Abstract
HIV-1 has high mutation rates and exists as mutant swarms within the host. Rapid evolution of HIV-1 allows the virus to outpace the host immune system, leading to viral persistence. Approaches to target immutable components are needed to clear HIV-1 infection. Here, we report that the CARD8 inflammasome senses HIV-1 protease activity. HIV-1 can evade CARD8 sensing because its protease remains inactive in infected cells prior to viral budding. Premature intracellular activation of the viral protease triggered CARD8 inflammasome-mediated pyroptosis of HIV-1-infected cells. This strategy led to the clearance of latent HIV-1 in patient CD4+ T cells after viral reactivation. Thus, our study identifies CARD8 as an inflammasome sensor of HIV-1, which holds promise as a strategy for clearance of persistent HIV-1 infection.
The adaptive immune system recognizes cognate epitopes based on amino acid sequences and associated conformations that are subject to mutation. Retroviruses such as human immunodeficiency virus type 1 (HIV-1) accumulate mutations at high rates due to the lack of proofreading activity of their reverse transcriptase coupled with high levels of virus replication in vivo (1). The within-host diversity of HIV-1 allows rapid selection of antibody and T cell escape variants (2-7). In patients who received antiretroviral therapy (ART), HIV-1 persists in a latent form primarily in quiescent CD4+ T cells (8-10) and possibly tissue macrophages (11). Immune escape variants achieved in the latent viral reservoirs present one of the major obstacles to HIV-1 eradication (12-15). In addition, host cells persistently infected with HIV-1 are long-lived and resistant to virus- and immune-mediated apoptotic cell death (16, 17). In this study, we evaluated whether the immune system could (1) sense the functions of an essential HIV-1 protein including those with viral enzymatic activities, which are highly immutable and (2) mediate robust target cell killing independent of apoptosis. The host immune system utilizes germline-encoded pattern-recognition receptors (PRRs) to detect microbial products. A set of intracellular PRRs characterized by the presence of a caspase recruitment domain (CARD) or a pyrin (PYD) domain co-oligomerize with pro-caspase-1 and form high-molecular weight inflammasome complexes upon sensing of their cognate ligands (18). Inflammasome assembly in response to cytoplasmic microbial or danger signals leads to caspase-1 (CASP1) activation and pyroptosis, an inflammatory form of programmed cell death. Murine nucleotide-binding domain leucine-rich repeat pyrin domain–containing 1b (NLRP1b) contains the FIIND and CARD domains. NLRP1b can be activated via direct proteolysis of its N-terminus by Bacillus anthracis lethal factor protease (LF) (19). Briefly, N-terminal cleavage of murine NLRP1b creates a neo-N-terminus, which is ubiquitylated by the N-end-rule pathway and targeted for proteasome degradation (20, 21). Due to the break in the polypeptide chain by FIIND auto-processing, the C-terminal bioactive subunit is liberated from the proteasome complex and can initiate CASP1-dependent inflammasome assembly (22, 23). Recently, two studies on human NLRP1 inflammasome reported that it senses viral infection in epithelial barrier tissues and its activation is also proteasome-dependent (24, 25). There is no evidence that HIV-1 infection can be sensed by innate sensors associated with inflammasome activation in CD4+ T cells, with the exception of the bystander effect triggered by abortive HIV-1 transcripts (26). The bystander cell death mechanism only applies to quiescent, lymphoid-resident CD4+ T cells that are non-permissive to HIV-1. It does not apply to the cells productively or latently infected with HIV-1, and thus has no impact on persistently infected cells. In the present study, we aimed to identify the inflammasome sensor(s) that recognize intracellular HIV-1 activity.
