Abstract
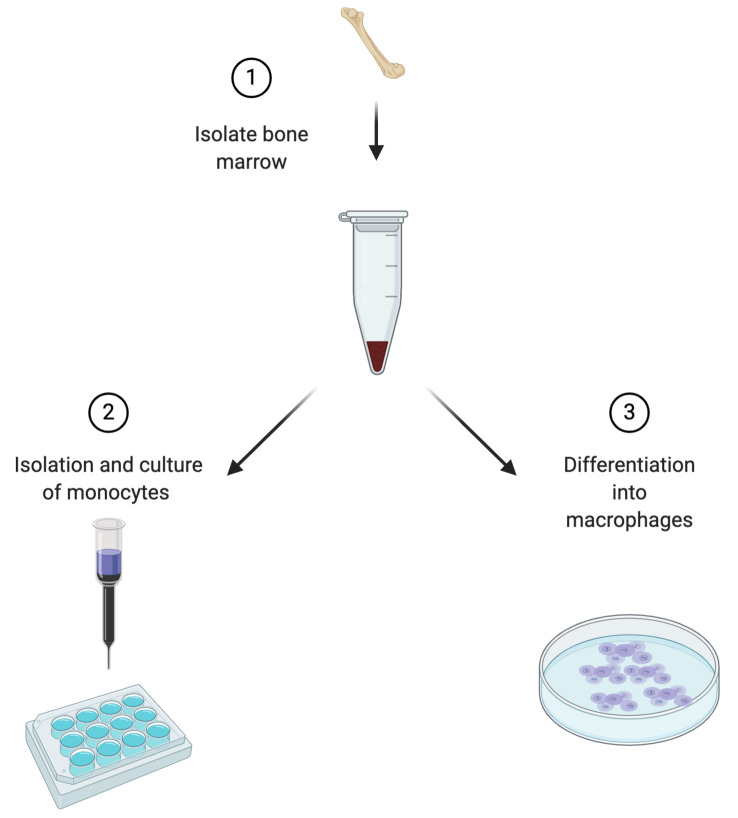
Myeloid progenitors in the bone marrow generate monocytes, macrophages, granulocytes and most dendritic cells. Even though these innate immune cells are part of the same lineage, each cell type plays a specific and critical role in tissue development, host defense and the generation of adaptive immunity. Protocols have been developed in the past to differentiate myeloid cell types from bone marrow cells, enabling functional investigation and furthering our understanding about their contribution to mammalian physiology. In this protocol, we describe a simple and rapid method to isolate monocytes from murine bone marrow, culture them for up to 5 days and lastly, differentiate them into bone marrow derived macrophages (Figure 1).
Graphic abstract:
Figure 1. Experimental outline depicting steps for murine monocyte and macrophage culture.
Keywords: Bone marrow isolation, Monocyte culture, BMDMs, Macrophage differentiation, Monocyte differentiation
Background
Innate immune cells within the hematopoietic compartment constitutes myeloid cells such as monocytes, macrophages, granulocytes as well as most dendritic cells (DCs). These cells are primary sensors of infection or tissue injury and provide a bridge to adaptive immunity via antigen presentation, lymphocyte priming, and cytokine production.
Monocytes exhibit remarkable plasticity in their capacity to patrol tissue, remodel local microenvironments and present antigens under specific conditions. These cells can differentiate into DC or macrophage subsets upon sustained tissue residence (Davies and Gordon, 2005). Macrophages, on the other hand, develop from yolk sac-derived or hematopoietic progenitors or monocytes and are critical for organ development and homeostasis ( Francke et al., 2011 ).
To study myeloid cell biology in primary cells, researchers have developed several protocols to either isolate monocytes or macrophages directly from primary tissue or generate bone marrow derived macrophages (BMDMs) from hematopoietic progenitors. BMDMs can be maintained in culture for several days and can be generated by culture of isolated bone marrow in media containing M-CSF/CSF1 ( Freund et al., 2020 ; Geissmann et al., 2010 ; Helft et al., 2015 ). Dendritic cell (DC) subsets can be generated in culture by differentiation in media containing GM-CSF/CSF2 or Fms-like tyrosine kinase 3 ligand (FLT3L). While both systems generated a mixed population of myeloid and DC subsets, FLT3L allows for differentiation of multiple physiologically relevant subsets such as plasmacytoid DCs (pDCs), CD8+ and CD8- DCs ( Jakubzick et al., 2017 ; Naik et al., 2005 ).
