Abstract
Horizontal gene transfer is a major force in bacterial evolution. Mobile genetic elements are responsible for much of horizontal gene transfer and also carry beneficial cargo genes. Uncovering strategies used by mobile genetic elements to benefit host cells is crucial for understanding their stability and spread in populations. We describe a benefit that ICEBs1, an integrative and conjugative element of Bacillus subtilis, provides to its host cells. Activation of ICEBs1 conferred a frequency-dependent selective advantage to host cells during two different developmental processes: biofilm formation and sporulation. These benefits were due to inhibition of biofilm-associated gene expression and delayed sporulation by ICEBs1-containing cells, enabling them to exploit their neighbors and grow more prior to development. A single ICEBs1 gene, devI (formerly ydcO), was both necessary and sufficient for inhibition of development. Manipulation of host developmental programs allows ICEBs1 to increase host fitness, thereby increasing propagation of the element.
Research organism: B. subtilis
eLife digest
Many bacteria can ‘have sex’ – that is, they can share their genetic information and trade off segments of DNA. While these mobile genetic elements can be parasites that use the resources of their host to make more of themselves, some carry useful genes which, for example, help bacteria to fight off antibiotics.
Integrative and conjugative elements (or ICEs) are a type of mobile segments that normally stay inside the genetic information of their bacterial host but can sometimes replicate and be pumped out to another cell. ICEBs1 for instance, is an element found in the common soil bacterium Bacillus subtilis. Scientists know that ICEBs1 can rapidly spread in biofilms – the slimly, crowded communities where bacteria live tightly connected – but it is still unclear whether it helps or hinders its hosts.
Using genetic manipulations and tracking the survival of different groups of cells, Jones et al. show that carrying ICEBs1 confers an advantage under many conditions. When B. subtilis forms biofilms, the presence of the devI gene in ICEBs1 helps the cells to delay the production of the costly mucus that keeps bacteria together, allowing the organisms to ‘cheat’ for a little while and benefit from the tight-knit community without contributing to it. As nutrients become scarce in biofilms, the gene also allows the bacteria to grow for longer before they start to form spores – the dormant bacterial form that can weather difficult conditions.
Mobile elements can carry genes that make bacteria resistant to antibiotics, harmful to humans, or able to use new food sources; they could even be used to artificially introduce genes of interest in these cells. The work by Jones et al. helps to understand the way these elements influence the fate of their host, providing insight into how they could be harnessed for the benefit of human health.
Introduction
Conjugative elements and phages are abundant mobile genetic elements in bacteria, capable of transferring DNA between cells (Frost et al., 2005). Integrative and conjugative elements (ICEs) appear to be the most widespread type of conjugative element (Guglielmini et al., 2011). ICEs are found integrated in a host genome. When activated, they excise and produce conjugation machinery that transfers the element DNA from the host cell to recipients (Carraro and Burrus, 2015; Johnson and Grossman, 2015; Wozniak and Waldor, 2010).
ICEs often carry ‘cargo’ genes that are not necessary for transfer but confer a phenotype to host cells. In fact, ICEs (conjugative transposons) were first identified because of the phenotypes conferred by cargo genes (Franke and Clewell, 1981). Cargo genes include those encoding antibiotic resistances, metabolic pathways, and determinants of pathogenesis and symbiosis (Johnson and Grossman, 2015). Transfer of mobile elements between cells contributes to rapid evolution and spread of associated cargo genes and phenotypes (Frost et al., 2005; Treangen and Rocha, 2011).
Despite the benefits cargo genes can provide, the maintenance and transfer of mobile genetic elements requires host cellular resources and in some cases is lethal (Baltrus, 2013). Maintenance of a mobile genetic element in host cells requires balancing the costs and benefits to the host or a sufficiently high transfer frequency. Many mobile elements, especially ICEs, have been identified bioinformatically (Bi et al., 2012; Guglielmini et al., 2011). Many of these ICEs contain putative cargo genes. However, the phenotypes conferred by these genes cannot be inferred from sequence nor are they easily detected experimentally (Cury et al., 2017).
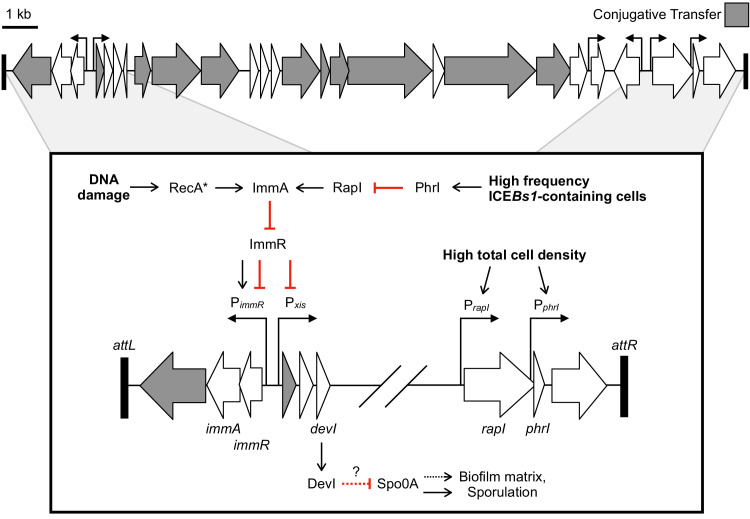
ICEBs1, a relatively small (~20 kb) ICE found in most strains of Bacillus subtilis, was identified bioinformatically (Burrus et al., 2002) and experimentally based on its regulation by cell-cell signaling (Auchtung et al., 2005). Most of the ICEBs1 genes needed for conjugation are grouped together in an operon that is repressed until activating signals are sensed (Figure 1). Two pathways activate ICEBs1, both of which lead to cleavage of the repressor ImmR by the protease and anti-repressor ImmA (Auchtung et al., 2007; Bose et al., 2008; Bose and Grossman, 2011). ICEBs1 contains the cell-cell signaling genes, rapI and phrI, which regulate ICEBs1 activation by sensing population density and the relative abundance of ICEBs1-containing host cells (Auchtung et al., 2005). RapI is produced at high cell density and during the transition to stationary phase and stimulates the proteolytic cleavage of the repressor ImmR by the protease ImmA (Bose and Grossman, 2011). Overproduction of RapI stimulates activation of ICEBs1 in >90% of cells (Auchtung et al., 2005). RapI activity (and therefore ICEBs1 activation) is inhibited by PhrI, a peptide that is secreted by cells that contain ICEBs1. PhrI levels indicate the relative abundance of ICEBs1-containing cells in the population, preventing the activation and possible redundant transfer of ICEBs1 if most nearby cells already contain the element. ICEBs1 is also activated during the RecA-dependent DNA damage response (Auchtung et al., 2005).
Figure 1. Genetic map and regulatory pathways of ICEBs1.
Genes are represented by horizontal block arrows indicating the direction of transcription. Vertical right-angle arrows mark the positions of promoters, and the arrowhead indicates the direction of transcription. Genes known to be involved in the conjugative life cycle of ICEBs1 are shaded in gray. The 60 bp direct repeats that mark the ends of ICEBs1 are shown as black rectangles. (Inset) A partial genetic map that highlights factors involved in the regulation of ICEBs1. The major promoter Pxis drives expression of most genes in ICEBs1. Pxis is repressed by the ICE-encoded repressor ImmR. Repression is relieved when ImmR is cleaved by the protease ImmA, and proteolytic cleavage is stimulated by activated RecA (RecA*) in response to DNA damage, or, independently by the cell signaling regulator RapI. RapI is made when cells are crowded by potential recipients, but repressed by the ICE-encoded secreted peptide PhrI if the neighboring cells already contain a copy of ICEBs1. devI (formerly ydcO) is the third open-reading frame downstream of Pxis. DevI inhibits sporulation and expression of biofilm matrix genes, likely by inhibiting Spo0A (directly or indirectly). In the genetic pathways, black arrows indicate activation and red T-bars indicate inhibition.
Biofilms appear to be hotspots of horizontal gene transfer for bacteria growing in natural settings (Madsen et al., 2012; Molin and Tolker-Nielsen, 2003). Undomesticated strains of B. subtilis form complex biofilms on agar plates and at the air-liquid interface in standing cultures (Vlamakis et al., 2013). There is also extensive spore formation in B. subtilis biofilms (Branda et al., 2001; Vlamakis et al., 2008). In addition, during growth in a biofilm, ICEBs1 is naturally activated and transfers efficiently, generating on the order of 10 new ICEBs1-containing host cells (transconjugants) per donor cell under appropriate conditions (Lécuyer et al., 2018). B. subtilis biofilms are held together by a matrix composed of secreted exopolysaccharides, protein fibers, and DNA (Vlamakis et al., 2013). This matrix reinforces cell-cell contacts, likely promoting rapid spread of ICEBs1 by conjugation. Additionally, the conditions that promote biofilm formation (high cell density) also promote activation and transfer of ICEBs1 and sporulation (Auchtung et al., 2005; Grossman and Losick, 1988). Although biofilm growth is clearly beneficial to conjugation, it is unknown how ICEBs1 impacts its host cells under these conditions.
In this study, we describe a selective advantage provided by ICEBs1 to its host cells during growth in biofilms. This fitness benefit was due to inhibition of host biofilm and spore development. We identified the ICEBs1 gene devI (formerly ydcO) as necessary and sufficient to inhibit host development and provide a selective advantage to ICEBs1-containing cells. We also provide evidence indicating that devI likely inhibits the key developmental transcription factor Spo0A, reducing its ability to stimulate biofilm and sporulation gene expression. devI (ydcO) is conserved in other ICEBs1-like elements, indicating that manipulation of host development may be a conserved strategy among this family of mobile genetic elements. We postulate that manipulation of host pathways may be a common function of many of the as yet uncharacterized cargo genes in ICEs.
Results
ICEBs1 spreads efficiently in biofilms by conjugation
Biofilm formation is characteristic of many bacteria growing in natural settings, including B. subtilis. We used biofilm growth to determine if ICEBs1 affected the fitness of its host cells under conditions that naturally promote its spread. We performed competition experiments in biofilms using strains of undomesticated B. subtilis (NCIB3610 plasmid-free) with or without ICEBs1.
We observed highly efficient spread of ICEBs1 at low donor to recipient ratios (Table 1) during growth in biofilms, similar to results reported previously (Lécuyer et al., 2018). To measure mating, we mixed ICEBs1-containing cells (potential donors) with cells that did not contain ICEBs1 (ICEBs10, potential recipients) and co-cultured the mix on standard biofilm-stimulating growth medium (MSgg agar) (Branda et al., 2001). Since ICEBs1 induction is regulated by cell-cell signaling, we varied the initial frequency of ICEBs1+ cells between approximately 0.01 and 0.9. We inserted unique selectable markers (antibiotic resistances) in the chromosomes of the donors and recipients as well as within ICEBs1. After 4 days of growth at 30°C (approximately 17 net doublings of the initial population), biofilms were disrupted and the number of transconjugants was determined by selective plating.
