ABSTRACT
As the most common histologic subtype of renal cancer, clear cell renal cell carcinoma (ccRCC) poses a serious threat to public health. However, there are no specific molecular-targeted drugs for ccRCC at present. Human ATP-binding cassette (ABC) transporter family plays an important role in homeostasis maintenance. This study aimed to evaluate the potential diagnostic value of ABC genes in ccRCC. A total of 952 samples of ccRCC patients (707) and controls (245) from three different datasets were included for analysis. Receiver operating characteristic analysis and t-test were used to analyze the differential expression of ABC genes in ccRCC patients and control samples at mRNA level during screening and validations. The Cancer Genome Atlas (TCGA-ccRCC) dataset was utilized to investigate the correlation between ABC genes expression and prognostic value in ccRCC. We then investigated the interactions between ABCG1 and proteins in the Comparative Toxicogenomics Database (CTD). Finally, we found that ATP-binding cassette transporter G member 1 (ABCG1) was over-expressed in ccRCC patients compared with healthy samples at mRNA level. Cox regression analysis and Kaplan–Meier analysis showed that ccRCC patients with high ABCG1 expression had better overall survival (OS) than those patients with low expression (hazard ratio (HR) = 0.662, p = 0.007). This study demonstrated that ABCG1 is a potential diagnostic and prognostic biomarker in ccRCC and discussed the molecular mechanisms underlying the relationship between ccRCC and ABCG1, which might provide guidance for better management and treatment of ccRCC in the future.
KEYWORDS: ABCG1, ccRCC, diagnosis, prognosis, cholesterol efflux
Introduction
Renal cell carcinoma (RCC) has become one of the most common malignant tumors in urology and accounts for 85% of primary renal cancer. It was estimated that almost 403,262 (2.2%) new cases of kidney cancer and 175,098 (1.8%) deaths worldwide occurred in 2018 [1]. Besides, the global morbidity and mortality of RCC are increasing by approximately 2–3% per decade [2]. Clear cell renal cell carcinoma (ccRCC) is the most common pathological type of RCC in adults. Surgery is recommended as the preferred option in local ccRCC [3], with five-year survival at more than 90% [4]. Then, due to the absence of obvious clinical symptoms at the early stage, cancer metastasis has occurred in 25–30% patients at the time of initial diagnosis of ccRCC [5]. Although there has been a significant progress in the management of advanced ccRCC, with improved knowledge of disease and the application of targeted drugs. Five-year survival drops to 12% for patients with metastatic ccRCC [4]. Therefore, identification and validation of biomarkers will be crucial for optimizing the management of ccRCC. In recent years, many molecular biomarkers for ccRCC have been discovered. C1q/tumor necrosis factor (C1QTNF) [6] and six-snoRNA (small nucleolar RNA) signature (SNORA2, SNORD12B, SNORA59B, SNORA70B, SNORD93, and SNORD116-2) [7] could serve as an independent diagnostic and prognostic indicator for ccRCC. Because of the lack of precise and effective molecular targets for the therapy of ccRCC, it is still important to explore new molecular markers or therapeutic targets for the diagnosis and prognosis [8].
Human ATP-binding cassette (ABC) transporter family contains 49 members that are divided into eight subfamilies [9]. ABC transporter family is a widespread membrane-bound protein, which is mainly distributed in liver, intestine, blood–brain barrier, blood–testosterone barrier, placenta, and kidney. ABC protein can transport various endogenous substrates, including inorganic anions, metal ions, peptides, amino acids, sugars, hydrophobic, and metabolites [10]. Abnormal changes of the ABC genes can lead to multiple diseases, such as cystic fibrosis and disorder of cholesterol metabolism [11]. It has been demonstrated that cholesterol metabolism disorders are related to various cancers, and the cholesterol level in cancer cells elevates obviously compared with normal tissues [12,13].
Recently, several studies had explored the role of 10 ABC family members in ccRCC. ABCA1 [14] and ABCD1 [15] were related to the occurrence and development of tumors. ABCA13 [16,17], ABCB1 [18], ABCC1 [19], and ABCC2 [20,21] were associated with drug resistance and treatment of tumors. ABCB2 and ABCB3 were found to be involved in tumor immune evasion [21,22]. ABCG2 was correlated with tumor progression, prognosis [23] and drug resistance [18], and ABCB10 was associated with tumor progression as well as prognosis [24]. In this study, we mainly focused on the expression of ABC genes in multiple datasets to assess their diagnostic and prognostic value in ccRCC.
Material and methods
Gene expression of ABC family members in 952 samples from three independent public datasets (GSE40435 dataset, GSE53757 dataset, and TCGA-ccRCC dataset) was analyzed by screening and verification. Transcriptional expression of ABC genes from the Oncomine database (http://www.oncomine.org) was also investigated. Then, a prognostic analysis of the validated gene was conducted on TCGA-ccRCC dataset from UCSC Xena (https://xenabrowser.net) (Figure 1). The procedure was similar to the previous studies [25].
Figure 1.
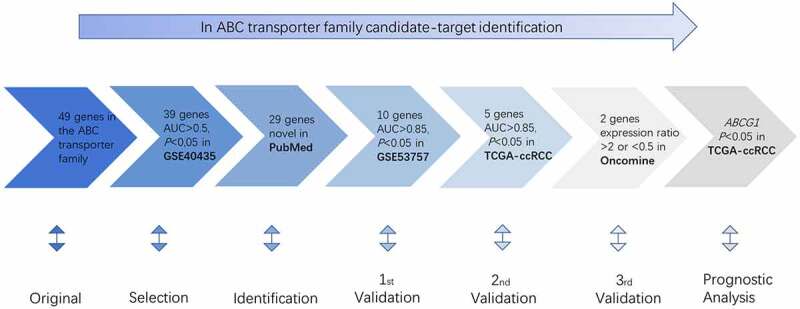
Procedure for the selection of the potential biomarker in ccRCC
Screening of ABC genes in the Gene Expression Omnibus (GEO) database – ccRCC vs normal samples
GSE40435 dataset containing 202 samples (101 ccRCC patients and 101 healthy controls) and the corresponding probe set GPL10558 was obtained from NCBI-GEO database (https://www.ncbi.nlm.nih.gov/geo/query/). T-test and receiver operating characteristic (ROC) analysis were carried out for analyzing the difference in gene expression on 202 samples. p < 0.05 of t-test and the area under curve (AUC)>0.5 of ROC analysis were considered to be significant statistically.