Our protocol describes the isolation of monocytes from the bone marrow and provides methods for culture and maintenance of these cells for several days. Additionally, we describe a rapid method to isolate total bone marrow from isolated tibia and femur, allowing for maximum cell recovery and survival. Our techniques have the advantages of circumventing the need of generating specific conditioned media for BMDM culture (e.g., from L929 cells) and shortcut the lengthy process of harvesting bone marrow through aspiration or flushing.
Taken together, these protocols allow for fast and simple isolation of bone marrow and provide the basis for growing primary monocytes and macrophages in high yields, which might be desired for downstream studies such as gene editing, functional genomics or adoptive cell transfer ( Zhang et al., 2008 ).
Materials and Reagents
Cell scraper (Corning/VWR, catalog number: 15621-010)
70 µm nylon sterile cell strainer (Corning, catalog number: 352350)
G18 needle (BD Bioscience, catalog number: 305196)
0.5 ml microfuge tube (Eppendorf, catalog number: 022363611)
1.5 ml microfuge tube (Eppendorf, catalog number: 022363204)
50 ml Falcon tube, screw top (Eppendorf, catalog number: 0030122178)
LC columns (Miltenyi, catalog number: 130-042-401)
Non-tissue culture treated 6-well plates (Corning, catalog number: 351146)
Serological pipettes (e.g., Fisher scientific catalog number: 10710810)
Low adherent plates (Petri-dish) (VWR, catalog number: 25384-326)
DMEM high glucose media (Gibco, catalog number: 11965092)
(Optional) EasySepTM (Stemcell Technologies, catalog number: 19861)
Penicillin/Streptomycin (Gibco, catalog number: 15070063)
Recombinant murine Macrophage Colony Stimulating Factor (rmM-CSF) (Genentech)
GlutaMAX (Gibco, catalog number: 35050061)
Fetal Calf Serum (FCS) (Thermo Fisher, catalog number: 26140)
Ammonium Chloride Potassium (ACK) lysis buffer (Thermo Fisher, catalog number: A1049201)
Sterile Phosphate Buffered Saline (PBS) (Genentech)
Monocyte isolation kit (Miltenyi, catalog number: 130-100-629)
Recombinant murine Granulocyte Macrophage Colony Stimulating Factor (rmGM-CSF) (R&D Systems, catalog number: 415-ML010)
Recombinant murine Interleukin 4 (rmIL-4) (R&D Systems, catalog number: 404-ML-010)
Macrophage media (see Recipes)
Equipment
Pipette boy (e.g., Integra, catalog number: 155017)
Tabletop centrifuge (e.g., Beckman, model: Allegra X14)
Microcentrifuge (e.g., Thermo Fisher Scientific, catalog number: 75002404)
Water-bath at 37 °C (Thermo Fisher Scientific, catalog number: TSCIP19)
Laminar Flow Hood (VWR)
Incubator at 37 °C with 5% CO2 (e.g., Heracell 150 CO2 incubator)
Vi-CELL XR Cell Viability Analyzer (Beckmann)
MACS Multi Stand magnet (Miltenyi, catalog number: 130-042-303)
Sterile Scissors (Thermo Fisher Scientific, catalog number: 08-950)
Sterile Forceps (Thermo Fisher Scientific, catalog number: 08-890)
Procedure
-
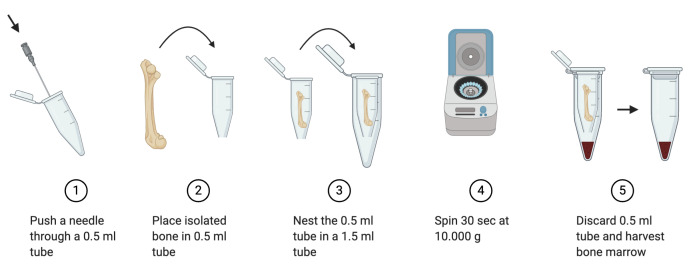
Bone marrow isolation (Figure 2)
Euthanize mouse using institutionally approved protocols.
Use sterile forceps and scissors to isolate femur and tibia.
Remove residual muscle or tissue attached to the bones.
Store isolated bones in a 6-well plated in PBS on ice until further processing.
Push a G 18 needle through the bottom of a 0.5 ml microcentrifuge tube to generate a small hole.
Place the 0.5 ml microcentrifuge tube in a larger 1.5 ml microcentrifuge tube.
