Abstract
Efforts to contain the spread of chronic wasting disease (CWD), a fatal, contagious prion disease of cervids, would be aided by the availability of additional diagnostic tools. RT-QuIC assays allow ultrasensitive detection of prion seeds in a wide variety of cervid tissues, fluids and excreta. The best documented antemortem diagnostic test involving RT-QuIC analysis targets lymphoid tissue in rectal biopsies. Here we have tested a more easily accessed specimen, ear pinna punches, using an improved RT-QuIC assay involving iron oxide magnetic extraction to detect CWD infections in asymptomatic mule and white-tailed deer. Comparison of multiple parts of the ear pinna indicated that a central punch spanning the auricular nerve provided the most consistent detection of CWD infection. When compared to results obtained from gold-standard retropharyngeal lymph node specimens, our RT-QuIC analyses of ear samples provided apparent diagnostic sensitivity (81%) and specificity (91%) that rivaled, or improved upon, those observed in previous analyses of rectal biopsies using RT-QuIC. These results provide evidence that RT-QuIC analysis of ear pinna punches may be a useful approach to detecting CWD infections in cervids.
Subject terms: Biological techniques, Microbiology, Biomarkers, Diseases of the nervous system, Diseases, Infectious diseases, Neurological disorders
Introduction
Chronic wasting disease (CWD) is a fatal, and contagious prion disease of cervids1. CWD is characterized by a prolonged incubation period, which can vary from 2 to 4 years2, after which animals develop clinical disease and die3. The long incubation period contributes to transmission as asymptomatic animals shed infectious prions in excreta and biological fluids4–7. Susceptible animals can be infected directly, through the contact with saliva, urine, feces or aerosols from an infected animal; or indirectly (environmental contamination), due to the ingestion of infectious prions bound to soil or plants, for instance4,8. CWD is characterized by the accumulation of the pathogenic form of prion protein, termed PrPSc. Following oral ingestion, the spread of PrPSc throughout the body follows a specific pattern, being first identified in the peripheral lymphoreticular system and later in the central nervous system9–11. Because retropharyngeal lymph nodes (RLN) are one of the first sites of prion replication, this tissue, along with a specific portion of the medulla oblongata (the obex), are the gold standard tissues approved for CWD detection by immunohistochemistry and ELISA (http://www.aphis.usda.gov/animal_health/animal_diseases/cwd/downloads/cwd-program-standards.pdf) (for review, see12). However, CWD surveillance and control in wildlife would be aided by the availability of accurate tests applicable to more accessible specimens that could be easily collected from live, as well as dead, cervids.
Toward this goal, ultrasensitive seed amplification assays13, such as real-time quaking-induced conversion (RT-QuIC)14,15, and protein misfolding cyclic amplification (PMCA)16, have been shown to be capable of detecting minute amounts of prions in CWD-infected animals through the analysis of certain peripheral tissues, biological fluids, and excreta4–7,15,17–21. In RT-QuIC reactions, PrPSc that may be present in a sample seeds the refolding and polymerization of the monomeric recombinant PrPC (the substrate), which is in vast excess in the reaction mix13. The resulting amyloid fibrils enhance the fluorescence of thioflavin T which is used to monitor the polymerization process over time. Cycles of shaking and rest accelerate the seeded polymerization reaction. Seed amplifications of a billion-fold or more can be achieved in RT-QuIC assays. Of particular interest with respect to antemortem diagnosis are multiple studies involving hundreds of white-tailed deer22 or elk23,24 showing that RT-QuIC analysis of rectal biopsy specimens yielded 65–83% sensitivity and 94–100% specificity. These RT-QuIC sensitivities were substantially higher than those obtained using immunohistochemistry for abnormal PrP deposits in recto-anal mucosa associated lymphoid tissue (RAMALT). Nonetheless, further improvements in diagnostic sensitivity, specificity, and practicality of antemortem CWD tests would facilitate the monitoring of CWD in cervids.
Previous studies have shown the accumulation of readily detectable prion seeding activity in the skin of humans with Creutzfeldt–Jakob disease25,26, and rodents with scrapie, even early in the course of infection27. Given that the outer ear could serve as an accessible source of both skin and nervous tissue, we sought to investigate the utility of RT-QuIC to diagnose CWD in pre-clinical white-tailed and mule deer using ear pinna punches. Here, by coupling iron oxide magnetic extraction to a modified RT-QuIC assay, we were able to diagnose CWD by analyzing ear samples collected from both clinical and asymptomatic animals with an accuracy at least as good as previous assays of RAMALT tissue, with the advantage herein of using a more easily collectable specimen.