Identification of genes that have not been reported in ccRCC
The genes selected in the screening stage were searched for ccRCC-related research in PubMed (https://www.ncbi.nlm.nih.gov/) on 5 December 2019. Our specific advanced search terms included “Renal clear cell carcinoma” OR “Clear cell renal cell carcinoma” OR “Kidney clear cell carcinoma” OR “KIRC” OR “ccRCC.” The genes that had not been studied before were considered new genes and then selected for the following validation.
Three rounds of validation
The first round of validation was performed by investigating the expression of the identified genes in the GSE53757 dataset from the NCBI-GEO database. The ROC analysis and t-test were carried out on 144 samples (72 ccRCC patients and 72 controls). Genes with p < 0.05 and AUC > 0.85 were selected for the following validation. The second round of validation was performed on TCGA-ccRCC dataset. ROC analysis and t-test were performed on 606 samples (72 healthy samples and 534 ccRCC patients). Genes with p < 0.05 and AUC > 0.85 were considered validated. The third round of validation was performed by analyzing transcriptional expression of ABC genes in 20 different tumors in the Oncomine database. The thresholds were as follows: p value: 0.05; multiple: 2; genetic rank: top 10%; data type: mRNA. Then, we found out the corresponding study on clear cell carcinoma of the kidney. The genes showing an expression ratio >2 or <0.5 were considered effective to be validated.
Prognostic analysis
To evaluate the prognostic value of the clinical characteristics in ccRCC patients, we analyzed the relationship between ABCG1 expression and clinical–pathological parameters including carcinoma in situ, expression, age, gender, survival outcome, overall survival (OS), stage, recurrence, survival after recurrence time (RFS), and smoking history from TCGA-ccRCC dataset. Five hundred and thirty-two patients with clinicopathologic information were equally divided into two groups on the basis of the gene expression. Univariate Cox regression analysis was carried out to find independent variables. Multivariate Cox regression analysis was performed for the parameters with p < 0.2 in univariate Cox regression analysis to assess the prognostic value.
Statistical analysis
ROC analysis and t-test were carried out with GraphPad 8.0 software during screening and validation. In the prognostic analysis step, univariate and multivariate analyses were performed on SPSS19.0. A chi-square test was performed and the OS curve of validated genes was also constructed using GraphPad 8.0 software.
Results
Screening and validation
The expression data of 49 ABC genes were collected from the GSE40435 dataset and samples were divided into two groups (patient group and control group). ROC analysis and t-test were used to evaluate the ability to discriminate ccRCC patients from control samples. The results showed that 39 genes have statistical significance with AUC > 0.5 and p < 0.05 (Table 1). A PubMed search was conducted on 5 December 2019. We found 29 genes that had not been reported to be associated with ccRCC (Table 1).
Table 1.
ROC analysis and t-test of ABC transporter family based on GSE40435 dataset
| NO. | Gene | Expression (ccRCC) | Expression (normal) | ccRCC/normal | AUC | p |
Gene and ccRCC co-occurrence |
||
|---|---|---|---|---|---|---|---|---|---|
| 1 | ABCA1 | 10.860 | 9.804 | 1.108 | ↑ | 0.961 | <0.0001 | ≥1 | |
| 2 | ABCA2a | 7.135 | 7.199 | 0.991 | ↓ | 0.604 | 0.0074 | 0 | |
| 3 | ABCA3 | 8.022 | 7.494 | 1.070 | ↑ | 0.901 | <0.0001 | 0 | |
| 4 | ABCA4 | 6.769 | 7.160 | 0.945 | ↓ | 0.883 | <0.0001 | 0 | |
| 5 | ABCA5 | 6.918 | 7.027 | 0.984 | ↓ | 0.742 | <0.0001 | 0 | |
| 6 | ABCA6 | 7.015 | 7.332 | 0.957 | ↓ | 0.845 | <0.0001 | 0 | |
| 7 | ABCA8 | 7.059 | 7.683 | 0.919 | ↓ | 0.946 | <0.0001 | 0 | |
| 8 | ABCA9 | 6.925 | 7.227 | 0.958 | ↓ | 0.929 | <0.0001 | 0 | |
| 9 | ABCA10 | 6.776 | 6.808 | 0.995 | ↓ | 0.646 | <0.0001 | 0 | |
| 10 | ABCA12 | 6.728 | 6.582 | 1.022 | ↑ | 0.883 | <0.0001 | 0 | |
| 11 | ABCA13 | 6.495 | 6.614 | 0.982 | ↓ | 0.740 | <0.0001 | ≥1 | |
| 12 | ABCB1 | 7.870 | 9.453 | 0.833 | ↓ | 0.982 | <0.0001 | ≥1 | |
