Abstract
Background
Antibody Fc-mediated functions, such as antibody-dependent cellular cytotoxicity, contribute to vaccine-induced protection against viral infections. Fc-mediated function of anti-Ebola glycoprotein (GP) antibodies suggest that Fc-dependent activation of effector cells, including natural killer (NK) cells, could play a role in vaccination against Ebola virus disease.
Methods
We analyzed the effect on primary human NK cell activation of anti-Ebola GP antibody in the serum of United Kingdom–based volunteers vaccinated with the novel 2-dose heterologous adenovirus type 26.ZEBOV, modified vaccinia Ankara–BN-Filo vaccine regimen.
Results
We demonstrate primary human NK cell CD107a and interferon γ expression, combined with down-regulation of CD16, in response to recombinant Ebola virus GP and post-vaccine dose 1 and dose 2 serum samples. These responses varied significantly with vaccine regimen, and NK cell activation was found to correlate with anti-GP antibody concentration. We also reveal an impact of NK cell differentiation phenotype on antibody-dependent NK cell activation, with highly differentiated CD56dimCD57+ NK cells being the most responsive.
Conclusions
These findings highlight the dual importance of vaccine-induced antibody concentration and NK cell differentiation status in promoting Fc-mediated activation of NK cells after vaccination, raising a potential role for antibody-mediated NK cell activation in vaccine-induced immune responses.
Keywords: antibody, Ebola, vaccine, natural killer cell
We found natural killer cell degranulation and interferon γ secretion in response to recombinant Ebola virus glycoprotein and postvaccination serum samples from healthy volunteers vaccinated with the novel 2-dose heterologous adenovirus type 26.ZEBOV, modified vaccinia Ankara–BN-Filo Ebola vaccine.
Determining correlates of protection for Ebola vaccines has proved difficult and ambiguous [1]. Anti-Ebola antibodies possess strong neutralizing capacity [2, 3]; moreover, antibodies with limited neutralizing activity were protective in animal models and human in vitro culture systems, suggesting that neutralization alone presents an incomplete mechanistic picture of in vivo protection [3–5]. Ebola-specific antibodies induce antibody-dependent cellular cytotoxicity (ADCC) in human peripheral blood natural killer (NK) cells and NK cell lines in vitro; in light of this, Fc-mediated function in anti-Ebola monoclonal antibody therapy and vaccine-induced protection is gaining in interest [5–7].
Analysis of the primary response to the candidate Ebola vaccine, rVSV-ZEBOV, revealed a correlation between early NK cell activation and anti-Ebola antibody titer [8]. Furthermore, protection of nonhuman primates against Ebola virus (EBOV) challenge is associated with a low immunoglobulin (Ig) G2/IgG1 antibody isotype ratio, compatible with ADCC as a major mechanism of protection [9]. In murine experimental filovirus vaccines, induction of anti–glycoprotein (GP) antibodies with robust ADCC function was critical for protection [10–12]. Taken together, these studies suggest that Fc functions of anti-Ebola antibodies may contribute to protection and may be exploited for improving vaccine and therapeutic monoclonal antibody efficacy and as markers of vaccine-induced immunity.
NK cells, like other innate immune effector cells, express Fc receptors (FcRs) on their surface, allowing activation of cell-mediated antibody-dependent antiviral functions [13]. Antibody-dependent phagocytosis of virus or virally infected cells by monocytes, macrophages, and neutrophils, along with ADCC mediated by NK cells, promotes the clearance of infected cells, reducing viral load and dissemination. NK cell ADCC is principally mediated by cross-linking of FcγRIIIa (CD16) by the Fc region of immunoglobulins—subclasses IgG1 and IgG3 in humans—which leads to phosphorylation of immunoreceptor tyrosine-based activation motifs and downstream pathway activation.
Killing proceeds by releasing lytic granules from activated NK cells, inducing apoptosis of virally infected cells. Cross-linking of CD16 by antibody induces its cleavage from the NK cell surface [14–16], but despite this, NK cells can move on to kill multiple targets, providing effective clearance of infected cells [17]. Fc functions of broadly neutralizing antibodies have been shown to be indispensable for protection against influenza virus infection [18, 19]; however, the role of Ebola vaccine–induced antibody-dependent NK cell functions is unknown.
The novel adenovirus type 26 (Ad26).ZEBOV, modified vaccinia Ankara (MVA)–BN-Filo 2-dose vaccine regimen has shown promising results in phase 1 and 2 studies; high levels of anti-Ebola GP–specific antibody are sustained for at least 360 days with high neutralizing activity and a strong correlation between binding and neutralizing antibody responses [20–23]. However, vaccine regimens based on Ad26.ZEBOV and MVA-BN-Filo and differing in order and interval between doses 1 and 2 induced substantially different serum antibody concentrations in United Kingdom–based volunteers at both post–dose 1 and post–dose 2 time points [21].
Therefore, the purpose of the current study was to assess the ability of post-Ad26.ZEBOV, MVA-BN-Filo vaccination serum samples (with differing regimens) to mediate antibody-dependent NK cell function in an in vitro ADCC assay targeting immobilized EBOV GP. We observed robust, antibody-dependent activation of NK cells in whole human peripheral blood mononuclear cell (PBMC) preparations cultured with EBOV GP in the presence of post-Ad26.ZEBOV, MVA-BN-Filo vaccination serum. NK cell activation varied depending on vaccine regimen and was positively correlated with antibody concentration. NK activity also varied between NK cell donors, consistent with the assumption that differentiation phenotype influences the potency of antibody-dependent NK cell responses.
METHODS
Study Participants and Samples
Eligible, healthy volunteers were recruited to take part in the EBL1001 (EBOVAC consortium) single-center, randomized, placebo-controlled, observer-blind Ebola vaccine trial held in Oxford, United Kingdom (ClinicalTrials.gov identifier NCT02313077). Another 15 volunteers were subsequently recruited for group 5 (see Milligan et al [21] for additional information on methods). Serum samples from 72 donors (age range, 18–50 years) were obtained for this study from nonplacebo arms (Table 1).
Table 1.
Vaccination Regimen of Each Trial Arm and Serum Samples Used in the Study
| Group | Vaccine Regimen | Timing of Serum Samples | ||
|---|---|---|---|---|
| Baseline (Visit 0) | After Dose 1 (Visit 1) | After Dose 2 (Visit 2) | ||
| 1 (n = 15) | MVA, Ad26 | d 1 | d 29 | d 50 |
| 2 (n = 15) | MVA, Ad26 | d 1 | d 57 | d 78 |
| 3 (n = 14) | Ad26, MVA | d 1 | d 29 | d 50 |
| 4 (n = 14) | Ad26, MVA | d 1 | d 57 | d 78 |
| 5 (n = 14)a | Ad26, MVA | d 1 | d 15 | d 36 |
Abbreviations: Ad26, adenovirus type 26.ZEBOV; MVA, modified vaccinia Ankara–BN-Filo.
a12 individuals in group 5 received dose 2, MVA at day 15.