Results
The CARD8 inflammasome senses HIV-1 protease activity
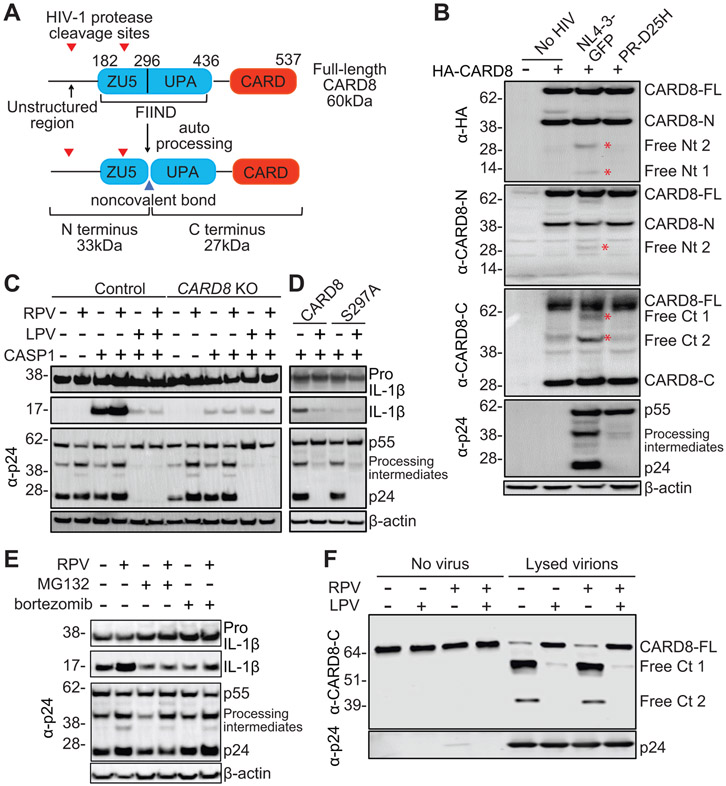
The caspase recruitment domain-containing protein 8 (CARD8) has been implicated in inflammasome activation and pyroptosis of CD4+ T cells and macrophages (27-29). A key question is whether CARD8 is an inflammasome sensor, and if so, which pathogens physiologically activate it. The C-terminus of CARD8 protein contains a “function-to-find” domain (FIIND) followed by a CARD domain. Human CARD8 shares structural similarity with murine NLRP1b and also undergoes autoprocessing (30), as evidenced by the detection of both the full length and the autoprocessed N- and C-terminal fragments by immunoblotting (Fig. 1A). To determine whether CARD8 is a sensor for HIV-1 protease, HEK293T cells were co-transfected with an HIV-1 reporter vector (pNL4-3-GFP) and human CARD8 vector with an N-terminal HA tag (HA-CARD8). The N-terminal tag added 43 amino acids to CARD8 protein. HIV-1 protease activation requires dimerization of the Gag–Pol polyprotein, which occurs during or soon after the viral budding process. Overexpression of HIV-1 Gag–Pol polyprotein in transfected cells resulted in its intracellular dimerization and premature protease activation (31). When the HIV-1 vector was co-transfected with HA-CARD8, two smaller N-terminal fragments were detected by an HA antibody, and one of them was also detected by a CARD8 N-terminus antibody (Fig. 1B). In addition, two neo-C-terminal fragments were detected by a CARD8 C-terminus antibody. These results suggested that the HA-tagged N-terminus was cleaved by HIV-1 protease. The first cleavage site is in the unstructured N-terminus, and the second site is likely in the ZU5 domain. By contrast, CARD8 was not cleaved by the viral vector when the viral protease was inactivated by a single mutation (D25H), as evidenced by the lack of HIV-1 Gag (p55) cleavage. To evaluate whether other viral proteins were required for CARD8 cleavage, we introduced various mutations to the viral vector (fig. S1). Cleavage of CARD8 was not affected when mutations were introduced to any viral genes except pol but was blocked by an HIV-1 protease inhibitor lopinavir (LPV) (fig. S2A). To test whether N-terminal cleavage of CARD8 by HIV-1 protease was sufficient to assemble a functional inflammasome complex, we co-transfected CASP1- and pro-IL1β-expressing plasmids together with pNL4-3-GFP into HEK293T cells. Although the level of endogenous expression of CARD8 in HEK293T cells was not sufficient for the detection of N-terminal cleavage, it was sufficient to trigger the downstream signaling cascade upon HIV-1 protease cleavage, as evidenced by the processing of pro-IL-1β into mature IL-1β (Fig. 1C). Some HIV-1-specific non-nucleoside reverse transcriptase inhibitors (NNRTI) such as rilpivirine (RPV) and efavirenz (EFV), but not nevirapine (NVP), can bind to HIV-1 Pol and enhance intracellular Gag–Pol polyprotein dimerization, which causes premature protease activation (32). As expected, IL-1β processing, as well as HIV-1 Gag processing, was enhanced by RPV but blocked by LPV (Fig. 1C and fig. S2B). By contrast, processing of pro-IL-1β was blocked in CARD8-KO HEK293T cells (Fig. 1C and fig. S2C). The autoproteolytic-deficient HA-CARD8S297A was not able to rescue inflammasome activation in CARD8-KO cells (Fig. 1D), suggesting that proteasome degradation of the N-terminus was required to release the bioactive UPA-CARD fragment. Thus, we added proteasome inhibitors MG132 or bortezomib together with RPV into transfected cells. The baseline level of IL-1β-p17 was not affected by proteasome inhibitors because the inhibitors were added 24 hours post transfection. RPV-induced processing of pro-IL-1β but not HIV-1 Gag cleavage was blocked by bortezomib and MG132 (Fig. 1E), which excluded the possibility that MG132 and bortezomib directly affected HIV-1 protease activity.
Fig. 1. The CARD8 inflammasome senses HIV-1 protease activity.
