Abstract
Background
Regional monitoring of the proportion of the population who have been infected by SARS-CoV-2 is important to guide local management of the epidemic, but is difficult in the absence of regular nationwide serosurveys. We aimed to estimate in near real time the proportion of adults who have been infected by SARS-CoV-2.
Methods
In this modelling study, we developed a method to reconstruct the proportion of adults who have been infected by SARS-CoV-2 and the proportion of infections being detected, using the joint analysis of age-stratified seroprevalence, hospitalisation, and case data, with deconvolution methods. We developed our method on a dataset consisting of seroprevalence estimates from 9782 participants (aged ≥20 years) in the two worst affected regions of France in May, 2020, and applied our approach to the 13 French metropolitan regions over the period March, 2020, to January, 2021. We validated our method externally using data from a national seroprevalence study done between May and June, 2020.
Findings
We estimate that 5·7% (95% CI 5·1–6·4) of adults in metropolitan France had been infected with SARS-CoV-2 by May 11, 2020. This proportion remained stable until August, 2020, and increased to 14·9% (13·2–16·9) by Jan 15, 2021. With 26·5% (23·4–29·8) of adult residents having been infected in Île-de-France (Paris region) compared with 5·1% (4·5–5·8) in Brittany by January, 2021, regional variations remained large (coefficient of variation [CV] 0·50) although less so than in May, 2020 (CV 0·74). The proportion infected was twice as high (20·4%, 15·6–26·3) in 20–49-year-olds than in individuals aged 50 years or older (9·7%, 6·9–14·1). 40·2% (34·3–46·3) of infections in adults were detected in June to August, 2020, compared with 49·3% (42·9–55·9) in November, 2020, to January, 2021. Our regional estimates of seroprevalence were strongly correlated with the external validation dataset (coefficient of correlation 0·89).
Interpretation
Our simple approach to estimate the proportion of adults that have been infected with SARS-CoV-2 can help to characterise the burden of SARS-CoV-2 infection, epidemic dynamics, and the performance of surveillance in different regions.
Funding
EU RECOVER, Agence Nationale de la Recherche, Fondation pour la Recherche Médicale, Institut National de la Santé et de la Recherche Médicale (Inserm).
Introduction
Little more than a year after the emergence of SARS-CoV-2 and a first pandemic wave that has had devastating consequences across the world, most European countries are now being confronted with an intense second or third wave of SARS-CoV-2. In this context, up-to-date regional estimates of the proportion of the population that has been infected with SARS-CoV-2 and might thus be temporarily protected against reinfection1, 2 constitute important information. Such estimates could help to characterise the burden of infection, epidemic dynamics, and the performance of surveillance in different regions of a country and inform local management of the scale of the epidemic. This information will become ever more important as the epidemic progresses and spatial heterogeneities in population immunity potentially increase.
In many European countries, serological studies have provided estimates of the proportion of the population infected during the first pandemic wave. For example, it was estimated that about 4–5% of the population in metropolitan France had developed antibodies against SARS-CoV-2 by May, 2020, with seroprevalences of the order of 10% in Grand Est and Île-de-France, the two most affected regions.3, 4, 5 Since then, the virus has continued to circulate. Unfortunately, up-to-date estimates of seroprevalence able to capture the most recent regional evolution of the epidemic are unavailable. This largely stems from the difficulty and cost of implementing large-scale nationwide representative serosurveys at regular intervals.6, 7 It is therefore important to develop methods that can track the proportion of the population that has been infected in different regions using the joint analysis of existing seroprevalence data and other surveillance data that are more readily available in real time. Such monitoring is difficult to perform from the analysis of case data, since testing practices have changed over both time and space. Joint analysis of serological and death data across different countries has been used to reconstruct the proportion of infected individuals and has allowed extrapolation to countries where serology was not available.8, 9 However, such an approach might have difficulties in capturing spread in younger age groups given low infection–fatality ratios in these groups, and might provide lagged estimates given the relatively long delays between infection and death.
Research in context.
Evidence before this study
To identify past analyses aiming to reconstruct in real time the number of infections from the joint analysis of serological and hospitalisation or death data, we searched PubMed for peer-reviewed articles published between Jan 1 and Dec 10, 2020, using the search query (“COVID-19” OR “SARS-CoV-2”) AND “sero*” AND ((“hosp*” OR “death*”) AND (“rate*” or “number*”)), with no language restrictions. The query returned 372 results. Among those, eight were relevant to our study and provided estimates for the number of infections using a combination of serological and death data. None of these studies combined hospitalisation data and serosurveys to estimate the cumulative number of infections, nor were they designed to map infections in near real time and at different spatial scales in a country.
Added value of this study
Here, we provide a simple approach to monitor in near real time the number of infections at regional and national scales, using a method that combines age-stratified hospitalisation and seroprevalence data in France. We determined the number of infections in the different regions of metropolitan France between March 1, 2020, and Jan 15, 2021, and also estimated the proportion of cases detected by surveillance.
Implications of all the available evidence
Our findings show how hospitalisation data can inform on the proportion of infected population even if a nationwide serological study is unavailable. In the absence of contemporary serosurveys, our study shows that the proportion infected by SARS-CoV-2 might be higher than 20% in some French regions.
