In a recent study published in Science Immunology,1 Kusnadi et al. performed single-cell RNA sequencing (scRNA-seq) of SARS-CoV-2-reactive CD8+ T cells and reported heterogeneity.
SARS-CoV-2 infection causes COVID-19, which is an ongoing pandemic disease threatening public health. The virology of SARS-CoV-2 and immune responses against the virus have been urgently investigated to develop effective measures against COVID-19. During viral infection, CD8+ T cells contribute to elimination of the virus by exerting cytotoxicity against virus-infected cells and producing effector cytokines, whereas neutralizing antibodies interfere with viral entry of host cells.
After the emergence of COVID-19, early studies examined the phenotypes and functions of various subtypes of immune cells from infected patients using high-dimensional techniques, including scRNA-seq and multi-parameter cytometry.2,3 These studies also revealed the profiles of CD8+ and CD4+ T cells in patients with COVID-19. However, the data did not include information regarding virus-specificity of T cells because these studies analyzed total CD8+ or CD4+ T cells, not SARS-CoV-2-reactive CD8+ or CD4+ T cells.
Other studies have detected and characterized SARS-CoV-2-reactive CD8+ and CD4+ T cells using ex vivo antigen stimulation-based assays, including interferon (IFN)-γ ELISpot assays, intracellular cytokine staining (ICS), and activation-induced marker (AIM) assays.4 Intriguingly, SARS-CoV-2-reactive CD8+ and CD4+ T cells have been detected not only in COVID-19 patients and convalescents, but also unexposed individuals. MHC class I (MHC-I) multimers were also used to directly detect SARS-CoV-2-specific CD8+ T cells without ex vivo stimulation, and their phenotypes were examined among COVID-19 patients and convalescents.5 Although these studies examined the phenotypes and functions of SARS-CoV-2-reactive T cells, high-dimensional techniques, such as scRNA-seq, could not be combined; thus, the deep profiles of SARS-CoV-2-reactive T cells have not been elucidated.
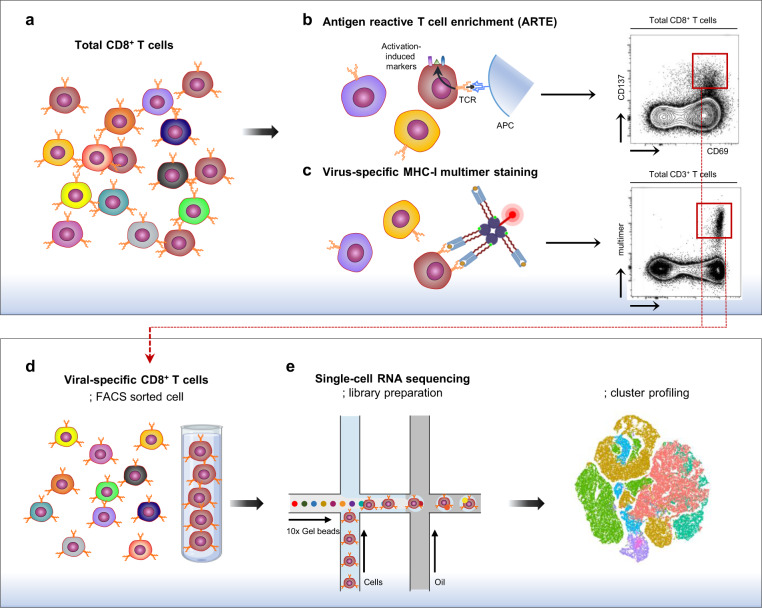
In a recent study, Kusnadi et al. examined landscapes of the SARS-CoV-2-reactive CD8+ T-cell population in a comparison with influenza A virus (IAV)-reactive and respiratory syncytial virus (RSV)-reactive CD8+ T-cell populations by scRNA-seq analysis.1 First, they isolated each virus-reactive CD8+ T-cell population from the peripheral blood mononuclear cells (PBMCs) of patients with COVID-19, or healthy donors via modified antigen-reactive T-cell enrichment (ARTE) (Fig. 1). In modified ARTE, PBMCs were stimulated ex vivo for 24 h with overlapping peptide pools for each viral protein, and responding CD8+ T cells were isolated based on the expression of activation markers CD137 and CD69. Next, they performed scRNA-seq analysis of each viral protein-reactive CD8+ T-cell population.
Fig. 1. Single-cell RNA sequencing of virus-specific CD8+ T cells.
