Abstract
In a systematic review, the authors explored genetic association studies of essential hypertension in African populations. Studies reporting on the association of polymorphism(s) with hypertension in African populations were included. Appropriate studies were pooled using random effects model meta‐analysis, under six potential inheritance models. In all, 46 polymorphisms in 33 genes were investigated for their association with hypertension or blood pressure levels. Meta‐analysis was possible for three single nucleotide polymorphisms: rs4340, rs699, and rs5186. An association was found between rs5186, rs699, and hypertension under allele contrast and homozygous codominant models (odds ratio, 1.63 [95% confidence interval, 1.04–2.54] and 4.01 [95% confidence interval, 1.17–13.80] for rs5186, respectively; and 1.80 [95% confidence interval, 1.13–2.87] for rs699). Findings were mostly robust in sensitivity analyses. According to the systematic review, there is currently insufficient evidence on the specific polymorphisms that pose the risk of hypertension in African populations. Large‐scale genetic studies are warranted to better understand susceptibility polymorphisms that may be specific to African populations.
Keywords: Africa, blood pressure, diastolic, genetics, hypertension, systolic
1. BACKGROUND
Hypertension was rarely reported in the early 20th century but is now recognized as the leading cardiovascular risk factor in African populations.1, 2 Africa has the highest prevalence of hypertension, particularly among persons 25 years and older, predominantly in sub‐Saharan Africa.1, 3 The increasing prevalence rate of hypertension has been particularly evident in this region, from approximately 80 million in 2000 to projections of 150 million by 2025.4 The burden associated with hypertension relates to cardiovascular5 and renal complications, which have also increased in African populations. The growing hypertension and related complications are attributed to environmental and behavioral changes including exposure to air pollution, increased alcohol consumption, tobacco use, sedentary lifestyle, adoption of a high‐salt diet,6 and consumption of refined sugar and unhealthy fats and oils.
Apart from environmental and lifestyle factors, genetics also play a major role in facilitating hypertension occurrence. According to family and twin studies, the heritability of hypertension ranges from 24% to 50%.7, 8 Hypertension exists not only as a monogenic trait but also as the essential type, which accounts for 95% of all cases of hypertension.9 While monogenic hypertension is well defined with 12 causative genes, essential hypertension involves a complex interaction of multiple polymorphisms in numerous genes. More than 60 single nucleotide polymorphisms (SNPs) have been reported in European populations,10, 11 African Americans,12 and Asians.13, 14 According to these studies, known loci account for only 2.5% of the phenotypic variance for systolic and diastolic blood pressure (BP). It is speculated that the missing heritability may be elucidated through search for rare and structural sequence variants, epigenetics, and investigating gene‐gene and gene‐environment interactions.15 While modifiable risk factors for hypertension are well established in African countries, the contribution of genetic factors remains elusive. The current systematic review focused on collating genetic association studies of hypertension conducted in African populations residing in their respective countries and critically assessing the extent to which these studies were performed.
2. METHODS
2.1. Data source and selection of studies
We conducted a systematic review of genetic association studies performed in populations residing in African countries, following the Systematic Reviews of Genetic Association Studies protocol.16 Four databases (PubMed, Embase, Scopus, and Web of Science) were searched using a combination of terms illustrated in Tables S1 and S2. This was supplemented by searching reference lists for relevant articles. Two investigators independently conducted the literature search to identify all potential studies related to polymorphisms associated with essential hypertension in populations residing in African countries. The last search date was July 21, 2017.
2.2. Selection criteria
Studies were included if they met the following criteria: (1) investigated the association between polymorphisms and essential hypertension in Africa‐based populations, (2) were original studies containing independent data, (3) were a case‐control or cohort design, and (4) contained sufficient data to calculate the odd ratio (OR) with a confidence interval (CI). Studies were excluded if they: (1) were duplicate publications, (2) reported selectively on migrant Africans outside Africa, (3) were family studies, (4) used linkage analysis, (5) were secondary hypertension studies, and (6) analyzed mixed populations of African descent without considering their country of residence.
2.3. Data extraction
Two reviewers independently extracted the following data from selected studies: first author and year of publication, study setting and design, population characteristics (number of cases and controls, and distribution of various genotypes), genetic models, Hardy‐Weinberg equilibrium (HWE) test, and measures of genetic association and adjustment for confounders. Disagreements were resolved by consensus. The presence of bias was assessed by considering the following: case definition, population stratification, and reporting of methods used (sample size, genotyping method and its reliability/accuracy, validation of results, and statistical analyses).
2.4. Statistical analyses
For polymorphisms assessed by more than two studies, we used random effects model meta‐analysis to derive the pooled OR and 95% CIs for the association with prevalent hypertension under six types of inheritance models: contrast, homozygote codominant, heterozygote codominant, dominant, overdominant, and recessive models. The departure from HWE of each study was retested, and credibility of the pooled association was assessed using the Venice interim criteria.17 Heterogeneity across studies was assessed using the Q‐statistic and quantified using the I 2 statistics.18 The Egger test was used to diagnose publication bias.19 Potential outliers were investigated in sensitivity analysis by dropping each study at a time. The Duval and Tweedie trim‐and‐fill method was used to adjust estimates for the effects of publication bias. Data analysis used R version 3.3.3 (2017‐03‐06; The R Foundation for Statistical Computing) and meta‐package. This systematic review is reported according to the recommendation of the HuGE Handbook and Preferred Reporting Items for Systematic Reviews and Meta‐Analyses statement (checklist available in Table S3).
3. RESULTS
3.1. Literature search
Figure 1 summarizes the studies selection process. We retrieved 409 records through searches: PubMed (n = 164), Embase (n = 136), Web of Science (n = 127), Scopus (n = 5), and reference lists (n = 1). After removing duplicates and screening titles and abstracts, the full text of 53 publications was assessed for eligibility. Of these, 15 were excluded for the following reasons: the study population was comprised of family members (n = 5), missing data (n = 1), different study design (eg, investigated the effect of polymorphisms in the plasma renin‐angiotensin‐aldosterone system in patients with hypertension, assessed the combined effect of multiple genes on hypertension, ecological variability and its relationship with fives genes involved in regulation of BP) (n = 3), the frequency of the heterozygous and minor homozygous genotypes were reported as a combined number (n = 2), African populations were analyzed together with African Americans (n = 1), and three articles were not accessible. Finally, 38 studies were included in this review.
