Abstract
Type I Interferons (IFN-I) mediate numerous immune interactions during viral infections, from the establishment of an antiviral state to invoking and regulating innate and adaptive immune cells that eliminate infection. While continuous IFN-I signaling plays critical roles in limiting virus replication during both acute and chronic infections, sustained IFN-I signaling also leads to chronic immune activation, inflammation and, consequently, immune exhaustion and dysfunction. Thus, an understanding of the balance between the desirable and deleterious effects of chronic IFN-I signaling will inform our quest for IFN-based therapies for chronic viral infections as well as other chronic diseases, including cancer. As such the factors involved in induction, propagation and regulation of IFN-I signaling, from the initial sensing of viral nucleotides within the cell to regulatory downstream signaling factors and resulting IFN-stimulated genes (ISGs) have received significant research attention. This review summarizes recent work on IFN-I signaling in chronic infections, and provides an update on therapeutic approaches being considered to counter such infections.
Keywords: Interferon, chronic virus infection, immune response, T cell, innate immunity, adaptive immunity, exhaustion, interferon regulatory factor, IRF, interferon stimulatory gene, ISG
Introduction
Viral infections induce an immune response that includes production of a family of pleiotropic cytokines that interfere with virus replication, interferons (IFNs). IFNs are broadly categorized into 3 groups – Type I IFN (IFN-I), IFN-II and the relatively newly identified IFN-III. IFN-I comprises a variety of genes including 13 IFNα and single copies of each of IFNβ, IFNω, IFNε, and IFNκ (1). IFN-II, also referred to as IFNγ, exists as a single copy gene structurally distinct from IFN-I. The IFN-III family, also referred to as lambda IFNs, represents members of the interleukin 10 (IL-10) superfamily, and include IFNλ1 (IL-29), IFNλ2 (IL-28A), IFNλ3 (IL-28B) and IFNλ4 genes. Since their discovery in 1957, IFN-Is have attracted attention, largely owing to their antiviral roles from generating an antiviral state to orchestrating both innate and adaptive immune responses (2). Indeed IFN-Is play important roles in inhibiting virus replication, initiating and enhancing antigen presentation, as well as directly and indirectly acting on T and B cells to trigger adaptive immune responses (3). Even though their functions are yet to be adequately elucidated, the IFN-III family also exerts antiviral activities and immune responses that overlap but at times are distinct from those of IFN-I (4, 5). While the protective roles of IFN-Is render IFN-Is the obvious main targets for antiviral therapies, recent studies have highlighted deleterious immunopathologies centred around the establishment of a dysfunctional immune state as a consequence of chronic IFN-I signaling (3). Chronic viral infections that lead to worsened disease outcomes are associated with a persisting IFN-I signaling signature and sustained inflammation and immune activation (3). This concept was nicely illustrated in monkey models of SIV infection where natural hosts (African green monkeys) that do not progress to AIDS despite high virus titers have a lower IFN-I signaling signature, whereas their AIDS-progressing non-natural hosts (rhesus macaques) had elevated levels of IFN-I signaling signatures despite comparable viral burdens (6–8). Thus, a deeper understanding of the intricacies of IFN-I signaling, from induction, modulation and the resulting gene products is necessary in order to delineate or tease apart the beneficial from the harmful effects of IFN-I signaling. This review, mainly focusing on chronic viral infections, explores the regulation of IFN-I signaling and recent works on the consequences of chronic IFN-I signaling. We then conclude with a discussion on the therapeutic potential of IFN-I.
Induction of IFN signaling
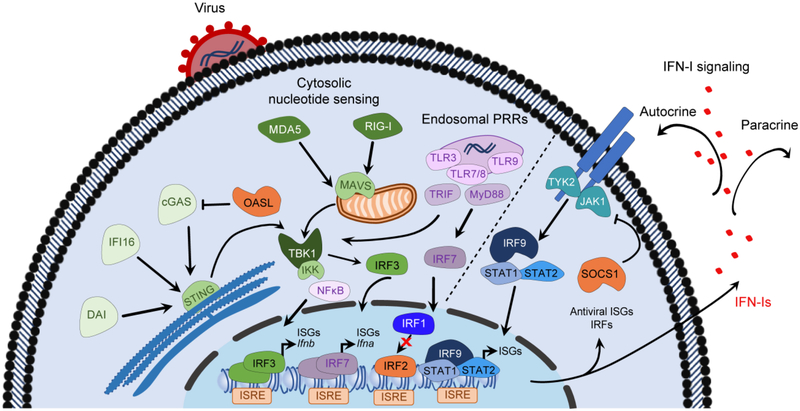
Induction of IFN signaling begins with recognition of pathogen-specific ligands including viral nucleic acids and viral proteins by pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), NOD-like receptors (NLRs) and DNA sensors (Figure 1). In humans, at least 10 functional TLRs (TLRs 1–10) recognizing distinct pathogen-associated molecular patterns (PAMPs) have been identified. These transmembrane receptors are expressed either at the cell surface or associated with intracellular vesicles. Viral nucleic acids are recognized by TLR3 (double-stranded RNAs), TLR7 and TLR8 (single-stranded RNAs) and TLR9 (unmethylated CpG DNA) expressed in endosomal compartments of primarily myeloid-lineage cells (9). Upon ligand engagement, TLRs signal through several adaptor molecules, including myeloid differentiation primary response gene 88 (MyD88) and TIR-domain containing adaptor protein-inducing IFNβ (TRIF), resulting in the recruitment and activation of members of the tumor necrosis factor receptor-associated factor (TRAF) family of proteins as well as the TRAF family member-associated NF-κB activator (TANK). In turn, this induces activation of tank-binding kinase-1 (TBK1) and inhibitor of NF-κB (IκB) kinase (IKK) family members. This then leads to the phosphorylation, activation and dimerization of interferon regulatory factor 3 (IRF3), as well as release of nuclear factor kappa B (NF-κB) from the IκB complex. IRF3 and NF-κB then translocate into the nucleus where they bind to promoter regions of IFNβ and other target genes thereby stimulating IFN-I and pro-inflammatory cytokine gene transcription (10). The resulting IFNβ also induces IRF-7 expression, in both autocrine and paracrine mechanisms, which in turn mediates a positive feedback loop that triggers synthesis of various members of the IFN-I family in a second burst of IFN-I production (Figure 1).
Figure 1: Induction, propagation and regulation of IFN-I signalling.
Viral nucleic acids and proteins are recognized by cytosolic DNA sensors, including DAI, IFI16 and cGAS, as well as by members of the RLR family (RIG-I and MDA5). Upon activation, the DNA sensors activate STING that then translocates to the Golgi where STING is phosphorylated by TBK1, allowing for the phosphorylation and activation of IRF3. Upon binding to their ligands, RIG-I and MDA5 engage MAVS leading to activation of TBK1 and members of the IKK family of kinases. Similarly, detection of endosomal viral nucleic acids is mediated by PRRs such as TLRs that then signal through MyD88 and TRIF adaptor molecules leading to activation of TBK1 and members of the IKK family. These kinases trigger phosphorylation, activation and dimerization of IRF3 and release of NF-κB from the IκB complex. IRF3 and NF-κB then translocate into the nucleus where they bind to promoter regions of IFNβ and other target genes thereby stimulating IFN-I and pro-inflammatory cytokine gene transcription. The resulting IFN genes then signal through a heterodimeric IFN-I receptor (IFNAR), triggering phosphorylation and activation of intracellular Jak1 and Tyr2 kinases. These kinases mediate recruitment and phosphorylation of STAT1 and STAT2 proteins that then recruit IRF9 to form the trimeric IFN-stimulated gene factor 3 (ISGF3) complex. The ISGF3 complex then translocates into the nucleus where it binds to ISRE regions within the promoter regions of IFNs or IFN-inducible genes. As a result, expression of a multitude of ISGs including antiviral and regulatory ISGs is induced, along with IRFs and other IFN-dependent genes. Both sensing of viral nucleic acids and IFN-I signaling can be reciprocally regulated by ISGs such as OASL and SOCS that respectively antagonize cGAS and the IFNAR-associated kinases.
DAI – DNA-dependent activator of IFN-regulatory factors; IFI16 – IFN-induced 16-kDa protein; cGAS – cyclic GMP-AMP synthase; RLR – RIG-I-like receptors; RIG-I – retinoic acid-inducible gene-I; MDA5 – melanoma differentiation-associated gene 5; STING – stimulator of IFN genes; TBK-1 – activation of tank-binding kinase-1; IRF – interferon regulatory factor; MAVS – mitochondrial antiviral signaling protein; IKK – inhibitor of NF-κB (IκB) kinase; NF-κB – nuclear factor kappa B; PRRs – pattern recognition receptors; TLRs – Toll-like receptors; MyD88 – myeloid differentiation primary response gene 88; TRIF – TIR-domain containing adaptor protein-inducing IFNβ; Jak1 – Janus kinase 1; Tyk2 – tyrosine kinase 2; ISRE – IFN-stimulated response elements; ISGs – IFN-stimulated genes; SOCS – Suppressor of cytokine signaling 1; OASL – 2′-5′-oligoadenylate synthetase-like.
Inside virus infected cells, members of the RLR family, including retinoic acid-inducible gene-I (RIG-I), melanoma differentiation-associated gene 5 (MDA5) and the laboratory of genetics and physiology 2 (LGP2) detect cytoplasmic viral RNAs and orchestrate production of both IFN-I and other pro-inflammatory cytokines (11). RIG-I binds short dsRNA and 5’-diphosphate or 5’tri-phosphate ssRNA whereas MDA5 binds long dsRNA and mRNA without the 2’-O-methylation (12). While these receptors are expressed in many cell types, they themselves are also products of IFN signaling resulting from a positive-feedback loop following viral infection (13). Upon their activation by viral RNA, RIG-I and MDA5 engage the mitochondrial antiviral signaling protein (MAVS, also referred to as VISA, IPS-1 and Cardif) adaptor that then activates the antiviral signaling (13) (Figure 1). MAVS signals much like MyD88 (discussed above), resulting in primarily IFNβ gene transcription. Interestingly, the interaction of RLRs with IRF3 not only mediates IFN-I signaling, but also induces apoptosis of infected cells via a RLR-induced IRF3-mediated pathway of apoptosis (RIPA) that is independent of and distinct from the transcriptional activities described here (14). It has also been suggested that IRF5 may utilize the MAVS adaptor to induce IFN-I responses in some subsets of dendritic cells (15).
DNA sensors, such as cyclic GMP-AMP synthase (cGAS), IFN-induced 16-kDa protein (IFI16), DNA-dependent activator of IFN-regulatory factors (DAI) and DEAD box polypeptide 41 (DDX41), can also detect viral DNA following infection with DNA viruses (16–19). In fact, cGAS can detect a variety of pathogens including RNA viruses, owing to its ability to recognize both PAMPs and DAMPs, as was illustrated in the detection of mitochondrial DNA from both herpes simplex virus (HSV) and dengue virus (20, 21). Interestingly, Schoggins and colleagues observed that cGAS exerted a broad inhibition of both DNA and RNA virus families independent of the canonical STAT1-based signaling pathway (22). Rather, these sensors activate the stimulator of IFN genes (STING) adaptor present on the membrane of the endoplasmic reticulum (23) (Figure 1). Activated STING then translocates to the Golgi, where it is phosphorylated by TBK1, allowing for the recruitment and phosphorylation of IRF3. As above, phosphorylated IRF3 then translocates into the nucleus where it drives expression of both IFNs and IFN-stimulated genes (ISGs). Beyond the IRF3 arm, STING also activates NF-κB dependent signaling, leading to expression of antiviral and pro-inflammatory cytokines (23). Of note, these DNA sensors are also ISGs produced in response to viral infections, serving to further amplify interferon production, the antiviral state, and the resulting immune response. Interestingly, the cGAS-STING axis also plays important roles in inducing IFN-I signaling in cancers, and can be triggered by cancer therapies such as radiation therapy and inhibitors of the DNA damage pathway (24, 25). Indeed, STING activation has emerged as a crucial regulator of anti-tumour immunity, and STING loss of function can occur in many cancers through missense mutation or epigenetic silencing, suggesting that it may contribute to malignant transformation (26, 27). Overall, the cGAS-STING pathway acts as a nexus on which many types of cellular perturbations converge.
IFN-I signaling
The IFN-I family signals through a heterodimeric IFN-I receptor (IFNAR) composed of two subunits, IFNAR1 (IFNα/β receptor α chain) and IFNAR2 (IFNα/β receptor β chain), that are expressed in all nucleated cells. Engagement of IFN-I to IFNAR induces dimerization of IFNAR1 and IFNAR2, leading to the phosphorylation and activation of intracellular receptor-associated kinases Janus kinase 1 (JAK1) and tyrosine kinase 2 (TYK2) (Figure 1). Upon activation, these kinases trigger recruitment and phosphorylation of members of the signal transducer and activator of transcription (STAT) family (mainly STAT1 and STAT2), forming a heterodimeric complex that then recruits IRF9 to form the trimeric IFN-stimulated gene factor 3 (ISGF3) complex. The ISGF3 complex then translocates into the nucleus where it binds to IFN-stimulated response elements (ISRE) within the promoter regions of IFNs or IFN-inducible or stimulated genes (ISGs) resulting in the induction of hundreds of ISGs that can further propagate the immune response against viral infection (Figure 1). How signaling via a ‘single’ dimeric receptor can induce such a diverse array of immunological outcomes likely depends on a number of factors including receptor surface expression, ligand binding affinities, ability of receptor to form dimers, regulatory mechanisms (both negative and positive), post-translational modifications, availability of co-factors, and epigenetic regulation of signaling components, among others.
While the canonical IFN signaling pathway is defined by heterodimerization of phosphorylated STAT1 (pSTAT1) and pSTAT2, IFNAR activation can also trigger formation of pSTAT1 homodimers that, upon translocating into the nucleus, bind to IFNγ response elements leading to expression of IFNγ-dependent proinflammatory genes (28). Of note, while STAT1/STAT2 phosphorylation is only transient, the induction of ISGs persists beyond the presence of IFN-I (29). IFN-Is stimulate overproduction of unphosphorylated STAT1 and 2, which can drive ISG gene activation in the absence of IFN-1 (30). Indeed, an unphosphorylated ISGF3 complex has been shown to exist in the overabundance of the unphosphorylated STATs, and drives a persistent IFN-I response (31).
