Abstract
Nutritional symbioses between bacteria and insects are prevalent and diverse, allowing insects to expand their feeding strategies and niches. A common consequence of long-term associations is a considerable reduction in symbiont genome size likely influenced by the radical shift in selective pressures as a result of the less variable environment within the host. While several of these cases can be found across distinct insect species, most examples provide a limited view of a single or few stages of the process of genome reduction. Stink bugs (Pentatomidae) contain inherited gamma-proteobacterial symbionts in a modified organ in their midgut and are an example of a long-term nutritional symbiosis, but multiple cases of new symbiont acquisition throughout the history of the family have been described. We sequenced the genomes of 11 symbionts of stink bugs with sizes that ranged from equal to those of their free-living relatives to less than 20%. Comparative genomics of these and previously sequenced symbionts revealed initial stages of genome reduction including an initial pseudogenization before genome reduction, followed by multiple stages of progressive degeneration of existing metabolic pathways likely to impact host interactions such as cell wall component biosynthesis. Amino acid biosynthesis pathways were retained in a similar manner as in other nutritional symbionts. Stink bug symbionts display convergent genome reduction events showing progressive changes from a free-living bacterium to a host-dependent symbiont. This system can therefore be used to study convergent genome evolution of symbiosis at a scale not previously available.
Subject terms: Molecular evolution, Bacterial evolution, Symbiosis
Introduction
Animal-microbe associations are prevalent throughout the tree of life and can greatly benefit and expand the host’s ability to survive in a variety of environments. Hosts can assimilate resources produced by their bacterial symbionts, thereby benefitting from entire metabolic pathways that they are unlikely to develop1. Insects, particularly hemipterans, have used these symbioses to expand their feeding strategies to include nutrient-poor or unbalanced resources2. Resource provisioning-based mutualisms between insects and bacteria have been well-documented in many insects, including aphids3, cockroaches4, armored scales5, cotton stainers6. Bacterial partners of insects can be members of diverse phyla, including Proteobacteria and Bacteroidetes, and comparative genomic analyses reveal that they functionally converge around the supplementation of essential amino acids and/or vitamins absent from the host’s diet7. Host-microbial mutualisms often involve modifications to the involved partners that are evident at genomic, transcriptomic, and physiological scales, and some common symptoms include the evolution of specialized host cells and organs to house symbionts8–11 and genomic streamlining in the symbiont7,12.
Some patterns of genomic modifications in bacterial symbionts have emerged across a wide range of symbioses. When these symbiont genomes are compared to those of free-living close relatives, the evolutionary pressures that may be unique to endosymbiotic lifestyles come into focus13. Dramatic gene losses are often observed in bacterial symbionts and this phenomenon is thought to be due, in part, to relatively stable environmental conditions within host tissues. Genome shrinkage is due primarily to relaxed selective pressures upon genes not essential for survival in the host and genetic bottlenecks resulting from vertical transmission14. For example, genes involved in cell wall biosynthesis and self-defense are consistently absent from symbiont genomes15. As a result, core symbiont genomes are consistently reduced relative to their free-living close relatives9. Pseudogenization of genes experiencing relaxed selection often precedes their loss16, however it is rare to find bacterial-insect symbioses within a single insect family undergoing radically different stages of gene loss, which would greatly facilitate the study of this process.
Members of the Pentatomomorpha often harbor an extracellular bacterial symbiont within posterior midgut crypts17–19. While this symbiotic organ is fairly similar across a wide variety of families, other traits of the symbiosis can be quite variable, including the acquisition strategy17,20–23, degree of cophylogeny between host and symbiont24, and the symbiont’s reliance on its host17. In the Pentatomidae, symbionts are typically vertically-inherited by nymphal consumption of maternal deposits of symbiont-enriched secretions during emergence from eggs. While symbionts can vary between cultivable and uncultivable, they are nonetheless transmitted extracellularly, unlike intracellularly-transmitted symbionts in other insects17,23,25. Some pentatomids can harbor both vertically and environmentally-acquired symbionts depending on which bacteria they were exposed to during their first instar26. For example, Plautia stali (Pentatomidae) can domicile up to six different symbiont ‘types’ (named A through F), that vary in origin and degree of host reliance27. However, the most common scenario is that of more specific host-symbiont associations, where a single host species is consistently associated with a single symbiont24,28–31, and cophylogeny is detected between genera29,32,33 but not necessarily at higher taxonomic levels34,35, which would indicate multiple associations at different points in time throughout the family.
Since the Pentatomidae have a conserved symbiotic organ but an imperfect and variable symbiont transmission mechanism and multiple symbiont acquisitions, different taxonomically distant species are likely to contain symbionts at different stages of the symbiosis. Previously, two cases of genome reduced symbionts in two distinct subfamilies within the Pentatomidae were identified with different degrees of genome reduction that originated separately35–37. This was identified as a potentially valuable system to understand the convergent evolution of nutritional symbioses from the bacterial symbiont’s perspective18. Therefore, sampling of known symbionts in this family by genome sequencing will shed light into the diversity and variability of these symbionts. A wide range of genome sizes across different species of the Pentatomidae were observed and the core genome of these symbionts and metabolic pathways were predicted to be retained or lost at different stages of genome shrinkage.
Methods
Genome sequencing and assembly
Collection permits for stink bugs were obtained from Costa Rican authorities (MINAET). Wild-caught stink bugs belonging to the subfamily Edessinae and Pentatominae were collected at La Selva Biological Station, Organization for Tropical Studies (coordinates 10.43028535, -84.007099) during June, 2017. These included Edessa oxcarti, E. sp, and Brachystethus rubromaculatus for the Edessinae and Arvelius albopunctatus, Sibaria englemani, Taurocerus edessoides, Mormidea sp., and an unidentified Pentatomidae species for the Pentatomidae. Additionally, individuals of the Brown Stink Bug Euschistus servus and Green Stink Bug Nezara viridula were donated by Michael Toews (from Tifton, GA) and individuals of the Harlequin Bug Murgantia histrionica were donated by Don Weber (from Beltsville, MD). Morphological identification was carried out with appropriate guides38–44 and confirmed by molecular analysis as described below. DNA extraction and genome sequencing were performed as in36. Briefly, insects were surface-sterilized with three washes of ethanol 70% and the symbiotic organ was dissected. DNA was extracted from the whole organ of individual specimens using the DNeasy Blood and Tissue kit (Qiagen) with RNAse treatment according to manufacturer instructions. Paired 300 bp Nextera Libraries were prepared and sequenced using Illumina MiSeq at the Ohio State MCIC with read lengths of 2 × 300 bp. Raw reads were corrected using Trimmomatic45 and assembly was performed using Unicycler 0.4.846. Resulting contigs were filtered based on coverage level and scaffold linkage using Bandage 0.8.047. Additionally, BLASTn48 searches were employed to separate host and symbiont sequences. Host mitochondria was obtained by searching resulting contigs for the 13 genes of stink bug mitochondria or performing de-novo assembly on a subset of reads obtained by mapping to these contigs using BWA49 and SAMtools50 until recovering a circularized mitochondrial genome.
Annotation and genome comparisons
Genomes were annotated using Prokka51, RAST52, and the NCBI’s PGAP53,54, and annotations were visualized using Geneious 8.1.9 (https://www.geneious.com). Genome completeness was assessed with Benchmarking Universal Single Copy Orthologs (BUSCO) 2.0.155 which compares gene content with a single copy ortholog set specific for gammaproteobacteria. Host mitochondria were annotated using the Geneious ‘Annotate’ feature with a database of available Pentatomomorpha mitochondria and manually curated. For analyses including shared gene content, Roary 3.11.056 was used on previously sequenced stink bug symbiont genomes and representative genomes of species of the genus Pantoea. Functional protein-coding gene categorization and completeness of metabolic pathways were determined by referring to KAAS57, eggNOG 4.5.158 and MetaCyc databases59.
Symbiont identification
It has been well documented that genome reduced symbionts are difficult to accurately place in a phylogeny due to their high mutation rates creating long branch attraction60, which can lead to erroneous predictions in tree-based identification methods. Additionally, this fast mutation rate significantly decreases the sequence identity between genome reduced symbionts and other bacteria. In order to identify sequenced genomes we used SINA61 which uses ribosomal RNA genes to classify sequences with the Least Common Ancestor method using available sequences. Additionally, we used a modified method for estimating a phylogenetic tree for these species as described in36 in which we estimated separate trees for each genome reduced bacteria with the other non-genome reduced members as well as members of the Erwiniaceae (Table S2) using FastTree 2.1.1162, which would prevent some of the long branch attraction artifacts, and evaluated the consensus of all trees. Alignments were produced from the 10 largest protein coding genes common to all strains by reciprocal best hit BLAST and aligned using TranslatorX63.
Host mitochondrial phylogeny
Cytochrome oxidase I (COX1) genes from the mitochondrial genomes sequenced were searched against the BOLD database64 to corroborate morphological identification (Table S1). Additionally, all 13 coding regions and two rRNA regions were aligned with MUSCLE65 using the default parameters. Protein sequences were manually inspected and corrected using the translation alignment feature from Geneious for the recovered mitochondrial genomes as well as for other complete pentatomid mitochondria. Coptosoma bifaria (Pentatomoidea; Plataspidae, NCBI accession number EU427334.1) was used as the outgroup. Trees were reconstructed using RAxML66 with separate partitions for each codon and the rDNA genes, and MrBayes67 with 4 heated chains and 10,000,000 iterations, with burn-in of 1,000,000 and sampling every 1,000.