| 13 | ABCB2(TAP1) | 11.140 | 8.970 | 1.242 | ↑ | 0.999 | <0.0001 | ≥1 | |
| 14 | ABCB3(TAP2) | 6.818 | 6.662 | 1.023 | ↑ | 0.774 | <0.0001 | ≥1 | |
| 15 | ABCB4 | 7.096 | 6.814 | 1.041 | ↑ | 0.744 | <0.0001 | 0 | |
| 16 | ABCB6 | 8.734 | 8.257 | 1.058 | ↑ | 0.809 | <0.0001 | 0 | |
| 17 | ABCB8 | 6.884 | 6.995 | 0.984 | ↓ | 0.739 | <0.0001 | 0 | |
| 18 | ABCB9 | 7.292 | 7.395 | 0.986 | ↓ | 0.735 | <0.0001 | 0 | |
| 19 | ABCB10 | 8.416 | 8.183 | 1.028 | ↑ | 0.762 | <0.0001 | ≥1 | |
| 20 | ABCC1 | 6.973 | 6.823 | 1.022 | ↑ | 0.881 | <0.0001 | ≥1 | |
| 21 | ABCC2 | 7.694 | 8.189 | 0.940 | ↓ | 0.738 | <0.0001 | ≥1 | |
| 22 | ABCC3 | 8.847 | 8.059 | 1.098 | ↑ | 0.918 | <0.0001 | 0 | |
| 23 | ABCC4 | 8.715 | 8.846 | 0.985 | ↓ | 0.596 | 0.0238 | 0 | |
| 24 | ABCC5 | 7.652 | 7.821 | 0.978 | ↓ | 0.737 | <0.0001 | 0 | |
| 25 | ABCC6 | 7.063 | 7.610 | 0.928 | ↓ | 0.911 | <0.0001 | 0 | |
| 26 | ABCC8 | 6.607 | 6.688 | 0.988 | ↓ | 0.755 | <0.0001 | 0 | |
| 27 | ABCC9 | 7.021 | 6.891 | 1.019 | ↑ | 0.752 | <0.0001 | 0 | |
| 28 | ABCC10 | 7.145 | 7.264 | 0.984 | ↓ | 0.687 | <0.0001 | 0 | |
| 29 | ABCC11 | 6.722 | 6.707 | 1.002 | ↑ | 0.579 | 0.0197 | 0 | |
| 30 | ABCC13 | 6.727 | 6.740 | 0.998 | ↓ | 0.593 | 0.02 | 0 | |
| 31 | ABCD1 | 7.406 | 7.179 | 1.032 | ↑ | 0.756 | <0.0001 | ≥1 | |
| 32 | ABCD3 | 7.983 | 8.618 | 0.926 | ↓ | 0.902 | <0.0001 | 0 | |
| 33 | ABCE1 | 8.864 | 8.598 | 1.031 | ↑ | 0.842 | <0.0001 | 0 | |
| 34 | ABCF1 | 9.295 | 9.626 | 0.966 | ↓ | 0.877 | <0.0001 | 0 | |
| 35 | ABCF2 | 7.334 | 7.283 | 1.007 | ↑ | 0.587 | 0.0026 | 0 | |
| 36 | ABCF3 | 7.363 | 7.502 | 0.981 | ↓ | 0.736 | <0.0001 | 0 | |
| 37 | ABCG1 | 7.591 | 7.139 | 1.063 | ↑ | 0.990 | <0.0001 | 0 | |
| 38 | ABCG2 | 7.234 | 7.088 | 1.021 | ↑ | 0.557 | 0.0036 | ≥1 | |
| 39 | ABCG4 | 7.032 | 7.074 | 0.994 | ↓ | 0.602 | 0.0035 | 0 | |
aNovel genes were marked in bold.
First round of validation
The 29 genes obtained from the above steps were then validated in the GSE53757 dataset. The 10 genes showing AUC > 0.85 and p < 0.05 were allowed to enter the second round of validation, namely ABCA3, ABCA8, ABCA9, ABCA12, ABCC3, ABCC6, ABCC8, ABCD3, ABCF1, and ABCG1 (Table S1). Notably, ABCA12 showed the most significant difference in expression in ccRCC patients vs healthy samples, and the ABCG1 gene with the highest AUC value.
Second round of validation
Ten genes selected from the first-round validation were analyzed on the TCGA-ccRCC dataset for the second round of validation. Five genes with AUC > 0.85 and p < 0.05 were statistically significant, namely, ABAC12, ABCC3, ABCD3, ABCF1, and ABCG1 (Table 2). ROC analysis was carried out to assess the diagnostic value of these five genes. The AUC values of the five genes indicated that they could identify ccRCC patients from normal samples effectively and independently (Figure 2).
Table 2.
T-test and ROC analysis of ABC transporter family members based on the TCGA-ccRCC dataset
| No. | Gene | Expression (ccRCC) | Expression (normal) | ccRCC/normal | AUC | p |
|---|---|---|---|---|---|---|
| 1 | ABCA12 | 6.50 | 3.42 | 1.90 | 0.88 | <0.0001 |
| 2 | ABCC3 | 11.56 | 9.14 | 1.26 | 0.93 | <0.0001 |
| 3 | ABCD3 | 10.43 | 11.49 | 0.91 | 0.95 | <0.0001 |
| 4 | ABCF1 | 10.24 | 10.67 | 0.96 | 0.87 | <0.0001 |
| 5 | ABCG1 | 10.44 | 9.09 | 1.15 | 0.91 | <0.0001 |
Figure 2.
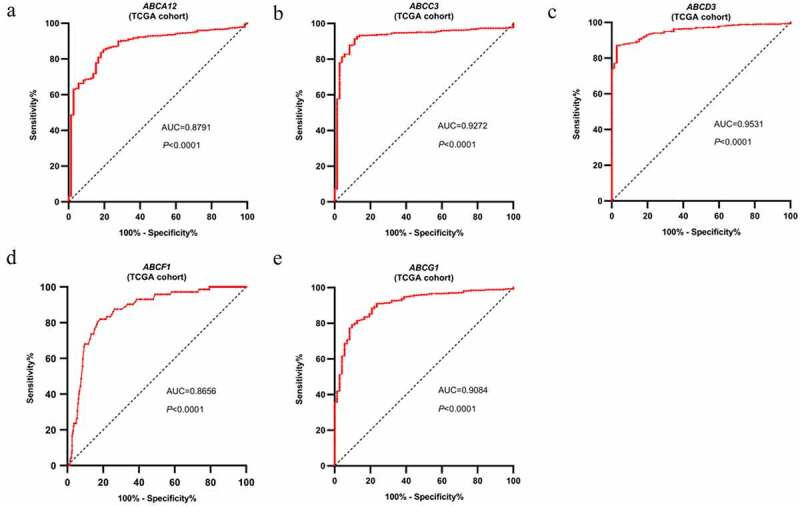
ROC analysis of the expression data for diagnostic assessment of five genes according to TCGA database. AUC statistics are used to evaluate the capacity to discriminate ccRCC samples from normal controls with specificity and sensitivity
Third round of validation
Transcriptional expression of the above five genes was verified in the Oncomine database. The expression of ABCC3, ABCF1, and ABCG1 in 20 types of cancers is shown in Figure S1. ABCA12 and ABCD3 had no available data. There are eight, five, and eight datasets of ABCC3, ABCF1, and ABCG1 for renal cancer, respectively. However, there is no clear cell renal cell carcinoma vs normal in the five datasets of ABCF1.The result showed that ABCG1 was highly overexpressed in all datasets (Table S2, Table S3).