Active vaccination comprised monovalent Ad26.ZEBOV, expressing the GP of the Ebola Zaire virus (Mayinga variant), and multivalent MVA-BN-Filo, expressing the GP of the Sudan and Zaire EBOVs and Marburg virus together with Taï Forest virus nucleoprotein (Janssen Vaccines and Prevention and Bavarian Nordic). Vaccination schedules were as follows; groups 1 and 2 received MVA-BN-Filo on day 1 and Ad26.ZEBOV on either day 29 or 57, respectively; and groups 3, 4, and 5 received Ad26.ZEBOV on day 1 and MVA-BN-Filo on days 29, 57, or 15, respectively.
Additional blood samples were obtained from nonvaccinated, nonstudy volunteers. PBMCs were isolated using Histopaque 1077 (Sigma-Aldrich) gradient centrifugation and cryopreserved in liquid nitrogen or used immediately. Ebola GP–specific IgG concentration and Ebola GP–specific pseudovirion virus neutralizing antibody (psVNA) titers were determined previously [21], and human cytomegalovirus (HCMV) serostatus was determined by means of IgG enzyme-linked immunosorbent assay (Demeditec) (36% HCMV seropositive). The trial protocol and study documents were approved by the National Research Ethics Service (reference no. 14/SC/1408) and the London School of Hygiene and Tropical Medicine Research Ethics Committee (reference no. 14383).
In Vitro Culture Assays
For antibody-dependent NK cell activation assays, 10 μg/mL purified EBOV GP, Mayinga variant (Janssen Vaccines and Prevention), was immobilized on 96-well flat-bottom tissue culture plates overnight at 4°C, washed, blocked with 5% fetal calf serum (FCS) in Roswell Park Memorial Institute 1640 medium supplemented with 100 U/mL penicillin/streptomycin and 20 mmol/L L-glutamine (Gibco; ThermoFisher), and washed again. Fresh PBMCs from a single individual donor (nonvaccinated) were washed in Roswell Park Memorial Institute 1640 medium supplemented as above and counted using a Countess II FL Automated Cell Counter (Invitrogen; ThermoFisher). PBMCs were seeded (3 × 105 per well) onto the antigen-coated plates together with pre- or postvaccination serum at various concentrations (with total serum concentration made up to 5% with FCS) and incubated for 6 hours at 37°C. Alternatively, cryopreserved PBMCs from multiple (nonvaccinated) donors were thawed, washed, and seeded onto the antigen-coated plates with pooled pre- or postvaccination serum from group 2 (regimen: MVA-BN-Filo on day 1 and Ad26.ZEBOV on day 57).
Anti-CD107a fluorescein isothiocyanate (clone H4A3; BD Biosciences) was added to the cultures for the entire culture period, and GolgiStop (Monensin; 1:1500 concentration; BD Biosciences) and GolgiPlug (Brefeldin A; 1:1000 final concentration; BD Biosciences) were added for the final 3 hours of culture. Positive control cultures comprised the CD20-expressing human Burkitt lymphoma cell line (Raji cells; European Collection of Authenticated Cell Cultures [ECACC]) with monoclonal anti-CD20, rituximab (Ritxan; Genentech), at varying concentrations. In all cases, cells were harvested into round-bottom plates by soaking and resuspension in fluorescence-activated cell sorting (FACS) buffer (phosphate-buffered saline, 0.5% FCS, 0.05% sodium azide, and 2 mmol/L ethylenediaminetetraacetic acid) for staining.
Flow Cytometry
Cells were stained in 96-well round-bottom plates, as described elsewhere [24]. Briefly, cells were blocked with FcR Blocking Reagent (Miltenyi Biotech) and stained with fluorophore-labeled antibodies for surface markers, including viability marker (Fixable Viability Stain 700; BD Biosciences) in FACS buffer. Cells were washed in FACS buffer, fixed, and permeabilized using Cytofix/Cytoperm Kit (BD Biosciences). Cells were then stained for intracellular markers with further FcR blocking, washed again, resuspended in FACS buffer, acquired using a BD LSRII flow cytometer and FACSDiva software, and analyzed using FlowJo V10 (Tree Star). FACS gates were set using unstimulated cells or Fluorescence Minus One (FMO) controls, a minimum cutoff was determined as the frequency of responding NK cells in the presence of FCS alone [21], and samples with <100 NK cell events were excluded from the analysis.
The fluorophore-labeled antibodies used were anti-CD3-V500 (clone UCHT1) (BD Biosciences), anti-CD56-BV605 (clone HCD56), anti-IFN-γ-BV785 (clone 45.B3) (Biolegendanti-CD16-APC (clone CB16), anti-CD57-e450 (clone TB01) (eBiosciences), and anti-NKG2C-PE (clone 134591) (R&D Systems).
Statistics
Statistical analysis was performed using GraphPad Prism software, version 7.04 (GraphPad). Functional responses were compared using Wilcoxon signed rank test or 1-way analysis of variance Friedman test with Dunn correction for multiple comparisons. Correlation analysis was performed using linear and nonlinear regression models, and the P value for the correlation of the 2 variables was determined using Pearson correlation analysis.
RESULTS
Ad26.ZEBOV, MVA-BN-Filo Ebola Vaccine–Induced Antibody-Dependent NK Cell Activation In Vitro
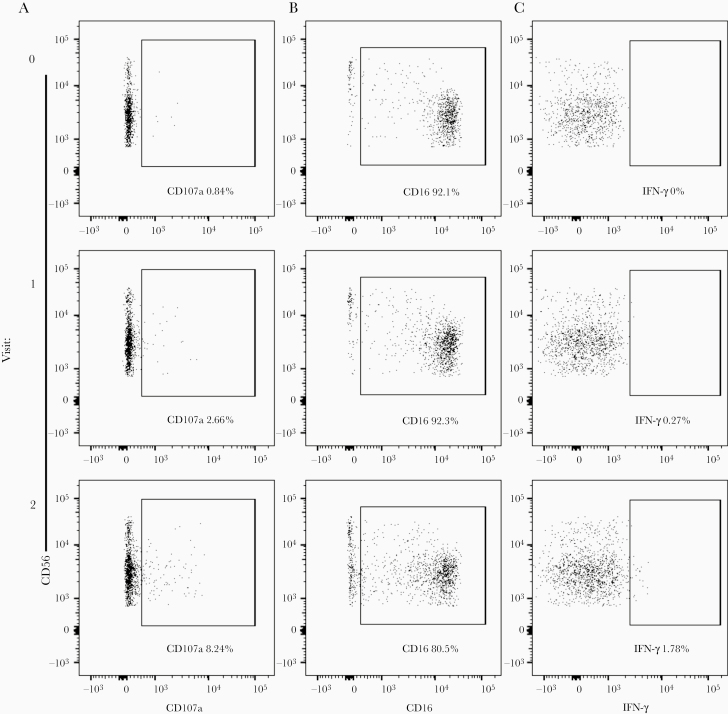
To assess the effect of Ad26.ZEBOV, MVA-BN-Filo vaccine–induced anti-GP antibody on NK cell activation, whole PBMCs from 1 nonvaccinated donor were cultured with plate-bound EBOV GP plus pre- or postvaccination serum samples. Optimal serum concentrations were established for CD3−CD56+ NK cell CD107a surface expression (gating strategy shown in Figure 1A and Supplementary Figure 1A; Supplementary Figure 1B). NK cell CD107a, CD16, and IFN-γ expression was then measured in response to 5% prevaccination (visit 0), post–dose 1 (visit 1), or post–dose 2 (visit 2) serum from each individual study participant (n = 72) (gating strategy shown in Figure 1A–1C). Initially, data from all 5 vaccination arms were combined for analysis.