Results
Genome assembly
Sequencing resulted in libraries containing between 8 and 12 million paired reads for the different samples. After removing adapters and low-quality bases, individual genome assemblies were performed and each resulted in few-to-many scaffolds (ranging between 9 and 285 per assembly, see Table 1), forming a single connected component of ambiguously connected scaffolds ranging from 0.82 to 5.6 Mb with few or no dead ends. Separately, between one and three contigs belonging to the host mitochondria or circular high coverage plasmids were found. Scaffolds of all the main components had BLAST matches to bacteria from the Enterobacteriaceae, namely Pantoea, Erwinia, and other genome-reduced insect symbionts. Circularizing the genomes was not possible using only the Illumina data due to most components converging multiple times on contigs containing the 5S, 16S, and 23S rRNA genes, which are repeated across the genome. Despite this, we are confident that the genomes described represent the complete gene set for each symbiont with few and negligible omissions given that: 1) the assemblies contained no additional contigs of similar or higher coverage, 2) the evidence that each contig was connected to the main component via the ribosomal gene operon (with the exception of circular plasmids) which is present multiple times in each genome, 3) the multiple assemblies for symbionts of the Edessa matched in contig number and connections to the repetitive sequences, and 4) in the case of the unreduced genomes, the BUSCO completeness was above 96%.
Table 1.
Bacterial symbiont genomes of the Pentatomidae. Genome characteristics were derived from the corresponding genomes deposited in public databases. Mean cov.: mean x-fold coverage of genome sequence as assessed by Geneious.
| Name | SILVA Classification | Host | Subfamily | Genome Size (Mb) | Busco Completeness (%) | G + C% | Coding density (%) | Genes | Mean Cov | Scaffolds | References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SoEE | Enterobacterales | Edessa eburatula | Edessinae | 0.85 | 73.4 | 26.9 | 84.3 | 750 | 86 | 12 | Otero-Bravo and Sabree29 |
| SoEL | Enterobacterales | Edessa loxdalii | Edessinae | 0.837 | 73 | 27.6 | 84.3 | 741 | 96 | 12 | Otero-Bravo and Sabree29 |
| SoEO | Enterobacterales | Edessa bella | Edessinae | 0.836 | 71.6 | 26.6 | 81.6 | 721 | 206 | 12 | Otero-Bravo and Sabree29 |
| SoET | Enterobacterales | Edessa sp. 2 | Edessinae | 0.841 | 70.7 | 27.8 | 82.7 | 733 | 172 | 11 | Otero-Bravo and Sabree29 |
| SoEOx | Enterobacterales | Edessa oxcartii | Edessinae | 0.827 | 71.7 | 26.8 | 84.4 | 731 | 940 | 12 | This paper |
| SoEF | Enterobacterales | Edessa F | Edessinae | 0.839 | 69.5 | 27.8 | 82.1 | 739 | 860 | 21 | This paper |
| SoBr | Enterobacterales | Brachystethus rubromaculatus | Edessinae | 0.843 | 69.9 | 25.5 | 80.7 | 761 | 430 | 30 | This paper |
| SoMh | Pantoea | Murgantia histrionica | Pentatominae | 1.025 | 75.9 | 27.1 | 70.8 | 794 | 110 | 17 | This paper |
| SoAa | Erwiniaceae | Arvelius albopunctatus | Pentatominae | 1.143 | 81.4 | 27.9 | 77.5 | 943 | 190 | 9 | This paper |
| SoNv | Pantoea | Nezara viridula | Pentatominae | 1.425 | 87.2 | 40.6 | 68.9 | 1045 | 70 | 11 | This paper |
| SoEus | Pantoea | Euschistus servus | Pentatominae | 3.928 | 96.7 | 55.2 | 81.8 | 3998 | 25 | 30 | This paper |
| SoPt | Pantoea | Pentatomidae sp. | Pentatominae | 4.314 | 98.9 | 57.7 | 86.7 | 4142 | 79 | 102 | This paper |
| SoSe | Pantoea | Sibaria englemanni | Pentatominae | 5.046 | 96.7 | 53.9 | 88.3 | 4764 | 5 | 49 | This paper |
| SoTe | Pantoea | Taurocerus edessoides | Pentatominae | 5.458 | 100 | 53.7 | 87.7 | 5099 | 35 | 70 | This paper |
| SoMo | Pantoea | Mormidea sp. | Pentatominae | 5.612 | 99.1 | 56.9 | 85 | 5497 | 5.5 | 285 | This paper |
| Can P. carbekii | Can ‘P. carbekii’ | Halyomorpha halys | Pentatominae | 1.151 | 75.8 | 30.4 | 69.1 | 877 | 71 | 1 | Kenyon et al.37 |
| SoPS-A | Pantoea | Plautia stali | Pentatominae | 4.093 | 97.8 | 57 | 84.8 | 5202 | 20.5 | 131 | Hosokawa et al.35 |
| SoPS-B | Pantoea | Plautia stali | Pentatominae | 2.429 | 92.9 | 55.9 | 72.5 | 3214 | 89 | 34 | Hosokawa et al.35 |
| SoPS-C | Pantoea | Plautia stali | Pentatominae | 5.138 | 99.1 | 57.4 | 86.7 | 4850 | 18 | 201 | Hosokawa et al.35 |
| SoPS-D | Pantoea | Plautia stali | Pentatominae | 5.531 | 97.1 | 53.8 | 86.4 | 5299 | 19 | 235 | Hosokawa et al.35 |
| SoPS-E | Pantoea | Plautia stali | Pentatominae | 5.406 | 98 | 53.7 | 87.2 | 5135 | 19 | 146 | Hosokawa et al.35 |
| SoPS-F | Pantoea | Plautia stali | Pentatominae | 4.665 | 98.9 | 56.7 | 86.2 | 4381 | 23 | 103 | Hosokawa et al.35 |
Symbiont genome annotation
The genomes of symbionts of stink bugs belonging to the Edessinae and Pentatominae subfamilies were sequenced and annotated. Symbionts of Edessinae stinkbugs, which included the symbionts of six Edessa species (hereafter called SoE) and the symbiont of Brachystethus rubromaculatus (henceforth SoBr), had genomes that were < 1 Mb in size, varying little in size, and taxonomically categorized by SINA as ‘unclassified Enterobacteriaceae’ (See Table 1, S3). Symbionts of the Pentatominae stink bugs were larger than 1 Mb, ranging from 1 to 5.6 Mb, and were assigned to the Pantoea genus.
The smallest genomes among the symbionts of the Pentatominae belong to the symbiont of the harlequin bug Murgantia histrionica (henceforth SoMh) at 1.02 Mb, followed by the symbiont of Arvelius albopunctatus (henceforth SoAa) at 1.14 Mb, and the symbiont of the brown marmorated stink bug (Halyomorpha halys), Candidatus ‘Pantoea carbekii’ (Kenyon, et al., 2014) (henceforth P. carbekii) at 1.15 Mb. The symbiont of the green stink Bug, Nezara viridula (henceforth SoNv) showed a slightly larger size, at 1.42 Mb. This is followed by a much larger genome, belonging to the B-type symbiont of Plautia stali (henceforth SoPs-B) with the smallest symbiont genome identified for this host at 2.4 Mb. Finally, the remaining symbionts contained genomes larger than 3.9 Mb, which is within the range of non-stink bug associated strains of the Pantoea genus.
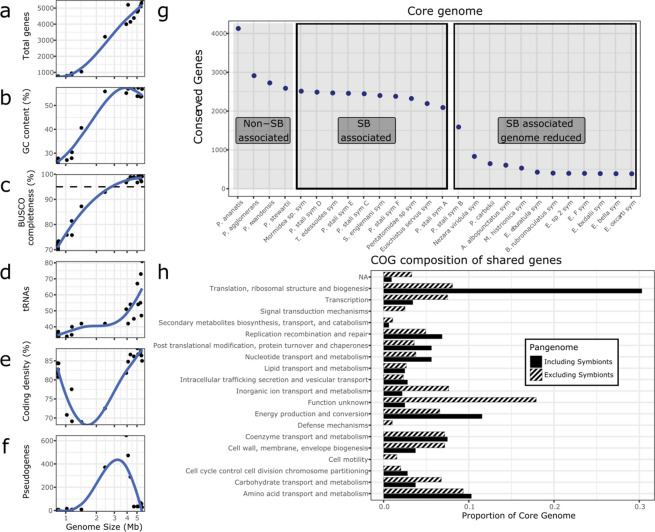
Metrics such as gene number, GC content, and genome completeness (Table 1, Fig. 1a-c) were also in accordance to expectations for genomes of given size: GC content showed little variation from the range between 53 and 57% among unreduced genomes and even the moderately reduced genome of SoPs-B (at 2.4 Mb). Reduced genomes on the other hand, presented a GC bias characteristic of genome reduction, ranging from 25 to 30%, while the intermediately sized SoNv showed a slightly lesser skew, with a GC content of 40% (Table 1, Fig. 1b). BUSCO completeness and number of total genes also decreased with greater genome reduction, with intermediate genomes of SoPs-B and SoNv also having intermediate values (Table 1, Fig. 1c,d). Coding density and pseudogene number followed a different trend, where both large and small genomes showed similar values (near 100% and 0, respectively), while intermediate sized genomes deviated significantly from this (Fig. 1e,f).