Carefully place the isolated femur and tibia (maximum 1 femur and 2 tibia) in the 0.5 ml microcentrifuge tube nested in the 1.5 ml tube and close the lid.
Centrifuge the tubes at 10,000 × g for 30 s.
After centrifugation confirm complete transfer of marrow into the bottom tube. If transfer was not successful, cut off epiphysis and repeat centrifugation step.
Discard the 0.5 ml microcentrifuge tube containing the bones. They should be empty of marrow.
Resuspend the obtained bone marrow in 1 ml ACK lysis buffer and incubate at room temperature for 2 min.
Place a 0.70 µm sterile filter on a 50 ml Falcon tube.
Filter the bone marrow suspension into tube and wash the filter with 10 ml PBS.
Add an additional volume of 30 ml PBS through the filter; discard the filter.
Close the lid on the tube and centrifuge at 350 × g for 4 min.
Remove PBS by gently decanting; retain bone marrow pellet.
If desired, repeat PBS wash by gently resuspending the pellet.
Count isolated bone marrow cells using a Vi-CELL or other standard method.
-
Isolation and culture of monocytes
Following bone marrow isolation, monocytes are enriched by negative selection using the monocyte isolation kit (Miltenyi) according to manufacturer’s instructions. Please note that other mouse monocyte isolation kits can be used for this step (e.g., EasySepTM, Stemcell Technologies); however, these have not been evaluated for this specific protocol.
Isolated monocytes can be cultured in macrophage media supplemented with 5 ng/ml rmGM-CSF and 2.5 ng/ml rmIL-4 for up to 5 days. Monocytes should be plated at a density of 1 × 106 cells in 2 ml final volume in a non-tissue culture treated 6-well plate.
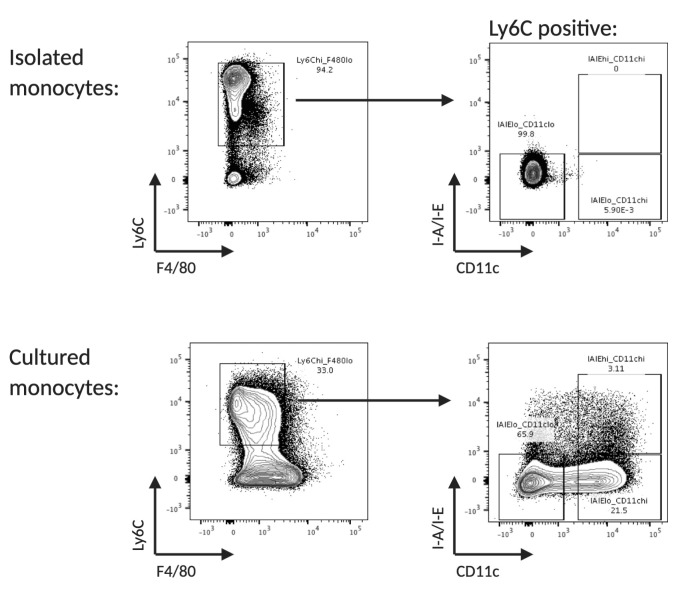
If desired, monocytes can be enriched from the day 5 culture by sorting for Ly6C positive, F4/80 negative, MHCII negative and CD11c negative cells at day 5 (Figure 3).
-
Differentiation into bone marrow derived macrophages (BMDMs)
Supplement macrophage media with 50 ng/ml rmM-CSF and pre-warm in a water bath to 37 °C.
Suspend 12 × 106 isolated total bone marrow cells (from Procedure A) or enriched monocytes (Procedure B) in 20 ml supplemented macrophage media and plate cells in 15 cm non-tissue culture treated plates.
Place dish in cell culture incubator at 37 °C and 5 % CO2 and culture for a total of 5 days.
At day 3 add 20 ml freshly supplemented macrophage media without removal of the original media.
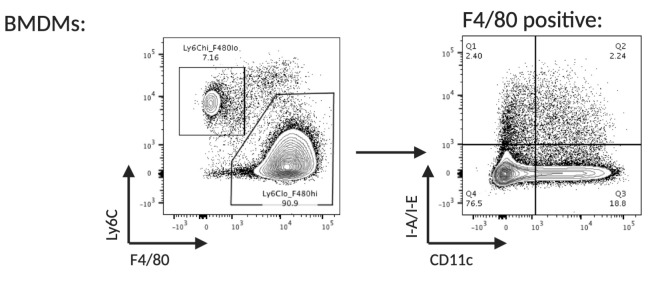
At day 5 confirm adherence of differentiated BMDMs under a microscope. Assess differentiation by flow cytometry (Figure 4).