Figure 1.
Flow cytometry gating strategy for natural killer (NK) cell CD107a, CD16, and interferon (IFN) γ expression. Flow cytometry plots show CD3−CD56+ NK cell (gating strategy shown in Supplementary Figure 1A) CD107a (A), CD16 (B) and IFN-γ (C) expression in response to 5% prevaccination (visit 0), post–dose 1 (visit 1), and post–dose 2 (visit 2) vaccination serum and plate-bound Ebola virus glycoprotein antigen. Whole human peripheral blood mononuclear cells from 1 nonvaccinated single donor were used for NK cell activation assays in Figures 1–4, and the NK cell differentiation phenotype of the donor is shown in Supplementary Figure 1D.
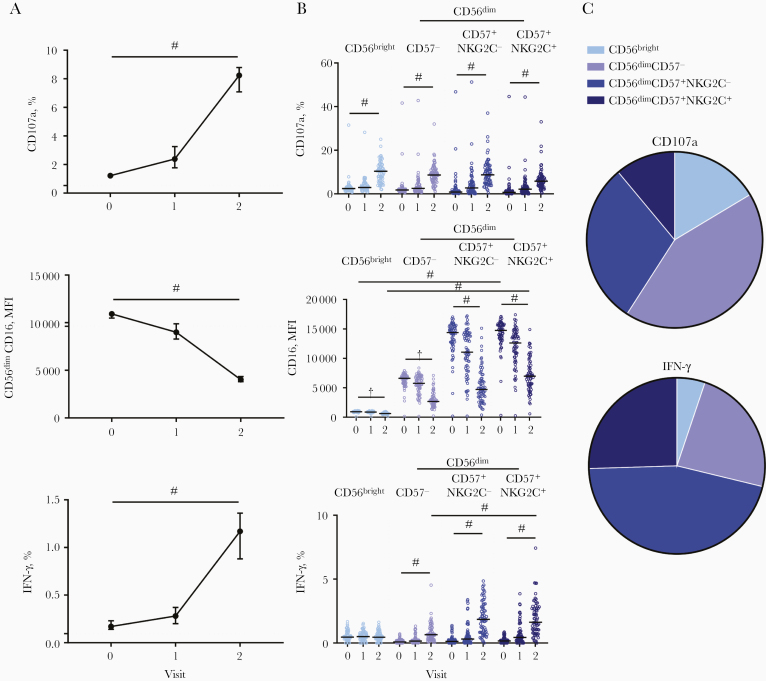
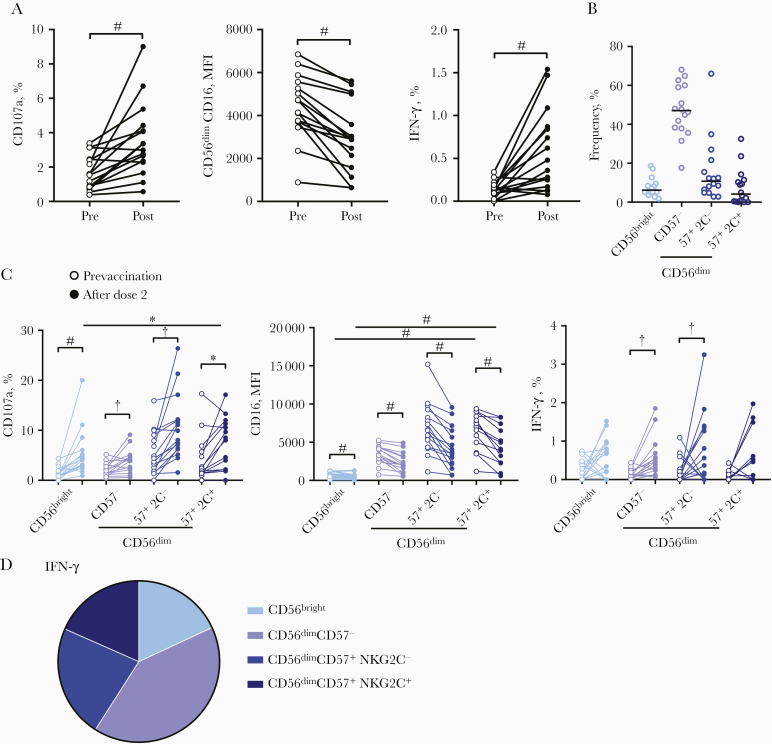
Significantly higher frequencies of CD107a+ NK cells were observed with post–dose 1 serum compared with prevaccination serum, and these frequencies were further enhanced with post–dose 2 serum (median, 2.39% and 8.24%, respectively, for post–dose 1 and post–dose 2 serum) (Figure 2A). CD56dim NK cell CD16 expression measured based on mean fluorescence intensity (MFI) decreased significantly in cells cultured with post–dose 1 serum, and there was a further decrease in cells cultured with post–dose 2 serum (median MFI, 8990 and 4020, respectively for post–dose 1 and post–dose 2 serum) (Figure 2A). Frequencies of NK cells producing IFN-γ in response to post-dose 1 serum were low but significantly higher than in response to prevaccination serum, and again, these were greatly increased with post–dose 2 serum (median, 0.28% for post–dose 1 and 1.17% for post–dose 2 serum) (Figure 2).
Figure 2.
