Abstract
The mRNA therapeutics is a new class of medicine to treat many various diseases. However, in vitro transcribed (IVT) mRNA triggers immune responses due to recognition by human endosomal and cytoplasmic RNA sensors, but incorporation of modified nucleosides have been shown to reduce such responses. Therefore, an assay signifying important aspects of the human immune system is still required. Here, we present a simple ex vivo method called ‘RNA ImmunoGenic Assay’ to measure immunogenicity of IVT-mRNAs in human whole blood. Chemically modified and unmodified mRNA are complexed with a transfection reagent (TransIT), and co-incubated in human whole blood. Specific cytokines are measured (TNF-α, INF-α, INF-γ, IL-6 and IL-12p70) using ELISAs. The qPCR analysis is performed to reveal the activation of specific immune pathways. The RNA ImmunoGenic Assay provides a simple and fast method to detect donor specific - immune response against mRNA therapeutics.
Graphic abstract:
Schematic representation of RNA ImmunoGenic Assay
Keywords: cmRNA, IVT-mRNA, Immunogenicity, Cytokines, Whole blood assay
Background
The mRNA therapeutics is an important class of gene therapy ( Sahin et al., 2014 ; Antony et al., 2015 ). Although no mRNA drugs are marketed, the efficacy demonstrated in ongoing clinical trials is remarkable. The in vitro transcribed mRNA induces immune responses due to its recognition by human endosomal and cytoplasmic RNA sensors including Toll-like receptors (TLRs), RIG-I like receptors (RLRs), and NOD-like receptors (NLRs). Therefore, the assessment of immunogenicity of mRNA therapeutics is pivotal for their possible use in humans.
Existing cell-based methods have been reported to study immunogenicity of IVT-mRNA including peripheral blood mononuclear cells (PBMCs) based assays, dendritic cells-based assays and THP-1 cell-based assays ( Kariko et al., 2011 ; Vaidyanathan et al., 2018 ). However, these assays are associated with laborious ficoll isolation together with isolation related immune cell activation, and artificial mono-immune cell systems that do not imitate the complexity of human immune cells. Therefore, a fast method that mimics the human immune system is desirable. Here, we report an adapted version of whole blood assay (WBA) as an ‘RNA ImmunoGenic Assay’ which is simple and fast method and eliminates the hurdles associated with PBMCs isolation and DCs enrichment ( Haque et al., 2020 ). Interestingly, the assay utilizes human whole blood and it provides the most physiologically relevant immune cell types, especially platelets, and white and red blood cells (RBCs) in an easily accessible ex vivo context.
The proposed assay is capable of sensing different immune responses caused by IVT-mRNA administration such as NF-kB activation and interferon stimulation. Notably, results of ‘RNA ImmunoGenic assay’ using whole blood can be compared to PBMC based assay. Since the assay requires small volumes of whole blood (< 10 ml), it is suitable for pediatric patients and other patients with severe illnesses. Moreover, the RNA ImmunoGenic assay is capable to detect personalized immune responses to specific mRNA compounds with or without formulation. Together, the presented RNA ImmunoGenic assay is easy to complete, fast to perform, and provides measurable immunogenicity results to be used in a clinical setting.