Results
RT-QuIC analysis of RLN samples
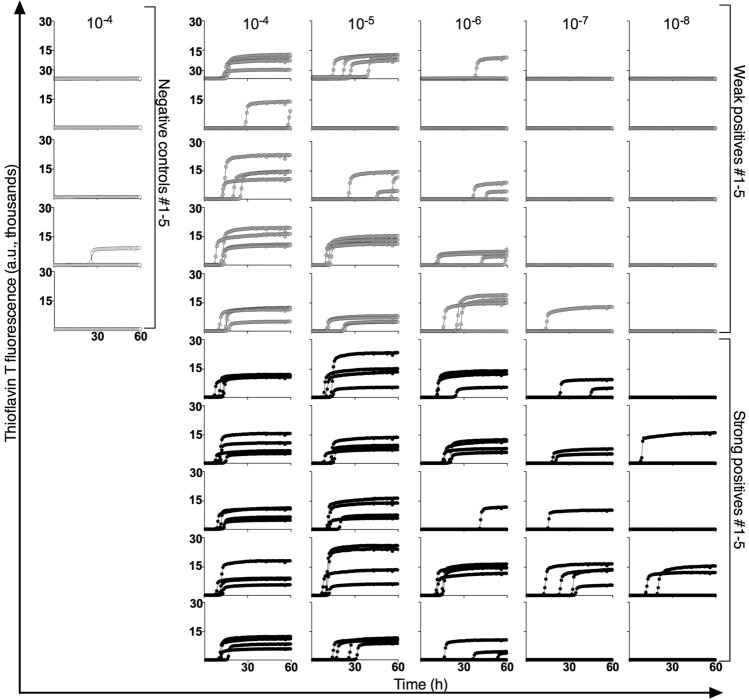
Analysis of postmortem RLN tissue for PrPSc by ELISA is an approved regulatory assay for identifying CWD-infected animals, as longitudinal studies have indicated that mule deer can become positive 3–6 months after oral inoculation, well prior to the onset of symptoms10. Based upon the optical density (OD) value obtained through ELISA of one of their RLNs, deer were classified as negative (OD < 0.1), weak positive (0.1 < OD < 1.0), and strong positive (OD > 3.0). Figure 1 shows that, consistent with previous studies28, prion seeding activity can be detected in the other RLNs collected from deer with weak (WP) as well as strong (SP) ELISA-positive RLN samples, but not in ELISA-negative controls (N1–N5). End point dilution RT-QuIC analysis of RLN from the weaker ELISA positive deer (OD = 0.539) had seed concentration that was ~ 100-fold lower than the deer with the strong ELISA positive RLN (OD = 3.999). Analysis of RLNs from a total of ten ELISA-positive deer indicated that the RLNs from weak positive deer (n = 5) had lower (p = 0.04, two tailed t test) mean (± SD) log 50% seeding units (doses)15, i.e. log SD50, per mg tissue (5.75 ± 0.91) than those from strong-positive deer (7.00 ± 0.64 log SD50 per mg; n = 5).
Figure 1.
RT-QuIC reactions seeded with RLN homogenates from deer with RLNs that gave either negative, relatively weak, or strong ELISA readings as described in the main text. Replicate aliquots (n = 4) of the designated dilution of each homogenate were independently assayed in microplate wells with the thioflavin T fluorescence indicated in arbitrary relative units (a.u.) as a function of reaction time. Reactions seeded with 10–4 RLN tissue dilutions (w/v) from negative control deer (n = 5) or the designated dilutions from weak ELISA-positive (n = 5) and strong ELISA-positive deer (n = 5).
IOME enhances the sensitivity of the RT-QuIC assay seeded with ear pinna homogenates
Aiming to develop an assay that would allow the use of a tissue sample that is more accessible than RLN or obex, we assessed the ability of RT-QuIC to detect prion seeding activity in post-mortem ear pinna punches from infected deer. We first used our previous CWD-adapted RT-QuIC assay to analyze a panel of 167 apparently asymptomatic hunter-killed deer that were designated as apparent CWD-positive and -negative deer according to ELISA assays of their RLN. However the diagnostic sensitivity provided by that assay was insufficient (< 50%). Therefore, to improve sensitivity, we added NaI in the RT-QuIC reactions based on previous findings29 and an initial iron oxide magnetic extraction (IOME) step that has been shown previously to aid in prion seed concentration and removal of potentially interfering tissue matrix components7. IOME extraction exploits the tendency of prion to bind to certain metals. Punches from tip, middle, and bottom of the ear apex (Fig. 2) were assayed by RT-QuIC, preceded or not by IOME. As shown in Fig. 3, the IOME-coupled RT-QuIC (IOME-RT-QuIC) provided more positive replicate reactions using samples from deer with both weakly and strongly ELISA-positive RLNs. Thus, we employed IOME-RT-QuIC in subsequent tests.
Figure 2.