Here, we present a method to reconstruct the proportion of the adult population infected by SARS-CoV-2 and the proportion of infections detected by surveillance from the joint analysis of age-stratified seroprevalence, hospitalisation, and case data. The method is applied to metropolitan France and makes it possible to track in near real time the underlying SARS-CoV-2 infections by region and age group.
Methods
Infection–hospitalisation ratios
Estimates of age-stratified infection–hospitalisation ratios (IHRs; ie, the proportion of infected individuals in an age group that require hospital admission for COVID-19) were derived from the joint analysis of hospitalisation and serological data documenting the impact of the first pandemic wave in Île-de-France and Grand Est, the two regions of metropolitan France that were most affected. This calculation has been described elsewhere.10 In short, seroprevalence estimates were obtained from the SAPRIS study,4 which gathered data from the large population-based French cohorts Constances, E3N-E4N, and NutriNet-Santé, and the numbers of hospital admissions were obtained from the SI-VIC database, the national exhaustive inpatient surveillance system used during the pandemic (appendix p 1). 9782 adult participants (aged ≥20 years) were recruited in the SAPRIS study in Île-de-France and Grand Est and sampling weights were used to adjust for selection and participation in the cohorts before random selection. Sociodemographic covariates were used to correct for selection and participation bias. A complete description of the SAPRIS study has been provided elsewhere.4
The median date of sample collection in the SAPRIS serosurvey was May 14, 2020 (IQR May 12 to May 19). Seropositive individuals were assumed to have been infected at least 19 days before that date (April 25). Assuming a delay of 11 days between infection and hospital admission,11, 12 these individuals would correspond to hospitalisations occurring up to May 6, 2020. The IHR was therefore obtained by dividing the cumulative numbers of hospital admissions up to May 6, 2020, by the number of infected people estimated from the SAPRIS serosurvey. Seroprevalence status of the participants was inferred using a series of tests (ELISA-S, ELISA-NP, and seroneutralisation). Participants were classified as being truly infected, truly negative, or as having inconsistent serological results. The serological status of those remaining participants was inferred using a multiple imputation method, details of which are given in the appendix (pp 2–3) and elsewhere.4 To adjust for the imperfect sensitivity observed in the serological tests used, we applied a correction of 85% to our multiple imputation estimates to obtain the IHR and to derive the proportion infected. We also considered test sensitivities of 80%, 90%, and 100% in sensitivity analyses.
To characterise uncertainty in seroprevalence estimates, 1000 values were drawn from Student's t distribution (the reference distribution for the multiple imputation inference), and 1000 values for the IHR were derived.
7934 patients from nursing homes had been admitted to hospital with SARS-CoV-2 infection by May 6, 2020, but because the dynamics of transmission in that population differ to those in the general population, and as they were not part of the cohort target population used for the estimation of the IHR, those patients were excluded from the calculation.
Reconstruction of the dynamics of infection
The curve of the daily number of infections was reconstructed from the daily number of hospital admissions and the distribution of the delay from infection to hospitalisation. For each age group, the number of infections was obtained as the deconvolution of the daily number of hospitalisations and the infection-to-hospitalisation delay distribution, divided by the IHR (appendix p 1). The infection-to-hospitalisation delay is discrete and parameterised with a gamma distribution with a mean of 11 days and SD of 3·2 days.12 The deconvolution approach used a Richardson-Lucy scheme that was adapted to account for right censoring in the hospitalisation curve (appendix pp 1, 4).13 The number of infections was reconstructed for all 13 regions of metropolitan France. Hospitalised individuals with missing age represented 0·7% (n=1480) of total hospital admissions and were not included in the study.
The heterogeneity of infections across regions was assessed using the coefficient of variation (CV). We report the estimated cumulative number of infections in the adult population on May 11, 2020 (after the first wave), on Oct 31, 2020 (during the second wave before the lockdown), and on Jan 15, 2021 (most recent estimate).
Internal and external validation of seroprevalence estimates
To internally validate our method, we compared seroprevalence estimates of the SAPRIS study in Grand Est, Ile-de-France and Nouvelle-Aquitaine with the seroprevalence predicted by our method on the median date of the study (May 14, 2020), reconstructed from the infections that happened up to April 25, to account for the 19-day delay between infection and seroconversion.
We validated our method externally using a separate national seroprevalence study3 done between May 2 and June 2, 2020, among individuals aged 15 years or older in 12 regions of metropolitan France. Serological results for SARS-CoV-2 were measured by the detection of IgG antibodies directed against the viral envelope using the ELISA-S method on 12 114 samples from throughout France. Corsica was excluded from this analysis since only 36 samples were available. We compared the results of this survey with the seroprevalence predicted by our method on May 17, 2020, the median date of sample collection, which we reconstructed from the predicted infections up to April 28 in those 12 regions (assuming a 19-day delay between infection and seroconversion).
Pearson's correlation was used to compare seroprevalence estimated with the model and from the external dataset.
Estimation of the proportion of infections detected by surveillance
Dates of infections of confirmed cases were reconstructed with the same deconvolution approach using the national virological surveillance database of confirmed cases (SI-DEP; appendix p 1) and assuming the infection-to-detection delay has a gamma distribution of mean 8·5 days and SD 2·8 days, which accounts for an incubation period of 5·5 days14 and a delay of 3 days to testing.15 Proportions of infections detected by surveillance were estimated over three periods (June 1 to Aug 31, 2020; Sept 1 to Oct 31, 2020; and Nov 1, 2020 to Jan 15, 2021) for all 13 regions as the ratio of the cumulative number of infections reconstructed from the confirmed cases recorded in SI-DEP over the cumulative number of infections, as estimated with the hospitalisation data.
