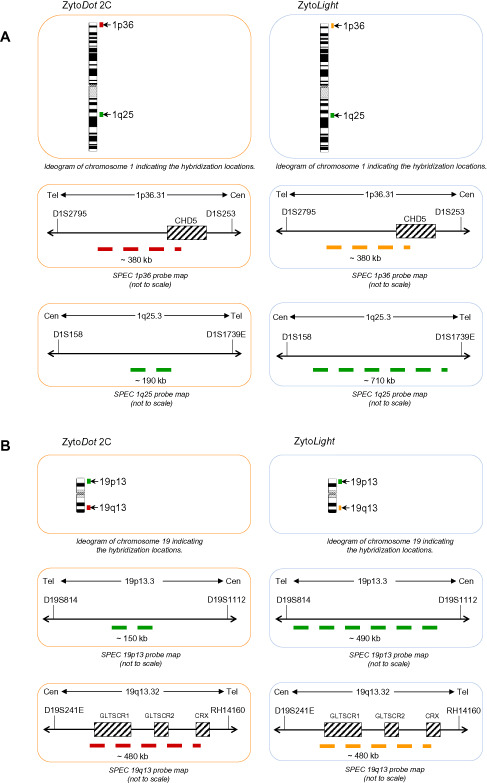
Comparison of locus‐specific CytoDot2C and CytoLight probes by Zytovision for 1p (A) and 19q (B). The target regions are depicted in the colors displayed by the respective detection systems. A. The probes for 1p36 deletion (red/orange) map to the smallest region of consistent deletion (SRD) and cover identical regions in chromogenic in situ hybridization (CISH) and fluorescence in situ hybridization (FISH). The reference probes target the same region in 1q25.3 with the CISH probe covering a shorter sequence localized in the centre of the FISH probe target sequence. B. The probes for 19q13 deletion (red/orange) map to the region of common deletion in gliomas at 19q13.32 and cover identical regions in CISH and FISH. The reference probes target the same region in 19p13.3 with the CISH probe covering a shorter sequence localized in the centre of the FISH probe target sequence.
In conclusion, the present study identified a novel CISH‐based assay as a promising and suitable alternative for FISH to analyze the 1p/19q status in gliomas. Our results indicate that 1p/19q CISH is a safe and easy to handle technique that is likely more sensitive to detect 1p19q status than FISH analysis.
Acknowledgments
We would like to thank Diana Rieker and Tanja Mandelkow for excellent technical assistance.
References
- 1. Ambros PF, Ambros IM (2001) Pathology and biology guidelines for resectable and unresectable neuroblastic tumors and bone marrow examination guidelines. Med Pediatr Oncol 37:492–504. [DOI] [PubMed] [Google Scholar]
- 2. Arnould L, Roger P, Macgrogan G, Chenard MP, Balaton A, Beauclair S, Penault‐Llorca F (2012) Accuracy of HER2 status determination on breast core‐needle biopsies (immunohistochemistry, FISH, CISH and SISH vs FISH). Mod Pathol 25:675–682. [DOI] [PubMed] [Google Scholar]
- 3. Asif M, Khadim MT, Mushtaq S, Mamoon N, Akhtar F, Ali Z (2011) Determination of her‐2/neu by chromogenic in situ hybridization on borderline (2+) immunohistochemistry cases in carcinoma breast. Asian Pac J Cancer Prev 12:211–214. [PubMed] [Google Scholar]
- 4. Bartlett JM, Campbell FM, Ibrahim M, O'Grady A, Kay E, Faulkes C et al (2011) A UK NEQAS ISH multicenter ring study using the Ventana HER2 dual‐color ISH assay. Am J Clin Pathol 135:157–162. [DOI] [PubMed] [Google Scholar]
- 5. Bernet L, Martinez Benaclocha M, Castera C, Cano Munoz R, Sevilla F, Alba J et al (2012) mRNA in situ hybridization (HistoSonda): a new diagnostic tool for HER2‐status in breast cancer—A multicentric Spanish study. Diagn Mol Pathol 21:84–92. [DOI] [PubMed] [Google Scholar]
- 6. Bigner SH, Matthews MR, Rasheed BK, Wiltshire RN, Friedman HS, Friedman AH et al (1999) Molecular genetic aspects of oligodendrogliomas including analysis by comparative genomic hybridization. Am J Pathol 155:375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bourdon V, Plessis G, Chapon F, Guarnieri J, Derlon JM, Jonveaux P (2004) Chromosome imbalances in oligodendroglial tumors detected by comparative genomic hybridization. Ann Genet 47:105–111. [DOI] [PubMed] [Google Scholar]
- 8. Burger PC, Minn AY, Smith JS, Borell TJ, Jedlicka AE, Huntley BK et al (2001) Losses of chromosomal arms 1p and 19q in the diagnosis of oligodendroglioma. A study of paraffin‐embedded sections. Mod Pathol 14:842–853. [DOI] [PubMed] [Google Scholar]