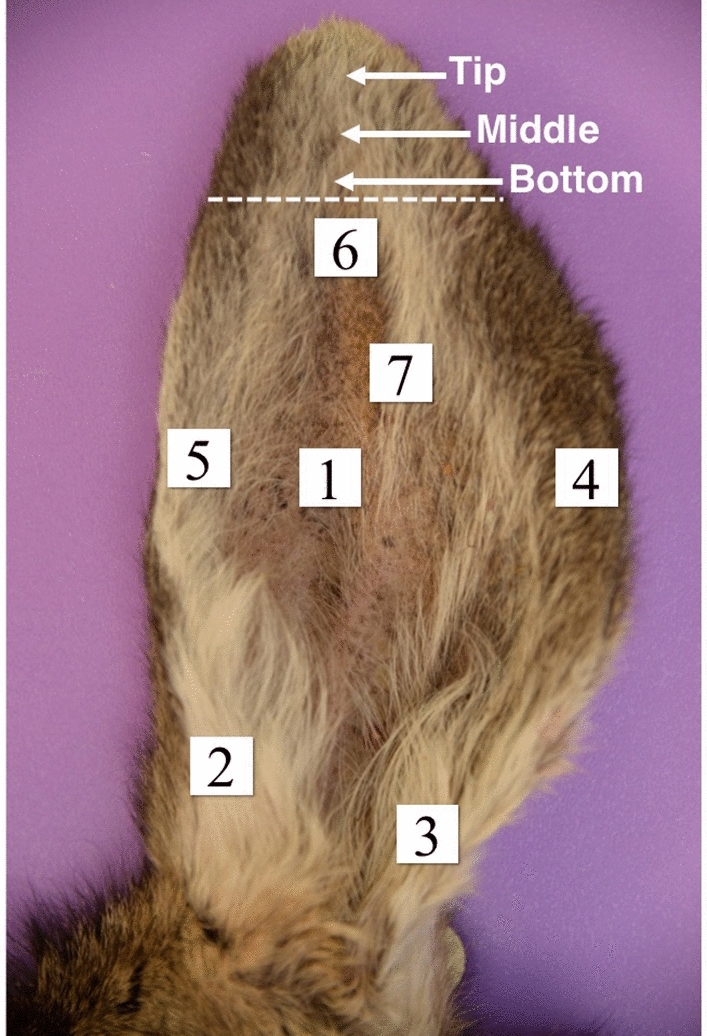
Representative picture of the ear parts from which the skin tissue was collected. The region of the apex of the ear above the dashed line shows areas samples for the initial survey of ear specimens using the original, suboptimal CWD RT-QuIC assay conditions. The numbers below the dashed line indicate areas from which 0.5 cm punches were taken for the analysis shown in Fig. 4.
Figure 3.
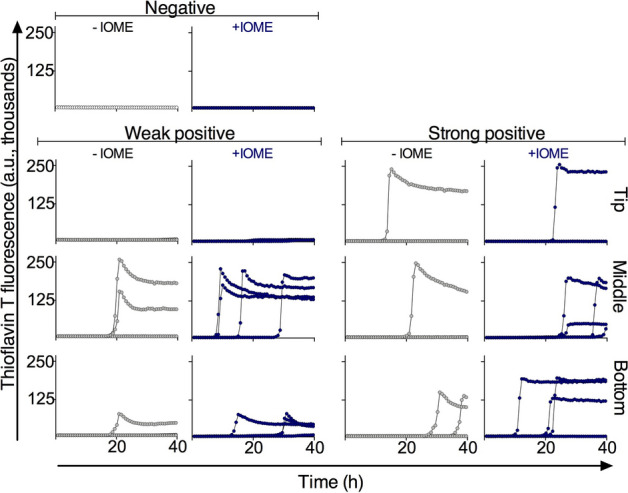
Fluorescence traces from individual wells (n = 4 per sample) of RT-QuIC reactions seeded with homogenates of ear punches collected from different regions of pinna apex (above the dashed line in Fig. 2). The RT-QuIC reactions (without IOME extraction, − IOME) were seeded with 10–2 dilution of ear homogenate in 0.1% SDS/PBS/N2. For the IOME-RT-QuIC reactions, a 1:10 dilution of iron oxide magnetic beads in 0.1% SDS/PBS/N2 was prepared, and the reactions were seeded with ear tissue equivalents comparable to the 10–2 dilution used in the reactions without IOME. ‘Weak positive’ and ‘strong positive’ labels refer to the ELISA positivity of RLN tissue from the same deer.