Prognostic analysis
To explore the prognostic value of ABCG1 expression in the TCGA-ccRCC dataset, Cox regression analysis and Kaplan–Meier analysis were performed. The relationship between ABCG1 expression and clinical characteristics in ccRCC is shown in Table 3. We found significant differences in living status between the high-expression group and the low-expression group (p = 0.01), but there was no statistical difference in gender, clinical stage, smoking history, and recurrence history between the two groups. Univariate Cox regression analysis showed that age, clinical stage, and ABCG1 expression were associated with OS (Table 4). Meanwhile, multivariate Cox regression analysis demonstrated that ABCG1 expression might be an independent prognostic factor for ccRCC patients. Kaplan–Meier analysis also showed that ccRCC patients with high ABCG1 expression was significantly associated with better OS than those patients with low ABCG1 expression (p = 0.0067, hazard ratio (HR) = 0.6621) (Figure 3).
Table 3.
Chi-square test of clinicopathologic parameters and ABCG1 mRNA expression in the TCGA-ccRCC cohort
| Parameters | Group |
ABCG1 mRNA expression |
|||
|---|---|---|---|---|---|
| High (n = 266) | Low (n = 266) | χ2 | p | ||
| Age (Mean + SD) | 60.80 ± 12.77 | 60.36 ± 12.03 | |||
| Gender | Female | 82 | 104 | 4.001 | 0.056 |
| Male | 184 | 162 | |||
| Clinical stage | I/II | 164 | 188 | 4.872 | 0.034 |
| III/IV | 101 | 77 | |||
| Discrepancy | 1 | 1 | |||
| Recurrence status | No | 68 | 53 | 0.325 | 0.655 |
| Yes | 15 | 9 | |||
| Null | 183 | 204 | |||
| Smoking history | 1 | 29 | 16 | 2.147 | 0.143 |
| 2/3/4/5 | 20 | 21 | |||
| Null | 229 | 217 | |||
| Living status | Living | 193 | 164 | 7.161 | 0.010 |
| Dead | 73 | 102 | |||
Table 4.
Univariate and multivariate Cox regression analysis of clinical pathologic features according to the TCGA-ccRCC dataset
| Parameters | Univariate analysis |
Multivariate analysis |
|||||
|---|---|---|---|---|---|---|---|
| HR | 95%CI | p | HR | 95%CI | p | ||
| Age | |||||||
| ≥60 vs <60 | 1.808 | 1.321–2.478 | 0.000 | 1.563 | 1.140–2.144 | 0.006 | |
| Gender | |||||||
| Female vs male | 1.060 | 0.779–1.442 | 0.710 | - | - | - | |
| Clinical stage | |||||||
| III/IV vs I/II | 3.838 | 2.799–5.292 | 0.000 | 3.675 | 2.676–5.046 | 0.000 | |
| Smoking history | |||||||
| 2/3/4/5 vs 1 | 0.778 | 0.254–2.383 | 0.661 | - | - | - | |
| ABCG1 expression | |||||||
| High vs low | 0.665 | 0.492–0.898 | 0.008 | 0.662 | 0.490–0.895 | 0.007 | |
Figure 3.
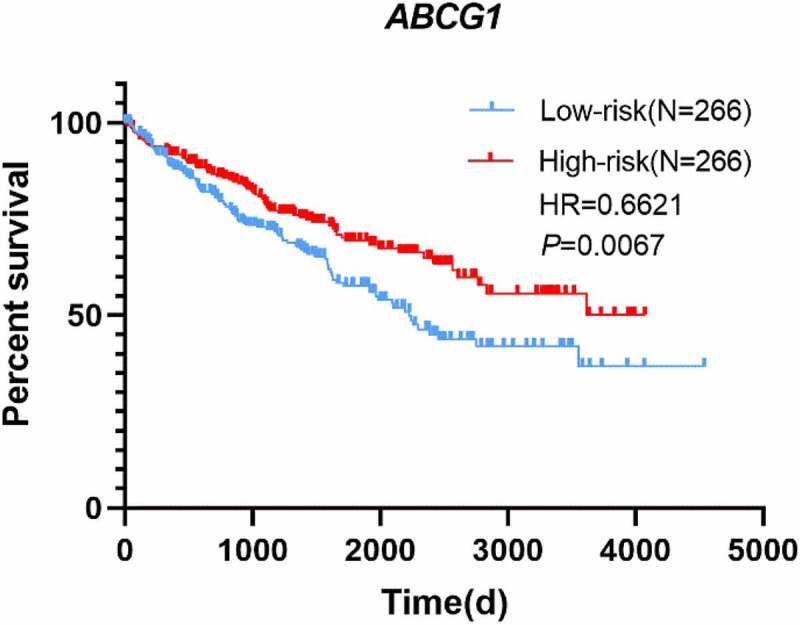
Kaplan–Meier survival curve of ABCG1 mRNA expression in ccRCC patients. Survival curves suggested that patients with decreased ABCG1 mRNA level significantly correlated with poorer OS (p = 0.0067)
Discussion
Many studies have demonstrated that ABC family genes play important roles in the maintaining cellular environment [26,27], cholesterol metabolism [9,10,28–31], disease occurrence [32–34], and tumor resistance [35,36]. ABC transporter genes could promote drug efflux and enhance chemical resistance of cancer cells [37]. Mutations in the ABC genes could affect the phenotypes of cancer cells such as proliferation, differentiation, migration, and invasion [38]. ABCG1 is involved in lipid balance and cholesterol efflux from macrophages [10]. ABCG1 is also able to transport sterols, which can regulate the expression of macrophage inflammatory cytokines, chemokines, and lymphocyte proliferation response [28]. ABCG1 was found to be a potential biomarker for lung cancer [39,40], head and neck squamous cell carcinoma [41], and prostate cancer [42]. However, it has not been studied in ccRCC. After screening, identification, and three rounds of verification, ABCG1 was selected from 49 ABC transporter genes. We first showed that ABCG1 has the diagnostic and prognostic value for ccRCC patients.