Table 1. Frequency of transconjugants generated in biofilm matings.
| Initial frequency ICEBs1 donors* | Final frequency transconjugants† |
|---|---|
| 0.008 ± 0.002 | 0.42 ± 0.13 |
| 0.10 ± 0.03 | 0.64 ± 0.15 |
| 0.47 ± 0.05 | 0.44 ± 0.07 |
| 0.89 ± 0.03 | 0.063 ± 0.008 |
*ICEBs1-containing cells (JMJ592) were mixed with ICEBs1-cured cells (JMJ550). The initial frequencies reported are the average ± standard deviation from three independent experiments.
†The final frequencies of transconjugants reported are the average ± standard deviation from a total of nine biofilms from three independent experiments.
We found that after four days of biofilm growth, the frequency of transconjugants ranged from ~0.4 to 0.6 of total cells in the biofilm for starting donor frequencies of ~0.01–0.5 (Table 1). The highest frequency of transconjugants was observed when the starting frequency of ICEBs1-containing cells was ~0.1. Enhanced conjugation at low donor to recipient ratios is likely due to regulation of ICEBs1 by cell-cell signaling (induction is inhibited by the presence of other potential donors) and the higher likelihood of contacting potential recipients at low frequencies of donors.
The high levels of ICEBs1 conjugation during growth in biofilms presented a challenge for quantifying the fitness of ICEBs1-containing host cells relative to ICEBs10 cells. Mating converts a large fraction of ICEBs10 cells to transconjugants (ICEBs1-containing), reducing the ICEBs10 proportion of the population in a manner unrelated to host fitness. To measure the effect of ICEBs1 on host fitness, we blocked conjugative DNA transfer using the conEK476E mutation (Berkmen et al., 2010). We then compared the proportion of ICEBs1-containing hosts to ICEBs10 cells without the confounding influence of conjugation.
ICEBs1 provides a frequency-dependent selective advantage in biofilms
We found that cells containing ICEBs1 that is incapable of conjugation {ICEBs1(conEK476E)} had a fitness advantage over cells lacking ICEBs1 during biofilm growth when they were initially present as a minority of the population. As before, we varied the initial frequency of ICEBs1-containing host cells in the inoculum between approximately 0.01 and 0.9. To measure fitness we determined the frequency of ICEBs1-containing cells (fICE) and ICEBs10 (fNULL) cells in the initial inoculum and in mature biofilms (4 days of growth at 30°C) by selective plating. The relative fitness of the ICEBs1-containing cells was calculated as the fold change in the ratio of fICE / fNULL over the course of the competition.
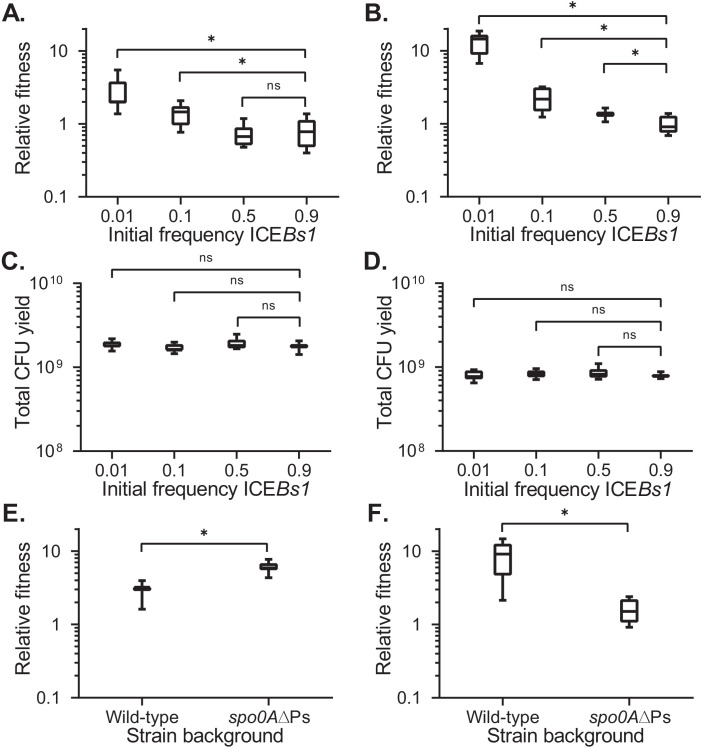
We found that the fitness of ICEBs1-containing cells was dependent on their initial frequency in the population (Figure 2A). The frequency-dependence was most likely due to regulation of ICEBs1 gene expression by the cell-cell signaling genes rapI-phrI or some other function of rapI. Cells with ICEBs1 had a selective advantage at low frequencies (0.01 or 0.1) when the element is most strongly activated. At high frequencies in the population (0.5 or 0.9), when there is little or no activation, fitness of ICEBs1-containing cells was approximately neutral (Figure 2A). The final growth yields of the populations were similar regardless of the frequency of ICEBs1-containing cells (Figure 2C).
Figure 2. Fitness of ICEBs1-containing cells relative to ICEBs1-cured cells during development.
The fitness of ICEBs1-containing cells (JMJ593) relative to ICEBs1-cured cells (JMJ550) was measured by competition during biofilm growth (A) or growth on sporulation medium (B). Total growth yields in biofilms (C) and on sporulation medium (D) were determined by counting CFUs derived from both cells and spores. The fitness of ICEBs1-containing cells relative to ICEBs1-cured cells was compared between the wild-type strain background (JMJ593 vs. JMJ550) and in the sporulation-deficient spo0A∆Ps background (JMJ788 vs. JMJ786) by competition during biofilm growth (E) or growth on sporulation medium (F). ICEBs1-containing cells were inoculated at an initial frequency of 0.01. Data shown are pooled from three independent experiments. A total of nine populations (biological replicates) were analyzed per condition. Boxes extend from the lower to upper quartiles of the data, and the middle line indicates the median fitness. Whiskers indicate the range of the fitness measurements. Asterisks indicate a p-value<0.05 (two-tailed T-test, unequal variance). Exact p-values: (A) 3.7 × 10−5, 8.8 × 10−3, 6.0 × 10−1; (B) 4.9 × 10−11, 1.6 × 10−4, 7.1 × 10−3; (C) 3.7 × 10−1, 4.2 × 10−1, 2.2 × 10−2; (D) 8.9 × 10−1, 1.9 × 10−1, 1.7 × 10−1; (E) 5.5 × 10−6; (F) 2.4 × 10−5.
We performed control competitions of two differentially marked ICEBs10 strains to verify that the enhanced fitness we observed was due to the presence of ICEBs1 rather than an inherent fitness difference associated with antibiotic resistances (see Materials and methods, Source data 1). There was a small cost associated with the marker used to select cells containing ICEBs1 (median relative fitness 0.7 ± 0.09), leading to a slight underestimate of the selective advantage to these cells.
There is a large amount of sporulation in B. subtilis biofilms (Branda et al., 2001; Vlamakis et al., 2008). Consistent with this, we found that approximately 80% of viable colony-forming units (CFUs) in a mature biofilm after 3 days were from spores. The selective advantage to cells containing ICEBs1 growing in biofilms could be due to sporulation and/or biofilm development.
ICEBs1 confers a selective advantage in biofilms without sporulation
We blocked sporulation using a mutation that causes a reduction in the amount of the transcription factor Spo0A that is required for spore formation. The spo0A∆Ps mutation is a deletion of the sigma-H-dependent promoter upstream of spo0A (Siranosian and Grossman, 1994). This mutation reduces production of Spo0A, and cells do not achieve the threshold concentration required to initiate sporulation (Chung et al., 1994). spo0A∆Ps mutant cells formed biofilms that were morphologically similar to those formed by wild-type cells.
In biofilms without sporulation, spo0A∆Ps mutant cells containing ICEBs1 (JMJ788) had a selective advantage compared to spo0A∆Ps mutant cells without ICEBs1 (JMJ786) (Figure 2E). Notably, the median fitness for spo0A∆Ps mutant cells containing ICEBs1 at a low frequency in the population was approximately six. Thus, sporulation was not required for a fitness benefit to ICEBs1-containing cells in biofilms.
Fitness of cells in biofilms can be affected by production of the biofilm matrix. For example, cells that ‘cheat’ by contributing less to biofilm matrix production reap the benefits of growing with other cells that bear the cost of matrix gene expression (Dragoš et al., 2018). We showed that cells containing ICEBs1 ‘cheat’ by decreasing expression of biofilm matrix genes compared to cells without ICEBs1 (see below).
ICEBs1 confers a selective advantage during sporulation, in the absence of biofilms
We found that cells containing ICEBs1 also had a frequency-dependent selective advantage during sporulation, in the absence of biofilms. We prepared mixtures of cells with and without ICEBs1 as described above. These mixtures were spotted onto a medium (DSM agar) that promotes high levels of sporulation. During growth on this medium, there are no complex colony features found in biofilms. As in the biofilm competitions, cells containing ICEBs1 had a frequency-dependent selective advantage during sporulation (Figure 2B). At an initial frequency of approximately 0.01, the median relative fitness of the ICEBs1-containing cells was approximately 14 (14.5 ± 4.3). As in biofilms, the total growth yields of the populations were similar regardless of ICEBs1 host frequency (Figure 2D). These results demonstrate that ICEBs1 confers a selective advantage to cells growing on DSM agar, outside the context of biofilms. This could be due to sporulation or growth under these specific conditions.
We found that sporulation was required for the strong selective advantage during growth on sporulation medium (DSM agar). The fitness benefit associated with the ICEBs1-containing cells at a low frequency in the population was greatly reduced in the spo0A∆Ps mutant (no sporulation) (Figure 2F). The sporulation mutant with ICEBs1 had a median fitness of approximately 1.5 compared to approximately nine for wild-type. Based on these results, we conclude that the presence of ICEBs1 confers a frequency-dependent selective advantage during sporulation.
Together, our results demonstrate that cells containing ICEBs1 have a frequency-dependent selective advantage in biofilms and during sporulation. This selective advantage is independent of the ability of ICEBs1 to actually transfer from one cell to another. Biofilm formation (Hamon and Lazazzera, 2001) and sporulation (Hoch, 1993; Sonenshein, 2000) are both regulated by the transcription factor Spo0A. Our results indicate that the presence of ICEBs1 could somehow be inhibiting the activity or activation of Spo0A.