Figure 1.
Genomic characteristics of pentatomid symbionts. (a–f) Genomic characteristics as a function of genome size for stink bug symbionts. (a) number of genes annotated, (b) GC % of the entire genome, (c) percentage of conserved genes found according to the BUSCO gammaproteobacterial set (Dotted line in indicates the recommended 95% threshold of BUSCO completeness for new species descriptions). (d) tRNAs annotated, (e) coding density of the genome, (f) number of pseudogenes annotated by Prokka. Blue line indicates the LOWESS fitted curve. (g) Number of conserved genes between each genome and all larger genomes (excluding pseudogenes). (h) COG composition of the shared genes for non-symbiotic and large genome symbionts (> 3.8 Mb, dashed bar) and for all genomes including genome reduced symbionts (< 1.4 Mb, solid bar).
The subset of genes that is common to all strains, was obtained for the four species of the Pantoea genus with complete representative genomes available (which are not associated with stink bugs) and the symbiont genomes used for previous analyses (Table 1). The number of shared genes plateaus for all genomes larger than 4 Mb, following the trend from the non-SB associated strains. This subset of approximately 2450 genes likely represents the core genome of the genus Pantoea, which most stink bug symbionts have been assigned to68. However, the number of shared genes begins decreasing with the addition of the symbiont of Euschistus servus (henceforth SoEus) which despite a relatively large 3.9 Mb genome has lost some of these conserved genes due to pseudogenization. Subsequently, the number of shared genes rapidly decays with the genomes of SoPs-B (2.4 Mb) and SoNv (1.4 Mb). The reduction between the next few genomes is modest due to their similar size of 1 Mb, followed by a larger decrease when the most reduced genomes (< 1 Mb) are added. The set of shared genes for the studied genomes, including all symbionts and non-stink bug associated Pantoea, includes 454 genes (Fig. 1g).
The differences between the two sets of shared genes can yield insights into the change in requirements for the symbiotic lifestyle (Fig. 1h). Primarily, the proportion of genes in the translation, ribosomal structure and biogenesis (J) and energy production (C) is much higher for the shared gene set including symbionts than the set excluding them. This difference comes at the expense of a large loss of proteins with poorly characterized function, in the category unknown function (S), general function prediction only (R), or no COG category assigned. Notably, the categories cell motility (N), defense mechanisms (V), and signal transduction mechanisms (T) contain genes shared between the large genomes, but are completely absent when including reduced genomes.
Host mitochondrial genome sequences and identification
Host mitochondrial scaffolds were recovered from the assembly as a single circular chromosome or in some cases two or three scaffolds that would map to the other stink bug mitochondrial genomes which allowed them to be circularized. All mitochondria ranged between 13 and 16 kbps in size and contained the 13 protein coding genes found in previously described pentatomid mitochondria69, two rRNA genes and 22 tRNAs. Gene order was conserved across all genomes. COX1 genes were searched against the BOLD database confirming species identification for 9 samples, while in 6 cases for specimens collected in the Neotropics a certain match was not found, likely due to the species not being present in the database (See Table S4).
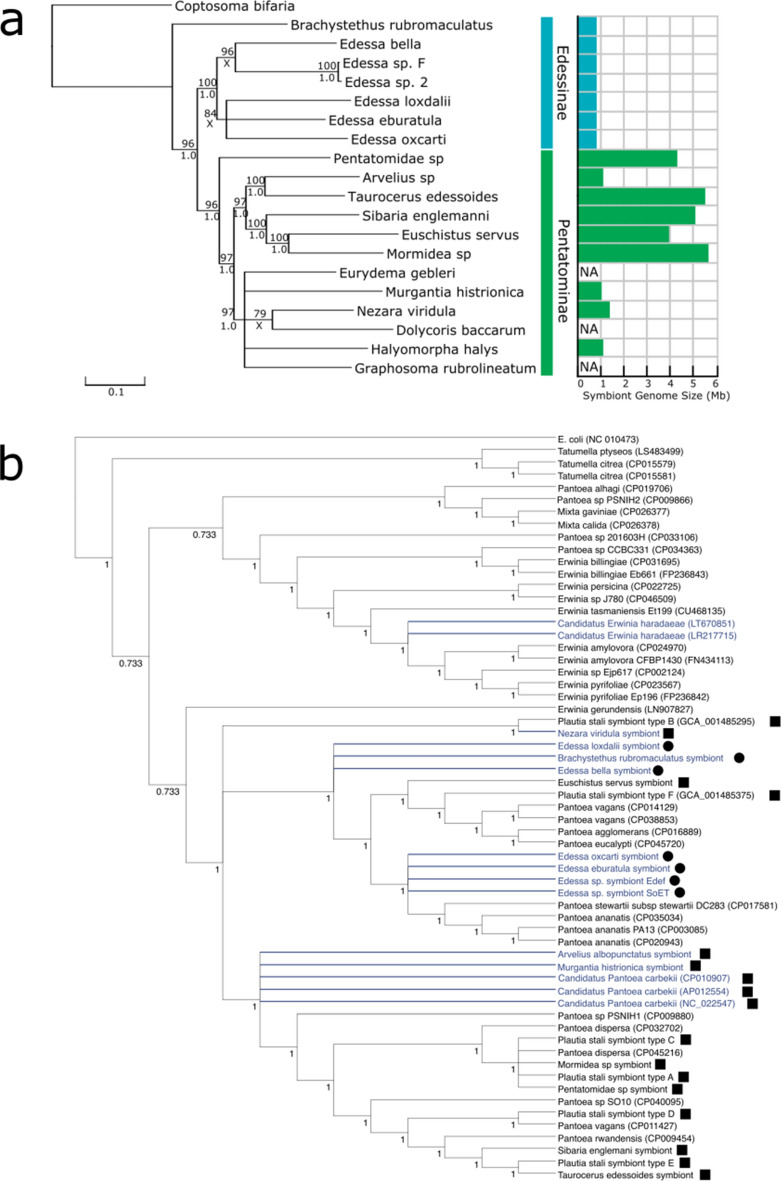
The reconstructed host mitochondrial tree showed good support for the genus Edessa (Fig. 2a, S1), containing the smaller symbiont genomes sequenced. The Brachystethus genus, also in the subfamily Edessinae, contains a symbiont of similar size, however the subfamily node was not well supported under this analysis (Fig. 2a). Additionally, despite similar sizes the genome of SoBr has several key differences from those of the SoE (See below). For the second subfamily, the Pentatominae, we see a large variation in symbiont genome sizes, ranging from moderately reduced (~ 1 Mb) to showing no signs of reduction (> 5 Mb).
Figure 2.
Host and symbiont relationships: (a) Stink bug mitochondrial tree with their respective symbiotic bacterial genome size. Nodes with lower than 60% bootstrap support were collapsed. Numbers above nodes represent bootstrap support while numbers underneath represent posterior probability from Bayesian Inference run. X indicates lower than 0.8 posterior probability. NA-genome size was not available. (b) Consensus cladogram of representative genomes of Erwiniaceae and stink bug symbionts. Individual FastTree reconstructions for the 10 longest protein sequences identified to be common for all taxa using reciprocal best hit Blast. E. coli was used as an outgroup. NCBI accession numbers for genomes are shown in parenthesis. Squares indicate a symbiont of a member of the Pentatominae while circles indicate symbionts of Edessinae. Numbers at nodes indicate Shimodaira-Hasegawa (SH)-like local support values. Taxa that were individually reconstructed are shown as polytomies in blue.
Placement of symbiotic strains within the Erwiniaceae
Traditional phylogenetic reconstruction approaches were not used because long branch attraction artifacts60 often confound accurate reconstruction of phylogenies of genome reduced species. We used a modified approach where separate trees were estimated for each genome reduced taxon and a consensus tree was created where all genomes were placed in the genus Pantoea and stink bug symbionts were shown to be paraphyletic within the genus (Fig. 2b). While these placements are tentative due to the lack of resolution on nodes with long branches, it provides evidence for the paraphyly of these symbionts.