For downstream assays, remove media and gently wash day 5 cells in 20 ml sterile PBS. Carefully scrape cells off the dish using a cell scraper in 5 ml macrophage media. If desired, wash the dish with an additional 5 ml of macrophage media to collect additional cells.
Count collected cells and re-suspend in macrophage media supplemented with 50 ng/ml rmM-CSF at desired cell density (typical density is 0.5 × 106 cells/ml). Plate cells onto tissue-culture treated dishes of desired formats.
If desired, resting macrophages can be enriched from the day 5 culture by sorting for Ly6C low, F4/80 high, CD11c low and I-A/I-E low.
Figure 2. Experimental outline depicting bone marrow isolation steps.
Figure 3. Flow cytometry analysis of freshly isolated and cultured monocytes.
Example of surface marker expression of Ly6C, F4/80, CD11b and I-A/I-E of freshly isolated and cultured monocytes. Monocytes are identified as Ly6C high, F4/80 low, MHCII low and CD11c low.
Figure 4. Flow cytometry analysis of monocytes differentiated into bone marrow derived macrophages (day 5).
Differentiated, resting macrophages can be identified as Ly6C low, F4/80 high, CD11c low and I-A/I-E low.
Recipes
-
Macrophage medium
DMEM high glucose media
10% FBS
1× GlutaMAX
1× Penicillin/Streptomycin
Acknowledgments
This study was funded by Genentech, Inc.
References 8 and 9 have utilized the procedure for murine BMDM isolation and culture.
Competing interests
SMH and AM are employees of Genentech, Inc. AM is a shareholder in Roche.
Ethics
All mice and experiments in this study were conducted following protocols approved by Genentech Institutional Animal Care and Use Committee (Protocol ID TH19-0370; valid until 02/14/2022).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Jakubzick C. V., Randolph G. J. and Henson P. M.(2017). Monocyte differentiation and antigen-presenting functions. Nat Rev Immunol 17(6): 349-362. [DOI] [PubMed] [Google Scholar]
- 2. Geissmann F., Manz M. G., Jung S., Sieweke M. H., Merad M. and Ley K.(2010). Development of monocytes, macrophages, and dendritic cells. Science 327(5966): 656-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davies J. Q. and Gordon S.(2005). Isolation and culture of murine macrophages. Methods Mol Biol 290: 91-103. [DOI] [PubMed] [Google Scholar]
- 4. Francke A., Herold J., Weinert S., Strasser R. H. and Braun-Dullaeus R. C.(2011). Generation of mature murine monocytes from heterogeneous bone marrow and description of their properties. J Histochem Cytochem 59(9): 813-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang X., Goncalves R. and Mosser D. M.(2008). The isolation and characterization of murine macrophages. Curr Protoc Immunol Chapter 14: Unit 14 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Helft J., Böttcher J., Chakravarty P., Zelenay S., Huotari J., Schraml B. U., Goubau D. and Reis e Sousa C.(2015). GM-CSF Mouse Bone Marrow Cultures Comprise a Heterogeneous Population of CD11c+MHCII+ Macrophages and Dendritic Cells . Immunity 42(6): 1197-1211. [DOI] [PubMed] [Google Scholar]
- 7. Naik S. H., Proietto A. I., Wilson N. S., Dakic A., Schnorrer P., Fuchsberger M., Lahoud M. H., O'Keeffe M., Shao Q. X., Chen W. F., Villadangos J. A., Shortman K. and Wu L.(2005). Cutting edge: generation of splenic CD8+ and CD8- dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures . J Immunol 174(11): 6592-6597. [DOI] [PubMed] [Google Scholar]
- 8. Freund E. C., Lock J. Y., Oh J., Maculins T., Delamarre L., Bohlen C. J., Haley B. and Murthy A.(2020). Efficient gene knockout in primary human and murine myeloid cells by non-viral delivery of CRISPR-Cas9. J Exp Med 217(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lim J., Park H., Heisler J., Maculins T., Roose-Girma M., Xu M., Mckenzie B., van Lookeren Campagne M., Newton K., and Murthy A.(2019). Autophagy regulates inflammatory programmed cell death via turnover of RHIM-domain proteins. Elife 8: e44452. [DOI] [PMC free article] [PubMed] [Google Scholar]