(A) Domain architecture of the CARD8 protein. CARD8 undergoes autoproteolytic processing in the FIIND domain at position 296, generating the N-terminal ZU5 and C-terminal UPA-CARD fragments that remain associated non-covalently. Two HIV-1 protease cleavage sites are in the non-structured N-terminus and ZU5 domain, respectively. (B) HIV-1 protease cleaves the N-terminus of CARD8. HEK293T cells were co-transfected with HA-CARD8-expressing plasmid together with pNL4-3-GFP or PR-D25H. Cells were collected 24 hours after transfection. Anti-HA, anti-CARD8-N, anti-CARD8-C, and anti-p24 antibodies were used sequentially on the same blot. Cleaved fragments are denoted with red asterisks. (C) HIV-1 protease triggers CARD8 inflammasome activation. Control or CARD8-KO HEK293T cells were co-transfected with plasmids encoding CASP1, pro-IL-1β, and pNL4-3-GFP. RPV and LPV were added immediately after transfection. (D) Autoproteolytic processing is required for HIV-1 protease triggered CARD8 inflammasome activation. CARD8-KO HEK293T cells were co-transfected with plasmids encoding CASP1, pro-IL-1β, and CARD8 or CARD8S297A, together with pNL4-3-GFP. LPV was added immediately after transfection. (E) HIV-1 protease triggered CARD8 inflammasome activation is proteasome-dependent. HEK293T cells were co-transfected with plasmids encoding CASP1, pro-IL-1β, and pNL4-3-GFP. Indicated drugs were added 24 hours after transfection. Cells were collected 6 hours after drug treatment. In B to E, cell lysates were evaluated by immunoblotting. (F) HIV-1 protease cleaves CARD8 in vitro. HA-tagged CARD8 was immunoprecipitated and incubated with lysed HIV-1 particles with the presence of indicated drugs. The eluate was collected for immunoblotting. Full-length CARD8 (CARD8-FL), N terminal CARD8 (CARD8-N), freed N terminus (free Nt), freed C terminus (free Ct). Data are representative of three or more independent experiments.
HIV-1 protease cleaves the N-terminus of CARD8
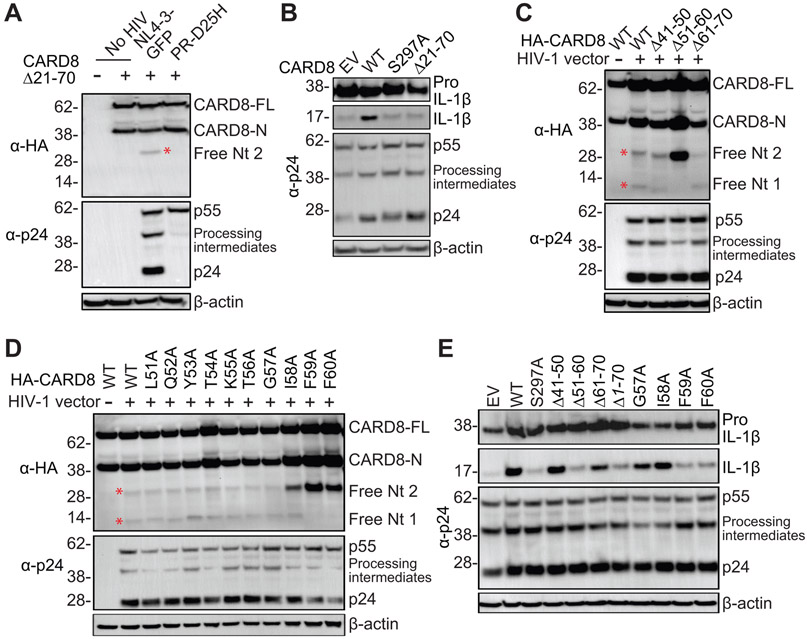
To examine whether CARD8 was directly cleaved by HIV-1 protease, HA-CARD8S297A was immunoprecipitated and incubated with purified and lysed HIV-1 particles. When the viral protease was functional, two additional bands were visualized using a CARD8 C-terminus antibody (Fig. 1F), which confirmed the two cleavage sites recognized by the viral protease. RPV did not further enhance CARD8 cleavage because the lysed HIV-1 particles already contained mature viral protease. To determine whether one or both cleavage sites were needed to activate CARD8, we first generated a truncated CARD8Δ21-70 which did not contain the first cleavage site. CARD8Δ21-70 was cleaved at the second site (Fig. 2A), but was unable to trigger inflammasome activation (Fig. 2B). To identify the first cleavage site, we tested a set of truncated CARD8 proteins and found that CARD8Δ51-60 was not cleaved (Fig. 2C). Next, we were able to determine that F59 and F60 were likely to be at the P1 and P1′ positions, respectively (Fig. 2D). The HIV-1 protease prefers to have large hydrophobic amino acids flanking the scissile bond. Phenylalanine is the most common residue at the P1 position, and its presence at both P1 and P1′ positions improved the cleavage rate of HIV-1 protease (33). We further demonstrated that the first cleavage site was required for the CARD8 inflammasome activation by HIV-1 protease (Fig. 2E). Thus, HIV-1 protease can cleave the N-terminus of CARD8 and that activation of the viral protease can lead to CARD8 inflammasome activation in the HEK293T transfection system.