Branched chain amino acid biosynthesis pathway
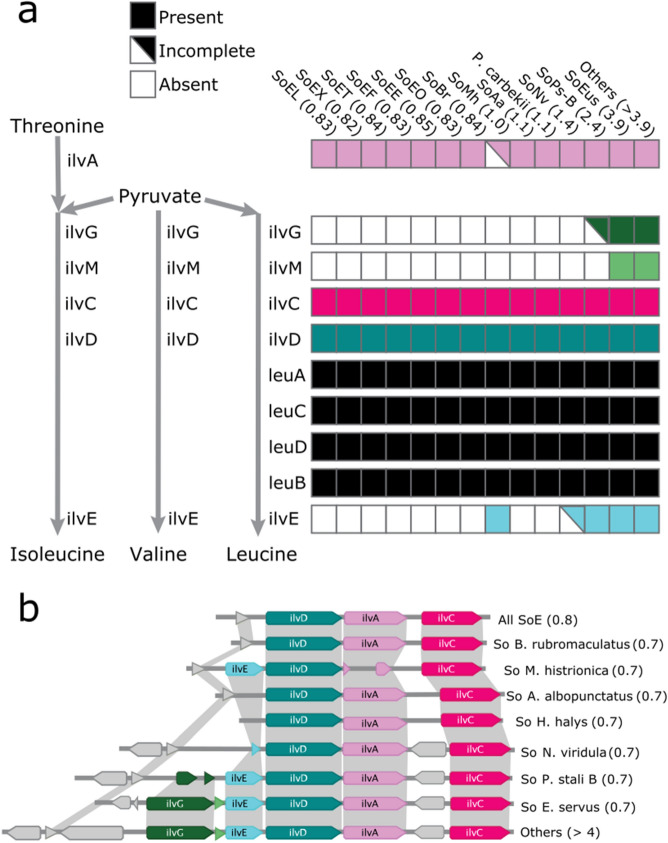
The supplementation of essential amino acids not present in phytophagous pentatomid host diets is one of the likely advantages of these inheritable symbionts18,70. We identified the canonical biosynthetic pathways for these amino acids, and found all genomes contained full pathways for the biosynthesis of all essential amino acids with the exception of the branched chain amino acid biosynthesis pathway. This is in contrast with nonessential amino acid pathways in which several key enzymes are missing from some of the stink bug symbiont genomes (for a more detailed description see36. The only loss in an essential amino acid pathway is in the ilv operon. The ilv operon encoding most of the pathway71 is present in all genomes with the particularity that the ilvE gene which encodes the branched-chain-amino-acid aminotransferase BCAT, the enzyme responsible for the final step in the valine, isoleucine and leucine biosynthesis pathway is missing in all symbionts with a genome under 2 Mb, with the exception of SoMh (Fig. 3a). In the SoNv, ilvE gene is in the process of being pseudogenized, being present as a small ORF that does not include any of the protein’s catalytic sites but retains nucleotide and protein identity to the functional genes in other genomes (Fig. 3b). ilvG and ilvM are also missing from these genomes, which when present are located adjacent to ilvE and in SoPs-B ilvG is in a similar process of pseudogenization as ilvE in SoNv.
Figure 3.
Presence of genes involved in branched chain amino acid biosynthesis pathway. The presence of genes in each genome according to the order of the pathway (a) and gene order and presence in the ilv operon region (b). Numbers in parentheses represent genome sizes in Mb. Colors are used to match genes in between panels for ease of visualization. Unlabeled grey pentagons denote genes not directly involved in the pathway.
While ilvE was present in the genome of the SoMh, being the only genome under 2 Mb to retain this gene, it is also the only genome of those sampled to contain a pseudogenized ilvA gene, which is completely conserved in all others. ilvA encodes L-threonine deaminase, which catalyzes the first step of the synthesis of isoleucine from threonine which shares the remaining steps with the other branch chain amino acid synthesis pathways72.
LPS and antigen biosynthesis gene loss during genome reduction
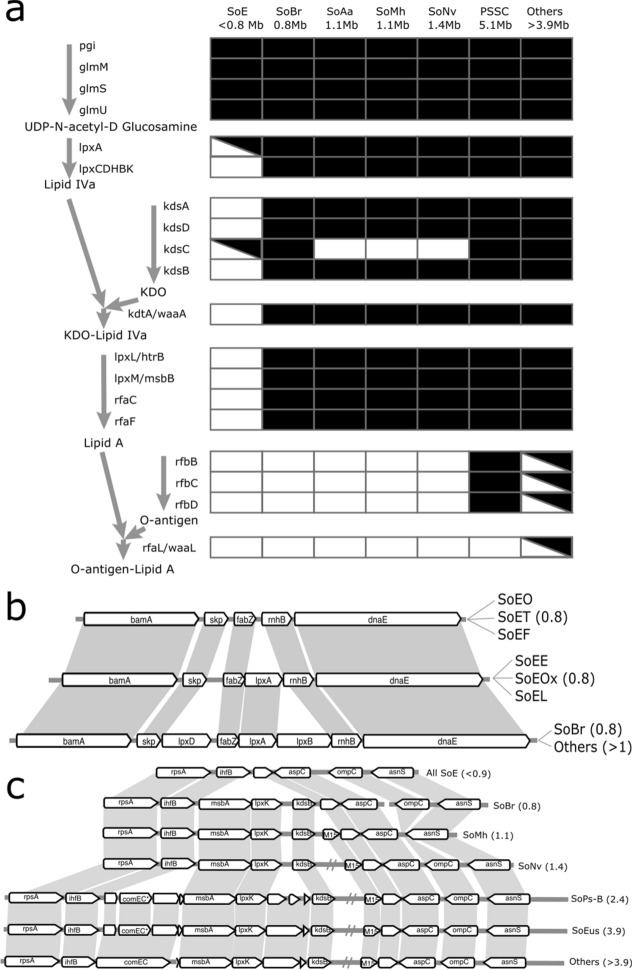
External cell wall components are responsible for protection of the cell in an outside environment but also are in direct contact with the host while in symbiosis. We compared the genes and pathways involved in some important and well conserved cell wall components: lipid A, peptidoglycan (PG), the O-antigen, and the enterobacterial common antigen (ECA). Lipid A is a precursor for the outer cell membrane lipopolysaccharide (LPS) present in Gram-negative bacteria including the Enterobacteria73. All genomes contained the necessary genes for the production of UDP-N-Acetyl-D-glucosamine (UDP-GlcNAc) pgi and the glm operon, which is a necessary precursor for both Lipid A and peptidoglycan74,75. The genes necessary for the production of peptidoglycan, murABCDEFGIJ, ddl, and mraY are also present in all genomes studied.
The canonical pathway encoding the production of lipid A from UDP-GlcNAc includes the genes lpxACDHBK (for the conversion to lipid IVA), kdtA (for the addition of KDO), lpxLM (conversion to KDO-Lipid A) and rfaCFGPQYBOJ (synonym waa) (addition of sugars to Lipid A). The pathway for the synthesis of lipid IVA is conserved in all genomes larger than 1 Mb but lost in smaller ones, with the exception of lpxA which is found in some SoE and SoBr and lpxB which is found in SoBr (Fig. 4a,b). In some cases, these genes are lost without any change to the adjacent genes (such as the case of lpxC, see Figure S2) while in others, regions containing several adjacent genes are lost (such as the case of lpxL, Fig. 4c).
Figure 4.
Genes involved in the production of Lipid A and attachment of O-antigen. (a) Intermediate metabolites are represented in bold and the presence or absence of a gene is represented in the table, with a crossed box indicating not present in all members. (b) Progressive elimination of lpxA, lpxB, and lpxD in the SoE. (c) Loss of lpxK kdsB, and adjacent genes. A double grey line indicates a region of approximately 15 kb containing several genes not shown here. Genes labeled ‘hypoth.’ or unlabeled indicate hypothetical proteins or interrupted CDS. Asterisk indicates a fragmented CDS. Numbers in parentheses correspond to genome sizes in Mb.
The genes lpxD, lpxA, and lpxB, are located syntenically in the Pantoea genomes along with fabZ, rnhB, and dnaE in a conserved pattern (Fig. 4b). This region is disrupted in the different genome reduced symbionts: The SoE lack lpxB and lpxD, and some (SoEO, SoEF) also lack lpxA. Despite this loss, the adjacent genes fabZ and rnhB are retained in these genomes. lpxC is found in all genomes between the ftsZ and secA genes, but it is lost in the SoE, where these two genes are adjacent to each other with no ORFs in the intergenic space. lpxH is also lost on all SoE along with the adjacent gene ppiB. However, genes flanking these two, cysS and purE, are conserved in the remaining genomes, the exception being P. carbekii, where ppiB and lpxH are retained but purEK are lost, which are part of the purine biosynthesis pathway. This loss is also present in SoEO. lpxK is also absent in all SoE genomes, in a gap where several genes are missing. In other genome reduced symbionts, lpxH is retained together with some adjacent genes that are absent in SoE. This region contains several gaps of multiple genes that are conserved in large genomes (Fig. 4c).
Adjacent to this region, we see evidence of pseudogenization of another gene, comEC, in the larger genomes that have begun the reduction process. In SoEus and SoPs-B, with genomes of 3.9 and 2.4 Mb respectively, rpsA and ihfB sit upstream of comEC while msbA and lpxK sit downstream. While in smaller genomes such as SoNv only a small intergenic region between ihfB and msbA remains, in SoEus and SoPs-B the region is roughly the same length as in unreduced genomes and contains several small, interrupted ORFs, some similar to the full protein, but likely not functional.
Another requirement for the production of lipid A are the kdsABCD genes in charge of the production of KDO. The first gene in the pathway, kdsC is the most conserved, present in all genomes except some SoE (SoEL & SoEE retain it). In SoEus, kdsC is split into two ORFs, indicating a possible pseudogenization occurring, and it is unsure if either of these have catalytic function. These results together show how the SoE have a complete or almost complete loss of the lipid A biosynthesis pathway, while others, including the similarly sized symbiont SoBr, conserve all genes.
The O-antigen consists of an oligosaccharide that can be attached to Lipid A to produce a mature LPS. Several glycosyltransferases can be involved in this process, in the case of Pantoea the rfbBCD genes encode enzymes involved in the production of dTDP-beta-L-rhamnose, a monomer of their O-antigen. This operon is absent in all genomes under 2 Mb, as well as some of the larger genomes, including those of the non-associated Pantoea species. The Enterobacterial common antigen, or ECA, is another antigen that can be found in the periplasmic space in a cyclic form, attached to PG or attached to LPS. The genes for its production wecABDEFG, and rffG, as well as the gene in charge of producing the cyclic form wzzE are lost in all genomes smaller than SoPs-B. rfaL, which binds both the O-antigen and the ECA to the LPS, and wzxE and wzyE which are in charge of the export of both antigens76,77 are also lost in smaller genomes.