In addition to STAT1/2, IFN-I also activates other members of the STAT family (STATs 3–6) resulting in a wide array of outcomes (32). STAT6 forms complexes with STAT2 in order to promote IFN-I dependent antiproliferative activities via upregulation of Sp1 and downregulation of Bcl6 (33). Intriguingly, STAT4, which can bind IFNAR, counteracts the IFN-I mediated inhibition of proliferation, and in so doing allows for enhanced expansion of antigen-specific CD8 T cells (34). This antagonizing of IFN-I mediated activities is also observed with STAT3, which limits the antiviral activity of IFN-Is, as well as counters the IFN-I mediated differentiation of CD4 T cells into T helper 1 (Th1) following viral infection (35, 36). STAT3 skews CD4 T cell differentiation towards a T follicular helper (Tfh) phenotype, which in turn promotes germinal center (GC) B cell development and virus-specific antibody responses (35, 36). Accordingly, STAT3-deficient Tfh cells show elevated IFN-inducible transcripts, suggesting that STAT3 downmodulates IFN-I signaling in CD4 T cells likely as a means to keep IFN-I signaling in check and avoid immunopathology. Perhaps not surprisingly, IFN-Is antagonize STAT3 in order to promote their antiviral responses, as was recently shown following HCV infection in vitro (37). This varied pairing of STATs offers an important negative feedback regulatory mechanism, which is especially important given that IFN-I signaling occurs in almost all cell types, raising the risk of immunopathology/cytokine storm associated with an uncontrolled or dysregulated IFN-I signaling. A tight regulation of the IFN-I mediated pro- and anti-inflammatory activities is indeed critical as further evidenced by the presence of various positive and negative regulatory mechanisms involving IRFs and ISGs, as discussed below.
In addition to the above, IFN-I leads to the activation of numerous STAT-independent signaling pathways, including the c-Jun N-terminal kinase (JNK) pathway, the extracellular signal regulated kinase (ERK) pathway, p38 mitogen-activated protein kinase (MAPK) pathway, and the mammalian target of rapamycin (mTOR) pathway. These pathways play essential roles in the ultimate expression of IFN-I dependent genes, both at the transcriptional and mRNA translation levels, and thus the ultimate immunologic outcome of IFN-I signaling. (See review by Platanais, pages _____)
Interferon regulatory factors in chronic infection
Induction of IFN-I is primarily controlled at the gene transcription level, a stage heavily modulated by IRFs. IRFs were originally identified as regulators of IFN-I gene expression following virus infections (38), and have since been implicated in the regulation of IFN-II and IFN-III as well (39). To date, nine members of the IRF family – IRF1, IRF2, IRF3, IRF4 (LSIRF, PIP or ICSAT), IRF5, IRF6, IRF7, IRF8 (also known as ICSBP), and IRF9 (ISGF3γ) have been identified in humans and mice (40). Structurally, all IRF family members share a highly conserved amino-terminal DNA binding domain (DBD) characterized by five tryptophan repeats, which is recognized by the consensus IRF recognition motif 5’- AANNGAAA-3’ (where N denotes any nucleotide) on DNA sequences including IRF elements (IRFEs) and ISREs (41). At the carboxyl-terminal end of all IRFs is an IRF association domain (IAD) that enables IRFs to form homomeric and heteromeric complexes with other IRF family members or members of other transcription factor families, including AP1 and various STATs. IRF1 and IRF2 share a fairly similar IAD (IAD2) that is distinct from the rest of the IRFs, which each contain an IAD1 that shares homology with the SMAD family of transcription factors (39). The IAD regions show considerable sequence variation among IRFs, a property believed to confer unique functional properties or selectivity for interactions.
Among the IRF family, IRF1, IRF3, IRF7 and IRF9 are considered positive regulators or activators of IFN-I gene transcription. The cytosolic IRF3 and IRF7 are the main antiviral transcription regulators following virus infection (39). As discussed above, the constitutively and ubiquitously expressed IRF3 facilitates the initial signaling following sensing of viral nucleic acids, leading to IFNβ production, which will in turn enhance IRF7 production and, consequently, gene transcription of various IFNα gene subtypes (39, 42, 43). IRF3 is also important for regulating several ISGs such as the interferon-induced protein with tetratricopeptide repeats (IFIT) family of proteins and ISG15, as well as various cytokines and chemokines including IL-23, CXCL10 and CCL5 (44–46). As is frequently observed in IRF signaling, while IRF3 enhances gene expression of many genes, it can also inhibit gene transcription in other contexts, as was demonstrated for TGFβ and IL-12β (47, 48). IRF1 and IRF7 are also constitutively expressed, albeit at low levels under naïve conditions. IRF1, IRF3 and IRF7 are capable of inducing ISGs in an IFN-independent manner as reported for the STAT independent expression of ISGs including beta 2- microglobulin (β2m), guanylate binding protein (GBP), Myxovirus protein A (MxA), CXCL10, ISG15 and MHC-I in immune cells (49–51). As well, IRF1 can interact with MyD88 and, in so doing, amplify TLR signaling (52). Upon IFN-I stimulation, the levels of IRF1 and IRF7 are significantly upregulated. In fact, all the IRFs, except IRF3, are cytokine-inducible. IRF1 and IRF7 further amplify the IFN response through a positive feedback loop (39, 53). IRF9, on the other hand, is the only member with a STAT2 binding site, forming part of the ISGF3 complex, to enable ISGF3-complex binding to ISRE on promoter regions of target genes (54). IRF9 was recently shown to interact with unphosphorylated STAT2 to drive expression of cytokines including IL-6, which in turn can activate STAT3 (55). Consistent with IRF9 being a key player in IFN-signaling, IRF9 deficient mice largely behave like IFN receptor knock out (IFNARKO) mice, and show increased susceptibility to various viral infections including HSV and vesicular stomatitis virus (VSV) (56). Moreover, as a driver of IRF7 and other ISG expression, IRF9 deficiency disrupts both the IFN-I amplification loop and the ability to control viral replication, leading to establishment of a chronic infection and T cell exhaustion (57).
IRF5 and IRF8 are frequently grouped together as their transcriptional activities can be positive or negative depending on the context. IRF5 has long been considered a primary mediator of MyD88-depedent TLR signaling, culminating in the induction of various inflammatory cytokines such as IFN-Is, TNFα and IL-6 (58–61). Perhaps testament to its important role in antiviral immunity, polymorphisms within IRF5 increase both susceptibility to and pathogenesis of hepatitis B virus (HBV) infection (62). Recently, TLR7-induced IRF5 activation was shown to upregulate the death receptor 5 (DR5) leading to enhanced death of protective CD4 T cells during chronic visceral leishmaniasis and, ultimately, allowed for establishment of a chronic infection (63). IRF5 has been shown to possess both transcriptional activator and repressor functions, as was illustrated in macrophages where it activated IL-12 and IL-23, but repressed IL-10 gene transcription (64). Similar to IRF5, the IFNγ-induced IRF8 also plays dual context-dependent roles as a transcriptional activator for genes such as IL-12p40 and repressor for the tripartite-motif 21 (TRIM21) (65, 66). It is interesting to note that TRIM-21 ubiquitinates and activates IRF8 transcriptional activity (66), which suggests that IRF8 uses its repressor function as a self-regulatory mechanism. With a weaker binding affinity to target ISREs, IRF8 works in concert with IRF1 and IRF2 to obtain optimal binding affinities (67). Whether these interactions with IRF1 or IRF2 determine whether IRF8 functions as a stimulator or repressor of gene expression remains to be elucidated.
IRF2 and IRF4 are generally considered inhibitory IRFs. IRF2, which shares significant sequence homology with IRF1, was originally identified as a factor antagonizing IRF1-mediated transcriptional activities largely through recognizing common DNA motifs on the promoter regions of target genes (68). IRF2 has now been shown to associate with NF-κB, IRF8 and STAT1 within the cytoplasm and, in so doing, prevents these factors from translocating into the nucleus where they would regulate gene transcription (67, 69–72). Under steady state conditions, IRF2 prevents tonic IFN-I signaling and its absence resulted in development of an ISGF3-driven autoimmune inflammatory skin disease in mice, pointing to a role of IRF2 in antagonizing the activities of the IRF9-STAT1-STAT2 complex (73). Of note, mice lacking IRF2 exhibit a heightened ISG signature without an accompanying elevation in IFN genes, implying that IRF2 regulates IFN signaling and not IFN gene production (73). In addition to its repressor functions, IRF2 has been shown to act, typically in concert with IRF1, as a transcriptional activator for several genes including the H4 histone gene and Tlr3 (74–76). On the other hand, IRF4 antagonizes IRF5 largely through binding to similar sequences on target genes as well as by competing for MyD88 interaction (77, 78). IRF4 regulates both CD4 and CD8 T cell differentiation, as well as B cell maturation [reviewed in (79)]. In addition, unlike IRFs 1, 5 and 8, which are associated with polarization towards M1 macrophage phenotype, IRF4 is involved in skewing towards the anti-inflammatory M2 phenotype (80). In an acute infection, IRF4 was shown to help sustain virus-specific CD8 T cell effector functions and prevent establishment of a chronic infection, but it induced immunopathology (81). However, in chronic LCMV infection, IRF4 suppressed effector cytokine production, increased inhibitory receptor expression and reduced anabolic metabolism in virus-specific CD8 T cells, and further repressed development of memory T cells (82). It did, however, promote expansion and maintenance of the antigen-specific T cells, as well as prevented immunopathology (82).
Overall, IRFs play important roles in regulating IFN-I signaling and, consequently, antiviral activities. It is therefore not surprising that viruses specifically target the functioning of these factors in order to prevent IFN-I induction. Viruses such as HSV, hepatitis C (HCV), dengue and HIV have dedicated some of their viral proteins to inhibit IKKε-, IKKα- and TBK1-mediated phosphorylation of IRFs, thereby dampening IFN-I signaling [reviewed in (83)]. HIV-I further uses its viral protein R (Vpr) and viral infectivity factor (Vif) to target IRF3 for degradation (84). Human papillomavirus (HPV) and paramyxoviruses (Measles virus, Newcastle disease virus) can interact with IRF3 and inhibit its transactivation potential or prevent its translocation into the nucleus (85). Furthermore, viruses have been shown to prevent the association of IRFs to the promoters of their target genes, as illustrated by HSV and Kaposi’s sarcoma-associated herpesvirus (KSHV) that prevent IRF3 binding to IFNβ promoter regions (86, 87). KSHV uses its viral homologs vIRF1 and vIRF2 to prevent IRF3-dependent gene transcription and vIRF3 to prevent IRF7 from binding to its target genes [reviewed in (88)]. As well, viruses have evolved to exploit IRFs for their own benefit. HIV-1 induces IRF1 expression, which it exploits to stimulate viral transcription of its genome in the early phases of infection as well as to increase T cell activation to favor virus replication. Once it achieves its goals, HIV-1 then directs its Tat protein to inhibit the IRF1 transcriptional activities and/or facilitates its proteasomal degradation [reviewed in (89)].
Diverse functions of interferon stimulated genes in viral infection
The activities of IFN-Is are potentiated by hundreds of ISGs that are directly stimulated downstream of IFNAR. ISGs include antimicrobial proteins, chemokines, cytokines and other immune mediators that induce recruitment of immune cells and inflammation. ISGs play diverse roles in many cellular processes such as migration, antigen processing and presentation, cellular activation, differentiation, mitosis and apoptosis. To add to this complexity, cell-type specificity, ISG expression kinetics, duration and context of signaling all likely contribute to the impact of IFN-Is on immune cell function.
Antiviral and anti-proliferative roles of ISGs
First discovered for their essential role in antiviral immunity, many studies have characterized the antiviral and anti-proliferative properties of ISGs. A productive virus infection requires virus entry, uncoating and RNA/DNA replication in the nucleus or cytoplasm of the infected cell. The viral genome is then transcribed into new viral RNA that is either translated into viral proteins or gets incorporated into new virions. The mature virus particles are then released in order to initiate infection of new target cells. IFN-Is stimulate expression of genes that will target each of these virus replication cycle steps, and in so doing prevent spread of infection. First, an ISG family of IFN inducible transmembrane proteins (IFITMs) interferes with membrane fusion thereby blocking cellular entry of a broad range of viruses including HIV, WNV, Ebola virus, Zika virus and influenza A virus, while the tripartite motif 5 alpha (TRIM5α) E3 ubiquitin ligase induces premature disassembly of the incoming virion capsid thus blocking synthesis of the viral complementary DNA (cDNA) [reviewed in (90, 91)]. Another ISG sterile alpha motif and histidine aspartic domain (HD) containing protein 1 (SAMHD1) not only inhibits HIV reverse transcription through mediating depletion of deoxynucleoside triphosphates (dNTPs) required for cDNA synthesis but also targets RNA for degradation (90). The reverse transcription step is also directly inhibited by an ISG family of apolipoprotein B mRNA editing enzyme catalytic-like 3 (APOBEC3). The APOBEC3 cytidine deaminases also hypermutates the viral genome by introducing G to A substitutions that are detrimental to virus replication [reviewed in (90, 92)]. Further down the replication cycle, the IFN inducible human myxovirus resistance proteins 1 (MxA) and 2 (MxB) inhibit nuclear import and/or integration of the viral DNA into the host cell chromosomes (90, 92, 93). Several ISGs including Schlafen proteins and Protein Kinase R (PKR) inhibit viral mRNA translation, whereas GBPs and ISG15 target post-translational modifications including processing and incorporation of viral proteins into assembling virions at the cell surface (92, 93). The ISGs serine incorporator 3 (SERINC3) and SERINC5 get incorporated into the forming virions, and they reduce infectivity of the nascent virion through mechanisms that ultimately inhibit virus fusion to the membranes of a new target cell (94, 95). At the cell surface, the cellular ISG restriction factor tetherin (also known as BST-2 or CD137) potently restricts release of a broad spectrum of enveloped viral particles by tethering or trapping them to the surface of the infected cell (92, 96, 97). While each of these ISGs appear sufficient to carry out their respective functions, there exist cases where ISGs further enhance functional activities of their fellow antiviral ISGs. To illustrate, in addition to its antiviral responsibilities, ISG15 also directly activates PKR to further promote or enforce inhibition of viral gene translation, and targets proteins being synthesized de novo during an active infection for eventual degradation (98, 99). As well, ISG15 stabilizes IRF3 transcriptional activity, leading to enhanced IFN-I signaling (100). The fact that IFNs greatly upregulate so many ISGs targeting essentially all the virus replication cycle steps underscores their absolute requirement in controlling virus infections. Consistent with this, viruses have evolved to antagonize these ISG-mediated activities. For example, HIV mainly uses its Vif, Nef and Vpu accessory proteins to respectively counter APOBEC3, SERINC3 and tetherin.
In addition to directly targeting steps of the replication cycle, IFN-Is mediate upregulation of RNA/DNA sensors such as MDA5 and RIG-I that further sensitize or enhance detection of viral nucleotides, resulting in expression of IFN genes, as discussed earlier. Importantly, MDA5 was found to be a major trigger of IFN-I production following both acute and chronic LCMV infections and its deficiency delayed antiviral CD8 T cell responses, which in turn resulted in viral persistence (101). Moreover, IFN-Is induce a wide array of ISGs including IRFs and STATs that can further stimulate expression of both IFN and IFN-inducible genes thereby propagating IFN-I signaling through positive feedback amplification mechanisms as well as sustaining an antiviral state not conducive for spread of infection. All considered, studies on IFN-Is and ISGs have provided information not only of how IFN-mediated activities contribute to control of virus replication and resolution of infection, but also how chronic IFN-I signaling is likely induced and sustained. Further studies are required in order to fully appreciate both the precise roles of these ISGs as well as the context-dependency of their functions (cell type and virus specificities). Indeed, specific ISGs may be selective for the viruses they inhibit. While some ISGs including IFI16, OASL, IFITM2, MOV10 and TREX1 show targeted specificity in their antiviral roles, ISGs such as IRF1, IFTIM3, RIG-I and MDA5 were broadly acting in their inhibitory roles (102). In fact IRF1, which induces a wide variety of ISGs in both a STAT1- dependent and independent manner, inhibited all viruses tested in a study by Schoggins and colleagues (102). Whether these pan effector functions of IRF1 are as a result of IFN signaling beyond the canonical STAT1-reliant IFN-I, IFN-II and IFN-III remains to be tested. Of note, in some virus infections STAT1 expression is downregulated, perhaps in a bid by the virus to dampen IFN-I responses, while expression of other STATs is enhanced (34). The ability of ISGs such as IRF1 to utilize multiple non-overlapping effectors (such as different STATs) to exert their function likely enables ISGs to inhibit a broad range of viruses, and is advantageous in counteracting virus-mediated antagonistic activities.