Sensitivity analyses
Several sensitivity analyses were done. We studied the impact of the delay distributions on the estimated proportion of infected individuals in the population and on the proportion of cases detected (appendix p 2). We changed the mean and variance of the gamma distribution of the time-to-hospitalisation delay, and varied the delay with age and over the course of the epidemic (appendix pp 12–14). In another series of sensitivity analyses, we varied the distribution of infection-to-detection delays and changed the delays during the course of the epidemic (appendix pp 15–16). We also varied the cutoff date chosen for the estimation of the IHR (appendix p 17). Finally, we evaluated the impact of a 10–30% decrease of the IHR during the second wave.
R (version 3.6.1) was used for all statistical analyses. The latest estimates are available online.
Ethical approval
Ethical approval and written or electronic informed consent were obtained from each participant before enrolment in the original cohort. The SAPRIS survey was approved by the Institut National de la Santé et de la Recherche Médicale ethics committee (approval number 20-672; March 30, 2020). The SAPRIS-SERO study was approved by the Sud-Mediterranée III ethics committee (approval number 20.04.22.74247) and electronic informed consent was obtained from all participants for dried blood spot testing.
Role of the funding source
The funders of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report.
Results
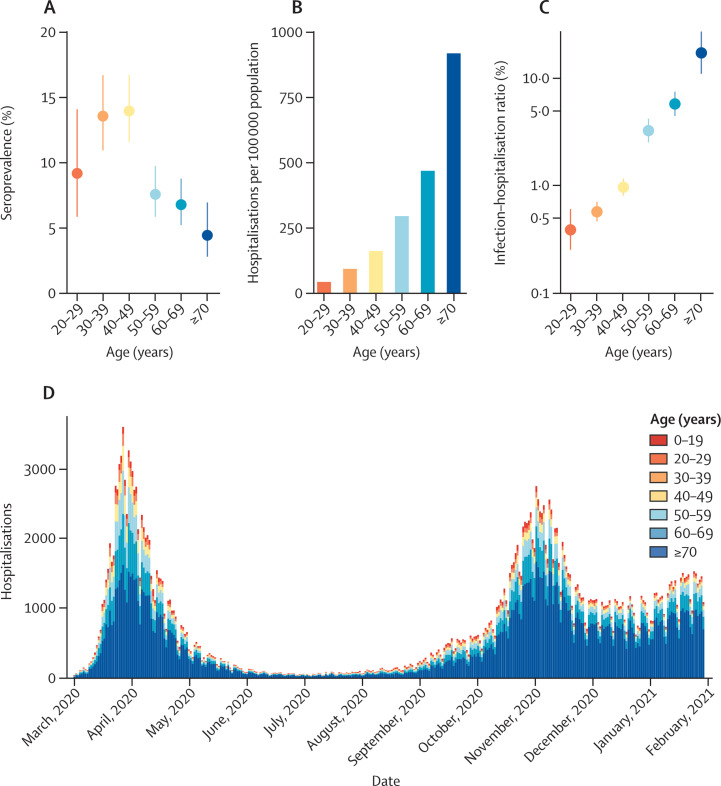
Observed seroprevalence in the Île-de-France and Grand Est regions in May to June, 2020, was highest among 40–49-year-olds (14·0%, 95% CI 11·6–16·7) and lower in older age groups, reaching a minimum among those aged 70 years or older (4·4%, 2·8–7·0; figure 1 ). Both the cumulative number of hospitalisations per 100 000 population and the IHR increased with age, with the IHR increasing from 0·4% (0·3–0·6) in 20–29-year-olds to 17·6% (11·2–27·8) in 70–89-year-olds (figure 1). The patterns of hospitalisations by age and the IHR were similar in Grand Est and Île-de-France (appendix p 5).
Figure 1.
Description of seroprevalence and hospitalisation data
(A) Estimates of seroprevalence by age group in the Île-de-France and Grand Est regions, in May to June, 2020 (median date May 14). (B) Cumulative number of hospitalisations per 100 000 population, in Île-de-France and Grand Est, from March 1 to May 6, 2020. (C) Estimates of infection–hospitalisation ratio by age group in Île-de-France and Grand Est. The y-axis is displayed in logarithmic scale. (D) Daily number of hospitalisations by age group in metropolitan France, from March 1, 2020, to Jan 30, 2021.