Discussion
Different stages of reduction among pentatomid stink bugs
There is a large variation in genome sizes of inhabitants of the M4 region of stink bugs in the family Pentatomidae. The placement of these symbionts is in the genus Pantoea consistent with previously discovered pentatomid symbionts30,35,68,78. The true relationships between bacteria with reduced genomes can be incredibly difficult to ascertain because accurate phylogenetic reconstructions are confounded by elevated mutation rates that can result in artifactual long branch attraction. However, the radically different genome size, different placement of taxa in the absence of long branches, as well as considerable differences in the gene order between these different organisms36 are all indications of separate association events at different times. The SoE have almost identical genome size and gene order amongst each other, while being considerably different from that of P. carbekii indicating these to be separately evolving groups. However, gene order, size, and independent phylogenetic placement between P. carbekii and SoMh or SoAa are not similar enough to ascertain that they belong to the same clade and not different enough to claim the contrary. Therefore, while largely different genome sizes are likely an indication of separate clades, similar genome sizes should not be taken as evidence of the same clade and comparisons must be cautious. For example, SoBr is of a similar genome size to the SoE and while B. rubromaculatus is the only other member of the Edessinae that is not in the genus Edessa, there are considerable differences between them, most prominently the latter retaining the full pathway for synthesis of lipid A. Gene order in SoBr is similar to that of the SoE, but to a lesser extent as within the genus, in which gene order is near identical.
While some of these genomes are considerably small for extracellular bacteria, undoubtedly at a late stage of genome reduction, examples such as the symbiont of P. stali-B and the symbiont of N. viridula allow us to see a glimpse of the intermediate stage of this process. Additionally, the larger genomes of symbionts, while retaining a total genome size similar to their non-host associated relatives, show several characteristics of an early stage genome reduction such as a considerable number of pseudogenes and a decrease in the coding density of the genome. This likely reflects a recent symbiont replacement event in which the host associated with a new clade, as SoEus is shown to be closely associated with P. vagans and P. agglomerans while SoPs-A and SoPt are more closely associated with P. dispersa (Fig. 2b). For SoPs-A, a shift in geography may have facilitated this replacement as different populations in separate islands contain different associates, including a more genome reduced SoPs-B27. The variation in genome sizes indicates that the replacement process can be relatively common and actively occurs in species of this family35. Given the extracellular nature of these bacteria and the host transmission and acquisition system, it is not difficult for other bacteria to invade. However, the high symbiont titers of genome reduced species show colonization must be favored for closer associates. In the case that genome reduction proceeds to an irreversible deletion within the symbiont that affects its utility to the host, it may more advantageous for the host to replace it12,79 with a large genome-bearing bacterium and start the process anew.
A wide variety of genome reduced insect symbionts has been described, covering the full range of genome sizes down to near organelle levels and with great diversity in the association to their host. Several of these symbionts have been hypothesized to be in a transitional state towards a stable symbiosis80, while others have severely reduced functions as in the case of intracellular bacteria with small genomes that are approaching organelle status81. Many of these cases require the host evolution of traits such as methods for intracellular vertical transmission82, a second symbiont’s complementation83, or even horizontal gene transfer of symbiont genes to the host84. However, most of these cases allow a limited view on an individual symbiosis without understanding the gradual steps required to achieve it, allowing us only to understand the process of symbiosis by comparing sometimes distant hosts with different organs, lifestyles, and diets. Here, we show how within a single family of stink bugs with little host change of the symbiotic organ we can evidence a range of steps in the development of the symbiosis from the bacterial perspective due to repeated establishment of the symbiosis.
While most of the extremely reduced genomes of symbionts are from intracellular symbionts, which are carefully protected within host cells, extracellular symbionts have additional constraints that likely impede further genome reduction. However, these can be overcome if there is significant investment from the host on structures that guarantee housing and transmission of its bacterium85–87. Stink bug symbionts are housed in separate gut compartments (or crypts) developed with a complex morphogenetic process from birth to adulthood26 and they are externally transmitted17,23,37, although it is unclear to what degree these symbionts are impacted by abiotic conditions outside of the host (i.e. temperature, dessication, UV light) given that they are often ensconced in maternal secretions. Some further strategies have been developed in other stink bug relatives such as a symbiont capsule in Plataspidae88 or transmission jelly in the Urostylididae20 which may have enhanced genome size reduction, but SoE species have similarly-reduced genomes but are not transmitted in either a capsule or jelly34,36. Additionally, some genome reduction may co-occur as a relationship with an amenable domiciling host emerges, as observed in deep sea anglerfish-associated luminescent symbionts with genomes that have undergone ‘extreme reduction’ that is likely impacted by association with anglerfish but independent of obligate intracellular incarceration89,90.
Cases where change is required in the physiology of the host to accommodate a symbiont require longer evolutionary time due to the difference in generation time and population size1. These traits can vary considerably, such as with the different symbiotic organs of Lygaeid bugs91–93, which makes it unlikely for the exact same structure to appear convergently. Thus, comparison of genome reduction for these cases can only be done with distantly related taxa, if at all. In the case of stink bugs, the midgut crypts are common to most of the Pentatomoidea (except in cases where they were subsequently lost such as for carnivorous groups or otherwise modified)17–19,94 and the nymphal probing behavior is also common across the superfamily23,95. Since host traits that enable the symbiosis are almost identical across the group, yet the symbionts appear at radically different stages of association, this system is invaluable for understanding the effects of varying constraints on genome composition.
Common genes after genome reduction
The core genome obtained for the unreduced genomes consists of 2450 genes, which includes between 46.6% and 61.4% of the total genes in the genome. Our estimate is slightly larger than previous core genome analyses of the Pantoea96,97 which estimate them at between 38.8–56% and 30% of the total genes, respectively. However, previous analyses included more distant strains of Pantoea and more samples for some of species which could contribute to their smaller core genome. When including all symbionts down to the SoE, the number of shared genes is reduced to 450 genes, which constitutes between 47.7% and 62% of the genes in the smaller genomes. This estimate is likely low given that these methods use sequence similarity to identify orthologous genes which can result in false negatives because the high mutation rate of genome reduced bacteria significantly lowers the sequence identity of orthologous proteins. We observed some cases where a group of shared genes was incorrectly counted as two non-overlapping groups due to insufficient sequence identity, even though annotations and genome position for the genes was identical. This is a considerable problem for organisms with such high mutation rates and should be carefully considered in comparative genomic frameworks.
Amino acid pathway loss
ilvE is the only gene in the valine, isoleucine, and leucine biosynthesis pathway lost between the core genome of Pantoea and the set of shared genes for all genomes. It encodes BCAT which catalyzes the last step for the production of valine, leucine and isoleucine. This gene is missing from multiple other nutritional symbionts5,98–100. In Buchnera it has been shown the host aphid upregulates its own BCAT in its bacteriocytes3 completing the pathway. BCAT is also present in the brown marmorated stink bug genome (LOC106691811) indicating stink bugs may also be completing this pathway in the symbiosis. The loss of ilvE in these cases also comes with the loss of ilvG and ilvM, which encode the two subunits of acetolactate synthase, which catalyzes the first step of the branched-chain amino acid biosynthesis pathway 101. However, there are three isozymes with this function in other Enterobacteria such as E. coli, among them IlvIH, encoded by ilvI and ilvH102, which is found in all genomes including those of the SoE. The exception to the loss of ilvE is SoMh, which retains a full copy of ilvE. However, it is unique among the stink bug symbionts in the loss of ilvA, a gene encoding L-threonine dehydratase [E.C:4.3.1.19] which is required for a previous step in the biosynthesis of isoleucine (Fig. 4a). This loss is also present in Buchnera and Wigglesworthia, and in aphids can be replaced with the host enzyme TcdB which is expressed and marginally upregulated in the bacteriocytes3. A similar protein is found in the genome of the closest pentatomid genome (H. halys, LOC106681826). This could be evidence of an alternative evolution of a shared pathway between host and symbiont, regulating exclusively the isoleucine pathway instead of all three branched-chain amino acids. This complementation is found in multiple genome reduced symbionts and their hosts, including other pathways such as tyrosine supplementation in weevils103. The loss of a vital step of the biosynthesis pathway for the symbiosis is likely beneficial for the host as it facilitates increased control over the production of the required nutrient and the growth of the symbiotic bacteria104.
The genomic region containing the previously mentioned ilv genes is a clear example of progressive gene decay: SoPs-B contains an identical gene order to large genome symbionts but has lost ilvM and ilvG is in the process of pseudogenization, the next smaller genome SoNv has lost ilvG altogether and ilvE is being pseudogenized, and finally in SoAa and P. carbekii ilvE is lost altogether. Since IlvGM is redundant it is likely one of the fastest genes to disappear, and ilvE or ilvA disappear in the next stage.
