Abstract
In late 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in Wahan, China and it causes disease which is known as COVID-19. This infection spreads everywhere in the world, and it leads to an enormous number of death among individuals. The mystery issue about SARS-CoV-2 that appears not to have functions of a hemagglutinin and neuraminidase like other coronaviruses. Angiotensin-converting enzyme 2 (ACE2) is the main surface receptor for entering SARS-CoV-2 into the host cell. This entry process is mediated by binding the SARS-CoV-2 spike receptor-binding domain (RBD) to ACE2. Recently, researchers discover a new receptor responsible for the SARS-CoV-2 entry which is neuropilin-1 (NRP1). So, this work provides afford a knowledge of how the initial interaction between SARS-CoV-2 spike RBD and NRP1 b1 domain may occur. Understanding this interaction would be very necessary for drug design against SARS-CoV-2.
Keywords: COVID-19, SARS-CoV-2, Microbiology, In silico, Coronavirus
1. Introduction
COVID-19 is disease caused by new emerged virus known as Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This virus is quickly spread to all world countries and causing high mortality and morbidity when compared to MERS-CoV and SARS-CoV (Datta et al., 2020). SARS-CoV-2 infects many cells such as pneumocytes in lungs and epithelial cells in kidney (Farkash et al., 2020). These cells are express angiotensin converting enzyme type 2 (ACE2) that facilitates SARS-CoV-2 entry (Harmer et al., 2002, Soler et al., 2013). The entry of this virus into the host cell is mediated by binding of SARS-CoV-2 spike receptor-binding domain (RBD) to ACE2 (Robson, 2020). This binding leads to the downregulation of ACE2 that may facilitate the progression of COVID-19 from slight to more severe illness such as huge stimulation of platelets and coagulation pathways (Datta et al., 2020). ACE2 may play a critical role in COVID-19 progression. ACE2 is expressed in several tissues such as heart, intestine and kidney (Zhou et al., 2020). SARS-CoV-2 use its surface spike protein to mediate the entry into host cells. This virus comprises a RBD that specifically recognized by its receptor, ACE2 (F. Li et al., 2005; W. Li et al., 2003). Besides ACE2 researchers found that neuropilin-1 (NRP1) plays as an alternative pathway for SARS-CoV-2 entry and also involved in the infection of SARS-CoV-2 (Cantuti-Castelvetri et al., 2020, Daly et al., 2020). NRP1 is a transmembrane receptor and works as a co-receptor for many ligands (Guo and Vander Kooi, 2015, Roy et al., 2017). It composes extracellular regions; CUB domains (a1 and a2), coagulation factor domains (b1 and b2), and a MAM domain (Guo & Vander Kooi, 2015). Of these, SARS-CoV-2 spike protein can bind to the b1 domain of NRP1 (Cantuti-Castelvetri et al., 2020, Daly et al., 2020). Based on these findings, there is no data about the virus-host interactions that identifying the potential binding sites of SARS-CoV-2 spike RBD on NRP1 b1 domain. In this work, an in silico analysis of interaction of SARS-CoV-2 spike RBD with b1 domain of NRP1 was generated for providing knowledge about the binding sites of this coronavirus spike protein on NRP1 b1 domain. So, this computational method would contribute in achieving the aim of understanding about the SARS-CoV-2-NRP1 interactions.
2. Materials and methods
2.1. Sequence information
The wild type of the SARS-CoV-2 genome corresponds to the NCBI reference sequence (NC_045512.2), isolated in December 2019 from a patient with COVID-19 in Wuhan, China (Lan et al., 2020).
2.2. Preparation
Structures of SARS-CoV-2 spike RBD and structure of the human neuropilin-1 b1 were downloaded from Protein Data Bank (PDB) (www.rcsb.org). The spike structure of SARS-CoV-2 with PDB code 6m0j (Lan et al., 2020) was used as the reference to align other structures of the protein to facilitate further analyses of the docking results. The structure 6m0j is provided as the complex between the spike RBD domain and ACE-2 (i.e., PDBcode_6m0j-ACE2) and without ACE-2 (i.e., PDB code_6m0j-noACE2, 6m0j:E). The structure of the human NRP1 b1 domain was found with PDB code_ 5JGQ. In the preparation process, we added the hydrogen atoms and optimized the hydrogen bond network with PROPKA (Olsson et al., 2011) considering pH = 7. None of the ionizable residues required manual adjustment of protonation states. The selenomethionine residues were converted to methionines and all non-protein molecules were deleted (waters, ions, cofactors and ligands).
2.3. Docking study and configuration
To perform docking studies, PDB files of 5JGQ and 6m0j:E were submitted to Cluspro 2.0 server (Kozakov et al., 2013, Kozakov et al., 2017, Vajda et al., 2017). Cluspro 2.0 was generated several docking models, then ProQdock was used to check the quality of the docking models (Basu et al., 2016).