Materials and Reagents
-
IVT mRNA production
Petri dish (Greiner Bio-One, catalog number: 633161)
Rattler plating beads (5 mm) (Zymo Research, catalog number: S1001, store at RT)
One ShotTM TOP10 chemical competent E. coli (Thermo Fisher, catalog number: C404003, store at -80 °C)
pVAX1.A120 vector based on pVAX1 (Life Technologies, catalog number: V26020, store at -20 °C)
XhoI (NEB, catalog number: R0146L), store at -20 °C
KpnI (NEB, catalog number: R0142L), store at -20 °C
NheI (NEB, catalog number: R0131L), store at -20 °C
NEBufferTM
1x CutSmart® Buffer (provided with the appropriate enzyme) , store at -20 °C
1x NEBufferTM 1.1 (provided with the appropriate enzyme), store at -20 °C
1x NEBufferTM 2.1 (provided with the appropriate enzyme)
T4 DNA ligase (NEB, catalog number: M0202L), store at -20 °C
1x T4 DNA Ligase Reaction Buffer (provided with the appropriate enzyme), store at -20 °C
S.O.C. medium (Thermo Fisher, catalog number: 15544034), store at RT
MEGAscriptTM T7 Transcription kit (Invitrogen, catalog number: AM1334), store at -20 °C
m27,3'-OGP3G (ARCA Cap Analog)–Solution (Jena Bioscience, catalog number: NU-855L), store at -20 °C
Modified Base 1, e.g., N1-Methylpseudo-UTP (Jena Bioscience, catalog number: NU-890L), store at -20 °C
Modified Base 1, e.g., 2-Thio-UTP (Jena Bioscience, catalog number: NU-1151L), store at -20 °C
MEGAclearTM Kit (Invitrogen, catalog number: AM1908), store at 4 °C
TURBOTM DNase (Thermo Fisher, catalog number: AM2239), store at -20 °C
Agilent RNA 6000 Nano Kit (Agilent, catalog number: 5067-1511), store reagents at 4 °C, chips at RT
Luria Broth Base (Miller's LB Broth Base) (Thermo Fisher, catalog number: 12795027), store at RT
Select Agar (Thermo Fisher, catalog number: 30391023), store at RT
Kanamycin Sulfate (Thermo Fisher, catalog number: 11815024), store at RT
GeneJET PCR Purification kit (Thermo Fisher, catalog number: K0702), store at RT
-
Blood Collection and ex vivo stimulation
12-well plate (Corning, catalog number: 3512)
EDTA containing vacutainer tubes (Sarstedt, catalog number: 02.1066.001)
Precellys bacterial/fungal RNA kit (PeqLab, catalog number: 12-7611-01, store at RT)
Precellys Tissue homogenizing CKMix–2 ml (Bertin Instruments, catalog number: P000918-LYSK0-A), store at RT
Resiquimod (R848) (Sigma-Aldrich, catalog number: SML0196), store at -20 °C
TransIT®-mRNA Transfection Kit (Mirus, catalog number: MIR 2225), store at 4 °C
-
Measurement of cytokines by ELISA
96-well plate (provided with the ELISA kit)
Absorbent paper
Acetat Foil for Microtesting (Sarstedt, catalog number: 82.1586)
Capture Antibody (provided with the ELISA kit)
Biotinylated Detection Ab (provided with the ELISA kit)
Washing buffer
DPBS, no calcium, no magnesium (Thermo Fisher, catalog number: 14190094), store at RT
TWEEN® 20 (Sigma-Aldrich, catalog number: P9416-50ML), store at RT
HRP Conjugate working solution (provided with the ELISA kit)
Substrate Reagent (provided with the ELISA kit)
TNF-alpha Human Uncoated ELISA Kit with Plates (Thermo Fisher, catalog number: 88-7346-22), store at 4 °C
InvitrogenTM eBioscienceTM Platin-ELISA-Kit for human IFN-alpha (Fisher Scientific, catalog number: 15561697), store at 4 °C
IFN gamma Human Uncoated ELISA Kit with Plates (Thermo Fisher, catalog number: 88-7316-22), store at 4 °C
IL-6 Human Uncoated ELISA Kit with Plates (Thermo Fisher, catalog number: 88-7066-22), store at 4 °C
IL-12 p70 Human Uncoated ELISA Kit with Plates (Thermo Fisher, catalog number: 88-7126-22), store at 4 °C