Figure 1.
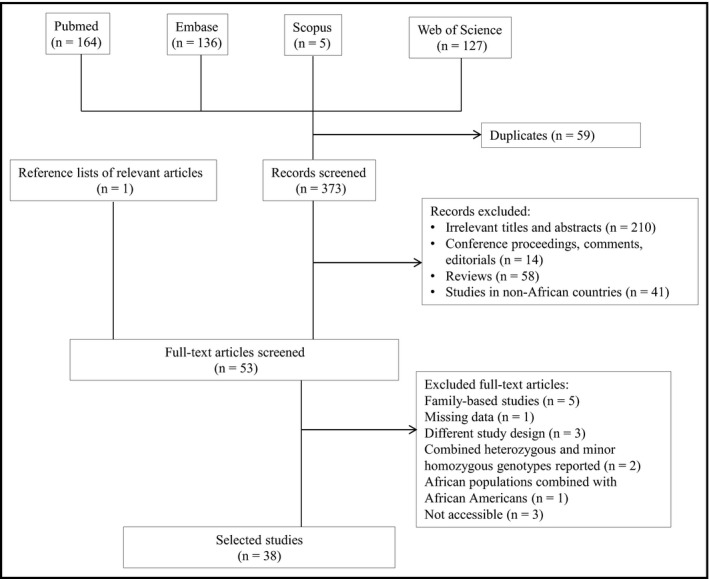
Flowchart for the studies selection process
3.2. Characteristics of studies
Study characteristics are presented in Table S4. The majority (n = 34, 92%) used a case‐control design. Nine studies were conducted in Tunisia; eight in South Africa; six in Egypt; five in Ghana; three in Algeria; and two in Cameroon, Nigeria, and Morocco each; and one in Burkina Faso. Twenty studies (53%) did not report on the ethnicity of the study population. Studies varied according to the number of included participants, which ranged from 65 to 1939. Most studies defined hypertension as systolic BP/diastolic BP (SBP/DBP) ≥140/90 mm Hg or the use of antihypertensive medications at inclusion. Other studies used either auscultatory DBP >90 mm Hg or 24‐hour ambulatory DBP >85 mm Hg20 and auscultatory DBP >95 mm Hg,21 SBP/DBP >139/89 mm Hg,22 SBP/DBP >140/90 mm Hg,23, 24 SBP/DBP ≥125/80 mm Hg,25 seated DBP >60 mm Hg,26 and SBP/DBP >159/94 mm Hg.27 Ranjith and colleages28 did not assign any values to define hypertensive status. On the other hand, Kooffreh and coworkers29 grouped patients with hypertension according to the severity of the disease as prehypertension (SBP/DBP: 120–139/80–89 mm Hg), stage 1 hypertension (SBP/DBP: 140–159/90–99 mm Hg), and stage 2 hypertension (SBP/DBP ≥160/100 mm Hg). AbdRaboh and colleagues30 included patients who had stage 1 hypertension (SBP/DBP > 140–145/90–95 mm Hg) or were using at least one antihypertensive medication.
3.3. Genetic assessment and associations
A total of 46 polymorphisms in 33 genes were investigated for their association with prevalent hypertension and/or BP. None of the studies conducted a genome‐wide scan; instead a candidate gene approach was used. The polymerase chain reaction–restriction fragment length polymorphism was a genotyping method of choice for most studies (55.3%). Five studies22, 28, 30, 31, 32 did not provide covariates adjustment, while six22, 27, 33, 34, 35, 36, 37 reported P values without effect estimates (Table S5). Some investigators included multiethnic populations, but participants were stratified by tribes or chiefdoms in only one study.38
The widely studied polymorphisms were those in genes of the renin‐angiotensin‐aldosterone system (RAAS) including angiotensin‐converting enzyme (ACE) I/D (n = 9 studies), angiotensinogen (AGT) M235T (n = 7), and angiotensin II type 1 receptor (AGTR1) 1166A>C (n = 7). The SNPs of these genes were included in the meta‐analysis. Four of the 30 polymorphisms (CPS1‐T1405N; BSND‐43V>I; SH2B3‐W262R; GNB3‐825C>T; AGT‐T174M) that were investigated by one to three studies deviated from HWE. We also observed variable associations of the 30 SNPs in relation to hypertension or its traits (Table S5). For example, AGT‐T147M was investigated in Algerian39 and Ghanaian33 populations, and demonstrated association with both SBP and DBP only in Ghanaians. On the other hand, MTFHR‐677C>T was studied in Algerians40 and Cameroonians37 with no evidence of association in both populations, while in Moroccans an increased risk of hypertension was found in TT carriers.41 One polymorphism (NPCR‐55C>A) had low minor allele frequency and could not be assessed for its role in hypertension.24 Seventeen of the remaining 27 polymorphisms demonstrated statistically significant associations with hypertension, SBP, or DBP (Table S5).
3.4. Meta‐analysis
Meta‐analysis was possible for only three SNPs within the ACE, AGT, and AGTR1, respectively, rs4340, rs699, and rs5186 (Figures 2, 3, 4). These are the SNPs that were investigated by more than three different studies (Table 1). Details of these studies are summarized in Table 1 and Table S4.
Figure 2.