Resolution of IFN-I signaling
Similar to many cytokine-receptor signaling pathways, excessive IFN-I signaling is prevented through multiple negative feedback mechanisms. For example, IFNAR is rapidly downregulated following engagement with IFN-I, immediately desensitizing cells that have responded to IFN-I from further stimulation (103, 104). As well, intracellular protein tyrosine phosphatases such as CD45, SH2 domain containing tyrosine phosphatase 1 (SHP-1) and SHP-2 inactivate JAK kinases and STAT proteins, further dampening IFN-I signaling (105). Further, a group of constitutively expressed proteins referred to as protein inhibitors of activated STAT (PIAS) suppresses STAT-mediated functions by blocking binding of STATs to their target DNAs (105). PIAS proteins also mediate SUMOylation of STAT proteins, ultimately channeling them for degradation via a SUMO-mediated degradation pathway (105). In addition to these cell-intrinsic desensitization mechanisms, some of the ISGs that are upregulated act at various points in the IFNAR signaling cascade to blunt or diminish signaling. Suppressor of cytokine signaling 1 (SOCS1), which is upregulated by IFN-I signaling, inhibits JAK-STAT signaling through associating with and inactivating JAK kinases (106, 107). As such, phosphorylation and activation of STATs is reduced, resulting in defective IFN-I dependent antiviral and antiproliferative activities. Another ISG, ubiquitin carboxy-terminal hydrolase 18 (USP18), plays an important role in inducing desensitization to IFN-I signaling. USP18 is an isopeptidase that counteracts ISG15-mediated activities by removing ISG15 conjugated (ISGylated) to target host and viral proteins, thus protecting these target proteins from being committed to or channelled for degradation (108). In addition to triggering de-ISGylation, USP18 also attenuates IFN-I signaling by displacing JAK1 bound to the IFNAR2 subunit (93, 109).
Anti-inflammatory and negative regulatory signals
Prolonged IFN-I signaling can lead to detrimental effects for the host. Immune stimulatory functions that lead to enhanced antigen cross-presentation and sensitivity can result in self-recognition and autoimmunity (110, 111). Therefore, the simultaneous induction of negative immune regulatory factors, such as indoleamine-2,3 dioxygenase (IDO), programmed cell death-1 ligand (PDL1) and IL-10, serves to protect the host from excessive stimulation by IFN-Is. Immune suppression by ISGs occurs at various levels: through signaling, altered metabolism, transcription and translation. IDO for example, catalyses the conversion of tryptophan to kynurenine, which then suppresses immunity by signaling through the aryl hydrocarbon receptor (112). In addition, tryptophan is required to support T cell proliferation, therefore redirecting tryptophan metabolism through IDO indirectly limits T cell function by depleting T cell resources (113). PDL1 and IL-10, on the other hand, directly attenuate T cell proliferation and function, allowing the resolution of immune responses after the infection is eliminated. In addition, cytosolic RNA/DNA sensors that are upregulated by IFN-Is can also paradoxically promote viral replication within host cells by interfering with other nucleotide sensing pathways. Whereas 2′-5′-oligoadenylate synthetase-like (OASL) limits viral replication by enhancing signaling through the RNA sensor RIG-I in the context of RNA viruses, induction of OASL inhibits cGAS and promotes viral replication in the context of infection by DNA viruses such as HSV (114). Similarly, chronic infections lead to induction of the ISG adenosine deaminase acting on RNA 1 (ADAR1), an RNA deaminase that counteracts the antiviral activity of PKR thereby promoting retroviral replication (102, 115). APOBEC3A, IDO1, LY6E are examples of other ISGs that promote virus replication in a broad range of viruses (102). Some ISGs have also been shown to target and inhibit IRFs. The interaction of GBP4 with IRF7 decreases the activation and transcriptional activity of IRF7, leading to defective IFNα expression (116). Thus, the myriad of ISGs induced by IFN-I signaling serve to both drive and then ultimately inhibit the immune responses to viral infection.
Chronic IFN signaling effects on innate and adaptive immunity
Much of the antiviral effects of IFN-I signaling were initially established in acute infections where IFN-I production is rapidly shut down with the elimination of infection (Figure 2). The same robust IFN-I dependent antiviral response is also initially observed in the case of chronic infections for initial virus control. Blockade of IFN-I signaling early during SIV infection decreased antiviral gene expression, and enabled increased virus replication, elevated viral reservoirs and led to an enhanced depletion of CD4 T cells and a faster progression to AIDS (117). Similarly, in LCMV infection, the administration of IFNAR blocking antibodies at the time of infection led to heightened viral loads while the receptor was being blocked and turned the otherwise acute LCMV infection into a chronic infection (118, 119). However, in persistent infections, this beneficial, heightened antiviral response is short-lived as the systemic IFN-I burst is reduced to levels observed in naïve hosts, even though the viral burden remains elevated (7, 118). Despite the undetectable levels of IFN-Is themselves, IFN-I signaling is ongoing as evidenced by the high continuing ISG production that is ablated by blocking the IFNAR (118). The cellular and/or environmental factors maintaining IFN-responsiveness (even when IFN-Is are systemically undetectable) are yet to be elucidated, but could represent smoldering levels of IFN-I production in distinct niches, or epigenetic restructuring that enables enhanced sensitivity to IFNAR signaling.
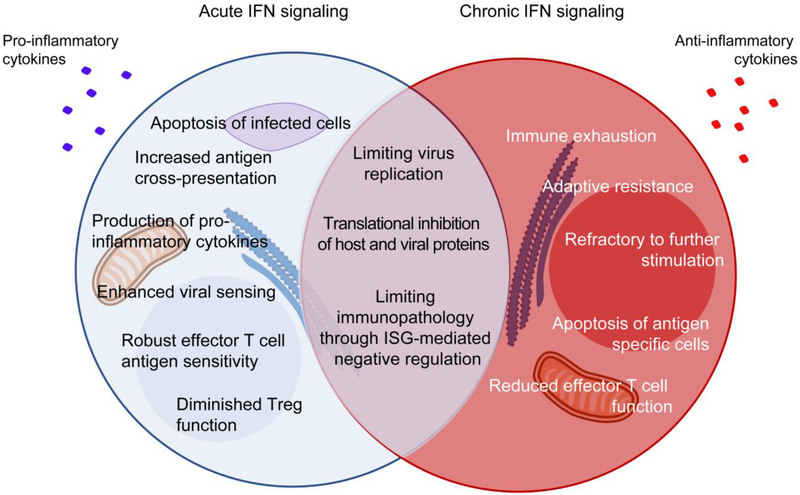
Figure 2: Chronic IFN-I signaling directly and indirectly induces immune activation and inflammation leading to an exhausted state.
Following infection, robust IFN-I signaling allows for strong antiviral responses characterized by inhibition of viral replication, apoptosis of infected cells, increased antigen cross-presentation, enhanced effector functions and overall control of infection. While chronic IFN-I signaling contributes towards controlling virus replication and limiting inflammation-induced immunopathology, it also leads to chronic immune stimulation and ultimately, immune dysfunction. The immunosuppressed or exhausted state resulting from chronic IFN-I signaling is characterized by several features including direct overexpression and accumulation of inhibitory factors (PDL1, IL-10, IDO, TRAIL, among others), and indirect effects due to chronic immune activation it drives (e.g., PD1, CD38, Tim3, CD38, among others). The ultimate outcome of these events is defective cytokine production, reduced metabolic fitness, adaptive resistance to therapy, failure to respond to further stimulation, contraction of antigen-specific cells, overall contributing to promote viral persistence.
PD-1 – programmed cell death-1; PDL1 – PD-1 ligand; Tim-3 – T-cell immunoglobulin and mucin-domain containing-3; IDO – indoleamine-2,3 dioxygenase; TRAIL – tumour-necrosis factor (TNF)-related apoptosis-inducing ligand.
Similarly, the downregulating mechanisms underlying diminished IFN-I expression despite sustained virus replication, remain ill-defined. Many studies have observed that corresponding to the downregulation of IFN-I production, antigen-presenting cells (and likely other immune subsets) become refractory to subsequent TLR signaling (117, 120, 121) and remain in a hypo-activated state throughout infection (122) (Figure 2). Interestingly, prolonging the strong antiviral response obtained during the acute stage may improve outcomes, suggesting that downregulation of IFN-I signaling may be premature against some viruses and enable virus persistence. Consistent with this, chronic LCMV (Cl-13)-infected mice given a recombinant form of IFN-I during the acute stage of infection or deletion of the IFN-I inhibitor OASL improved virus-specific CD8 T cell responses and accelerated virus control (101). Interestingly, attempts to recapitulate these beneficial effects obtained at the acute stage of infection (Figure 2) were unsuccessful when IFN-I levels were boosted during the chronic stage of LCMV infection (101). In fact, sustained IFN-I administration in SIV-infected rhesus macaques revealed an expanded SIV reservoir, accelerated loss of CD4 T cells (even though this may have been in part due to CD4 T cells being a target of SIV) and a faster progression to AIDS, much like the case in which IFN-I was blocked during the acute phase (117). Similarly positive correlations between a sustained IFN-I signature and disease progression were reported in HIV-infected patients (123, 124) and defective immune restoration following combination anti-retroviral therapy (cART) was associated with higher levels of IFN-I and immune activation (125, 126).
In contrast to their antiviral and stimulatory roles in acute viral infections, our group and Michael Oldstone’s group identified IFN-I signaling during chronic LCMV infection to drive many of the immune dysfunctions associated with viral persistence (118, 119). Inhibiting IFN-I signaling reduced chronic inflammation and apoptosis, reduced expression of multiple negative regulatory factors, including PDL1 and IL-10, and improved functional properties of T cells (Figure 2) (118, 119, 127). Paradoxically, blocking IFNAR at the onset of infection or during the established infection accelerated elimination of the chronic infection in a CD4 T cell and IFNγ dependent manner (118, 119). Thus, early IFNAR blockade directly or indirectly ‘programs’ the course of immune exhaustion. IFNAR blockade in the middle of a persisting infection restores antiviral functional properties of T cells (118, 119). The underlying mechanisms via which IFN-Is exert their effect in chronic infections still remain elusive, but likely involve various immune cell types associated with both innate and adaptive immunity.
Upon sensing of viral nucleotides through TLRs, plasmacytoid dendritic cells (pDCs) respond by producing massive amounts of IFN-Is, with contributions from other myeloid cells (128). The importance of pDC-driven IFN-Is in particular is highlighted by the failure of pDC-deficient mice to control chronic LCMV (128). Proliferation and functionality of virus-specific T cells were impaired in these pDC-deficient animals; however, this impairment was not due to lack of antigen-presentation since pDCs deficient in MHC II elicited functional T cell responses (128). Exposure of both pDCs and conventional DCs (cDCs) to IFN-Is enhances their maturation and activation (129). Interfering with IFNAR signaling reduces expression of MHC, costimulatory and adhesion molecules, and interferes with the ability of the DCs to stimulate naïve T cells (129). In chronic LCMV infection, IFN-I signaling induces expression of TNF receptor superfamily ligands such as the glucocorticoid-induced tumor necrosis factor receptor related protein ligand (GITRL) specifically on monocyte-derived antigen presenting cells (APCs) and not on cDCs (130). Triggering the GITRL-GITR axis helps control virus infection by promoting post-priming accumulation of CD4 T cells, which in turn provides help necessary for accumulation and functional properties of CD8 T cells (130). Additionally, in chronic infections, IFN-Is protect antiviral T cells from NK cell-mediated elimination, allowing for robust T cell responses and suppression of virus loads (131–133). In agreement, when given early following infection, IFN-Is rescue CD8 T cell function and promote virus clearance (101). Of note, Le Bon et al provided evidence that IFN-Is facilitate APC-mediated crosspriming of CD8 T cells in a manner independent of CD4 T cell help (134). The authors observed that while MHC II and CD40 were dispensable, IFN-I signaling was imperative for optimal priming and differentiation of CD8 T cells into effector cells in LCMV infected mice (134). Exogenous administration of IFNα yielded similar results, further strengthening the notion that this effect is IFN-I dependent (101, 134). In addition, IFN-I signaling by innate immune cells mediates expression of various ISGs that can either inhibit virus replication and/or induce expression of antiviral cytokines or IRFs that can further propagate IFN-mediated antiviral responses. However, consistent with the complexities of IFN-I mediated outcomes, IFN-I signaling also dampens immune functions, resulting in impaired control of infections. As alluded to earlier, some of the ISGs induced actually abrogate further IFN-I signaling. In accordance, mice deficient in the negative regulatory ISG, OASL1, produce enhanced levels of IFNs that potentiate antiviral CD8 T cells responses leading to efficient virus control (135). Moreover, IFN-I signaling was shown to trigger expression of pro-apoptotic factors such as Bax, Bid, Bim in pDCs, that activate caspases leading to pDC cell death (136). IFN-I signaling is also associated with reduction in size of the DC, macrophage and NK cell pools in the spleen. Further, IFN-I signaling enhances expression of multiple negative regulatory factors, including PDL1 and IL-10 in myeloid cells (118, 137–139). Our group has further shown that sustained IFN-I signaling in chronic infections not only mediates generation of immunosuppressive DCs, but also represses generation of cDCs capable of stimulating T cells (137). Additionally, in chronic HIV infection, IFN-Is desensitize monocytes such that they fail to respond to activating microbial factors (140). Taken together, these studies associate chronic IFN-I signaling with heightened immune activation and immunosuppression, as well as defective antigen presentation in persisting viral infections. As such, blocking of IFN-I signaling can restore immune function and help control viral infections (118, 119). We observe that IFNAR blockade during the chronic stage of infection suppresses not only production of proinflammatory cytokines IL-1 and IL-18, but also expression of caspases in DCs and macrophages, together implying that both inflammation and activation of the inflammasome are prevented in these innate immune cells (118). It is worth mentioning that IFNAR blockade at the onset of a would-be persistent viral infection initially results in high virus titres, but this appears to be transient as the number of immune cells, especially virus-specific CD4 T cells, increase along with production of the effector cytokine IFNγ, ultimately resulting in enhanced virus control (118).