2 N HCl (Stop solution) (Carl Roth, catalog number: T134.1), store at RT
-
Real-time RT-PCR
PAXgene 96 Blood RNA Kit (Qiagen, catalog number: 762331), store at RT
iScriptTM cDNA Synthesis Kit (Bio-Rad, catalog number: 1708891), store at -20 °C
Power SYBR Green PCR Master mix (Thermo Fisher, catalog number: 4367659), store at -20 °C
Erythrocyte Lysis Buffer (PAN Biotech, Catalog number: P10-90100), store at RT
-
Primers:
hTNFAlpha forward: 5′- GGA GAA GGG TGA CCG ACT CA-3′
hTNFAlpha reverse: 5′-CTG CCC AGA CTC GGC AA-3′
hIFNAlpha forward: 5′-GAC TCC ATC TTG GCT GTG A-3′
hIFNAlpha reverse: 5′-TGA TTT CTG CTC TGA CAA CCT-3′
hIFNgamma forward: 5′-CCA ACG CAA AGC AAT ACA TGA-3′
hIFNgamma reverse: 5′-CCT TTT TCG CTT CCC TGT TTT A-3′
hIL-12 forward: 5′- CAA GAA CTT GCA GCT GAA G-3′
hIL-12 reverse: 5′- TGG GTC TAT TCC GTT GTG TC-3′
IL‐6 forward: 5′-AGACAGCCACTCACCTCTTCAG-3′
IL-6 reverse: 5′-TTCTGCCAGTGCCTCTTTGCTG-3′
IRF7 forward: 5′-CCACGCTATACCATCTACCTGG-3′
IRF7 reverse: 5′-GCTGCTATCCAGGGAAGACACA-3′
IRF3 forward: 5′-TCTGCCCTCAACCGCAAAGAAG-3′
IRF3 reverse: 5′-TACTGCCTCCACCATTGGTGTC-3′
eIF 2A forward: 5′-CTGGACCTCATGCAGCTTTAGCm-3′
eIF 2A reverse: 5′-CTCCATAGTAGGAAGCTCCTGTC -3′
RNase L forward: 5′-AAGGCTGTTCAAGAACTACACTTG-3′
RNase L reverse: 5′-TGGATCTCCAGCCCACTTGATG-3′
-
Flow cytometry analysis
DBPS, no calcium, no magnesium (Thermo Fisher, catalog number: 14190094), store at RT
10% FBS; FBS Superior (Sigma-Aldrich, catalog number: S0615), store at -20 °C
Erythrocyte Lysis Buffer (PAN Biotech, Catalog number: P10-90100), store at RT
Equipment
ThermoShaker Univortemp MT 100 (Universal Labortechnik)
Centrifuge 5430R (Eppendorf, catalog number: 5427000015)
2100 Bioanalyzer Instrument (Agilent, catalog number: G2939BA)
2100 Bioanalyzer Laptop (Agilent Original Bundle PC, catalog number: G2953CA)
Cell culture hood HeraSafe KS (Thermo Fisher, catalog number: 10110910)
CO2 incubator HeraCell VIOS 160i (Thermo Fisher, catalog number: 15381075)
Precellys evolution homogenizer (Bertin Technologies, catalog number: P000062-PEVO0-A)
EnsightTM multimode plate reader (Perkin Elmer, catalog number: HH34000000)
Flow Cytometer, BD LSRFortessaTM X20 (BD Biosciences)
ViiA 7 Real-Time PCR System (Thermo Fisher, catalog number: 4453545)
Software
2100 Expert for Bioanalyzer (Agilent)
Graphpad Prism v.6.0d software (www.graphpad.com)
KaleidoTM 2.0 Data Acquisition and Analysis software for EnsightTM plate reader (PerkinElmer)
BD FACSDivaTM Software for Flow Cytometry Analysis (BD Biosciences)
ViiA7 software v.1.2 (Applied Biosystems by Life Technologies)
NEBioCalculator v.1.12.0 (New England Biolabs)
Procedure
-
IVT mRNA production
-
Gene cloning
-
PCR amplify open reading frame of the gene of interest (hCFTR/ZFN/Cas9/RFP) from pcDNA3 (50 ng) using 2x AmpliTaq Gold mastermix (25 µl) with each primers adding (1 µl each, 1.0 µM) NheI (Fwd: 5′-TTAGCTAGATGCAGAGGTCGCCTC-3′) and KpnI (Rev: 5′-GCGGGTACCTATCTTGCATCTCTTCT-3′) restriction site to each end respectively in 50 µl reaction volume in 8 different reactions.
Holding: 95 °C for 5 min
-
40 cycle of
Denaturing 95 °C 15 s
Annealing 60 °C 30 s
Extending 72 °C for 5 min (1 min/Kb)
Final extension 72 °C for 7 min.
Pool the eight different PCR reactions and verify the amplification for theoretical size in agarose gel electrophoresis and purify the PCR product using GeneJET PCR Purification kit following manufacturer instructions.