ICEBs1-containing cells have a frequency-dependent delay in sporulation
We hypothesized that some ICEBs1-encoded gene(s) inhibit host cell development. This inhibition could delay development and enable cells to continue growth for a small number of generations. This model derives from analogous phenotypes of mutants that do not enter the sporulation pathway (Dawes and Mandelstam, 1970). Mutants that delay the start of sporulation have a growth advantage as they are able to divide one or a few more times while other cells in the population stop growing and start to sporulate.
We found that in mixed populations, sporulation was delayed in cells containing ICEBs1 compared to cells without ICEBs1, in a frequency-dependent manner. As above, we used an ICEBs1 mutant that is incapable of conjugation {ICEBs1(conEK476E)}. We started several replicate populations, each of which we sampled once at different times to create a time-course. (This was done because sampling to quantify CFUs [spores and cells] disrupts and prevents monitoring a single population over time.) Spore frequency was determined by measuring heat-resistant CFUs as a fraction of total CFUs for ICEBs1-containing and ICEBs1-cured strains that contained different antibiotic resistance markers to distinguish the strains.
Sporulation is delayed in ICEBs1 host cells during biofilm formation
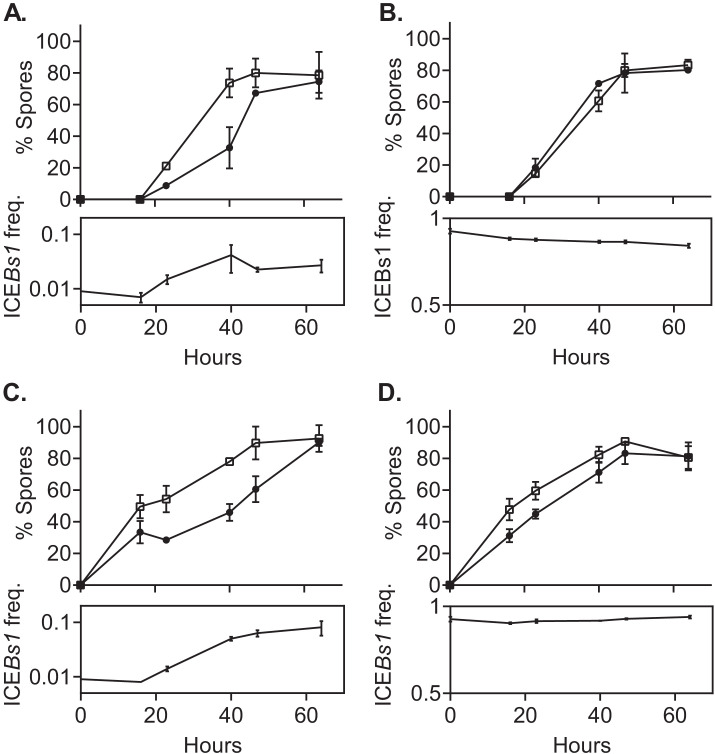
We found that sporulation of ICEBs1-containing cells was delayed in a frequency-dependent manner during growth in biofilms (Figure 3A and B). When ICEBs1-containing cells were started at a frequency of approximately 0.01, they reached their maximum sporulation frequency (>80% spores) roughly 17 hr later than the cells without ICEBs1 (Figure 3A). After 3 days of biofilm growth, the sporulation frequencies of ICEBs1-containing and ICEBs1-cured cells were indistinguishable. Over this period of time the total frequency of ICEBs1-containing cells in the population typically rose from ~0.01 to ~0.03, giving a relative fitness (~3) consistent with results above (Figure 2A). When the ICEBs1-containing cells were the majority in the population (initial frequency ~0.9) the timing and sporulation frequencies of the ICEBs1-containing and ICEBs1-cured cells were indistinguishable (Figure 3B).
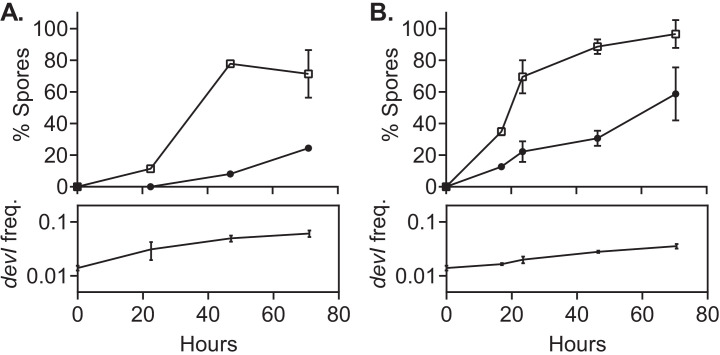
Figure 3. ICEBs1-containing cells delay sporulation in a frequency-dependent manner.
ICEBs1-containing cells (JMJ593, black circles) were mixed with ICEBs1-cured cells (JMJ550, open squares) at an initial frequency of 0.01 and spotted onto biofilm growth medium (A) or sporulation medium (C). Mixtures with ICEBs1-containing cells at an initial frequency of 0.9 were also spotted onto biofilm medium (B) and sporulation medium (D). Biofilms and colonies were harvested at the indicated times to determine the fraction of CFUs derived from heat-resistant spores for both the ICEBs1-containing and the ICEBs1-cured cells. Boxes below each graph indicate the frequency of ICEBs1-containing CFUs at each timepoint. Data shown are the average from two populations (biological replicates) per timepoint with error bars indicating the standard deviation. A representative experiment is shown.
Sporulation is delayed in ICEBs1 host cells during sporulation in the absence of biofilms
We also found that sporulation of ICEBs1-containing cells was delayed in a frequency-dependent manner during sporulation in the absence of biofilm formation (Figure 3C and D). When the ICEBs1 containing cells were inoculated at a low frequency (approximately 0.01), the delay in sporulation was qualitatively similar to that observed in biofilms (Figure 3C). However, the increase in the total frequency of ICEBs1-containing cells in the population was approximately 10-fold, giving a relative fitness of approximately 10, consistent with results described above (Figure 2B). This increase was much greater than the approximately threefold increase during biofilm formation.
We suspect that the stronger selective advantage of ICEBs1-containing cells on sporulation medium is due to the earlier onset of sporulation. By 16 hr of growth on sporulation medium, spores made up about 40% of the total CFUs. By the same time in biofilms, spores were undetectable (limit of detection ~0.03% spores).
rapI-phrI are necessary but not sufficient for enhanced fitness
The fitness benefits provided by ICEBs1 were dependent on the relative abundance of ICEBs1-containing cells, indicating that the cell-cell signaling genes rapI-phrI in ICEBs1 were likely involved, either directly or indirectly. Other Rap proteins in B. subtilis are known to regulate development by inhibiting phosphorylation (activation) of the transcription factor Spo0A (Sonenshein, 2000). RapI, like other Rap proteins in B. subtilis, can inhibit the pathway needed to phosphorylate (activate) the transcription factor Spo0A, and overexpression of rapI in vivo inhibits sporulation (Even-Tov et al., 2016; Parashar et al., 2013; Singh et al., 2013). However, it was unknown whether RapI regulates development in vivo under physiological conditions. Results described below demonstrate that the rapI-phrI system is required for the fitness advantage of ICEBs1-containing cells, but that this requirement is by virtue of causing induction of ICEBs1 gene expression. Another gene in ICEBs1 is both necessary and sufficient for the selective advantage of ICEBs1-containing cells during development.
rapI-phrI are required for the fitness benefit of cells containing ICEBs1
We deleted rapI-phrI (∆rapI-phrI) in ICEBs1 and compared the fitness conferred by this mutant to that conferred by ICEBs1 with rapI-phrI. Because loss of rapI prevents induction of gene expression, excision, and replication of ICEBs1, we used ICEBs1 mutants (‘locked-in’) that are incapable of excision or replication (see Materials and methods), regardless of the presence or absence of rapI. Preventing excision and replication of ICEBs1 allowed us to compare the fitness of wild-type ICEBs1 to ICEBs1 ∆rapI-phrI (and other mutants), which would otherwise have a lower gene copy number due to a lower frequency of induction.
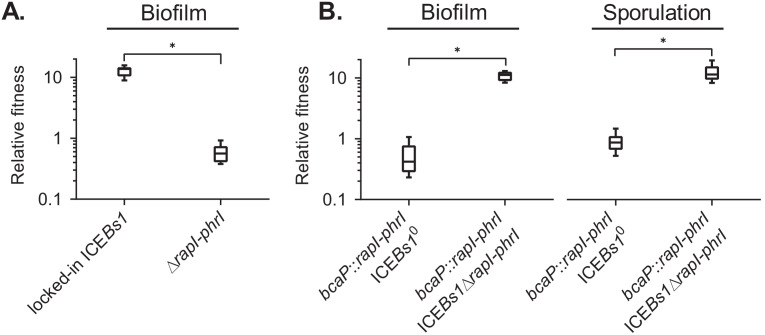
We verified that locked-in ICEBs1 still conferred a fitness benefit to host cells. During sporulation in biofilms (MSgg agar), cells containing locked-in ICEBs1 had a relative fitness of approximately 14 when they were started at a low frequency in the population (~0.01) (Figure 4A). This benefit was much greater than that conferred by wild-type ICEBs1 that can excise and replicate. We suspect that replication of ICEBs1 incurs a fitness cost to the host cell that reduces the apparent benefit. The sources of this burden could include use of host resources, the host’s response to single-stranded DNA produced by rolling-circle-replication of ICEBs1, and increases in ICEBs1 gene expression due to increased copy number.
Figure 4. The ICEBs1 cell-cell signaling genes, rapI-phrI, are necessary but not sufficient to confer a selective advantage.
(A) The fitness of cells containing locked-in ICEBs1 (JMJ646) or an isogenic rapI-phrI mutant (JMJ686) was measured relative to ICEBs1-cured cells (JMJ550) during biofilm competitions. The ICEBs1-containing cells were started at a frequency of 0.01. (B) The fitness of cells containing rapI-phrI alone (JMJ576) or cells containing both rapI-phrI and locked-in ICEBs1∆rapI-phrI (JMJ785) was measured relative to ICEBs1-cured cells (JMJ714) during biofilm and sporulation medium competitions. JMJ576 and JMJ785 were started at a frequency of 0.01. Data shown are pooled from three independent experiments with a total of nine populations (biological replicates) analyzed per strain mixture. Boxes extend from the lower to upper quartiles, and the middle line indicates the median fitness. Whiskers indicate the range of the fitness measurements. Asterisks indicate a p-value<0.05 (two-tailed T-test, unequal variance). Exact p-values: (A) 1.3 × 10−12; (B) 2.1 × 10−8, 4.6 × 10−2.