T cell specific effects of chronic IFN-I signaling CD4 T cells
In activated CD4 T cells, IFN-Is can either trigger antiproliferative and proapoptotic responses or survival and proliferative responses depending on the STATs they activate [reviewed in (50)]. While the mechanisms regulating each of these opposing pathways are incompletely understood, STAT1 activation has been shown to mediate CD4 T cell antiproliferation and apoptosis through inducing IRF1, activating caspase-7 and activating TRAIL (50). In contrast, signaling through STATs 3 and 5 generates antiapoptotic CD4 T cell responses, and is frequently observed in low amounts of STAT1 (50). During chronic viral infection, a sustained IFN-I signature leads to CD4 T cell attrition, exhaustion and, consequently, suboptimal control of infection (127, 141, 142). Unlike treatment during an acute infection, treatment of SIV chronically-infected macaques with a recombinant IFNα2a led to accelerated depletion of CD4 T cells that correlated with rapid progression to AIDS, potentially through enhanced cellular activation generating a larger target pool for SIV infection (117). In chronic HIV infection, sustained IFN-I signaling was a key driver of virus-induced apoptosis, TRAIL- and TRAIL-DR5-dependent apoptosis, Bak- and Fas-mediated apoptosis, as well as elevated caspase-3 activity in CD4 T cells (142–145). Further, expression of PDL1 and IL-10 driven by IFN-Is prevents CD4 T cell priming and Th1 polarization during chronic infection (146). Taken together, these mechanisms of CD4 T cell loss or exhaustion deprive CD8 T cells of the help the CD4 T cells would otherwise provide, hence compounding poor virus control.
The effect of IFN-I signaling on regulatory T cells (Tregs) remains controversial. IFN-Is have been shown to promote Treg development and survival, as well as to maintain FoxP3 expression and Treg functions under inflammatory conditions (147, 148). However, in acute virus infections IFN-I signaling in Tregs inhibited both proliferation and activation of Tregs, enabling efficient antiviral T cell responses and leading to efficient virus clearance (149). With the exception of the IFN-I mediated depletion of Tregs, similar results were obtained in a chronic LCMV infection wherein IFN-I signaling in Tregs reduced their activation and expression of genes associated with an enhanced suppressive potential (150). In this system, IFN-I signaling in Tregs was associated with increased numbers of effector cytokine-producing virus-specific CD8 T cells, resulting in reduced virus burden and limited expression of inhibitory receptors PD1 and CD39, among others (150). Interestingly, these authors were able to extend their findings into tumor models where abrogation of IFNAR signaling in Tregs led to an increase in the frequency of tumor infiltrating PD1+ Tregs, repressed activation and cytokine production by both conventional CD4 and CD8 tumor infiltrating T cells, and ultimately poor control of tumor growth (150). While IFN-I signaling in Tregs appears to potentiate antiviral and antitumor T cell functions, it is worth noting that IFN-I signaling within the tumor microenvironment is associated with Treg expansion and enhanced activation of their immune suppressive potential, both of which favor tumor progression (151). Further, Treg depletion during the chronic stage of LCMV infection restored antiviral functions of virus-specific CD8 T cells albeit with an increase in PDL1 expression that secondarily limited viral control (152).
CD8 T cells
IFN-Is play important roles in potentiating antiviral responses by cytotoxic CD8 T cells, which are essential for elimination of chronic viral infections. IFN-Is enhance T cell priming, allowing for T cell differentiation that is associated with expression of key transcription factors T-bet and eomesodermin (Eomes) and acquisition of effector functions (129, 134, 153, 154). Additionally, IFN-Is directly enhance the functionality of antigen-specific CD8 T cell responses including their cytotoxic capacity (155, 156). Abrogation of direct interaction between virus-specific CD8 T cells and IFN-Is almost entirely (>99%) abolished their ability to expand and generate memory cells following LCMV infection (156). In a way, this emphasizes IFN-Is as costimulatory factors necessary for antigen-specific cell expansion. In this case, IFN-Is promoted cell survival during the antigen-driven proliferation phase, without affecting the CD8 T cell proliferative potential per se. In another study, polyinosinic-polycytidylic acid [poly(I:C)]-mediated IFN-I signaling was critical in augmenting virus-specific CD4 and CD8 T cell responses and reducing both the virus burden and risk of virus-induced leukemia in Friend retrovirus (FV)-infected mice (157). This poly(I:C) effect was again entirely IFN-dependent as the effector cytokine-production and cytotoxicity of virus-specific CD8 T cells were not observed in infected IFNAR-deficient mice (157). As discussed earlier, IFN-Is can either activate STAT1-dependent signaling that inhibits CD8 T cell proliferation or trigger other STATs such as STAT4 that favor antiviral CD8 T cell expansion (50, 155). In LCMV infection, virus- activated CD8 T cells (and intriguingly not CD4 T cells) were shown to downregulate STAT1, with a concomitant increase in STAT4 levels, suggesting that this could be a mechanism through which the IFN- mediated block in proliferation is circumvented (34). It is interesting that several studies have observed that in the earlier stages of an infection, memory CD8 T cells, and to a lesser extent naïve CD8 T cells, are preferentially vulnerable to IFN-mediated apoptosis [reviewed in (153)]. It is indeed tempting to think of this as yet another mechanism the immune system uses to ensure that there is ‘room’ for expansion of antigen-specific CD8 T cells. These studies support a beneficial role of IFN-I signaling in enhancing the antiviral roles of virus-specific CD8 T cells. In agreement, administration of exogenous IFN-Is early following chronic LCMV infection immediately enhanced both numbers and antiviral functions of virus-specific CD8 T cells (101). Importantly, the authors further observed that this early IFN-I administration maintained the enhanced numbers and preserved polyfunctionality of the LCMV-specific CD8 T cells at later time points, ultimately resulting in an accelerated resolution of the infection (101). In accordance with other studies (117), the “window of opportunity” during which IFN-I administration is beneficial is rather short, around one week in the case of chronic LCMV infection (101).
An emerging theme, however, is that during chronic viral infections, virus-specific CD8 T cells are continuously exposed to antigens and chronic IFN-I signaling leading to their differentiation into an exhausted state in which their effector functions, memory potential, and proliferative capabilities are significantly impaired (158). The exhausted cells are characterized by several features including overexpression and accumulation of inhibitory receptors (PD1, Tim-3, TIGIT, CTLA-4, LAG-3 and several others), defective cytokine production, reduced metabolic fitness, among others (158). Somewhat perplexingly, IFNAR blockade reduces the expression of these exhaustion markers and can restore functional properties in virus-specific CD8 T cells (118, 143, 159, 160). The identification of IFN-Is as mediators of CD8 T cell exhaustion during the chronic stage of infections helps explain, at least in part, the observations indicating that delayed exogenous IFN-I administration in chronically infected animals neither enhanced numbers nor functionality of antiviral CD8 T cells (101, 117). Instead, IFN-I administration during the chronic stage was associated with worsened outcomes culminating in poor virus control. Even though ongoing IFN-I signaling during a chronic viral infection induces an exhausted state among CD8 T cells, several recent studies have highlighted the heterogeneity among the virus-specific CD8 T cells. In particular, these studies identified a population of PD-1+ CD8 T cells that exhibit stem cell- and memory-like properties, capable of self-renewal and terminal differentiation, whose generation is dependent on TCF1 (161, 162). Even though this population lacked an effector T cell signature, it sustained an antiviral T cell response and had greater proliferative potential following PDL1 blockade during the chronic stage of infection (161, 162). Whether IFN-Is play a role in maintenance of this population or affect its functional and/or proliferative capabilities remain to be tested. Notably, a recent study found that TLR9-mediated induction of IFN-Is using biodegradable nanoparticles functionalized with CpG and retroviral T cell epitopes of FV during the chronic stage of infection efficiently reactivated CTL antiviral activities leading to a pronounced decrease in virus burden (154). Interestingly, this immunization strategy was also shown effective in cancers where the tumor antigens and CpG drove IFN production that was associated with enhanced numbers of CTLs infiltrating the tumor, resulting in the control of tumor growth (163).
Overall, IFN-I signaling early following infection offers critical benefits for both innate and adaptive immunity, and contributes to generation of an antiviral state that limits virus infections. The direct antiviral roles of IFN-Is have seen IFN-Is become central in therapies to treat chronic HBV and HCV infections. However, sustained IFN-I signaling during the chronic stage of infection leads to exhaustion of immune cells and other immunosuppressive mechanisms that appear to temper the IFN-I mediated antiviral benefits, ultimately leading to failure to control the infection.
IFN therapy
IFN therapy during chronic infections remains controversial in the realm of therapeutic approaches aimed at controlling and eliminating viral infections. This underscores the dual, opposing roles IFN-Is play during the chronic stage of infection. IFNs continue to play a critical beneficial role throughout the course of infection, which led to the hypothesis that enhancing IFN-signaling in chronic patients would improve disease outcomes. While IFNα has been successfully used to treat HBV and HCV, IFNα administration to HIV-1 chronically infected patients failed to offer significant long-term benefits in curtailing disease progression, even though the viral loads and HIV-I cell-associated RNA levels were reduced in some cases (164–167). Consistent with this, several reports indicated that IFNα correlated with immune activation, enhanced CD4 T cells loss and disease progression (142, 168).
However, recent advances in the field suggest a therapeutic potential of IFN-I administration. This is largely due to the emerging understanding that antiviral potencies of IFN-I subtypes vary (169, 170). This variation is largely due to distinct IFNα subtypes and IFNβ exhibiting differential affinity for IFNAR, thereby eliciting differential signal transduction and, consequently, different immune responses (171–173). As such, the perceived failure of earlier IFNα boosting therapies could be as a result of administering IFNα subtypes that are ineffective in virus control during the chronic stage of infection. Of particular note, IFN therapies in HIV-I infected patients have primarily focused on IFNα2 (164). Intriguingly, IFNα2 was recently shown to be ineffective in potentiating anti-HIV immunity in HIV-infected humanized mouse models (169, 174), while instead, IFNβ and IFNα14 were individually potent against HIV. During chronic LCMV infection, blockade of IFNβ, but not multiple IFNα subtypes, increased the antiviral immune responses and virus clearance, albeit not as well as IFNAR blockade (175), suggesting that distinct IFN-I subtypes have different roles in modulating immunity to chronic viruses. This raises the question of whether, and if so which, specific IFN-I subtypes should be targeted in these IFN administration approaches. It is interesting to note that while IFNα2a treatment may not have been efficacious in anti-HIV therapies, it enhanced clearance of HIV-infected cells via antibody-dependent cellular cytotoxicity mediated by broadly neutralizing antibodies (176), underscoring the diverse roles of IFN-Is in viral infections.
IFN-I blockade
An approach that has yielded consistent positive outcomes involves blocking IFN-I signaling during the chronic stage of infection. As detailed above, this was first performed in the chronic LCMV model, leading to enhanced control of viral infection (118, 119). In a humanized mouse model of HIV, we showed that blocking IFNAR signaling during the chronic stage of infection lowered ongoing ISG expression, reversed the HIV-induced immune activation, enhanced anti-HIV T cell immune responses, and reduced viral titers and the size of the reactivatable viral reservoir, particularly in conjunction with concurrent antiretroviral therapy (159). In a separate study, HIV infected humanized mice with cART-suppressed viral replication were treated with anti-IFNAR blocking antibodies leading to “blips” in virus reactivation (even while on continued cART) and delayed virus rebound when cART was discontinued (160), suggesting that continued IFN-I signaling during cART suppresses smoldering reservoirs or prevents virus reactivation from latency and in so doing helps to maintain the latent reservoir. These observations in mouse models have generated interest in the potential of blocking IFN-I in the middle of a chronic infection. However, a recent study did not reveal any differences in T cell activation and size of the reservoir following treatment with an engineered IFNα antagonist (177). The treatment did reduce (albeit not completely) the previously elevated IFN-I signature in the infected animals. While reasons for this discrepancy between these animal models are not immediately clear, it is worth pointing out that the studies in humanized mice used a highly effective anti-IFNAR antibody, whereas the SIV study antagonized the frequently targeted IFNα2. It would be informative to test the antiviral role(s) of IFNα2 against SIV in these non-human primate models, to understand the suitability or potential of IFNα2 as a therapeutic target, particularly considering the ineffective antiviral contributions of IFNα2 in models of HIV infection (169, 174). Importantly, this highlights the necessity for greater understanding of the specific role and therapeutic potential of different IFN-I subtypes, which can then be targeted in single or combinatorial blockade approaches.
It must be emphasized though that IFN-I signaling, although having some detrimental effects during chronic infections, still plays critical antiviral roles throughout the infection. Overall, this argues that IFN-Is do not convert from being beneficial to becoming ‘entirely detrimental’, rather it is a delicate balance of the ‘good’ versus the ‘bad’ and further research is required in order to understand the temporal and cell type specific effects of IFN-I signaling that will likely enable us to tease apart these seemingly conflicting roles of IFN-Is in chronic infections.
Selective inhibitors
Our increasing understanding of the variable effects of IFN-I subtypes that drive the different IFN-I induced effects enables targeting of specific stages or molecules within the IFN-I signaling cascade. Two FDA-approved inhibitors of the JAK1/2 kinases (ruxolitinib and tofacitinib approved for myelofibrosis and arthritis, respectively) were shown to potently suppress HIV replication in vitro (178) and ruxolitinib was further demonstrated to inhibit virus replication in a murine model of HIV encephalitis (179). Additionally, ruxolitinib treatment of HIV infected PBMCs in combination with a latency reversing agent (protein kinase C agonist) that specifically induces T cell activation, limited production of multiple inflammatory cytokines IL-6, IL-1α/β, TNFα and IFNγ (180). These JAK inhibitors have also been shown to not only reduce HIV virus production but also reduce the size of the HIV reservoir (181). Further, these inhibitors limit IL-15-induced viral reactivation, which overall resulted in reduced immune activation, and therefore infection of bystander target CD4 T cells in a pSTAT5-dependent manner (181). It is interesting that the potency of these inhibitors improves when they are combined, suggesting they can be manipulated to achieve the desirable affinities or combinatorial effects. However, given that these JAK inhibitors affect very early stages of the JAK/STAT pathway, there remains concern that shutting down IFN-I signaling altogether may dampen responses to opportunistic infections. Indeed, a JAK3 inhibitor, which also targets JAK1/2, was recently shown to lead to increased SIV viral loads in chronically infected rhesus macaques (182, 183). Nonetheless, the therapeutic potential of these inhibitors has generated interest, and they are currently being explored in clinical trials.
In addition to JAK inhibitors, inhibitors of the cGAS-STING pathway have emerged as critical potential therapeutic targets to dampen chronic IFN-I signaling. While potent small molecules targeting cGAS activity have been synthesized, their safety profiles or off-target effects and reduced functionality in cells have dissuaded their further consideration (184, 185). The lessons learnt from these compounds will undoubtedly inform the future design of more potent and specific cGAS inhibitors. In contrast, blocking STING has yielded very promising results, as the compounds are believed to be STING-specific largely due to their covalent association with the transmembrane domain of STING (186). These STING antagonists are potent in human cells, effectively attenuating expression of STING-dependent proinflammatory cytokines (186). While this is an important advance, it still remains to be shown whether other off-target effects for these antagonists exist. Notwithstanding their therapeutic potential, caution must be exercised when inhibiting the cGAS-STING pathway, as defective DNA sensing and STING-mediated antiviral/immune responses may increase susceptibility to infections. It has been argued that targeting the cGAS-STING pathway poses an acceptable risk relative to inhibiting fundamental components of the IFN-I signaling pathway such as IFNAR, JAK/TYK kinases and TBK1. In line with this, cGAS-STING pathway deactivates only a fraction of the PRR sensing pathway, suggesting that non STING-dependent viral sensing and immune responses would remain intact.
