Abstract
To gain a better understanding of the transcriptional response of Aspergillus fumigatus during invasive pulmonary infection, we used a NanoString nCounter to assess the transcript levels of 467 A. fumigatus genes during growth in the lungs of immunosuppressed mice. These genes included ones known to respond to diverse environmental conditions and those encoding most transcription factors in the A. fumigatus genome. We found that invasive growth in vivo induces a unique transcriptional profile as the organism responds to nutrient limitation and attack by host phagocytes. This in vivo transcriptional response is largely mimicked by in vitro growth in Aspergillus minimal medium that is deficient in nitrogen, iron, and/or zinc. From the transcriptional profiling data, we selected 9 transcription factor genes that were either highly expressed or strongly up-regulated during in vivo growth. Deletion mutants were constructed for each of these genes and assessed for virulence in mice. Two transcription factor genes were found to be required for maximal virulence. One was rlmA, which is required for the organism to achieve maximal fungal burden in the lung. The other was sltA, which regulates of the expression of multiple secondary metabolite gene clusters and mycotoxin genes independently of laeA. Using deletion and overexpression mutants, we determined that the attenuated virulence of the ΔsltA mutant is due in part to decreased expression aspf1, which specifies a ribotoxin, but is not mediated by reduced expression of the fumigaclavine gene cluster or the fumagillin-pseruotin supercluster. Thus, in vivo transcriptional profiling focused on transcription factors genes provides a facile approach to identifying novel virulence regulators.
Author summary
Although A. fumigatus causes the majority of cases of invasive aspergillosis, the function of most genes in its genome remains unknown. To identify genes encoding transcription factors that may be important for virulence, we used a NanoString nCounter to measure the mRNA levels of A. fumigatus transcription factor genes in the lungs of mice with invasive aspergillosis. The transcriptional profiling data indicate that the organism is exposed to nutrient limitation and stress during growth in the lungs, and that it responds by up-regulating genes that encode mycotoxins and secondary metabolites. In vitro, this response was most closely mimicked by growth in medium that was deficient in nitrogen, iron and/or zinc. Using the transcriptional profiling data, we identified two transcription factors that govern A. fumigatus virulence. These were RlmA, which is governs factors that enables the organism to proliferate maximally in the lung and SltA, which controls the production of mycotoxins and secondary metabolites.
Introduction
The fungus Aspergillus fumigatus is the major cause of invasive aspergillosis, a progressive pulmonary infection that may disseminate [1–3]. Risk factors for invasive aspergillosis include chemotherapy, corticosteroids, HIV infection, anti-TNF therapy, and solid organ or stem cell transplantation. Because of the growing population of patients at risk of invasive aspergillosis, the annual incidence of this infection has more than tripled since 1990 [2,4]. Moreover, resistance has emerged to azoles, the front-line therapy for invasive aspergillosis [5,6]. Therefore, there is an urgent need to understand A. fumigatus pathogenicity mechanisms to develop new therapeutic and diagnostic approaches.
Gene expression during infection can provide deep insight into virulence determinants. However, among 10,180 predicted genes in the A. fumigatus genome, over 95% are uncharacterized, and fewer than 100 genes have demonstrated roles in virulence. There have been three genome-wide studies of A. fumigatus gene expression during in vivo infection in the mouse model of pulmonary infection [7–9]. These studies revealed that in early germlings there is up-regulation of respiration, central metabolism, and amino acid biosynthesis genes. At later times, as tissue invasion is initiated, there is up-regulation of cation transport, secondary metabolism, and iron metabolism genes. The authors noted consistent up-regulation of secreted protein genes throughout the infection time-course. These gene expression results are mirrored by functional analysis indicating that defects in iron acquisition, amino acid biosynthesis regulation, and secondary metabolite synthesis all lead to reduced virulence in mouse infection models [1,10].
These foundational studies investigated gene expression in A. fumigatus cells recovered by bronchoalveolar lavage. However, it was not possible to investigate gene expression more than 16 h post-infection because the fungal cells had invaded the lung tissue. To investigate A. fumigatus gene expression during tissue invasion, we have taken a different approach, using NanoString technology to assay fungal gene expression in whole lung homogenates. The NanoString nCounter measures RNA levels through probe-based technology and is generally more practical for focused gene set assays than for genome-wide analysis. Here, we used this approach to assay expression of predicted transcription factor genes during invasive growth in the lungs of immunosuppressed mice. Using these data, we selected a panel of transcription factor genes for functional analysis. We found two transcription factor genes, RlmA and SltA, with distinct roles in pathogenicity. RlmA is required for proliferation in the lung, whereas SltA is required for production of mycotoxins, especially Asp f1, that mediate pathogenicity. Furthermore, we determined that in vitro growth of A. fumigatus in Aspergillus minimal medium with low zinc and low nitrogen induced a transcriptional response that was similar to the response induced during invasive growth in the lung of immunosuppressed mice.
Results and discussion
Invasive infection induces expression of genes involved in nutrient acquisition, stress response, and secondary metabolite biosynthesis
To determine the transcriptional response of A. fumigatus during invasive infection, we developed two NanoString probesets. The first was a pilot set that contained probes for 97 genes with functional annotations that included metabolism, iron acquisition, hypoxia, cell wall, and stress response, as described previously [11,12]. The second contained probes for 400 genes that specify virtually all predicted transcription factors in the A. fumigatus genome. We focused on transcription factor genes (TF genes) for three reasons. First, a single transcription factor often controls many functionally related genes, so transcription factor mutants frequently have more prominent phenotypes than mutations in individual target genes [13–17]. Thus, our dataset might be useful for selection of TF genes for functional analysis. Second, there are many methods to identify the indirect or direct targets of a transcription factor, including expression profiling and chromatin immunoprecipitation. Thus, a transcription factor defect can be linked to its target genes to provide physiological and mechanistic insight. Third, transcription factors are prospective drug targets [18], so their analysis can lead to therapeutic benefit.
The transcriptional profiling was performed on RNA isolated from the lungs of mice with invasive aspergillosis, a model that mimics many aspects of human disease [19,20]. In this model, the mice were immunosuppressed with cortisone acetate and then infected with A. fumigatus Af293 via an aerosol chamber, which delivered approximately 5 x103 conidia to the lungs of each mouse. RNA was prepared from whole lung samples at 2, 4 and 5 days post-infection. In this infection model, mortality begins by day 6 post-infection. There was some overlap between the two sets of probes so the final dataset contained expression data for 467 genes. Data quality was excellent at days 4 and 5 post-infection (S1 Fig); the majority of probes gave detectable signals, and R2 values for independent determinations were all >0.95. Data quality was weaker at day 2 post-infection, a reflection of the low A. fumigatus titer at that time.
We expected that once A. fumigatus achieved steady-state invasive growth, its gene expression profile would be similar in successive time points. Our data support this hypothesis, indicating that there are similar gene expression states at days 4 and 5 post-infection (S1 Table). The mean normalized probe counts for all genes assayed at days 4 and 5 were very similar, with an R2 value of 0.96, and no statistical support for differential expression of 88% of genes. The genes that showed most extreme variation between samples were close to the lower detection limits on both days 4 and 5, and thus small changes in the numbers of probe counts lead to large differences in calculated expression ratios. Therefore, our data indicate that gene expression states are very similar on days 4 and 5, shortly before mortality begins.
Growth in the mouse lung resulted in an extensive change in the A. fumigatus transcriptional profile compared to growth in Aspergillus minimal medium (AMM). Of the 467 genes analyzed, 125 (27%) were up-regulated by at least 2-fold and 85 (18%) were down-regulated by at least 2-fold at 5 days post-infection (S1 Table). There was significant up-regulation of genes involved in iron acquisition, including fre2, hapX, sidA, sidD, mirB, and sit1 (Table 1). Concomitantly, there was down-regulation of sreA, whose product represses iron uptake and siderophore synthesis [21]. Other up-regulated genes include zrfC, zrfA, aspf2, and zafA, which govern zinc uptake, nrtB and areA, which control nitrogen uptake, and Afu5g00710, which specifies a GABA permease (Table 1). We infer that growth in the lung imposes limitation for key nutrients, including iron, zinc, and nitrogen, on A. fumigatus. Our results are consistent with prior studies of A. fumigatus mutants with defects in iron acquisition (ΔhapX, ΔsidA), zinc homeostasis (ΔzafA), and nitrogen uptake (ΔareA), all of which have attenuated virulence in mice [22–25].
Table 1. Fold-change and probe counts for selected A. fumigatus genes during growth in Aspergillus minimal medium (AMM) and during invasive growth in the mouse lung on days 2, 4, and 5.