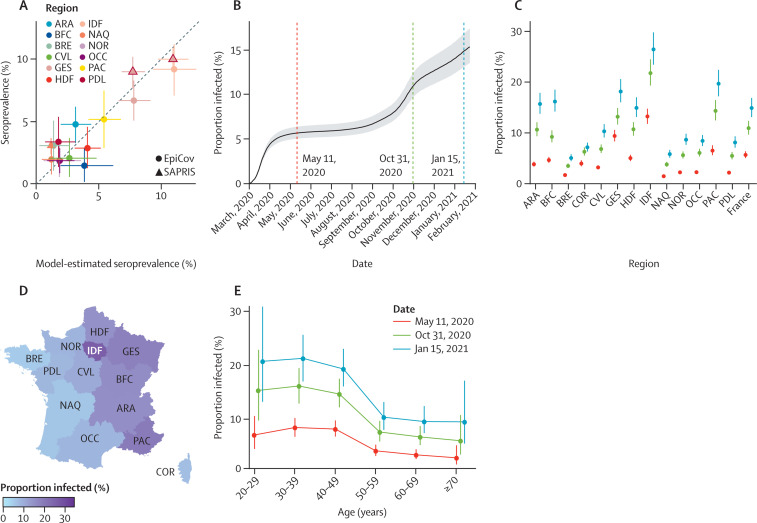
Our model is calibrated to serological data collected in two regions in May, 2020, but can be used to reconstruct the seroprevalence and proportion infected in all regions and over time (figure 2 ). Consistent with national seroprevalence from the dataset for external validation,3 we estimated that 4·8% (95% CI 4·3–5·4) of adults were seropositive to SARS-CoV-2 in May, 2020, in metropolitan France. Our regional estimates of seroprevalence in May were strongly correlated with the external validation dataset (coefficient of correlation 0·89), with ten of the 12 estimates contained in the 95% CI of the serosurvey (figure 2A). After correcting for the imperfect sensitivity of the serological assay, we found that 5·7% (5·1–6·4) of the adult population had been infected by SARS-CoV-2 by May 11, 2020, in metropolitan France, with important regional variations (figure 2B, C). The proportion of the adult population that had been infected remained stable during the summer months in 2020 and increased in September to reach 14·9% (13·2–16·9) by Jan 15, 2021 (appendix p 9). On that date, the proportion infected was highest in Île-de-France (ie, Paris area; 26·5%, 23·4–29·8), followed by Provence-Alpes-Côte d'Azur (19·7%, 17·2–22·4), Grand Est (18·2%, 16·1–20·6), Bourgogne-Franche-Comté (16·2%, 14·2–18·5), and Auvergne-Rhône-Alpes (15·7%, 13·8–17·9; figure 2D; appendix p 9). The lowest proportion was in Brittany (5·1%, 4·5–5·8). The proportion infected was more homogeneous across regions in January, 2021 (CV 0·50) than in May, 2020 (0·74).
Figure 2.
Reconstruction of the proportion infected in metropolitan France
(A) Scatter plot of the seroprevalence in regions estimated with our model on May 11, 2020 (x-axis) and in seroprevalence studies in May, 2020 (y-axis), obtained from the SAPRIS serosurvey and EpiCov database. Data from the SAPRIS serosurvey in Île-de-France and Grand Est (triangles contoured in red) were used to calibrate the model. Bars represent the 95% CIs of the seroprevalence estimated by the model. (B) Proportion infected among adults in metropolitan France between March 1, 2020, and Jan 24, 2021. Timing of infection was reconstructed from the daily number of hospitalisations for COVID-19 and the delay from infection to hospital admission. The grey area represents the 95% CI. (C) Proportion infected in metropolitan France and in the 13 regions of metropolitan France, by date. (D) Geographical distribution of the proportion infected on Jan 15, 2021. (E) Proportion infected by age group and date. ARA=Auvergne-Rhône-Alpes. BFC=Bourgogne-Franche-Comté. BRE=Bretagne. COR=Corsica. CVL=Centre-Val de Loire. GES=Grand Est. HDF=Hauts-de-France. IDF=Île-de-France. NAQ=Nouvelle-Aquitaine. NOR=Normandie. OCC=Occitanie. PAC=Provence-Alpes-Côte d'Azur. PDL=Pays de la Loire.
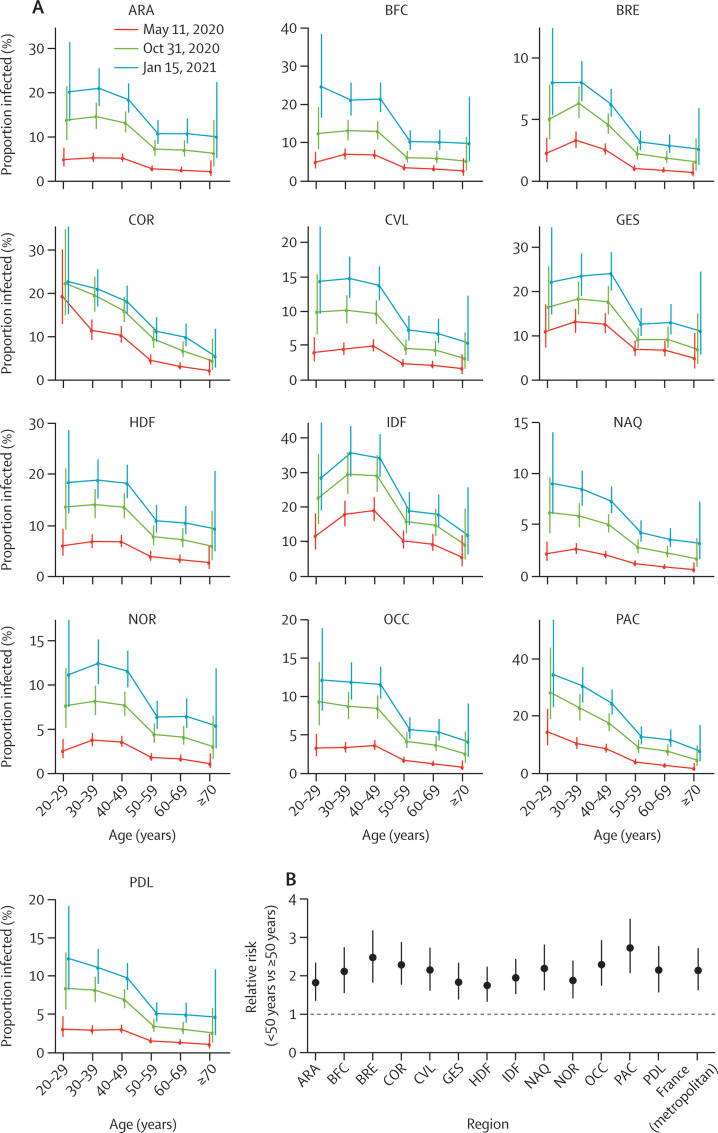
The proportion infected in metropolitan France was highest in those aged 20–49 years (20·4%, 95% CI 15·6–26·3), with lower rates of 9·7% (6·9–14·1) in those aged 50 years or older (figure 2E; appendix p 10). The same pattern by age was seen in most regions, with the risk of infection in those aged 20–49 years being 2–3 times higher than that in those aged 50 years or older, depending on the region (figure 3 ).