Small molecule inhibitors of the inflammatory TBK1 and IKKε kinases have been developed and proven effective against various inflammation-related disease conditions (187–190). Moreover, functional benefits of these inhibitors have also been reported in cancers where the inhibitors blocked oral squamous cell carcinoma xenograft growth, potentiated responses to PD-1 blockade therapies, and sensitized melanomas that were resistant to clinical MEK inhibitors (191–194). However, these inhibitors have been associated with toxic side effects likely stemming from their lack of specificity as well as the fact that the TBK1/IKKε kinases regulate a plethora of critical cellular processes including modulation of Akt-mTOR signaling, DC function, autophagy, apoptosis, IRF activation, NF-kB functions and overall IFN-I signaling events (195–198). In addition, inhibition of TBK1 and IKKε kinases will likely negate the antiviral effects of IFN-Is during the chronic stage of infections, leading to both impairment in virus clearance and enhanced risk of opportunistic viral or bacterial infections given that sensing of multiple pattern recognition pathways and the initial immune defense mechanisms would be compromised.
Consistent with emerging realization that the effects of IFN-Is in chronic viral infections also occur in cancers, some of the strategies being explored in chronic infections to modulate IFN-I signaling are garnering interest in the cancer field. Indeed, targeting IFN-I induced expression of negative regulatory proteins that limit viral and tumor control has been associated with improved outcomes (199). Importantly, activating TLR signaling in combination with checkpoint inhibition (such as PD-1 blockade) during a chronic virus infection was very recently shown to increase the number of virus-specific T cells and restored immune function of exhausted virus-specific T cells, with a corresponding decrease in virus burden (200). Similarly, superior results were obtained from combining checkpoint inhibitors with TLR activation in various cancers (201–203). Fu et al reported that inclusion of STING agonists improved control of tumors that were otherwise resistant to PD-1 blockade monotherapy (204). Very importantly, a recent study reported that blockade of IFN signaling significantly improved responses to either anti-PD1 or anti-CTLA4 monotherapies in melanoma and breast cancer models that originated from tumors that had initially responded to a combination of radiotherapy and anti-CTLA4 but later on relapsed (199). Interestingly, blockade of IFN receptors for both IFN-I and IFN-II led to complete responses to anti-CTLA4, which was remarkable considering that such superior results could not be obtained even when quadruple immune check point inhibitors were combined (199). These striking results were CD8 T cell dependent. Of particular interest, blocking IFN-signaling using JAK inhibitors was also effective, but only when the inhibitors were administered following the immune checkpoint therapy, underscoring the idea that IFNs play important roles at the onset of CD8-mediated antitumor functions, but become less beneficial at later time points (199). Taken together, these studies support the notion that therapies modulating IFN-I signaling have potential in overcoming both chronic virus infections and cancer.
Adaptive Resistance
One challenge that limits the effectiveness of many therapies against chronic viral infection and cancer is adaptive resistance – a phenomenon where an initial response to therapy triggers suppressive mechanisms that then curtails the induced immune response and can limit the long-term functionality required to continue to control virus replication or tumor growth. Genetic mutation is a major mechanism of adaptive resistance common to both chronic infection and cancer (205, 206). Therapeutic intervention can therefore favor the outgrowth or survival of mutants that do not respond to therapy, exacerbating disease or preventing clearance (207). Mutations that promote resistance to therapy include loss of function of IFNAR-associated JAK1/2, rendering infected or tumor cells unresponsive to IFN-Is and IFN-IIs (206). As well, truncating mutations in β2m, an ISG protein associated with antigen presentation, resulted in loss of MHCI surface expression, with possible negative implications in CD8 T cell recognition and therefore resistance to T cell-based therapies (206). Recently, Gao et al demonstrated that resistance to anti-CTLA4 therapy was associated with loss or significant mutations on several genes that regulate IFN-signaling, including IRF1 (207). Moreover, as discussed earlier, chronic infections can downregulate STAT1 proteins, thereby attenuating beneficial antiviral STAT1-dependent signaling outcomes (34). In agreement, in HIV-infected patients, monocytes downregulate IFNAR expression, resulting in impaired IFN-I signaling that associates with increased expression of CD38 by memory CD8 T cells - a measure of disease progression (208). Thus, viral infections appear to induce adaptive resistance measures that dampen or promote desensitization to IFN-I signaling. Interestingly, recent evidence also suggests that human cancers actually mediate deletion of IFN-I genes, which is associated with resistance to checkpoint immunotherapies (209). Such a resistance strategy would shield tumor cells from immune surveillance and IFN-I dependent antitumor effects, ultimately favoring tumorigenesis.
In addition to genetic adaptation, upregulation of host-based immune suppressive pathways is a mechanism of adaptive resistance (199). This occurs when the host activates negative regulatory factors and receptors in response to therapy induced activation. It should be noted that although bad for therapy, the counter-regulation of inflammation with subsequent increase in suppressive pathways is an appropriate immune response to prevent excessive pathology and to ultimately downregulate the immune response against the invading pathogen. Although more widely studied in terms of cancer treatment, the concept of adaptive resistance is analogous to the persistence of HIV despite ART (210). Indeed, expression of exhaustion markers can be predictive of failure of ART to restore immune function (210).
Further, immune checkpoint blockade is currently being explored as a potential treatment to cure and clear HIV infection alongside ART (211). In both chronic viral infections and cancer IFN-Is promote immune escape by facilitating the expression of anti-inflammatory mediators. While it is known that particular IFN signatures can be predictive of resistance to therapy, the mechanisms by which IFN-Is interfere with cancer treatment are areas of active research. Recent studies have shown that IFN-Is induced by radiation therapy are critical for immune activation and tumor control but that they also can lead to the expression of PDL1 (199). Blocking PDL1 after radiation therapy can mitigate the IFN-driven adaptive resistance that occurs (199, 212). The heightened expression of immunoregulatory molecules such as PDL1 on APCs as a result of chronic IFN-I signaling during persisting viral infections is one adaptive resistance mechanism that engages activated (PD1+) antiviral CD8 T cells resulting in subdued or impaired cytotoxic and antiviral functions of these virus-specific CD8 T cells (137). Similarly, the IFN-associated expression of PDL1 in tumors appears positively correlated with infiltrating T cell frequencies, suggesting that PDL1 expression is once again adaptively upregulated so as to evade cytotoxic T cell-mediated immune functions (213). Further, although most of the focus has been on IFN-I induced activation of PDL1, sustained IFN-I signaling can trigger other negative regulatory factors such as IL-10 and IDO and suppressive cell types, which can also serve to dampen T cell responses in both chronic infections and cancers (137, 199, 214, 215). It will be worth pursuing these immunosuppressive factors and cell types as secondary targets for combination checkpoint inhibition therapy to allow for continued immune function in order to promote or sustain virus and/or tumor control.
Following radiation therapy, IFN-Is promote recruitment of lymphocytes to tumor sites, in part by inducing levels of the chemokine receptor CXCR3 that regulates T cell trafficking and function (216). Lim et al showed that IFN-Is were necessary for DC-mediated activation of tumor-reactive CD8 T cells and that a combination of exogenous IFN-I administration and radiation therapy significantly enhanced the potency of tumor-reactive CD8 T cells (216). Lastly, radiotherapy is increasingly being shown to function synergistically with immune-based treatments, such as immune checkpoint inhibition, to mediate “spreading” antitumor effects following localized therapy, in a process referred to as the abscopal effect (217). Given the roles of IFN-Is in immune-based treatments, their roles in these abscopal effects deserve further examination.
In addition to the immune response, it is becoming clear that adaptive resistance triggers unique vulnerabilities in cancer cells that can subsequently be targeted. For example, ER+ breast tumors that become resistant to endocrine treatment simultaneously acquire a sensitivity to TRAIL-mediated cell death (218). Given the strong link between IFN-I signaling and TRAIL-mediated cell death, at least in chronic infections, this suggests that IFN-based therapies may be effective in targeting these otherwise resistant tumors (145). As well, non-small cell lung carcinomas that express high levels of ISGs and are otherwise resistant to therapy respond well to JAK2 inhibition (219). Finally, two recent studies have shown that tumor-derived IFN-Is trigger a unique sensitivity within tumor cells to loss of ADAR1 (214, 220). Ishizuka and colleagues found that loss of function mutations in ADAR1 prevents tumor resistance to checkpoint immunotherapy (214). Meanwhile, Liu et al examined tumor-intrinsic ISG signatures to discover that tumors expressing high levels of ISGs were exquisitely sensitive to loss of ADAR1 (215). These studies suggest that further targeting of ADAR1 could be a strategy to overcome resistance to immunotherapy and a path to explore in chronic infections.
Concluding remarks
IFN-Is play crucial roles in preventing infections, regulating viral replication and eliminating viruses. However, chronic IFN-I signaling sustains immune activation and inflammation which induce immune exhaustion with a resulting inability to clear infection. Thus, as infection progresses, IFN-Is continue to provide critical antiviral functions throughout infection, but simultaneously adapt counterregulatory functions that limit the immune response to prevent excessive immunopathology, but with the long-term effect of sustaining suppressive factors that limit the ability to control chronic infection. Indeed, excessive and prolonged IFN-I signaling is associated with autoimmunity and potentially the autoimmune adverse events observed in cancer following checkpoint blockade (221, 222). It is therefore imperative to dissociate the factors mediating positive outcomes from those driving worsened disease outcomes. To this end, it has become clear that reinforcing IFN-I signaling early during infection is beneficial whereas it is detrimental during the chronic stage of infection. This temporal effect of IFN-I administration therapy warrants further investigation in order to understand the precise governing mechanisms. Identifying the IFN-I subtypes that modulate virus control during the early and late stages of chronic infection and those potentially associated with various “positive” and “negative” functions during viral persistence will be key in harnessing the antiviral activities of IFN-Is while inhibiting the immunosuppressive functions. As well, selectively targeting IFN-I subtypes will circumvent the need to inhibit the entire or global IFN-I signaling pathway, which would otherwise abolish the IFN-I mediated suppression of virus replication during the chronic stage and be critical for preventing secondary infections. Moreover, our increasing understanding of IFN-I signal transduction factors (components of IFN-I signaling pathway, IRFs, ISGs) has identified potential therapeutic targets that can be explored in order to modulate IFN-I signaling during the chronic stage of infections and in cancer. Distinguishing the roles of distinct IRFs and ISGs and their interactions in different contexts will likely improve the specificity of IFN-Is and immune checkpoint-based treatments and may permit individualized treatments. In addition, recent progress identifying de novo vulnerabilities in the face of resistance to therapies ushers a new therapeutic area of focus in order to extend the immune stimulatory functions that may lead to viral and tumor clearance. Further research is needed in this area so as to ensure that these positive immune functions are not achieved at the expense of immune regulatory functions aimed to prevent host tissue damage.
In summary, our understanding of factors mediating IFN-I signaling during chronic infections has significantly advanced IFN-I based approaches towards restoring immune function and ultimately clearing infection or tumors. Notwithstanding this encouraging progress, there are still critical gaps in our appreciation of the potential therapeutic targets, from identification of the targets (IFN-I subtypes, IFN-I signaling modulatory factors, cellular sources and targets of IFN-I, etc) to the molecular structures (IRFs and TBK1- and IKKε-interacting partners) and mechanisms of action (exhaustion markers and the resistance-induced sensitivities) of the potential targets.
Outstanding questions.
What are the specific roles for IFN-I subtypes, and can they be selectively targeted to achieve favorable outcomes in chronic IFN-I signaling, while suppressing the detrimental immunosuppressive functions.
Are IRFs therapeutic targets? Dysregulation of IRFs can lead to either suppression or hyperactivation of immune responses. Can we identify means to target the activities of these factors?
What roles do different IRFs and ISGs play in the context of a chronic infection? Can specific transcriptional networks and/or ISGs be harnessed to modulate IFN-I signaling during the chronic stage of infection to obtain desired outcomes?
Can RNA/DNA sensors or other factors involved in IFN-I signaling be selectively targeted to induce specific, measured effects of IFN-I signaling? Can such an approach selectively extend the strong antiviral responses observed during the early stage of infections into the chronic stage?
How does IFN-I signaling mediate resistance to therapeutic approaches? Can we specifically target these mechanisms while retaining the beneficial roles IFN-Is play in chronic diseases?