Cell membrane component loss during symbiosis
Lipopolysaccharides (LPS) are the main component of the exterior cell membrane of Gram-negative bacteria acting as a protective barrier for the cell. It is composed of Lipid A, which binds to the cell membrane, a core oligosaccharide, and an external polysaccharide chain sometimes referred to as the O-antigen. There are both conserved regions as well as considerable variation in the composition of LPS, and particularly the O-antigen region is a target of host immune responses105. We identified two major changes in the biosynthetic capability of stink bug symbionts with regards to LPS: the first being the loss of addition of O-antigen and ECA and the second being the complete loss of LPS. We found that the genes responsible for the production and attachment of the ECA and O-antigens to lipid A are absent in stink bug symbionts with reduced genomes which would render them unable to produce smooth-type (antigen containing) LPS. In other species of the genus Pantoea and symbionts with regular genome size the rfb operon can be complete (P. rwandensis, SoEus, SoPs-A, SoSe and SoPd) or incomplete (P. ananatis, SoMo, SoTe, and other SoPs) which indicates the addition of the O-antigen is not essential for the survival of the bacteria. E. coli mutants are able to survive without the addition of O-antigen, however, their membrane is considerably more permeable making the it hypersusceptible106. The ECA behaves similarly, while it is not essential for the survival of strains in culture, outer membrane permeability is considerably reduced76.
In the Burkholderia and bean bug Riptortus pedestris symbiosis, symbiont Burkholderia cells produce LPS with O-antigen when grown in culture media but the O-antigen is absent in symbiotic cells. Additionally, symbiotic Burkholderia do not induce host antimicrobial peptides (AMPs), and the expression of host AMPs in the M4 region is lower than the basal expression in the fat bodies. It has been shown that cells without the O-antigen are much more susceptible to cell lysis and host immune responses. However, this downregulation of AMPs in the symbiotic organ likely allows the survival of a weakened symbiotic cell107. It is likely that as in the bean bug-Burkholderia symbiosis, the weakened membrane due to the loss of O-antigen evidenced in the stink bug symbiont genomes is compensated by the host protection and/or changes in its immune reaction. The loss of these genes in stink bug symbionts is likely due to increased pressure to prevent the activation of the host’s immune system or as a different adaptation to the host environment108. Since these symbionts need to travel through the host gut during its first instar in order to colonize, as well as proliferate throughout the host’s development, the loss of this antigen is likely helpful if not necessary for the consistent establishment of symbiont populations, however, a there is a trade-off involved of increased cell permeability which may be related to the inability of genome reduced symbionts to grow in vitro. Additionally, as with Burkholderia, unreduced genome bacteria may be able to stop production of these antigens when in a symbiotic state.
Furthermore, we found that all SoE lacked the complete pathway to produce lipid A (with the exception of the gene lpxA encoding UDP-GlcNAc acyltransferase which catalyzes the first step in this pathway, which was present in SoEL, SoEE, and SoOX). Under normal circumstances the disruption of lipid A is fatal for most gram negative bacteria, with few exceptions109,110. While the SoE lacked the ability to produce lipid A, the production of UDP-GlcNAc remained intact in all genomes. The only other pathway that this metabolite has been associated with is in the biosynthesis of peptidoglycan. All genomes of stink bug symbionts, including I. capsulata and T. gelatinosa, contained all genes necessary for the production of peptidoglycan, which is found in the periplasmic space. Only the most reduced genome symbionts in other systems are able to lose production of peptidoglycan, and these are restricted to intracellular symbionts and those that can rely on host production of peptidoglycan through horizontally transferred genes83,111. Given that stink bug symbionts are extracellular bacteria, we hypothesize that maintenance of LPS in the outer membrane is not necessary for an extracellular symbiosis but peptidoglycan production in the periplasmic space must be conserved likely for cell wall integrity. This hypothesis could be tested in systems such as the extracellular Stammera – tortoise leaf beetle symbiosis, where some symbionts retain some, all, or almost none of the genes required for peptidoglycan biosynthesis86.
Conclusion
Genome reduction has been widely studied across insect symbionts, yet comparative genomic methods studying convergence in genome evolution have been limited to either too distant or too similar instances of symbiosis. Here we show how the extracellular symbionts of stink bugs can be used as a system of similar and convergent instances of genome reduction, as well as covering the range of symbiosis from free living bacteria to highly specialized symbionts. We identify convergence in gene loss in multiple pathways associated with symbiosis and different stages of loss including partial gene losses and ongoing pseudogenization. We identify a convergence in the loss of a single gene in branched chain amino acid biosynthesis, with one example displaying the loss of a different step in the pathway (ilvA as opposed to ilvE) found in other nutritional symbionts, as well as the selective loss of genes involved in the production and attachment of antigens to the cell wall which likely influences interactions with the host. Further study of the diversity and evolution of these symbionts will likely elucidate key factors in the process of genome reduction.
Supplementary Information
Acknowledgements
The authors thank the Organization of Tropical Studies (OTS), La Selva Biological Research Station and staff, and CONAGEBio. This work was supported by The US National Science Foundation (IOS1656786) to Z.L.S., the Ohio Agricultural Research Development Center SEEDS Research Enhancement- Interdisciplinary Grant to Z.L.S., The Ohio State University College of Arts and Sciences, the computation resources of the Ohio Biodiversity Conservation Partnership (OBCP) cluster, and the Colombian Administrative Department of Science, Technology and Innovation COLCIENCIAS to A.O. All field work was approved by the Costa Rican authorities (CONAGEBIO commission from MINAET Ministerio del Ambiente, Energia y Telecomunicaciones) permit number: R-026–2017-OT.
Author contributions
A.O. and Z.L.S. conceived and designed the study. A.O. collected specimens and performed the data analysis. Z.L.S. supervised all data analysis and interpretation. The manuscript was written and edited by A.O. and Z.L.S. All authors approved the manuscript.
Funding
Stink bug mitochondrial genomes have been deposited at GenBank under the NCBI accession numbers MN783643-MN783657 and bacterial genomes have been deposited under BioProject PRJNA413893.
Data availability
These Whole Genome Shotgun projects have been deposited at GenBank under the NCBI accessions SZZU00000000, VOQV00000000, VOQW00000000, SZZT00000000, SZZY00000000, VOQX00000000, SZZX00000000, SZZW00000000, VOQY00000000, SZZV00000000, PDKT00000000, PDKU00000000, PDKR00000000, PDKS00000000, SZZZ00000000.
Data deposition
Stink bug mitochondrial genomes have been deposited at GenBank under the NCBI accession numbers MN783643-MN783657 and bacterial genomes have been deposited under BioProject PRJNA413893.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-86574-8.
References
- 1.Sudakaran S, Kost C, Kaltenpoth M. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 2017 doi: 10.1016/j.tim.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 2.Giron D, et al. Influence of microbial symbionts on plant-insect interactions. Adv. Bot. Res. 2017;81:225–257. doi: 10.1016/bs.abr.2016.09.007. [DOI] [Google Scholar]
- 3.Hansen AK, Moran NA. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2849–2854. doi: 10.1073/pnas.1013465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabree ZL, Kambhampati S, Moran NA. Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc. Natl. Acad. Sci. 2009;106:19521–19526. doi: 10.1073/pnas.0907504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabree ZL, Huang CY, Okusu A, Moran NA, Normark BB. The nutrient supplying capabilities of Uzinura, an endosymbiont of armoured scale insects. Environ. Microbiol. 2013;15:1988–1999. doi: 10.1111/1462-2920.12058. [DOI] [PubMed] [Google Scholar]
- 6.Salem H, et al. Vitamin supplementation by gut symbionts ensures metabolic homeostasis in an insect host. Proc. Biol. Sci. 2014;281:20141838. doi: 10.1098/rspb.2014.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 8.Okude G, et al. Novel bacteriocyte-associated pleomorphic symbiont of the grain pest beetle Rhyzopertha dominica (Coleoptera: Bostrichidae) Zool. Lett. 2017;3:13. doi: 10.1186/s40851-017-0073-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moran NA, Bennett GM. The tiniest tiny genomes. Annu. Rev. Microbiol. 2014 doi: 10.1146/annurev-micro-091213-112901. [DOI] [PubMed] [Google Scholar]
