Abstract
Virus infection can alter immune regulatory activity, and thus may be involved in the occurrence of autoimmune diseases. Recently, the pandemic of COVID-19 has posed a huge threat to public health and emerging evidence suggests that coronavirus may be implicated in the development and pathogenesis of autoimmune diseases. However, how coronavirus infection impacts the risk of autoimmune disease remains largely unknown. In this review, we focused on the association between coronavirus and autoimmunity, and elucidated the molecular mechanisms linking coronavirus exposure to autoimmunity. Additionally, we briefly introduced the role that coronavirus plays in several autoimmune diseases including multiple sclerosis (MS), rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), and idiopathicthrombocytopenic purpura (ITP).
Key Words: Coronavirus, Autoimmune disease, Autoimmunity; COVID-19
Introduction
The pandemic of coronavirus disease 19 (COVID-19) which started in December 2019 has posed a huge threat to public health. The outbreaks of Severe Acute Respiratory Syndrome (SARS) in 2003 (1) and Middle East Respiratory Syndrome (MERS) in 2012 (2), both caused by coronavirus, had also raised concerns from governments, public health officials, clinicians, and the general public.
Coronaviruses (CoVs) (order Nidovirales, family Coronaviridae, subfamily Coronavirinae) have the largest genomes of all RNA viruses. Coronaviruses can be divided into three genera: α-coronavirus, β-coronavirus, and γ-coronavirus (3). A recent study has found that there is also a new genus-δ-coronavirus (4). The viral nucleocapsid is composed of RNA and nucleocapsid proteins. For coronaviruses, there are several important structural proteins: spike (S) protein, membrane (M) protein, nucleocapsid (N) protein and envelope (E) protein. Group A β-coronaviruses also express an additional structural protein, hemagglutinin esterase (HE) (2,3,5). Among these proteins, S protein plays a crucial role in promoting the process of viral infection by binding to cellular receptors and mediating membrane fusion (6).
Autoimmune diseases are a group of diseases in which the body reacts immune responses to autoantigens and causes damage to its own tissues. In recent years, the incidence of autoimmune diseases shows tendency to ascend (7). It is a good proof that the incidence of childhood type 1 diabetes has been rising for 50 years (8). Common autoimmune diseases include rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), idiopathic thrombocytopenic purpura (ITP), multiple sclerosis (MS), and autoimmune hepatitis, etc. There is epidemiological evidence suggesting that the occurrence of autoimmune diseases is associated with viral infection. For example, Epstein-Barr virus (EBV)-negative individuals do not acquire MS unless they acquire virus for the first time, and individuals who have not been infected with virus are also less likely to have SLE (9).
Nevertheless, there are few studies on the relationship between coronavirus and autoimmune diseases. This review attempts to summarize the structure and immunity of coronavirus, several common autoimmune diseases (including RA, SLE, ITP and MS) and the relationship between them.
Coronavirus and Immune Responses
Coronavirus are enveloped viruses with a positive-sense single-stranded RNA genome and a nucleocapsid of helical symmetry, which can cause the infections of upper respiratory tract, gastrointestinal tract and nervous system. The genome size of coronaviruses ranges from approximately 26–32 kilobases, one of the largest among RNA viruses (3). The average diameter of the virus particles is around 125 nm (0.125 mmol). Presently, there are seven coronaviruses that can infect humans, including HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, SARS-CoV, MERS-CoV and COVID-19 (10). Under the electron microscope, the spikes of the coronavirus look like the corona, which is the origin of the name. The severe acute respiratory syndrome (SARS) that ravaged the world from the winter of 2002 to the spring of 2003 was caused by the SARS-CoV. The virus caused 916 deaths at a mortality rate of 11% (11). MERS-Cov with a mortality rate of 35% was prevalent in Saudi Arabia since September 2012 (12). COVID-19 broke out in Wuhan, China, in December 2019, and has spread rapidly all over the world.
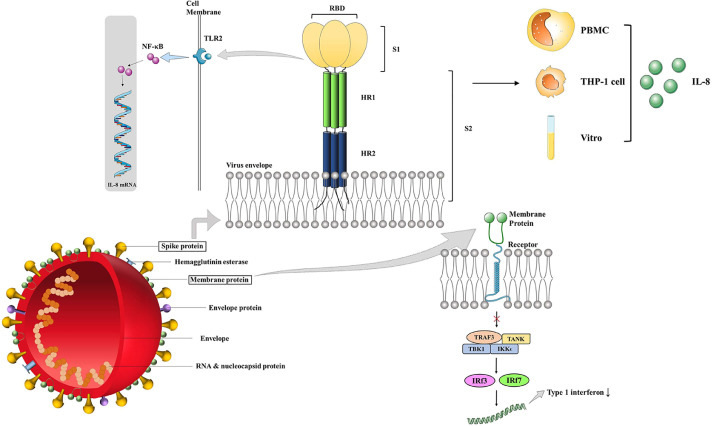
The relationship between these four structural proteins and immunity is introduced in the following section (Figure 1 ).