Antibody-dependent natural killer (NK) cell responses to plate-bound Ebola virus glycoprotein after adenovirus type 26.ZEBOV, modified vaccinia Ankara (MVA)–BN-Filo vaccination. The median and 95% confidence interval of NK cell CD107a, CD56dimCD16 mean fluorescence intensity (MFI), and interferon (IFN) γ responses to prevaccination (visit 0), post–dose 1 (visit 1) and post–dose 2 (visit 2) vaccination serum sample are shown. A, All vaccine arms combined (n = 72). NK cell CD107a, CD16, and IFN-γ responses were analyzed according to NK cell differentiation subset, defined by CD56, CD57, and NKG2C expression (gating strategy shown in Supplementary Figure 1C). B, Each individual serum donor is represented by a dot with a line at the median. The proportion of total NK cell CD107a and IFN-γ expression (after dose 2) attributed to each subset is shown as a pie graph, with each slice representing the median. C, Comparisons across vaccination visits and between subsets were performed using 1-way analysis of variance with Holm-Sidak test for multiple comparisons. ‡P < .001.
The effect of Ad26.ZEBOV, MVA-BN-Filo–induced anti-GP antibody on antibody-dependent NK cell activation was analyzed according to NK cell differentiation subset (gating strategy shown in Supplementary Figure 1C; Figure 1Dshows the NK cell subset distribution for the single donor used in this assay). NK cell CD107a expression was induced equally in less differentiated CD56bright and more differentiated CD56dim NK cell subsets and in subsets further subdivided into moderately and highly differentiated CD56dimCD57− and CD56dimCD57+ (NKG2C− and NKG2C+) cells (Figure 2B). This was consistent with significant CD16 down-regulation observed in all NK cell subsets (Figure 2B).
Basal CD16 expression increased with increasing differentiation status (CD56dimCD57− < CD56dimCD57+NKG2C− < CD56dimCD57+NKG2C+), and CD16 expression was maintained at higher MFI after dose 2 in the most differentiated subsets (Figure 2B). CD107a was induced within the CD56bright NK cell population in response to postvaccination serum, but its overall contribution was <14% of the total expression (P < .001) (Figure 2C). IFN-γ expression in response to postvaccination serum was attributed to CD56dim NK cells, with no increase in expression observed within the least differentiated CD56bright NK cell subset (Figure 2B).
The most highly differentiated CD56dimCD57+NKG2C− and CD56dimCD57+NKG2C+ NK cell subsets showed the most extensive CD16 down-regulation and the highest frequencies of IFN-γ producing cells (Figure 2B); 71.2% of all the NK cells producing IFN-γ in response to post–dose 2 serum were CD56dimCD57+ (NKG2C+/−) NK cells, with 25.5% of IFN-γ + cells being CD56dimCD57+NKG2C+ (Figure 2C). Consistent with antibody-dependent activation of more differentiated NK cell subsets, anti-CD20 (rituximab) and CD20-expressing Raji cells also preferentially induced NK cell degranulation and IFN-γ expression in CD56dimCD57+ (NKG2C+/−) cells (Supplementary Figure 2). These data suggest that EBOV GP–specific antibody induces antibody-dependent NK cell activation, including IFN-γ secretion, in more differentiated NK cell subsets.
Variation in Antibody-Dependent NK Cell Activation by Vaccine Regimen
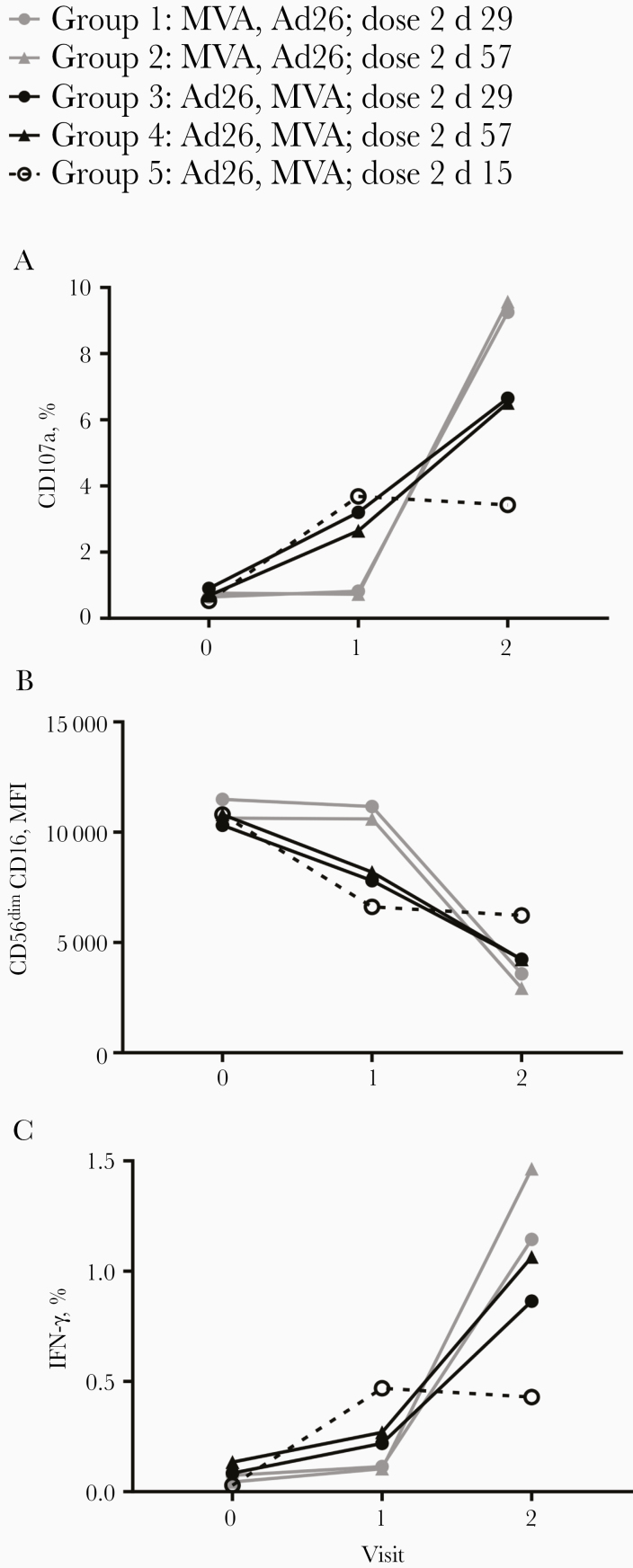
The Ebola GP–specific IgG concentration in the serum samples of Ad26.ZEBOV, MVA-BN-Filo–vaccinated individuals varied depending on the vaccination regimen [21]. We therefore analyzed the impact of vaccine regimen on antibody-dependent NK cell activation. There was significant up-regulation of CD107a and IFN-γ and down-regulation of CD16 with post–dose 2 serum in all groups compared with prevaccination serum responses, but responses differed significantly between study arms (Figure 3A–3C).
Figure 3.