Figure 3.
Proportion infected in the regions by age group and over time
(A) Estimates for the 13 regions of metropolitan France are shown on three dates. (B) Relative risk of infection of younger (<50 years) versus older (≥50 years) individuals. ARA=Auvergne-Rhône-Alpes. BFC=Bourgogne-Franche-Comté. BRE=Bretagne. COR=Corsica. CVL=Centre-Val de Loire. GES=Grand Est. HDF=Hauts-de-France. IDF=Île-de-France. NAQ=Nouvelle-Aquitaine. NOR=Normandie. OCC=Occitanie. PAC=Provence-Alpes-Côte d'Azur. PDL=Pays de la Loire.
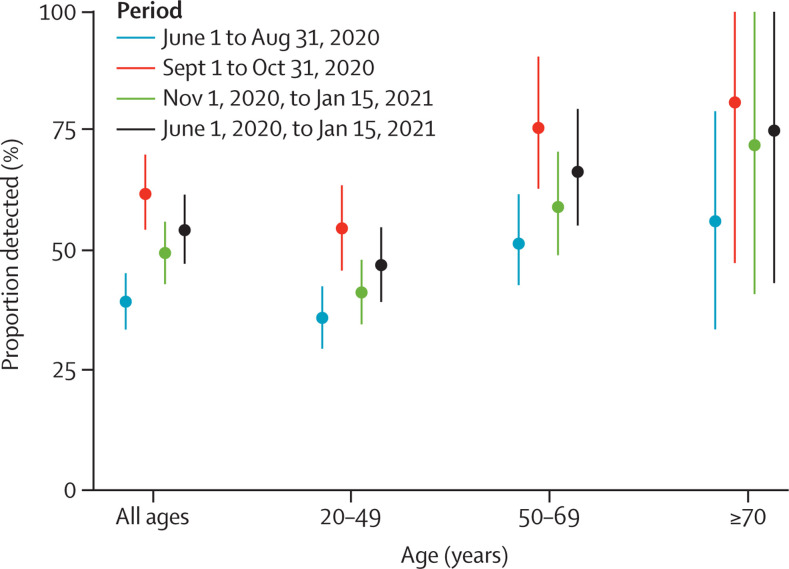
We estimated that 54·5% (95% CI 47·4–61·9) of SARS-CoV-2 infections in the adult population were detected by surveillance between June, 2020, and January, 2021, with a probability of detection of 40·2% (34·3–46·3) in June to August, 2020; 62·3% (54·7–70·5) in September to October, 2020; and 49·3% (42·9–55·9) in November, 2020, to January, 2021 (figure 4 ; appendix p 11). The probability of detection between June, 2020, and January, 2021, was higher in those aged 50 years or older (68·7%, 54·4–82·6) than in those aged 20–49 years (47·1%, 39·4–55·1; figure 4). These estimates are consistent with a simple analysis of the raw data from the SI-VIC and SI-DEP databases: between June 1 and Nov 30, 2020, approximately 170 000 adults were hospitalised and 2 400 000 cases were detected by surveillance in metropolitan France, leading to a proportion detected of about 46% for an average estimated IHR of 3·3%.
Figure 4.
Proportion of infections detected by surveillance over different periods between June, 2020, and January, 2021
Bars represent 95% CIs.