-
Clone the PCR product into polyA-120 containing pVAX (pVAX.A120) by sticky-end ligation in below steps.
Perform double digestion of both PCR (5 µg) product and pVAX.A120 (10 µg) with KpnI and NheI (Reaction Condition in 50 µl volume-10x NEBufferTM 1.1 (5 µl), 10 Units of KpnI and NheI) and incubate at 37 °C overnight. Verify the digestion of pVAX.A120 vector in agarose gel electrophoresis and purify both digested PCR product and digested pVAX.A120 vector backbone using GeneJET PCR Purification kit following manufacturer instructions.
For optimal ligation, the concentration of digested pVAX.A120 vector backbone and digested PCR product from gene of interest are calculated in NEBiocalculator to achieve 3:1 of insert:vector ratio. The calculated amount of insert and vector are added to the 20 µl ligation mix consisting of 20 Units of of T4 DNA ligase and 2 µl 10x DNA ligase buffer. Incubate the ligation complex at 22 °C for 10 min.
-
Cell transformation
Use 50 µl Top10 E. coli competent cells and thaw it on ice.
Chill approximately 10 µl of the ligation mixture.
Add 50 µl of competent cells to the 4 µl of ligation mixture and mix gently. Do not exceed 10% volume of the competent cells, i.e., not more than 5 µl of ligation mixture.
Incubate the complex on ice for 30 min.
Heat shock at 42 °C for 45 s.
Add 950 µl of SOC media stored at room temperature.
Place tube at 37 °C for 60 min. Shake vigorously at 250 rpm.
Warm selection plates (Kanamycin for pVAX.A120) at 37 °C.
Spin the incubated SOC media at 2,650 x g for 10 min.
Carefully discard 900 µl without disturbing the pellet and resuspend the same.
Spread 100 µl of the cells and ligation mixture onto the plates.
Incubate overnight at 37 °C.
-
-
mRNA production
Linearize 10 µg of pVAX.A120 containing the open reading frame of interested gene using XhoI enzyme [10 units (1 µl) with10x CutsmartTM buffer (5 µl) in 50 µl reaction volume and incubate at 37 °C overnight). Verify the linearization of pVAX.A120 vector with desired gene in agarose gel electrophoresis and purify it using GeneJET PCR Purification kit following manufacturer instructions.
Use the linearized product to produce mRNA using MEGAscript kit (Use 1 µg Linear template DNA and anti-reverse CAP analog (ARCA; [m7G(5’)G])). The reaction volume is 20 µl (1x). For co-transcriptional capping of ARCA, the concertation of GTP is reduced to 5 mM instead of 10 mM as with other nucleosides. To increase the chance of ARCA being first base to be transcribed, 6 mM of ARCA is added to the reaction.
Choose the bases (ATP, CTP, GTP and UTP) for unmodified mRNA production.
The chemically modified nucleosides (Example: 100% Pseudo-UTP, 100% N1-methylpseudo-UTP, 25% s2-thio-UTP/5-methyl-CTP and 100% N1-methylpseudo-UTP/100% 5-methyl-CTP) are used to generate chemically modified mRNA. Table 1 shows an example to generate both modified and unmodified mRNA.
Mix the solution by flicking the tube and use parafilm to cover the lid.
Incubate at 37 °C for 2 h.
Add 1 μl turbo DNase and incubate at 37 °C for 15 min.
Clean up the reaction using MEGAclear kit following manufacturer instructions and quantify them using Nanaophotometer.
Dilute specific amount of the purified mRNA (ng)with DEPC water provided in MEGAclear kit in 1:5 ratio to get 100 ng in 1 µl and denature the solution at 95 °C for 2 min.
Load 100 ng (1 µl) it in Agilent 2100 Bioanalyzer to check the size (identity) and purity.
Store the mRNA at -80 °C until further use.
-
-
Preparation of bacterial total RNA
Grow E. coli cells in 50 ml culture at 37 °C for overnight in LB media.
Collect pellet by spinning at 2,650 x g for 15 min.
Homogenize the bacterial pellet with the Precellys Evolution Homogenizer at 2,650 x g for 20 s using 0.5 mm glass beads tube provided in a Precellys Bacterial/Fungal RNA isolation Kit.