| Fold-change relative to growth in AMM | Normalized Probe Counts | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Process | Gene ID | Gene Name | Function | Day 2 | Day 4 | Day 5 | AMM | Day 2 | Day 4 | Day 5 |
| Iron homeostasis | ||||||||||
| Afu1g17270 | fre2 | Metalloreductase involved in response to iron starvation | 1.09 | 23.14 | 21.64 | 232 | 253 | 5369 | 5019 | |
| Afu5g03920 | hapX | bZIP transcription factor required for adaption to both iron depletion and excess and for transcriptional activation of the siderophore system | 1.18 | 4.19 | 3.51 | 948 | 1123 | 3974 | 3326 | |
| Afu2g07680 | sidA | L-ornithine N5-oxygenase; first committed step in siderophore biosynthesis | 3.06 | 10.93 | 9.07 | 9646 | 29495 | 105444 | 87529 | |
| Afu3g03420 | sidD | Nonribosomal peptide synthetase 4; involved in extracellular siderophore biosynthesis | 4.98 | 15.17 | 17.36 | 1000 | 4981 | 15166 | 17351 | |
| Afu3g03640 | mirB | Putative siderophore iron transporter | 5.27 | 31.94 | 22.07 | 1231 | 6480 | 39307 | 27157 | |
| Afu7g06060 | sit1 | Putative siderophore transporter | 0.44 | 10.50 | 6.88 | 1126 | 498 | 11822 | 7751 | |
| Afu5g11260 | sreA | GATA transcription factor that regulates iron uptake | 0.00 | 0.14 | 0.08 | 2310 | 0 | 316 | 177 | |
| Zinc homeostasis | ||||||||||
| Afu4g09560 | zrfC | Zinc transporter that functions in neutral or alkaline environments | 58.86 | 158.24 | 143.41 | 248 | 14598 | 39244 | 35566 | |
| Afu1g01550 | zrfA | Putative plasma membrane zinc transporter | 18.82 | 10.41 | 8.15 | 19 | 354 | 196 | 153 | |
| Afu4g09580 | aspf2 | Allergen Asp f 2; expressed in alkaline zinc-limiting conditions | 47.91 | 218.08 | 136.52 | 170 | 8141 | 37057 | 23198 | |
| Afu1g10080 | zafA | Putative C2H2 zinc-responsive transcriptional activator | 3.99 | 2.06 | 2.40 | 1010 | 4034 | 2084 | 2429 | |
| Nitrogen uptake | ||||||||||
| Afu1g17470 | nrtB | Putative high-affinity nitrate transporter | 461.99 | 16.95 | 32.20 | 13 | 6162 | 226 | 429 | |
| Afu6g01970 | areA | Putative GATA-like transcription factor; required for growth on numerous nitrogen sources | 0.00 | 1.71 | 2.08 | 1891 | 0 | 3234 | 3934 | |
| Afu5g00710 | GABA permease | 9.63 | 45.45 | 39.14 | 168 | 1616 | 7633 | 6572 | ||
| Stress response | ||||||||||
| Afu2g01260 | srbA | Sterol regulatory element binding protein (SREBP); basic helix-loop-helix leucine zipper DNA binding domain | 0.85 | 1.81 | 1.73 | 9403 | 7991 | 16995 | 16299 | |
| Afu3g04070 | hacA | bZIP transcription factor, major regulator of the unfolded protein response | 0.54 | 0.46 | 0.46 | 12752 | 6930 | 5895 | 5926 | |
| Afu3g11970 | pacC | C2H2 finger domain transcription factor; required for response to alkaline pH | 0.59 | 1.73 | 1.72 | 1864 | 1100 | 3230 | 3202 | |
| Afu4g09080 | sebA | Putative transcription factor; localizes to the nucleus in response to oxidative stress and heat shock | 4.56 | 6.42 | 5.33 | 868 | 3959 | 5571 | 4623 | |
| Afu1g05800 | mkk2 | Putative mitogen-activated protein kinase kinase; essential for cell wall integrity signaling | 0.11 | 2.91 | 2.62 | 1600 | 175 | 4662 | 4193 | |
| Afu5g08420 | sho1 | Putative transmembrane osmosensor | 0.44 | 2.30 | 2.42 | 2990 | 1309 | 6864 | 7233 | |
| Afu5g04170 | hsp90 | Heat shock protein; allergen Asp f 12; | 0.01 | 0.18 | 0.19 | 7795 | 90 | 1406 | 1518 | |
| Afu8g01670 | cat2 | Putative bifunctional catalase-peroxidase | 0.22 | 0.45 | 0.22 | 1450 | 319 | 655 | 312 | |
| Afu4g11580 | sod2 | Putative manganese-superoxide dismutase | 0.10 | 0.20 | 0.23 | 932 | 90 | 185 | 211 | |
| Afu4g00860 | dprA | Dehydrin-like protein; plays a role in oxidative, osmotic and pH stress responses | 0.43 | 0.50 | 0.44 | 210 | 90 | 105 | 92 | |
| Secondary metabolite production | ||||||||||
| Afu6g09690 | gliG | Glutathione S-transferase encoded in the gliotoxin biosynthetic gene cluster | 5469.48 | 6614.27 | 9370.72 | 0 | 882 | 1066 | 1510 | |
| Afu6g09660 | gliP | Non-ribosomal peptide synthetase encoded in the gliotoxin biosynthetic gene cluster | 805.63 | 649.15 | 657.75 | 47 | 38108 | 30706 | 31113 | |
| Afu6g09630 | gliZ | Zn2Cys6 binuclear transcription factor, regulates genes required for gliotoxin biosynthesis | 31.18 | 50.79 | 39.87 | 114 | 3552 | 5786 | 4542 | |
| Afu6g02690 | mtfA | Transcription factor involved in regulation of morphogenesis, gliotoxin production and virulence | 2.47 | 8.05 | 9.08 | 351 | 867 | 2822 | 3184 | |
| Afu7g00130 | nscR | Pathway-specific Zn(II)2Cys6 transcriptional factor; role in neosartoricin and fumicycline A biosynthesis | 21.31 | 209.35 | 308.36 | 12 | 263 | 2582 | 3803 | |
| Afu3g00590 | aspHS | Asp-hemolysin; hemolytic toxin | 26.33 | 0.15 | 4.07 | 103 | 2707 | 15 | 419 | |
| Afu5g02330 | aspf1 | Allergen Asp f 1; ribonuclease mitogillin family of cytotoxins | 0.00 | 0.01 | 0.00 | 99853 | 468 | 951 | 320 | |
| Afu8g00420 | fumR | C6 zinc finger domain protein required for expression of fumagillin and pseurotin gene clusters | 0.10 | 0.15 | 0.12 | 2416 | 237 | 365 | 290 | |
| Afu3g12890 | hasA | C6 transcription factor; hexadehydroastechrome biosynthesis | 0.00 | 0.21 | 0.15 | 8234 | 0 | 1712 | 1231 | |
| Afu4g14540 | tpcE | Putative Zn2Cys6 transcription factor involved in trypacidin biosynthesis | 0.00 | 0.23 | 0.16 | 172 | 0 | 40 | 27 | |
Some TF genes that are known to govern virulence were not strongly up-regulated by invasive growth in the lung. These include srbA, which is required for growth under hypoxia [26], hacA which governs the unfolded protein response [27], and pacC, which is required for growth under alkaline pH [7]. The expression of these TF genes was increased by less than 2-fold in vivo (srbA and pacC) or even reduced (hacA) (Table 1). However, the NanoString counts indicated that all three genes were among the 30 most highly expressed genes in vivo. This result suggests that in A. fumigatus, TF genes that are highly expressed in vivo are likely to govern virulence, even if their expression in vivo does not increase relative to growth in vitro. We have observed a similar relationship between in vivo gene expression levels and virulence regulation in C. albicans [13].
In corticosteroid-treated mice with invasive aspergillosis, the invading hyphae stimulate an influx of neutrophils into the lung that accumulate around the organisms [19,20]. These neutrophils are almost certainly responsible for the up-regulation of genes that are required for normal stress response and virulence in A. fumigatus, such as sebA, mkk2, and sho1 [28–30] (Table 1). Surprisingly, some genes involved in stress response were actually down-regulated during in vivo growth. These genes included hsp90, cat2, sod2, and dprA (Table 1). Collectively, these results indicate that in vivo growth induces expression of only a subset of stress response genes.
Growth in the lung also altered the expression of genes involved in the production of specific secondary metabolites. As compared to organisms grown in AMM, organisms in the mouse lung had a 700–10,000-fold increase in expression of gliG and gliP, which specify enzymes in the gliotoxin biosynthesis pathway [31–33] (Table 1). Furthermore, there was 40-fold up-regulation of gliZ¸ which encodes the transcription factor that governs gliotoxin synthesis [34], and 9-fold up-regulation of mtfA, which induces synthesis of both gliotoxin and extracellular proteases [35] (Table 1). In vivo growth also up-regulated expression of nscR whose product governs synthesis of neosartoricin [36] and of aspHS, which specifies a hemolysin (Table 1). However, in vivo growth repressed the expression of aspf1 which encodes a ribotoxin [12,37], fumR, which governs production of fumagillin and pseurotin [38,39], hasA, which controls production of hexadehydroastechrome [40], and tpcE, which regulates production of trypacidin and questin [41] (Table 1). Thus, growth in the mouse lung induces production of a distinct subset of mycotoxins. We speculate that the mycotoxin genes whose expression was down-regulated in the lung must be expressed in other environmental niches, possibly including other anatomic sites within the host.
Comparisons among gene expression profiles to identify in vitro conditions that mimic in vivo growth
Similarities among gene expression profiles can reveal parallels among diverse genetic or environmental regulatory inputs. To assess similarity, we adapted our Fisher’s Exact Test (FET) comparison tool for use with A. fumigatus gene expression data. We created a database from 129 published microarray or RNA-seq datasets and generated NanoString profiles of A. fumigatus during growth in standard AMM and in AMM modified to mimic aspects of in vivo growth: limitation for nitrogen, iron, zinc, or oxygen; presence of serum; limitation for combinations of nitrogen, iron, and zinc. Datasets with significant similarity among genes up-regulated on day 5 postinfection (compared to growth in AMM) are listed in Table 2. A heat map clustering of some of these datasets is shown in Fig 1.
Table 2. Comparisons among datasets of differentially expressed genes.
| Dataset | Similarity to lung day 5 vs AMM (P-value) |
|---|---|
| lung day 2 vs AMM | <1.00E-10 |
| lung day 4 vs AMM | <1.00E-10 |
| AMM low N Fe vs AMM | <1.00E-10 |
| AMM low N Fe Zn vs AMM | <1.00E-10 |
| AMM low Zn vs AMM | <1.00E-10 |
| AMM low Fe Zn vs AMM | <1.00E-10 |
| AMM low N Zn vs AMM | <1.00E-10 |
| AMM low Fe vs AMM | 2.38E-10 |
| AMM low N vs AMM | 6.20E-08 |
| lung germling 12–14 hr vs YPD 37° [8] | 4.57E-07 |
| lung germling 12 hr vs spores [7] | 7.30E-07 |
| lung germling 16 hr vs spores [7] | 9.00E-07 |
| AMM+serum vs AMM | 1.35E-06 |
Description: Genes up-regulated ≥2 fold in our lung day 5 vs AMM dataset were compared to genes up-regulated ≥2 fold in each comparison dataset listed above. The probability that a correlation was due to chance alone was calculated using Fisher’s Exact Test. The calculated threshold of significance is a P-value of 0.00017.
Fig 1. Hierarchical clustering of gene expression datasets.
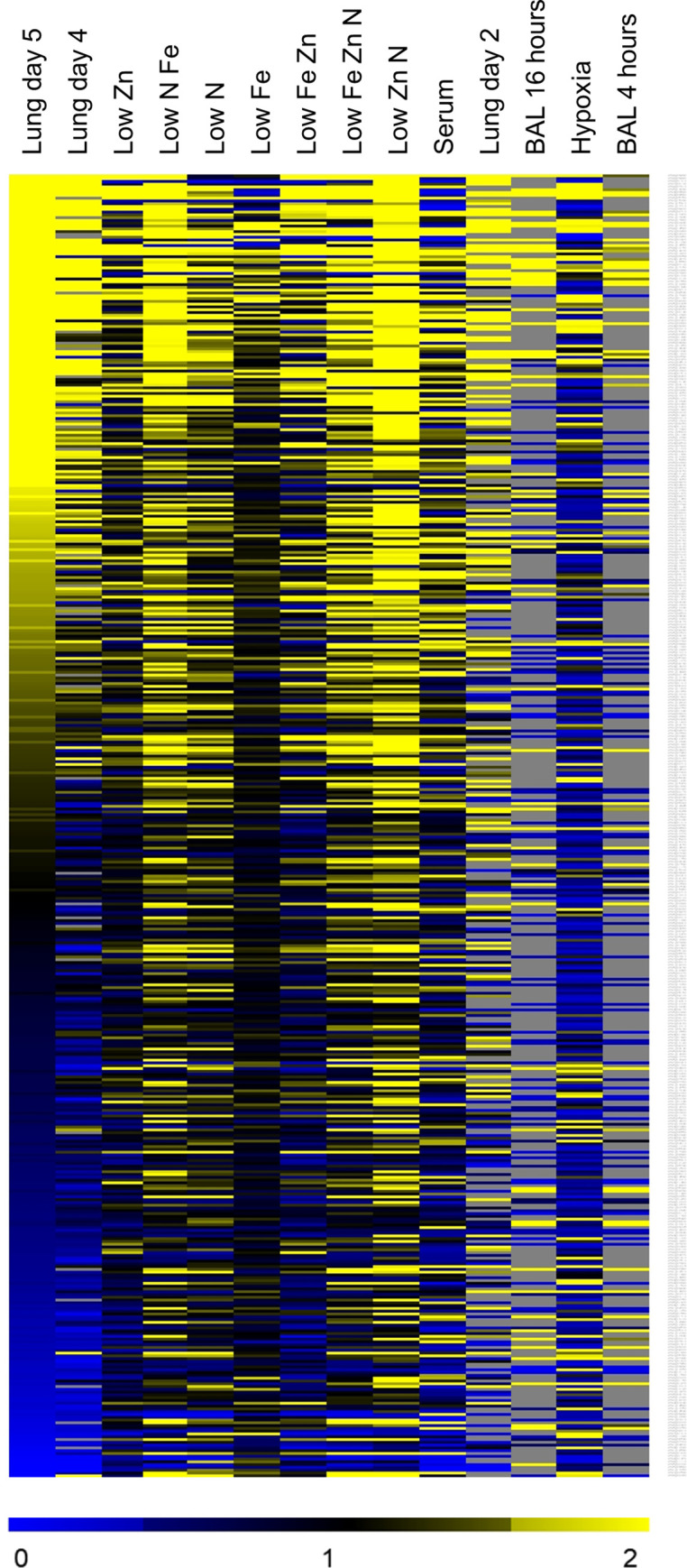
The NanoString datasets and published datasets were compared by hierarchical clustering based on the 467 genes in the NanoString datasets. Select dataset are indicated, including lung germlings [7,8], invasive infection (current data), growth in low zinc or low nitrogen (current in vitro data for Aspergillus minimal medium (AMM) lacking zinc or nitrate alone, or in combination with each other and with limiting iron), and low iron alone [21]. Grey areas indicated genes with undetectable expression.