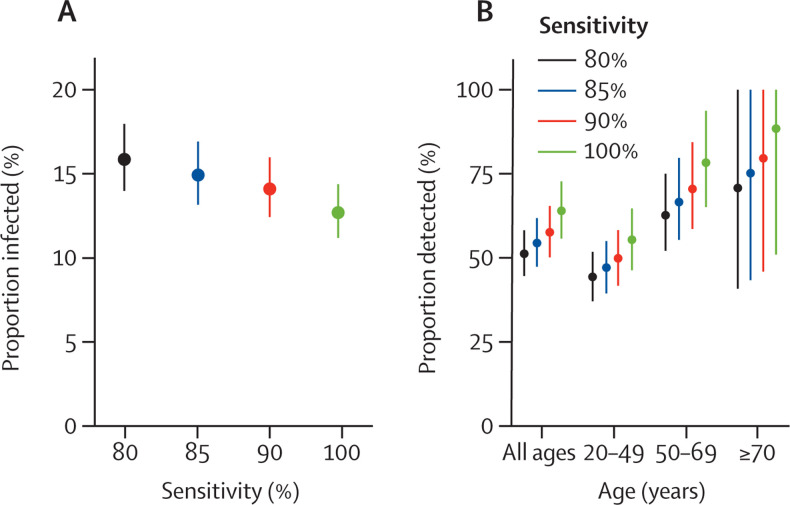
In our baseline scenario, we assumed that the sensitivity of the serological test was 85%. In a sensitivity analysis, we found that estimates of the proportion infected by Jan 15, 2021, increased from 12·7% (95% CI 11·2–14·3) for a sensitivity of 100% to 15·8% (14·0–18·0) for a sensitivity of 80% (figure 5A ). The proportion of infections that were detected varied from 51·2% (44·5–58·1) for 80% sensitivity to 63·9% (55·7–72·7) for 100% sensitivity (figure 5B).
Figure 5.
Sensitivity analysis
(A) Proportion infected on Jan 15, 2021, assuming different sensitivities of the serological tests. (B) Proportion of infections detected by surveillance between June, 2020, and January, 2021, assuming different sensitivities of the serological tests. In our baseline analysis, we consider a sensitivity of the test of 85%.
In a simulation study, we checked that our deconvolution approach could correctly retrieve the daily numbers of infections if delay distributions (from infection to hospitalisation and from infection to detection) were known (appendix pp 1, 4). However, this approach is in principle sensitive to uncertainty in these distributions. We therefore conducted a series of sensitivity analyses and showed that our results on the proportion infected and proportion detected were robust to misspecification of the delay distributions (appendix pp 2, 12–16). This is because we were not aiming to precisely estimate the number of infections on a given day, but only the cumulative number of infections since the start of the pandemic. We also showed that our results are robust to changes in the cutoff date used to compute the IHR (May 6, 2020), because epidemic activity was low in France in May, 2020 (appendix pp 2, 17).
In a sensitivity analysis, we found that a 10–30% reduction in the IHR during the second wave would have little impact on estimates of the proportion infected overall (15·8% [95% CI 13·9–17·9] for 10% reduction and 18·4% [16·2–20·9] for 30% reduction; appendix p 6) but would reduce the proportion of infections being detected to 49·6% (43·2–56·4) for 10% reduction in the IHR and to 39·3% (34·3–44·7) for 30% reduction (appendix p 6).
Discussion
We have presented a method to reconstruct the proportion of the adult population infected by SARS-CoV-2 by region and age group from the joint analysis of readily available hospital surveillance data and existing serological surveys. This approach offers a simple way to track the number of infections in the population with a lag of a few weeks (ie, from infection to hospitalisation), which is challenging in the absence of regular, large-scale, representative serosurveys.
After accounting for the imperfect sensitivity of serology, we estimate that the proportion infected by SARS-CoV-2 in metropolitan France increased by two to three times from about 6% in May, 2020, to about 15% in mid-January, 2021. There are important differences between the two waves. First, the first wave occurred over a much shorter time period than the second wave that is still ongoing (figure 2). Second, while the first wave was mostly concentrated in two regions, all regions were impacted by the second wave. As a consequence, the proportion infected was more homogeneous in January, 2021, than in May, 2020. However, substantial heterogeneities remain. For example, the proportion infected in Île-de-France (Paris area) was about twice the national average. Overall, relatively similar patterns of infection by age were reconstructed in the different regions, with individuals aged 20-49 years being at substantially higher risk of infection.
Assuming that those infected are immunised against reinfection, the estimated 27% immunity could contribute to slowing down the spread of the virus in Île-de-France. Consider, for example, a situation in which control measures are such that, in a naive population, a case infects on average 1·6 people (reproduction number R 0=1·6). In such a scenario, we would expect the number of cases to double about every 10 days. With 27% immunity, the effective reproduction number R eff would be reduced to 0·73 × 1·6 = 1·2, leading to a substantially longer doubling time of about 26 days. However, given the very high transmissibility of SARS-CoV-2 (estimated at R 0=3),12 about 70% herd protection is likely to be needed for viral circulation to stop if all control measures were lifted.16 In the absence of control measures, 27% immunity would be insufficient to avoid a major crisis in hospitals since R eff would then be 0·73 × 3 = 2·32, with the number of cases expected to double about every 6 days. This does not take into account the potential waning of natural immunity.
For the period between June and August, 2020, we estimated that 40·2% (95% CI 34·3–46·3) of infections were detected, which is consistent with another modelling study15 that reported a detection rate of 38% (35–44) at the end of June. In our baseline scenario, we assumed a sensitivity of 85% for our assay, consistent with existing estimates.17 Higher sensitivities would lead to slightly lower estimates of the proportion infected and would inflate the proportion of infections being detected by surveillance at surprisingly high levels in some age groups.