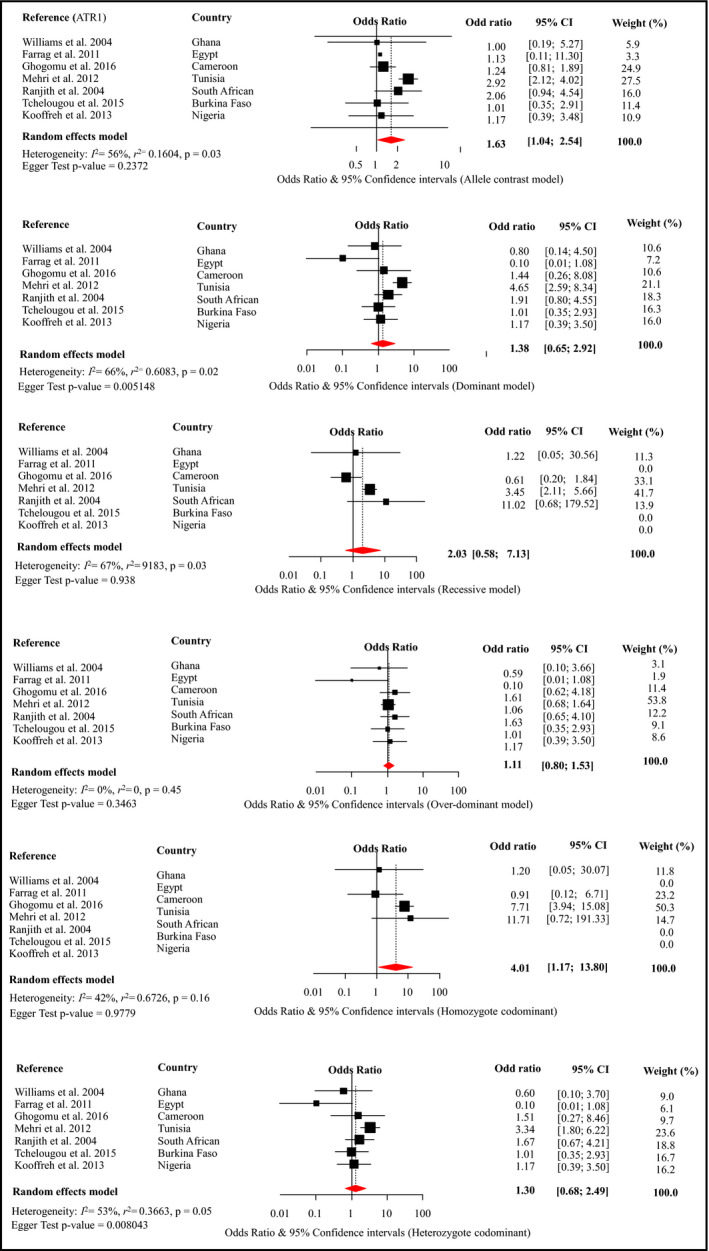
Forest plot for the effects of rs5186 from different genetic models on the risk of hypertension across studies in Africa. Figures panels (top to bottom) are for the “alleles contrast,” “dominant,” and “recessive” models of genetic associations, respectively, for the first column, and “overdominant,” “homozygote codominant,” and “heterozygote codominant” models for the second column. For each figure panel, the black boxes are for the effect estimates (odd ratio) of the association, and the horizontal bars are for the 95% confidence interval (CI). The sizes of the boxes are proportional to the inverse variance of the effect estimates. The diamond beneath the black boxes is for the overall effect estimates across studies, from random effects model meta‐analysis. A dotted vertical line centered on the diamond has been added to assist visual interpretation. For each contributing study, the odd ratio and 95% CI are also shown, together with the weight (in percentage), reflecting the contribution of the study to the overall estimates. The horizontal bar (x‐axis) is on log scale to allow a balanced distribution of the CIs around the effect estimates. The heterogeneity statistics are also shown. ATR1 indicates angiotensin II type 1 receptor
Figure 3.
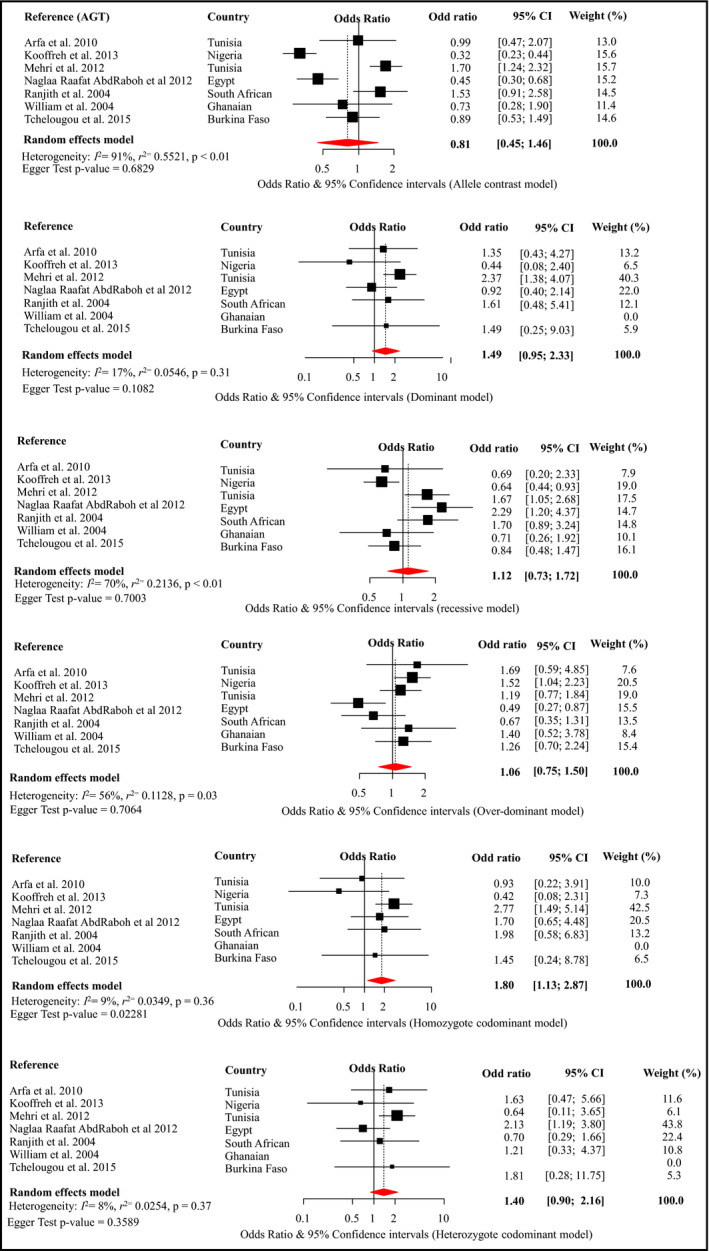
Forest plot for the effects of rs699 from different genetic models on the risk of hypertension across studies in Africa. Conventions are as per Figure 2
Figure 4.