Antibody-dependent natural killer (NK) cell activation varies with vaccine regimen. NK cell CD107a (A), CD56dimCD16 mean fluorescence intensity (MFI) (B), and interferon (IFN) γ (C) expression was plotted according to vaccine regimen (groups 1–5) for prevaccination (visit 0), post–dose 1 (visit 1), and post–dose 2 (visit 2) time points; graphs show median only. Comparisons between visits within each group were performed using 1-way analysis of variance with Dunn correction for multiple comparisons and summarized in Supplementary Table 1.
Groups 1 and 2 (MVA-BN-Filo followed by Ad26.ZEBOV) resulted in the strongest induction of CD107a and IFN-γ expression and the lowest CD16 MFI of all 5 groups (Figure 3A–3C). Serum collected after MVA-BN-Filo (dose 1) did not induce NK cell activation, but significant induction of CD107a and IFN-γ and down-regulation of CD16 was seen with post-Ad26.ZEBOV (post–dose 2) serum (compared with both prevaccination and post–dose 1 vaccination serum) (Figure 3A–3C and Supplementary Table 1). By contrast, in groups 3, 4, and 5 (Ad26.ZEBOV followed by MVA-BN-Filo) there was induction of NK cell responses in post-Ad26.ZEBOV (post–dose 1) serum that was further enhanced by post–MVA-BN-Filo (post–dose 2) serum (Figure 3A–3C and Supplementary Table 1).
However, earlier MVA-BN-Filo dose 2 in groups 3 (day 29) and 5 (day 15) did not result in further significant NK activation (1-way analysis of variance; Supplementary Table 1) compared with the first dose of Ad26.ZEBOV (except weak boosting of IFN-γ in group 3), and group 5 resulted in the weakest overall response (Figure 3A–3C and Supplementary Table 1). This suggests that Ad26.ZEBOV as the first dose induces sufficient concentrations of antibody for a robust NK cell response that is further increased by the MVA-BN-Filo second dose, whereas MVA-BN-Filo alone does not induce sufficient antibody (or antibody of the correct isotype or subclass) to mediate ADCC.
Correlation of NK Cell Function With Anti-GP Antibody Concentration and psVNA Titers
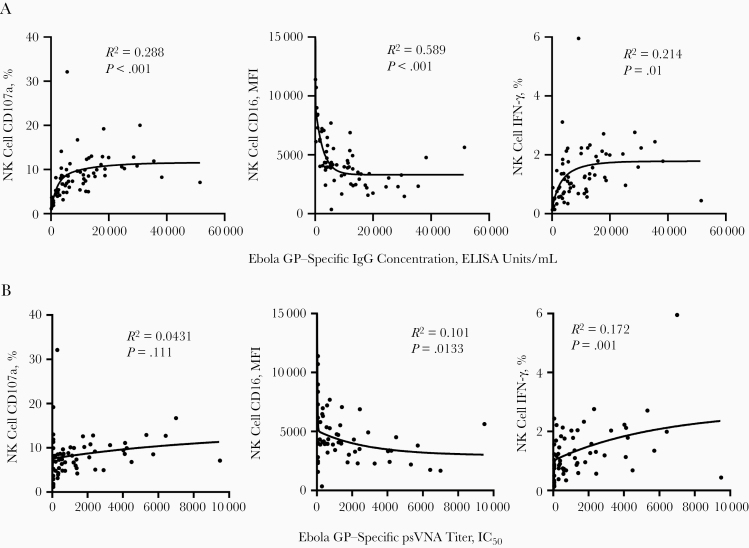
Variation in NK cell function according to vaccine regimen is consistent with data on the effect of vaccine regimen on anti-GP antibody concentration [21]. Therefore, we next analyzed the relationship between individual Ebola GP–specific IgG concentration (determined by Milligan et al [21]) and antibody-dependent NK cell activation. With all groups combined, there was a significant positive correlation between post–dose 2 antibody concentration and frequencies of NK cell CD107a and IFN-γ and a negative correlation with CD16 MFI (Figure 4A).
Figure 4.
Natural killer (NK) cell activation correlates with anti–glycoprotein (GP) antibody concentration and pseudovirion virus neutralizing antibody (psVNA) titers. Post–dose 2 anti-GP antibody concentrations (A) or Ebola GP–specific pseudovirion virus neutralizing antibody (psVNA) titers (B) (determined previously by Milligan et al [21]) were correlated with post–dose 2 NK cell CD107a, CD56dimCD16 mean fluorescence intensity (MFI) and interferon (IFN) γ expression, all vaccination groups combined. A 2-phase nonlinear regression model was fitted in prism, and R2 goodness-of-fit analysis is shown; P values were determined by Pearson correlation, with significance defined as P < .05. Abbreviations: ELISA, enzyme-linked immunosorbent assay; IC50, median inhibitory concentration; IgG, immunoglobulin G.
Groups 1 and 2 (MVA-BN-Filo followed by Ad26.ZEBOV) demonstrated the highest median NK cell functional responses after dose 2 (CD107a+ NK cell frequency, P = .048 for group 1 vs 3 and P = .02 for group 2 vs 4; Wilcoxon paired t test). However, when analyzed according to vaccination group, only groups 3–5 (Ad26.ZEBOV followed by MVA-BN-Filo) had a significant correlation between post–dose 2 antibody concentration and NK cell function (Table 2). Antibody concentration and NK cell function were also significantly correlated after dose 1 when all groups were combined (Supplementary Figure 3A–3C), but when groups were analyzed separately, this relationship was significant only for group 3 (Ad26.ZEBOV followed by MVA-BN-Filo at day 29) (Table 2). There was no correlation between antibody concentration and antibody-dependent NK cell function after dose 1 or dose 2 in individuals vaccinated by MVA-BN-Filo followed by Ad26.ZEBOV (groups 1 and 2) (Table 2). Therefore, in vaccine regimens inducing the highest post–dose 2 responses (groups 1 and 2), the association between the 2 variables is lost.
Table 2.
Correlation Between Natural Killer Cell CD107a, CD16, and Interferon γ Expression in Response to Plate-Bound Ebola Virus Glycoprotein (GP) Plus Post-Dose 1 (Visit 1) and Post-Dose 2 (Visit 2) Serum and Anti-GP Antibody Concentration According to Vaccine Regimen Groupa
| Visit by Group | R 2 (P Value)b | ||
|---|---|---|---|
| CD107a (%) | CD16 (MFI) | IFN-γ (%) | |
| Visit 1 | |||
| Group 1 (d 29) | 0.129 (.21)c | 0.00292 (.85)c | 0.0214 (.60)c |
| Group 2 (d 57) | 0.00469 (.82)c | 0.0193 (.62)c | 0.00638 (.78)c |
| Group 3 (d 29) | 0.480 (.006) | 0.550 (.002) | 0.553 (.002) |
| Group 4 (d 57) | 0.0924 (.29)c | 0.312 (.04) | 0.248 (.07)c |
| Group 5 (d 15) | 0.394 (.052)c | 0.221 (.17)c | 0.397 (.051)c |
| Visit 2 | |||
| Group 1 (d 50) | 0.0209 (.61)c | 0.00775 (.76)c | 0.00144 (.89)c |
| Group 2 (d 78) | 0.0639 (.36)c | 0.0339 (.51)c | 0.0895 (.28)c |
| Group 3 (d 50) | 0.660 (<.001) | 0.554 (.002) | 0.531 (.003) |
| Group 4 (d 78) | 0.364 (.02) | 0.612 (<.001) | 0.327 (.03) |
| Group 5 (d 36) | 0.859 (<.001) | 0.690 (.003) | 0.276 (.12)c |
Abbreviations: IFN, interferon; MFI, mean fluorescence intensity.
aAntiglycoprotein antibody concentrations were determined previously by Milligan et al [21].
b R 2 values determined by means of linear regression. Significance was defined as P < .05.
cNonsignificant correlations.