Isolate total RNA from the homogenized pellet using Precellys Bacterial/Fungal RNA isolation Kit.
Store the bacterial RNA at -80 °C until further use.
-
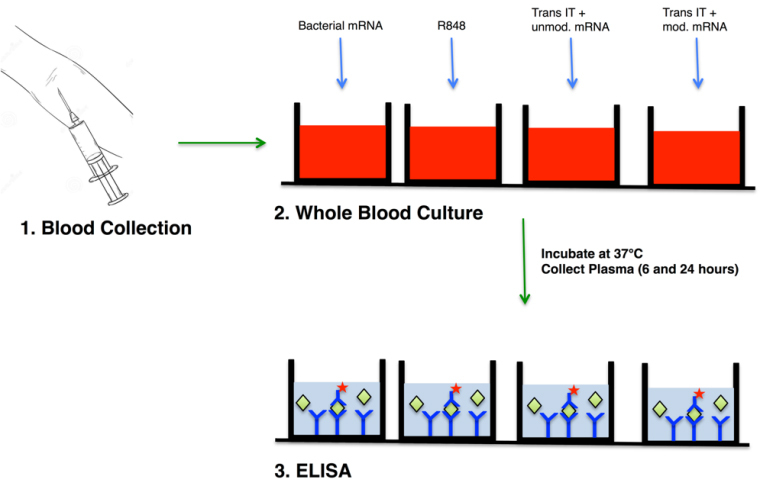
Blood collection and ex vivo stimulation
Day 1
Collect peripheral blood from healthy adult volunteers or patients in EDTA containing vacutainer tubes (all blood has to be used immediately).
Add 2 ml of blood in a well of a 12-well plate.
Add 15 µg of IVT-mRNA or total bacterial RNA/R848 (as positive control) with or without TransIT transfection complex to 2 ml blood and mix it gently.
Incubate the blood for 6 to 24 h at 37 °C in a humidified incubator containing 5% CO2.
After 6 h collect 1 ml of blood and spin it at 425 x g for 10 min at 4 °C.
Collect the separated blood plasma from coagulated blood. The PBMCs are isolated from rest part of the blood after erythrocyte lysis followed by centrifugation 500 x g for 10 min at 4 °C .
Store the plasma at -20 °C. PBMCs are processed into RNA isolation for qPCR analysis.
Day 2
After 24 h collect 1 ml remaining blood and spin it at 425 x g for 10 min.
Collect the separated blood plasma from coagulated blood.
Store it at -20 °C.
-
Measurement of cytokines by ELISA (of TNF-α, INF-α, INF-γ, IL-6 and IL-12p70)
Thaw the plasma on ice.
Use 50 µl of collected plasma and dilute it in the appropriate diluent for ELISA kit.
Load the diluted plasma on a pre coated 96-well plate.
Load the appropriate standards made by serial dilution (1:2 dilution).
Incubate for 2 h at 37 °C or overnight at 4 °C.
Add biotinylated detection antibody (prepared following instruction for specific ELISA).
Wash the plate 5 times with wash buffer (appropriate for specific ELISA and included in the kit). Otherwise, wash buffer consists of PBS and 0.05% Tween-20. Make sure for each wash all the wells are covered with wash buffer and soaked for 1 min.
Decant the wash buffer and dry the wells using absorbent paper.
Add HRP conjugate working solution (prepared following instruction for specific ELISA) to each well and incubate for 30 min at 37 °C.
Wash the plate 7 times with washing buffer (appropriate for specific ELISA). Make sure for each wash all the wells are covered with wash buffer and soaked for 1 min.
Decant the wash buffer and dry the wells using absorbent paper.
Add 90 μl of Substrate Reagent to each well. Cover with a new plate sealer. Incubate for about 15 min at 37 °C in the dark.
Add 50 µl stop solution and determine the optical density of each well using a plate reader at the specific wavelength (normally 450 nm).
Cytokine levels are assessed by interpolating unknown values from standard curve using the Michaelis-Menten model in the Graphpad Prism v.6.0 software (www.graphpad.com).
-
Real-time RT-PCR
-
Collect total RNA
For RT-PCR collect and stimulate the blood as mentioned above.