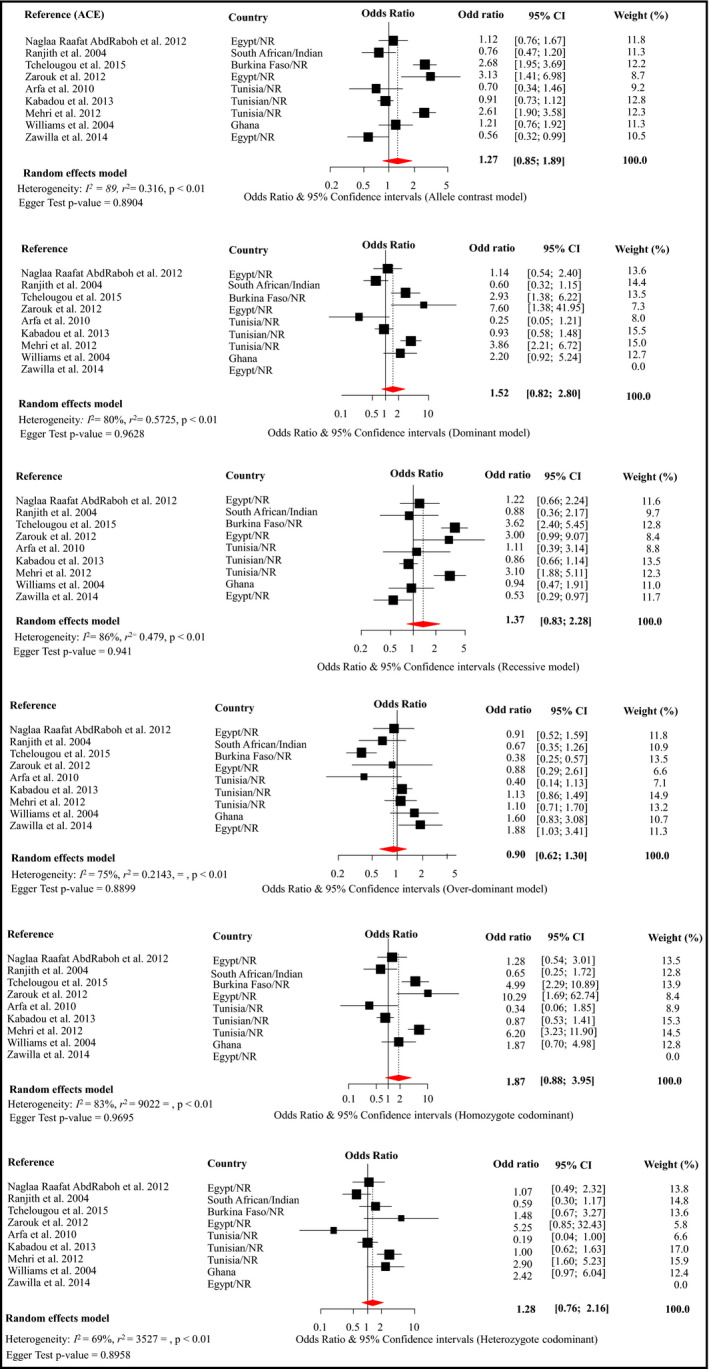
Forest plot for the effects of rs4340 from different genetic models on the risk of hypertension across studies in Africa. Conventions are as per Figure 2
Table 1.
Genotype frequency distribution of the rs4340, rs5186, and rs699 in African populations
| Reference | Country | Genotypes, hypertension, No. | HWE (P value) | Genotypes, no hypertension, No. | HWE (P value) | ||||
|---|---|---|---|---|---|---|---|---|---|
| rs5186 | AA | AC | CC | AA | AC | CC | |||
| Williams et al27 | Ghana | 113 | 3 | 1 | <.0001 | 45 | 2 | 0 | .882 |
| Farrag et al34 | Egypt | 39 | 1 | 0 | .936 | 12 | 3 | 0 | .667 |
| Ghogomu et al36 | Cameroon | 2 | 67 | 5 | <.0001 | 4 | 89 | 11 | <.0001 |
| Mehri et al42 | Tunisia | 17 | 63 | 62 | .871 | 74 | 82 | 35 | .151 |
| Ranjith et al28 | South African | 35 | 6 | 1 | .268 | 410 | 42 | 1 | .950 |
| Tchelougou et al58 | Burkina Faso | 195 | 7 | 0 | .802 | 197 | 7 | 0 | .803 |
| Kooffreh et al59 | Nigeria | 605 | 7 | 0 | .887 | 606 | 6 | 0 | .906 |
| rs699 | MM | MT | TT | MM | MT | TT | |||
|---|---|---|---|---|---|---|---|---|---|
| Arfa et al32 | Tunisia | 10 | 21 | 8 | .618 | 7 | 9 | 6 | .398 |
| Kooffreh et al29 | Nigeria | 4 | 67 | 541 | .232 | 2 | 52 | 642 | .391 |
| Mehri et al60 | Tunisia | 23 | 67 | 52 | .856 | 60 | 82 | 49 | .056 |
| AbdRaboh et al30 | Egypt | 14 | 57 | 39 | .330 | 11 | 64 | 18 | .0002 |
| Ranjith et al28 | South African | 3 | 14 | 25 | .599 | 50 | 193 | 210 | .573 |
| William et al27 | Ghana | 0 | 18 | 92 | .350 | 0 | 6 | 43 | .648 |
| Tchelougou et al58 | Burkina Faso | 2 | 29 | 171 | .541 | 3 | 24 | 177 | .051 |
| rs4340 | II | ID | DD | DD | DI | II | |||
|---|---|---|---|---|---|---|---|---|---|
| AbdRaboh et al30 | Egypt | 17 | 59 | 34 | .299 | 25 | 52 | 16 | .214 |
| Ranjith et al28 | South African | 18 | 18 | 6 | .666 | 72 | 240 | 141 | .071 |
| Tchelougou et al58 | Burkina Faso | 10 | 57 | 135 | .225 | 73 | 104 | 27 | .289 |
| Zarouk et al61 | Egypt | 2 | 14 | 24 | .982 | 7 | 8 | 6 | .279 |
| Arfa et al32 | Tunisia | 14 | 16 | 19 | .017 | 8 | 12 | 2 | .402 |
| Kabadou et al23 | Tunisian | 39 | 176 | 173 | .553 | 205 | 180 | 40 | .957 |
| Mehri et al60 | Tunisia | 20 | 65 | 57 | .832 | 34 | 83 | 74 | .208 |
| William et al27 | Ghana | 14 | 74 | 38 | .014 | 16 | 24 | 11 | .723 |
| Zawilla et al62 | Egypt | NA | 21 | 78 | .238 | 244 | 35 | NA | .264 |
HWE, Hardy‐Weinberg equilibrium.