Analysis of antibody-dependent NK cell responses and Ebola GP–specific psVNA titers revealed a significant positive correlation across the entire cohort with the frequency of NK cell IFN-γ and a negative correlation with CD16 expression, although no association was observed with CD107a expression (Figure 4B). Consistent with the association with anti-GP antibody concentration, we observed the strongest correlations between all NK cell functions and psVNA titers for groups 3 and 4 (Ad26.ZEBOV followed by MVA-BN-Filo) (Table 3).
Table 3.
Correlation Between Natural Killer Cell CD107a, CD16, and Interferon γ Expression in Response to Plate-bound Ebola Virus Glycoprotein (GP) Plus Post–Dose 2 (Visit 2) Serum and Ebola GP–Specific Pseudovirion Virus Neutralizing Antibody Titers According to Vaccine Regimen Groupa
| Visit 2 by Group | R 2 (P Value) | ||
|---|---|---|---|
| CD107a (%) | CD16 (MFI) | IFN-γ (%) | |
| Group 1 (d 50) | 0.0001 (.97)c | 0.006 (.79)c | 0.425 (.01) |
| Group 2 (d 78) | 0.063 (.36)c | 0.034 (.51)c | 0.089 (.28)c |
| Group 3 (d 50) | 0.446 (.02) | 0.331 (.050) | 0.352 (.04) |
| Group 4 (d 78) | 0.485 (.006) | 0.548 (.003) | 0.503 (.005) |
| Group 5 (d 36) | 0.182 (.29)c | 0.306 (.16)c | 0.380 (.10)c |
Abbreviations: IFN, interferon; MFI, mean fluorescence intensity.
aTiters determined previously by Milligan et al l [21].
b R 2 values determined by means of linear regression. Significance was defined as P < .05.
cNonsignificant correlations.
Variation in Antibody-dependent NK Cell Function by NK Cell Donor
To analyze the effect of donor variation, PBMCs from nonvaccinated donors (n = 16) were cultured with plate-bound EBOV GP and pooled prevaccination or post–dose 2 serum samples from group 2 (MVA-BN-Filo followed by Ad26.ZEBOV at day 57). NK cell CD107a (14 of 16 donors responding), CD16 down-regulation (all 16 responding), and IFN-γ (13 of 16 responding) was induced with pooled post–dose 2 serum compared with pooled prevaccination serum (Figure 5A), suggesting that the majority of donors respond to Ad26.ZEBOV, MVA-BN-Filo vaccine–induced antibody.
Figure 5.
Natural killer (NK) cell activation varies with NK cell donor. A, NK cell CD107a, CD56dimCD16 mean fluorescence intensity (MFI), and interferon (IFN) γ expression (multiple nonvaccinated donors; n = 16) in response to 5% pooled prevaccination (Pre) and post–dose 2 (Post) vaccination serum (group 2) and plate-bound Ebola virus glycoprotein. B, NK cell subset frequency distribution is shown for each donor. C, D, NK cell CD107a, CD16, and IFN-γ responses were also analyzed according to NK cell differentiation subset (C) and the proportion of total NK cell IFN-γ expression (after dose 2) attributed to each subset is shown as a pie graph, with each slice representing the median (D). Graphs show before-and-after plots with a line connecting each donor or 1 dot per donor with a line representing the median. Comparisons between pre- and postvaccination serum responses were performed using Wilcoxon signed rank test and comparison between subsets using 1-way analysis of variance with Dunn correction for multiple comparisons. *P < .05; †P < .01; ‡P < .001.
We next analyzed NK cell activation in response to pooled post-dose 2 serum according to NK cell differentiation subset. Among the individuals tested, frequencies of the most highly differentiated CD56dimCD57+NKG2C+ NK cells varied widely (with frequencies >10% in 5 of 16 donors), with a wide range of subset frequency (Figure 5B). Overall, NK cell CD107a expression was apparent in all NK cell subsets, as was CD16 down-regulation (Figure 5C). IFN-γ was significantly up-regulated with post–dose 2 serum in CD56dimCD57− and CD56dimCD57+NKG2C− subsets and with the highest frequency of IFN-γ expression observed within the CD56dim NK cell subsets (Figure 5C). Almost half (41.0%) of total NK cell IFN-γ production was attributed to CD56dimCD57+ NK cells (Figure 5D). These data demonstrate that differences in NK cell differentiation status introduce additional interdonor variation in NK cell ADCC responses.
DISCUSSION
We have shown that antibodies to EBOV GP induced by the Ad26.ZEBOV, MVA-BN-Filo vaccine regimen activate robust in vitro NK cell degranulation and IFN-γ secretion on coculture with Ebola GP antigen. These NK cell responses are highly variable depending on vaccine regimen and interval and are correlated with anti-GP IgG concentration and are markedly enriched in (though not limited to) more differentiated NK cell subsets. Variation in NK cell differentiation status between donors contributes to the heterogeneity of postvaccination ADCC responses.
The positive correlation between post–dose 2 antibody concentration and NK cell activation demonstrates a good readout of antibody-dependent effector function. Interestingly, MVA-BN-Filo followed by Ad26.ZEBOV vaccine regimen (groups 1 and 2), giving rise to both the highest numerical geometric mean Ebola GP–specific IgG concentration and median NK cell function, did not result in significant correlations between the 2 factors on an individual level. NK cell CD107a expression did not increase further with the higher antibody concentrations induced by a delayed second dose (day 57) compared with an earlier second dose (day 29), suggesting that sufficient antibody concentrations can be achieved with the early dose 2 (day 29) to obtain optimal responses [21]. NK cell IFN-γ expression was higher with a later second dose (day 57) than with the earlier second dose (day 29), suggesting that increasing levels of anti-GP antibody are associated with stronger NK cell cytokine secretion.