Metabolic change is the main feature of tumors [43], and ccRCC is also considered as metabolic disease [44], which is characterized by the accumulation of cholesterol, cholesterol esters, other neutral lipids, and glycogen [45]. The total cholesterol content in the ccRCC tissues is eight times higher than that of the normal kidney, and the esterified cholesterol content is 35 times higher than that of the normal kidney [46]. The abnormalities in cholesterol metabolism in ccRCC cells may affect the physiological and biochemical functions of cells and produce pathological changes. Many studies have shown that serum cholesterol levels are associated with ccRCC invasion and prognosis [47–49]. Patients with low preoperative cholesterol levels have lower OS than patients with high cholesterol levels [47], and cholesterol metabolism may be involved in ccRCC metastasis [50]. The function of the ABCG1 gene is mainly related to cell cholesterol outflow [10], which indicates that ABCG1 may play an important role in the tumorigenesis and progress of ccRCC.
Our study showed that ABCG1 was over-expressed among patients with ccRCC compared with normal people (Table 1–2, Table S1, Figure 3). It may be hypothesized that when normal cells mutate into cancer cells, the energy demand increases, which activates a certain cholesterol transport mechanism and begins to take cholesterol from the outside. The decrease in serum cholesterol levels is correlated with the uptake of low-density lipoprotein in serum by tumor cells [49]. Yang et al. found that the accumulation of cholesterol is one of the characteristics of ccRCC [50]. As serum cholesterol decreased, ABCG1 began to be highly expressed and promoted the efflux of cellular cholesterol to maintain serum cholesterol level.
Moreover, we found that ccRCC patients with high expression level had longer OS time than those with low expression level. It may be that the high expression of ABCG1 can inhibit the growth of cancer cells and affect the survival of cancer cells by reducing the cholesterol content in cancer cells. Wu et al. found that liver X receptor 623 (LXR623) downregulates low-density lipoprotein receptor (LDLR) expression while upregulating ABCA1, leading to a decrease in intracellular cholesterol content and the occurrence of apoptosis [14]. They speculated that LXR623 could kill tumor cells by promoting cholesterol outflow [14]. ABCG1 and ABCA1 have many similarities: both of them are regulated by LXR623 [51–54] and they have 101 common interacting chemicals (Figure S2), promoting the outflow of cholesterol from macrophages [51–54] and regulating the expression of macrophage inflammatory cytokines [28], etc. Hence, it may be possible to kill or suppress tumor cells by upregulating ABCG1 with LXR623.
In addition, ABCG1 could affect tumor growth by regulating macrophages. Macrophages participate in the formation of tumor microenvironment [55,56], tumor growth, and metastasis [57–60], apoptosis [61], and play an important role in tumor immunity [62]. There are two types of it: M1 cells can produce a large number of inflammatory cytokines, which can activate the immune response and play an anti-tumor role; M2 cells promote angiogenesis, remodeling, and tumor growth [63]. In most tumor models, most of the macrophages in the tumor are shown as tumor promoting M2 phenotype [62]. Researches have shown that the deficiency of ABCG1 increases the signaling of Toll-like receptors in macrophages, leading to an enhanced inflammatory response of macrophages to LPS or other TLR ligands [64–67], and also reduces the number and proportion of M2 phenotype [29,68]. This means that the upregulation of ABCG1 leads to an increase in M2 macrophages, which is conducive to tumor growth. However, this is inconsistent with the result that the high ABCG1 expression group can have a longer survival time, so more work is needed to explain the problem.
Furthermore, we identified 35 potential active drugs from 165 interplay substances with ABCG1 in the Comparative Toxicogenomics Database (CTD) (http://ctdbase.org), of which 18 drugs can upregulate ABCG1 expression at mRNA level, while 17 drugs can downregulate ABCG1 expression (Table S4). In addition, six ABCG1 target genes were identified from 60 ABCG1 interacting genes: ACSL4 [69], AP1S2 [69], ELAVL1 [70], HNRNPL [71], SGO1 [69], and UBC [72] (Table S5).
There are still several limitations in this study. In the screening and the first rounds of validation analysis, ABCA12 and ABCD3 were significantly different between ccRCC and normal tissues. Unfortunately, there was no data available for ABCA12 and ABCD3 in the Oncomine database. But it could not exclude that ABCA12 may be associated with the diagnosis and prognosis in ccRCC. Protein expression of ABC genes was analyzed before prognostic analysis in the HPA database (https://www.proteinatlas.org/). Grayscale conversion analysis was carried out on a total of 57 histological sections (11 normal tissue sections and 46 pathological sections of ccRCC patients) using ImageJ (Table S6). The t-test was then performed and a scatter plot was constructed according to the area percentage of histological sections (Figure S3). As shown in Figure S3, ABCC3 was not statistically significant in t-test, and in the protein expression level of ABCG1 was reduced in ccRCC patients, while positively expressed in normal tissue sections. It is not consistent with the results of mRNA level, which may be related to the type and specificity of antibody and the small sample size. Additional works and experiments need to be performed to validate them.