Fig. 2. HIV-1 protease cleaves the N-terminus of CARD8.
(A) CARD8Δ21-70 is cleaved at the second site. HEK293T cells were transfected with plasmids encoding CARD8 or CARD8Δ21-70, and pNL4-3-GFP or PR-D25H. (B) Cleavage in the ZU5 domain does not activate the CARD8 inflammasome. CARD8-KO HEK293T cells were co-transfected with plasmids encoding CASP1, pro-IL-1β, and WT or mutant CARD8, together with pNL4-3-GFP. (C and D) Mapping of the first cleavage site. HEK293T cells were transfected with plasmids encoding truncated or mutated CARD8, and pNL4-3-GFP. (E) Cleavage in the N-terminus is required to activate the CARD8 inflammasome. CARD8-KO HEK293T cells were co-transfected with plasmids encoding CASP1, pro-IL-1β, and WT or mutant CARD8, together with pNL4-3-GFP. Cell lysates were evaluated by immunoblotting. Data are representative of three or more independent experiments. Empty vector (EV).
HIV-1 triggers CARD8-depedent pyroptosis of infected macrophages
Since CARD8 is expressed in HIV-1 target cells, including primary CD4+ T cells and macrophages (fig. S3), the next question was whether induction of premature intracellular activation of HIV-1 protease could trigger CARD8-dependent pyroptosis of infected cells. When treated with EFV or RPV, HIV-1-infected (GFP+) macrophages rapidly underwent pyroptotic cell death as evidenced by membrane swelling and rupture (Fig. 3, A to D and movie S1 and S2), as well as secretion of IL-1β (Fig. 3E). The cell death triggered by EFV or RPV was rapid and dose-dependent and completely blocked by LPV (Fig. 3, B and C). CASP1 activation was evidenced by detection of its active subunits p10 and p20 (Fig. 3F). In addition, we tested the CASP1-specific inhibitor VX765 and pan-caspase inhibitor Z-VAD-FMK. Although most of the infected macrophages were killed by RPV, blocking of CASP1 activity reduced the loss of infected cells to 20% (Fig. 3G). The N-end-rule pathway mediates proteasome degradation of mouse NLRP1b (34). To test whether it was required for CARD8 activation, we added MG132 and bortezomib or an N-end-rule pathway inhibitor bestatin methyl ester (Me-Bs) together with RPV to HIV-1-infected macrophages. MG132, bortezomib, and Me-Bs effectively blocked RPV-mediated killing of infected macrophages and secretion of IL-1β (Fig. 3, H and I). HIV-1-infected THP-1 cells were also susceptible to NNRTI-triggered killing (Fig. 4, A and B). As chemical inhibitors often have off-target effects, we confirmed that the CARD8 inflammasome was responsible for HIV-1 sensing by generating bulk or single clone CARD8-, ASC-, CASP1- and NLRP3-knockout THP-1 cells (Fig. 4C and fig. S4B). Deletion of CARD8 or CASP1, but not ASC or NLRP3, inhibited HIV-1 protease-mediated pyroptosis and IL-1β secretion (Fig. 4, D and E, and fig. S4, C to G), consistent with the inhibitor experiments. This finding also confirms studies showing that CARD8 can form an ASC-independent inflammasome complex (28, 35).
Fig. 3. HIV-1 protease triggers pyroptosis of infected macrophages upon NNRTI treatment.
(A to E) HIV-1 protease activation by NNRTIs induced rapid pyroptosis of infected monocyte-derived macrophages (MDMs). MDMs were infected with replication-competent HIVNL4-3/BaL. On day 4, RAL and T-20 were added to block new infection. Cells were then treated with RPV, EFV, LPV, or combinations for 24 hours or as indicated. GFP+ cells were detected by flow cytometry. DMSO controls were used to determine cell killing by NNRTIs (A to C). Representative images of infected MDMs were taken at 0 and 3 hours post RPV treatment. Scale bars represent 100 μm (D). Culture supernatant was collected after NNRTI treatment for IL-1β ELISA (E). (F and G) Pyroptosis of HIV-1-infected MDMs is CASP1-dependent. MDMs were infected and treated as described above. Cleavage of pro-CASP1 and cleaved CASP1 (p10 and p20) in infected MDMs after RPV treatment for one hour (F). Infected MDMs were pre-treated with CASP1 inhibitor VX-765 or pan-Caspase inhibitor Z-VAD-FMK for 3 hours then treated with RPV for 4 hours. Flow cytometry analysis was performed to determine cell killing (G). (H and I) HIV-1 protease mediated inflammasome activation is proteasome-dependent. MDMs were infected and treated as described above. Infected MDMs were pretreated with proteasome inhibitors MG132, bortezomib, or Me-Bs for 30 min and then treated with RPV for 4 hours. Cell killing was determined by flow cytometry (H). Culture supernatant from (H) was collected for IL-1β measurement by ELISA (I). In B, G, and H, P-values were calculated using one-way ANOVA and Dunnett's test. In C, E, and I, P-values were calculated using two-way ANOVA and Tukey’s multiple comparison tests. *P<0.05, ****P<0.0001. In each graph, n≥3. Error bars show mean values with SEM. Data are representative of three or more independent experiments.