- 10.Nakabachi A, et al. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5477–5482. doi: 10.1073/pnas.0409034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirota B, et al. A novel, extremely elongated, and endocellular bacterial symbiont supports cuticle formation of a grain pest beetle. MBio. 2017;8:e01482–e1517. doi: 10.1128/mBio.01482-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett GM, Moran NA. Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. Proc. Natl. Acad. Sci. 2015;2015:201421388. doi: 10.1073/pnas.1421388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 2011;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 14.Rispe C, Moran NA. Accumulation of deleterious mutations in endosymbionts: Muller’s Ratchet with two levels of selection. Am. Nat. 2000;156:425–441. doi: 10.1086/303396. [DOI] [PubMed] [Google Scholar]
- 15.Wilson ACC, Duncan RP. Signatures of host/symbiont genome coevolution in insect nutritional endosymbioses. Proc. Natl. Acad. Sci. 2015;112:201423305. doi: 10.1073/pnas.1423305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lo W-S, Huang Y-Y, Kuo C-H. Winding paths to simplicity: genome evolution in facultative insect symbionts. FEMS Microbiol. Rev. 2016;2:158–161. doi: 10.1093/femsre/fuw028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salem H, Florez L, Gerardo N, Kaltenpoth M. An out-of-body experience: the extracellular dimension for the transmission of mutualistic bacteria in insects. Proc. Biol. Sci. 2015;282:20142957. doi: 10.1098/rspb.2014.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otero-Bravo A, Sabree ZL. Inside or out? Possible genomic consequences of extracellular transmission of crypt-dwelling stinkbug mutualists. Front. Ecol. Evol. 2015;3:1–7. doi: 10.3389/fevo.2015.00064. [DOI] [Google Scholar]
- 19.Kikuchi Y, Hosokawa T, Fukatsu T. Diversity of bacterial symbiosis in stinkbugs. In: Van Dijk T, editor. Microbial Ecology Research Trends. Hauppauge: Nova Science Publishers, Inc.,; 2008. pp. 39–63. [Google Scholar]
- 20.Kaiwa N, et al. Symbiont-supplemented maternal investment underpinning Host’s ecological adaptation. Curr. Biol. 2014;24:1–6. doi: 10.1016/j.cub.2014.08.065. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi Y, Hosokawa T, Fukatsu T. Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 2007;73:4308–4316. doi: 10.1128/AEM.00067-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olivier-Espejel S, Sabree ZL, Noge K, Becerra JX. Gut microbiota in nymph and adults of the giant mesquite bug (Thasus neocalifornicus) (Heteroptera: Coreidae) is dominated by Burkholderia acquired de novo every generation. Environ. Entomol. 2011;40:1102–1110. doi: 10.1603/EN10309. [DOI] [PubMed] [Google Scholar]
- 23.Hosokawa T, Fukatsu T. Relevance of microbial symbiosis to insect behavior. Curr. Opin. Insect Sci. 2020;39:91–100. doi: 10.1016/j.cois.2020.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Karamipour N, et al. Gammaproteobacteria as essential primary symbionts in the striped shield bug, Graphosoma lineatum (Hemiptera: Pentatomidae) Sci. Rep. 2016;6:33168. doi: 10.1038/srep33168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scopel W, Cônsoli FL. Culturable symbionts associated with the reproductive and digestive tissues of the Neotropical brown stinkbug Euschistus heros. Antonie Van Leeuwenhoek. 2018 doi: 10.1007/s10482-018-1130-9. [DOI] [PubMed] [Google Scholar]
- 26.Oishi S, Moriyama M, Koga R, Fukatsu T. Morphogenesis and development of midgut symbiotic organ of the stinkbug Plautia stali (Hemiptera: Pentatomidae) Zool. Lett. 2019;5:16. doi: 10.1186/s40851-019-0134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosokawa T, et al. Obligate bacterial mutualists evolving from environmental bacteria in natural insect populations. Nat. Microbiol. 2016;1:15011. doi: 10.1038/nmicrobiol.2015.11. [DOI] [PubMed] [Google Scholar]
- 28.Tada A, et al. Obligate association with gut bacterial symbiont in Japanese populations of the southern green stinkbug Nezara viridula (Heteroptera: Pentatomidae) Appl. Entomol. Zool. 2011;46:483–488. doi: 10.1007/s13355-011-0066-6. [DOI] [Google Scholar]
- 29.Otero-Bravo A, Sabree ZL. Comparing the utility of host and primary endosymbiont loci for predicting global invasive insect genetic structuring and migration patterns. Biol. Control. 2018;116:10–16. doi: 10.1016/j.biocontrol.2017.04.003. [DOI] [Google Scholar]
- 30.Bansal R, Michel A, Sabree Z. The crypt-dwelling primary bacterial symbiont of the polyphagous pentatomid pest Halyomorpha halys (Hemiptera: Pentatomidae) Environ. Entomol. 2014;43:617–625. doi: 10.1603/EN13341. [DOI] [PubMed] [Google Scholar]
- 31.Itoh H, Matsuura Y, Hosokawa T, Fukatsu T, Kikuchi Y. Obligate gut symbiotic association in the sloe bug Dolycoris baccarum (Hemiptera: Pentatomidae) Appl. Entomol. Zool. 2016 doi: 10.1007/s13355-016-0453-0. [DOI] [Google Scholar]
- 32.Kikuchi Y, et al. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol. 2009;7:2. doi: 10.1186/1741-7007-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kikuchi Y, Hosokawa T, Nikoh N, Fukatsu T. Gut symbiotic bacteria in the cabbage bugs Eurydema rugosa and Eurydema dominulus (Heteroptera: Pentatomidae) Appl. Entomol. Zool. 2012;47:1–8. doi: 10.1007/s13355-011-0081-7. [DOI] [Google Scholar]
- 34.Bistolas KSI, Sakamoto RI, Fernandes JAM, Goffredi SK. Symbiont polyphyly, co-evolution, and necessity in pentatomid stinkbugs from Costa Rica. Front. Microbiol. 2014;5:1–15. doi: 10.3389/fmicb.2014.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hosokawa T, Matsuura Y, Kikuchi Y, Fukatsu T. Recurrent evolution of gut symbiotic bacteria in pentatomid stinkbugs. Zool. Lett. 2016;2:24. doi: 10.1186/s40851-016-0061-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otero-Bravo A, Goffredi S, Sabree ZL. Cladogenesis and genomic streamlining in extracellular endosymbionts of tropical stink bugs. Genome Biol. Evol. 2018;10:680–693. doi: 10.1093/gbe/evy033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenyon LJ, Meulia T, Sabree ZL. Habitat visualization and genomic analysis of ‘Candidatus Pantoea carbekii’, the primary symbiont of the brown marmorated stink bug. Genome Biol. Evol. 2015;7:620–635. doi: 10.1093/gbe/evv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Duzee EP. Annotated list of the pentatomidæ recorded from America North of Mexico, with descriptions of some new species. Trans. Am. Entomol. Soc. 1904;30:1–80. [Google Scholar]
- 39.Rolston LH, McDonald FJD. Keys and diagnoses for the families of Western Hemisphere Pentatomoidea, subfamilies of Pentatomidae and Tribes of Pentatominae (Hemiptera) J. N. Y. Entomol. Soc. 1979;87:189–207. [Google Scholar]
- 40.Rolston LH, McDonald FJD, Thomas DBJ. A Conspecuts of Pentatomini genera of the Western Hemisphere. Part I (Hemiptera: Pentatomidae) J. N. Y. Entomol. Soc. 1980;88:120–132. [Google Scholar]
- 41.Rolston LH, McDonald FJD. Conspectus of Pentatomini genera of the western hemisphere: part 2 (Hemiptera: Pentatomidae) J. N. Y. Entomol. Soc. 1980;88:257–272. [Google Scholar]
- 42.Rolston LH, McDonald FJD. A conspectus of Pentatomini of the western hemisphere. Part 3 (Hemiptera: Pentatomidae) J. N. Y. Entomol. Soc. 1984;92:69–86. [Google Scholar]
- 43.Barcellos A, Grazia J. Revision of Brachystethus (Heteroptera, Pentatomidae, Edessinae) Iheringia. Série Zool. 2003;93:413–446. doi: 10.1590/S0073-47212003000400008. [DOI] [Google Scholar]
- 44.Do Nascimento DA, Da Silva Mendonça MT, Fernandes JAM. Description of a new group of species of Edessa (Hemiptera: Pentatomidae: Edessinae) Zootaxa. 2017;4254:136–150. doi: 10.11646/zootaxa.4254.1.10. [DOI] [PubMed] [Google Scholar]
- 45.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 52.Aziz RK, et al. The RAST server: rapid annotations using subsystems technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tatusova T, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haft DH, et al. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018;46:D851–D860. doi: 10.1093/nar/gkx1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waterhouse RM, et al. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2018;35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huerta-Cepas J, et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caspi R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016;44:D471–D480. doi: 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Husník F, Chrudimský T, Hypša V. Multiple origins of endosymbiosis within the Enterobacteriaceae (γ-Proteobacteria): convergence of complex phylogenetic approaches. BMC Biol. 2011;9:87. doi: 10.1186/1741-7007-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pruesse E, Peplies J, Glöckner FO. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Price MN, Dehal PS, Arkin AP. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38:W7–W13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ratnasingham, S. & Hebert, P. D. N. bold: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes7, 355–364 (2007). [DOI] [PMC free article] [PubMed]
- 65.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duron O, Noël V. A wide diversity of Pantoea lineages are engaged in mutualistic symbiosis and cospeciation processes with stinkbugs. Environ. Microbiol. Rep. 2016;8:715–727. doi: 10.1111/1758-2229.12432. [DOI] [PubMed] [Google Scholar]
- 69.Yuan M-L, Zhang Q-L, Guo Z-L, Wang J, Shen Y-Y. Comparative mitogenomic analysis of the superfamily Pentatomoidea (Insecta: Hemiptera: Heteroptera) and phylogenetic implications. BMC Genom. 2015;16:460. doi: 10.1186/s12864-015-1679-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taylor C, Johnson V, Dively G. Assessing the use of antimicrobials to sterilize brown marmorated stink bug egg masses and prevent symbiont acquisition. J. Pest Sci. 2017;2004(90):1287–1294. doi: 10.1007/s10340-016-0814-z. [DOI] [Google Scholar]