Estimates of the proportion of the population infected by SARS-CoV-2 constitute useful contextual information to better characterise the burden of infection and epidemic dynamics in regions, as well as to ascertain the performance of surveillance. Such estimates are also important to ensure that mathematical models used to support policy making are correctly calibrated. However, it would be premature to use them as a basis to design differential control strategies in regions. First, although most people infected by SARS-CoV-2 appear to acquire protection against reinfection for at least 6 months,1, 2 we still lack data to document waning of immunity over longer time periods. Immunity might also be less important for asymptomatic infections that constitute a substantial proportion of infections.18 If there is waning of immunity, in the absence of vaccines, the estimated number of people infected by SARS-CoV-2 will be an upper bound of the number of people that are protected against infection. The interpretation of contemporary seroprevalence estimates might be equally challenging since antibody decay following infection is not necessarily synonymous with a loss of protection.19 Therefore, in the long run, in the absence of vaccines, the proportion protected against SARS-CoV-2 could fall between the proportion seropositive estimated from seroprevalence studies and the proportion infected estimated with an approach such as ours. Obviously, as the vaccine roll-out progresses, it will be essential to track the level of immunity acquired through vaccination. Second, regions with the highest proportions of infected population might also be those that have larger transmission rates for example because of larger population densities. Third, we need to remain cautious in a context of emergence of new variants that are more transmissible and appear to partly escape the immune response.20 As more recent seroprevalence studies and data on the duration of immunity become available, this information could easily be integrated into our statistical framework to estimate the proportion of the population that is currently immunised from the time series of infections over time that we reconstructed and an assumption about the distribution of the duration of immunity.
Our estimates rely on the assumption that age-specific IHRs remained constant over time and across regions. However, it is possible that IHRs changed during the course of the pandemic—eg, as a function of the stress on the health-care system. In a sensitivity analysis, assuming a reduced IHR during the second wave had little impact on the proportion infected. Our IHR estimates are calculated during the first pandemic wave and therefore constitute averages over a time period during which the stress on the health-care system changed rapidly. They are nonetheless in line with national estimates of the IHR for other countries.21 IHRs might also vary with regional hospitalisation policies. For example, if there is higher propensity to hospitalise young adults in some regions, we might overestimate the proportion of infected individuals in this age group and therefore in the overall population. However, despite these possible regional and contextual variations in IHRs, our regional estimates of seroprevalence were strongly correlated with our external validation dataset, despite using IHR estimates only for Île-de-France and Grand Est, the two regions that were the most affected during the first wave. IHRs might also have changed if the population of those infected changed between the first and second wave. Our results should be robust to variations in the age distribution of those infected since our approach controls for age. However, if in a given age group, the proportion of infected individuals with higher IHR (eg, because of comorbidities) decreased between the first and second wave owing to improved protective measures, we might underestimate the proportion infected in that age group. Patients with undiagnosed COVID-19 admitted to hospital at the very beginning of the pandemic might have led to overestimation of the IHR. However, any such effect would probably be small since most COVID-19-related hospitalisations are likely to have been detected once hospital surveillance was in place from mid-March, 2020, given the exponential nature of the first wave. Since our framework relies on the analysis of hospitalisation data and very few children were hospitalised, our approach would be likely to generate large CIs for that age group. We therefore decided to focus on adults.
In conclusion, we have presented a simple framework to track the proportion of the population infected with a lag of a few weeks using the joint analysis of age-stratified hospitalisation and serological data. Age-specific IHRs might vary by country given the different health-care systems. However, it should be easy to recalibrate our model to data from countries in which hospital surveillance and results of serosurveys are available.
This online publication has been corrected. The corrected version first appeared at thelancet.com/public-health on Month DD, 2021
Data sharing
The code and data will be made available online.
Declaration of interests
FC reports personal fees from Imaxio and Sanofi, outside of the submitted work. The other authors declare no competing interests.
Acknowledgments
Acknowledgments
Support for this study was provided by Agence Nationale de la Recherche (ANR; #ANR-20-COVI-000, #ANR-10-COHO-06), Fondation pour la Recherche Médicale (#20RR052-00), Institut National de la Santé et de la Recherche Médicale (Inserm; #C20-26). We acknowledge financial support from the Investissement d'Avenir programme, the Laboratoire d'Excellence Integrative Biology of Emerging Infectious Diseases programme (grant ANR-10-LABX-62-IBEID), Santé Publique France, the INCEPTION project (PIA/ANR-16-CONV-0005), the EU Horizon 2020 research and innovation programme under grants 101003589 (RECOVER) and 874735 (VEO), AXA, and Groupama. The CONSTANCES Cohort Study is supported by the Caisse Nationale d'Assurance Maladie, the French Ministry of Health, the Ministry of Research, and Inserm. CONSTANCES benefits from a grant from the French National Research Agency (grant number ANR-11-INBS-0002) and is also partly funded by Merck Sharp & Dohme, AstraZeneca, Lundbeck, and L'Oreal. The E3N-E4N cohort is supported by the following institutions: Ministère de l'Enseignement Supérieur, de la Recherche et de l'Innovation; Inserm; University Paris-Saclay; Gustave Roussy; the Mutuelle générale de l'Éducation nationale; and the French League Against Cancer. The NutriNet-Santé study is supported by the following public institutions: Ministère de la Santé, Santé Publique France, Inserm, Institut National de la Recherche Agronomique, Conservatoire National des Arts et Métiers, and Sorbonne Paris Nord. The CEPH-Biobank is supported by the Ministère de l'Enseignement Supérieur, de la Recherche et de l'Innovation. NH, JP, and SC acknowledge Rachel Torchet, Rémi Planel, Thomas Ménard, and Hervé Ménager from Institut Pasteur, Paris, for their technical contributions.