These comparisons indicated that our transcriptional profiles of invasive A. fumigatus were significantly similar to the published profiles of germlings isolated from the lungs by lavage [7,8] (Table 2). These correlations make sense, because we would expect common regulatory pathways that govern growth in the lung environment. It is probable that the incomplete overlap between these profiles is due to changes in the microenvironment that the organism experiences as it invades the lung parenchyma and is attacked by phagocytes.
The FET comparison also identified some in vitro growth conditions that induced transcriptional profiles that were similar to what was induced by invasive growth in the lungs. While in vitro growth in the presence of serum correlated with invasive growth in vivo, the most significant correlations were with gene expression in organisms grown in AMM that was limited in nitrogen, iron, zinc, or combinations of the three (Table 2). These correlations also make sense, because all invasive pathogens must combat nutritional immunity imposed by the host. Our data thus fit well with current understanding of A. fumigatus infection biology. In addition, these data indicate growing A. fumigatus in AMM that is deficient in nitrogen, iron and/or zinc induces a transcriptional response that is quite similar to that induced by invasive growth in the lungs.
Analysis of deletion mutants to assess transcription factor function in invasive aspergillosis
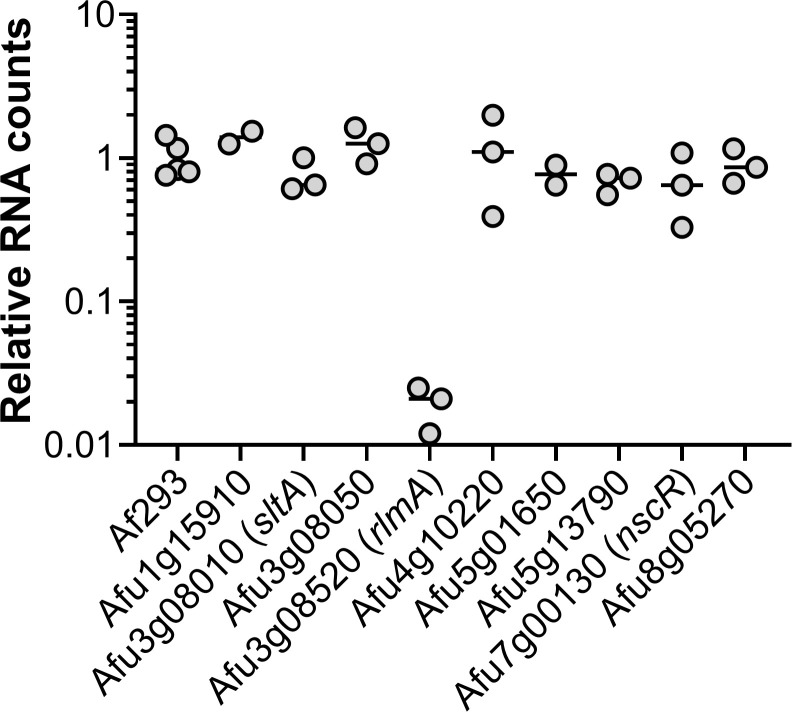
We sought to use our expression profiling data to prioritize TF genes for functional analysis during invasive aspergillosis. We chose a set of 9 A. fumigatus genes that were either highly expressed or highly upregulated during invasive growth in the lung, and whose functions had not been reported at the time of experimentation (Table 3). We created a deletion mutant for each gene and tested the mutants for proliferation in the mouse model of invasive aspergillosis. In this screen, proliferation was assayed by NanoString measurement of A. fumigatus rRNA levels relative to mouse housekeeping gene RNA (ACTB, GAPDH, and PPIA) levels in whole lung homogenates. The ΔrlmA mutant displayed 10-fold lower levels of A. fumigatus rRNA than the wild-type strain or any other mutant (Fig 2). The ΔsltA mutant displayed rRNA levels comparable to those of the wild-type strain, but the lung tissue of the mice infected with the ΔsltA mutant appeared to be notably healthier compared to that of mice infected with the wild-type strain. Also, the mass of lungs of the mice infected with the ΔsltA mutant was significantly less than those of mice infected with the wild-type strain (S2 Fig), suggesting that this mutant induced less inflammation or tissue damage. Therefore, we chose to pursue analysis of rlmA and sltA during infection.
Table 3. Genes encoding potential transcription factors that were selected for construction of deletion mutants.
| Gene ID | Gene Name | Fold-change (relative to growth in AMM) | Mean lung probe counts (day 5) | |||
|---|---|---|---|---|---|---|
| AMM 24 h | Lung (day 2) | Lung (day 4) | Lung (day 5) | |||
| Afu1g15910 | 1 | 26.4 | 35.1 | 43.1 | 11138 | |
| Afu3g08010 | SltA | 1 | 0.8 | 1.0 | 0.9 | 10221 |
| Afu3g08050 | 1 | 0.4 | 3.9 | 6.2 | 4186 | |
| Afu3g08520 | RlmA | 1 | 2.2 | 2.1 | 2.1 | 4134 |
| Afu4g10220 | 1 | 0.9 | 2.0 | 1.8 | 21511 | |
| Afu5g01650 | 1 | 1.6 | 2.5 | 2.3 | 27841 | |
| Afu5g13790 | 1 | 29.2 | 1.9 | 2.6 | 815 | |
| Afu7g00130 | NscR | 1 | 21.3 | 209.4 | 308.4 | 3803 |
| Afu8g05270 | 1 | 0.5 | 2.2 | 2.0 | 3969 | |
Fig 2. RlmA is required for growth in the lungs during invasive aspergillosis.
Pulmonary fungal burden of mice after 5 days of infection with either A. fumigatus strain Af293 or mutants deleted for the indicated genes. Lung fungal burden was determined by NanoString measurement of A. fumigatus rRNA levels relative to mouse ACTB, GAPDH, and PPIA levels. Results are from 2–3 mice per strain and are normalized data from mice infected with strain Af293.
RlmA is required for maximal lung fungal burden during invasive aspergillosis
RlmA is a putative MADS-box transcription factor whose orthologs in many ascomycetes function in cell wall integrity. rlmA RNA levels were down-regulated in the lung germling datasets at 4, 8, and 12 hours post-infection, then began to increase slightly at 16 hours [7]. We found that rlmA was up-regulated in our invasive infection datasets at 2, 4, and 5 days post-infection (Table 3). These results led us to the simple hypothesis that RlmA may be required specifically for invasive infection. It is neither up- nor down-regulated 2-fold in any of the in vitro published datasets we collected. This observation suggested to us that rlmA may be regulated by a signal that is distinctive of the invasive infection environment.
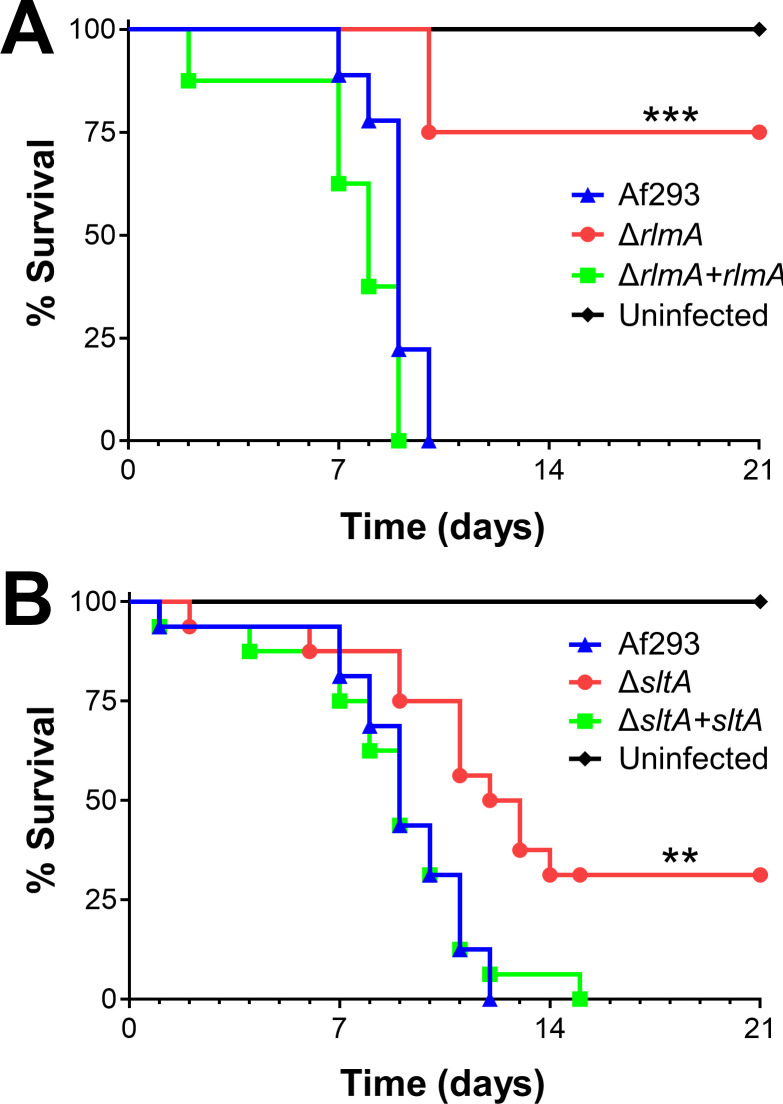
To test RlmA function during invasive aspergillosis, we characterized a ΔrlmA deletion mutant in the Af293 background. Mice infected with the ΔrlmA mutant survived significantly longer than those infected with the wild-type or ΔrlmA+rlmA complemented strains (Fig 3A). This result indicates that RlmA is required for pathogenicity in mice immunosuppressed with corticosteroids. Recently, another group reported RlmA is a member of the cell wall integrity pathway in A. fumigatus and required for virulence in neutropenic mice [42].
Fig 3. ΔrlmA and ΔsltA mutants have attenuated virulence.
Survival of corticosteroid-immunosuppressed mice infected with the indicated strains. Results are the combined data from two independent experiments, each using 8 mice per strain. **, p < 0.01; ***, p < 0.001.