Figure 1.
Immune Effects of Coronavirus Structural Proteins S protein and M protein are two important structural proteins of coronavirus, which cause a variety of immune responses. S protein can cause the increase of IL-8 level through the following two ways: a) S protein is composed of S1 and S2. The former subunit can bind to Toll-like receptor 2, activate NF-κB, and increase the level of IL-8 mRNA. b) Coronavirus causes an increase in IL-8 in PBMC and THP-1 cells. These reactions can also be induced in vitro. M protein prevents the formation of TRAF3•TANK•TBK1/IKKϵ complex which leads to the activation of IRF3 and IRF7. Therefore, the level of type 1 interferon decreases in M protein infected cells. IL-8, interleukin-8; NF-κB, nuclear factor κ-B; PBMC, peripheral blood mononuclear cell; IRF, interferon regulatory factor; IKK, IκB kinase; TRAF, tumor necrosis factor receptor-associated factor; TANK, TRAF-associated NFκB activator; TBK1, TANK binding kinase.
Spike Protein
The ectodomain of the coronavirus spike protein consists of S1 and S2, and the former is responsible for receptor binding and the latter is associated with membrane fusion. The S1 subunit includes two domains, in which the N-terminal domain (S1-NTD) binds to sugar and the C-terminal domain (S1-CTD) binds to ACE2 (6). A study showed that S protein increased IL-8 in PBMC, in THP-1, and in vitro. S protein activates NF-κB through TLR2 ligand, which increases IL-8mRNA and may be inhibited by NF-κB inhibitor. The increase in the level of proinflammatory mediators is the result of the interaction of the S protein with monocytes and leads to an increase in NK, neutrophils, and monocytes at the site of infection. TPCK increases the secretion of IL-8 and IL-6 can indicate these (13).
Membrane Protein
The M protein, a transmembrane protein, is the largest number of structural protein and gives the virion envelope a special shape (14). Recently, emerging evidence showed that the M protein of SARS-CoV could suppress the production of Interferon-1 (IFN-1) by preventing the formation of TRAF3·TANK·TBK1/IKKϵ (15).
Envelope Protein
The E protein, a small polypeptide, is a minor structure of virions, ranging from 8.4–12 kDa. Due to its small size and limited number, E is considered to be a structural protein much later than others, first in IBV (14). A study showed that SARS-CoV E protein played an essential role in inducing inflammation. The amino terminus, transmembrane and carboxy terminus domains of E protein enhance the virulence. The activity of ion channel of E protein and the binding motif of PDZ domain participate in the occurrence of edema, ARDS (16).
Nucleocapsid Protein
The N protein, ranging from 43–50 kDa, constitutes the viral nucleocapsid. N protein exerts biological activity by combining with viral RNA (14). However, a study showed that the N protein of SARS-CoV inhibited dsRNA-induced IFN-β expression and participates in the suppression of the inception of IFN induction in the innate immune pathway (17).
Molecular Mechanisms Linking Coronavirus Exposure and Autoimmunity
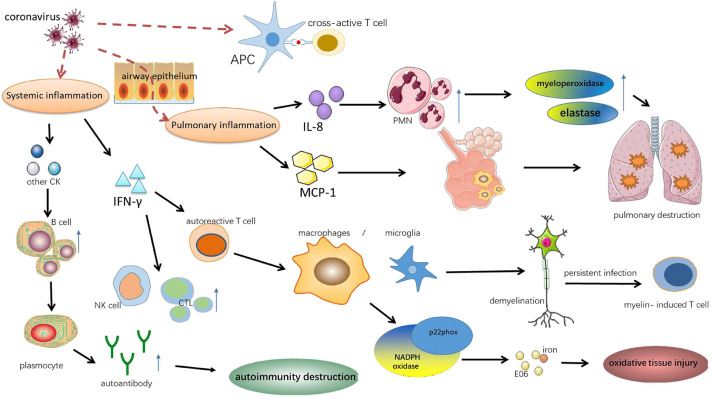
Mounting evidence indicates that coronavirus invasion could cause cross reaction, bystander activation, persistent viral infection and hyper innate inflammatory response, through which augmenting autoimmune responses and affecting autoimmunity (18., 19., 20., 21.) (Figure 2 ).