Higher prion seeding activity is found around the central nerve in the ear pinna
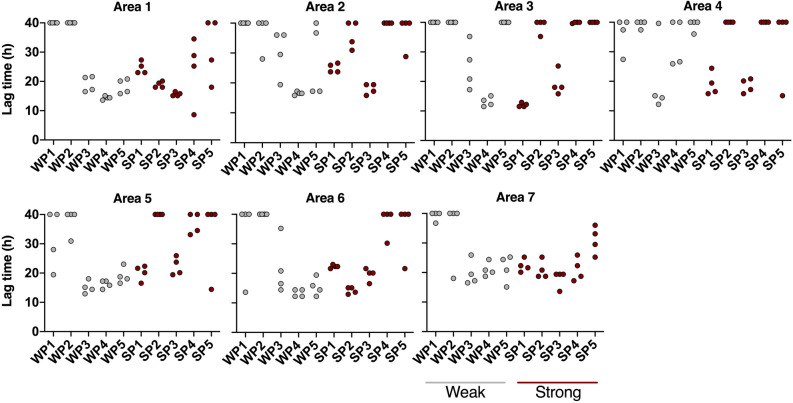
Considering that prion seeding activity might not be evenly distributed throughout the ear pinna, we analyzed punches from seven different pinna areas as shown in Fig. 2 from 5 each of weak (WP) and strong (SP) ELISA-positive deer. After homogenization, samples were subjected to IOME-RT-QuIC. We found that prion seeding activity was more concentrated in areas 1, 6, and 7, which are located at (area 7) or adjacent to (1 and 6) the auricular nerve, with area 7 giving the highest proportion of positive reactions (Fig. 4).
Figure 4.
Lag times of RT-QuIC reactions seeded with ear pinna homogenates prepared from different areas (see Fig. 2) of five weak ELISA-positive (WP, grey) and five strong positive (SP, maroon) deer. Samples were subjected to IOME-RT-QuIC. Lag time (time to reach 10% maximum ThT fluorescence on the plate) within the cutoff time of the assay (40 h). Reactions with lag times of ≥ 40 h are aligned at the top of each graph.
IOME-RT-QuIC analysis of a panel of ear pinna samples
Based on these results, we focused on sampling area 7 in a subsequent analysis of 58 ears collected from deer killed by hunters (n = 56) or submitted for diagnostic examination (n = 2). None of the hunter-killed deer had been reported as appearing unhealthy. Deer were deemed positive or negative for CWD infection based on both ELISA and RT-QuIC (without IOME) assays of their RLNs. These analyses were 100% concordant giving a total of 26 CWD-positive deer in the panel (Table 1). IOME-RT-QuIC analyses of pinna area 7 were performed by an operator who was blinded to the CWD status of the donors. Using the criteria described in the Materials and Methods for calling the samples positive or negative overall, 24 of the ear samples were positive (Table 2). Twenty-one of those samples were from deer with positive RLNs, giving an apparent diagnostic sensitivity of 81% based on the RLN diagnosis. The 3 remaining IOME-RT-QuIC-positive ear samples were from the group of 32 total deer with negative RLNs, giving 29 true IOME-RT-QuIC negatives out 32, or an apparent diagnostic specificity of 91%.
Table 1.
Comparison of RLN ELISA (optical density; OD) and RT-QuIC results with ear (area 7) IOME-RT-QuIC results.
| Sample ID#,* | CWD status** | OD | RLN RT-QuIC*** | EAR RT-QuIC*** |
|---|---|---|---|---|
| 3681 | Positive | 3.999 | 4/4 | 4/4 |
| 3806w | Positive | 3.999 | 4/4 | 4/4 |
| 5685w | Positive | 0.490 | 2/4 | 1/4 and 1/4 |
| 7424 | Positive | 3.999 | 4/4 | 1/4 and 4/4 |
| 0482 | Positive | 0.389 | 4/4 | 4/4 |
| 0504 | Not detected | – | 0/4 | 0/4 |
| 0902 | Not detected | – | 0/4 | 0/4 |
| 0913 | Not detected | – | 0/4 | 0/4 |
| 0924 | Not detected | – | 0/4 | 0/4 |
| 1042**** | Positive | 0.689 | 4/4 | 4/4 |
| 1403 | Not detected | – | 0/4 | 0/4 |
| 2136 | Positive | 0.372 | 4/4 | 4/4 |
| 3422w | Positive | 0.315 | 4/4 | 3/4 and 4/4 |
| 0262**** | Not detected | – | 0/4 | 0/4 |
| 0623 | Not detected | – | 0/4 | 0/4 |
| 0881 | Not detected | – | 0/4 | 0/4 |
| 0903 | Not detected | – | 0/4 | 2/4 and 2/4 |
| 1010 | Not detected | – | 0/4 | 0/4 |
| 1194 | Positive | 2.468 | 4/4 | 4/4 |
| 1356 | Not detected | – | 0/4 | 0/4 |
| 1894 | Not detected | – | 0/4 | 0/4 |
| 1920 | Not detected | – | 0/4 | 0/4 |
| 1953 | Not detected | – | 0/4 | 1/4 and 3/4 |
| 2546 | Not detected | – | 0/4 | 1/4 and 1/4 |
| 2550 | Not detected | – | 0/4 | 0/4 |
| 2561 | Not detected | – | 0/4 | 0/4 |
| 2594 | Not detected | – | 0/4 | 0/4 |
| 2616 | Not detected | – | 0/4 | 0/4 |
| 2863 | Not detected | – | 0/4 | 0/4 |