- 71.Smith JM, Smolin DE, Umbarger HE. Polarity and the regulation of the ilv gene cluster in Escherichia coli strain K-12. MGG Mol. Gen. Genet. 1976;148:111–124. doi: 10.1007/BF00268374. [DOI] [PubMed] [Google Scholar]
- 72.Favre R, Iaccarino M, Levinthal M. Complementation between different mutations in the ilvA gene of Escherichia coli K-12. J. Bacteriol. 1974;119:1069–1071. doi: 10.1128/JB.119.3.1069-1071.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X, Quinn PJ. Lipopolysaccharide: biosynthetic pathway and structure modification. Prog. Lipid Res. 2010;49:97–107. doi: 10.1016/j.plipres.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 74.Heijenoort JV. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology. 2001;11:25R–36R. doi: 10.1093/glycob/11.3.25R. [DOI] [PubMed] [Google Scholar]
- 75.Opiyo SO, Pardy RL, Moriyama H, Moriyama EN. Evolution of the Kdo2-lipid A biosynthesis in bacteria. BMC Evol. Biol. 2010;10:362. doi: 10.1186/1471-2148-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mitchell AM, Srikumar T, Silhavy TJ. Cyclic enterobacterial common antigen maintains the outer membrane permeability barrier of Escherichia coli in a manner controlled by YhdP. MBio. 2018 doi: 10.1128/mBio.01321-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Danese PN, et al. Accumulation of the enterobacterial common antigen lipid II biosynthetic intermediate stimulates degP transcription in Escherichia coli. J. Bacteriol. 1998;180:5875–5884. doi: 10.1128/JB.180.22.5875-5884.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Prado SS, Almeida RPP. Phylogenetic placement of pentatomid stink bug gut symbionts. Curr. Microbiol. 2009;58:64–69. doi: 10.1007/s00284-008-9267-9. [DOI] [PubMed] [Google Scholar]
- 79.McCutcheon JP, Boyd BM, Dale C. The life of an insect endosymbiont from the cradle to the grave. Curr. Biol. 2019;29:R485–R495. doi: 10.1016/j.cub.2019.03.032. [DOI] [PubMed] [Google Scholar]
- 80.Clayton AL, Jackson DG, Weiss RB, Dale C. Adaptation by deletogenic replication slippage in a nascent symbiont. Mol. Biol. Evol. 2016;33:1957–1966. doi: 10.1093/molbev/msw071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tamames J, et al. The frontier between cell and organelle: genome analysis of Candidatus Carsonella ruddii. BMC Evol. Biol. 2007;7:181. doi: 10.1186/1471-2148-7-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dan H, Ikeda N, Fujikami M, Nakabachi A. Behavior of bacteriome symbionts during transovarial transmission and development of the Asian citrus psyllid. PLoS ONE. 2017;12:e0189779. doi: 10.1371/journal.pone.0189779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bublitz DC, et al. Peptidoglycan production by an insect-bacterial mosaic. Cell. 2019;179:703–712.e7. doi: 10.1016/j.cell.2019.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Husnik F, et al. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell. 2013;153:1567–1578. doi: 10.1016/j.cell.2013.05.040. [DOI] [PubMed] [Google Scholar]
- 85.Salem H, et al. Drastic genome reduction in an herbivore’s pectinolytic symbiont. Cell. 2017 doi: 10.1016/j.cell.2017.10.029. [DOI] [PubMed] [Google Scholar]
- 86.Salem H, et al. Symbiont digestive range reflects host plant breadth in herbivorous beetles. Curr. Biol. 2020;30:2875–2886.e4. doi: 10.1016/j.cub.2020.05.043. [DOI] [PubMed] [Google Scholar]
- 87.Reis F, et al. Bacterial symbionts support larval sap feeding and adult folivory in (semi-)aquatic reed beetles. Nat. Commun. 2020;11:2964. doi: 10.1038/s41467-020-16687-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fukatsu T, Hosokawa T. Capsule-transmitted gut symbiotic bacterium of the Japanese common plataspid stinkbug, Megacopta punctatissima. Appl. Environ. Microbiol. 2002;68:389–396. doi: 10.1128/AEM.68.1.389-396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hendry TA, et al. Ongoing transposon-mediated genome reduction in the luminous bacterial symbionts of deep-sea ceratioid anglerfishes. MBio. 2018;9:e01033–e1118. doi: 10.1128/mBio.01033-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Baker LJ, et al. Diverse deep-sea anglerfishes share a genetically reduced luminous symbiont that is acquired from the environment. Elife. 2019 doi: 10.7554/eLife.47606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Matsuura Y, Kikuchi Y, Meng XY, Koga R, Fukatsu T. Novel clade of alphaproteobacterial endosymbionts associated with stinkbugs and other arthropods. Appl. Environ. Microbiol. 2012;78:4149–4156. doi: 10.1128/AEM.00673-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kuechler SM, Renz P, Dettner K, Kehl S. Diversity of symbiotic organs and bacterial endosymbionts of: Lygaeoid bugs of the families blissidae and lygaeidae (Hemiptera: Heteroptera: Lygaeoidea) Appl. Environ. Microbiol. 2012;78:2648–2659. doi: 10.1128/AEM.07191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kuechler SM, Fukatsu T, Matsuura Y. Repeated evolution of bacteriocytes in lygaeoid stinkbugs. Environ. Microbiol. 2019;21:4378–4394. doi: 10.1111/1462-2920.14804. [DOI] [PubMed] [Google Scholar]
- 94.Kikuchi Y, Hosokawa T, Fukatsu T. An ancient but promiscuous host-symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J. 2011;5:446–460. doi: 10.1038/ismej.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hosokawa T, Kikuchi Y, Shimada M, Fukatsu T. Symbiont acquisition alters behaviour of stinkbug nymphs. Biol. Lett. 2008;4:45–48. doi: 10.1098/rsbl.2007.0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang L, Wang J, Jing C. Comparative genomic analysis reveals organization, function and evolution of ars genes in Pantoea spp. Front. Microbiol. 2017;8:471. doi: 10.3389/fmicb.2017.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Palmer M, Steenkamp ET, Coetzee MPA, Blom J, Venter SN. Genome-based characterization of biological processes that differentiate closely related bacteria. Front. Microbiol. 2018;9:113. doi: 10.3389/fmicb.2018.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wilson ACC, et al. Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Insect Mol. Biol. 2010;19:249–258. doi: 10.1111/j.1365-2583.2009.00942.x. [DOI] [PubMed] [Google Scholar]
- 99.Rio RVM, et al. Insight into the transmission biology and species-specific functional capabilities of tsetse (Diptera: glossinidae) obligate symbiont Wigglesworthia. MBio. 2012;3:e00240–e311. doi: 10.1128/mBio.00240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mccutcheon, J. P., Moran, N. A. & Greenberg, E. P. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. www.pnas.org/cgi/content/full/ (2007). [DOI] [PMC free article] [PubMed]
- 101.Hill MC, Pang SS, Duggleby GR. Purification of Escherichia coli acetohydroxyacid synthase isoenzyme II and reconstitution of active enzyme from its individual pure subunits. Biochem. J. 1997;327(3):891–898. doi: 10.1042/bj3270891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lawther RP, Calhoun DH, Adams CW, Hauser CA, Gray J, Hatfield GW. (1981) Molecular basis of valine resistance in Escherichia coli K-12. Proc Natl Acad Sci. 1981;78(2):922–925. doi: 10.1073/pnas.78.2.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Anbutsu H, et al. Small genome symbiont underlies cuticle hardness in beetles. Proc. Natl. Acad. Sci. 2017 doi: 10.1073/pnas.1712857114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ankrah NYD, Chouaia B, Douglas AE. The cost of metabolic interactions in symbioses between insects and bacteria with reduced genomes. MBio. 2018 doi: 10.1128/mBio.01433-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Carlsson A, Nystrom T, de Cock H, Bennich H. Attacin an insect immune protein binds LPS and triggers the specific inhibition of bacterial outer membrane protein synthesis. Microbiology. 1998;144:2179–2188. doi: 10.1099/00221287-144-8-2179. [DOI] [PubMed] [Google Scholar]
- 106.Murata T, Tseng W, Guina T, Miller SI, Nikaido H. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar typhimurium. J. Bacteriol. 2007;189:7213–7222. doi: 10.1128/JB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim JK, Park HY, Lee BL. The symbiotic role of O-antigen of Burkholderia symbiont in association with host Riptortus pedestris. Dev. Comp. Immunol. 2016;60:202–208. doi: 10.1016/j.dci.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 108.Kobayashi H, Kawasaki K, Takeishi K, Noda H. Symbiont of the stink bug Plautia stali synthesizes rough-type lipopolysaccharide. Microbiol. Res. 2011;167:48–54. doi: 10.1016/j.micres.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 109.Steeghs L, et al. Meningitis bacterium is viable without endotoxin. Nature. 1998;392:449–449. doi: 10.1038/33046. [DOI] [PubMed] [Google Scholar]
- 110.Zhang G, Meredith TC, Kahne D. On the essentiality of lipopolysaccharide to Gram-negative bacteria. Curr. Opin. Microbiol. 2013;16:779–785. doi: 10.1016/j.mib.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Skidmore IH, Hansen AK. The evolutionary development of plant-feeding insects and their nutritional endosymbionts. Insect Sci. 2017;24:910–928. doi: 10.1111/1744-7917.12463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
These Whole Genome Shotgun projects have been deposited at GenBank under the NCBI accessions SZZU00000000, VOQV00000000, VOQW00000000, SZZT00000000, SZZY00000000, VOQX00000000, SZZX00000000, SZZW00000000, VOQY00000000, SZZV00000000, PDKT00000000, PDKU00000000, PDKR00000000, PDKS00000000, SZZZ00000000.
Stink bug mitochondrial genomes have been deposited at GenBank under the NCBI accession numbers MN783643-MN783657 and bacterial genomes have been deposited under BioProject PRJNA413893.