Figure 2.
Molecular mechanisms linking coronavirus exposure and autoimmunity. There are several molecular mechanisms that explain the link between coronavirus and autoimmunity. Frist, molecular mimicry can active cross-reactive T cells because viral antigens mimic host antigens. Second, coronavirus infection can result in local pulmonary inflammation. Hyper innate inflammatory response can accumulate alveolar macrophages, stimulate PMN to secrete the myeloperoxidase and elastase, which is accompanied with increased MCP-1 and IL-8, eventually lead to pulmonary destruction. Third, the production of inflammatory cytokines activate B cells, resulting autoimmunity destruction by autoantibodies. And IFN-γ activates autoreactive T cell, NK cell and stimulates cytotoxic lymphocyte, causing the oxidative tissue injury and demyelination by macrophages/microglia. IL-8, interleukin-8; CK, cytokines; CTL, cytotoxic lymphocyte; MCP-1, monocyte chemotactic protein 1; IFN-γ, interferon-γ.
Autoimmune diseases have been suggested to be associated with cross reaction, which is defined as reactions of antibodies or activated lymphocytes to different antigens which own the same or similar epitopes. In terms of activated antibodies, SARS-CoV spike protein domain 2 (S2) owns the cross-linked epitopes like A549 cells, the type 2 lung cells. Autoantibodies (mainly IgG) induced by SARS-CoV (S2) can bind to A549 cells, enhance the adhesion of immune cells to epithelial cells and then trigger cytotoxicity which is positively correlated with anti-epithelial cell IgG levels (21). In addition, studies have demonstrated that the disorder of the immune system on the host's antigens may exert a role in the diffuse alveolar damage in SARS (22). When it comes to activated lymphocytes, molecular mimicry can active cross-reactive T cells due to the fact that viral antigens mimic host antigens (19). The cross reactions between human coronavirus 229E antigen and the myelin basic protein are revealed in MS patients (23), and the CDR3 region in Vβ chains of the cross-reactive T cell lines (TCL) is identified by sequencing (24). Then HCoV serotypes (HCoV 229E and HCoV OC43) and their myelin antigen (MBP and PLP) cross-reactive clones are obtained (25).
Bystander activation is a widely accepted virus-induced initiating in autoimmune diseases and occurs when the release of cytokines triggers the autoreactive T cells during a virus-induced immune response (18,19). This mechanism also provides a theoretical basis for the construction of animal models of autoimmune diseases (e.g., MS). The process of bystander T-cell-mediated demyelination induced by mouse hepatitis virus, strain JHM (JHM), is dependent on interferon-gamma (IFN-γ), which in the surrounding of infection might activate autoreactive T cells. These T cells may move to chronic inflammation areas and interact with macrophages/microglia to induce bystander pathology (26).
Besides, the activation of microglia and macrophage is followed by the expression of the NADPH oxidase subunit p22phox. Oxidized lipids (E06) and iron deposition can be detected in oxidative tissue injury induced by MHV-JHM (27).
Coronavirus may cause autoimmune diseases by continually infecting the body. Most animal models can recover from this active disease phase to chronic infection phase, and viral antigens and antibody components can be detected in autopsy samples. Not only can anti-viral antibodies contribute a lot in autoimmune disease but also persistent T cell reaction against infected cells lead to chronic inflammation. For example, during JHMv2.2-1 infection, both the SR T cell and myelin-specific CD4+ T cells contribute to persistent viral infection. Although demyelination is initially associated with a spike in SR T cells, the severity of demyelination during the chronic infection phase is consistent with the trend toward myelin- induced T cell rather than with the SR T cells in bystander activation (28).
Coronavirus invasion is followed by the hyper innate inflammatory response featured by a significant elevation in proinflammatory cytokines and chemokines in serum of the infected (20,29,30). IFN-γ, a Th1-related cytokine, can stimulate NK cells and cytotoxic T lymphocytes, activating the Th1 cell‐mediated immunity (20,29). Moreover, hyper innate inflammatory response can accumulate alveolar macrophages, stimulate polymorphonuclear neutrophil (PMN) to secrete the myeloperoxidase and elastase, which is accompanied with increased MCP-1 and IL-8. Through these ways, hyper innate inflammatory response augments the immune imbalance and leads to the pulmonary destruction (20).