SltA governs production of secondary metabolites and toxins during invasive aspergillosis
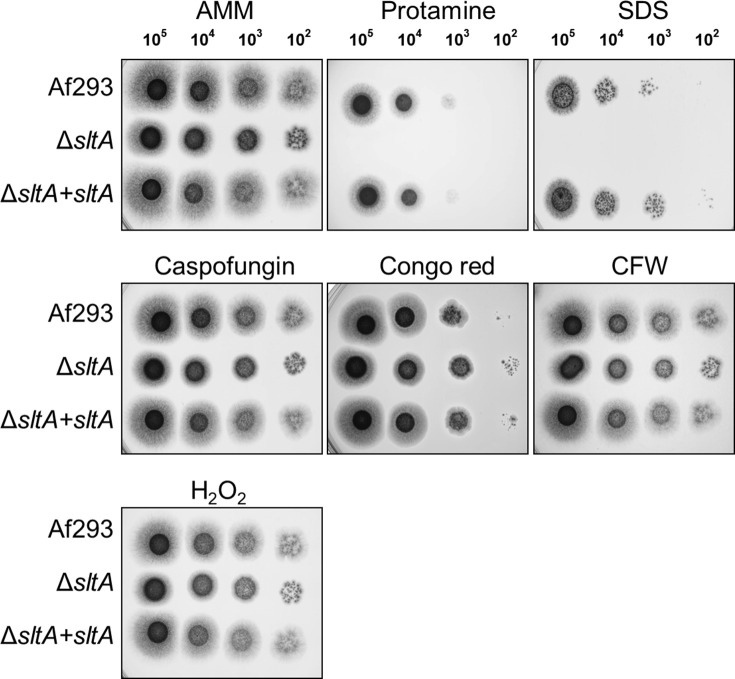
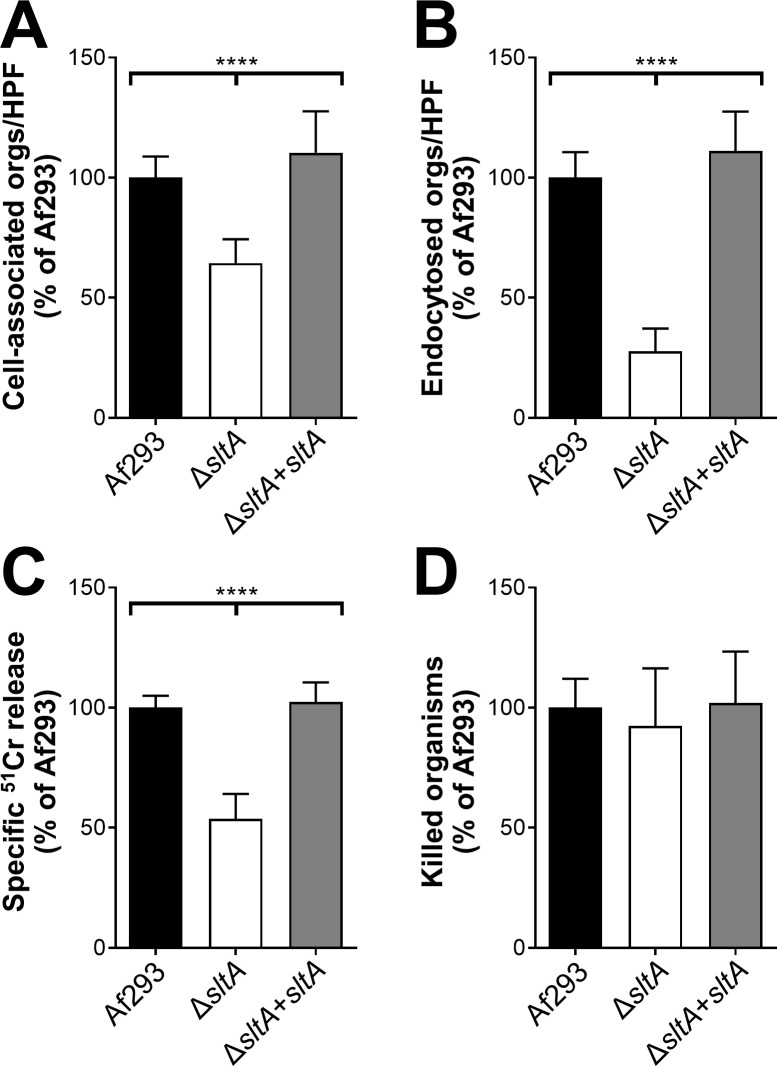
SltA is a C2H2 zinc finger protein whose A. nidulans ortholog, SltA, governs ion homeostasis, growth under alkaline pH, and sporulation [43,44]. Recently, SltA has been found to control azole resistance in A. fumigatus, but its role in virulence was previously unknown [45]. We constructed an A. fumigatus ΔsltA mutant and found that it grew comparably to the wild-type strain in the presence of high cations and at pH 8 (S3 Fig). The ΔsltA mutant also sporulated similarly to the wild-type strain. Although the ΔsltA mutant had increased susceptibility to cell membrane stress caused by protamine and SDS, it had wild-type susceptibility to Congo red, caspofungin, calcofluor white, and hydrogen peroxide (Fig 4). The ΔsltA mutant had reduced capacity to adhere to, invade, and damage the A549 pulmonary epithelial cell line, but had wild-type susceptibility to macrophage killing (Fig 5). These results suggest that SltA governs the response to cell membrane stress and the capacity of A. fumigatus to invade and damage host cells.
Fig 4. Increased susceptibility of the ΔsltA mutant to protamine and SDS.
Serial 10-fold dilutions of the indicated strains of A. fumigatus were spotted onto Aspergillus minimal medium (AMM) containing the indicated stressors. The plates were imaged after incubation at 37°C for 2 d. CFW, calcofluor white.
Fig 5. The ΔsltA mutant is defective in pulmonary epithelial cell adherence, invasion, and damage.
(A-B) The indicated strains of A. fumigatus were incubated with the A549 pulmonary epithelial cell line for 2.5 h, after which the number of cell-associated (A; a measure of adherence) and endocytosed (B) organisms was determined by a differential fluorescence assay. (C) The extent of epithelial cell damage induced by the indicated strains after 16 h of infection. (D) The percentage of cells of the indicated A. fumigatus strains that were killed by mouse bone marrow-derived macrophages after 8 h of infection. Results are mean ± SD of 3 experiments, each performed in triplicate. Orgs/HPF, organisms per high-powered field; ****, p < 0.0001.
In the mouse model of invasive aspergillosis, our rRNA-based titer measurement described above indicated that the ΔsltA and wild-type strains proliferated to similar levels in the lung at day 5 post-infection (Fig 2). However, gross inspection of the ΔsltA infected lungs and their reduced mass suggested that there was less fungal-induced damage. This observation led to the hypothesis that SltA may be required for specific pathogenicity functions during invasive aspergillosis rather than for proliferation.
We tested that hypothesis by monitoring mouse survival post-infection in our non-neutropenic invasive aspergillosis model (Fig 3B). We observed that ΔsltA-infected mice survived significantly longer than mice infected with the wild-type strain or the ΔsltA+sltA complemented strain. The finding that mice infected with the ΔsltA mutant maintained a high pulmonary fungal burden yet had reduced mortality is similar to what has been found with A. fumigatus mutants with defects in secondary metabolite or mycotoxin production [12,31], suggesting that SltA may govern the expression of secondary metabolite or mycotoxin genes.
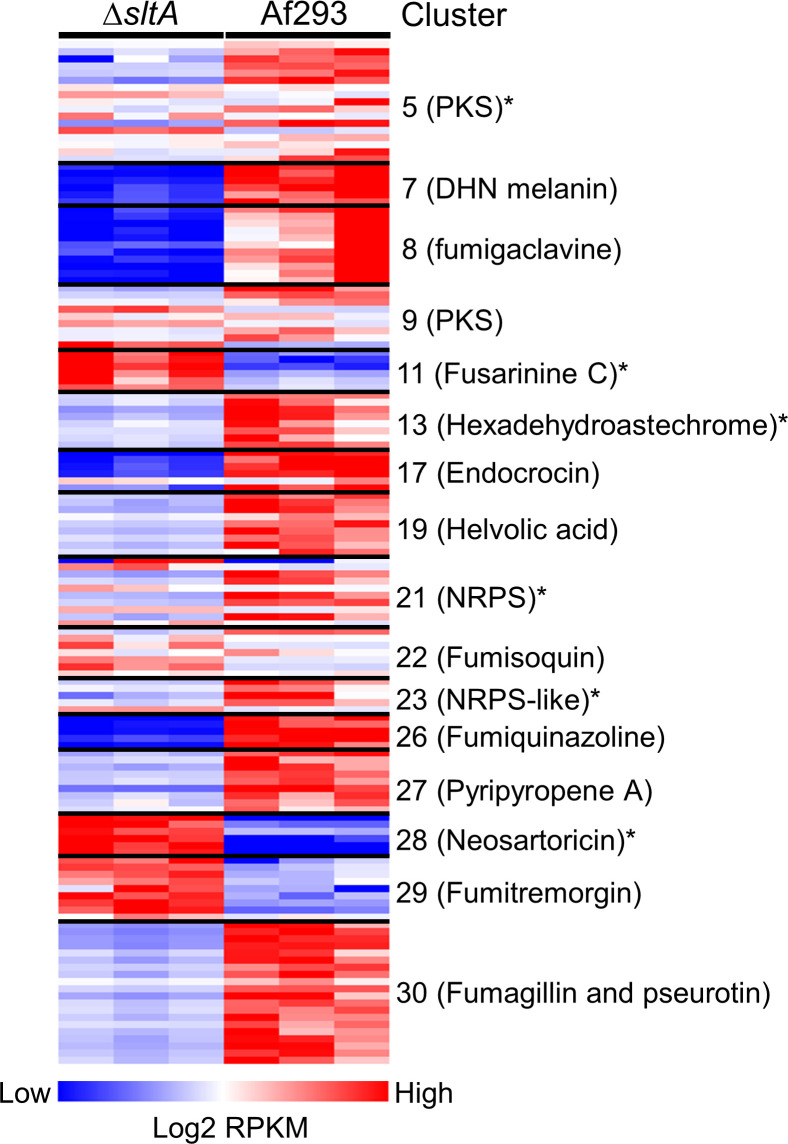
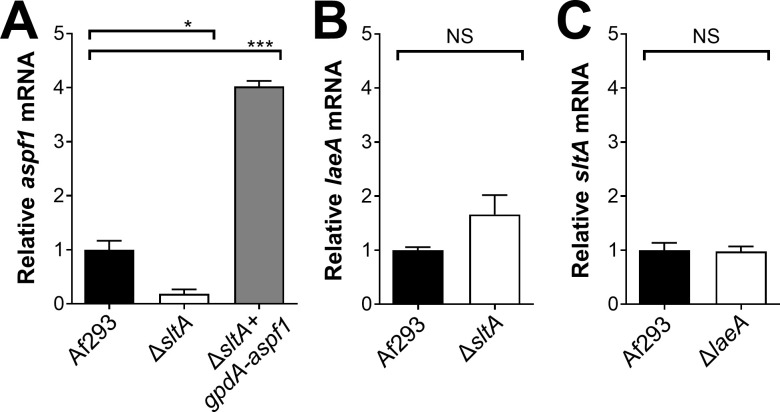
To identify SltA target genes that might be responsible for the virulence, we performed RNA-seq analysis of the wild-type and ΔsltA mutant strains grown in AMM with low nitrogen and low zinc to mimic the conditions during invasive infection in the lung. In Aspergillus spp., genes encoding proteins involved in the biosynthesis of secondary metabolites are frequently located in contiguous clusters in the genome. A total of 33 non-overlapping secondary metabolite gene clusters have been identified in A. fumigatus Af293 [46]. We found that SltA governs the expression of at least 50% of the genes in 16 of these biosynthetic gene clusters (Fig 6 and S2 Table). Of these gene clusters, 10 had reduced mRNA expression in the ΔsltA mutant, 3 had increased mRNA expression, and 3 had both increased and decreased mRNA expression. In the ΔsltA mutant, there was also reduced expression of aspf1 (Afu5g02330), which encodes a ribotoxin that enhances A. fumigatus virulence [12,37] (S2 Table). We verified the low levels of aspf1 mRNA in the ΔsltA mutant by qPCR (Fig 7A). These results indicate that a principal function of SltA is the regulation of production of secondary metabolites and mycotoxins.
Fig 6. SltA governs the expression of secondary metabolite gene clusters.
Heat map showing secondary metabolite gene clusters in which the expression of at least 50% of genes were altered in the ΔsltA mutant relative to strain Af293. The transcript levels were assessed by RNA-seq analysis of organisms that were grown in liquid AMM with low nitrogen and low zinc in biological triplicate. Secondary metabolite cluster numbers are from [46]. *, gene clusters that were not found to be regulated by LaeA by microarray analysis [48]; NRPS, non-ribosomal peptide synthase; PKS, polyketide synthase.
Fig 7. qPCR verification of transcriptional profiling results.
Real-time PCR analysis of the relative transcript levels of aspf1 (A), laeA (B), and sltA (C) in the indicates strains. The organisms were grown in AMM with low nitrogen and low zinc. Results are the mean ± SD of 3 biological replicates. *, P < 0.05; ***, P <0.001; NS, not significant.
To determine if SltA has a similar function in another strain of A. fumigatus, we obtained a ΔsltA mutant constructed in the A1160 strain background [47] and used real-time PCR to assess the expression levels of 19 SltA-dependent genes that specified mycotoxins and secondary metabolites. We found that 15 of these genes were regulated by SltA similarly in both Af293 and A1160 (S4 Fig). The expression of two genes, AFUA_2g18040 (AFUB_033730) and AFUA_3g01480 (AFUB_046920) was not significantly influenced by deletion of sltA in strain A1160. The expression of AFUA_8g00250 (AFUB_086290) was increased in the ΔsltA mutant in strain Af293, but decreased in the corresponding A1160 mutant. By contrast, the expression of AFUA_5g12780 (AFUB_060450) was decrease in the ΔsltA mutant in strain Af293, but increased in the A1160 mutant. Thus, although there is some strain-to-strain variation in the SltA regulon, this transcriptional regulator governs mycotoxins and secondary metabolites in at least two strains of A. fumigatus.