Conclusion
Excluding the ABC family members that have been studied, through multiple rounds of validation, a novel diagnostic and prognostic biomarker of ccRCC – ABCG1 – was found. According to the high expression of ABCG1 in ccRCC and its correlation with better prognosis, it may be helpful for the diagnosis and provides new ideas for the development of molecular-targeted drugs for ccRCC.
Supplementary Material
Acknowledgments
This study was supported by Program for Innovative Talents of Science and Technology in Henan Province (No. 202102310205), China Postdoctoral Science Foundation (No. 2017M62237), and Henan Postdoctoral Foundation (No. 001702052).
Funding Statement
This work was supported by the Program for Innovative Talents of Science and Technology in Henan Province [202102310205]; Henan Postdoctoral Foundation [001702052]; China Postdoctoral Science Foundation [2017M62237].
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018. November;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- [2].Gupta K, Miller JD, Li JZ, et al. Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): a literature review. Cancer Treat Rev. 2008. May;34(3):193–205. [DOI] [PubMed] [Google Scholar]
- [3].Escudier B, Porta C, Schmidinger M, et al. Renal cell carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2019. May 1;30(5):706–720. [DOI] [PubMed] [Google Scholar]
- [4].Atkins MB, Tannir NM.. Current and emerging therapies for first-line treatment of metastatic clear cell renal cell carcinoma. Cancer Treat Rev. 2018. November;70:127–137. [DOI] [PubMed] [Google Scholar]
- [5].Motzer RJ, Bander NH, Nanus DM. Renal-cell carcinoma. N Engl J Med. 1996. September 19;335(12):865–875. [DOI] [PubMed] [Google Scholar]
- [6].Lin W, Chen X, Chen T, et al. C1QTNF6 as a novel diagnostic and prognostic biomarker for clear cell renal cell carcinoma. DNA Cell Biol. 2020. April 13;39(6):1000–1011. [DOI] [PubMed] [Google Scholar]
- [7].Zhao Y, Yan Y, Ma R, et al. Expression signature of six-snoRNA serves as novel non-invasive biomarker for diagnosis and prognosis prediction of renal clear cell carcinoma. J Cell Mol Med. 2020. February;24(3):2215–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Xu D, Xu Y, Lv Y, et al. Identification of four pathological stage-relevant genes in association with progression and prognosis in clear cell renal cell carcinoma by integrated bioinformatics analysis. Biomed Res Int. 2020;2020:2137319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dean M, Hamon Y, Chimini G. The human ATP-binding cassette (ABC) transporter superfamily. J Lipid Res. 2001. July;42(7):1007–1017. [PubMed] [Google Scholar]
- [10].Vasiliou V, Vasiliou K, Nebert DW. Human ATP-binding cassette (ABC) transporter family. Hum Genomics. 2009. April 3;3(3):281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lam F, Hussain S, Sinha J. An unusual cause of a limp in a child: developmental coxa vara. Emerg Med J. 2001. July;18(4):314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rudling M, Collins VP. Low density lipoprotein receptor and 3-hydroxy-3-methylglutaryl coenzyme A reductase mRNA levels are coordinately reduced in human renal cell carcinoma. Biochim Biophys Acta. 1996. January 5;1299(1):75–79. [DOI] [PubMed] [Google Scholar]
- [13].Dessi S, Batetta B, Pulisci D, et al. Cholesterol content in tumor tissues is inversely associated with high-density lipoprotein cholesterol in serum in patients with gastrointestinal cancer. Cancer. 1994. January 15;73(2):253–258. [DOI] [PubMed] [Google Scholar]
- [14].Wu G, Wang Q, Xu Y, et al. Targeting the transcription factor receptor LXR to treat clear cell renal cell carcinoma: agonist or inverse agonist? Cell Death Dis. 2019. May 28;10(6):416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hour TC, Kuo YZ, Liu GY, et al. Downregulation of ABCD1 in human renal cell carcinoma. Int J Biol Markers. 2009. Jul-Sep;24(3):171–178. [DOI] [PubMed] [Google Scholar]
- [16].Yun EJ, Zhou J, Lin CJ, et al. The network of DAB2IP-miR-138 in regulating drug resistance of renal cell carcinoma associated with stem-like phenotypes. Oncotarget. 2017. September 15;8(40):66975–66986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Arai E, Sakamoto H, Ichikawa H, et al. Multilayer-omics analysis of renal cell carcinoma, including the whole exome, methylome and transcriptome. Int J Cancer. 2014. September 15;135(6):1330–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Reustle A, Fisel P, Renner O, et al. Characterization of the breast cancer resistance protein (BCRP/ABCG2) in clear cell renal cell carcinoma. Int J Cancer. 2018. December 15;143(12):3181–3193. [DOI] [PubMed] [Google Scholar]
- [19].Li S, Yang J, Wang J, et al. Down-regulation of miR-210-3p encourages chemotherapy resistance of renal cell carcinoma via modulating ABCC1. Cell Biosci. 2018;8(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Haenisch S, Zimmermann U, Dazert E, et al. Influence of polymorphisms of ABCB1 and ABCC2 on mRNA and protein expression in normal and cancerous kidney cortex. Pharmacogenomics J. 2007. February 7;7(1):56–65. [DOI] [PubMed] [Google Scholar]
- [21].Saleeb RM, Farag M, Lichner Z, et al. Modulating ATP binding cassette transporters in papillary renal cell carcinoma type 2 enhances its response to targeted molecular therapy. Mol Oncol. 2018. October;12(10):1673–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Seliger B, Atkins D, Bock M, et al. Characterization of human lymphocyte antigen class I antigen-processing machinery defects in renal cell carcinoma lesions with special emphasis on transporter-associated with antigen-processing down-regulation. Clin Cancer Res. 2003. May;9(5):1721–1727. [PubMed] [Google Scholar]