- 9. Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR et al (1998) Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst 90:1473–1479. [DOI] [PubMed] [Google Scholar]
- 10. Chaturbedi A, Yu L, Linskey ME, Zhou YH (2012) Detection of 1p19q deletion by real‐time comparative quantitative PCR. Biomark Insights 7:9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Claes A, Schuuring J, Boots‐Sprenger S, Hendriks‐Cornelissen S, van der Dekkers M, Kogel AJ et al (2008) Phenotypic and genotypic characterization of orthotopic human glioma models and its relevance for the study of anti‐glioma therapy. Brain Pathol 18:423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elliott K, Hamilton PW, Maxwell P (2008) Fluorescence (FISH) and chromogenic (CISH) in situ hybridisation in prostate carcinoma cell lines: comparison and use of virtual microscopy. Br J Biomed Sci 65:167–171. [DOI] [PubMed] [Google Scholar]
- 13. Felsberg J, Erkwoh A, Sabel MC, Kirsch L, Fimmers R, Blaschke B et al (2004) Oligodendroglial tumors: refinement of candidate regions on chromosome arm 1p and correlation of 1p/19q status with survival. Brain Pathol 14:121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Franco‐Hernandez C, de Martinez‐Glez V, Campos JM, Isla A, Vaquero J, Gutierrez M et al (2009) Allelic status of 1p and 19q in oligodendrogliomas and glioblastomas: multiplex ligation‐dependent probe amplification versus loss of heterozygosity. Cancer Genet Cytogenet 190:93–96. [DOI] [PubMed] [Google Scholar]
- 15. Giannini C, Burger PC, Berkey BA, Cairncross JG, Jenkins RB, Mehta M et al (2008) Anaplastic oligodendroglial tumors: refining the correlation among histopathology, 1p 19q deletion and clinical outcome in Intergroup Radiation Therapy Oncology Group Trial 9402. Brain Pathol 18:360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Griffin CA, Burger P, Morsberger L, Yonescu R, Swierczynski S, Weingart JD, Murphy KM (2006) Identification of der(1;19)(q10;p10) in five oligodendrogliomas suggests mechanism of concurrent 1p and 19q loss. J Neuropathol Exp Neurol 65:988–994. [DOI] [PubMed] [Google Scholar]
- 17. Hartmann C, Mueller W, Lass U, von Kamel‐Reid S, Deimling A (2005) Molecular genetic analysis of oligodendroglial tumors. J Neuropathol Exp Neurol 64:10–14. [DOI] [PubMed] [Google Scholar]
- 18. Hatanpaa KJ, Burger PC, Eshleman JR, Murphy KM, Berg KD (2003) Molecular diagnosis of oligodendroglioma in paraffin sections. Lab Invest 83:419–428. [DOI] [PubMed] [Google Scholar]
- 19. Idbaih A, Marie Y, Lucchesi C, Pierron G, Manie E, Raynal V et al (2008) BAC array CGH distinguishes mutually exclusive alterations that define clinicogenetic subtypes of gliomas. Int J Cancer 122:1778–1786. [DOI] [PubMed] [Google Scholar]
- 20. Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M et al (2006) A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res 66:9852–9861. [DOI] [PubMed] [Google Scholar]
- 21. Jeuken J, Cornelissen S, Boots‐Sprenger S, Gijsen S, Wesseling P (2006) Multiplex ligation‐dependent probe amplification: a diagnostic tool for simultaneous identification of different genetic markers in glial tumors. J Mol Diagn 8:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jeuken JW, Sijben A, Bleeker FE, Boots‐Sprenger SH, Rijntjes J, Gijtenbeek JM et al (2011) The nature and timing of specific copy number changes in the course of molecular progression in diffuse gliomas: further elucidation of their genetic “life story”. Brain Pathol 21:308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jeuken JW, Sprenger SH, von Boerman RH, Deimling A, van Teepen HL, Overbeeke JJ, Wesseling P (2001) Subtyping of oligo‐astrocytic tumours by comparative genomic hybridization. J Pathol 194:81–87. [DOI] [PubMed] [Google Scholar]