Coronavirus and Autoimmune Diseases
Multiple Sclerosis. Is an autoimmune disease featured by chronic inflammation, axonal damage, and demyelination (31). It has been reported that a virus or virus-triggered immunopathology may promote demyelinating disorder, such as MS (32).Data from some epidemiological studies in MS patients and animal models verify the hypothesis that coronaviruses can initiate the immune pathological reactions in MS. For example, the neurotropic strains of the coronavirus mouse hepatitis virus (JHMV) infection results in demyelination and intracranial axonal degeneration (33), serving as one of the few recognized mouse models for MS (Table 1) (Figure 3).
Table 1.
Links of several autoimmune diseases with coronavirus
| Subjects | Links to coronavirus | References |
|---|---|---|
| MS patients | Positivities for HCV-229E were found in multiple sclerosis cerebrospinal fluids | Cristallo, A et al. (35) |
| MS patients | Activate myelin-reactive T cells | Boucher, A. et al. (24) |
| MS Mice | MS mouse model was triggered by JHMV | Mangale, V. et al. (33) |
| RA patients | Associated with an increasing incidence of RA | Joo, Y. B. et al. (19) |
| SLE patients | Cytoplasmic myxovirus-like renal tubule structure appears in the tissues of SLE patients | Hurd, E. R. et al. (42) |
| SLE patients | Oxidative stress caused by the infection of SARS-CoV-2 will aggravate DNA methylation defect in patients with lupus | Sawalha, A. H. et al. (44) |
| SLE patients | The incidence of certain viruses was significantly higher than that of the general population | Chen CJ, et al. (30); Li TH, et al. (45) |
| SLE patients | CD38 increasing expression in SLE patients can suppress the function of CD8 T cells | Katsuyama, E et al. (47) |
| SARS patients and ITP | Directly infects hematopoietic stem cells and megakaryocytes, Induces antibodies or immune complexes and causes lung damage | Yang, M et al. (49) |
| ITP patients | Severe ITP can be triggered by CoV.HKU145 | Magdi, M et al. (48) |
Abbreviation: MS, multiple sclerosis; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; ITP, idiopathicthrombocytopenic purpura; SARS, severe acute respiratory syndrome; HCV, hepatitis C virus; JHMV, JHM strain of mouse hepatitis virus (mouse coronavirus); Cov.HKU145, coronavirus HKU145.
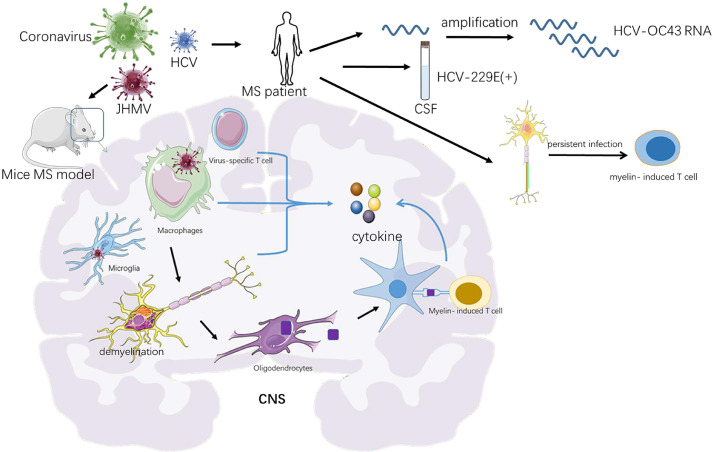
Figure 3.
Potential relationship between multiple sclerosis and coronavirus infections. There is some evidence to reveal the relationship between multiple sclerosis and coronavirus infections. a) The neurotropic strains of the coronavirus mouse hepatitis virus (JHMV) can serve as a recognized mouse models for MS. It is the interaction between T cells, macrophages and microglia that lead to MS pathology featured by axonal damage and demyelination. With the long-term infection of the HMV, myelin-reactive T cells can be active by myelin debris. These cells participate in chronic inflammation by releasing cytokines. b) Previous research has detected polyadenylated RNA sequences of HCV-OC43. c) HCV-229E Positivity were also observed in multiple sclerosis cerebrospinal fluids CNS, central nervous system; MS, multiple sclerosis; CSF, cerebrospinal fluid; JHMV, mouse hepatitis virus
Based on research of experimental coronavirus infections in mice, it has been confirmed that natural infection of coronavirus can lead to pathological features like MS. A study with massive autopsy samples of human brain reported a statistically significant higher morbidity rate of HCV OC43 in MS patients (35.9%; 14 of 39) than in general group(13.7%; 7 of 51) (34). Previous research has revealed that polyadenylated RNA sequences of human coronavirus can be found in cerebrospinal fluid of MS patients and seven positivities for HCV-229E were observed in 20 multiple sclerosis cerebrospinal fluids (35). Hence, two serotypes of human coronavirus, OC43 and 229E, are able to activate myelin-reactive T cells, which add evidence to the possible effect of coronaviruses that drives MS pathogenesis (25). Emerging evidences suggest that it is the interaction between T cells, macrophages and microglia, following with astrocytes that lead to MS pathological features (36).