| 2922 | Not detected | – | 0/4 | 0/4 |
| 3353 | Positive | 1.252 | 4/4 | 0/4 |
| 3386 | Positive | 3.999 | 4/4 | 4/4 |
| 3482 | Not detected | – | 0/4 | 0/4 |
| 3493 | Positive | 3.999 | 4/4 | 3/4 and 4/4 |
| 3810w | Positive | 3.999 | 4/4 | 0/4 |
| 4624 | Not detected | – | 0/4 | 3/4 and 1/4 |
| 4716 | Positive | 2.137 | 4/4 | 4/4 |
| 4720 | Positive | 3.999 | 4/4 | 4/4 |
| 4753 | Positive | 3.999 | 4/4 | 4/4 |
| 5630 | Positive | 1.565 | 4/4 | 4/4 |
| 5652 | Not detected | – | 0/4 | 0/4 |
| 5755 | Not detected | – | 0/4 | 0/4 |
| 5770 | Not detected | – | 0/4 | 0/4 |
| 5781 | Not detected | – | 0/4 | 0/4 |
| 5803 | Positive | 0.539 | 4/4 | 1/4 and 3/4 |
| 5851 | Positive | 3.999 | 4/4 | 2/4 and 4/4 |
| 7376 | Positive | 2.371 | 4/4 | 2/4 and 2/4 |
| 7472w | Not detected | – | 0/4 | 0/4 |
| 7542 | Positive | 3.999 | 4/4 | 4/4 |
| 7995 | Not detected | – | 0/4 | 0/4 |
| 8264 | Not detected | – | 0/4 | 0/4 |
| 8360 | Positive | 3.999 | 4/4 | 4/4 |
| 0390w | Positive | 0.453 | 4/4 | 4/4 |
| 0456 | Not detected | – | 0/4 | 0/4 |
| 2556 | Not detected | – | 0/4 | 0/4 |
| 2582 | Positive | 0.604 | 4/4 | 0/4 |
| 3385 | Positive | 0.373 | 4/4 | 0/4 |
| 5285 | Positive | 3.333 | 4/4 | 4/4 |
*Sample numbers with a “w” suffix were from white-tailed deer; all others were from mule deer.
**Based on RLN ELISA and RT-QuIC results in 3rd and 4th columns, which were 100% concordant.
***Positive/total replicate reactions. Two fractions for the same sample indicate results from repeated assays in quadruplicate.
****Carcass submitted for diagnostic examination. Samples not marked came from apparently healthy hunter-killed deer.
Table 2.
Summary of RLN ELISA, RLN RT-QuIC and ear (area 7) IOME-RT-QuIC results from 58 deer (53 mule and 5 white-tailed).
| Assay results | Concordance of ELISA and RT-QuIC of RLN | Concordance of ear positives with RLN | App.* sensitivity or ear vs RLN | Concordance of ear negatives with RLN | App.* specificity of ear vs RLN | ||
|---|---|---|---|---|---|---|---|
| + | − | ||||||
| RLN (ELISA) | 26 | 32 | 100% | ||||
| RLN (RT-QuIC) | 26 | 32 | |||||
| Ear (RT-QuIC) | 24 | 34 | 21/26 | 81% | 29/32 | 91% | |
*Apparent; we use this adjective to indicate that these sensitivity and specificity values are based on analysis of RLN tissues, which might not be a completely accurate indication of the overall CWD infection status of the deer.
Discussion
Given concerns about the growing distribution of CWD and uncertainty about the potential for transmission to non-cervid species, it remains critical to improve the practicality and accuracy of detection of infection in both wild and farmed populations. One key challenge is the simple and cost-effective collection of diagnostic specimens from both live and dead animals in the field. Often this is done under less-than-optimal circumstances by people who may lack specific training in the relevant anatomy and the prevention of sample cross-contamination. Cross contamination can be a significant concern when the samples are being tested using ultra-sensitive prion seed amplification assays such as RT-QuIC. The collection of diagnostic specimens from live cervids presents particular problems in that sample collection usually requires restraint. As RLN tissue cannot be biopsied practically, several studies have focused on RT-QuIC analysis of RAMALT that can be obtained by rectal biopsy of cervids by trained personnel once the animal is restrained. Although these studies have yielded promising results as noted above, we hoped in undertaking this current study that the outer ear of cervids might be a useful specimen for RT-QuIC analysis due to ease of collection.