We found that rapI-phrI were required for the fitness benefit conferred by ICEBs1. During sporulation in biofilms (MSgg agar), the relative fitness of the ∆(rapI-phrI) host strain was approximately neutral (Figure 4A), in contrast to the high fitness (median ~14) provided by ICEBs1 containing rapI-phrI when the ICEBs1-containing cells were started at a low frequency (~0.01). The requirement for rapI-phrI could be due to a direct role for one of these, likely RapI, or an indirect role in activating expression of ICE genes.
rapI-phrI are not sufficient in the absence of other ICEBs1 genes to provide a fitness benefit
We found that rapI-phrI alone were not sufficient to provide a fitness benefit during sporulation or during sporulation in biofilms. We cloned rapI-phrI and their native promoters and inserted them in an ectopic locus (bcaP) in a strain that did not contain ICEBs1. When this strain was started at a low frequency (~0.01), fitness of this strain was neutral relative to a control strain without rapI-phrI (Figure 4B). To verify that rapI-phrI were functional, we added back ICEBs1 that was missing rapI-phrI. Adding the rest of ICEBs1 restored the fitness advantage during sporulation and in biofilms, indicating that the ectopic copy of rapI-phrI was functional (Figure 4B). The requirement for rapI-phrI and some other ICEBs1 gene(s) indicated that the selective advantage was likely dependent on induction of ICEBs1 by RapI.
Activation of ICEBs1 is required for the fitness benefit
We found that expression of one or more ICEBs1 genes controlled by the promoter Pxis was required for the selective advantage in biofilms with sporulation. Pxis drives most of the genes in ICEBs1 and is indirectly activated by RapI in a frequency-dependent manner (Auchtung et al., 2005; Bose and Grossman, 2011). We deleted Pxis in a strain in which ICEBs1 was unable to excise or replicate (locked-in-ICEBs1). In this strain, only genes not dependent on Pxis could be expressed, including rapI-phrI. Fitness of this strain was approximately neutral during sporulation in biofilms (Figure 5B). This indicated that expression of one or more genes controlled by Pxis, either alone or in combination with rapI, was required for the fitness benefit conferred by ICEBs1.
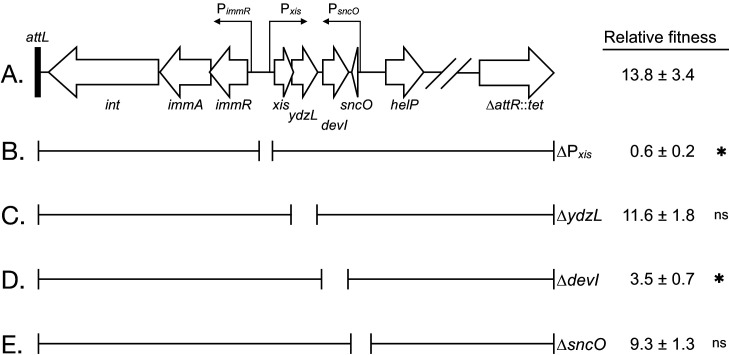
Figure 5. Effects of deletions in ICEBs1 on host fitness.
(A) Abbreviated genetic map of locked-in-ICEBs1 showing genes as open block arrows, promoters as thin right-angle arrows, and the left attachment site (attL) as a black bar. (B to E) Brackets under the map of ICEBs1 indicate regions contained in isogenic derivatives of locked-in-ICEBs1. Open spaces represent regions deleted. The fitness of strains containing locked-in-ICEBs1 (JMJ646) and its derivatives (∆Pxis, JMJ662; ∆ydzL, JMJ704; ∆devI, JMJ703; ∆sncO, JMJ688) is indicated at the right. Fitness was measured relative to ICEBs1-cured cells (JMJ550) during competitions in biofilms. ICEBs1-containing cells were started at a frequency of 0.01. Data shown are the median ± standard deviation from at least three independent experiments (at least nine total biological replicates per strain mixture), with the exception of JMJ688 which was measured only in one experiment (three total biological replicates). Asterisks indicate a statistically significant difference in fitness compared to JMJ646 (p-value<0.05, two-tailed T-test, unequal variance). Exact p-values: (B) 2.3 × 10−16; (C) 4.5 × 10−1; (D) 6.3 × 10−12; (E) 6.2 × 10−2.
Since most of the genes controlled by Pxis have known roles in the conjugative life cycle, we focused our search on genes without a known function, starting with the genes near Pxis. Of these, we found that a deletion of devI (ydcO) reduced fitness. Results described below demonstrate that a single ICEBs1 gene, devI (ydcO), is both necessary and sufficient to inhibit development and provide a selective advantage. The primary role of rapI in the fitness benefit appears to be the induction of devI expression.
devI is necessary for the fitness benefit conferred by ICEBs1
We found that an ICEBs1 gene of unknown function, devI (ydcO), was necessary for the fitness advantage of ICEBs1 host cells. We constructed a deletion of devI in the locked-in-ICEBs1 strain. When started at a low frequency in the population (~0.01) the relative fitness of the devI mutant was approximately 3.5 (Figure 5D), much less than that of the isogenic devI+ cells (median fitness ~14) in biofilms with sporulation (Figure 5A). Interestingly, the deletion of devI did not reduce fitness fully to neutral, indicating a possible role for other ICEBs1 genes. devI (ydcO) is predicted to encode an 86 amino acid protein. A search for conserved motifs and structural similarity between DevI (YdcO) and other proteins did not significantly inform our understanding of DevI function. However, devI (ydcO) homologs are found in other Bacillus species (see below).
devI is sufficient to inhibit sporulation and provide a fitness benefit
We found that when expressed constitutively, devI alone, in the absence of all other ICEBs1 genes, was sufficient to inhibit sporulation and provide a selective advantage. We cloned devI under the control of Pxis at an ectopic locus (lacA) in a strain without ICEBs1. In the absence of ICEBs1 (and its repressor ImmR), Pxis is constitutively active (Auchtung et al., 2007). Fitness was measured relative to a control strain that had Pxis with no gene downstream.
Sporulation of the Pxis-devI strain was strongly inhibited under conditions that normally support robust sporulation, including in biofilms (Figure 6). During sporulation either with (Figure 6A) or without biofilm formation (Figure 6B), the frequency of the Pxis-devI strain in the population rose from ~0.01 to ~0.05, giving a relative fitness of ~5. This is greater than the typical fitness conferred by ICEBs1 in biofilms, but less than that observed during sporulation without biofilms. We suspect these differences are due to constitutive expression of devI in the absence of ICEBs1's regulatory systems and the earlier onset of starvation on DSM agar compared to MSgg agar; cells that are unable to sporulate eventually die.
Figure 6. devI alone is sufficient to inhibit sporulation and provide a selective advantage.
ICEBs1-cured cells that constitutively express devI (JMJ725, black circles) were mixed at an initial frequency of 0.01 with ICEBs1-cured cells containing an empty expression construct (JMJ727, open squares). The mixture was spotted onto biofilm growth medium (A) or sporulation medium (B). Biofilms and colonies were harvested at the indicated times to determine the fraction of CFUs derived from heat-resistant spores for both the devI-containing and control cells. Boxes below each graph indicate the frequency of devI-containing CFUs at each timepoint. Data shown are the average from two populations (biological replicates) per timepoint with error bars indicating the standard deviation.
DevI likely targets the developmental transcription factor Spo0A
Results described above demonstrated that devI is a robust inhibitor of sporulation. Sporulation is controlled by the transcription factor Spo0A (reviewed in Hoch, 1993; Sonenshein, 2000}) which both directly and indirectly regulates the expression of many genes needed for development, including biofilm formation (Hamon and Lazazzera, 2001). The results described below indicate that DevI most likely targets Spo0A, either directly or indirectly.
devI inhibits early sporulation gene expression
We found that devI inhibits expression of genes normally activated early during sporulation. Sporulation is initiated when Spo0A~P directly stimulates transcription of several genes, including the three sporulation operons, spoIIA, spoIIE, and spoIIG (Sonenshein, 2000). Using lacZ fusions to the promoters of each of these operons, we found that Pxis-devI inhibited activity of each promoter compared to wild-type during sporulation in liquid sporulation medium (Figure 7A). This indicates that DevI inhibits the initiation of sporulation, perhaps by affecting the activity or accumulation of Spo0A~P.
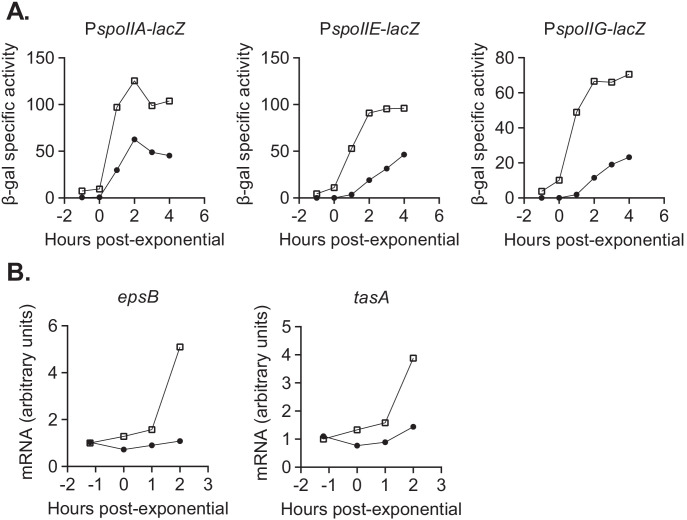
Figure 7. devI inhibits expression of genes associated with sporulation initiation and biofilm formation.