Fig. 4. The CARD8 inflammasome is required for pyroptosis of HIV-1-infected THP-1 cells.
(A and B) NNRTIs induce death of HIV-1-infected THP-1 cells. THP-1 cells were infected with VSV-G pseudotyped HIV-1 reporter virus NL4-3-GFP. On day 4 post infection, cells were treated with NNRTIs for another 2 days before flow cytometry analysis. (C to E) NNRTI-triggered cell death is CARD8 inflammasome-dependent. Bulk populations of knockout THP-1 cells were used. Knockouts of CARD8, ASC, CASP1 or NLRP3 in THP-1 cells were confirmed by immunoblotting (C). Infected THP-1 cells were pre-treated with LPS (100 ng/ml) for 3 hours before RPV treatment. GFP expression was analyzed by flow cytometry 24 hours post RPV treatment. Data were normalized to the control group (D). Culture supernatant was collected 48 hours post RPV treatment for IL-1β detection (E). In B and D, P-values were calculated using one-way ANOVA and Dunnett's test. In E, P-values were calculated using two-way ANOVA and Tukey’s multiple comparison tests. **P<0.01, ****P<0.0001. In each bar graph, n=3. Error bars show mean values with SEM. Data are representative of three or more independent experiments.
HIV-1 triggers CARD8-depedent pyroptosis of infected CD4+ T cells
Several studies have reported that NNRTIs can induce killing of HIV-1-infected CD4+ T cells through an unknown mechanism (36-38). We hypothesized that the cell killing observed in those studies was due to NNRTI-triggered HIV-1 protease activation which led to CARD8 inflammasome activation. Since resting CD4+ T cells are the most well characterized cellular reservoirs for HIV-1, we examined the expression levels of key components of the CARD8 inflammasome in different subsets of primary CD4+ T cells. CARD8 is expressed in both activated and unstimulated blood CD4+ T cells, as well as in memory and naïve CD4+ T cells in lymphoid tissues (Fig. 5A and fig. S3). Both activated (Fig. 5B and fig. S5, B and C) and unstimulated (fig. S5D) CD4+ T cells were susceptible to HIV-1 protease-triggered cell death when treated with NNRTIs including EFV, RPV, and Etravirine (ETR), but not NVP. Since several HIV-1 proteins can induce death of primary CD4+ T cells, we produced different reporter viruses carrying mutations in vif, vpr, vpu, env, and nef by transfecting HEK293T cells with different viral plasmids (fig. S1). Reporter viruses without a functional protease (ΔGag–Pol and PR-D25H) or deficient in Gag–Pol dimerization (RT-L234A and RT-W401A) were unable to trigger cell death. None of the other viral proteins were required for cell killing (Fig. 5C). CASP1 activation was induced by EFV or RPV but blocked by LPV (Fig. 5, D and E). Both VX765 and Z-VAD-FMK blocked killing of HIV-1-infected CD4+ T cells (Fig. 5F). Similar to infected macrophages, HIV-1 protease triggered CASP1 activation and cell death was also blocked by MG132, bortezomib, and Me-Bs (fig. S6). NNRTI-induced CASP1 activation and pyroptosis of HIV-1-infected primary CD4+ T cells was abrogated when CARD8 was knocked out (Fig. 5, G to I). Similarly, a CASP1 knockout or knockdown in primary CD4+ T cells also conferred resistance to HIV-1 protease-mediated pyroptosis (Fig. 5J and fig. S7). In addition to the pseudotyped reporter virus, we also observed cell killing with a clinical isolate HIVBaL. Since HIVBaL is a replication-competent virus, all classes of antiretroviral drugs blocked viral replication; NNRTIs could further reduce viral infection by clearing cells already infected with HIV-1 (fig. S8, A to C). NNRTI-mediated killing of primary CD4+ T cells infected with HIVBaL was also CARD8-dependent (fig. S8, D to F).
Fig. 5. HIV-1 Protease induces CARD8 inflammasome activation and subsequent cell death in CD4+ T cells.