Our results show that IOME-RT-QuIC testing of deer pinna punch samples can provide sensitivity (81%) and specificity (91%) relative to RLN-based diagnosis (Table 2) that was better than, or comparable to, those previously reported for RT-QuIC analysis of RAMALT biopsy tissue (69.8% and > 93.9%, respectively)22. We note that whereas this previous RAMALT study sampled deer depopulated from a CWD-contaminated farm, the majority of samples we analyzed came from apparently asymptomatic deer killed by hunters. In neither study was the duration of the infection in the CWD-positive animals clear, but this same uncertainty will apply to most, if not all, naturally exposed cervids. Further experimental infection studies will be required to ascertain how early prion seeding activity can be detected in cervid ear pinna tissue after infections with various doses by different routes of inoculation. It is not clear why we failed to detect prion seeding activity in the area 7 ear samples of all deer that were positive by RLN analysis. However, one possible explanation is that the primary mechanism of spread to the ear pinna is via anterograde spread through nerves from the central nervous system. Indeed, the fact that we more frequently obtained positive IOME-RT-QuIC reactions from parts of the ear (i.e. area 7) that span the auricular nerve compared to other areas is consistent with this possibility. Given that evidence of infection is often seen in the lymphoreticular system of deer prior to detection in nervous tissues10, it seems likely that prion seeds might often appear in the ear pinna well after infection of RLNs.
Clearly, from a diagnostic perspective, there is room for improvement in the diagnostic performance of IOME-RT-QuIC analysis of outer ear tissue relative to standard postmortem RLN analysis. Sensitivity might be improved by exploring new methods of sample handling and RT-QuIC testing, unless the false negativity rate that we obtained was due entirely to deer being infected without yet having any CWD prions in their outer ears. There may also exist other cutaneous sites that are amenable to punch biopsy and IOME-RT-QuIC analysis that may harbor higher doses of PrPSc, beyond what is found in the ear pinna during pre-clinical CWD infection. The apparently less-than-perfect specificity of IOME-RT-QuIC might be due to inadvertent cross-contamination of the sample, or occasional non-CWD components of ear pinna tissue that accelerate spontaneous nucleation and fibrillization of our recombinant PrP substrate. Such confounding factors might be more effectively removed from samples by as yet unknown improvements in sample handling and prion seed capture. On the other hand, the possibility remains that apparent false positive tests of ear samples were actually real positives obtained from deer that were CWD-infected in a way that was not yet manifest in their RLNs. For example, rare cases of CNS infection without lymphoid involvement have been observed3. Such cases might not only be a manifestation of infection route, but also CWD strain. Again, such possibilities would have to be addressed by deliberate, controlled experimental infection studies. Hopefully such studies will be encouraged by our current findings.
Methods
Samples collection
No animal-use-protocol approval was required for this study. The samples analyzed herein were collected opportunistically from heads or carcasses of deer that had been killed by licensed hunters (n = 56) or submitted for postmortem examination after dying as a result of injury or illness (n = 2). Entire ears were collected in addition to RNLs and biological data (species, sex, approximate age). Fresh RLN tissues were screened for evidence of PrPSc by ELISA30 at the Colorado State University Veterinary Diagnostic Laboratory (Fort Collins, Colorado USA). Ears and remaining RLN tissue samples were stored frozen until assayed.
Recombinant PrP
Recombinant prion protein was expressed and purified as described previously15. Briefly, Syrian golden hamster prion protein sequence encompassing residues 90-231 was inserted into the pET41 vector (EMD Biosciences) and then transformed into Rosetta (DE3) E. coli. Protein expression was induced by the autoinduction system31,32. Protein was purified from inclusion bodies in an AKTA fast protein liquid chromatographer (GE Healthcare Life Sciences) using nickel-nitrilotriacetic acid (NTA) Superflow resin (Qiagen). The refolding process was performed under a Gdn-HCl reduction gradient and elution was achieved with an imidazole gradient. Protein was then dialyzed against 10 mM Na2HPO4 buffer (pH 5.8).
Skin homogenization
The tissue was sprayed with 70% ethanol and then shaved with a scalpel. The shaved area was washed with PBS and the tissue was cut in 0.5 cm square pieces. The small pieces of tissue were placed in a tube with 1 mm glass beads, weighed, and homogenization buffer (2 mM CaCl2, 2.5 mg/mL collagenase A [0.25% w/v] in PBS) was added in order to make a 10% ear homogenate. The samples were homogenized in a bead beater (max power for 3 min) and incubated overnight at 37 °C in a thermomixer under mild agitation (650 rpm). Samples were agitated in a bead beater again (max power for 1 min), centrifuged for 5 min at 2000g to remove gross debris, and the supernatant was transferred into a new tube. Samples were stored at − 80 °C.
Retropharyngeal lymph nodes homogenization
Retropharyngeal lymph nodes (RLN) were homogenized following the same protocol described above, except for the omission of shaving and washing with ethanol.