(A) ICEBs1-cured cells that constitutively express devI (black circles) or contain an empty expression construct (open squares) were grown in liquid sporulation medium. Cells were harvested at the indicated times and β-galactosidase specific activity was measured. Strains: PspoIIA-lacZ Pxis-devI (JMJ732), PspoIIA-lacZ WT (JMJ735), PspoIIE-lacZ Pxis-devI (JMJ731), PspoIIE-lacZ WT (JMJ734), PspoIIG-lacZ Pxis-devI (JMJ733), PspoIIG-lacZ WT (JMJ736). A representative experiment is shown. (B) ICEBs1-cured cells that constitutively express devI (JMJ725, black circles) and ICEBs1-cured cells containing an empty expression construct (JMJ727, open squares) were grown in liquid biofilm medium, and cells were harvested at the indicated times. cDNA was synthesized using reverse transcriptase and RT-qPCR was used to measure expression of biofilm-associated genes epsB and tasA. The transcript copy numbers of these genes were measured relative to a housekeeping gene gyrA. The data reported are the average of three technical replicates from one experiment. The relative expression levels are normalized to wild-type at T-1. A representative experiment is shown.
devI inhibits biofilm gene expression
We also found that devI inhibits expression of genes needed for extracellular matrix production during biofilm formation. We measured expression of biofilm matrix genes epsB and tasA (Hahn et al., 1995; Hamon et al., 2004; Bai et al., 1993; Kearns et al., 2005) by RT-qPCR with primers internal to each gene. In early stationary phase in liquid biofilm medium, transcript levels of epsB and tasA were reduced by about 5-fold and 3-fold, respectively, in the Pxis-devI strain compared to wild-type (Figure 7B). We suspect that inhibition of biofilm matrix genes, in addition to delaying sporulation, is an important mechanism of selection for ICEBs1 host cells during growth in a biofilm. This is consistent with the selective advantage of ICEBs1 host cells in biofilms without sporulation described earlier. Inhibition of biofilm and early sporulation genes is consistent with DevI functioning as an inhibitor of Spo0A or its activation by phosphorylation.
devI is conserved among ICEs homologous to ICEBs1
We found that devI (ydcO) is conserved among Bacillus species and in many cases is located within what appear to be ICEs similar to ICEBs1. We used NCBI BLAST to search for homologous protein sequences using both pBLAST (protein database) and tBLASTn (translated nucleotide database). Homologs with 100% sequence coverage and greater than 70% identity to YdcO from B. subtilis NCIB3610 were found in dozens of other B. subtilis strains and in closely related species including B. licheniformis, B. atrophaeus, and B. amyloliquefaciens. Excluding Bacillus species from the searches to possibly identify more distantly related proteins with known functions produced no hits.
We analyzed the sequence surrounding the devI (ydcO) homologs identified to determine if there is similarity to ICEBs1. All of the devI (ydcO) homologs appear to be within mobile element regions similar to ICEBs1, though some are clearly missing genes present in ICEBs1. Although we cannot infer whether any of these regions are functional mobile elements, we suspect that the ability to inhibit host development may be a conserved strategy among ICEBs1-like elements and possibly other ICEs with cargo genes of unknown function.
Discussion
Our work demonstrates that ICEBs1 confers a selective advantage on its host cells by delaying biofilm and spore development, enabling the host to grow more than cells without ICEBs1. When ICEBs1-containing cells are the minority in a mixed population, ICEBs1 genes are induced. One of these genes, devI, is necessary and sufficient to inhibit biofilm- and sporulation-associated gene expression, likely by inhibiting the key developmental regulator Spo0A, either directly or indirectly. Together with previous findings we conclude that ICEBs1 encodes at least three distinct strategies to benefit its host cells. (1) Inhibition of development (described here) provides a growth advantage in biofilms and during sporulation. (2) Exclusion, mediated by yddJ, blocks the conjugation machinery and protects the host cell from lethal excessive transfer (Avello et al., 2019). (3) An abortive infection mechanism, mediated by spbK (yddK) protects populations of ICEBs1 host cells from predation by the lysogenic phage SPβ (Johnson et al., 2020). We propose that all three strategies provide a competitive advantage for ICEBs1 and its host cells in different conditions.
Expression of devI reduces biofilm matrix expression and delays the initiation of sporulation. Production of the biofilm matrix is a public good, benefiting the whole community (Dragoš et al., 2018). Avoidance of matrix production can therefore be considered an exploitative behavior. Exploitation can be detrimental to the population as a whole (Smith and Schuster, 2019), but we did not observe any negative effects on populations under conditions where ICEBs1 host cells had an advantage. This is in agreement with the facultative nature of ICEBs1 cheating (Even-Tov et al., 2016; Pollak et al., 2016). Quorum-sensing by rapI-phrI ensures that ICEBs1 cheats only as a minority, where its impact on total public goods levels is negligible. Interestingly, the pBS32 plasmid utilizes direct regulation of biofilm formation by a Rap receptor to its benefit (Omer Bendori et al., 2015; Pollak et al., 2015), while in ICEBs1 this regulation was moved from the Rap receptor to one of its regulated genes.
The fitness consequences of sporulation inhibition are complicated (Mutlu et al., 2018). Delaying sporulation too long would result in a loss of viability of the starved cells. Inhibition of sporulation by ICEBs1 appears to be transient; ICEBs1 host cells eventually sporulate and do not lose significant viability as a consequence of delaying sporulation. Regulation of devI expression by the cell-cell signaling genes rapI-phrI is likely critical for transient developmental inhibition. Because commitment to sporulation is irreversible, sporulating too early is detrimental if nutrient deprivation is short-lived. B. subtilis cells with activated Spo0A that have not yet committed to sporulate also delay commitment to sporulation by killing sibling cells to liberate nutrients (‘cannibalism’) (González-Pastor et al., 2003). Cannibalism is regulated by Spo0A and the subpopulation of cannibal cells (those doing the killing) overlaps with those producing the biofilm matrix (López et al., 2009). Because of this overlap, it seems unlikely that devI delays sporulation by stimulating cannibalism.
ICEBs1 stability during sporulation
We hypothesize that in addition to providing a fitness advantage to its host cell, delaying sporulation may also improve stability of ICEBs1 in the host during development. Sporulation involves the formation of an asymmetric division septum generating the larger mother cell and the smaller forespore (Errington, 2001; Higgins and Dworkin, 2012). Sporulation is induced when cells are at a high population density and running out of nutrients, conditions that also activate ICEBs1 (Auchtung et al., 2005; Grossman and Losick, 1988). The plasmid form of ICEBs1 that is generated after excision from the chromosome is not known to have a mechanism for active partitioning and is more likely to remain in the larger mother cell if the cells do enter the sporulation pathway and divide asymmetrically. Therefore, the ability of ICEBs1 to delay the initiation of sporulation under conditions where the element is activated could help prevent loss of the element and maintain ICEBs1 in host cells.
Mobile genetic elements employ various strategies to promote their maintenance during sporulation. Rates of curing during sporulation for various plasmids in Bacillus species vary widely and do not necessarily correlate with their stability during normal cell division (Tokuda et al., 1993; Turgeon et al., 2008). Mechanisms encoded by plasmids to promote their stability during growth and sporulation include the production of dynamic cytoskeletal filaments (Becker et al., 2006) and post-segregational killing of plasmid-cured pre-spores with toxin-antitoxin systems (Short et al., 2015). Interestingly, even lytic phage genomes can be incorporated into spores (first described in the 1960 s) by co-opting the host’s chromosomal partitioning system (Meijer et al., 2005).
Diversity of cargo genes and associated phenotypes
Mobile genetic elements, especially ICEs, are widespread in bacteria (Frost et al., 2005; Guglielmini et al., 2011). Many known mobile genetic elements encode cargo genes that confer easily recognizable phenotypes, notably antibiotic resistance. Other cargo genes provide less obvious phenotypes but still fundamentally alter the physiology of the host cell. A large (500 kb) ICE was discovered in Mesorhizobium loti because its horizontal transfer conferred the ability to form nitrogen-fixing symbiotic rood nodules on Lotus plant species (Sullivan and Ronson, 1998). In many pathogens, cargo genes in mobile elements are largely responsible for virulence. For example, Vibrio cholerae is capable of a pathogenic lifestyle in human hosts due to the toxin-encoding phage CTXΦ (Waldor and Mekalanos, 1996). In the sporulating pathogen Bacillus anthracis, mobile genetic elements regulate both virulence and host development. Two plasmids, pXO1 and pXO2, provide the genes for toxin synthesis and production of a protective capsule, respectively (Green et al., 1985; Mikesell et al., 1983). pXO1 also contains a regulatory gene, atxA, that regulates virulence factor production and inhibits host cell sporulation (Dale et al., 2018). Co-regulation of virulence factors and sporulation is likely important during infection, as B. anthracis spores are thought to be more susceptible than vegetative cells to eradication by the immune system (Mock and Fouet, 2001).
Mobile elements are also known to alter the host’s interaction with other horizontally acquired DNA, which has implications for the fitness and evolvability of the host. For example, the plasmid pBS32 in B. subtilis encodes an inhibitor of the host’s DNA uptake machinery, blocking natural transformation (Konkol et al., 2013). Interestingly, genes with roles in defense against foreign DNA, CRISPR-Cas systems, are also identified within mobile elements (Faure et al., 2019; McDonald et al., 2019; Millen et al., 2012). Competition between mobile elements not only shapes the repertoire of cargo genes in a given cell, but it may also protect the host from harmful elements.
Many mobile genetic elements have been identified bioinformatically from genome sequences or discovered by means other than the phenotypes they provide (Bi et al., 2012; Guglielmini et al., 2011; Johnson and Grossman, 2015). Many elements lack obvious cargo genes, or at least lack cargo genes that have recognizable functions (Cury et al., 2017). We suspect that many elements with uncharacterized cargo genes provide important traits to their hosts beyond the scope of the phenotypes currently attributed to mobile elements. Although mobile genetic elements can have remarkably broad host ranges, such as the Tn916-Tn1545 group of ICEs (Clewell et al., 1995; Roberts and Mullany, 2009) and the IncP-1 group of plasmids (Popowska and Krawczyk-Balska, 2013), cargo genes and their associated functions could be highly specific to certain hosts.
Characterization of unknown cargo genes is likely to expand the diversity of traits currently attributed to mobile genetic elements. We speculate that many of these genes modulate normal host functions rather than provide entirely new phenotypes. Understanding cargo gene function is critical for understanding interactions between and co-evolution of mobile elements and their hosts.