Rheumatoid Arthritis. Is a chronic autoimmune disease characterized by inflammatory synovitis, and it is hard to cure. Despite the high burden of rheumatoid arthritis, the etiology of this disease remains unclear. An analysis of data from the Korean Centers for Disease Control and Prevention on a population close to the whole country found that respiratory viral infections were associated with the incident of rheumatoid arthritis, and coronavirus was one of these viruses (19). At present, there are some popular pathogenesis mechanisms.
Molecular Mimicry Hypothesis
The structure of some microbial antigens is like that of host autoantigens, so specific antibodies or effector T cells against microbial antigens can react with corresponding host antigens, thus causing autoimmune diseases. Chinese scholars found that the level of interleukin-2 receptor increased with the severity of the disease by analyzing the serum inflammatory cytokines in 29 patients suffering from 2019 novel coronavirus pneumonia (37). American scholars have found that virus-specific memory CD8 T cells could produce a variety of effector cytokines including interleukin-2 while facing SARS virus (38). IL-2 plays a vital role in the activation of regulatory T cells (Treg). Some studies have shown that if this activation pathway is destroyed, autoimmune diseases related to Th1 will occur, including rheumatoid arthritis. The results showed that anti-IL-2 receptor induced Treg's injury, which resulted in systemic inflammation and changes of autoimmune process (39). We can speculate that when coronavirus infects human body, it will lead to the production of many IL-2, which may lead to autoimmunity, and then produce many anti-IL-2, changing the autoimmune process and leading to rheumatoid arthritis. In this process, IL-2 plays a role of mimetic autologous molecule. However, we have not found the direct effect of coronavirus by molecular mimicry mechanism.
Epitope Spreading Hypothesis
Epitope expansion means that the body can respond to the cryptic epitopes one after another, usually by the antigen presenting cells (APC) to provide the cryptic epitopes of their own antigens to the self-reactive lymphocyte clones. Studies have found that rheumatoid arthritis is characterized by the persistence and sustained affinity maturation of the IgA-ACPA response. This may be due to the presence of a persistent mucosal antigen that promotes IgA production, affinity maturation and epitope diffusion, leading to the production of ACPAs, which bind to ACPA and promote TNF production by macrophages (40). Serological analysis of 88 SARS patients in Taiwan showed that the level of TNF-β and other cytokines increased significantly in the acute phase, leading to a series of cytokine storms (41). We hypothesized that coronavirus infection would lead to an elevation of TNF, which may be associated with RA caused by the above epitope expansion leading to an elevation of TNF.
Systemic Lupus Erythematosus. Is a chronic autoimmune disease featured by the formation of pathogenic autoantibodies and immune complexes, which mediates multi-system damages. The cytoplasmic myxovirus-like renal tubule structure appears in the tissues of SLE patients, which is most often reported in kidney biopsies of patients with lupus nephritis (42). Emerging evidence suggests that viral infections are involved in the development of autoimmune diseases (43). Moreover, recent studies have explained the reasons for the increased sensitivity of lupus patients to viral infections, as well as their underlying mechanisms. The increasing oxidative stress caused by the infection of SARS-CoV-2 will aggravate DNA methylation defect in patients with lupus, which will lead to the overexpression of ACE2 and further cause the susceptibility to COVID-19 (44). Besides, Chinese scholars have found that the incidence of certain viruses, such as Herpes Simplex virus (HSV) and Epstein-Barr virus (EBV), in SLE patients was significantly higher than that of the general population (45,46).One of involved mechanisms is that CD38 increasing expression in SLE patients can depress the function of CD8 T cells, resulting in decreased cytotoxic capacity and high susceptibility to infection (47).
Idiopathicthrombocytopenic Purpura. Is an autoimmune disease characterized by isolated low levels of circulating platelets secondary to autoimmune destruction of platelets or inhibition of synthesis (48). The etiology of ITP remains incompletely understood, but recently emerging evidence suggests that coronavirus infection is associated with the prevalence of ITP.
Yang M, et al showed that SARS-CoV could cause thrombocytopenia in patients who got severe acute respiratory syndrome. Three main hypotheses may explain the relationship between coronavirus and ITP. First, coronavirus may directly infect hematopoietic stem cells and megakaryocytes, thus destructing the platelets. Second, antibodies or immune complexes induced by coronavirus may contribute to thrombocytopenia. Third, damaged lung would result in thrombocytopenia for activating platelets to form the thrombi (49). Moreover, a case report in 2019 observed that severe immune thrombocytopenic purpura can be triggered by CoV.HKU1 (48).