A major regulator of secondary metabolite production in A. fumigatus is LaeA, which removes heterochromatic marks from the promoters of numerous genes, enabling the transcription of secondary metabolite genes [46,48]. Microarray analysis indicates that LaeA governs the expression of at least 16 secondary metabolite gene clusters [46,48]. Comparison of the microarray analysis of the ΔlaeA mutant with the current RNA-seq analysis of the ΔsltA mutant indicates that SltA governs the expression of 6 biosynthetic gene clusters that are not known to be regulated by LaeA (Fig 6). LaeA and SltA differ in additional respects. All gene clusters that are regulated by LaeA are down-regulated in the ΔlaeA mutant [48], whereas some gene clusters that are regulated by SltA, such as fusarinine C, neosartoricin, and fumitremorgin, are up-regulated in the ΔsltA mutant. Also, LaeA governs asexual development and conidiation [46,49], whereas we found no evidence that SltA governs these processes. These results suggest that SltA regulates the expression of secondary metabolite gene clusters by a different mechanism than LaeA.
To further investigate the relationship between SltA and LaeA, we used qPCR to measure the transcript levels of TF genes in the ΔsltA and ΔlaeA mutant. The transcript levels of laeA were slightly higher in the ΔsltA mutant than in the wild-type strain, but this difference was not significant (Fig 7B). Also, sltA was expressed at wild-type levels in the ΔlaeA mutant (Fig 7C). Overall, these data indicate that SltA regulates secondary metabolite gene clusters independently of LaeA.
SltA governs virulence via Asp f1
The RNA-seq data suggested that the ΔsltA mutant had reduced virulence because of decreased production of mycotoxins. We investigated whether ergot alkaloids produced by the fumigaclavine biosynthesis cluster play a role in A. fumigatus virulence. The first enzyme in the fumigaclavine biosynthesis pathway is DmaW [50] and deletion of dmaW results in absent production of all detectable ergot alkaloids and attenuated virulence in Galleria mellonella [51]. We constructed a ΔdmaW mutant and analyzed its virulence in corticosteroid treated mice. The survival of mice infected with this mutant was similar to that of mice infected with the wild-type strain (Fig 8A), indicating that the reduced virulence of the ΔsltA mutant was not due to the absence of ergot alkaloid production.
Fig 8. Forced expression of aspf1 rescues the virulence defect of the ΔsltA mutant.
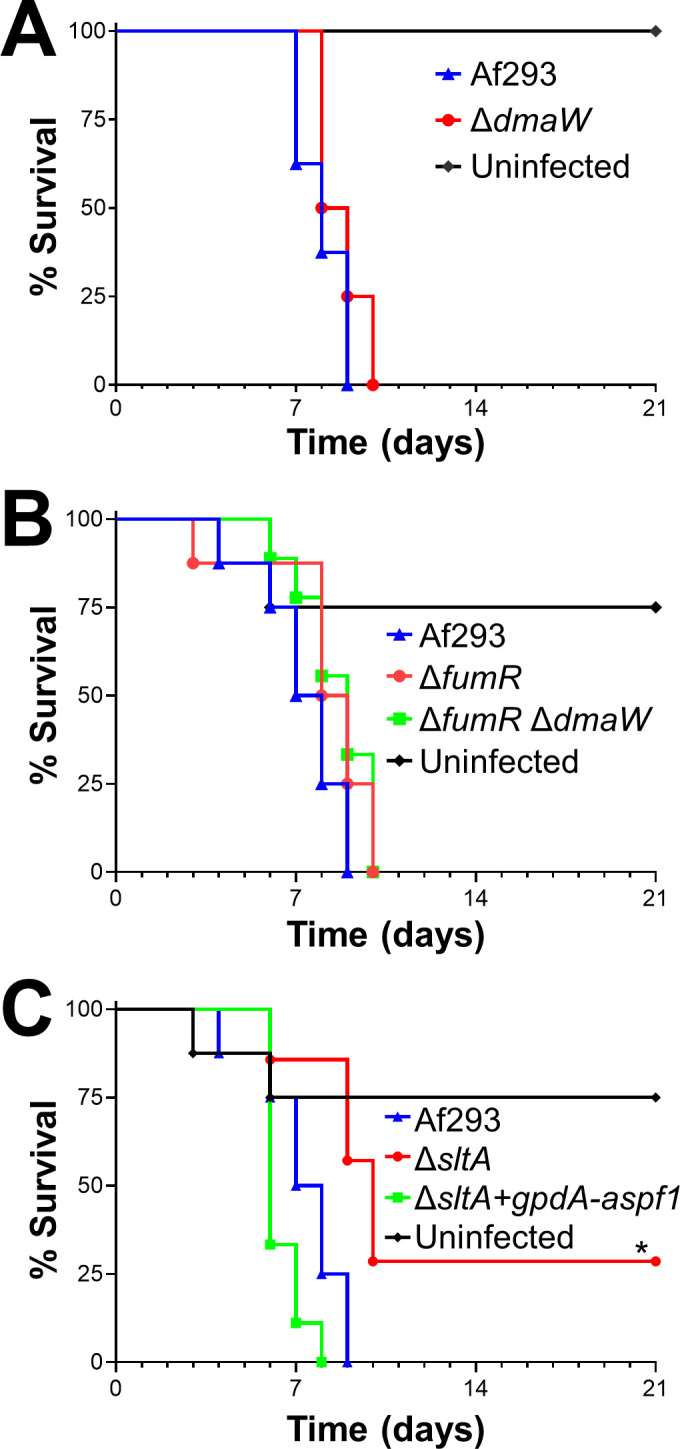
Survival of mice infected with the indicated strains of A. fumigatus. Results are from 8 mice per strain. *, p < 0.05.
Another gene cluster that was down-regulated in the ΔsltA mutant was the large fumagillin and pseruotin supercluster. Fumagillin inhibits neutrophil function [52] and is required for A. fumigatus to cause maximal damage to the A549 pulmonary epithelial cell line [53]. Within the fumagillin biosynthetic gene cluster is fumR, which specifies a putative C6 type transcription factor that is required for fumagillin and pseurotin synthesis [38,39]. We constructed a ΔfumR mutant and found that it had wild-type virulence in mice (Fig 8B). A ΔfumR ΔdmaW double mutant also had no detectable reduction in virulence (Fig 8B). Collectively, these data suggest that both fumagillin and the fumigaclavine ergot alkaloids are dispensable for virulence in the corticosteroid treated mouse model of invasive aspergillosis. Thus, the decreased production of these secondary metabolites does not explain the reduced virulence of the ΔsltA mutant.
Next, we investigated whether the attenuated virulence of the ΔsltA mutant was due to decreased expression of aspf1, which encodes a ribotoxin [37]. Previously, we have determined that Asp f1 is required for the maximal virulence of A. fumigatus in corticosteroid treated mice [12]. We constructed a variant of the ΔsltA mutant in which the expression of aspf1 was driven by the constitutive gpdA promoter (Fig 7A). The forced expression of aspf1 restored the virulence of the ΔsltA mutant to wild-type levels (Fig 8C). Thus, the reduced expression of aspf1 likely contributes to the decreased virulence of the ΔsltA mutant. Because SltA governs the expression of multiple secondary metabolite genes, it remains possible that the products of other SltA-dependent genes also mediate A. fumigatus virulence.
Collectively, our results indicate that invasive growth in the lungs of corticosteroid treated mice induces a unique transcription profile in A. fumigatus as the organism responds to nutrient limitation and attack by host phagocytes. Also, growth in AMM with low zinc and low nitrogen in vitro induces a transcriptional response that largely mimics that induced by growth in vivo. This set of conditions can be used for RNA-seq analysis of A. fumigatus TF gene mutants to identify potential downstream target genes whose products mediate virulence. NanoString profiling of A. fumigatus during invasive growth in the lungs identified RlmA as a transcription factor that governs lung fungal burden in vivo. It also identified SltA as a transcription factor that governs pathogenicity by regulating the expression of multiple secondary metabolite gene clusters and aspf1 independently of LaeA. In our NanoString dataset are additional TF genes that are either up-regulated or highly expressed during invasive growth in vivo. Determining the roles of these genes in governing A. fumigatus virulence is currently in progress.
Methods
Ethics statement
All mouse studies were conducted in compliance with the NIH Guide for the Care and Use of Laboratory Animals. The experimental procedures were approved in advance by the Institutional Animal Care and Use Committee at the Lundquist Institute. The mice were group-housed according to experimental group in HEPA-filtered laminar flow cages with unrestricted access to food and water. The vivarium is managed by the Lundquist Institute in compliance with all policies and regulations of the Office of Laboratory Animal Welfare of the Public Health Service. The facility is fully accredited by the American Association for Laboratory Animal Care.
Strains, media and growth conditions
The A. fumigatus strains used in this study are listed in Table 4. All strains were grown on Sabouraud dextrose agar (Difco) at 37°C for 7 d prior to use. Conidia were harvested with phosphate-buffered saline (PBS) containing 0.1% Tween 80 (Sigma-Aldrich) and enumerated with a hemacytometer.
Table 4. A. fumigatus strains used in the current study.
| Strain | Genotype | Reference |
|---|---|---|
| Af293 | Wild-type | [69] |
| ΔrlmA | Af293; rlmA::hph | Present study |
| ΔsltA | Af293; sltA::hph | Present study |
| Δafu1g15910 | Af293; Afu1g15910::hph | Present study |
| Δafu3g08050 | Af293; Afu3g08050::hph | Present study |
| Δafu4g10220 | Af293; Afu4g10220::hph | Present study |
| Δafu5g01650 | Af293; Afu5g01650::hph | Present study |
| Δafu5g13790 | Af293; Afu5g13790::hph | Present study |
| Δafu7g00130 | Af293; Afu7g00130::hph | Present study |
| Δafu8g05270 | Af293; Afu8g05270::hph | Present study |
| ΔrlmA+rlmA | ΔrlmA; rlmA; ble | Present study |
| ΔsltA+sltA | ΔsltA; sltA; ble | Present study |
| ΔlaeA | Af293.1; laeA::pyrG1 | [48] |
| ΔdmaW | Af293; dmaW::hph | Present study |
| ΔfumR | Af293; fumR::hph | Present study |
| ΔfumRΔdmaW | ΔdmaW; fumR::ble | Present study |
| ΔsltA+gpdA-aspf1 | ΔsltA; gpdA-aspf1; ble | Present study |
| Af293+gpdA-GFP | Af293; gpdA-GFP; ble | [57] |
| ΔsltA+gpdA-GFP | ΔsltA; gpdA-GFP; ble | Present study |
| ΔsltA+sltA+gpdA-GFP | ΔsltA+sltA; gpdA-GFP; ptrA | Present study |
For in vitro gene expression profiling analysis, 1.5 X108 conidia of A. fumigatus were added to 300 ml liquid standard AMM or modified AMM (no added iron, no added zinc, or the addition of 10% fetal bovine serum) and incubated for 24 h at 37°C in a shaking incubator. For growth under low nitrogen conditions, the A. fumigatus cells were grown in either AMM or modified AMM for 20 h, after which the hyphae were collected by filtration, washed with water, then added to AMM or modified AMM without nitrate and incubated for an additional 4 h. At the end of the incubation period, the resulting hyphae were collected by filtration and the RNA was extracted using the RNeasy Plant Minikit (Qiagen) following the manufacturer’s instructions.
For RNA-seq analysis, 1.5 X 108 conidia of A. fumigatus were incubated in 300 ml of AMM without zinc for 20 h and then incubated in AMM without zinc and nitrate for 4 additional h prior to RNA extraction.