Instead of collecting plasma after 6 h and 24 h use the blood to collect total RNA.
Collect total RNA from blood using PAXgene Blood RNA kit.
-
Check the concentration and purity of total RNA (RNA integrity number-RIN score should be above five).
Note: For qPCR application RIN score above 5 (partially degraded RNA)works moderately well. However for RNA-seq application RIN score above 7 (pure RNA) is needed.
Store the RNA -80 °C.
-
cDNA synthesis
-
Use 200 ng total RNA to perform cDNA synthesis using iScriptTM cDNA kit.
Priming: 25 °C for 7 min.
Reverse Transcription: 45 °C for 20 min.
RT inactivation: 95 °C for 2 min.
Dilute the cDNA 1:20.
Store at -20 °C.
-
-
SYBR-Green based quantitative Real-time PCR
Use 5 µl of diluted cDNA with 2x Power-SYBR green PCR master mix (7.5 µl) and both forward and reverse primer (1 µl each, 1.0 µM) in 15 µl reaction volume.
-
Use the following program in ViiA7.
Initial denaturation for 10 min at 95 °C.
40 cycles of 15 s at 95 °C and 2 min at 60 °C (annealing and extension).
Collect and analyze melting curve and Ct values.
-
-
Flow cytometry analysis of DsRed transfected blood samples
In order to assess whether TransIT complexed DsRed mRNA transfected any cells in whole blood, stimulate the blood as mentioned above.
Instead of collecting plasma or total RNA after 6 h and 24 h use the whole blood for flow cytometry.
Add 2 ml of Erythrocyte lysis buffer to 2 ml whole blood mix well.
Incubate at room temperature for 10 min (Do not exceed the time of incubation).
Spin the tube at 500 x g for 5 min.
Decant the supernatant.
Wash the cells with 1 ml PBS.
Spin it at 500 x g for 5 min and remove PBS and resuspend the pellet at FACS buffer (10% FBS in PBS).
-
Measure DsRed expression using appropriate laser and red channel (558 ex/583 em for DsRed).
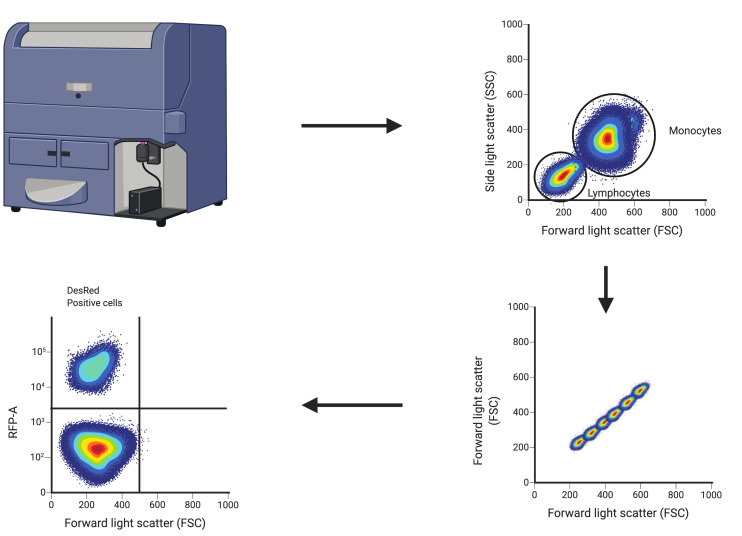
The gating strategy to detect DsRed positive cells illustrated in Figure 1.
-
Count minimum of 50,000 gated cells per tube with flow rate of 100 cells per second.
The RNA ImmunoGeneic Assay protocol uses R848 (Resiquimod) TLR7/8 agonist as a positive control that mimic the endosomal TLR7/8 activation upon RNA endosomal entry. The blood only sample will serve as a background control whereas TransIT treated samples will be the appropriate negative control.
Table 1. 1x unmodified or modified mRNA.
| Reagents | Volume (µl) | Stock (mM) | Final concentration (mM) |
|---|---|---|---|
| ATP | 2 | 100 | 10 |
| CTP/Modified CTP | 2 | 100 | 10 |
| UTP/Modified UTP | 2 | 100 | 10 |
| GTP | 1 | 100 | 5 |
| ARCA | 1.2 | 100 | 6 |
| 10x reaction buffer | 2 | ||
| Plasmid DNA | XX.X | ||
| DEPC-Water | XX.X | ||
| Enzyme | 5.8 | ||
| Total-volume | 20 |
Figure 1. Gating strategy of whole blood cells.