Conclusion
Current evidence indicates that coronavirus exerts an indispensable role in the development of autoimmune diseases. Coronavirus can cause diret-infection, elevation of proinflammatory cytokines and chemokines, production of SR T cells and autoantibodies as well as oxidative tissue injury to reinforce the immunity and aggravate autoimmune diseases. The past decades have witnessed many outbreaks of infectious diseases caused by coronavirus. A retrospective study has suggested production of autoimmune antibodies as well as lymphocytopenia (87%) was found in severe and critical cases with COVID-19 (50). Lymphocytopenia is also a characteristic manifestation of SLE and SARS (26), which assumes increase in lymphocyte apoptosis may be a mechanism involved in both coronavirus pneumonia and active lupus. Additional clinical and laboratory data from cases with COVID-19 are needed to better evaluate the exact role of coronavirus in the pathophysiology of autoimmune diseases.
Encouragingly, studies have also indicated the existence of autoimmunity response and provided a theoretical basis to optimize their immune treatment (51). Two cross-sectional studies both observe that patients with autoimmune diseases do not seem to carry a higher risk of SARS-CoV-2 compared with the general population (52,53), while Jose et al. observed a significant elevation in the SARS-CoV-2 infection rate in patients with systemic autoimmune or immune-mediated disease, comparing with controls (54). Additionally, it is widely acknowledged that immunosuppressive treatments will undoubtedly increase the risk of opportunistic infections. How to apply the immunosuppressive treatments and whether to continue it still worth our consideration and research (53,55). Therefore, it is of vital significance to take actions to utilize advanced molecular biology techniques to further confirm and study the mechanism of coronavirus leading to autoimmune diseases at molecular and cellular levels, aiming to formulate a reasonable treatment strategy to optimize immunosuppression, find the appropriate application time and better maintaining dosage and ultimately reduce the incidence of autoimmune diseases caused by coronavirus.
Conflicts of Interest
The authors report no conflicts of interest.
References
- 1.Chan KS, Zheng JP, Mok YW, et al. SARS: prognosis, outcome and sequelae. Respirology (Carlton, Vic) 2003;8(Suppl 1):S36–S40. [DOI] [PMC free article] [PubMed]
- 2.Zheng J, Yamada Y, Fung TS, et al. Identification of N-linked glycosylation sites in the spike protein and their functional impact on the replication and infectivity of coronavirus infectious bronchitis virus in cell culture. Virology. 2018;513:65–74. doi: 10.1016/j.virol.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo PC, Lau SK, Lam CS, et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saghazadeh A, Rezaei N. Immune-epidemiological parameters of the novel coronavirus - a perspective. Expert Rev Clin Immunol. 2020;16:465–470. doi: 10.1080/1744666X.2020.1750954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirchdoerfer RN, Cottrell CA, Wang N, et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Wang FS, Gershwin ME. Human autoimmune diseases: a comprehensive update. J Intern Med. 2015;278:369–395. doi: 10.1111/joim.12395. [DOI] [PubMed] [Google Scholar]
- 8.Gillespie KM, Bain SC, Barnett AH, et al. The rising incidence of childhood type 1 diabetes and reduced contribution of high-risk HLA haplotypes. Lancet (London, England) 2004;364:1699–1700. doi: 10.1016/S0140-6736(04)17357-1. [DOI] [PubMed] [Google Scholar]
- 9.Ascherio A, Munger KL. EBV and Autoimmunity. Curr Top Microbiol Immunol. 2015;390(Pt 1):365–385. doi: 10.1007/978-3-319-22822-8_15. [DOI] [PubMed] [Google Scholar]
- 10.Malik YA. Properties of Coronavirus and SARS-CoV-2. Malays J Pathol. 2020;42:3–11. [PubMed] [Google Scholar]
- 11.Chan-Yeung M, Xu RH. SARS: epidemiology. Respirology (Carlton, Vic) 2003;8(Suppl 1):S9–S14. doi: 10.1046/j.1440-1843.2003.00518.x. Suppl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chafekar A, MERS-CoV Fielding BC. Understanding the Latest Human Coronavirus Threat. Viruses. 2018;10:93. doi: 10.3390/v10020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dosch SF, Mahajan SD, Collins AR. SARS coronavirus spike protein-induced innate immune response occurs via activation of the NF-kappaB pathway in human monocyte macrophages in vitro. Virus research. 2009;142:19–27. doi: 10.1016/j.virusres.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siu KL, Kok KH, Ng MH, et al. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J Biol Chem. 2009;284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeDiego ML, Nieto-Torres JL, Jimenez-Guardeño JM, et al. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 2014;194:124–137. doi: 10.1016/j.virusres.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu X, Pan J, Tao J, et al. SARS-CoV nucleocapsid protein antagonizes IFN-β response by targeting initial step of IFN-β induction pathway, and its C-terminal region is critical for the antagonism. Virus genes. 2011;42:37–45. doi: 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olson JK, Croxford JL, Miller SD. Virus-induced autoimmunity: potential role of viruses in initiation, perpetuation, and progression of T-cell-mediated autoimmune disease. Viral Immunol. 2001;14:227–250. doi: 10.1089/088282401753266756. [DOI] [PubMed] [Google Scholar]
- 19.Joo YB, Lim Y-H, Kim K-J, et al. Respiratory viral infections and the risk of rheumatoid arthritis. Arthritis Res Ther. 2019;21:199. doi: 10.1186/s13075-019-1977-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong CK, Lam CW, Wu AK, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin YS, Lin CF, Fang YT, et al. Antibody to severe acute respiratory syndrome (SARS)-associated coronavirus spike protein domain 2 cross-reacts with lung epithelial cells and causes cytotoxicity. Clin Exp Immunol. 2005;141:500–508. doi: 10.1111/j.1365-2249.2005.02864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo AW, Tang NL, To KF. How the SARS coronavirus causes disease: host or organism? J Pathol. 2006;208:142–151. doi: 10.1002/path.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talbot PJ, Paquette JS, Ciurli C, et al. Myelin basic protein and human coronavirus 229E cross-reactive T cells in multiple sclerosis. Ann Neurol. 1996;39:233–240. doi: 10.1002/ana.410390213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boucher A, Desforges M, Duquette P, et al. Long-term human coronavirus-myelin cross-reactive T-cell clones derived from multiple sclerosis patients. Clin Immunol. 2007;123:258–267. doi: 10.1016/j.clim.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boucher A, Denis F, Duquette P, et al. Generation from multiple sclerosis patients of long-term T-cell clones that are activated by both human coronavirus and myelin antigens. Adv Exp Med Biol. 2001;494:355–362. doi: 10.1007/978-1-4615-1325-4_53. [DOI] [PubMed] [Google Scholar]
- 26.Dandekar AA, Anghelina D, Perlman S. Bystander CD8 T-cell-mediated demyelination is interferon-gamma-dependent in a coronavirus model of multiple sclerosis. Am J Pathol. 2004;164:363–369. doi: 10.1016/S0002-9440(10)63126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuh C, Wimmer I, Hametner S, et al. Oxidative tissue injury in multiple sclerosis is only partly reflected in experimental disease models. Acta Neuropathol. 2014;128:247–266. doi: 10.1007/s00401-014-1263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savarin C, Bergmann CC. Viral-induced suppression of self-reactive T cells: Lessons from neurotropic coronavirus-induced demyelination. J Neuroimmunol. 2017;308:12–16. doi: 10.1016/j.jneuroim.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahallawi WH, Khabour OF, Zhang Q, et al. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine. 2018;104:8–13. doi: 10.1016/j.cyto.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X, Zhao B, Qu Y, et al. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clin Infect Dis. 2020;71:1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owens GP, Gilden D, Burgoon MP, et al. Viruses and multiple sclerosis. Neuroscientist. 2011;17:659–676. doi: 10.1177/1073858411386615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng Y, Skinner DD, Lane TE. Innate Immune Responses and Viral-Induced Neurologic Disease. J Clin Med. 2018;8:3. doi: 10.3390/jcm8010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mangale V, McIntyre LL, Walsh CM, et al. Promoting remyelination through cell transplantation therapies in a model of viral-induced neurodegenerative disease. Dev Dyn. 2019;248:43–52. doi: 10.1002/dvdy.24658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arbour N, Day R, Newcombe J, et al. Neuroinvasion by human respiratory coronaviruses. J Virol. 2000;74:8913–8921. doi: 10.1128/jvi.74.19.8913-8921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cristallo A, Gambaro F, Biamonti G, et al. Human coronavirus polyadenylated RNA sequences in cerebrospinal fluid from multiple sclerosis patients. New Microbiol. 1997;20:105–114. [PubMed] [Google Scholar]