Strain construction
The TF gene mutant strains used in this research were constructed using a split marker strategy. For each gene, approximately 1.5 kb of the 5’-flanking sequence upstream of the protein coding region was PCR-amplified from genomic DNA of strain Af293 using primers F3 and F4. The PCR primers used in the experiments are listed in S3 Table. The resulting fragment was cloned into plasmid pNLC106 [54]. Using the plasmid as a template, the 5’-flanking sequence region was linked to the 5’ portion of the hph hygromycin resistance gene by fusion PCR using primers F4 and HY. Next, about 1.5 kb of the 3’-flanking sequence downstream of the protein coding region was PCR-amplified from genomic DNA of strain A293 using primers F2 and F1 and the sequence of hph was amplified from pAN7 [55] using primers HYG-F and HYG-R. Using a mixture of the two fragments as the template, the 3’-flanking sequence region of target gene linked to 3’ portion of hph was amplified by fusion PCR with primers F1 and YG. Finally, the two fragments were used to transform Af293 protoplasts. The hygromycin-resistant clones were screened for the deletion of target gene by colony PCR using primers Screen-F and Screen-R.
To construct the ΔrlmA + rlmA complemented strain, a 4481 bp fragment containing the rlmA protein coding sequence and approximately 2 kb of 5’ flanking sequence and 0.5 kb of 3’ flanking sequence was PCR-amplified from Af293 genomic DNA using primers 3g08520-Com-F and 3g08520-Com-R. Similarly, to construct the ΔsltA+sltA complemented strain, a 4997 bp fragment containing the sltA protein coding sequence and flanking regions was PCR-amplified from using primers 3g08010-Com-F and 3g08010-Com-R. Each fragment was cloned into the NotI/XbaI sites of plasmid p402 [56]. The resulting plasmids were used to transform the ΔrlmA and ΔsltA strains. To confirm the presence of the complementation plasmids, the phleomycin-resistant colonies were screened by colony PCR using primers 3g08520-Com-F and 3g08520-Com-R to detect rlmA or 3g08010-Com-F and 3g08010-Com-R to detect sltA. The transcript levels of rlmA or sltA in various clones were quantified by real-time RT-PCR using primers RT-F and RT-R. The clones in which the transcript levels of rlmA or sltA was most similar to that of the wild-type strain was used in all experiments.
For use in the epithelial cell invasion assays, strains of A. fumigatus that expressed GFP were constructed. The ΔsltA mutant was transformed with plasmid GFP-Phleo and the ΔsltA+sltA complemented strain was transformed with plasmid GFP-pPTRI [57].
To construct the ΔdmaW (Afu2g18040) and ΔfumR (Afu8g00420) deletion mutants, a transient CRISPR-Cas9 gene deletion system was used [58,59]. The Cas9 expression cassette was amplified from plasmid pFC331 [59], using primers Cas9-F and Cas9-R. To construct the sgRNA expression cassette, two DNA fragments were amplified from plasmid pFC334 [59] using primers sgRNA-F and sgRNA-ss-R, and sgRNA-R, sgRNA-ss-F. Next, the sgRNA expression cassette was amplified by fusion PCR from the two DNA fragments, using primers sgRNA-F and sgRNA-R. The hygromycin resistance (HygR) repair template was amplified from plasmid pVG2.2-hph [60] using primers Hyg-F and Hyg-R, which had about 50 bp of homology to the 5’ end of the protein coding sequence of the gene and the 3’ end of the protein coding sequence, respectively. The HygR repair template was mixed with the Cas9 cassette and the two sgRNA cassettes and then used for protoplast transformation. Hygromycin resistant clones were screened for deletion of the target gene by colony PCR using primers RT-F and RT-R. The positive clones were also confirmed for absence of integration of DNA encoding Cas9 or the gRNA, using primers Cas9RT-F and Cas9RT-R, and sgRT-F, sgRT-R.
The ΔfumR ΔdmaW double mutant was constructed using the above CRISPR-Cas9 approach starting with the ΔdmaW mutant strain. The phleomycin (PhlR) repair template was amplified from plasmid p402 using primers Phleo-F and Phleo-R, which contained regions that were homologous to the 5’ and 3’ ends of the fumR protein coding sequence. Protoplasts were transformed with the repair template along with the Cas9 and sgRNA cassettes that were used to construct the ΔfumR deletion mutant. Phleomycin resistant clones were screened by colony PCR to identify ones with deletion of both genes and that lacked Cas9 and sgRNA sequences.
A strain of the ΔsltA mutant in which aspf1 expression was driven by the gpdA promoter was constructed using the CRISPR-Cas9 system. The protein coding region of aspf1 was amplified from genomic DNA using primers Aspf1-F and Aspf1-R. The resulting fragment was cloned into the BamHI-NcoI sites of plasmid pGFP-phleo using the NEBuilder DNA assembly kit (New England Biolabs). In this plasmid, the expression of aspf1 was driven by the A. nidulans gpdA promoter. The gpdA-aspf1-phleomycin template was PCR amplified from this plasmid using primers Phleo-OE-F and Phleo-OE-R (S1 Table), which had about 50 bp of homology to the safe haven region of the A. fumigatus genome [61]. The ΔsltA mutant was transformed with the gpdA-aspf1-phleomycin template, the Cas9 cassette and the two safe haven sgRNA cassettes [61]. In the resulting phleomycin resistant clones, the aspf1 transcript levels were quantified by real-time RT-PCR using primers Aspf1-RT-F and Aspf1-RT-R. A clone in which the aspf1 mRNA expression was approximately 4-fold higher than the wild-type strain was used in all subsequent experiments.
For all deletion mutants that were constructed, we performed real-time PCR to verify that there was no mRNA expression of the deleted gene (S5A Fig). Because the mRNA of nssR was not detectable when the organisms were grown in vitro, we used standard PCR to verify the presence of the HYG hygromycin resistance gene at the nscR locus (S5B Fig).
Mouse model of invasive pulmonary aspergillosis
A non-neutropenic, immunosuppressed mouse model of invasive aspergillosis was used to assess the transcriptional profile and virulence of the various strains [19]. Briefly, 6 week old, male Balb/c mice (Taconic Laboratories) were immunosuppressed with 7.5 mg cortisone acetate (Sigma-Aldrich) administered subcutaneously every other day starting at day -4 before infection for a total of 5 doses. To prevent bacterial infections, enrofloxacin (Baytril, Western Medical Supply) was added to the drinking water at a final concentration of 0.005% on day -5 relative to infection. The mice were infected by placing them for 1 h in an acrylic chamber into which 12 ml of 1x109 conida/ml were aerosolized. Control mice were immunosuppressed, but not infected.
For the transcriptional profiling experiments, 3 mice infected with each strain were sacrificed after 2, 4, and 5 days infection. Their lungs were harvested and snap frozen in liquid nitrogen for RNA extraction. To isolate fungal RNA from the infected mouse lungs, the RNeasy minikit (Qiagen) was used with modifications [12]. Approximately 2.4 ml of buffer RLT with 1% β-mercaptoethanol was added to the lungs from each mouse and the tissue was homogenized in an M tube (Miltenyi Biotec) using a gentleMACS dissociator (Miltenyi Biotec) on setting RNA_02.01. Next, the homogenate was mixed with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) and a half volume of zirconium beads (Ambion) and then vortexed with a Mini-Beadbeater (Biospec Products) for 3 min. After centrifugation, the aqueous phase was collected and mixed with an equal volume of 70% ethanol. The RNA was isolated from this mixture using an RNeasy spin column (Qiagen) following the manufacturer’s instructions.
To assess the virulence of the various A. fumigatus strains using survival as the end point, 11 mice were infected with each strain. Shortly after infection, 3 mice from each group were sacrificed, and their lungs were harvested, homogenized and quantitatively cultured to verify conidia delivery to the lung. The remaining mice were monitored twice daily for survival. 5 mice that were immunosuppressed, but not infected were included as a negative control.
NanoString analysis
A NanoString nCounter digital analyzer was used for transcriptional profiling of A. fumigatus Af293 both in vivo and in vitro as previously described [12]. For each condition, the adjusted data were normalized to total probe counts. However, when we compared 18 genes that were in common between our two probe sets (called TF and ER), we observed poor agreement in fold changes. The ER genes had been chosen because they respond dramatically to environmental changes, and we reasoned that large expression changes may make total counts unreliable for normalization. The TF probe set, representing 400 different putative transcription factor genes, is extremely diverse and thus total counts are more reliable for normalization. With that point in mind, the ER datasets were renormalized to TF dataset measurements as follows. For each of the 18 common genes in both ER and TF probe sets, we calculated the ratio of mean TF counts/mean ER counts for each growth condition. Then the median ratio for the 18 genes was used to calculate a normalization factor for each growth condition. The normalization factor for each growth condition was applied to all ER genes. Finally, the TF and ER datasets were combined, with counts for common genes from the TF datasets, and 10 TF genes with the lowest counts removed. Each condition was tested in 3 biological replicates and the expression ratios were calculated using the mean values. Genes were considered differentially expressed when there was at least a 2-fold change in the transcript levels and an unpaired, two-tailed student’s t-test p-value ≤ 0.05.
RNA-seq
RNA-seq libraries (strand-agnostic, 150 bp paired-end) were generated from total fungal RNA by Novogene Corporation Inc. Sequencing reads were aligned to the reference A. fumigatus Af293 genome using HISAT2 [62] and alignment files were used to generate read counts for each gene using HTseq [63]. We obtained an average of 46.4 million aligned reads per sample. Statistical analysis of differential gene expression was performed using the DEseq package from Bioconductor [64]. A gene was considered differentially expressed if the FDR value for differential expression was below 0.05. The RNA-seq analysis was performed in biological triplicate.
Stress assays
To test the susceptibility of the various strains to cell wall, cell membrane, oxidant, and ionic stress, serial 10-fold dilutions of conidia ranging from 105 to 102 cells in a volume of 5 μl were spotted onto AMM agar plates supplemented with 5 mM protamine (Sigma-Aldrich), 0.01% SDS (Sigma), 40 μg/ml caspofungin (Bellavida Pharmacy), 200 ug/ml Congo red (Sigma-Aldrich), 300 μg/ml Calcofluor White (Sigma-Aldrich), 4 mM H2O2, 200 mM KCl, 200 mM MgCl2, 200 mM NaCl, or 50 mM CaCl2. To determine growth at alkaline pH, the conidia were spotted onto AMM agar adjusted to pH 8.0 with NaOH. Fungal growth was analyzed after incubation at 37°C for 2 d.
Real-time PCR
The total RNA was reverse transcribed into cDNA using Moloney murine leukemia virus reverse transcriptase (Promega). Real-time PCR was performed using the POWER SYBR green PCR master mix (Applied Biosystems) and an ABI 7000 thermocycler (Applied Biosystems). Gene transcript levels were quantified by ΔΔCt method, using GAPDH as the endogenously expressed gene [65].
Host cell interaction assays
The capacity of the various strains to adhere to, invade, and damage the A549 pulmonary epithelial cell line (American Type Culture Collection) was determined using our previously described methods [11,57,66]. To measure adherence and invasion, 105 germlings of the various GFP-expressing strains of A. fumigatus in F12k medium (American Type Culture Collection) were added to A549 cells that had been grown to confluency in 24-well tissue culture plates containing fibronectin coated circular glass coverslips in each well. After incubation for 2.5 h, the cells were rinsed with 1 ml HBSS in a standardized manner and then fixed with 3% paraformaldehyde. The noninternalized portions of the organisms were stained with a polyclonal rabbit anti-A. fumigatus primary antibody (Meridian Life Science, Inc.) followed by an AlexaFluor 568-labeled secondary antibody (Life Technologies). After the coverslips were mounted inverted on microscope slides, they were viewed by epifluorescence. The number of cell-associated organisms was determined by counting the number of GFP-expressing organisms per high-powered field (HPF). The number of endocytosed organisms was determined by subtracting the number of non-internalized organisms (which fluoresced red) from the number of cell-associated organisms. At least 100 organisms per coverslip were scored and each strain was tested in triplicate in three independent experiments.