ACKNOWLEDGEMENTS
These researchers are supported by the Canadian Institutes of Health Research (CIHR) Foundation Grant FDN148386, the National Institutes of Health (NIH) grant AI085043, the Scotiabank Research Chair to D.G.B, and the Helena Lam Fellowship in Cancer Research Fellowship (S.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, Fisher PB. Interleukin-10 and related cytokines and receptors. Annu Rev Immunol. 2004;22:929–79. [DOI] [PubMed] [Google Scholar]
- 2.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147(927):258–67. [DOI] [PubMed] [Google Scholar]
- 3.Snell LM, McGaha TL, Brooks DG. Type I Interferon in Chronic Virus Infection and Cancer. Trends Immunol. 2017;38(8):542–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, et al. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 2015;347(6219):269–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lazear HM, Nice TJ, Diamond MS. Interferon-lambda: Immune Functions at Barrier Surfaces and Beyond. Immunity. 2015;43(1):15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacquelin B, Mayau V, Targat B, Liovat AS, Kunkel D, Petitjean G, et al. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest. 2009;119(12):3544–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosinger SE, Li Q, Gordon SN, Klatt NR, Duan L, Xu L, et al. Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J Clin Invest. 2009;119(12):3556–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris LD, Tabb B, Sodora DL, Paiardini M, Klatt NR, Douek DC, et al. Downregulation of robust acute type I interferon responses distinguishes nonpathogenic simian immunodeficiency virus (SIV) infection of natural hosts from pathogenic SIV infection of rhesus macaques. J Virol. 2010;84(15):7886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1–20. [DOI] [PubMed] [Google Scholar]
- 10.Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347(6227):aaa2630. [DOI] [PubMed] [Google Scholar]
- 11.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38(5):855–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fensterl V, Chattopadhyay S, Sen GC. No Love Lost Between Viruses and Interferons. Annu Rev Virol. 2015;2(1):549–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–20. [DOI] [PubMed] [Google Scholar]
- 14.Chattopadhyay S, Kuzmanovic T, Zhang Y, Wetzel JL, Sen GC. Ubiquitination of the Transcription Factor IRF-3 Activates RIPA, the Apoptotic Pathway that Protects Mice from Viral Pathogenesis. Immunity. 2016;44(5):1151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazear HM, Lancaster A, Wilkins C, Suthar MS, Huang A, Vick SC, et al. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog. 2013;9(1):e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448(7152):501–5. [DOI] [PubMed] [Google Scholar]
- 17.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11(11):997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12(10):959–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520(7548):553–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguirre S, Luthra P, Sanchez-Aparicio MT, Maestre AM, Patel J, Lamothe F, et al. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat Microbiol. 2017;2:17037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505(7485):691–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding L, Kim H-J, Wang Q, Kearns M, Jiang T, Ohlson CE, et al. PARP Inhibition Elicits STING-Dependent Antitumor Immunity in Brca1-Deficient Ovarian Cancer. Cell Reports. 2018;25(11):2972–80.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 2017;548:466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konno H, Yamauchi S, Berglund A, Putney RM, Mule JJ, Barber GN. Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production. Oncogene. 2018;37(15):2037–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Queiroz N, Xia T, Konno H, Barber GN. Ovarian Cancer Cells Commonly Exhibit Defective STING Signaling Which Affects Sensitivity to Viral Oncolysis. Mol Cancer Res. 2019;17(4):974–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5(5):375–86. [DOI] [PubMed] [Google Scholar]
- 29.Jaitin DA, Schreiber G. Upregulation of a small subset of genes drives type I interferon-induced antiviral memory. J Interferon Cytokine Res. 2007;27(8):653–64. [DOI] [PubMed] [Google Scholar]
- 30.Cheon H, Stark GR. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci U S A. 2009;106(23):9373–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheon H, Holvey-Bates EG, Schoggins JW, Forster S, Hertzog P, Imanaka N, et al. IFNbeta-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 2013;32(20):2751–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uddin S, Lekmine F, Sassano A, Rui H, Fish EN, Platanias LC. Role of Stat5 in type I interferon-signaling and transcriptional regulation. Biochem Biophys Res Commun. 2003;308(2):325–0. [DOI] [PubMed] [Google Scholar]
- 33.Hsu YA, Huang CC, Kung YJ, Lin HJ, Chang CY, Lee KR, et al. The anti-proliferative effects of type I IFN involve STAT6-mediated regulation of SP1 and BCL6. Cancer Lett. 2016;375(2):303–12. [DOI] [PubMed] [Google Scholar]
- 34.Gil MP, Ploquin MJ, Watford WT, Lee SH, Kim K, Wang X, et al. Regulating type 1 IFN effects in CD8 T cells during viral infections: changing STAT4 and STAT1 expression for function. Blood. 2012;120(18):3718–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang WB, Levy DE, Lee CK. STAT3 negatively regulates type I IFN-mediated antiviral response. J Immunol. 2011;187(5):2578–85. [DOI] [PubMed] [Google Scholar]
- 36.Ray JP, Marshall HD, Laidlaw BJ, Staron MM, Kaech SM, Craft J. Transcription factor STAT3 and type I interferons are corepressive insulators for differentiation of follicular helper and T helper 1 cells. Immunity. 2014;40(3):367–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao LJ, He SF, Wang W, Ren H, Qi ZT. Interferon alpha antagonizes STAT3 and SOCS3 signaling triggered by hepatitis C virus. Cytokine. 2016;80:48–55. [DOI] [PubMed] [Google Scholar]
- 38.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, et al. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-beta gene regulatory elements. Cell. 1988;54(6):903–13. [DOI] [PubMed] [Google Scholar]
- 39.Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25(3):349–60. [DOI] [PubMed] [Google Scholar]
- 40.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–84. [DOI] [PubMed] [Google Scholar]
- 41.Fujii Y, Shimizu T, Kusumoto M, Kyogoku Y, Taniguchi T, Hakoshima T. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO J. 1999;18(18):5028–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13(4):539–48. [DOI] [PubMed] [Google Scholar]
- 43.Marie I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17(22):6660–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grandvaux N, Servant MJ, tenOever B, Sen GC, Balachandran S, Barber GN, et al. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76(11):5532–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith S, Gabhann JN, Higgs R, Stacey K, Wahren-Herlenius M, Espinosa A, et al. Enhanced interferon regulatory factor 3 binding to the interleukin-23p19 promoter correlates with enhanced interleukin-23 expression in systemic lupus erythematosus. Arthritis Rheum. 2012;64(5):1601–9. [DOI] [PubMed] [Google Scholar]
- 46.Brownell J, Bruckner J, Wagoner J, Thomas E, Loo YM, Gale M Jr., et al. Direct, interferon-independent activation of the CXCL10 promoter by NF-kappaB and interferon regulatory factor 3 during hepatitis C virus infection. J Virol. 2014;88(3):1582–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu P, Bailey-Bucktrout S, Xi Y, Xu D, Du D, Zhang Q, et al. Innate antiviral host defense attenuates TGF-beta function through IRF3-mediated suppression of Smad signaling. Mol Cell. 2014;56(6):723–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koshiba R, Yanai H, Matsuda A, Goto A, Nakajima A, Negishi H, et al. Regulation of cooperative function of the Il12b enhancer and promoter by the interferon regulatory factors 3 and 5. Biochem Biophys Res Commun. 2013;430(1):95–100. [DOI] [PubMed] [Google Scholar]
- 49.Porritt RA, Hertzog PJ. Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol. 2015;36(3):150–60. [DOI] [PubMed] [Google Scholar]
- 50.van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25(3):361–72. [DOI] [PubMed] [Google Scholar]
- 51.Ashley CL, Abendroth A, McSharry BP, Slobedman B. Interferon-Independent Upregulation of Interferon-Stimulated Genes during Human Cytomegalovirus Infection is Dependent on IRF3 Expression. Viruses. 2019;11(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Negishi H, Fujita Y, Yanai H, Sakaguchi S, Ouyang X, Shinohara M, et al. Evidence for licensing of IFN-gamma-induced IFN regulatory factor 1 transcription factor by MyD88 in Toll-like receptor-dependent gene induction program. Proc Natl Acad Sci U S A. 2006;103(41):15136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levy DE, Marie I, Smith E, Prakash A. Enhancement and diversification of IFN induction by IRF-7-mediated positive feedback. J Interferon Cytokine Res. 2002;22(1):87–93. [DOI] [PubMed] [Google Scholar]
- 54.Rengachari S, Groiss S, Devos JM, Caron E, Grandvaux N, Panne D. Structural basis of STAT2 recognition by IRF9 reveals molecular insights into ISGF3 function. Proc Natl Acad Sci U S A. 2018;115(4):E601–E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nan J, Wang Y, Yang J, Stark GR. IRF9 and unphosphorylated STAT2 cooperate with NF-kappaB to drive IL6 expression. Proc Natl Acad Sci U S A. 2018;115(15):3906–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kimura T, Kadokawa Y, Harada H, Matsumoto M, Sato M, Kashiwazaki Y, et al. Essential and non-redundant roles of p48 (ISGF3 gamma) and IRF-1 in both type I and type II interferon responses, as revealed by gene targeting studies. Genes Cells. 1996;1(1):115–24. [DOI] [PubMed] [Google Scholar]
- 57.Huber M, Suprunenko T, Ashhurst T, Marbach F, Raifer H, Wolff S, et al. IRF9 Prevents CD8(+) T Cell Exhaustion in an Extrinsic Manner during Acute Lymphocytic Choriomeningitis Virus Infection. J Virol. 2017;91(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryzhakov G, Eames HL, Udalova IA. Activation and function of interferon regulatory factor 5. J Interferon Cytokine Res. 2015;35(2):71–8. [DOI] [PubMed] [Google Scholar]
- 59.Steinhagen F, McFarland AP, Rodriguez LG, Tewary P, Jarret A, Savan R, et al. IRF-5 and NF-kappaB p50 co-regulate IFN-beta and IL-6 expression in TLR9-stimulated human plasmacytoid dendritic cells. Eur J Immunol. 2013;43(7):1896–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ouyang X, Negishi H, Takeda R, Fujita Y, Taniguchi T, Honda K. Cooperation between MyD88 and TRIF pathways in TLR synergy via IRF5 activation. Biochem Biophys Res Commun. 2007;354(4):1045–51. [DOI] [PubMed] [Google Scholar]
- 61.Schoenemeyer A, Barnes BJ, Mancl ME, Latz E, Goutagny N, Pitha PM, et al. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. J Biol Chem. 2005;280(17):17005–12. [DOI] [PubMed] [Google Scholar]
- 62.Sy BT, Hoan NX, Tong HV, Meyer CG, Toan NL, Song LH, et al. Genetic variants of interferon regulatory factor 5 associated with chronic hepatitis B infection. World J Gastroenterol. 2018;24(2):248–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fabie A, Mai LT, Dagenais-Lussier X, Hammami A, van Grevenynghe J, Stager S. IRF-5 Promotes Cell Death in CD4 T Cells during Chronic Infection. Cell Rep. 2018;24(5):1163–75. [DOI] [PubMed] [Google Scholar]
- 64.Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol. 2011;12(3):231–8. [DOI] [PubMed] [Google Scholar]
- 65.Sjostrand M, Ambrosi A, Brauner S, Sullivan J, Malin S, Kuchroo VK, et al. Expression of the immune regulator tripartite-motif 21 is controlled by IFN regulatory factors. J Immunol. 2013;191(7):3753–63. [DOI] [PubMed] [Google Scholar]
- 66.Kong HJ, Anderson DE, Lee CH, Jang MK, Tamura T, Tailor P, et al. Cutting edge: autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J Immunol. 2007;179(1):26–30. [DOI] [PubMed] [Google Scholar]
- 67.Sharf R, Azriel A, Lejbkowicz F, Winograd SS, Ehrlich R, Levi BZ. Functional domain analysis of interferon consensus sequence binding protein (ICSBP) and its association with interferon regulatory factors. J Biol Chem. 1995;270(22):13063–9. [DOI] [PubMed] [Google Scholar]
- 68.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, et al. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58(4):729–39. [DOI] [PubMed] [Google Scholar]
- 69.Chae M, Kim K, Park SM, Jang IS, Seo T, Kim DM, et al. IRF-2 regulates NF-kappaB activity by modulating the subcellular localization of NF-kappaB. Biochem Biophys Res Commun. 2008;370(3):519–24. [DOI] [PubMed] [Google Scholar]
- 70.Drew PD, Franzoso G, Carlson LM, Biddison WE, Siebenlist U, Ozato K. Interferon regulatory factor-2 physically interacts with NF-kappa B in vitro and inhibits NF-kappa B induction of major histocompatibility class I and beta 2-microglobulin gene expression in transfected human neuroblastoma cells. J Neuroimmunol. 1995;63(2):157–62. [DOI] [PubMed] [Google Scholar]
- 71.Bovolenta C, Driggers PH, Marks MS, Medin JA, Politis AD, Vogel SN, et al. Molecular interactions between interferon consensus sequence binding protein and members of the interferon regulatory factor family. Proc Natl Acad Sci U S A. 1994;91(11):5046–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rouyez MC, Lestingi M, Charon M, Fichelson S, Buzyn A, Dusanter-Fourt I. IFN regulatory factor-2 cooperates with STAT1 to regulate transporter associated with antigen processing-1 promoter activity. J Immunol. 2005;174(7):3948–58. [DOI] [PubMed] [Google Scholar]
- 73.Hida S, Ogasawara K, Sato K, Abe M, Takayanagi H, Yokochi T, et al. CD8(+) T cell-mediated skin disease in mice lacking IRF-2, the transcriptional attenuator of interferon-alpha/beta signaling. Immunity. 2000;13(5):643–55. [DOI] [PubMed] [Google Scholar]
- 74.Vaughan PS, Aziz F, van Wijnen AJ, Wu S, Harada H, Taniguchi T, et al. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature. 1995;377(6547):362–5. [DOI] [PubMed] [Google Scholar]
- 75.Ren G, Cui K, Zhang Z, Zhao K. Division of labor between IRF1 and IRF2 in regulating different stages of transcriptional activation in cellular antiviral activities. Cell Biosci. 2015;5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun L, Jiang Z, Acosta-Rodriguez VA, Berger M, Du X, Choi JH, et al. HCFC2 is needed for IRF1- and IRF2-dependent Tlr3 transcription and for survival during viral infections. J Exp Med. 2017;214(11):3263–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Negishi H, Ohba Y, Yanai H, Takaoka A, Honma K, Yui K, et al. Negative regulation of Toll-like-receptor signaling by IRF-4. Proc Natl Acad Sci U S A. 2005;102(44):15989–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu D, Meyer F, Ehlers E, Blasnitz L, Zhang L. Interferon regulatory factor 4 (IRF-4) targets IRF-5 to regulate Epstein-Barr virus transformation. J Biol Chem. 2011;286(20):18261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huber M, Lohoff M. IRF4 at the crossroads of effector T-cell fate decision. Eur J Immunol. 2014;44(7):1886–95. [DOI] [PubMed] [Google Scholar]
- 80.Chistiakov DA, Myasoedova VA, Revin VV, Orekhov AN, Bobryshev YV. The impact of interferon-regulatory factors to macrophage differentiation and polarization into M1 and M2. Immunobiology. 2018;223(1):101–11. [DOI] [PubMed] [Google Scholar]
- 81.Grusdat M, McIlwain DR, Xu HC, Pozdeev VI, Knievel J, Crome SQ, et al. IRF4 and BATF are critical for CD8(+) T-cell function following infection with LCMV. Cell Death Differ. 2014;21(7):1050–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Man K, Gabriel SS, Liao Y, Gloury R, Preston S, Henstridge DC, et al. Transcription Factor IRF4 Promotes CD8(+) T Cell Exhaustion and Limits the Development of Memory-like T Cells during Chronic Infection. Immunity. 2017;47(6):1129–41 e5. [DOI] [PubMed] [Google Scholar]