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Bacillus subtilis NCIB3610) | DS2569 |
Konkol et al., 2013. PMID:23836866 |
NCIB3610 cured of pBS32 plasmid. Gift of Daniel Kearns to Avigdor Eldar. | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ550 | This paper | ICEBs10 lacA::{Ppen-mApple2 kan} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ574 | This paper | ICEBs10 lacA::{Pveg-mTagBFP mls} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ576 | This paper | ICEBs10 bcaP::{PrapI-rapIphrI kan} lacA::{Pveg-mTagBFP mls} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ592 | This paper | ICEBs1 yddJ-cat-yddK lacA::{Pveg-mTagBFP mls} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ593 | This paper | ICEBs1 conEK476E yddJ-cat-yddK lacA::{Pveg-mTagBFP mls} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ646 | This paper | ICEBs1 oriT* attR::tet lacA::{Pveg-mTagBFP mls} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ662 | This paper | ICEBs1 ∆Pxis oriT* attR::tet lacA::{Pveg-mTagBFP mls} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ686 | This paper | ICEBs1 oriT* ∆rapIphrI attR::tet lacA::{Pveg-mTagBFP mls} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ688 | This paper | ICEBs1 oriT* ∆sncO attR::tet lacA::{Pveg-mTagBFP mls} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ703 | This paper | ICEBs1 oriT* ΔdevI attR::tet lacA::{Pveg-mTagBFP mls} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ704 | This paper | ICEBs1 oriT* ∆ydzL attR::tet lacA::{Pveg-mTagBFP mls} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ714 | This paper | ICEBs10 lacA::spec bcaP::kan | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ725 | This paper | ICEBs10 lacA::{Pxis-devI mls} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ727 | This paper | ICEBs10 lacA::{Pxis-empty mls} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ731 | This paper | ICEBs10 lacA::{Pxis-devI mls} amyE::{PspoIIE-lacZ cat} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ732 | This paper | ICEBs10 lacA::{Pxis-devI mls} amyE::{PspoIIA-lacZ cat} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ733 | This paper | ICEBs10 lacA::{Pxis-devI mls} amyE::{PspoIIG-lacZ cat} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ734 | This paper | ICEBs10 lacA::{Pxis-empty mls}amyE::{PspoIIE-lacZ cat} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ735 | This paper | ICEBs10 lacA::{Pxis-empty mls} amyE::{PspoIIA-lacZ cat} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ736 | This paper | ICEBs10 lacA::{Pxis-empty mls} amyE::{PspoIIG-lacZ cat} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ785 | This paper | ICEBs1 oriT* ∆rapIphrI attR::tet bcaP::{PrapI-rapIphrI kan} lacA::{Pveg-mTagBFP mls} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ786 | This paper | ICEBs10 spo0A∆Ps lacA::{Ppen-mApple2 kan} | |
| Strain, strain background (Bacillus subtilis DS2569) | JMJ788 | This paper | ICEBs1 conEK476E yddJ-cat-yddK spo0A∆Ps lacA::{Pveg-mTagBFP mls} | |
| Strain, strain background (Escherichia coli MC1061) | AG1111 | Ireton et al., 1993 PMID:8436298 | E. coli strain for cloning and maintaining plasmids. MC1061 with F’ proAB+ lacIq lacZM15 Tn10. | |
| Recombinant DNA reagent | pJMJ196 (plasmid) | This paper | For generating oriT* nick; derived from pCAL1422. | |
| Recombinant DNA reagent | pJMJ430 (plasmid) | This paper | For generating unmarked rapI-phrI deletion; derived from pCAL1422. | |
| Recombinant DNA reagent | pJMJ199 (plasmid) | This paper | For generating unmarked Pxis deletion; derived from pCAL1422. | |
| Recombinant DNA reagent | pELS5 (plasmid) | Other | For generating unmarked ydzL deletion; derived from pCAL1422. From Grossman lab collection. | |
| Recombinant DNA reagent | pELS1 (plasmid) | Other | For generating unmarked devI deletion; derived from pCAL1422. From Grossman lab collection. | |
| Recombinant DNA reagent | pELC815 (plasmid) | Other | For generating unmarked sncO deletion; derived from pCAL1422. From Grossman lab collection. | |
| Recombinant DNA reagent | pJT245 (plasmid) | Other | Source of oriT* nicK allele. From Grossman lab collection. | |
| Recombinant DNA reagent | pCAL1422 (plasmid) | Thomas et al., 2013. PMID:23326247 | For generating markerless deletions/mutations. | |
| Recombinant DNA reagent | pMMH253 (plasmid) | Other | Vector for integration of constructs at bcaP. From Grossman lab collection. | |
| Recombinant DNA reagent | pJMJ354 (plasmid) | This paper | Native rapI-phrI expression construct for integration at bcaP; derived from pMMH253 | |
| Sequence-based reagent | oJJ363 | Sigma-Aldrich | qPCR primer | CGGAACAATATCGCACCATTC |
| Sequence-based reagent | oJJ364 | Sigma-Aldrich | qPCR primer | CGCTGCACTGAACGATTTAC |
| Sequence-based reagent | oJJ367 | Sigma-Aldrich | qPCR primer | GGATCACTTGCGATCAAAGAAG |
| Sequence-based reagent | oJJ368 | Sigma-Aldrich | qPCR primer | CTTCAAACTGGCTGAGGAAATC |
| Sequence-based reagent | oMEA128 | Sigma-Aldrich | qPCR primer | TGGAGCATTACCTTGACCATC |
| Sequence-based reagent | oMEA129 | Sigma-Aldrich | qPCR primer | AGCTCTCGCTTCTGCTTTAC |
| Commercial assay or kit | RNeasy PLUS | Qiagen | Cat No. 74136 | |
| Commercial assay or kit | iScript Reverse Transcription Supermix | Bio-Rad | Cat No. 1708840 | |
| Commercial assay or kit | SsoAdvanced SYBR master mix | Bio-Rad | Cat No. 1725270 |
Media and growth conditions
Prior to competition experiments, cells were grown as light lawns for approximately 20 hr at room temperature on 1.5% agar plates containing 1% w/v glucose, 0.1% w/v monopotassium glutamate, and 1x Spizizen’s salts (2 g/l (NH4)SO4, 14 g/l K2HPO4, 6 g/l KH2PO4, 1 g/l Na3citrate-2H2O, and 0.2 g/l MgSO4-7H2O) (Harwood and Cutting, 1990). Cells were resuspended from light lawns and grown at 37°C with shaking in S750 defined minimal medium (Jaacks et al., 1989) with 1% w/v glucose and 0.1% w/v monopotassium glutamate. Biofilms were grown at 30°C on MSgg agar plates (as defined in Branda et al., 2001 with the exception of tryptophan and phenylalanine, which we did not include). The sporulation medium used was DSM (in liquid form or as plates solidified with 1.5% agar) (Harwood and Cutting, 1990). MSgg agar and DSM agar plates were dried for 20–24 hr at 37°C prior to use. Antibiotics were used at the following concentrations for selection on LB agar plates: chloramphenicol (5 μg/ml), kanamycin (5 μg/ml), spectinomycin (100 μg/ml), tetracycline (12.5 μg/ml), and a combination of erythromycin (0.5 μg/ml) and lincomycin (12.5 μg/ml) to select for macrolide-lincosamide-streptogramin (MLS) resistance.
Strains and alleles
The B. subtilis strains used are listed in Table 2. The strain background used in all experiments was a derivative of the undomesticated strain NCIB3610 lacking its endogenous plasmid pBS32 (NCIB3610 plasmid-free). ICEBs10 indicates the strain is cured of ICEBs1. Standard techniques were used for cloning and strain construction (Harwood and Cutting, 1990). Some alleles were previously described and are summarized below. Variants of ICEBs1 that were blocked for transfer contained the conEK476E mutation derived from MMB1118 (Berkmen et al., 2010). The spo0A∆Ps allele was derived from AG1242 (Siranosian and Grossman, 1994). The amyE::PspoIIA-lacZ cat allele was derived from KI938 (Chung et al., 1994). Essentially identical alleles with PspoIIE and PspoIIG were also used.
Table 2. B. subtilis strains used*.
| Strain | Relevant genotype |
|---|---|
| JMJ550 | ICEBs10 lacA::{Ppen-mApple2 kan} |
| JMJ574 | ICEBs10 lacA::{Pveg-mTagBFP mls} |
| JMJ576 | ICEBs10 bcaP::{PrapI-rapIphrI kan} lacA::{Pveg-mTagBFP mls} |
| JMJ592 | ICEBs1 yddJ-cat-yddK lacA::{Pveg-mTagBFP mls} |
| JMJ593 | ICEBs1 conEK476E yddJ-cat-yddK lacA::{Pveg-mTagBFP mls} |
| JMJ646 | ICEBs1 oriT* attR::tet lacA::{Pveg-mTagBFP mls} |
| JMJ662 | ICEBs1 ∆Pxis oriT* attR::tet lacA::{Pveg-mTagBFP mls} |
| JMJ686 | ICEBs1 oriT* ∆rapIphrI attR::tet lacA::{Pveg-mTagBFP mls} |
| JMJ688 | ICEBs1 oriT* ∆sncO attR::tet lacA::{Pveg-mTagBFP mls} |
| JMJ703 | ICEBs1 oriT* ΔdevI attR::tet lacA::{Pveg-mTagBFP mls} |
| JMJ704 | ICEBs1 oriT* ∆ydzL attR::tet lacA::{Pveg-mTagBFP mls} |
| JMJ714 | ICEBs10 lacA::spec bcaP::kan |
| JMJ725 | ICEBs10 lacA::{Pxis-devI mls} |
| JMJ727 | ICEBs10 lacA::{Pxis-empty mls} |
| JMJ731 | ICEBs10 lacA::{Pxis-devI mls} amyE::{PspoIIE-lacZ cat} |
| JMJ732 | ICEBs10 lacA::{Pxis-devI mls} amyE::{PspoIIA-lacZ cat} |
| JMJ733 | ICEBs10 lacA::{Pxis-devI mls} amyE::{PspoIIG-lacZ cat} |
| JMJ734 | ICEBs10 lacA::{Pxis-empty mls} amyE::{PspoIIE-lacZ cat} |
| JMJ735 | ICEBs10 lacA::{Pxis-empty mls} amyE::{PspoIIA-lacZ cat} |
| JMJ736 | ICEBs10 lacA::{Pxis-empty mls} amyE::{PspoIIG-lacZ cat} |
| JMJ785 | ICEBs1 oriT* ∆rapIphrI attR::tet bcaP::{PrapI-rapIphrI kan} lacA::{Pveg-mTagBFP mls} |
| JMJ786 | ICEBs10 spo0A∆Ps lacA::{Ppen-mApple2 kan} |
| JMJ788 | ICEBs1 conEK476E yddJ-cat-yddK spo0A∆Ps lacA::{Pveg-mTagBFP mls} |
1All strains derived from NCIB3610 plasmid-free.
Construction of selective markers for mating and competition experiments
ICEBs1 was marked with the cat gene (conferring chloramphenicol resistance) between the divergently transcribed genes yddJ and spbK (yddK). Markers used to select ICEBs1-containing and ICEBs10 cells were all integrated at lacA and contained spec (spectinomycin resistance), mls (macrolide-lincosamide-streptogramin resistance), or kan (kanamycin resistance). The mls and kan constructs also contained constitutively expressed fluorescent proteins BFP and RFP, respectively. The plating efficiency of all markers was verified, and control competitions (described below) were performed to measure marker-associated fitness effects.