a Total CD8+ T cells include CD8+ T cells with various antigen specificity. b, c Virus-specific CD8+ T cells are fluorescently detected by activation-induced markers, such as CD69 and CD137, following ex vivo stimulation with viral antigens (b) or MHC-I multimer staining (c). d Virus-specific CD8+ T cells are enriched by sorting fluorescently stained cells. The procedure enriching activation-induced marker+ cells is called antigen-reactive T-cell enrichment (ARTE). e Enriched virus-specific CD8+ T cells are analyzed by single-cell RNA sequencing, and single-cell heterogeneity is revealed
They analyzed the single-cell transcriptome and T cell receptor (TCR) sequence of >84,000 virus-reactive CD8+ T cells from 49 subjects in total, including patients with COVID-19 and healthy donors. Virus-reactive CD8+ T cells created seven clusters according to gene expression profiles, indicating heterogeneity among virus-reactive CD8+ T cells. They then described distinct characteristics of SARS-CoV-2-reactive CD8+ T cells compared to IAV-reactive or RSV-reactive CD8+ T cells. SARS-CoV-2-reactive CD8+ T cells from patients with COVID-19 and healthy donors were mainly composed of clusters enriched with T-cell exhaustion signature genes, IFN-stimulated genes, and cytotoxicity-related genes. In contrast, IAV-reactive or RSV-reactive CD8+ T cells were mainly composed of clusters enriched with inflammatory cytokine genes. They concluded that SARS-CoV-2-reactive CD8+ T cells exhibit exhausted phenotypes with type I IFN stimulation, and have a decreased capacity to secrete inflammatory cytokines.
Focusing on the transcriptome and TCR sequence data of SARS-CoV-2-reactive CD8+ T cells from patients with mild and severe COVID-19, they attempted to differentiate mild and severe COVID-19. SARS-CoV-2-reactive CD8+ T cells from patients with severe COVID-19 had a significantly lower frequency of the exhausted cluster than mild patients. When the analysis was narrowed down to the exhausted cluster, severe COVID-19-specific upregulated genes were highly enriched with cytotoxicity-related genes, pro-inflammatory cytokine genes, and genes for T-cell activation-associated transcription factors and negatively enriched with IFN response genes. These findings suggest that SARS-CoV-2-reactive CD8+ T cells are less exhausted, and more functional with an impaired type I IFN response in severe compared to mild COVID-19.
They also analyzed the non-exhausted cluster. Severe COVID-19-specific upregulated genes were enriched with genes related to co-stimulation and NF-κB activation, suggesting that SARS-CoV-2-reactive CD8+ T cells are more activated in patients with severe disease than those with mild disease. In the analysis of TCR clonality, clonal expansion was increased in SARS-CoV-2-reactive CD8+ T cells from patients with severe disease compared to those with mild disease. Collectively, SARS-CoV-2-reactive CD8+ T cells present a robust response in severe patients.
Kusnadi et al. reported a valuable resource for understanding the heterogeneity of the host immune response against SARS-CoV-2 infection by investigating SARS-CoV-2 reactive CD8+ T cells with the modified ARTE assay and scRNA-seq analysis. Unlike previous studies investigating total CD8+ T cells, this study described a landscape of SARS-CoV-2-reactive CD8+ T cells isolated by modified ARTE for the first time.
ARTE is a useful technique for enriching T cells reactive to specific antigens. However, it has inherent limitations for the proper characterization of antigen-reactive CD8+ T cells. Because the process for ARTE includes ex vivo stimulation of T cells with overlapping peptide antigens, the phenotypes and transcriptomes of antigen-reactive T cells can be changed by stimulation. In addition, ARTE cannot capture antigen-specific, non-functioning T cells. This is critical because a considerable proportion of antigen-specific CD8+ T cells detected by MHC-I multimer staining do not exert effector functions.5 These limitations can be overcome by using DNA barcode-tagged MHC-I multimers in scRNA-seq analysis (Fig. 1). MHC-I multimer staining enables the detection of virus-specific CD8+ T cells without stimulation regardless of their functions. However, MHC-I multimer combined scRNA-seq analysis has not yet been reported in the study of SARS-CoV-2-specific CD8+ T cells in patients with COVID-19.
One of the main findings by Kusnadi et al. is that SARS-CoV-2-reactive CD8+ T cells are mainly clustered in the exhausted subset. However, a recent study demonstrated that, among SARS-CoV-2-specific CD8+ T cells detected by MHC-I multimer staining, PD-1+ cells, as well as PD-1− cells, produced IFN-γ in patients with COVID-19 regardless of disease severity, indicating that SARS-CoV-2-specific CD8+ T cells are not exhausted, but functional.5 Further studies are required to examine the functional characteristics of CD8+ T cells in the exhausted cluster identified by Kusnadi et al. In addition, further studies are required to reveal a possible association between co-morbidities of COVID-19 patients and T cell functions.
The COVID-19 pandemic has urged us to investigate host immune responses, including the SARS-CoV-2-specific T-cell response. High-dimensional analysis adopting ARTE or MHC-I multimers will uncover the molecular characteristics, functions, and heterogeneity of SARS-CoV-2-specific CD8+ T cells in COVID-19 patients.
Acknowledgements
This research was supported by the 2020 Joint Research Project of Institutes of Science and Technology.
Competing interests
The authors declare no competing interests.
References
- 1.Kusnadi A, et al. Severely ill COVID-19 patients display impaired exhaustion features in SARS-CoV-2-reactive CD8(+) T cells. Sci. Immunol. 2021;6:eabe4782. doi: 10.1126/sciimmunol.abe4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathew D, et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JS, et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020;5:eabd1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grifoni A, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed Individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rha MS, et al. PD-1-expressing SARS-CoV-2-specific CD8(+) T cells are not exhausted, but functional in patients with COVID-19. Immunity. 2021;54:44–52. doi: 10.1016/j.immuni.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]