(A) Analysis of CARD8, caspase 1, and caspase 3 expression levels in unstimulated (U) and activated (A) primary CD4+ T cells by immunoblotting. (B and C) HIV-1 protease activation by NNRTIs leads to killing of infected primary CD4+ T cells. Activated primary CD4+ T cells were infected with HIV-1 reporter virus NL4-3-Pol (B) or different HIV-1 reporter viruses (C) for 3 days before treatment with RPV or indicated NNRTIs. Cells were analyzed for GFP expression by flow cytometry 48 hours post NNRTI treatment. (D and E) HIV-1 protease activation by NNRTIs induced CASP1 activation in infected primary CD4+ T cells. Activated primary CD4+ T cells were infected with the NL4-3-Pol. On day 3 post infection, cells were treated with EFV, RPV, LPV or combinations for 3 hours before staining for active CASP1. (F) Chemical inhibition of caspase-1 blocks NNTRI-induced cell death. Activated primary CD4+ T cells were infected with the NL4-3-Pol. On day 3 post infection, cells were pre-treated with CASP1 inhibitor VX765 or pan-caspase inhibitor Z-VAD-FMK for 3 hours before adding RPV. GFP expression was measured by flow cytometry. (G) Knockout of CARD8 in primary CD4+ T cells was confirmed by immunoblotting. (H to J) The CARD8 inflammasome is required for pyroptosis of HIV-1-infected primary CD4+ T cells. CARD8-, ASC- or CASP1-knockout primary CD4+ T cells were co-stimulated and then infected with NL4-3-Pol. On day 3 post infection, cells were treated with RPV. Staining of active CASP1 3 hours post RPV treatment (H). GFP expression was measure by flow cytometry 24 hours after RPV treatment (I and J). In C, E, I, and J, P-values were calculated using one-way ANOVA and Dunnett's test. In F and H, P-values were calculated using two-way ANOVA and Tukey’s multiple comparison tests. **P<0.01, ***P<0.001, and ****P<0.0001. In each bar graph, n≥3. Error bars show mean values with SEM. Data are representative of five or more independent experiments.
Activation of the CARD8 inflammasome clears latent HIV-1 in patient CD4+ T cells
To determine whether HIV-1 protease function in activating the CARD8 inflammasome is conserved, we tested a panel of HIV-1 virus isolates from chronically infected individuals of subtypes A, B, C, and D (39). Subtype B is the dominant subtype in Europe and North America, whereas A, C, and D are more prevalent worldwide. T-20 and Raltegravir (RAL) were used to completely block new infection but had no killing effect or cellular toxicity (fig. S9). The addition of EFV and RPV but not NVP effectively cleared primary CD4+ T cells infected with all HIV-1 subtypes. Additionally, the killing efficiency did not correlate with viral replication fitness (Fig. 6A and fig. S10), suggesting that the enzymatic activity of HIV-1 protease with regard to CARD8 activation is well conserved across major HIV-1 subtypes. Next, we confirmed that EFV and RPV treatment induced CARD8-dependent caspase-1 activation and pyroptosis of primary CD4+ T cells infected with clinical viral isolates (Fig. 6, B and C, and figs. S11 and 12). To test whether strategies involving targeted activation of the CARD8 inflammasome could be used for the clearance of latent HIV-1, we obtained blood CD4+ T cells from patients under suppressive ART to measure the size of viral reservoirs (40). The control antiretroviral (ARV) combination containing T-20, RAL, and NVP had no killing effect. The median IUPM in control and RPV groups was 2.61 and 0.16, respectively, suggesting a rapid clearance of 93.9% of the latent HIV-1 reservoirs (Fig. 6, D and E). Notably, three out of eight patient samples had no detectable viral replication after RPV treatment. In our “shock and kill” assay, cells were only treated with RPV for the first 2-3 days. It is possible that the residual viruses in the RPV group came from delayed virus reactivation which occurred after removal of RPV.
Fig. 6. Induction of CARD8 inflammasome activation clears infection of clinical HIV-1 isolates.
(A) Killing of primary CD4+ T cells infected with different subtypes of clinical HIV-1 isolates. Activated CD4+ T cells were infected with a panel of international HIV-1 isolates. RAL and T-20 were added with or without EFV, RPV or NVP on day 6 post infection. Viral infection was measured by intracellular p24 staining on day 8 and normalized to the DMSO controls. (B) Killing of infected primary CD4+ T cells is proteasome-dependent. Activated primary CD4+ T cells were infected with different subtypes of clinical HIV-1 isolates. RAL, T-20, and RPV were added with or without bortezomib on day 6 post infection. Viral infection was measured by intracellular p24 staining 6 hours after drug treatment. (C) The CARD8 inflammasome is required for the killing of infected primary CD4+ T cells. Activated CARD8-KO or control primary CD4+ T cells were infected with different subtypes of clinical HIV-1 isolates. RAL, T-20, and RPV were added on day 6 post infection. Viral infection was measured by intracellular p24 staining 24 hours after drug treatment. In A to C, n=3. Error bars show mean values with SEM. (D) Scheme of shock and kill strategy and quantitative viral outgrowth assay (QVOA) using patient CD4+ T cells. (E) Clearance of latent HIV-1 by RPV treatment. Frequency of latent HIV-1 was determined by QVOA. Red: ARV for 2 days and RPV at 2.5 μM. Blue: ARV for 3 days and RPV at 5 μM. Open circle: no detectable HIV-1 infection by p24 ELISA. In B and C, P-values were calculated using multiple t test. In E, P-value was calculated by ratio paired t test. *P<0.01, **P<0.01, ***P<0.001, and ****P<0.001.