- 36.Savarin C, Dutta R, Bergmann CC. Distinct Gene Profiles of Bone Marrow-Derived Macrophages and Microglia During Neurotropic Coronavirus-Induced Demyelination. Front Immunol. 2018;9:1325. doi: 10.3389/fimmu.2018.01325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L, Liu HG, Liu W, et al. [Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia]. Zhonghua jie he he hu xi za zhi = Zhonghua jiehe he huxi zazhi = Chinese journal of tuberculosis and respiratory diseases 2020;43:203–208. [DOI] [PubMed]
- 38.Channappanavar R, Fett C, Zhao J, et al. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J Virol. 2014;88:11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bo M, Niegowska M, Erre GL, et al. Rheumatoid arthritis patient antibodies highly recognize IL-2 in the immune response pathway involving IRF5 and EBV antigens. Sci Rep. 2018;8:1789. doi: 10.1038/s41598-018-19957-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elliott SE, Kongpachith S, Lingampalli N, et al. Affinity Maturation Drives Epitope Spreading and Generation of Proinflammatory Anti-Citrullinated Protein Antibodies in Rheumatoid Arthritis. Arthritis Rheumatol. 2018;70:1946–1958. doi: 10.1002/art.40587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang KJ, Su IJ, Theron M, et al. An interferon-gamma-related cytokine storm in SARS patients. J Med Virol. 2005;75:185–194. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hurd ER, Dowdle W, Casey H, et al. Virus antibody levels in systemic lupus erythematosus. Arthritis Rheum. 1972;15:267–274. doi: 10.1002/art.1780150308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horwitz MS, Sarvetnick N. Viruses, host responses, and autoimmunity. Immunol Rev. 1999;169:241–253. doi: 10.1111/j.1600-065X.1999.tb01319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sawalha AH, Zhao M, Coit P, et al. Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clin Immunol. 2020;215 doi: 10.1016/j.clim.2020.108410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li TH, Lai CC, Wang WH, et al. Risk of severe herpes simplex virus infection in systemic lupus erythematosus: analysis of epidemiology and risk factors analysis in Taiwan. Ann Rheum Dis. 2019;78:941–946. doi: 10.1136/annrheumdis-2018-214844. [DOI] [PubMed] [Google Scholar]
- 46.Chen CJ, Lin KH, Lin SC, et al. High prevalence of immunoglobulin A antibody against Epstein-Barr virus capsid antigen in adult patients with lupus with disease flare: case control studies. J Rheumatol. 2005;32:44–47. [PubMed] [Google Scholar]
- 47.Katsuyama E, Suarez-Fueyo A, Bradley SJ, et al. The CD38/NAD/SIRTUIN1/EZH2 Axis Mitigates Cytotoxic CD8 T Cell Function and Identifies Patients with SLE Prone to Infections. Cell Rep. 2020;30:112–123.e4. doi: 10.1016/j.celrep.2019.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Magdi M, Rahil A. Severe Immune Thrombocytopenia Complicated by Intracerebral Haemorrhage Associated with Coronavirus Infection: A Case Report and Literature Review. Eur J Case Rep Intern Med. 2019;6 doi: 10.12890/2019_001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang M, Ng MH, Li CK. Thrombocytopenia in patients with severe acute respiratory syndrome (review) Hematology. 2005;10:101–105. doi: 10.1080/10245330400026170. [DOI] [PubMed] [Google Scholar]
- 50.Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Y, Han T, Chen J, et al. Clinical and Autoimmune Characteristics of Severe and Critical Cases with COVID-19. Clin Transl Sci. 2020;13:1077–1086. doi: 10.1111/cts.12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zen M, Fuzzi E, Astorri D, et al. SARS-CoV-2 infection in patients with autoimmune rheumatic diseases in northeast Italy: A cross-sectional study on 916 patients. J Autoimmun. 2020;112 doi: 10.1016/j.jaut.2020.102502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emmi G, Bettiol A, Mattioli I, et al. SARS-CoV-2 infection among patients with systemic autoimmune diseases. Autoimmun Rev. 2020;19 doi: 10.1016/j.autrev.2020.102575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pablos JL, Abasolo L, Alvaro-Gracia JM, et al. Prevalence of hospital PCR-confirmed COVID-19 cases in patients with chronic inflammatory and autoimmune rheumatic diseases. Ann Rheum Dis. 2020 doi: 10.1136/annrheumdis-2020-217763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarzi-Puttini P, Marotto D, Antivalle M, et al. How to handle patients with autoimmune rheumatic and inflammatory bowel diseases in the COVID-19 era: An expert opinion. Autoimmun Rev. 2020;19 doi: 10.1016/j.autrev.2020.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]