Meta‐analysis of rs5186 included seven studies totaling the number of cases and controls to 1229 and 1626, respectively. There was evidence of an association between rs5186 and hypertension under allele contrast (OR, 1.63; 95% CI, 1.04–2.54) and homozygous codominant (OR, 4.01; 95% CI, 1.17–13.80) models (Figure 2). Sensitivity analyses suggested that these significant pooled estimates were driven by the effects of Mehri42 and Ranjith28 and colleagues since the pooled estimates for both models were nonsignificant each time one of the two studies was omitted (Figure S1). There was evidence of substantial heterogeneity for the alleles contrast (P = .03) and dominant models (P < .01), driven by the study by Mehri and associates42 (Figure S1), and for the recessive model (P = .03), driven by the study by Goghomu and coworkers36 The Egger test was in favor of significant publication bias for the dominant model (P = .005) and heterozygote codominant model (P = .008). For both models, the trim‐and‐fill method imputed four studies with unrealistic effect estimates (OR ranging from 5.65 to 106.2) (Figure S4). There was a lack of information about genotyping quality control and controlling for population stratification and therefore likelihood of bias. The significant deviation from HWE of the distribution of genotypes (P < .0001 in Ghanaian and Cameroonian populations) indicates an existence of genotyping error. We provided Venice grading ABB or moderate epidemiological credibility for this meta‐analysis (Table 2).
Table 2.
Venice interim assessments of the credibility of each meta‐analysis
| SNP | Phenotype | Studies, No. | Pooled sample size | Genotyping quality control | Deviation from HWE | Risk of population stratification | Venice interim rating | Overall rating |
|---|---|---|---|---|---|---|---|---|
| rs5186 | Hypertension (SBP >140 mm Hg and/or DBP >90 mm Hg, doctor diagnosis) | 7 | 2855 | Not reported | Yes (two studies) | Yes | ABB | Moderate |
| rs699 | Hypertension (SBP >140 mm Hg and/or DBP >90 mm Hg, doctor diagnosis) | 7 | 2967 | Not reported | No | Yes | AAB | Moderate |
| rs4340 | Hypertension (SBP >140 mm Hg and/or DBP >90 mm Hg, doctor diagnosis) | 9 | 2938 | Not reported | Yes (two studies) | Yes | ACA | Weak |
DBP, diastolic blood pressure; HWE, Hardy‐Weinberg equilibrium, SBP, systolic blood pressure.
The rs699 meta‐analysis included seven studies with a total of 1257 cases and 1710 controls. An association between rs699 and hypertension was observed for the homozygous codominant model (OR, 1.80; 95% CI, 1.13–2.87), with no evidence of statistical heterogeneity between studies (I 2 = 9%) (Figure 3). In sensitivity analyses, this effect estimate was enhanced when findings reported by Arfa32 or Kooffreh29 and colleagues were omitted (Figure S2). In similar analyses, omitting AbdRaboh and colleagues30 led to a significant pooled estimate for the dominant model (OR, 1.83; 95% CI, 1.20–2.81 [I 2 = 0]), for the overdominant model (OR, 1.26; 95% CI, 1.00–1.58 [I 2 = 0]), and for the heterozygote codominant model (OR, 1.75; 95% CI, 1.11–2.75 [I 2 = 0]) (Figure S2). There was significant publication bias (P = .023) for the homozygous codominant model. The trim‐and‐fill methods imputed three studies with effect size (OR) ranging from 3.76 to 12.94 and a resulting pooled estimate of 2.27 (1.33–3.85, heterogeneity P = .14)) (Figure S5). The significant P value of the Egger test and lack of reporting on adjusting for population stratification prompted us to grade this meta‐analysis AAB or moderate epidemiological credibility (Table 2). No deviation from HWE was found in studies included in this meta‐analysis (P > .2).
Pooled data on rs4340 included 1198 cases and 1740 controls and suggested no evidence of association with hypertension across the six genetic models (Figure 4). There was substantial heterogeneity across studies in all models (all heterogeneity P < .01), not accounted for by a particular study in sensitivity analysis (Figure S3). For the homozygote codominant models, the pooled estimate reached statistical significance when Kabadou and colleagues23 was dropped, with an OR of 2.14 (95% CI, 1.01–4.77; I 2 = 0.84) (Figure S3). There was no evidence of publication bias (all P > .890 for the Egger test). The trim‐and‐fill methods imputed two studies (OR ranging from 3.13 to 3.35) for the overdominant model, and one study (OR, 0.28) for the homozygote codominant model, but the resulting pooled estimates were always nonsignificant (Figure S6). Although there was no evidence of publication bias, these studies failed to report on the quality control of genotyping methods, and the population stratification was not accounted for, thus the weak (ACA) Venice interim grading. Deviation from HWE was found in the Tunisian and Ghanaian hypertension groups (P = .017) that were included in the studies by Arfa32 and Williams27 and colleagues, respectively.
4. DISCUSSION
Heterogeneous susceptibility to hypertension among different populations has been largely documented. This has been particularly noted in the United States, with studies demonstrating that African Americans generally have higher BP levels and are characterized by early‐onset hypertension compared with other ethnic groups.43 Susceptibility to hypertension is influenced, in part, by variations in regulation of body salt and water among many population groups. One of the well‐studied pathways involved in regulation of thirst and sodium homeostasis is the RAAS, and genetic variants have been extensively investigated in relation to hypertension.44, 45
Studies conducted in Africa so far generally lack the power to detect statistically significant association between SNPs and complex diseases, as we observed in this review. We conducted meta‐analyses of three SNPs in three genes of the RAAS: ACE I/D (rs4340), AGT M235T (rs699), and AGTR1 1166A>C (rs5186) to achieve an optimum power. We observed an association between rs5186 and hypertension, which is in contrast to findings from a meta‐analysis by Liu and coworkers46 in African Americans. Instead, Liu and associates46 observed the rs5186‐hypertension association in Asians and Caucasians in support of findings reported elsewhere.45, 47 Our meta‐analysis also suggests an association of rs699 with hypertension, similar to studies conducted in Asians,48, 49 while no association between any of the common SNPs found in RAAS genes and hypertension has been reported by other authors.50 The inconsistent findings may be attributable to differences in study design, specifically generalization of hypertension by some studies as hypertension,45, 46, 47 while Sun and colleagues50 investigated the salt‐sensitive hypertension phenotype. Regulation of BP in the presence or absence of salt sensitivity varies considerably among individuals within the same or in different ethnic population groups, and this may also contribute to the lack of consistent associations reported by many studies.