Discussion
Due to rapid viral evolution, it is very difficult for the host immune system to control HIV-1 infection and clear residual viral reservoirs without targeting immutable components of the virus. In this study, we found that CARD8 is a sensor for HIV-1 protease activity to trigger inflammasome activation and pyroptosis of infected cells. This work demonstrates that the CARD8 and NLRP1 inflammasomes share similar mechanisms of activation, which involves their N-terminal cleavage by microbial proteases, followed by proteasome-mediated release of the bioactive C-terminal fragment to trigger inflammasome assembly and CASP1 activation. Interestingly, HIV-1 protease cleaves CARD8 at two different sites. Cleavage of the unstructured N-terminus but not the FIIND domain leads to inflammasome activation. We also observed that a deletion (Δ51-60) or mutations (F59A or F60A) of the first cleavage site increased the cleavage efficiency at the second site (Fig. 2C and D), suggesting a competition between the two sites. Cleavage of HIV-1 Gag and Gag–Pol by the viral protease is a sequential process regulated by the rate of cleavage at individual site (41). Since CARD8 activation requires a cleavage within the unstructured N-terminus, the cleavage site preference may influence the CARD8 inflammasome activation. In HIV-1-infected cells, the CARD8 inflammasome cannot detect the virus because the viral protease remains inactive as a subunit of unprocessed viral Gag–Pol polyprotein. Surprisingly, some NNRTIs which have been used to treat HIV-1 infection for more than two decades can facilitate CARD8 sensing by mediating premature intracellular activation of HIV-1 protease. NNRTIs bind to HIV-1 RT and act as enhancers of Gag–Pol dimerization to activate Pol-embedded viral protease (32). Additional investigations are needed to better understand the mechanism of the NNRTI-mediated Gag–Pol dimerization process. Although NNRTI-containing treatment regimens cannot eliminate HIV-1 infection in patients because the viral latent reservoirs are rapidly established prior to treatment initiation, inclusion of NNRTIs without protease inhibitors in the initial ARV regimen may partially reduce the seeding of latent viral reservoirs. In addition, inclusion of NNRTIs in HIV-1 cure strategies should facilitate the elimination of infected cells after viral latency reversal. Intriguingly, CARD8 is preferentially and highly expressed in blood and lymphoid tissues (42) as well as in many hematopoietic-derived cells (27), suggesting that targeting the CARD8 inflammasome may be effective in lymphoid tissues, the most important anatomical sites for persistent HIV-1 infection. Notably, the cell-killing IC50 of EFV and RPV is approximately 1-2 μM (Figs. 2C and 3B), which is about 100-fold higher than the infection-blocking IC50. The plasma EFV concentration in patients receiving EFV-containing regiments (1-4 μg/ml or 3-12 μM) is within the therapeutic range for cell killing. This strategy is unlikely to be effective in tissues with markedly lower drug concentration such as central nervous system. Importantly, HIV-1 Pol that confer resistance to NNRTIs also abrogate NNRTI-triggered cell killing (38) likely because the resistant viral variants can avoid drug binding. Thus, the identification of more potent chemical compounds that promote Gag–Pol dimerization regardless of viral inhibition is warranted. Taken together, this work reveals a mechanism of innate sensing of HIV-1 infection that has immediate implications for HIV-1 cure strategies.
Supplementary Material
Acknowledgements
We thank the volunteers for participating in this study. We thank L. Kessels, M. Klebert, A. Haile, T. Spitz, and T. Minor for study subject recruitment. We thank M. Cella for the tonsil biopsies. We thank M. Diamond and R. Jackson for comments and suggestions on this manuscript. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: rilpivirine, efavirenz, lopinavir, etravirine, nevirapine, maraviroc, T-20, tenofovir, raltegravir, pNL4-3-GFP, international HIV-1 isolates, MOLT-4 CCR5+ cells and HIV-1 p24 antibodies.
Funding: This work was supported by NIH grants R00AI125065 (to L. Shan), AI150470 (S. Kutluay), and 5DP5OD026427 (to C. DeSelm).
Footnotes
Competing interests: the authors declare no competing financial interests.
Data and materials availability: All data is available in the main text or the supplementary materials.