Construction of ICEBs1 mutants
Fitness measurements of ICEBs1 mutants were performed in a version of ICEBs1 unable to excise and replicate (locked-in ICEBs1). All mutants were isogenic to JMJ646 (ICEBs1 oriT* ∆attR::tet), which is unable to excise due to the attR::tet deletion (Lee and Grossman, 2007). The origin of transfer was mutated (oriT*) to prevent ICEBs1 replication while integrated, which is detrimental (Lee and Grossman, 2007; Menard and Grossman, 2013). The markerless oriT* mutation was constructed by cloning nicK(oriT*) from pJT245 and ~1 kb of upstream sequence into pCAL1422 (a plasmid that contains E. coli lacZ) by isothermal assembly (Gibson et al., 2009), essentially as previously described (Thomas et al., 2013). The resulting plasmid, pJMJ196, was integrated into the chromosome by single-crossover recombination. Transformants were screened for loss of lacZ, indicating loss of the integrated plasmid, and PCR was used to identify a clone containing the oriT* allele. Markerless deletions of ICEBs1 sequence were also generated using pCAL1422-derived plasmids. The rapI-phrI deletion was generated using pJMJ430 and removes the rapI and phrI ORFs. The Pxis deletion was generated using pJMJ199 and removes sequence from 149 bp to 27 bp upstream of the xis ORF. This removes the presumed −35 and −10 of the promoter but does not remove the known regulatory sites at the neighboring immR promoter (Auchtung et al., 2007). The ydzL deletion was generated using pELS5 and fuses the first four and last two codons of ydzL. The devI deletion was generated using pELS1 and fuses the first four and last two codons of devI. The sncO deletion was generated using pELC815 and removes the sncO gene and 44 bp of upstream sequence.
Construction of ectopic rapI-phrI construct
The rapI-phrI ORFs plus the promoter region (352 bp upstream of rapI) and 112 bp of downstream sequence were cloned into pMMH253 (vector for integration at bcaP). The resulting plasmid (pJMJ354) was linearized and introduced to B. subtilis by transformation and selection for kanamycin resistance. The corresponding empty control construct was generated by transforming linearized pMMH253.
Construction of lacA::Pxis-devI
We expressed devI from the Pxis promoter by cloning existing elements from a Pxis gene expression construct marked with mls at lacA and inserting the devI ORF by isothermal assembly. The resulting product was introduced to the chromosome by transformation and selection for MLS resistance. The Pxis-empty control strain contains an identical construct lacking an ORF fused to Pxis.
Biofilm mating experiments
Cultures were started from resuspended light lawns (described above) diluted to an initial OD600 of 0.05 in S750 minimal medium. Cultures were grown to mid-exponential phase (OD600 ~0.5) at 37°C with shaking. Cells were pelleted, resuspended in 1x Spizizen’s salts, and diluted to an OD600 of 0.01. Donor and recipient strains were mixed at the indicated frequencies and 50 μl of the mixture was spotted onto the center of MSgg agar plates. Spots were allowed to dry at 30°C before flipping the plates. Plates were incubated at 30°C for 4 days. At the time of inoculation, the strain mixes were diluted and plated in duplicate on LB agar plates containing the appropriate antibiotics to determine the initial CFU/ml of the donor and recipient strains. After 4 days, the mature biofilms were scraped from the agar surface with sterile wooden sticks and resuspended in 5 ml 1x Spizizen’s salts, followed by mild sonication to disperse the cells. Cells were diluted and selectively plated to determine the final CFU/ml of transconjugants.
Competition experiments
Cells were prepared for competition experiments as described above for biofilm mating experiments. Strain mixtures at the indicated frequencies were spotted onto MSgg agar plates for biofilm competitions and DSM agar plates for sporulation competitions. Plates were incubated at 30°C for 4 days unless otherwise indicated. Biofilms/colonies were collected, disrupted, and plated as described above. For time-course competitions, two replicate biofilms/colonies were collected at each of the indicated times. Sporulation frequency was determined by selective plating before and after a heat treatment at 85°C for 20 min to enumerate total CFUs and CFUs derived from heat-resistant spores. Relative fitness of ICEBs1-containing cells over ICEBs10 cells was determined as (pf/(1-pf))/(pi/(1-pi)), where pf, pi are the frequencies of ICEBs1-containing cells in the final and initial populations, respectively. Control competitions between ICEBs1-cured cells were performed to determine the fitness associated with the lacA::{Pveg-mTagBFP mls} marker (JMJ574) used in ICEBs1-containing cells relative to the lacA::{Ppen-mApple2 kan} marker used in ICEBs1-null cells (JMJ550). When the mls-marked cells were started at a frequency of 0.01, their relative fitness was 0.7 ± 0.09 (average and standard deviation from three independent experiments and a total of 9 biofilms).
Gene expression assays
Expression of sporulation genes was measured in cultures grown from single colonies in liquid DSM at 37° with shaking. Cells were harvested at the indicated timepoints. For β-galactosidase assays, growth was stopped by the addition of toluene (~1.5% final concentration). β-galactosidase specific activity ([∆A420 per minute per ml of culture per OD600] x 1000) was measured as described (Miller, 1972) after pelleting cell debris.
Biofilm gene expression was measured in cultures grown from single colonies in liquid MSgg at 37° with shaking. For RT-qPCR assays, cells were harvested directly into ice-cold methanol (1:1 methanol to culture volume) and pelleted. RNA was isolated using Qiagen RNeasy PLUS kit with 10 mg/ml lysozyme. iScript Supermix (Bio-Rad) was used for reverse transcriptase reactions to generate cDNA. Control reactions without reverse transcriptase were performed to assess the amount of DNA present in the RNA samples. RNA was degraded by adding 75% vol of 0.1 M NaOH and incubating at 70°C for 10 min, followed by neutralization with an equal volume of 0.1 M HCl. qPCR was done using SSoAdvanced SYBR master mix and CFX96 Touch Real-Time PCR system (Bio-Rad). Primers used to measure epsB were oJJ363 (5’-CGGAACAATATCGCACCATTC-3’) and oJJ364 (5’-CGCTGCACTGAACGATTTAC-3’). Primers used to quantify tasA were oJJ367 (5’-GGATCACTTGCGATCAAAGAAG-3’) and oJJ368 (5’-CTTCAAACTGGCTGAGGAAATC-3’). Primers used to measure the control locus gyrA were oMEA128 (5’-TGGAGCATTACCTTGACCATC-3’) and oMEA129 (5’-AGCTCTCGCTTCTGCTTTAC-3’). The relative transcript copy numbers (as indicated by the Cp values measured by qPCR) of epsB and tasA were normalized to gyrA after subtracting the signal from control reactions without reverse transcriptase.
Acknowledgements
We thank Mary Anderson and Janet Smith for helpful comments on the manuscript. Research reported here is based upon work supported, in part, by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM050895 and R35 GM122538 to ADG. Any opinions, findings, and conclusions or recommendations expressed in this report are those of the authors and do not necessarily reflect the views of the National Institutes of Health. Funding for travel was provided by the MIT International Science and Technology Initiatives (MISTI) MIT-Israel Seed Fund.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Avigdor Eldar, Email: avigdor@gmail.com.
Alan D Grossman, Email: adg@mit.edu.
Petra Anne Levin, Washington University in St. Louis, United States.
Gisela Storz, National Institute of Child Health and Human Development, United States.
Funding Information
This paper was supported by the following grants:
National Institute of General Medical Sciences R01 GM050895 to Alan D Grossman.
National Institute of General Medical Sciences R35 GM122538 to Alan D Grossman.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Formal analysis, Validation, Investigation, Visualization, Methodology, Writing - original draft, Writing - review and editing.
Investigation, Methodology.
Conceptualization, Formal analysis, Supervision, Funding acquisition, Writing - review and editing.
Conceptualization, Supervision, Funding acquisition, Visualization, Project administration, Writing - review and editing.
Additional files
Counts of two ICEBs1-cured strains each bearing an antibiotic resistance marker (used to distinguish strains) at the start and end of competitions.
Data availability
All data generated or analysed during this study are included in the manuscript and supporting files. Source data files have been provided for all figures.
References
- Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. PNAS. 2005;102:12554–12559. doi: 10.1073/pnas.0505835102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung JM, Lee CA, Garrison KL, Grossman AD. Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis. Molecular Microbiology. 2007;64:1515–1528. doi: 10.1111/j.1365-2958.2007.05748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avello M, Davis KP, Grossman AD. Identification, characterization and benefits of an exclusion system in an integrative and conjugative element of Bacillus subtilis. Molecular Microbiology. 2019;112:1066–1082. doi: 10.1111/mmi.14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai U, Mandic-Mulec I, Smith I. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes & Development. 1993;7:139–148. doi: 10.1101/gad.7.1.139. [DOI] [PubMed] [Google Scholar]
- Baltrus DA. Exploring the costs of horizontal gene transfer. Trends in Ecology & Evolution. 2013;28:489–495. doi: 10.1016/j.tree.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Becker E, Herrera NC, Gunderson FQ, Derman AI, Dance AL, Sims J, Larsen RA, Pogliano J. DNA segregation by the bacterial actin AlfA during Bacillus subtilis growth and development. The EMBO Journal. 2006;25:5919–5931. doi: 10.1038/sj.emboj.7601443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkmen MB, Lee CA, Loveday EK, Grossman AD. Polar positioning of a conjugation protein from the integrative and conjugative element ICEBs1 of Bacillus subtilis. Journal of Bacteriology. 2010;192:38–45. doi: 10.1128/JB.00860-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi D, Xu Z, Harrison EM, Tai C, Wei Y, He X, Jia S, Deng Z, Rajakumar K, Ou HY. ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Research. 2012;40:D621–D626. doi: 10.1093/nar/gkr846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose B, Auchtung JM, Lee CA, Grossman AD. A conserved anti-repressor controls horizontal gene transfer by proteolysis. Molecular Microbiology. 2008;70:570–582. doi: 10.1111/j.1365-2958.2008.06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose B, Grossman AD. Regulation of horizontal gene transfer in Bacillus subtilis by activation of a conserved site-specific protease. Journal of Bacteriology. 2011;193:22–29. doi: 10.1128/JB.01143-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. PNAS. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrus V, Pavlovic G, Decaris B, Guédon G. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid. 2002;48:77–97. doi: 10.1016/S0147-619X(02)00102-6. [DOI] [PubMed] [Google Scholar]
- Carraro N, Burrus V. The dualistic nature of integrative and conjugative elements. Mobile Genetic Elements. 2015;5:98–102. doi: 10.1080/2159256X.2015.1102796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JD, Stephanopoulos G, Ireton K, Grossman AD. Gene expression in single cells of Bacillus subtilis: evidence that a threshold mechanism controls the initiation of sporulation. Journal of Bacteriology. 1994;176:1977–1984. doi: 10.1128/JB.176.7.1977-1984.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell DB, Flannagan SE, Jaworski DD. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends in Microbiology. 1995;3:229–236. doi: 10.1016/S0966-842X(00)88930-1. [DOI] [PubMed] [Google Scholar]