- [23].Kuo CY, Yoo TJ. In vitro inhibition of tritiated thymidine uptake in Morris hepatoma cells by normal rat liver extract: a possible liver chalone. J Natl Cancer Inst. 1977. December;59(6):1691–1695. [DOI] [PubMed] [Google Scholar]
- [24].Huang Y, Zhang Y, Jia L, et al. Circular RNA ABCB10 promotes tumor progression and correlates with pejorative prognosis in clear cell renal cell carcinoma. Int J Biol Markers. 2019. June;34(2):176–183. [DOI] [PubMed] [Google Scholar]
- [25].D’Arcangelo D, Giampietri C, Muscio M, et al. WIPI1, BAG1, and PEX3 autophagy-related genes are relevant melanoma markers. Oxid Med Cell Longev. 2018;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chai AB, Ammit AJ, Gelissen IC. Examining the role of ABC lipid transporters in pulmonary lipid homeostasis and inflammation. Respir Res. 2017. February 28;18(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhu L, Tang F, Lei Z, et al. Antiapoptotic properties of MALT1 protease are associated with redox homeostasis in ABC-DLBCL cells. Mol Carcinog. 2019. September 26;58(12):2340–2352. . [DOI] [PubMed] [Google Scholar]
- [28].Yvan-Charvet L, Wang N, Tall AR. Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler Thromb Vasc Biol. 2010. February;30(2):139–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sag D, Cekic C, Wu R, et al. The cholesterol transporter ABCG1 links cholesterol homeostasis and tumour immunity. Nat Commun. 2015. February 27;6(1):6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kober AC, Manavalan APC, Tam-Amersdorfer C, et al. Implications of cerebrovascular ATP-binding cassette transporter G1 (ABCG1) and apolipoprotein M in cholesterol transport at the blood-brain barrier. Biochim Biophys Acta Mol Cell Biol Lipids. 2017. June;1862(6):573–588. [DOI] [PubMed] [Google Scholar]
- [31].Vaughan AM, Oram JF. ABCA1 and ABCG1 or ABCG4 act sequentially to remove cellular cholesterol and generate cholesterol-rich HDL. J Lipid Res. 2006. November;47(11):2433–2443. [DOI] [PubMed] [Google Scholar]
- [32].Pereira CD, Martins F, Wiltfang J, et al. ABC transporters are key players in Alzheimer’s disease. J Alzheimers Dis. 2018;61(2):463–485. [DOI] [PubMed] [Google Scholar]
- [33].Schumacher T, Benndorf RA, Transport ABC. Proteins in cardiovascular disease—a brief summary. Molecules. 2017. April 6;22(4):589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tarling EJ, De Aguiar Vallim TQ, Edwards PA. Role of ABC transporters in lipid transport and human disease. Trends Endocrinol Metab. 2013. July;24(7):342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Harandi NM, Stavness I, Woo J. Subject-specific biomechanical modelling of the oropharynx: towards speech production. Comput Methods Biomech Biomed Eng Imaging Vis. 2017;5(6):416–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liu X. ABC family transporters. Adv Exp Med Biol. 2019;1141:13–100. [DOI] [PubMed] [Google Scholar]
- [37].Briz O, Perez-Silva L, Al-Abdulla R, et al. What “The Cancer Genome Atlas” database tells us about the role of ATP-binding cassette (ABC) proteins in chemoresistance to anticancer drugs. Expert Opin Drug Metab Toxicol. 2019. July;15(7):577–593. [DOI] [PubMed] [Google Scholar]
- [38].Fletcher JI, Williams RT, Henderson MJ, et al. ABC transporters as mediators of drug resistance and contributors to cancer cell biology. Drug Resist Updat. 2016. May;26:1–9. [DOI] [PubMed] [Google Scholar]
- [39].Tian C, Huang D, Yu Y, et al. ABCG1 as a potential oncogene in lung cancer. Exp Ther Med. 2017. June;13(6):3189–3194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [40].Wang Y, Liu H, Ready NE, et al. Genetic variants in ABCG1 are associated with survival of nonsmall-cell lung cancer patients. Int J Cancer. 2016. June 1;138(11):2592–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gonzalez HE, Gujrati M, Frederick M, et al. Identification of 9 genes differentially expressed in head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2003. July;129(7):754–759. [DOI] [PubMed] [Google Scholar]
- [42].Demidenko R, Razanauskas D, Daniunaite K, et al. Frequent down-regulation of ABC transporter genes in prostate cancer. BMC Cancer. 2015. October 12;15(1):683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jiang L, Zhao L, Bi J, et al. Glycolysis gene expression profilings screen for prognostic risk signature of hepatocellular carcinoma. Aging (Albany NY). 2019. December 2;11(23):10861–10882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Massari F, Ciccarese C, Santoni M, et al. Metabolic alterations in renal cell carcinoma. Cancer Treat Rev. 2015. November;41(9):767–776. [DOI] [PubMed] [Google Scholar]
- [45].Drabkin HA, Gemmill RM. Cholesterol and the development of clear-cell renal carcinoma. Curr Opin Pharmacol. 2012. December;12(6):742–750. [DOI] [PubMed] [Google Scholar]
- [46].Gebhard RL, Clayman RV, Prigge WF, et al. Abnormal cholesterol metabolism in renal clear cell carcinoma. J Lipid Res. 1987. October;28(10):1177–1184. [PubMed] [Google Scholar]
- [47].Kang HW, Seo SP, Kim WT, et al. Low preoperative serum cholesterol level is associated with aggressive pathologic features and poor cancer-specific survival in patients with surgically treated renal cell carcinoma. Int J Clin Oncol. 2018. February;23(1):142–150. [DOI] [PubMed] [Google Scholar]