The role of AGTR1 and AGT in hypertension has also been highlighted in patients with renal cell cancer, in whom a significant interaction with hypertension was observed.51 Similar to many studies,52 our meta‐analysis found no association between rs4340 and hypertension. In contrast, other studies have reported the association of rs4340 with hypertension.53 Decker and coworkers51 on the other hand found an interaction between ACE and sodium intake, an association that may also be involved in hypertension occurrence. In light of these findings, the role of ACE in hypertension may not be ruled out in African populations, as our meta‐analysis considered one ACE SNP but not its interaction with sodium intake.
The mechanism underlying rs5186‐hypertension association is unclear as the SNP is located within the 3′ untranslated region of AGTR1. It is possible that rs5186 is in linkage disequilibrium with other functional polymorphisms. Alternatively, this SNP may be involved in regulation of AGTR1 expression. Despite the elusive function of the rs5186 SNP, the AGTR1 has been selected as the potential treatment target for hypertension because of its evident association with hypertension in certain population groups46 and effectiveness of its antagonists as antihypertensive therapy.54 However, BP‐lowering efficacy of RAAS inhibitors is not evident in African Americans, probably because of their low renin profile and sensitivity to salt intake.55 Similarly, black populations in Africa have a characteristic salt‐sensitive hypertension56 and may be less responsive to RAAS inhibitors. It is also important to consider that antihypertensive therapy may be influenced by the presence or absence of polymorphisms in RAAS genes. For example, Woodiwiss and coworkers57 demonstrated that genotypes of the functional SNPs in AGT contributed to the variability of antihypertensive responses to ACE inhibitors in individuals of African ancestry. This study, therefore, highlights the importance of investigating sequence variations in the RAAS genes that may be involved in hypertension occurrence and those that may affect response to treatments in African populations.
5. STUDY LIMITATIONS AND STRENGTHS
We found moderate heterogeneity among studies included in our meta‐analysis, particularly for the rs5186 SNP. This may be attributable to heterogeneous populations of the studies included in the meta‐analysis, which included Arabs, Indians, and black and Caucasian Africans. Furthermore, deviation from HWE in two population groups might have contributed to the existing heterogeneity. The studies included and our meta‐analysis did not address the presence of population stratification. Our review has other limitations, such as selective reporting of methodological aspects. Not all of the studies included reported sample size calculation, HWE, and adjustment of confounders. The sample sizes of selected studies were relatively small. Furthermore, the definition of hypertension varied among studies. This may have hampered identification of small effects of other polymorphisms investigated. Our meta‐analysis was also hindered by the small number of genetic association studies conducted in Africa. Only three SNPs have been investigated by more than three African studies. Our study also has several strengths. This is the first systematic review and meta‐analysis of genetic association studies conducted in African populations residing in Africa. We extensively appraised the existing studies using appropriate protocols designed for genetic association studies. We increased the likelihood of detecting significant effects across studies by conducting a meta‐analysis under several genetic models, using the Venice interim criteria to assess the quality of studies included.
6. CONCLUSIONS
Our study findings suggest that, to date, only two SNPs of the AGT (rs699) and AGTR1 (rs5186) are likely to predispose African populations to hypertension, necessitating further investigation in larger‐scale genetic studies so that more susceptibility polymorphisms specific to African populations can be discovered.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
Supporting information
Yako YY, Balti EV, Matsha TE, et al. Genetic factors contributing to hypertension in African‐based populations: A systematic review and meta‐analysis. J Clin Hypertens. 2018;20:485–495. 10.1111/jch.13225
Funding information
South African Medical Research Council.
REFERENCES
- 1. van de Vijver S, Akinyi H, Oti S, et al. Status report on hypertension in Africa––consultative review for the 6th Session of the African Union Conference of Ministers of Health on NCD's. Pan Afr Med J. 2013;16:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ataklte F, Erqou S, Kaptoge S, et al. Burden of undiagnosed hypertension in sub‐saharan Africa: a systematic review and meta‐analysis. Hypertension. 2015;65:291‐298. [DOI] [PubMed] [Google Scholar]
- 3. Danaei G, Finucane MM, Lin JK, et al. National, regional, and global trends in systolic blood pressure since 1980: systematic analysis of health examination surveys and epidemiological studies with 786 country‐years and 5.4 million participants. Lancet. 2011;377:568‐577. [DOI] [PubMed] [Google Scholar]
- 4. Opie LH, Seedat YK. Hypertension in sub‐Saharan African populations. Circulation. 2005;112:3562‐3568. [DOI] [PubMed] [Google Scholar]
- 5. Franklin SS, Wong ND. Hypertension and cardiovascular disease: contributions of the framingham heart study. Glob Heart. 2013;8:49‐57. [DOI] [PubMed] [Google Scholar]
- 6. Rodrigues SL, Baldo MP, Machado RC, et al. High potassium intake blunts the effect of elevated sodium intake on blood pressure levels. J Am Soc Hypertens. 2014;8:232‐238. [DOI] [PubMed] [Google Scholar]
- 7. Kupper N, Ge D, Treiber FA, et al. Emergence of novel genetic effects on blood pressure and hemodynamics in adolescence: The Georgia Cardiovascular Twin Study. Hypertension. 2006;47:948‐954. [DOI] [PubMed] [Google Scholar]