- 83.Marsili G, Perrotti E, Remoli AL, Acchioni C, Sgarbanti M, Battistini A. IFN Regulatory Factors and Antiviral Innate Immunity: How Viruses Can Get Better. J Interferon Cytokine Res. 2016;36(7):41–32. [DOI] [PubMed] [Google Scholar]
- 84.Okumura A, Alce T, Lubyova B, Ezelle H, Strebel K, Pitha PM. HIV-1 accessory proteins VPR and Vif modulate antiviral response by targeting IRF-3 for degradation. Virology. 2008;373(1):85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Irie T, Kiyotani K, Igarashi T, Yoshida A, Sakaguchi T. Inhibition of interferon regulatory factor 3 activation by paramyxovirus V protein. J Virol. 2012;86(13):7136–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cloutier N, Flamand L. Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen inhibits interferon (IFN) beta expression by competing with IFN regulatory factor-3 for binding to IFNB promoter. J Biol Chem. 2010;285(10):7208–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang M, Liu Y, Wang P, Guan X, He S, Luo S, et al. HSV-2 immediate-early protein US1 inhibits IFN-beta production by suppressing association of IRF-3 with IFN-beta promoter. J Immunol. 2015;194(7):3102–15. [DOI] [PubMed] [Google Scholar]
- 88.Rustagi A, Gale M, Jr. Innate antiviral immune signaling, viral evasion and modulation by HIV-1. J Mol Biol. 2014;426(6):1161–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sgarbanti M, Marsili G, Remoli AL, Stellacci E, Mai A, Rotili D, et al. IkappaB kinase epsilon targets interferon regulatory factor 1 in activated T lymphocytes. Mol Cell Biol. 2014;34(6):1054–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Doyle T, Goujon C, Malim MH. HIV-1 and interferons: who’s interfering with whom? Nat Rev Microbiol. 2015;13(7):403–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bailey CC, Zhong G, Huang IC, Farzan M. IFITM-Family Proteins: The Cell’s First Line of Antiviral Defense. Annu Rev Virol. 2014;1:261–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Soper A, Kimura I, Nagaoka S, Konno Y, Yamamoto K, Koyanagi Y, et al. Type I Interferon Responses by HIV-1 Infection: Association with Disease Progression and Control. Front Immunol. 2017;8:1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Usami Y, Wu Y, Gottlinger HG. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature. 2015;526(7572):218–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, et al. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature. 2015;526(7572):212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451(7177):425–30. [DOI] [PubMed] [Google Scholar]
- 97.Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3(4):245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Durfee LA, Lyon N, Seo K, Huibregtse JM. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol Cell. 2010;38(5):722–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Okumura F, Okumura AJ, Uematsu K, Hatakeyama S, Zhang DE, Kamura T. Activation of double-stranded RNA-activated protein kinase (PKR) by interferon-stimulated gene 15 (ISG15) modification down-regulates protein translation. J Biol Chem. 2013;288(4):2839–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shi HX, Yang K, Liu X, Liu XY, Wei B, Shan YF, et al. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol Cell Biol. 2010;30(10):2424–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang Y, Swiecki M, Cella M, Alber G, Schreiber RD, Gilfillan S, et al. Timing and magnitude of type I interferon responses by distinct sensors impact CD8 T cell exhaustion and chronic viral infection. Cell Host Microbe. 2012;11(6):631–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472(7344):481–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Constantinescu SN, Croze E, Wang C, Murti A, Basu L, Mullersman JE, et al. Role of interferon alpha/beta receptor chain 1 in the structure and transmembrane signaling of the interferon alpha/beta receptor complex. Proc Natl Acad Sci U S A. 1994;91(20):9602–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Constantinescu SN, Croze E, Murti A, Wang C, Basu L, Hollander D, et al. Expression and signaling specificity of the IFNAR chain of the type I interferon receptor complex. Proc Natl Acad Sci U S A. 1995;92(23):10487–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bohmer FD, Friedrich K. Protein tyrosine phosphatases as wardens of STAT signaling. JAKSTAT. 2014;3(1):e28087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fenner JE, Starr R, Cornish AL, Zhang JG, Metcalf D, Schreiber RD, et al. Suppressor of cytokine signaling 1 regulates the immune response to infection by a unique inhibition of type I interferon activity. Nat Immunol. 2006;7(1):33–9. [DOI] [PubMed] [Google Scholar]
- 107.Song MM, Shuai K. The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins inhibit interferon-mediated antiviral and antiproliferative activities. J Biol Chem. 1998;273(52):35056–62. [DOI] [PubMed] [Google Scholar]
- 108.Villarroya-Beltri C, Guerra S, Sanchez-Madrid F. ISGylation - a key to lock the cell gates for preventing the spread of threats. J Cell Sci. 2017;130(18):2961–9. [DOI] [PubMed] [Google Scholar]
- 109.Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KG, Fuchs SY, et al. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25(11):2358–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Arakura F, Hida S, Ichikawa E, Yajima C, Nakajima S, Saida T, et al. Genetic control directed toward spontaneous IFN-alpha/IFN-beta responses and downstream IFN-gamma expression influences the pathogenesis of a murine psoriasis-like skin disease. J Immunol. 2007;179(5):3249–57. [DOI] [PubMed] [Google Scholar]
- 111.Gough DJ, Messina NL, Clarke CJ, Johnstone RW, Levy DE. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 2012;36(2):166–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39(2):372–85. [DOI] [PubMed] [Google Scholar]
- 113.Fallarino F, Grohmann U, You S, McGrath BC, Cavener DR, Vacca C, et al. The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J Immunol. 2006;176(11):6752–61. [DOI] [PubMed] [Google Scholar]
- 114.Ghosh A, Shao L, Sampath P, Zhao B, Patel NV, Zhu J, et al. Oligoadenylate-Synthetase-Family Protein OASL Inhibits Activity of the DNA Sensor cGAS during DNA Virus Infection to Limit Interferon Production. Immunity. 2019;50(1):51–63 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nie Y, Hammond GL, Yang JH. Double-stranded RNA deaminase ADAR1 increases host susceptibility to virus infection. J Virol. 2007;81(2):917–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hu Y, Wang J, Yang B, Zheng N, Qin M, Ji Y, et al. Guanylate binding protein 4 negatively regulates virus-induced type I IFN and antiviral response by targeting IFN regulatory factor 7. J Immunol. 2011;187(12):6456–62. [DOI] [PubMed] [Google Scholar]
- 117.Sandler NG, Bosinger SE, Estes JD, Zhu RT, Tharp GK, Boritz E, et al. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511(7511):601–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wilson EB, Yamada DH, Elsaesser H, Herskovitz J, Deng J, Cheng G, et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 2013;340(6129):202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Teijaro JR, Ng C, Lee AM, Sullivan BM, Sheehan KC, Welch M, et al. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 2013;340(6129):207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zuniga EI, Liou LY, Mack L, Mendoza M, Oldstone MB. Persistent virus infection inhibits type I interferon production by plasmacytoid dendritic cells to facilitate opportunistic infections. Cell Host Microbe. 2008;4(4):374–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wonderlich ER, Barratt-Boyes SM. SIV infection of rhesus macaques differentially impacts mononuclear phagocyte responses to virus-derived TLR agonists. J Med Primatol. 2013;42(5):247–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Snell LM, MacLeod BL, Law JC, Osokine I, Elsaesser HJ, Hezaveh K, et al. CD8(+) T Cell Priming in Established Chronic Viral Infection Preferentially Directs Differentiation of Memory-like Cells for Sustained Immunity. Immunity. 2018;49(4):678–94 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rotger M, Dalmau J, Rauch A, McLaren P, Bosinger SE, Martinez R, et al. Comparative transcriptomics of extreme phenotypes of human HIV-1 infection and SIV infection in sooty mangabey and rhesus macaque. J Clin Invest. 2011;121(6):2391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Herbeuval JP, Nilsson J, Boasso A, Hardy AW, Kruhlak MJ, Anderson SA, et al. Differential expression of IFN-alpha and TRAIL/DR5 in lymphoid tissue of progressor versus nonprogressor HIV-1-infected patients. Proc Natl Acad Sci U S A. 2006;103(18):7000–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fernandez S, Tanaskovic S, Helbig K, Rajasuriar R, Kramski M, Murray JM, et al. CD4+ T-cell deficiency in HIV patients responding to antiretroviral therapy is associated with increased expression of interferon-stimulated genes in CD4+ T cells. J Infect Dis. 2011;204(12):1927–35. [DOI] [PubMed] [Google Scholar]
- 126.Stylianou E, Aukrust P, Bendtzen K, Muller F, Froland SS. Interferons and interferon (IFN)-inducible protein 10 during highly active anti-retroviral therapy (HAART)-possible immunosuppressive role of IFN-alpha in HIV infection. Clin Exp Immunol. 2000;119(3):479–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Osokine I, Snell LM, Cunningham CR, Yamada DH, Wilson EB, Elsaesser HJ, et al. Type I interferon suppresses de novo virus-specific CD4 Th1 immunity during an established persistent viral infection. Proc Natl Acad Sci U S A. 2014;111(20):7409–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cervantes-Barragan L, Lewis KL, Firner S, Thiel V, Hugues S, Reith W, et al. Plasmacytoid dendritic cells control T-cell response to chronic viral infection. Proc Natl Acad Sci U S A. 2012;109(8):3012–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Montoya M, Schiavoni G, Mattei F, Gresser I, Belardelli F, Borrow P, et al. Type I interferons produced by dendritic cells promote their phenotypic and functional activation. Blood. 2002;99(9):3263–71. [DOI] [PubMed] [Google Scholar]
- 130.Chang YH, Wang KC, Chu KL, Clouthier DL, Tran AT, Torres Perez MS, et al. Dichotomous Expression of TNF Superfamily Ligands on Antigen-Presenting Cells Controls Post-priming Anti-viral CD4(+) T Cell Immunity. Immunity. 2017;47(5):943–58 e9. [DOI] [PubMed] [Google Scholar]
- 131.Xu HC, Grusdat M, Pandyra AA, Polz R, Huang J, Sharma P, et al. Type I interferon protects antiviral CD8+ T cells from NK cell cytotoxicity. Immunity. 2014;40(6):949–60. [DOI] [PubMed] [Google Scholar]
- 132.Waggoner SN, Cornberg M, Selin LK, Welsh RM. Natural killer cells act as rheostats modulating antiviral T cells. Nature. 2011;481(7381):394–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Crouse J, Bedenikovic G, Wiesel M, Ibberson M, Xenarios I, Von Laer D, et al. Type I interferons protect T cells against NK cell attack mediated by the activating receptor NCR1. Immunity. 2014;40(6):961–73. [DOI] [PubMed] [Google Scholar]
- 134.Le Bon A, Etchart N, Rossmann C, Ashton M, Hou S, Gewert D, et al. Cross-priming of CD8+ T cells stimulated by virus-induced type I interferon. Nat Immunol. 2003;4(10):1009–15. [DOI] [PubMed] [Google Scholar]
- 135.Lee MS, Park CH, Jeong YH, Kim YJ, Ha SJ. Negative regulation of type I IFN expression by OASL1 permits chronic viral infection and CD8(+) T-cell exhaustion. PLoS Pathog. 2013;9(7):e1003478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Swiecki M, Wang Y, Vermi W, Gilfillan S, Schreiber RD, Colonna M. Type I interferon negatively controls plasmacytoid dendritic cell numbers in vivo. J Exp Med. 2011;208(12):2367–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cunningham CR, Champhekar A, Tullius MV, Dillon BJ, Zhen A, de la Fuente JR, et al. Type I and Type II Interferon Coordinately Regulate Suppressive Dendritic Cell Fate and Function during Viral Persistence. PLoS Pathog. 2016;12(1):e1005356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Boasso A, Hardy AW, Landay AL, Martinson JL, Anderson SA, Dolan MJ, et al. PDL-1 upregulation on monocytes and T cells by HIV via type I interferon: restricted expression of type I interferon receptor by CCR5-expressing leukocytes. Clin Immunol. 2008;129(1):132–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wilson EB, Kidani Y, Elsaesser H, Barnard J, Raff L, Karp CL, et al. Emergence of distinct multiarmed immunoregulatory antigen-presenting cells during persistent viral infection. Cell Host Microbe. 2012;11(5):481–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rempel H, Sun B, Calosing C, Pillai SK, Pulliam L. Interferon-alpha drives monocyte gene expression in chronic unsuppressed HIV-1 infection. AIDS. 2010;24(10):1415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sedaghat AR, German J, Teslovich TM, Cofrancesco J Jr., Jie CC, Talbot CC Jr., et al. Chronic CD4+ T-cell activation and depletion in human immunodeficiency virus type 1 infection: type I interferon-mediated disruption of T-cell dynamics. J Virol. 2008;82(4):1870–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fraietta JA, Mueller YM, Yang G, Boesteanu AC, Gracias DT, Do DH, et al. Type I interferon upregulates Bak and contributes to T cell loss during human immunodeficiency virus (HIV) infection. PLoS Pathog. 2013;9(10):e1003658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cheng L, Yu H, Li G, Li F, Ma J, Li J, et al. Type I interferons suppress viral replication but contribute to T cell depletion and dysfunction during chronic HIV-1 infection. JCI Insight. 2017;2(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Herbeuval JP, Hardy AW, Boasso A, Anderson SA, Dolan MJ, Dy M, et al. Regulation of TNF-related apoptosis-inducing ligand on primary CD4+ T cells by HIV-1: role of type I IFN-producing plasmacytoid dendritic cells. Proc Natl Acad Sci U S A. 2005;102(39):13974–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Herbeuval JP, Grivel JC, Boasso A, Hardy AW, Chougnet C, Dolan MJ, et al. CD4+ T-cell death induced by infectious and noninfectious HIV-1: role of type 1 interferon-dependent, TRAIL/DR5-mediated apoptosis. Blood. 2005;106(10):3524–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Snell LM, Osokine I, Yamada DH, De la Fuente JR, Elsaesser HJ, Brooks DG. Overcoming CD4 Th1 Cell Fate Restrictions to Sustain Antiviral CD8 T Cells and Control Persistent Virus Infection. Cell Rep. 2016;16(12):3286–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Metidji A, Rieder SA, Glass DD, Cremer I, Punkosdy GA, Shevach EM. IFN-alpha/beta receptor signaling promotes regulatory T cell development and function under stress conditions. J Immunol. 2015;194(9):4265–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lee SE, Li X, Kim JC, Lee J, Gonzalez-Navajas JM, Hong SH, et al. Type I interferons maintain Foxp3 expression and T-regulatory cell functions under inflammatory conditions in mice. Gastroenterology. 2012;143(1):145–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Srivastava S, Koch MA, Pepper M, Campbell DJ. Type I interferons directly inhibit regulatory T cells to allow optimal antiviral T cell responses during acute LCMV infection. J Exp Med. 2014;211(5):961–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gangaplara A, Martens C, Dahlstrom E, Metidji A, Gokhale AS, Glass DD, et al. Type I interferon signaling attenuates regulatory T cell function in viral infection and in the tumor microenvironment. PLoS Pathog. 2018;14(4):e1006985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kawano Y, Zavidij O, Park J, Moschetta M, Kokubun K, Mouhieddine TH, et al. Blocking IFNAR1 inhibits multiple myeloma-driven Treg expansion and immunosuppression. J Clin Invest. 2018;128(6):2487–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Penaloza-MacMaster P, Kamphorst AO, Wieland A, Araki K, Iyer SS, West EE, et al. Interplay between regulatory T cells and PD-1 in modulating T cell exhaustion and viral control during chronic LCMV infection. J Exp Med. 2014;211(9):1905–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Welsh RM, Bahl K, Marshall HD, Urban SL. Type 1 interferons and antiviral CD8 T-cell responses. PLoS Pathog. 2012;8(1):e1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Knuschke T, Rotan O, Bayer W, Kollenda S, Dickow J, Sutter K, et al. Induction of Type I Interferons by Therapeutic Nanoparticle-Based Vaccination Is Indispensable to Reinforce Cytotoxic CD8(+) T Cell Responses During Chronic Retroviral Infection. Front Immunol. 2018;9:614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Curtsinger JM, Valenzuela JO, Agarwal P, Lins D, Mescher MF. Type I IFNs provide a third signal to CD8 T cells to stimulate clonal expansion and differentiation. J Immunol. 2005;174(8):4465–9. [DOI] [PubMed] [Google Scholar]