3. Results
3.1. Docking model of interaction of SARS-CoV-2 spike RBD and human neuropilin-1 b1 domain
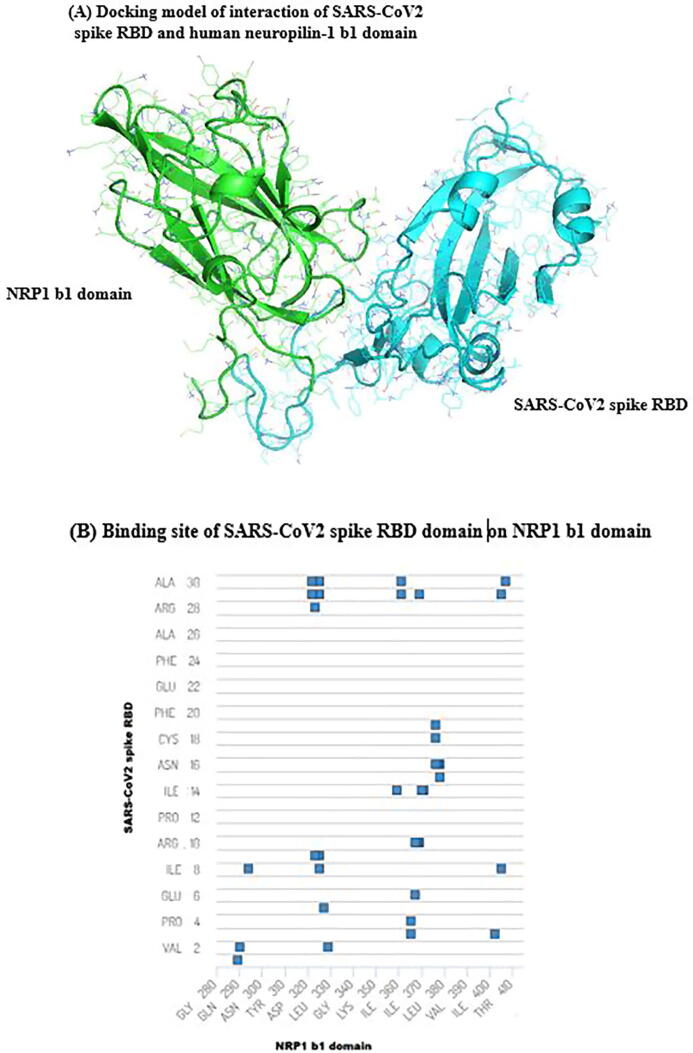
The database of PDB was explored for the 3D structure of SARS-CoV-2 spike RBD, the accession number 6m0j:E was found. The structure of 6m0j:E was revealed by X-Ray diffraction (Lan et al., 2020). After that, PDB database was also searched for the 3D structure of NRP1 b1 domain, PDB file with code 5JGQ was found. Therefore, work was carried out to identify binding sites of SARS-CoV-2 spike RBD on NRP1 b1 domain. Cluspro 2.0 was created several docking models. These models were ranked based on their quality score by ProQdock. The top-scored docking model was selected in this work (Fig. 1A). The score of modeling confidence was used to judge the reliability of this docking model, and its overall global quality score is 0.92520. This score showed that this docking has a more confident model. This docking model showed that there are potential sites of binding of SARS-CoV-2 spike RBD on NRP1 b1 domain (Fig. 1B). In this model, many locations of interaction were identified as potential binding sites for SARS-CoV-2 spike RBD on the b1 domain of NRP1. GLN280, ASP289, TYR322, ARG323, TRP325, GLN327, ASP329, LYS359, ASP361 residues were identified as potential binding sites on NRP1 b1 domain for SARS-CoV-2 spike RBD domain (Fig. 1B). Also, there is a contact between GLN3, ILE8, PHE29, ALA30 of SARS-CoV-2 spike RBD domain and ARG402, ARG405, LYS407 of NRP1 b1 domain (Fig. 1B). This docking study showed several important binding sites for SARS-CoV-2 spike RBD on human host NRP1 b1 domain. These data could be contribute in reaching the goal for treatment development against SARS-CoV-2 through providing more information about binding sites of the interaction of SARS-CoV-2 spike RBD with human host NRP1 b1 domain.
Fig. 1.
Overall three-dimensional view of interaction between SARS-CoV-2 spike RBD and NRP1 b1 domain. (A) Docking model of interaction of SARS-CoV-2 spike RBD with NRP1 b1 domain. The overall global quality score is 0.92520. This docking model was generated by Cluspro 2.0, then ProQdock was used to check its quality. This model was visualized by PyMOL1.3. (B) Binding site of SARS-CoV-2 spike RBD on NRP1 b1 domain. Residues of SARS-CoV-2 spike RBD on the Y-axis and NRP1 b1 domain residues on X-axis. Blue squares represents for residues that involves in interaction of SARS-CoV-2 spike RBD with NRP1 b1 domain.