The lack of correlation between antibody concentration and NK cell responses after the MVA-BN-Filo followed by Ad26.ZEBOV regimen highlights a requirement for a 2-dose vaccine regimen or primary vaccination with Ad26.ZEBOV to induce robust NK cell responses. This may also reflect effects of antibody affinity maturation and isotype/subclass switching. Of note, the Ad26.ZEBOV followed by MVA-BN-Filo vaccine regimen is being further evaluated in phase 2 and 3 clinical studies.
NK cell activation after the Ad26.ZEBOV, MVA-BN-Filo vaccination regimen required relatively high serum concentrations, with similar levels of NK cell degranulation occurring with up to 40-fold lower concentrations of post–seasonal influenza vaccination serum [16]. Importantly, only antibodies binding to EBOV GP contributed to the response; antibodies specific for the nucleoprotein (contained in MVA-BN-Filo) were not assessed. Significant correlations between NK cell function and psVNA were also observed after dose 2, most significantly for the Ad26.ZEBOV, MVA-BN-Filo vaccine regimen. This is consistent with a previously reported direct temporal and quantitative relationship between specific IgG concentrations and neutralizing activity and with a subset of vaccine-induced antibodies having both NK cell activation and virus neutralizing properties [20, 22].
Our data highlight substantial variation in frequencies of activated NK cells subsets both within a single donor and between donors in response to postvaccination antibody. Many variables affect NK cell ADCC function, including FcR polymorphisms [25], antibody glycosylation [26], and cytokine-mediated regulation [27]. Antibody-binding epitopes can also affect the ADCC function of vaccine-induced antibodies [3]; neutralizing antibodies bind Ebola GP core epitopes, potentially inhibiting virion cell entry, whereas antibodies with Fc function bind epitopes on the exposed upper domains of GP presented on the surface of infected cells [6, 28]. Ebola GP returning to the surface of the infected cell and liberation of soluble GP for immune complex formation could promote NK cell ADCC. However, GP shed from infected cells can also bind anti-GP antibodies and block, rather than facilitate, their activity [29].
HCMV has seropositivity rates up to 60% in adults in developed countries and is near universal in developing countries [30]. HCMV infection strongly influences NK cell function in response to viral antigens and promotes accumulation of NK cells expressing NKG2C with a mature (CD56dimCD57+) and “adaptive” (FcεRγ −) phenotype [31–33] and with enhanced IFN-γ secretion in response to antibody-coated targets [34–36]. HCMV serostatus may affect antibody-dependent NK cell activation after Ad26.ZEBOV, MVA-BN-Filo vaccination; therefore, measuring NK cell function may help evaluate vaccine responses across different human populations.
Future use of CD16-transfected NK cell lines for standardization of these assays could potentially enable comparison across multiple vaccine studies [37]. However, NK cell tumor lines, such as NK-92, are largely derived from pre-NK cells and do not reflect the range of FcR expression, activation requirements, or functional differentiation of primary human NK cells, factors important in African settings where NK cells are enriched for highly differentiated subsets. Alternatively, Wines et al [38] have described a system using soluble dimeric ectodomains of human FcγRIII or FcγRII (CD32), which facilitate evaluation of antibody isotype specificity and binding to low- and high-affinity variants of these FcRs.
In summary, Ad26.ZEBOV, MVA-BN-Filo vaccine–induced antibody promotes strong antibody-dependent NK cell activation that is correlated with antibody concentration. Our findings suggest that NK cells are potential mediators of immunity after Ebola vaccination, wherein responses are determined by both the level of antibody and effector NK cells differentiation status. Antibody-dependent NK cell function may help define the effector capacity of vaccine-induced antibodies when combined with antibody level or neutralization assays.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Carolynne Stanley for recruiting and obtaining consent from London School of Hygiene and Tropical Medicine study subjects and for blood sample collection.
Author contributions. H. R. W. and M. R. G. designed and performed the experiments and analyzed the data. H. R. W., M. R. G., and E. M. R. wrote the manuscript. V. B., J. N. S., and K. L. helped analyze the data. M. D. and G. S. helped conceive and design the work described. E. A. C. and A. J. P. were coinvestigators on the current trial. M. D. S. was chief investigator for the phase 1 clinical trial of Ad26.ZEBOV, MVA-BN-Filo. E. A. C., V. B., J. N. S., K. L., M. D., G. S., M. D. S., A. J. P., and E. M. R. advised on the manuscript.
Disclaimer. The views expressed in this publication are those of the authors and not necessarily those of the UK Department of Health and Social Care, the National Institute for Health Research (NIHR), the National Health Service, the Medical Research Council, the World Health Organization, or the European Union.
Financial support. This work was supported by a UK Medical Research Council Studentship in Vaccine Research (H.R.W). This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking, EBOVAC (Grant 115861) and Crucell Holland (now Janssen Vaccines and Prevention B.V.). This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation programme and European Federation of Pharmaceutical Industries and Associations (EFPIA). A.J.P. is supported by the NIHR Oxford Biomedical Research Centre and is an NIHR Senior Investigator. M.D.S. is supported by the NIHR Oxford Biomedical Research Centre.