- 8. van Rijn MJ, Schut AF, Aulchenko YS, et al. Heritability of blood pressure traits and the genetic contribution to blood pressure variance explained by four blood‐pressure‐related genes. J Hypertens. 2007;25:565‐570. [DOI] [PubMed] [Google Scholar]
- 9. Padmanabhan S, Caulfield M, Dominiczak AF. Genetic and molecular aspects of hypertension. Circ Res. 2015;116:937‐959. [DOI] [PubMed] [Google Scholar]
- 10. Wellcome Trust Case Control Consortium . Genome‐wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661‐678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. International Consortium for Blood Pressure Genome‐Wide Association Studies , Ehret GB, Munroe PB, Rice KM, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Franceschini N, Fox E, Zhang Z, et al. Genome‐wide association analysis of blood‐pressure traits in African‐ancestry individuals reveals common associated genes in African and non‐African populations. Am J Hum Genet. 2013;93:545‐554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kato N, Takeuchi F, Tabara Y, et al. Meta‐analysis of genome‐wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011;43:531‐538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kelly TN, Takeuchi F, Tabara Y, et al. Genome‐wide association study meta‐analysis reveals transethnic replication of mean arterial and pulse pressure loci. Hypertension. 2013;62:853‐859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747‐753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sagoo GS, Little J, Higgins JP. Systematic reviews of genetic association studies. Human Genome Epidemiology Network. PLoS Med. 2009;6:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ioannidis JP, Boffetta P, Little J, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol. 2008;37:120‐132. [DOI] [PubMed] [Google Scholar]
- 18. Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta‐analyses. BMJ. 2003;327:557‐560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Egger M, Davey Smith G, Schneider M, et al. Bias in meta‐analysis detected by a simple, graphical test. BMJ. 1997;315:629‐634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Candy G, Samani N, Norton G, et al. Association analysis of beta2 adrenoceptor polymorphisms with hypertension in a Black African population. J Hypertens. 2000;18:167‐172. [DOI] [PubMed] [Google Scholar]
- 21. Barlassina C, Norton GR, Samani NJ, et al. α‐Adducin polymorphism in hypertensives of South African ancestry. Am J Hypertens. 2000;13:719‐723. [DOI] [PubMed] [Google Scholar]
- 22. Jones ES, Patricia Owen E, Rayner BL. The association of the R563Q genotype of the ENaC with phenotypic variation in southern Africa. Am J Hypertens. 2012;25:1286‐1291. [DOI] [PubMed] [Google Scholar]
- 23. Kabadou IA, Soualmia H, Jemaa R, et al. G protein β3 subunit gene C825T and angiotensin converting enzyme gene insertion/deletion polymorphisms in hypertensive Tunisian population. Clin Lab. 2013;59:85‐92. [DOI] [PubMed] [Google Scholar]
- 24. Nkeh B, Tiago A, Candy GP, et al. Association between an atrial natriuretic peptide gene polymorphism and normal blood pressure in subjects of African ancestry. Cardiovasc J S Afr. 2002;13:97‐101. [PubMed] [Google Scholar]
- 25. van Deventer CA, Louw R, van der Westhuizen FH, et al. The contribution of the C‐824T tyrosine hydroxylase polymorphism to the prevalence of hypertension in a South African cohort: the SABPA study. Clin Exp Hypertens. 2013;35:614‐619. [DOI] [PubMed] [Google Scholar]
- 26. Sile S, Velez DR, Gillani NB, et al. CLCNKB‐T481S and essential hypertension in a Ghanaian population. J Hypertens. 2009;27:298‐304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Williams SM, Ritchie MD, Phillips JA 3rd, et al. Multilocus analysis of hypertension: a hierarchical approach. Hum Hered. 2004;57:28‐38. [DOI] [PubMed] [Google Scholar]
- 28. Ranjith N, Pegoraro RJ, Rom L, et al. Renin‐angiotensin system and associated gene polymorphisms in myocardial infarction in young South African Indians. Cardiovasc J S Afr. 2004;15:22‐26. [PubMed] [Google Scholar]
- 29. Kooffreh ME, Anumudu CI, Akpan EE, et al. A study of the M235T variant of the angiotensinogen gene and hypertension in a sample population of Calabar and Uyo, Nigeria. EJMHG. 2013;14:13‐19. [Google Scholar]
- 30. AbdRaboh NR, El Din Hemimi NS, Louka ML, et al. Association of angiotensin converting enzyme insertion/deletion and angiotensinogen T235 polymorphisms with risk of essential hypertension in egyptian patients. Int J Cancer Res. 2012;8:69‐82. [Google Scholar]
- 31. Saidi S, Mahjoub T, Almawi WY. Aldosterone synthase gene (CYP11B2) promoter polymorphism as a risk factor for ischaemic stroke in Tunisian Arabs. J Renin Angiotensin Aldosterone Syst. 2010;11:180‐186. [DOI] [PubMed] [Google Scholar]
- 32. Arfa I, Nouira S, Abid A, et al. Lack of association between renin‐angiotensin system (RAS) polymorphisms and hypertension in Tunisian type 2 diabetics. Tunis Med. 2010;88:38‐41. [PubMed] [Google Scholar]
- 33. Robinson M, Williams SM. Role of two angiotensinogen polymorphisms in blood pressure variation. J Hum Hypertens. 2004;18:865‐869. [DOI] [PubMed] [Google Scholar]
- 34. Farrag W, Eid M, El‐Shazly S, et al. Angiotensin II type 1 receptor gene polymorphism and telomere shortening in essential hypertension. Mol Cell Biochem. 2011;351:13‐18. [DOI] [PubMed] [Google Scholar]
- 35. Fisher DL, Plange‐Rhule J, Moreton M, et al. CYP3A5 as a candidate gene for hypertension: no support from an unselected indigenous West African population. J Hum Hypertens. 2016;30:778‐782. [DOI] [PubMed] [Google Scholar]
- 36. Ghogomu SM, Atanga R, Mungwa ST, et al. Lack of association of the A1166C polymorphism in the angiotensin II type 1 receptor (ATR1) gene and essential hypertension in the south west Region of Cameroon. Int J Clin Exp Med. 2016;9:4071‐4076. [Google Scholar]
- 37. Ghogomu SM, Ngolle NE, Mouliom RN, Asa BF. Association between the MTHFR C677T gene polymorphism and essential hypertension in South West Cameroon. Genet Mol Res. 2016;15:(1). [DOI] [PubMed] [Google Scholar]