4. Discussion
The SARS-CoV-2 is causative agent for the COVID-19 that emerged in Wahan, China in December 2019 and leading to an uncontrollable global outbreak. COVID-19 is associated with severe respiratory illness, fever, and pneumonia (Lai et al., 2020). The mechanism of SARS-CoV-2 entry into host has been extensively studied with ACE2 (Datta et al., 2020, Lan et al., 2020). Recently, scientists found that NRP1 is potential another pathway for SARS-CoV-2 entry and is implicated in the progression of COVID-19 (Cantuti-Castelvetri et al., 2020, Daly et al., 2020). The studying of the interaction of SARS-CoV-2 spike RBD with NRP1 b1 domain is conducted in this work due to there are limited studied about this interaction. So, docking study is carried out for this purpose. Binding sites of SARS-CoV-2 spike RBD with its NRP1 receptor that presents on surface with interacting amino acids; GLN280, ASP289, TYR322, ARG323, TRP325, GLN327, ASP329, LYS359, ASP361. Bound structure of SARS-CoV-2 spike RBD with NRP1 b1 domain is considered as target for drug discovery to treat SARS-CoV-2 infection.
5. Conclusions
The bioinformatics analyses indicated the possible binding specificity of SARS-CoV-2 spike RBD and NRP1 b1 domain. Data predict how the initial binding of SARS-CoV-2 spike RBD to NRP1 b1 domain may happen. These data are necessary for drug-design perspective.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
References
- Basu Sankar, Wallner Björn. Finding correct protein-protein docking models using ProQDock. Bioinformatics (Oxford, England) 2016;32(12):i262–i270. doi: 10.1093/bioinformatics/btw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri Ludovico, Ojha Ravi, Pedro Liliana D., Djannatian Minou, Franz Jonas, Kuivanen Suvi, Simons Mikael. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;eabd2985 doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly James L., Simonetti Boris, Klein Katja, Chen Kai-En, Williamson Maia Kavanagh, Antón-Plágaro Carlos, Yamauchi Yohei. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;eabd3072 doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, Prasun K., Liu, Fengming, Fischer, Tracy, Rappaport, Jay, & Qin, Xuebin, 2020. SARS-CoV-2 pandemic and research gaps: Understanding SARS-CoV-2 interaction with the ACE2 receptor and implications for therapy. Theranostics, 10(16), 7448-7464. doi: 10.7150/thno.48076 [DOI] [PMC free article] [PubMed]
- Farkash E.A., Wilson A.M., Jentzen J.M. Ultrastructural evidence for direct renal infection with SARS-CoV-2. J. Am. Soc. Nephrol. 2020;31(8):1683–1687. doi: 10.1681/asn.2020040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H.F., Vander Kooi C.W. Neuropilin functions as an essential cell surface receptor. J. Biol. Chem. 2015;290(49):29120–29126. doi: 10.1074/jbc.R115.687327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer D., Gilbert M., Borman R., Clark K.L. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532(1–2):107–110. doi: 10.1016/s0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- Kozakov D., Beglov D., Bohnuud T., Mottarella S.E., Xia B., Hall D.R., Vajda S. How good is automated protein docking? Proteins. 2013;81(12):2159–2166. doi: 10.1002/prot.24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C.…Vajda S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017;12(2):255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, Chih-Cheng, Ko, Wen-Chien, Lee, Ping-Ing, Jean, Shio-Shin, & Hsueh, Po-Ren, 2020. Extra-respiratory manifestations of COVID-19. Int. J. Antimicrobial Agents, 56(2), 106024-106024. doi: 10.1016/j.ijantimicag.2020.106024 [DOI] [PMC free article] [PubMed]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson, Mats H.M., Søndergaard, Chresten R., Rostkowski, Michal, Jensen, Jan H., 2011. PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput., 7(2), 525-537. doi: 10.1021/ct100578z [DOI] [PubMed]
- Robson, B., 2020. COVID-19 Coronavirus spike protein analysis for synthetic vaccines, a peptidomimetic antagonist, and therapeutic drugs, and analysis of a proposed achilles' heel conserved region to minimize probability of escape mutations and drug resistance. Comput. Biology Med., 121, 103749-103749. doi: 10.1016/j.compbiomed.2020.103749 [DOI] [PMC free article] [PubMed]
- Roy S., Bag A.K., Singh R.K., Talmadge J.E., Batra S.K., Datta K. Multifaceted role of neuropilins in the immune system: potential targets for immunotherapy. Front. Immunol. 2017;8:1228. doi: 10.3389/fimmu.2017.01228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler M.J., Wysocki J., Batlle D. ACE2 alterations in kidney disease. Nephrol. Dial. Transplant. 2013;28(11):2687–2697. doi: 10.1093/ndt/gft320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajda Sandor, Yueh Christine, Beglov Dmitri, Bohnuud Tanggis, Mottarella Scott E., Xia Bing.…Kozakov Dima. New additions to the ClusPro server motivated by CAPRI. Proteins. 2017;85(3):435–444. doi: 10.1002/prot.25219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W.…Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]