Our standard 51Cr release assay was used to evaluate the capacity of the various strains to damage the A549 cell line [11,67]. The A549 cells were grown to confluency in a 24-well tissue culture plate and then loaded with 51Cr overnight. After rinsing the cells to remove the unincorporated 51Cr, the cells were infected with 5x105 conidia of each strain in F12K medium. After 16 h of infection, the medium above the cells was collected and the cells were lysed with 6 N NaOH. The lysed cells were collected by rinsing the wells twice with RadiacWash (Biodex Medical Systems). The amount of 51Cr in the medium and the cell lysate was measured using a gamma counter. The spontaneous release of 51Cr was determined using uninfected A549 cells that were processed in parallel. The specific release of 51Cr was calculated using our previous described formula [11,67]. Each experiment was performed in triplicate and repeated three times.
The susceptibility of the various A. fumigatus strains to phagocyte killing was determined using bone marrow-derived macrophages (BMDMs), which were isolated from 6-week-old mice (Taconic Laboratories). The cells were differentiated into macrophages by incubation with 50 ng/ml macrophage colony–stimulating factor (M-CSF) (BioLegend) in Dulbecco’s Modified Eagle’s Medium (DMEM) (American Type Culture Collection) with 10% fetal bovine serum (Gemini Bio-Products), 1% streptomycin and penicillin for 10 d [68]. The day before the experiment, the adherent cells were harvested and 106 cells were seeded into each well of a 6-well tissue culture plate. The next day, 5 x 104 conidia were added to each well and incubated for 8 h. A similar number of conidia was added to a second 6-well tissue culture plate without BMDMs as a control. At the end of the incubation period, the BMDMs were lysed with distilled water and sonication. The contents of the wells were aspirated and quantitatively cultured on Sabouraud dextrose agar. For each strain, the percentage of A. fumigatus cells killed was calculated by the formula: 1—number of colonies in the wells containing BMDMs/number of colonies in the wells without BMDMs. Each experiment was performed in triplicates and repeated three times.
Statistical analysis
The data from the in vitro experiments were analyzed by the two-tailed Student’s t-test assuming unequal variance or one way analysis of variance followed by the Dunnett’s test for multiple comparisons. The survival data were analyzed using the Log-Rank test. A P-value of ≤ 0.05 was considered to be significant.
Supporting information
Shown are NanoString probe counts for 97 environmentally responsive A. fumigatus genes in lung samples from 3 mice after 5 d of infection and a single immunosuppressed, uninfected mouse.
(PDF)
Shown are the lung masses of mice after 5 d of infection with the indicated strains of A. fumigatus. Lines indicate the median and interquartile ranges of 5 mice per strain. Symbols indicate the results of individual mice. *P < 0.05.
(PDF)
Serial 10-fold dilutions of the indicated strains of A. fumigatus were spotted onto Aspergillus minimal medium (AMM) containing the indicated stressors. The plates were imaged after incubation at 37°C for 2 d.
(PDF)
Values represent Log2 expression ratios for each gene in the ΔsltA strain divided by the wild-type strain as determined by RNA-seq (Af293) or qRT-PCR (AF1160).
(PPTX)
(A) qRT-PCR results of relative gene expression in the indicated strains. (B) The replacement of nscR (Afu7g00130) by the hygromycin resistance cassette (HYG) was verified by PCR using two different sets of primers.
(PDF)
Results are from the lungs of mice infected with A. fumigatus strain Af293 and from the same strain grown in standard Aspergillus minimal medium (AMM) in normoxia or hypoxia, in AMM deficient in specific nutrients, or in AMM containing 10% bovine serum. Results are the mean of 3 biological replicates or mice per condition.
(XLSX)
Results are the mean of 3 biological replicates of organisms grown in AMM with low zinc and low nitrogen. Inf indicates no detectable reads in the ΔsltA mutant samples.
(XLSX)
(XLSX)
Acknowledgments
We thank the members of the Filler, Bruno, and Mitchell labs for helpful discussions and suggestions.
Data Availability
The raw sequencing reads from this study have been submitted to the NCBI sequence read archive (SRA) under BioProject accession number PRJNA693756. The remaining relevant data are within the paper and the Supporting Information files.
Funding Statement
This work was supported by NIH grants R01AI124566 and R01DE026600 to SGF and APM, U19AI110820 to SGF and VMB, and R01AI141360 to VMB. MJB was supported in part by Wellcome Trust grant 208396/Z/17/Z. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dagenais TR, Keller NP. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin Microbiol Rev. 2009;22(3):447–65. Epub 2009/07/15. 10.1128/CMR.00055-08 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Segal BH. Aspergillosis. N Engl J Med. 2009;360(18):1870–84. Epub 2009/05/01. 360/18/1870 [pii] 10.1056/NEJMra0808853. 10.1056/NEJMra0808853 . [DOI] [PubMed] [Google Scholar]
- 3.Sherif R, Segal BH. Pulmonary aspergillosis: clinical presentation, diagnostic tests, management and complications. Curr Opin Pulm Med. 2010;16(3):242–50. Epub 2010/04/09. 10.1097/MCP.0b013e328337d6de [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh TJ, Anaissie EJ, Denning DW, Herbrecht R, Kontoyiannis DP, Marr KA, et al. Treatment of aspergillosis: clinical practice guidelines of the Infectious Diseases Society of America. Clin Infect Dis. 2008;46(3):327–60. Epub 2008/01/08. 10.1086/525258 . [DOI] [PubMed] [Google Scholar]
- 5.Georgiadou SP, Kontoyiannis DP. The impact of azole resistance on aspergillosis guidelines. Ann N Y Acad Sci. 2012;1272:15–22. Epub 2012/12/13. 10.1111/j.1749-6632.2012.06795.x . [DOI] [PubMed] [Google Scholar]
- 6.Pfaller MA. Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am J Med. 2012;125(1 Suppl):S3–13. Epub 2012/01/04. S0002-9343(11)00913-2 [pii] 10.1016/j.amjmed.2011.11.001 . [DOI] [PubMed] [Google Scholar]
- 7.Bertuzzi M, Schrettl M, Alcazar-Fuoli L, Cairns TC, Munoz A, Walker LA, et al. The pH-responsive PacC transcription factor of Aspergillus fumigatus governs epithelial entry and tissue invasion during pulmonary aspergillosis. PLoS Pathog. 2014;10(10):e1004413. Epub 2014/10/21. 10.1371/journal.ppat.1004413 ; PubMed Central PMCID: PMC4199764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonagh A, Fedorova ND, Crabtree J, Yu Y, Kim S, Chen D, et al. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog. 2008;4(9):e1000154. 10.1371/journal.ppat.1000154 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kale SD, Ayubi T, Chung D, Tubau-Juni N, Leber A, Dang HX, et al. Modulation of immune signaling and metabolism highlights host and fungal transcriptional responses in mouse models of invasive pulmonary aspergillosis. Sci Rep. 2017;7(1):17096. Epub 2017/12/08. 10.1038/s41598-017-17000-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abad A, Fernandez-Molina JV, Bikandi J, Ramirez A, Margareto J, Sendino J, et al. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev Iberoam Micol. 2010;27(4):155–82. 10.1016/j.riam.2010.10.003 [DOI] [PubMed] [Google Scholar]
- 11.Pongpom M, Liu H, Xu W, Snarr BD, Sheppard DC, Mitchell AP, et al. Divergent targets of Aspergillus fumigatus AcuK and AcuM transcription factors during growth in vitro versus invasive disease. Infect Immun. 2015;83(3):923–33. 10.1128/IAI.02685-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Xu W, Solis NV, Woolford C, Mitchell AP, Filler SG. Functional convergence of gliP and aspf1 in Aspergillus fumigatus pathogenicity. Virulence. 2018;9(1):1062–73. 10.1080/21505594.2018.1482182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu W, Solis NV, Ehrlich RL, Woolford CA, Filler SG, Mitchell AP. Activation and alliance of regulatory pathways in C. albicans during mammalian infection. PLoS Biol. 2015;13(2):e1002076. Epub 2015/02/19. 10.1371/journal.pbio.1002076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fanning S, Xu W, Solis N, Woolford CA, Filler SG, Mitchell AP. Divergent targets of Candida albicans biofilm regulator Bcr1 in vitro and in vivo. Eukaryot Cell. 2012;11:896–904. Epub 2012/05/01. 10.1128/EC.00103-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nobile CJ, Solis N, Myers CL, Fay AJ, Deneault JS, Nantel A, et al. Candida albicans transcription factor Rim101 mediates pathogenic interactions through cell wall functions. Cell Microbiol. 2008;10(11):2180–96. 10.1111/j.1462-5822.2008.01198.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, et al. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006;2(7):e63. 10.1371/journal.ppat.0020063 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis DA, Bruno V, Loza L, Filler SG, Mitchell AP. C. albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics. 2002;162(4):1573–81. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bahn YS. Exploiting fungal virulence-regulating transcription factors as novel antifungal drug targets. PLoS Pathog. 2015;11(7):e1004936. Epub 2015/07/17. 10.1371/journal.ppat.1004936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiang LY, Sheppard DC, Gravelat FN, Patterson TF, Filler SG. Aspergillus fumigatus stimulates leukocyte adhesion molecules and cytokine production by endothelial cells in vitro and during invasive pulmonary disease. Infect Immun. 2008;76(8):3429–38. 10.1128/IAI.01510-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheppard DC, Rieg G, Chiang LY, Filler SG, Edwards JE, Jr., Ibrahim AS. Novel inhalational murine model of invasive pulmonary aspergillosis. Antimicrob Agents Chemother. 2004;48(5):1908–11. 10.1128/aac.48.5.1908-1911.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schrettl M, Kim HS, Eisendle M, Kragl C, Nierman WC, Heinekamp T, et al. SreA-mediated iron regulation in Aspergillus fumigatus. Mol Microbiol. 2008;70(1):27–43. 10.1111/j.1365-2958.2008.06376.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schrettl M, Beckmann N, Varga J, Heinekamp T, Jacobsen ID, Jochl C, et al. HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 2010;6(9):1001124. 10.1371/journal.ppat.1001124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schrettl M, Bignell E, Kragl C, Joechl C, Rogers T, Arst HN Jr., et al. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J Exp Med. 2004;200(9):1213–9. 10.1084/jem.20041242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno MA, Ibrahim-Granet O, Vicentefranqueira R, Amich J, Ave P, Leal F, et al. The regulation of zinc homeostasis by the ZafA transcriptional activator is essential for Aspergillus fumigatus virulence. Mol Microbiol. 2007;64(5):1182–97. Epub 2007/06/05. 10.1111/j.1365-2958.2007.05726.x . [DOI] [PubMed] [Google Scholar]
- 25.Hensel M, Arst HN, Jr., Aufauvre-Brown A, Holden DW. The role of the Aspergillus fumigatus areA gene in invasive pulmonary aspergillosis. Mol Gen Genet. 1998;258(5):553–7. Epub 1998/07/21. 10.1007/s004380050767 . [DOI] [PubMed] [Google Scholar]
- 26.Willger SD, Puttikamonkul S, Kim KH, Burritt JB, Grahl N, Metzler LJ, et al. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 2008;4(11):e1000200. 10.1371/journal.ppat.1000200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richie DL, Hartl L, Aimanianda V, Winters MS, Fuller KK, Miley MD, et al. A role for the unfolded protein response (UPR) in virulence and antifungal susceptibility in Aspergillus fumigatus. PLoS Pathog. 2009;5(1):e1000258. 10.1371/journal.ppat.1000258 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dinamarco TM, Almeida RS, de Castro PA, Brown NA, dos Reis TF, Ramalho LN, et al. Molecular characterization of the putative transcription factor SebA involved in virulence in Aspergillus fumigatus. Eukaryot Cell. 2012;11(4):518–31. 10.1128/EC.00016-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dirr F, Echtenacher B, Heesemann J, Hoffmann P, Ebel F, Wagener J. AfMkk2 is required for cell wall integrity signaling, adhesion, and full virulence of the human pathogen Aspergillus fumigatus. Int J Med Microbiol. 2010;300(7):496–502. 10.1016/j.ijmm.2010.03.001 [DOI] [PubMed] [Google Scholar]