- 24. Jeuken JW, Sprenger SH, Wesseling P, von Macville MV, Deimling A, Teepen HL et al (1999) Identification of subgroups of high‐grade oligodendroglial tumors by comparative genomic hybridization. J Neuropathol Exp Neurol 58:606–612. [DOI] [PubMed] [Google Scholar]
- 25. Jha P, Sarkar C, Pathak P, Sharma MC, Kale SS, Gupta D et al (2011) Detection of allelic status of 1p and 19q by microsatellite‐based PCR versus FISH: limitations and advantages in application to patient management. Diagn Mol Pathol 20:40–47. [DOI] [PubMed] [Google Scholar]
- 26. Johnson MD, Vnencak‐Jones CL, Toms SA, Moots PM, Weil R (2003) Allelic losses in oligodendroglial and oligodendroglioma‐like neoplasms: analysis using microsatellite repeats and polymerase chain reaction. Arch Pathol Lab Med 127:1573–1579. [DOI] [PubMed] [Google Scholar]
- 27. Kraus JA, Koopmann J, Kaskel P, Maintz D, Brandner S, Schramm J et al (1995) Shared allelic losses on chromosomes 1p and 19q suggest a common origin of oligodendroglioma and oligoastrocytoma. J Neuropathol Exp Neurol 54:91–95. [DOI] [PubMed] [Google Scholar]
- 28. Kros JM, van Run PR, Alers JC, van den Beverloo HB, Bent MJ, van Avezaat CJ, Dekken H (1999) Genetic aberrations in oligodendroglial tumours: an analysis using comparative genomic hybridization (CGH). J Pathol 188:282–288. [DOI] [PubMed] [Google Scholar]
- 29. Kumagai A, Motoi T, Tsuji K, Imamura T, Fukusato T (2010) Detection of SYT and EWS gene rearrangements by dual‐color break‐apart CISH in liquid‐based cytology samples of synovial sarcoma and Ewing sarcoma/primitive neuroectodermal tumor. Am J Clin Pathol 134:323–331. [DOI] [PubMed] [Google Scholar]
- 30. Li S, Yan C, Huang L, Qiu X, Wang Z, Jiang T (2012) Molecular prognostic factors of anaplastic oligodendroglial tumors and its relationship: a single institutional review of 77 patients from China. Neuro Oncol 14:109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Louis DN, Ohgaki H, Wiestler OD, Cavenee KC (2007) WHO Classification of Tumours of the Central Nervous System. International Agency for Research on Cancer (IARC): Lyon. [Google Scholar]
- 32. Magnani I, Moroni RF, Roversi G, Beghini A, Pfundt R, Schoenmakers EF, Larizza L (2005) Identification of oligodendroglioma specific chromosomal copy number changes in the glioblastoma MI‐4 cell line by array‐CGH and FISH analyses. Cancer Genet Cytogenet 161:140–145. [DOI] [PubMed] [Google Scholar]
- 33. Maintz D, Fiedler K, Koopmann J, Rollbrocker B, Nechev S, Lenartz D et al (1997) Molecular genetic evidence for subtypes of oligoastrocytomas. J Neuropathol Exp Neurol 56:1098–1104. [DOI] [PubMed] [Google Scholar]