Iron oxide magnetic extraction (IOME) coupled with RT-QuIC analysis
The protocol described here was developed based on a previously described IOME method7 with a couple of modifications. Superparamagnetic iron oxide beads (~ 9 μm, Bangs Laboratories, Indiana) were vortexed and 10 μL were transferred into a tube and washed with 0.5 mL phosphate buffered saline (PBS). After washing, beads were transferred to a tube containing 0.5 mL 0.1% SDS/PBS/N2 and 0.5 mL of 10% ear homogenate. The sample was incubated at room temperature in an end-over-end rotator for 2 h. Then, the beads were separated from the supernatant by placing the tube in the magnetic particle separator. Supernatant was removed and beads were resuspended in 10 μL of 0.1% SDS/PBS/N2. The suspension was sonicated for 1 min and a tenfold dilution of IOM beads was done in a 0.1% SDS/PBS/N2 solution. Two μL of this final dilution were added to single wells of a 96-well plate previously loaded with 98 μL of the RT-QuIC reaction mix (300 mM NaI, 10 mM Hepes, 1 mM EDTA [ethylenediamine tetraacetic acid], 10 μM thioflavin T, 0.1 mg/mL rPrP hamster 90-231). Each sample was assessed in quadruplicate, and following the first analyses, every sample which showed partial positive reactions (1–3 out of 4) was reanalyzed. A sample was considered positive when at least half of total replicates surpassed the threshold (10% maximum ThT fluorescence on the plate). Determination of log SD50 values by end-point dilution RT-QuIC analysis was performed as described previously15.
Acknowledgements
We thank Drs. Cathryn Haigh, Moses Leavens, and Ankit Srivastava for their in-house review of this manuscript at RML. This work is supported in part by the Intramural Research Program of the NIAID, Colorado Parks and Wildlife and US Fish and Wildlife Service Federal Aid in Wildlife Restoration funds, Basque Grant EP 2018_1_0036 “Ayudas para la realización de estancias en centros distintos al de aplicación del programa predoctoral de formación del personal investigador”, Spanish Grant RTI2018-098515-B-I00 (MICIU/FEDER), NIH Grants R01-NS061902 and P01-AI-077774 (to EAH) and Mary Hilderman Smith, Zoë Smith Jaye, and Jenny Smith Unruh in memory of Jeffrey Smith. MAM is supported by the NIH-Oxford/Cambridge Scholars Program.
Author contributions
B.C., M.W.M., and T.A.N. conceived the project. N.C.F., J.M.C., J.P., C.D.O., N.D.D., M.A.M., A.G.H., and B.R. designed and/or performed experiments. J.C. and E.A.H. provided project oversight. M.W.M., T.A.N. and K.A.G. provided samples. B.C. and N.C.F. wrote the first draft and M.W.M., T.A.N., J.C., J.P., C.D.O., A.G.H., B.R., and M.A.M. helped edit the manuscript.
Funding
Open Access funding provided by the National Institutes of Health (NIH).
Competing interests
NCF, EAH, JMC, JC, NDD, TAN, MAM, MWM, CDO, AGH, BR, JP, and KAG declare no conflict of interest in any of the contexts or instances stated. BC is an inventor on patents relating to RT-QuIC technology.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Williams ES, Young S. Chronic wasting disease of captive mule deer: A spongiform encephalopathy. J. Wildl. Dis. 1980;16:89–98. doi: 10.7589/0090-3558-16.1.89. [DOI] [PubMed] [Google Scholar]
- 2.Miller MW, Wild MA, Williams ES. Epidemiology of chronic wasting disease in captive Rocky Mountain elk. J. Wildl. Dis. 1998;34:532–538. doi: 10.7589/0090-3558-34.3.532. [DOI] [PubMed] [Google Scholar]
- 3.Williams ES. Chronic wasting disease. Vet. Pathol. 2005;42:530–549. doi: 10.1354/vp.42-5-530. [DOI] [PubMed] [Google Scholar]
- 4.Miller MW, Williams ES, Hobbs NT, Wolfe LL. Environmental sources of prion transmission in mule deer. Emerg. Infect. Dis. 2004;10:1003–1006. doi: 10.3201/eid1006.040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haley NJ, et al. Detection of chronic wasting disease prions in salivary, urinary, and intestinal tissues of deer: Potential mechanisms of prion shedding and transmission. J. Virol. 2011;85:6309–6318. doi: 10.1128/JVI.00425-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henderson DM, et al. Quantitative assessment of prion infectivity in tissues and body fluids by real-time quaking-induced conversion. J. Gen. Virol. 2015;96:210–219. doi: 10.1099/vir.0.069906-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denkers ND, Henderson DM, Mathiason CK, Hoover EA. Enhanced prion detection in biological samples by magnetic particle extraction and real-time quaking-induced conversion. J. Gen. Virol. 2016;97:2023–2029. doi: 10.1099/jgv.0.000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gough KC, Maddison BC. Prion transmission: Prion excretion and occurrence in the environment. Prion. 2010;4:275–282. doi: 10.4161/pri.4.4.13678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller MW, Williams ES. Detection of PrP(CWD) in mule deer by immunohistochemistry of lymphoid tissues. Vet. Rec. 2002;151:610–612. doi: 10.1136/vr.151.20.610. [DOI] [PubMed] [Google Scholar]