- 38. Nkeh B, Samani NJ, Badenhorst D, et al. T594M variant of the epithelial sodium channel β‐subunit gene and hypertension in individuals of African ancestry in South Africa. Am J Hypertens. 2003;16:847‐852. [DOI] [PubMed] [Google Scholar]
- 39. Amrani A, Baba Hamed MB, Mesli Talebbendiab F. Association study between some renin‐angiotensin system gene variants and essential hypertension in a sample of Algerian population: case control study. Ann Biol Clin (Paris). 2015;73:557‐563. [DOI] [PubMed] [Google Scholar]
- 40. Amrani‐Midoun A, Kiando SR, Treard C, et al. The relationship between MTHFR C677T gene polymorphism and essential hypertension in a sample of an Algerian population of Oran city. Int J Cardiol. 2016;225:408‐411. [DOI] [PubMed] [Google Scholar]
- 41. Nassereddine S, Kassogue Y, Korchi F, et al. Association of methylenetetrahydrofolate reductase gene (C677T) with the risk of hypertension in Morocco. BMC Res Notes. 2015;8:775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mehri S, Mahjoub S, Hammami S, et al. Renin‐angiotensin system polymorphisms in relation to hypertension status and obesity in a Tunisian population. Mol Biol Rep. 2012;39:4059‐4065. [DOI] [PubMed] [Google Scholar]
- 43. Go AS, Bauman MA, Coleman King SM, et al. An effective approach to high blood pressure control: a science advisory from the American Heart Association, the American College of Cardiology, and the Centers for Disease Control and Prevention. Hypertension. 2014;63:878‐885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rudnichi A, Safar ME, Lajemi M, et al. Gene polymorphisms of the renin‐angiotensin system and age‐related changes in systolic and diastolic blood pressure in subjects with hypertension. Am J Hypertens. 2004;17:321‐327. [DOI] [PubMed] [Google Scholar]
- 45. Ji LD, Zhang LN, Shen P, et al. Association of angiotensinogen gene M235T and angiotensin‐converting enzyme gene I/D polymorphisms with essential hypertension in Han Chinese population: A meta‐analysis. J Hypertens. 2010;28:419‐428. [DOI] [PubMed] [Google Scholar]
- 46. Liu DX, Zhang YQ, Hu B, et al. Association of AT1R polymorphism with hypertension risk: an update meta‐analysis based on 28,952 subjects. J Renin Angiotensin Aldosterone Syst. 2015;16:898‐909. [DOI] [PubMed] [Google Scholar]
- 47. Charita B, Padma G, Sushma P, et al. Estimation of risk and interaction of single nucleotide polymorphisms at angiotensinogen locus causing susceptibility to essential hypertension: a case control study. J Renin Angiotensin Aldosterone Syst. 2012;13:461‐471. [DOI] [PubMed] [Google Scholar]
- 48. Xi B, Shen Y, Yan Y, et al. Association of polymorphisms in the AGT gene with essential hypertension in the Chinese population. J Renin Angiotensin Aldosterone Syst. 2012;13:282‐288. [DOI] [PubMed] [Google Scholar]
- 49. Cheng JL, Wang AL, Wan J. Association between the M235T polymorphism of the AGT gene and cytokines in patients with hypertension. Exp Ther Med. 2012;3:509‐512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sun J, Zhao M, Miao S, et al. Polymorphisms of three genes (ACE, AGT and CYP11B2) in the renin‐angiotensin‐aldosterone system are not associated with blood pressure salt sensitivity: a systematic meta‐analysis. Blood Press. 2016;25:117‐122. [DOI] [PubMed] [Google Scholar]
- 51. Deckers IA, van den Brandt PA, van Engeland M, et al. Polymorphisms in genes of the renin‐angiotensin‐aldosterone system and renal cell cancer risk: interplay with hypertension and intakes of sodium, potassium and fluid. Int J Cancer. 2015;136:1104‐1116. [DOI] [PubMed] [Google Scholar]
- 52. Zaman MM, Yoshiike N, Tanaka H. Dissecting the contradictory findings of angiotensin converting enzyme genetic polymorphism with blood pressure and hypertension. Bangladesh Med Res Counc Bull. 2001;27:90‐95. [PubMed] [Google Scholar]
- 53. Niu W, Qi Y, Gao P, et al. Angiotensin converting enzyme D allele is associated with an increased risk of type 2 diabetes: evidence from a meta‐analysis. Endocr J. 2010;57:431‐438. [DOI] [PubMed] [Google Scholar]
- 54. Te Riet L, van Esch JH, Roks AJ, et al. Hypertension: renin‐angiotensin‐aldosterone system alterations. Circ Res. 2015;116:960‐975. [DOI] [PubMed] [Google Scholar]
- 55. Cooper‐DeHoff RM, Johnson JA. Hypertension pharmacogenomics: in search of personalized treatment approaches. Nat Rev Nephrol. 2016;12:110‐122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rayner BL, Spence JD. Hypertension in blacks: insights from Africa. J Hypertens. 2017;35:234‐239. [DOI] [PubMed] [Google Scholar]
- 57. Woodiwiss AJ, Nkeh B, Samani NJ, et al. Functional variants of the angiotensinogen gene determine antihypertensive responses to angiotensin‐converting enzyme inhibitors in subjects of African origin. J Hypertens. 2006;24:1057‐1064. [DOI] [PubMed] [Google Scholar]
- 58. Tchelougou D, Kologo JK, Karou SD, et al. Renin‐angiotensin system genes polymorphisms and essential hypertension in Burkina Faso, West Africa. Int J Hypertens. 2015;2015:979631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kooffreh ME, Anumudu CI, Duke R, et al. Angiotensin II type 1 receptor A1166C gene polymorphism and essential hypertension in Calabar and Uyo cities, Nigeria. Indian J Hum Genet. 2013;19:213‐218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mehri S, Baudin B, Mahjoub S, et al. Angiotensin‐converting enzyme insertion/deletion gene polymorphism in a Tunisian healthy and acute myocardial infarction population. Genet Test Mol Biomarkers. 2010;14:85‐91. [DOI] [PubMed] [Google Scholar]
- 61. Zarouk WA, Hussein IR, Esmaeil NN, et al. Association of angiotensin converting enzyme gene (I/D) polymorphism with hypertension and type 2 diabetes. Bratisl Lek Listy. 2012;113:14‐18. [DOI] [PubMed] [Google Scholar]
- 62. Zawilla N, Shaker D, Abdelaal A, et al. Angiotensin‐converting enzyme gene polymorphisms and hypertension in occupational noise exposure in Egypt. Int J Occup Environ Health. 2014;20:194‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