- 34. Mollerup J, Henriksen U, Muller S, Schonau A (2012) Dual color chromogenic in situ hybridization for determination of HER2 status in breast cancer: a large comparative study to current state of the art fluorescence in situ hybridization. BMC Clin Pathol 12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nigro JM, Takahashi MA, Ginzinger DG, Law M, Passe S, Jenkins RB, Aldape K (2001) Detection of 1p and 19q loss in oligodendroglioma by quantitative microsatellite analysis, a real‐time quantitative polymerase chain reaction assay. Am J Pathol 158:1253–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reifenberger J, Reifenberger G, Liu L, James CD, Wechsler W, Collins VP (1994) Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am J Pathol 145:1175–1190. [PMC free article] [PubMed] [Google Scholar]
- 37. Reisenbichler ES, Horton D, Rasco M, Andea A, Hameed O (2012) Evaluation of dual immunohistochemistry and chromogenic in situ hybridization for HER2 on a single section. Am J Clin Pathol 137:102–110. [DOI] [PubMed] [Google Scholar]
- 38. Ritland SR, Ganju V, Jenkins RB (1995) Region‐specific loss of heterozygosity on chromosome 19 is related to the morphologic type of human glioma. Genes Chromosomes Cancer 12:277–282. [DOI] [PubMed] [Google Scholar]
- 39. Rodriguez FJ, Mota RA, Scheithauer BW, Giannini C, Blair H, New KC et al (2009) Interphase cytogenetics for 1p19q and t(1;19)(q10;p10) may distinguish prognostically relevant subgroups in extraventricular neurocytoma. Brain Pathol 19:623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Scheie D, Meling TR, Cvancarova M, Skullerud K, Mork S, Lote K et al (2011) Prognostic variables in oligodendroglial tumors: a single‐institution study of 95 cases. Neuro Oncol 13:1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smith JS, Alderete B, Minn Y, Borell TJ, Perry A, Mohapatra G et al (1999) Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene 18:4144–4152. [DOI] [PubMed] [Google Scholar]
- 42. Terry J, Barry TS, Horsman DE, Hsu FD, Gown AM, Huntsman DG, Nielsen TO (2005) Fluorescence in situ hybridization for the detection of t(X;18)(p11.2;q11.2) in a synovial sarcoma tissue microarray using a break‐apart‐style probe. Diagn Mol Pathol 14:77–82. [DOI] [PubMed] [Google Scholar]
- 43. Tsiambas E, Karameris A, Stamatelopoulos A, Baltayiannis N, Manaios L, Gerontopoulos K et al (2006) Chromogenic in situ hybridization analysis of chromosomes 7, 9, and 17 in pancreatic ductal adenocarcinoma based on tissue microarrays. J BUON 11:205–211. [PubMed] [Google Scholar]
- 44. von Deimling A, Bender B, Louis DN, Wiestler OD (1993) A rapid and non‐radioactive PCR based assay for the detection of allelic loss in human gliomas. Neuropathol Appl Neurobiol 19:524–529. [DOI] [PubMed] [Google Scholar]
- 45. von Deimling A, von Louis DN, Ammon K, Petersen I, Wiestler OD, Seizinger BR (1992) Evidence for a tumor suppressor gene on chromosome 19q associated with human astrocytomas, oligodendrogliomas, and mixed gliomas. Cancer Res 52:4277–4279. [PubMed] [Google Scholar]
- 46. Woehrer A, Sander P, Haberler C, Kern S, Maier H, Preusser M et al (2011) FISH‐based detection of 1p 19q codeletion in oligodendroglial tumors: procedures and protocols for neuropathological practice—A publication under the auspices of the Research Committee of the European Confederation of Neuropathological Societies (Euro‐CNS). Clin Neuropathol 30:47–55. [DOI] [PubMed] [Google Scholar]
- 47. Zhang SK, Lu DH, Piao YS, Cai YN, Xu QZ (2006) Study of loss of heterozygosity in oligodendroglial tumors by real‐time quantitative polymerase chain reaction‐based microsatellite analysis. Zhonghua Bing Li Xue Za Zhi 35:731–734. [PubMed] [Google Scholar]