- 30.Ma Y, Qiao J, Liu W, Wan Z, Wang X, Calderone R, et al. The sho1 sensor regulates growth, morphology, and oxidant adaptation in Aspergillus fumigatus but is not essential for development of invasive pulmonary aspergillosis. Infect Immun. 2008;76(4):1695–701. 10.1128/IAI.01507-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spikes S, Xu R, Nguyen CK, Chamilos G, Kontoyiannis DP, Jacobson RH, et al. Gliotoxin production in Aspergillus fumigatus contributes to host-specific differences in virulence. J Infect Dis. 2008;197(3):479–86. 10.1086/525044 . [DOI] [PubMed] [Google Scholar]
- 32.Balibar CJ, Walsh CT. GliP, a multimodular nonribosomal peptide synthetase in Aspergillus fumigatus, makes the diketopiperazine scaffold of gliotoxin. Biochemistry. 2006;45(50):15029–38. 10.1021/bi061845b [DOI] [PubMed] [Google Scholar]
- 33.Scharf DH, Remme N, Habel A, Chankhamjon P, Scherlach K, Heinekamp T, et al. A dedicated glutathione S-transferase mediates carbon-sulfur bond formation in gliotoxin biosynthesis. J Am Chem Soc. 2011;133(32):12322–5. 10.1021/ja201311d [DOI] [PubMed] [Google Scholar]
- 34.Bok JW, Chung D, Balajee SA, Marr KA, Andes D, Nielsen KF, et al. GliZ, a transcriptional regulator of gliotoxin biosynthesis, contributes to Aspergillus fumigatus virulence. Infect Immun. 2006;74(12):6761–8. 10.1128/IAI.00780-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith TD, Calvo AM. The mtfA transcription factor gene controls morphogenesis, gliotoxin production, and virulence in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot Cell. 2014;13(6):766–75. 10.1128/EC.00075-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chooi YH, Fang J, Liu H, Filler SG, Wang P, Tang Y. Genome mining of a prenylated and immunosuppressive polyketide from pathogenic fungi. Org Lett. 2013;15(4):780–3. 10.1021/ol303435y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arruda LK, Platts-Mills TA, Fox JW, Chapman MD. Aspergillus fumigatus allergen I, a major IgE-binding protein, is a member of the mitogillin family of cytotoxins. J Exp Med. 1990;172(5):1529–32. 10.1084/jem.172.5.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dhingra S, Lind AL, Lin HC, Tang Y, Rokas A, Calvo AM. The fumagillin gene cluster, an example of hundreds of genes under veA control in Aspergillus fumigatus. PLoS ONE. 2013;8(10). 10.1371/journal.pone.0077147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiemann P, Guo CJ, Palmer JM, Sekonyela R, Wang CC, Keller NP. Prototype of an intertwined secondary-metabolite supercluster. Proc Natl Acad Sci U S A. 2013;110(42):17065–70. 10.1073/pnas.1313258110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin WB, Baccile JA, Bok JW, Chen Y, Keller NP, Schroeder FC. A nonribosomal peptide synthetase-derived iron(III) complex from the pathogenic fungus Aspergillus fumigatus. J Am Chem Soc. 2013;135(6):2064–7. 10.1021/ja311145n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Throckmorton K, Lim FY, Kontoyiannis DP, Zheng W, Keller NP. Redundant synthesis of a conidial polyketide by two distinct secondary metabolite clusters in Aspergillus fumigatus. Environ Microbiol. 2016;18(1):246–59. Epub 2015/08/06. 10.1111/1462-2920.13007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rocha MC, Fabri JH, Franco de Godoy K, Alves de Castro P, Hori JI, Ferreira da Cunha A, et al. Aspergillus fumigatus MADS-Box Transcription Factor rlmA Is Required for Regulation of the Cell Wall Integrity and Virulence. G3 (Bethesda). 2016;6(9):2983–3002. Epub 2016/07/31. 10.1534/g3.116.031112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shantappa S, Dhingra S, Hernandez-Ortiz P, Espeso EA, Calvo AM. Role of the zinc finger transcription factor SltA in morphogenesis and sterigmatocystin biosynthesis in the fungus Aspergillus nidulans. PLoS One. 2013;8(7):e68492. Epub 2013/07/11. 10.1371/journal.pone.0068492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spielvogel A, Findon H, Arst HN, Araujo-Bazan L, Hernandez-Ortiz P, Stahl U, et al. Two zinc finger transcription factors, CrzA and SltA, are involved in cation homoeostasis and detoxification in Aspergillus nidulans. Biochem J. 2008;414(3):419–29. Epub 2008/05/13. 10.1042/BJ20080344 . [DOI] [PubMed] [Google Scholar]
- 45.Du W, Zhai P, Wang T, Bromley MJ, Zhang Y, Lu L. The C2H2 transcription factor SltA contributes to azole resistance by coregulating the expression of the drug target Erg11A and the drug efflux pump Mdr1 in Aspergillus fumigatus Antimicrob Agents Chemother. 2021. Epub 2021/01/13. 10.1128/AAC.01839-20 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lind AL, Lim FY, Soukup AA, Keller NP, Rokas A. An LaeA- and BrlA-dependent cellular network governs tissue-specific secondary metabolism in the human pathogen Aspergillus fumigatus. mSphere. 2018;3(2). Epub 2018/03/23. 10.1128/mSphere.00050-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furukawa T, van Rhijn N, Fraczek M, Gsaller F, Davies E, Carr P, et al. The negative cofactor 2 complex is a key regulator of drug resistance in Aspergillus fumigatus. Nat Commun. 2020;11(1):427. Epub 2020/01/24. 10.1038/s41467-019-14191-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perrin RM, Fedorova ND, Bok JW, Cramer RA, Wortman JR, Kim HS, et al. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007;3(4):e50. 10.1371/journal.ppat.0030050 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bok JW, Balajee SA, Marr KA, Andes D, Nielsen KF, Frisvad JC, et al. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot Cell. 2005;4(9):1574–82. 10.1128/EC.4.9.1574-1582.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coyle CM, Panaccione DG. An ergot alkaloid biosynthesis gene and clustered hypothetical genes from Aspergillus fumigatus. Appl Environ Microbiol. 2005;71(6):3112–8. Epub 2005/06/04. 10.1128/AEM.71.6.3112-3118.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Panaccione DG, Arnold SL. Ergot alkaloids contribute to virulence in an insect model of invasive aspergillosis. Sci Rep. 2017;7(1):8930. Epub 2017/08/23. 10.1038/s41598-017-09107-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fallon JP, Reeves EP, Kavanagh K. Inhibition of neutrophil function following exposure to the Aspergillus fumigatus toxin fumagillin. J Med Microbiol. 2010;59(Pt 6):625–33. Epub 2010/03/06. 10.1099/jmm.0.018192-0 . [DOI] [PubMed] [Google Scholar]
- 53.Guruceaga X, Ezpeleta G, Mayayo E, Sueiro-Olivares M, Abad-Diaz-De-Cerio A, Aguirre Urizar JM, et al. A possible role for fumagillin in cellular damage during host infection by Aspergillus fumigatus. Virulence. 2018;9(1):1548–61. Epub 2018/09/27. 10.1080/21505594.2018.1526528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Catlett NL, Lee B-N, Yoder OC, Turgeon BG. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet News. 2002;50:9–11. [Google Scholar]
- 55.Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, van den Hondel CA. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene. 1987;56(1):117–24. 10.1016/0378-1119(87)90164-8 . [DOI] [PubMed] [Google Scholar]
- 56.Richie DL, Miley MD, Bhabhra R, Robson GD, Rhodes JC, Askew DS. The Aspergillus fumigatus metacaspases CasA and CasB facilitate growth under conditions of endoplasmic reticulum stress. Mol Microbiol. 2007;63(2):591–604. 10.1111/j.1365-2958.2006.05534.x . [DOI] [PubMed] [Google Scholar]
- 57.Liu H, Lee MJ, Solis NV, Phan QT, Swidergall M, Ralph B, et al. Aspergillus fumigatus CalA binds to integrin α5β1 and mediates host cell invasion. Nat Microbiol. 2016;2:16211. 10.1038/nmicrobiol.2016.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Min K, Ichikawa Y, Woolford CA, Mitchell AP. Candida albicans gene deletion with a transient CRISPR-Cas9 system. mSphere. 2016;1(3). Epub 2016/06/25. 10.1128/mSphere.00130-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nødvig CS, Nielsen JB, Kogle ME, Mortensen UH. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS One. 2015;10(7):e0133085. Epub 2015/07/16. 10.1371/journal.pone.0133085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Macheleidt J, Scherlach K, Neuwirth T, Schmidt-Heck W, Straßburger M, Spraker J, et al. Transcriptome analysis of cyclic AMP-dependent protein kinase A-regulated genes reveals the production of the novel natural compound fumipyrrole by Aspergillus fumigatus. Mol Microbiol. 2015;96(1):148–62. Epub 2015/01/15. 10.1111/mmi.12926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pham T, Xie X, Lin X. An intergenic "safe haven" region in Aspergillus fumigatus. Med Mycol. 2020. Epub 2020/03/15. 10.1093/mmy/myaa009 . [DOI] [PubMed] [Google Scholar]
- 62.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–15. Epub 2019/08/04. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. Epub 2014/09/28. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. Epub 2010/10/29. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu H, Gravelat FN, Chiang LY, Chen D, Vanier G, Ejzykowicz DE, et al. Aspergillus fumigatus AcuM regulates both iron acquisition and gluconeogenesis. Mol Microbiol. 2010;78(4):1038–54. Epub 2010/11/11. 10.1111/j.1365-2958.2010.07389.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ejzykowicz DE, Cunha MM, Rozental S, Solis NV, Gravelat FN, Sheppard DC, et al. The Aspergillus fumigatus transcription factor Ace2 governs pigment production, conidiation and virulence. Mol Microbiol. 2009;72(1):155–69. 10.1111/j.1365-2958.2009.06631.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ejzykowicz DE, Solis NV, Gravelat FN, Chabot J, Li X, Sheppard DC, et al. Role of Aspergillus fumigatus DvrA in host cell interactions and virulence. Eukaryot Cell. 2010;9(10):1432–40. Epub 2010/08/03. EC.00055-10 [pii] 10.1128/EC.00055-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Curr Protoc Immunol. 2008;Chapter 14:Unit 14 1. Epub 2008/11/20. 10.1002/0471142735.im1401s83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438(7071):1151–6. 10.1038/nature04332 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shown are NanoString probe counts for 97 environmentally responsive A. fumigatus genes in lung samples from 3 mice after 5 d of infection and a single immunosuppressed, uninfected mouse.
(PDF)
Shown are the lung masses of mice after 5 d of infection with the indicated strains of A. fumigatus. Lines indicate the median and interquartile ranges of 5 mice per strain. Symbols indicate the results of individual mice. *P < 0.05.
(PDF)
Serial 10-fold dilutions of the indicated strains of A. fumigatus were spotted onto Aspergillus minimal medium (AMM) containing the indicated stressors. The plates were imaged after incubation at 37°C for 2 d.
(PDF)
Values represent Log2 expression ratios for each gene in the ΔsltA strain divided by the wild-type strain as determined by RNA-seq (Af293) or qRT-PCR (AF1160).
(PPTX)
(A) qRT-PCR results of relative gene expression in the indicated strains. (B) The replacement of nscR (Afu7g00130) by the hygromycin resistance cassette (HYG) was verified by PCR using two different sets of primers.
(PDF)
Results are from the lungs of mice infected with A. fumigatus strain Af293 and from the same strain grown in standard Aspergillus minimal medium (AMM) in normoxia or hypoxia, in AMM deficient in specific nutrients, or in AMM containing 10% bovine serum. Results are the mean of 3 biological replicates or mice per condition.
(XLSX)
Results are the mean of 3 biological replicates of organisms grown in AMM with low zinc and low nitrogen. Inf indicates no detectable reads in the ΔsltA mutant samples.
(XLSX)
(XLSX)
Data Availability Statement
The raw sequencing reads from this study have been submitted to the NCBI sequence read archive (SRA) under BioProject accession number PRJNA693756. The remaining relevant data are within the paper and the Supporting Information files.