- Cury J, Touchon M, Rocha EPC. Integrative and conjugative elements and their hosts: composition, distribution and organization. Nucleic Acids Research. 2017;45:8943–8956. doi: 10.1093/nar/gkx607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale JL, Raynor MJ, Ty MC, Hadjifrangiskou M, Koehler TM. A dual role for the Bacillus anthracis Master Virulence Regulator AtxA: Control of Sporulation and Anthrax Toxin Production. Frontiers in Microbiology. 2018;9:482. doi: 10.3389/fmicb.2018.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes IW, Mandelstam J. Sporulation of Bacillus subtilis in continuous culture. Journal of Bacteriology. 1970;103:529–535. doi: 10.1128/JB.103.3.529-535.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragoš A, Kiesewalter H, Martin M, Hsu C-Y, Hartmann R, Wechsler T, Eriksen C, Brix S, Drescher K, Stanley-Wall N, Kümmerli R, Kovács Á. Division of labor during biofilm matrix production. Current Biology. 2018;28:1903–1913. doi: 10.1016/j.cub.2018.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J. Septation and chromosome segregation during sporulation in Bacillus subtilis. Current Opinion in Microbiology. 2001;4:660–666. doi: 10.1016/S1369-5274(01)00266-1. [DOI] [PubMed] [Google Scholar]
- Even-Tov E, Bendori SO, Valastyan J, Ke X, Pollak S, Bareia T, Ben-Zion I, Bassler BL, Eldar A. Social evolution selects for redundancy in bacterial quorum sensing. PLOS Biology. 2016;14:e1002386. doi: 10.1371/journal.pbio.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure G, Shmakov SA, Yan WX, Cheng DR, Scott DA, Peters JE, Makarova KS, Koonin EV. CRISPR-Cas in mobile genetic elements: counter-defence and beyond. Nature Reviews Microbiology. 2019;17:513–525. doi: 10.1038/s41579-019-0204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke AE, Clewell DB. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of "conjugal" transfer in the absence of a conjugative plasmid. Journal of Bacteriology. 1981;145:494–502. doi: 10.1128/JB.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nature Reviews Microbiology. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- González-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- Green BD, Battisti L, Koehler TM, Thorne CB, Ivins BE. Demonstration of a capsule plasmid in Bacillus anthracis. Infection and Immunity. 1985;49:291–297. doi: 10.1128/IAI.49.2.291-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AD, Losick R. Extracellular control of spore formation in Bacillus subtilis. PNAS. 1988;85:4369–4373. doi: 10.1073/pnas.85.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmini J, Quintais L, Garcillán-Barcia MP, de la Cruz F, Rocha EP. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLOS Genetics. 2011;7:e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J, Roggiani M, Dubnau D. The major role of Spo0A in genetic competence is to downregulate abrB, an essential competence gene. Journal of Bacteriology. 1995;177:3601–3605. doi: 10.1128/JB.177.12.3601-3605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon MA, Stanley NR, Britton RA, Grossman AD, Lazazzera BA. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Molecular Microbiology. 2004;52:847–860. doi: 10.1111/j.1365-2958.2004.04023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon MA, Lazazzera BA. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Molecular Microbiology. 2001;42:1199–1209. doi: 10.1046/j.1365-2958.2001.02709.x. [DOI] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM. Molecular Biological Methods for Bacillus. John Wiley & Sons; 1990. [Google Scholar]
- Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiology Reviews. 2012;36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch JA. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annual Review of Microbiology. 1993;47:441–465. doi: 10.1146/annurev.mi.47.100193.002301. [DOI] [PubMed] [Google Scholar]
- Ireton K, Rudner DZ, Siranosian KJ, Grossman AD. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes & Development. 1993;7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- Jaacks KJ, Healy J, Losick R, Grossman AD. Identification and characterization of genes controlled by the sporulation-regulatory gene spo0H in Bacillus subtilis. Journal of Bacteriology. 1989;171:4121–4129. doi: 10.1128/JB.171.8.4121-4129.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, Harden MM, Grossman AD. An integrative and conjugative element encodes an abortive infection system to protect host cells from predation by a bacteriophage. bioRxiv. 2020 doi: 10.1101/2020.12.13.422588. [DOI]
- Johnson CM, Grossman AD. Integrative and conjugative elements (ICEs): What they do and how they work. Annual Review of Genetics. 2015;49:577–601. doi: 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Molecular Microbiology. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- Konkol MA, Blair KM, Kearns DB. Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. Journal of Bacteriology. 2013;195:4085–4093. doi: 10.1128/JB.00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lécuyer F, Bourassa JS, Gélinas M, Charron-Lamoureux V, Burrus V, Beauregard PB. Biofilm formation drives transfer of the conjugative element ICEBs1 in Bacillus subtilis. mSphere. 2018;3:e00473. doi: 10.1128/mSphere.00473-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CA, Grossman AD. Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis. Journal of Bacteriology. 2007;189:7254–7261. doi: 10.1128/JB.00932-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López D, Vlamakis H, Losick R, Kolter R. Cannibalism enhances biofilm development in Bacillus subtilis. Molecular Microbiology. 2009;74:609–618. doi: 10.1111/j.1365-2958.2009.06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen JS, Burmølle M, Hansen LH, Sørensen SJ. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunology & Medical Microbiology. 2012;65:183–195. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- McDonald ND, Regmi A, Morreale DP, Borowski JD, Boyd EF. CRISPR-Cas systems are present predominantly on mobile genetic elements in Vibrio species. BMC Genomics. 2019;20:105. doi: 10.1186/s12864-019-5439-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer WJ, Castilla-Llorente V, Villar L, Murray H, Errington J, Salas M. Molecular basis for the exploitation of spore formation as survival mechanism by virulent phage phi29. The EMBO Journal. 2005;24:3647–3657. doi: 10.1038/sj.emboj.7600826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard KL, Grossman AD. Selective pressures to maintain attachment site specificity of integrative and conjugative elements. PLOS Genetics. 2013;9:e1003623. doi: 10.1371/journal.pgen.1003623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikesell P, Ivins BE, Ristroph JD, Dreier TM. Evidence for plasmid-mediated toxin production in Bacillus anthracis. Infection and Immunity. 1983;39:371–376. doi: 10.1128/IAI.39.1.371-376.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen AM, Horvath P, Boyaval P, Romero DA. Mobile CRISPR/Cas-mediated bacteriophage resistance in Lactococcus lactis. PLOS ONE. 2012;7:e51663. doi: 10.1371/journal.pone.0051663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Mock M, Fouet A. Anthrax. Annual Review of Microbiology. 2001;55:647–671. doi: 10.1146/annurev.micro.55.1.647. [DOI] [PubMed] [Google Scholar]
- Molin S, Tolker-Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Current Opinion in Biotechnology. 2003;14:255–261. doi: 10.1016/S0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- Mutlu A, Trauth S, Ziesack M, Nagler K, Bergeest JP, Rohr K, Becker N, Höfer T, Bischofs IB. Phenotypic memory in Bacillus subtilis links dormancy entry and exit by a spore quantity-quality tradeoff. Nature Communications. 2018;9:69. doi: 10.1038/s41467-017-02477-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer Bendori S, Pollak S, Hizi D, Eldar A. The RapP-PhrP quorum-sensing system of Bacillus subtilis strain NCIB3610 affects biofilm formation through multiple targets, due to an atypical signal-insensitive allele of RapP. Journal of Bacteriology. 2015;197:592–602. doi: 10.1128/JB.02382-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar V, Jeffrey PD, Neiditch MB. Conformational change-induced repeat domain expansion regulates rap phosphatase quorum-sensing signal receptors. PLOS Biology. 2013;11:e1001512. doi: 10.1371/journal.pbio.1001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak S, Omer Bendori S, Eldar A. A complex path for domestication of B. subtilis sociality. Current Genetics. 2015;61:493–496. doi: 10.1007/s00294-015-0479-9. [DOI] [PubMed] [Google Scholar]
- Pollak S, Omer-Bendori S, Even-Tov E, Lipsman V, Bareia T, Ben-Zion I, Eldar A. Facultative cheating supports the coexistence of diverse quorum-sensing alleles. PNAS. 2016;113:2152–2157. doi: 10.1073/pnas.1520615113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popowska M, Krawczyk-Balska A. Broad-host-range IncP-1 plasmids and their resistance potential. Frontiers in Microbiology. 2013;4:44. doi: 10.3389/fmicb.2013.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AP, Mullany P. A modular master on the move: the Tn916 family of mobile genetic elements. Trends in Microbiology. 2009;17:251–258. doi: 10.1016/j.tim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Short FL, Monson RE, Salmond GP. A type III protein-RNA toxin-antitoxin system from Bacillus thuringiensis promotes plasmid retention during spore development. RNA Biology. 2015;12:933–937. doi: 10.1080/15476286.2015.1073438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PK, Ramachandran G, Ramos-Ruiz R, Peiró-Pastor R, Abia D, Wu LJ, Meijer WJ. Mobility of the native Bacillus subtilis conjugative plasmid pLS20 is regulated by intercellular signaling. PLOS Genetics. 2013;9:e1003892. doi: 10.1371/journal.pgen.1003892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siranosian KJ, Grossman AD. Activation of spo0A transcription by sigma H is necessary for sporulation but not for competence in Bacillus subtilis. Journal of Bacteriology. 1994;176:3812–3815. doi: 10.1128/JB.176.12.3812-3815.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P, Schuster M. Public goods and cheating in microbes. Current Biology. 2019;29:R442–R447. doi: 10.1016/j.cub.2019.03.001. [DOI] [PubMed] [Google Scholar]
- Sonenshein AL. Control of sporulation initiation in Bacillus subtilis. Current Opinion in Microbiology. 2000;3:561–566. doi: 10.1016/S1369-5274(00)00141-7. [DOI] [PubMed] [Google Scholar]
- Sullivan JT, Ronson CW. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. PNAS. 1998;95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Lee CA, Grossman AD. A conserved helicase processivity factor is needed for conjugation and replication of an integrative and conjugative element. PLOS Genetics. 2013;9:e1003198. doi: 10.1371/journal.pgen.1003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda Y, Ano T, Shoda M. Characteristics of plasmid stability in Bacillus subtilis NB22, an antifungal-antibiotic iturin producer. Journal of Fermentation and Bioengineering. 1993;75:319–321. doi: 10.1016/0922-338X(93)90159-6. [DOI] [Google Scholar]
- Treangen TJ, Rocha EP. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLOS Genetics. 2011;7:e1001284. doi: 10.1371/journal.pgen.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon N, Laflamme C, Ho J, Duchaine C. Evaluation of the plasmid copy number in B. cereus spores, during germination, bacterial growth and sporulation using real-time PCR. Plasmid. 2008;60:118–124. doi: 10.1016/j.plasmid.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes & Development. 2008;22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nature Reviews Microbiology. 2013;11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nature Reviews Microbiology. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]