Post Erythrocyte lysis buffer treatment and isolation of white blood cells, the cells were subjected to flow cytometry analysis and analyzed. The gating strategies are detailed in the above figure. Briefly, singlets were gated based on their location in the FSC/SSC plot. From viable cells, lymphocytes and monocytes were gated based on their size and granularity in the FSC/SSC plot. DsRed positive cells were gated from the cell pool choosing RFP channel.
Data analysis
Cytokine levels are assessed by interpolating unknown values from standard curve using the Michaelis–Menten model in the Graphpad Prism v.6.0 software (www.graphpad.com). Mann-Whitney U rank sum tests orKruskal-Wallis test (one-way ANOVA) are applied to analyze differences in cytokine levels between or among the IVT-mRNA modifications in appropriate places. The relative quantification of each target genes in technical triplicates per sample referenced to non-stimulated whole blood is used to detect specific immune response of IVT mRNA using in ViiA7 software v1.2. The similarity in several cytokine expressions in same experimental set-up is tested by the correlation analyses between different cytokines levels by non-parametric Spearman’s rank coefficient tested as implemented in Graphpad Prism v.6.0d (www.graphpad.com). Flow cytometry data are analyzed with BD FACSDivaTM software. For detailed data please refer to the original article ( Haque et al., 2020 ).
Acknowledgments
This work was supported by an ERC Starting Grant (to MSDK, No. 637752) and HMZ Private Foundation (to MSDK). JSA was financially supported by UKT-fortüne grant (to JSA, No. 2485-0-0) MM was supported by a UKT-fortüne grant (to MM, No. 2412-0-0). The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Wiley press and the journal of Advances in Cell and Gene therapy for permission to submit the protocol of the original article of RNA immunoGenic Assay in Bio-protocol ( Haque et al., 2020 ). We thank all voluntary blood donors for our study.
Competing interests
MSDK holds a patent on RNA modification licensed to the biopharmaceutical company, Ethris GmbH (EP2459231B1), and is listed as main inventor on a patent application related to Nuclease encoding modified mRNA. MSDK, AH, and JSA hold a European patent on delivery of chemically modified mRNA complexed to nanoparticles for the treatment of pulmonary diseases (EP3398963A1).
Ethics
Human blood samples were collected from healthy volunteers and two adult cystic fibrosis patients. Written informed consent was obtained from all participants. Ethical approval was obtained from institutional review board, Children’s University Hospital Tuebingen, Germany (No: 349/2013BO2 and 829/2016BO2).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Antony J. S., Dewerth A., Haque A., Handgretinger R. and Kormann M. S.(2015). Modified mRNA as a new therapeutic option for pediatric respiratory diseases and hemoglobinopathies. Mol Cell Pediatr 2(1): 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Haque A. A., Weinmann P., Biswas S., Handgretinger R., Mezger M., Kormann M. S. D. and Antony J. S.(2020). RNA ImmunoGenic Assay: Simple method for detecting immunogenicity of in vitro transcribed mRNA. Adv Cell Gene Ther 3(2): e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kariko K., Muramatsu H., Ludwig J. and Weissman D.(2011). Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res 39(21): e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sahin U., Karikó K. and Türeci Ö.(2014). mRNA-based therapeutics— developing a new class of drugs. Nat Rev Drug Discov 13(10): 759-780. [DOI] [PubMed] [Google Scholar]
- 5. Vaidyanathan S., Azizian K. T., Haque A. K. M. A., Henderson J. M., Hendel A., Shore S., Antony J. S., Hogrefe R. I., Kormann M. S. D., Porteus M. H. and McCaffrey A. P.(2018). Uridine Depletion and Chemical Modification Increase Cas9 mRNA Activity and Reduce Immunogenicity without HPLC Purification. Mol Ther Nucleic Acids 12: 530-542. [DOI] [PMC free article] [PubMed] [Google Scholar]