Potential conflicts of interest. V. B., J. N. S., K. L., M. D., and G. S. are employees and potential stockholders of Janssen Pharmaceuticals. A. J. P. chairs the UK Department of Health and Social Care’s Joint Committee on Vaccination and Immunisation and the European Medicines Agency (EMA) Scientific Advisory Group on vaccines, and he is a member of the World Health Organization’s Strategic Advisory Group of Experts. M. D. S. is an investigator for the University of Oxford on clinical research studies funded by vaccine manufacturers, including Janssen, Pfizer, GlaxoSmithKline, Novavax, Medimmune, and MCM Vaccine; he receives no personal financial benefit for this work. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Meyer M, Malherbe DC, Bukreyev A. Can Ebola virus vaccines have universal immune correlates of protection? Trends Microbiol 2019; 27:8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Corti D, Misasi J, Mulangu S, et al. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science 2016; 351:1339–42. [DOI] [PubMed] [Google Scholar]
- 3. Saphire EO, Schendel SL, Fusco ML, et al. Systematic analysis of monoclonal antibodies against Ebola virus GP defines features that contribute to protection. Cell 2018; 174:938–52.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Qiu X, Wong G, Audet J, et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 2014; 514:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu Q, Fan C, Li Q, et al. Antibody-dependent-cellular-cytotoxicity-inducing antibodies significantly affect the post-exposure treatment of Ebola virus infection. Sci Rep 2017; 7:45552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gunn BM, Yu WH, Karim MM, et al. A role for Fc function in therapeutic monoclonal antibody-mediated protection against Ebola virus. Cell Host Microbe 2018; 24:221–33.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singh K, Marasini B, Chen X, Spearman P. A novel Ebola virus antibody-dependent cell-mediated cytotoxicity (Ebola ADCC) assay. J Immunol Methods 2018; 460:10–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rechtien A, Richert L, Lorenzo H, et al. ; VEBCON Consortium . Systems vaccinology identifies an early innate immune signature as a correlate of antibody responses to the Ebola vaccine rVSV-ZEBOV. Cell Rep 2017; 20:2251–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blaney JE, Marzi A, Willet M, et al. Antibody quality and protection from lethal Ebola virus challenge in nonhuman primates immunized with rabies virus based bivalent vaccine. PLoS Pathog 2013; 9:e1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Warfield KL, Swenson DL, Olinger GG, Kalina WV, Aman MJ, Bavari S. Ebola virus-like particle-based vaccine protects nonhuman primates against lethal Ebola virus challenge. J Infect Dis 2007; 196(suppl 2):S430–7. [DOI] [PubMed] [Google Scholar]
- 11. Marzi A, Murphy AA, Feldmann F, et al. Cytomegalovirus-based vaccine expressing Ebola virus glycoprotein protects nonhuman primates from Ebola virus infection. Sci Rep 2016; 6:21674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Keshwara R, Hagen KR, Abreu-Mota T, et al. A recombinant rabies virus expressing the Marburg virus glycoprotein is dependent upon ADCC for protection against Marburg virus disease in a murine model. J Virol 2019; 93:e01865–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bruhns P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood 2012; 119:5640–9. [DOI] [PubMed] [Google Scholar]
- 14. Bowles JA, Wang SY, Link BK, et al. Anti-CD20 monoclonal antibody with enhanced affinity for CD16 activates NK cells at lower concentrations and more effectively than rituximab. Blood 2006; 108:2648–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Romee R, Foley B, Lenvik T, et al. NK cell CD16 surface expression and function is regulated by a disintegrin and metalloprotease-17 (ADAM17). Blood 2013; 121:3599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goodier MR, Lusa C, Sherratt S, Rodriguez-Galan A, Behrens R, Riley EM. Sustained immune complex-mediated reduction in CD16 expression after vaccination regulates NK cell function. Front Immunol 2016; 7:384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Srpan K, Ambrose A, Karampatzakis A, et al. Shedding of CD16 disassembles the NK cell immune synapse and boosts serial engagement of target cells. J Cell Biol 2018; 217:3267–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DiLillo DJ, Palese P, Wilson PC, Ravetch JV. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J Clin Invest 2016; 126:605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jegaskanda S, Luke C, Hickman HD, et al. Generation and protective ability of influenza virus-specific antibody-dependent cellular cytotoxicity in humans elicited by vaccination, natural infection, and experimental challenge. J Infect Dis 2016; 214:945–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Anywaine Z, Whitworth H, Kaleebu P, et al. Randomized clinical trial examining safety and immunogenicity of heterologous prime-boost Ebola vaccines, Ad26.ZEBOV and MVA-BN-Filo: 12-month data from Uganda and Tanzania. J Infect Dis 2019; 220:46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Milligan ID, Gibani MM, Sewell R, et al. Safety and immunogenicity of novel adenovirus type 26- and modified vaccinia Ankara-vectored Ebola vaccines: a randomized clinical trial. JAMA 2016; 315:1610–23. [DOI] [PubMed] [Google Scholar]
- 22. Mutua G, Anzala O, Luhn K, et al. Randomized clinical trial examining safety and immunogenicity of heterologous prime-boost Ebola vaccines, Ad26.ZEBOV and MVA-BN-Filo: 12-month data from Nairobi, Kenya. J Infect Dis 2019; 220:57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Winslow RL, Milligan ID, Voysey M, et al. Immune responses to novel adenovirus type 26 and modified vaccinia virus Ankara-vectored Ebola vaccines at 1 year. JAMA 2017; 317:1075–7. [DOI] [PubMed] [Google Scholar]
- 24. Wagstaffe HR, Nielsen CM, Riley EM, Goodier MR. IL-15 promotes polyfunctional NK cell responses to influenza by boosting IL-12 production. J Immunol 2018; 200:2738–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oboshi W, Watanabe T, Matsuyama Y, et al. The influence of NK cell-mediated ADCC: structure and expression of the CD16 molecule differ among FcγRIIIa-V158F genotypes in healthy Japanese subjects. Hum Immunol 2016; 77:165–71. [DOI] [PubMed] [Google Scholar]
- 26. Shields RL, Lai J, Keck R, et al. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. J Biol Chem 2002; 277:26733–40. [DOI] [PubMed] [Google Scholar]
- 27. Jegaskanda S, Vanderven HA, Tan HX, et al. Influenza infection enhances antibody-mediated NK cell functions via type I interferon dependent pathways. J Virol 2019; 93:e02090–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saphire EO, Schendel SL, Gunn BM, Milligan JC, Alter G. Antibody-mediated protection against Ebola virus. Nat Immunol 2018; 19:1169–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mohan GS, Li W, Ye L, Compans RW, Yang C. Antigenic subversion: a novel mechanism of host immune evasion by Ebola virus. PLoS Pathog 2012; 8:e1003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Darboe A, Danso E, Clarke E, et al. Enhancement of cytokine-driven NK cell IFN-γ production after vaccination of HCMV infected Africans. Eur J Immunol 2017; 47:1040–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goodier MR, Jonjić S, Riley EM, Juranić Lisnić V. CMV and natural killer cells: shaping the response to vaccination. Eur J Immunol 2018; 48:50–65. [DOI] [PubMed] [Google Scholar]
- 32. Griffiths P, Baraniak I, Reeves M. The pathogenesis of human cytomegalovirus. J Pathol 2015; 235:288–97. [DOI] [PubMed] [Google Scholar]
- 33. Nielsen CM, White MJ, Bottomley C, et al. Impaired NK cell responses to pertussis and H1N1 influenza vaccine antigens in human cytomegalovirus-infected individuals. J Immunol 2015; 194:4657–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luetke-Eversloh M, Hammer Q, Durek P, et al. Human cytomegalovirus drives epigenetic imprinting of the IFNG locus in NKG2Chi natural killer cells. PLoS Pathog 2014; 10:e1004441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schlums H, Cichocki F, Tesi B, et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 2015; 42:443–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hart GT, Tran TM, Theorell J, et al. Adaptive NK cells in people exposed to Plasmodium falciparum correlate with protection from malaria. J Exp Med 2019; 216:1280–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jochems C, Hodge JW, Fantini M, et al. An NK cell line (haNK) expressing high levels of granzyme and engineered to express the high affinity CD16 allele. Oncotarget 2016; 7:86359–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wines BD, Vanderven HA, Esparon SE, Kristensen AB, Kent SJ, Hogarth PM. Dimeric FcγR ectodomains as probes of the Fc receptor function of anti-influenza virus IgG. J Immunol 2016; 197:1507–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.