- [48].Lee H, Kim YJ, Hwang EC, et al. Preoperative cholesterol level as a new independent predictive factor of survival in patients with metastatic renal cell carcinoma treated with cyto-reductive nephrectomy. BMC Cancer. 2017. May 25;17(1):364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Peng D, He ZS, Li XS, et al. A novelpredictor of survival with renal cell carcinoma after nephrectomy. J Endourol. 2017. April;31(4):397–404. [DOI] [PubMed] [Google Scholar]
- [50].Yang H, Li W, Lv Y, et al. Exploring the mechanism of clear cell renal cell carcinoma metastasis and key genes based on multi-tool joint analysis. Gene. 2019. December 15;720:144103. [DOI] [PubMed] [Google Scholar]
- [51].Frambach S, De Haas R, Smeitink JAM, et al. Brothers in arms: ABCA1- and ABCG1-mediated cholesterol efflux as promising targets in cardiovascular disease treatment. Pharmacol Rev. 2020. January;72(1):152–190. [DOI] [PubMed] [Google Scholar]
- [52].Ren K, Li H, Zhou HF, et al. Mangiferin promotes macrophage cholesterol efflux and protects against atherosclerosis by augmenting the expression of ABCA1 and ABCG1. Aging (Albany NY). 2019. December 2;11(23):10992–11009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Huesca-Gomez C, Torres-Paz YE, Martinez-Alvarado R, et al. Association between the transporters ABCA1/G1 and the expression of miR-33a/144 and the carotid intima media thickness in patients with arterial hypertension. Mol Biol Rep. 2020. February;47(2):1321–1329. [DOI] [PubMed] [Google Scholar]
- [54].Koga M, Kanaoka Y, Okamoto M, et al. Varenicline aggravates atherosclerotic plaque formation in nicotine-pretreated ApoE knockout mice due to enhanced oxLDL uptake by macrophages through downregulation of ABCA1 and ABCG1 expression. J Pharmacol Sci. 2020. January;142(1):9–15. [DOI] [PubMed] [Google Scholar]
- [55].Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006. January 15;66(2):605–612. [DOI] [PubMed] [Google Scholar]
- [56].Movahedi K, Laoui D, Gysemans C, et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 2010. July 15;70(14):5728–5739. [DOI] [PubMed] [Google Scholar]
- [57].Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004. January 4;4(1):71–78. [DOI] [PubMed] [Google Scholar]
- [58].Lin EY, Li JF, Gnatovskiy L, et al. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res. 2006. December 1;66(23):11238–11246. [DOI] [PubMed] [Google Scholar]
- [59].Sica A, Larghi P, Mancino A, et al. Macrophage polarization in tumour progression. Semin Cancer Biol. 2008. October;18(5):349–355. [DOI] [PubMed] [Google Scholar]
- [60].Hiraoka K, Zenmyo M, Watari K, et al. Inhibition of bone and muscle metastases of lung cancer cells by a decrease in the number of monocytes/macrophages. Cancer Sci. 2008. August;99(8):1595–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Li Y, Schwabe RF, DeVries-Seimon T, et al. Free cholesterol-loaded macrophages are an abundant source of tumor necrosis factor-alpha and interleukin-6: model of NF-kappaB- and map kinase-dependent inflammation in advanced atherosclerosis. J Biol Chem. 2005. June 10;280(23):21763–21772. [DOI] [PubMed] [Google Scholar]
- [62].Solinas G, Germano G, Mantovani A, et al. Tumor-associated macrophages (TAM) as major players of the cancer-related inflammation. J Leukoc Biol. 2009. November;86(5):1065–1073. [DOI] [PubMed] [Google Scholar]
- [63].Wei H, Tarling EJ, McMillen TS, et al. ABCG1 regulates mouse adipose tissue macrophage cholesterol levels and ratio of M1 to M2 cells in obesity and caloric restriction. J Lipid Res. 2015. December;56(12):2337–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Buczek M, Escudier B, Bartnik E, et al. Resistance to tyrosine kinase inhibitors in clear cell renal cell carcinoma: from the patient’s bed to molecular mechanisms. Biochim Biophys Acta. 2014. January;1845(1):31–41. [DOI] [PubMed] [Google Scholar]
- [65].Van Der Mijn JC, Mier JW, Broxterman HJ, et al. Predictive biomarkers in renal cell cancer: insights in drug resistance mechanisms. Drug Resist Updat. 2014. Oct-Dec;17(4–6):77–88. [DOI] [PubMed] [Google Scholar]
- [66].Randall JM, Millard F, Kurzrock R. Molecular aberrations, targeted therapy, and renal cell carcinoma: current state-of-the-art. Cancer Metastasis Rev. 2014. December;33(4):1109–1124. [DOI] [PubMed] [Google Scholar]
- [67].Garcia-Donas J, Rodriguez-Moreno JF, Romero-Laorden N, et al. Renal carcinoma pharmacogenomics and predictors of response: steps toward treatment individualization. Urol Oncol. 2015. April;33(4):179–186. [DOI] [PubMed] [Google Scholar]
- [68].Yvan-Charvet L, Welch C, Pagler TA, et al. Increased inflammatory gene expression in ABC transporter-deficient macrophages: free cholesterol accumulation, increased signaling via toll-like receptors, and neutrophil infiltration of atherosclerotic lesions. Circulation. 2008. October 28;118(18):1837–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hein MY, Hubner NC, Poser I, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015. October 22;163(3):712–723. [DOI] [PubMed] [Google Scholar]
- [70].Abdelmohsen K, Srikantan S, Yang X, et al. Ubiquitin-mediated proteolysis of HuR by heat shock. Embo J. 2009. May 6;28(9):1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Fei T, Chen Y, Xiao T, et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci U S A. 2017. June 27;114(26):E5207–E5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Na CH, Jones DR, Yang Y, et al. Synaptic protein ubiquitination in rat brain revealed by antibody-based ubiquitome analysis. J Proteome Res. 2012. September 7;11(9):4722–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