- 156.Kolumam GA, Thomas S, Thompson LJ, Sprent J, Murali-Krishna K. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J Exp Med. 2005;202(5):637–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gibbert K, Dietze KK, Zelinskyy G, Lang KS, Barchet W, Kirschning CJ, et al. Polyinosinic-polycytidylic acid treatment of Friend retrovirus-infected mice improves functional properties of virus-specific T cells and prevents virus-induced disease. J Immunol. 2010;185(10):6179–89. [DOI] [PubMed] [Google Scholar]
- 158.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15(8):486–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhen A, Rezek V, Youn C, Lam B, Chang N, Rick J, et al. Targeting type I interferon-mediated activation restores immune function in chronic HIV infection. J Clin Invest. 2017;127(1):260–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cheng L, Ma J, Li J, Li D, Li G, Li F, et al. Blocking type I interferon signaling enhances T cell recovery and reduces HIV-1 reservoirs. J Clin Invest. 2017;127(1):269–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016;537(7620):417–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, et al. T Cell Factor 1-Expressing Memory-like CD8(+) T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity. 2016;45(2):415–27. [DOI] [PubMed] [Google Scholar]
- 163.Hesse C, Kollenda S, Rotan O, Pastille E, Adamczyk A, Wenzek C, et al. A tumor-peptide based nanoparticle vaccine elicits efficient tumor growth control in anti-tumor immunotherapy. Mol Cancer Ther. 2019. [DOI] [PubMed] [Google Scholar]
- 164.Bosinger SE, Utay NS. Type I interferon: understanding its role in HIV pathogenesis and therapy. Curr HIV/AIDS Rep. 2015;12(1):41–53. [DOI] [PubMed] [Google Scholar]
- 165.Tavel JA, Huang CY, Shen J, Metcalf JA, Dewar R, Shah A, et al. Interferon-alpha produces significant decreases in HIV load. J Interferon Cytokine Res. 2010;30(7):461–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Azzoni L, Foulkes AS, Papasavvas E, Mexas AM, Lynn KM, Mounzer K, et al. Pegylated Interferon alfa-2a monotherapy results in suppression of HIV type 1 replication and decreased cell-associated HIV DNA integration. J Infect Dis. 2013;207(2):213–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Moron-Lopez S, Gomez-Mora E, Salgado M, Ouchi D, Puertas MC, Urrea V, et al. Short-term Treatment With Interferon Alfa Diminishes Expression of HIV-1 and Reduces CD4+ T-Cell Activation in Patients Coinfected With HIV and Hepatitis C Virus and Receiving Antiretroviral Therapy. J Infect Dis. 2016;213(6):1008–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hardy GA, Sieg S, Rodriguez B, Anthony D, Asaad R, Jiang W, et al. Interferon-alpha is the primary plasma type-I IFN in HIV-1 infection and correlates with immune activation and disease markers. PLoS One. 2013;8(2):e56527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lavender KJ, Gibbert K, Peterson KE, Van Dis E, Francois S, Woods T, et al. Interferon Alpha Subtype-Specific Suppression of HIV-1 Infection In Vivo. J Virol. 2016;90(13):6001–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Harper MS, Guo K, Gibbert K, Lee EJ, Dillon SM, Barrett BS, et al. Interferon-alpha Subtypes in an Ex Vivo Model of Acute HIV-1 Infection: Expression, Potency and Effector Mechanisms. PLoS Pathog. 2015;11(11):e1005254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Thomas C, Moraga I, Levin D, Krutzik PO, Podoplelova Y, Trejo A, et al. Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell. 2011;146(4):621–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jaks E, Gavutis M, Uze G, Martal J, Piehler J. Differential receptor subunit affinities of type I interferons govern differential signal activation. J Mol Biol. 2007;366(2):525–39. [DOI] [PubMed] [Google Scholar]
- 173.Lavoie TB, Kalie E, Crisafulli-Cabatu S, Abramovich R, DiGioia G, Moolchan K, et al. Binding and activity of all human alpha interferon subtypes. Cytokine. 2011;56(2):282–9. [DOI] [PubMed] [Google Scholar]
- 174.Abraham S, Choi JG, Ortega NM, Zhang J, Shankar P, Swamy NM. Gene therapy with plasmids encoding IFN-beta or IFN-alpha14 confers long-term resistance to HIV-1 in humanized mice. Oncotarget. 2016;7(48):78412–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ng CT, Sullivan BM, Teijaro JR, Lee AM, Welch M, Rice S, et al. Blockade of interferon Beta, but not interferon alpha, signaling controls persistent viral infection. Cell Host Microbe. 2015;17(5):653–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pham TN, Lukhele S, Dallaire F, Perron G, Cohen EA. Enhancing Virion Tethering by BST2 Sensitizes Productively and Latently HIV-infected T cells to ADCC Mediated by Broadly Neutralizing Antibodies. Sci Rep. 2016;6:37225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Nganou-Makamdop K, Billingsley JM, Yaffe Z, O’Connor G, Tharp GK, Ransier A, et al. Type I IFN signaling blockade by a PASylated antagonist during chronic SIV infection suppresses specific inflammatory pathways but does not alter T cell activation or virus replication. PLoS Pathog. 2018;14(8):e1007246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gavegnano C, Detorio M, Montero C, Bosque A, Planelles V, Schinazi RF. Ruxolitinib and tofacitinib are potent and selective inhibitors of HIV-1 replication and virus reactivation in vitro. Antimicrob Agents Chemother. 2014;58(4):1977–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Haile PA, Votta BJ, Marquis RW, Bury MJ, Mehlmann JF, Singhaus R Jr., et al. The Identification and Pharmacological Characterization of 6-(tert-Butylsulfonyl)-N-(5-fluoro-1H-indazol-3-yl)quinolin-4-amine (GSK583), a Highly Potent and Selective Inhibitor of RIP2 Kinase. J Med Chem. 2016;59(10):4867–80. [DOI] [PubMed] [Google Scholar]
- 180.Spivak AM, Larragoite ET, Coletti ML, Macedo AB, Martins LJ, Bosque A, et al. Janus kinase inhibition suppresses PKC-induced cytokine release without affecting HIV-1 latency reversal ex vivo. Retrovirology. 2016;13(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gavegnano C, Brehm JH, Dupuy FP, Talla A, Ribeiro SP, Kulpa DA, et al. Novel mechanisms to inhibit HIV reservoir seeding using Jak inhibitors. PLoS Pathog. 2017;13(12):e1006740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Takahashi Y, Mayne AE, Khowawisetsut L, Pattanapanyasat K, Little D, Villinger F, et al. In vivo administration of a JAK3 inhibitor to chronically siv infected rhesus macaques leads to NK cell depletion associated with transient modest increase in viral loads. PLoS One. 2013;8(7):e70992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Takahashi Y, Byrareddy SN, Albrecht C, Brameier M, Walter L, Mayne AE, et al. In vivo administration of a JAK3 inhibitor during acute SIV infection leads to significant increases in viral load during chronic infection. PLoS Pathog. 2014;10(3):e1003929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Vincent J, Adura C, Gao P, Luz A, Lama L, Asano Y, et al. Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice. Nat Commun. 2017;8(1):750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hall J, Brault A, Vincent F, Weng S, Wang H, Dumlao D, et al. Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay. PLoS One. 2017;12(9):e0184843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Haag SM, Gulen MF, Reymond L, Gibelin A, Abrami L, Decout A, et al. Targeting STING with covalent small-molecule inhibitors. Nature. 2018;559(7713):269–73. [DOI] [PubMed] [Google Scholar]
- 187.Reilly SM, Chiang SH, Decker SJ, Chang L, Uhm M, Larsen MJ, et al. An inhibitor of the protein kinases TBK1 and IKK-varepsilon improves obesity-related metabolic dysfunctions in mice. Nat Med. 2013;19(3):313–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Oral EA, Reilly SM, Gomez AV, Meral R, Butz L, Ajluni N, et al. Inhibition of IKKvarepsilon and TBK1 Improves Glucose Control in a Subset of Patients with Type 2 Diabetes. Cell Metab. 2017;26(1):157–70 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Louis C, Ngo D, D’Silva DB, Hansen J, Phillipson L, Jousset H, et al. Therapeutic Effects of a TANK-Binding Kinase 1 Inhibitor in Germinal Center-Driven Collagen-Induced Arthritis. Arthritis Rheumatol. 2019;71(1):50–62. [DOI] [PubMed] [Google Scholar]
- 190.Bodewes ILA, Huijser E, van Helden-Meeuwsen CG, Tas L, Huizinga R, Dalm V, et al. TBK1: A key regulator and potential treatment target for interferon positive Sjogren’s syndrome, systemic lupus erythematosus and systemic sclerosis. J Autoimmun. 2018;91:97–102. [DOI] [PubMed] [Google Scholar]
- 191.Bai LY, Chiu CF, Kapuriya NP, Shieh TM, Tsai YC, Wu CY, et al. BX795, a TBK1 inhibitor, exhibits antitumor activity in human oral squamous cell carcinoma through apoptosis induction and mitotic phase arrest. Eur J Pharmacol. 2015;769:287–96. [DOI] [PubMed] [Google Scholar]
- 192.Jenkins RW, Aref AR, Lizotte PH, Ivanova E, Stinson S, Zhou CW, et al. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer Discov. 2018;8(2):196–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Eskiocak B, McMillan EA, Mendiratta S, Kollipara RK, Zhang H, Humphries CG, et al. Biomarker Accessible and Chemically Addressable Mechanistic Subtypes of BRAF Melanoma. Cancer Discov. 2017;7(8):832–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547(7664):413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Xiao Y, Zou Q, Xie X, Liu T, Li HS, Jie Z, et al. The kinase TBK1 functions in dendritic cells to regulate T cell homeostasis, autoimmunity, and antitumor immunity. J Exp Med. 2017;214(5):1493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Yu J, Zhou X, Chang M, Nakaya M, Chang JH, Xiao Y, et al. Regulation of T-cell activation and migration by the kinase TBK1 during neuroinflammation. Nat Commun. 2015;6:6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37(2):223–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Verhelst K, Verstrepen L, Carpentier I, Beyaert R. IkappaB kinase epsilon (IKKepsilon): a therapeutic target in inflammation and cancer. Biochem Pharmacol. 2013;85(7):873–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell. 2016;167(6):1540–54 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang Y, Chung YR, Eitzinger S, Palacio N, Gregory S, Bhattacharyya M, et al. TLR4 signaling improves PD-1 blockade therapy during chronic viral infection. PLoS Pathog. 2019;15(2):e1007583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sato-Kaneko F, Yao S, Ahmadi A, Zhang SS, Hosoya T, Kaneda MM, et al. Combination immunotherapy with TLR agonists and checkpoint inhibitors suppresses head and neck cancer. JCI Insight. 2017;2(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sagiv-Barfi I, Czerwinski DK, Levy S, Alam IS, Mayer AT, Gambhir SS, et al. Eradication of spontaneous malignancy by local immunotherapy. Sci Transl Med. 2018;10(426). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wang D, Jiang W, Zhu F, Mao X, Agrawal S. Modulation of the tumor microenvironment by intratumoral administration of IMO-2125, a novel TLR9 agonist, for cancer immunotherapy. Int J Oncol. 2018;53(3):1193–203. [DOI] [PubMed] [Google Scholar]
- 204.Fu J, Kanne DB, Leong M, Glickman LH, McWhirter SM, Lemmens E, et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci Transl Med. 2015;7(283):283ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Martinez-Picado J, DePasquale MP, Kartsonis N, Hanna GJ, Wong J, Finzi D, et al. Antiretroviral resistance during successful therapy of HIV type 1 infection. Proceedings of the National Academy of Sciences. 2000;97(20):10948–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. New England Journal of Medicine. 2016;375(9):819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, et al. Loss of IFN-gamma Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell. 2016;167(2):397–404 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hardy GA, Sieg SF, Rodriguez B, Jiang W, Asaad R, Lederman MM, et al. Desensitization to type I interferon in HIV-1 infection correlates with markers of immune activation and disease progression. Blood. 2009;113(22):5497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ye Z, Dong H, Li Y, Ma T, Huang H, Leong HS, et al. Prevalent Homozygous Deletions of Type I Interferon and Defensin Genes in Human Cancers Associate with Immunotherapy Resistance. Clin Cancer Res. 2018;24(14):3299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Shive CL, Clagett B, McCausland MR, Mudd JC, Funderburg NT, Freeman ML, et al. Inflammation Perturbs the IL-7 Axis, Promoting Senescence and Exhaustion that Broadly Characterize Immune Failure in Treated HIV Infection. Journal of acquired immune deficiency syndromes (1999). 2016;71(5):483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wykes MN, Lewin SR. Immune checkpoint blockade in infectious diseases. Nature Reviews Immunology. 2017;18:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Twyman-Saint Victor C, Rech AJ, Maity A, Rengan R, Pauken KE, Stelekati E, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015;520(7547):373–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell. 2017;168(4):707–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ishizuka JJ, Manguso RT, Cheruiyot CK, Bi K, Panda A, Iracheta-Vellve A, et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature. 2019;565(7737):43–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Liu H, Golji J, Brodeur LK, Chung FS, Chen JT, deBeaumont RS, et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat Med. 2019;25(1):95–102. [DOI] [PubMed] [Google Scholar]
- 216.Lim JY, Gerber SA, Murphy SP, Lord EM. Type I interferons induced by radiation therapy mediate recruitment and effector function of CD8(+) T cells. Cancer Immunol Immunother. 2014;63(3):25–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Reynders K, Illidge T, Siva S, Chang JY, De Ruysscher D. The abscopal effect of local radiotherapy: using immunotherapy to make a rare event clinically relevant. Cancer Treat Rev. 2015;41(6):503–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Piggott L, Silva A, Robinson T, Santiago-Gomez A, Simoes BM, Becker M, et al. Acquired Resistance of ER-Positive Breast Cancer to Endocrine Treatment Confers an Adaptive Sensitivity to TRAIL through Posttranslational Downregulation of c-FLIP. Clin Cancer Res. 2018;24(10):2452–63. [DOI] [PubMed] [Google Scholar]
- 219.Pitroda SP, Stack ME, Liu GF, Song SS, Chen L, Liang H, et al. JAK2 Inhibitor SAR302503 Abrogates PD-L1 Expression and Targets Therapy-Resistant Non-small Cell Lung Cancers. Mol Cancer Ther. 2018;17(4):732–9. [DOI] [PubMed] [Google Scholar]
- 220.Liu H, Golji J, Brodeur LK, Chung FS, Chen JT, deBeaumont RS, et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nature Medicine. 2019;25(1):95–102. [DOI] [PubMed] [Google Scholar]
- 221.Sanmamed MF, Chen L. A Paradigm Shift in Cancer Immunotherapy: From Enhancement to Normalization. Cell. 2018;175(2):313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Walsh SR, Bastin D, Chen L, Nguyen A, Storbeck CJ, Lefebvre C, et al. Type I IFN blockade uncouples immunotherapy-induced antitumor immunity and autoimmune toxicity. The Journal of Clinical Investigation. 2018;129(2). [DOI] [PMC free article] [PubMed] [Google Scholar]