- 10.Fox KA, Jewell JE, Williams ES, Miller MW. Patterns of PrPCWD accumulation during the course of chronic wasting disease infection in orally inoculated mule deer (Odocoileus hemionus) J. Gen. Virol. 2006;87:3451–3461. doi: 10.1099/vir.0.81999-0. [DOI] [PubMed] [Google Scholar]
- 11.Rivera NA, Brandt AL, Novakofski JE, Mateus-Pinilla NE. Chronic wasting disease in cervids: Prevalence, impact and management strategies. Vet. Med. (Auckl.) 2019;10:123–139. doi: 10.2147/VMRR.S197404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haley NJ, Richt JA. Evolution of diagnostic tests for chronic wasting disease, a naturally occurring prion disease of cervids. Pathogens. 2017;6:35. doi: 10.3390/pathogens6030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira NDC, Caughey B. Proteopathic seed amplification assays for neurodegenerative disorders. Clin. Lab. Med. 2020;40:257–270. doi: 10.1016/j.cll.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atarashi R, et al. Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking-induced conversion. Nat. Med. 2011;17:175–178. doi: 10.1038/nm.2294. [DOI] [PubMed] [Google Scholar]
- 15.Wilham JM, et al. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 2010;6:e1001217. doi: 10.1371/journal.ppat.1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411:810–813. doi: 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- 17.Henderson DM, et al. Longitudinal detection of prion shedding in saliva and urine by chronic wasting disease-infected deer by real-time quaking-induced conversion. J. Virol. 2015;89:9338–9347. doi: 10.1128/JVI.01118-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haley NJ, et al. Prion-seeding activity in cerebrospinal fluid of deer with chronic wasting disease. PLoS ONE. 2013;8:e81488. doi: 10.1371/journal.pone.0081488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson DM, et al. Detection of chronic wasting disease prion seeding activity in deer and elk feces by real-time quaking-induced conversion. J. Gen. Virol. 2017;98:1953–1962. doi: 10.1099/jgv.0.000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davenport KA, et al. Assessment of chronic wasting disease prion shedding in deer saliva with occupancy modeling. J. Clin. Microbiol. 2018;56:e01243-17. doi: 10.1128/JCM.01243-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davenport KA, Hoover CE, Denkers ND, Mathiason CK, Hoover EA. Modified protein misfolding cyclic amplification overcomes real-time quaking-induced conversion assay inhibitors in deer saliva to detect chronic wasting disease prions. J. Clin. Microbiol. 2018;56:e00947-18. doi: 10.1128/JCM.00947-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haley NJ, et al. Antemortem detection of chronic wasting disease prions in nasal brush collections and rectal biopsy specimens from white-tailed deer by real-time quaking-induced conversion. J. Clin. Microbiol. 2016;54:1108–1116. doi: 10.1128/JCM.02699-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haley NJ, et al. Cross-validation of the RT-QuIC assay for the antemortem detection of chronic wasting disease in elk. Prion. 2020;14:47–55. doi: 10.1080/19336896.2020.1716657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haley NJ, et al. Chronic wasting disease management in ranched elk using rectal biopsy testing. Prion. 2018;12:93–108. doi: 10.1080/19336896.2018.1436925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orru CD, et al. Prion seeding activity and infectivity in skin samples from patients with sporadic Creutzfeldt–Jakob disease. Sci. Transl. Med. 2017;9:eaam7785. doi: 10.1126/scitranslmed.aam7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mammana A, et al. Detection of prions in skin punch biopsies of Creutzfeldt–Jakob disease patients. Ann. Clin. Transl. Neurol. 2020;7:559–564. doi: 10.1002/acn3.51000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, et al. Early preclinical detection of prions in the skin of prion-infected animals. Nat. Commun. 2019;10:247. doi: 10.1038/s41467-018-08130-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haley NJ, et al. Detection of chronic wasting disease in the lymph nodes of free-ranging cervids by real-time quaking-induced conversion. J. Clin. Microbiol. 2014;52:3237–3243. doi: 10.1128/JCM.01258-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metrick MA, 2nd, et al. Million-fold sensitivity enhancement in proteopathic seed amplification assays for biospecimens by Hofmeister ion comparisons. Proc. Natl. Acad. Sci. U.S.A. 2019 doi: 10.1073/pnas.1909322116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hibler, C. P. et al. Field validation and assessment of an enzyme-linked immunosorbent assay for detecting chronic wasting disease in mule deer (Odocoileus hemionus), white-tailed deer (Odocoileus virginianus), and Rocky Mountain elk (Cervus elaphus nelsoni). J. Vet. Diagn. Invest.15(4), 311–319 (2003). [DOI] [PubMed]
- 31.Fox BG, Blommel PG. Autoinduction of protein expression. Curr. Protoc. Protein Sci. 2009;Chapter 5:Unit 5 23. doi: 10.1002/0471140864.ps0523s56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]