Contributors
NH, JP, NL, DL-B, FC, and SC designed and planned the study. NH, JP, NL, CTK, and HS contributed to the statistical analysis. GS, MT, MZ, XdL, DL-B, and FC contributed to data collection. NH, JP, and SC wrote the original draft. All authors critically edited the manuscript. NH, NL, and JP directly accessed and verified the data. All authors had access to all the data reported in the study and had final responsibility to submit for publication.
Supplementary Material
References
- 1.Dan JM, Mateus J, Kato Y, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371 doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lumley SF, O'Donnell D, Stoesser NE, et al. Antibody status and incidence of SARS-CoV-2 infection in health care workers. N Engl J Med. 2021;384:533–540. doi: 10.1056/NEJMoa2034545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.EpiCov En mai 2020, 4,5 % de la population en France métropolitaine a développé des anticorps contre le SARS-CoV-2. Sept 10, 2020. https://drees.solidarites-sante.gouv.fr/publications/etudes-et-resultats/en-mai-2020-45-de-la-population-vivant-en-france-metropolitaine
- 4.Carrat F, de Lamballerie X, Rahib D, et al. Seroprevalence of SARS-CoV-2 among adults in three regions of France following the lockdown and associated risk factors: a multicohort study. SSRN. 2020 doi: 10.2139/ssrn.3696820. published online Oct 23. (preprint) [DOI] [Google Scholar]
- 5.Le Vu S, Jones G, Anna F, et al. Prevalence of SARS-CoV-2 antibodies in France: results from nationwide serological surveillance. medRxiv. 2020 doi: 10.1101/2020.10.20.20213116. published online Oct 21. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ward H, Cooke G, Atchison C, et al. Declining prevalence of antibody positivity to SARS-CoV-2: a community study of 365,000 adults. medRxiv. 2020 doi: 10.1101/2020.10.26.20219725. published online Oct 27. (preprint) [DOI] [Google Scholar]
- 7.Pouwels KB, House T, Pritchard E, et al. Community prevalence of SARS-CoV-2 in England from April to November, 2020: results from the ONS Coronavirus Infection Survey. Lancet Public Health. 2021;6:e30–e38. doi: 10.1016/S2468-2667(20)30282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Driscoll M, Dos Santos GR, Wang L, et al. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2020;590:140–145. doi: 10.1038/s41586-020-2918-0. [DOI] [PubMed] [Google Scholar]
- 9.Brazeau N, Verity R, Jenks S, et al. Imperial College London; London: 2020. Report 34: COVID-19 infection fatality ratio: estimates from seroprevalence. [Google Scholar]
- 10.Lapidus N, Paireau J, Levy-Bruhl D, et al. Do not neglect SARS-CoV-2 hospitalization and fatality risks in the middle-aged adult population. Infect Dis Now. 2021 doi: 10.1016/j.idnow.2020.12.007. published online Jan 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. SSRN. 2020 doi: 10.2139/ssrn.3546052. published online March 3. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salje H, Tran Kiem C, Lefrancq N, et al. Estimating the burden of SARS-CoV-2 in France. Science. 2020;369:208–211. doi: 10.1126/science.abc3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstein E, Dushoff J, Ma J, Plotkin JB, Earn DJD, Lipsitch M. Reconstructing influenza incidence by deconvolution of daily mortality time series. Proc Natl Acad Sci USA. 2009;106:21825–21829. doi: 10.1073/pnas.0902958106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lauer SA, Grantz KH, Bi Q, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pullano G, Di Domenico L, Sabbatini CE, et al. Underdetection of COVID-19 cases in France threatens epidemic control. Nature. 2021;590:134–139. doi: 10.1038/s41586-020-03095-6. [DOI] [PubMed] [Google Scholar]
- 16.Fontanet A, Cauchemez S. COVID-19 herd immunity: where are we? Nat Rev Immunol. 2020;20:583–584. doi: 10.1038/s41577-020-00451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stringhini S, Wisniak A, Piumatti G, et al. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): a population-based study. Lancet. 2020;396:313–319. doi: 10.1016/S0140-6736(20)31304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynolds CJ, Swadling L, Gibbons JM, et al. Discordant neutralizing antibody and T cell responses in asymptomatic and mild SARS-CoV-2 infection. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abf3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyllie DH, Mulchandani R, Jones HE, et al. SARS-CoV-2 responsive T cell numbers are associated with protection from COVID-19: a prospective cohort study in keyworkers. medRxiv. 2020 doi: 10.1101/2020.11.02.20222778. published online Nov 4. (preprint) [DOI] [Google Scholar]
- 20.Zucman N, Uhel F, Descamps D, Roux D, Ricard JD. Severe reinfection with South African SARS-CoV-2 variant 501Y.V2: a case report. Clin Infect Dis. 2021 doi: 10.1093/cid/ciab129. published online Feb 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knock E, Whittles L, Lees J, et al. Imperial College London; London: 2020. Report 41—The 2020 SARS-CoV-2 epidemic in England: key epidemiological drivers and impact of interventions. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code and data will be made available online.