References
- 1.Preston BD, Dougherty JP, Mechanisms of retroviral mutation. Trends in microbiology 4, 16–21 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Phillips RE et al. , Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354, 453–459 (1991). [DOI] [PubMed] [Google Scholar]
- 3.Borrow P et al. , Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nature medicine 3, 205–211 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Wei X et al. , Antibody neutralization and escape by HIV-1. Nature 422, 307–312 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Kwong PD et al. , HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Caskey M et al. , Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522, 487–491 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caskey M et al. , Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nature medicine 23, 185–191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finzi D et al. , Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–1300 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Wong JK et al. , Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278, 1291–1295 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Chun TW et al. , Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 94, 13193–13197 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganor Y et al. , HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nat Microbiol 4, 633–644 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Richman DD et al. , The challenge of finding a cure for HIV infection. Science 323, 1304–1307 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Deng K et al. , Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 517, 381–385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bar KJ et al. , Effect of HIV Antibody VRC01 on Viral Rebound after Treatment Interruption. N Engl J Med 375, 2037–2050 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheid JF et al. , HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 535, 556–560 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang SH et al. , Latent HIV reservoirs exhibit inherent resistance to elimination by CD8+ T cells. J Clin Invest 128, 876–889 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clayton KL et al. , Resistance of HIV-infected macrophages to CD8(+) T lymphocyte-mediated killing drives activation of the immune system. Nat Immunol 19, 475–486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broz P, Dixit VM, Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol 16, 407–420 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Chavarria-Smith J, Vance RE, The NLRP1 inflammasomes. Immunol Rev 265, 22–34 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Sandstrom A et al. , Functional degradation: A mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 364, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chui AJ et al. , N-terminal degradation activates the NLRP1B inflammasome. Science 364, 82–85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finger JN et al. , Autolytic proteolysis within the function to find domain (FIIND) is required for NLRP1 inflammasome activity. J Biol Chem 287, 25030–25037 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frew BC, Joag VR, Mogridge J, Proteolytic processing of Nlrp1b is required for inflammasome activity. PLoS Pathog 8, e1002659 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson KS et al. , Enteroviral 3C protease activates the human NLRP1 inflammasome in airway epithelia. Science 370, (2020). [DOI] [PubMed] [Google Scholar]
- 25.Bauernfried S, Scherr MJ, Pichlmair A, Duderstadt KE, Hornung V, Human NLRP1 is a sensor for double-stranded RNA. Science, (2020). [DOI] [PubMed] [Google Scholar]
- 26.Doitsh G et al. , Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 505, 509–514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson DC et al. , DPP8/DPP9 inhibitor-induced pyroptosis for treatment of acute myeloid leukemia. Nature medicine 24, 1151–1156 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linder A et al. , CARD8 inflammasome activation triggers pyroptosis in human T cells. The EMBO journal 39, e105071 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson DC et al. , DPP8/9 inhibitors activate the CARD8 inflammasome in resting lymphocytes. Cell death & disease 11, 628 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D'Osualdo A et al. , CARD8 and NLRP1 undergo autoproteolytic processing through a ZU5-like domain. PloS one 6, e27396 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park J, Morrow CD, Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. Journal of virology 65, 5111–5117 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Figueiredo A et al. , Potent nonnucleoside reverse transcriptase inhibitors target HIV-1 Gag-Pol. PLoS Pathog 2, e119 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potempa M et al. , HIV-1 Protease Uses Bi-Specific S2/S2' Subsites to Optimize Cleavage of Two Classes of Target Sites. Journal of molecular biology 430, 5182–5195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wickliffe KE, Leppla SH, Moayeri M, Killing of macrophages by anthrax lethal toxin: involvement of the N-end rule pathway. Cell Microbiol 10, 1352–1362 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ball DP et al. , Caspase-1 interdomain linker cleavage is required for pyroptosis. Life Sci Alliance 3, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jochmans D et al. , Selective killing of human immunodeficiency virus infected cells by non-nucleoside reverse transcriptase inhibitor-induced activation of HIV protease. Retrovirology 7, 89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zerbato JM, Tachedjian G, Sluis-Cremer N, Nonnucleoside Reverse Transcriptase Inhibitors Reduce HIV-1 Production from Latently Infected Resting CD4(+) T Cells following Latency Reversal. Antimicrob Agents Chemother 61, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trinite B, Zhang H, Levy DN, NNRTI-induced HIV-1 protease-mediated cytotoxicity induces rapid death of CD4 T cells during productive infection and latency reversal. Retrovirology 16, 17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown BK et al. , Biologic and genetic characterization of a panel of 60 human immunodeficiency virus type 1 isolates, representing clades A, B, C, D, CRF01_AE, and CRF02_AG, for the development and assessment of candidate vaccines. Journal of virology 79, 6089–6101 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siliciano JD, Siliciano RF, Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol 304, 3–15 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Erickson-Viitanen S et al. , Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS research and human retroviruses 5, 577–591 (1989). [DOI] [PubMed] [Google Scholar]
- 42.Fagerberg L et al. , Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Molecular & cellular proteomics : MCP 13, 397–406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, Mateo JL, CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PloS one 10, e0124633 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mochizuki H, Schwartz JP, Tanaka K, Brady RO, Reiser J, High-titer human immunodeficiency virus type 1-based vector systems for gene delivery into nondividing cells. Journal of virology 72, 8873–8883 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Potempa M et al. , A Direct Interaction with RNA Dramatically Enhances the Catalytic Activity of the HIV-1 Protease In Vitro. Journal of molecular biology 427, 2360–2378 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.