Abstract
Macrolide-resistant Streptococcus suis is highly prevalent worldwide. The acquisition of the erm(B) gene mediated by mobile genetic elements (MGEs) in particular integrative and conjugative elements (ICEs) is recognized as the main reason for the rapid spread of macrolide-resistant streptococcal strains. However, knowledge about different erm(B)-carrying elements responsible for the widespread of macrolide resistance and their transferability in S. suis remains poorly understood. In the present study, two erm(B)- and tet(O)-harboring putative ICEs, designated as ICESsuYSB17_rplL and ICESsuYSJ15_rplL, and a novel erm(B)- and aadE-spw-like-carrying genomic island (GI), named GISsuJHJ17_rpsI, were identified to be excised from the chromosome and transferred among S. suis strains with different serotypes. ICESsuYSB17_rplL and ICESsuYSJ15_rplL were integrated downstream the rplL gene, a conserve locus of the ICESa2603 family. GISsuJHJ17_rpsI, with no genes belonging to the conjugation module, was integrated into the site of rpsI. All transconjugants did not exhibit obvious fitness cost by growth curve and competition assays when compared with the recipient. The results demonstrate that different erm(B)-carrying elements were presented and highlight the role of these elements in the dissemination of macrolide resistance in S. suis.
Keywords: erm(B), ICEs, GIs, horizontal transfer, S. suis
Introduction
The rapid increase of macrolide resistance in Streptococcus has been reported worldwide from both pig and human isolates during the past two decades (Princivalli et al., 2009; Palmieri et al., 2011; Vela et al., 2017). Although numerous resistance genes have been reported since the early 1980’s1 (Roberts, 2008), macrolide resistance in streptococci is primarily due to the ribosomal alteration of the 23S rRNA target site by methylases encoded by the erm genes, predominantly erm(B), which mediate resistance to macrolides, lincosamides, and streptogramin B (MLSB) antimicrobials, and active efflux by the mef and msr genes (Wilson, 2014; Fyfe et al., 2016). These resistance genes are frequently carried by mobile genetic elements (MGEs), such as plasmids, transposons, prophages, and more recently, integrative and conjugative elements (ICEs) (Horaud et al., 1985; Woodbury et al., 2008; Varaldo et al., 2009; Huang et al., 2016b,c; Feßler et al., 2018; Libante et al., 2019). ICEs primarily reside in the bacterial chromosome and can excise from the donor chromosome to form a circular molecule that can be horizontally self-transferred to a recipient cell by conjugation (Bellanger et al., 2014). Other chromosomal elements, including integrative and mobilizable elements (IMEs), which encode a recombinase and only some conjugation proteins, and some genomic islands (GIs), which encode a recombinase but do not encode any conjugation proteins, were recently found to be mobilized in trans by ICEs (Daccord et al., 2010) and might have played crucial roles in bacterial evolution.
The erm(B) gene was originally identified on a 5,266 bp transposon Tn917 from Enterococcus faecalis (Tomich et al., 1979). In human streptococci strains, the erm(B)-containing Tn917 was usually integrated into Tn916 (designated as Tn3872), which also carries the tetracycline resistance gene tet(M) (Brenciani et al., 2007; Varaldo et al., 2009). Further, two other erm(B)-containing elements, erm(B) element and macrolide–aminoglycoside–streptothricin element, were frequently inserted into tet(M)-carrying Tn916-like structure (e.g., Tn6002/Tn6003, Tn1545, Tn2009/Tn2010) (Varaldo et al., 2009; Marosevic et al., 2017). This genetic linkage between erm(B) and tet(M) on different MGEs was considered to be the primary mechanism for the spread of streptococcal bacteria that are resistant to both macrolide and tetracycline antimicrobials (Brenciani et al., 2007; Cochetti et al., 2008; Xu et al., 2010). However, in the zoonotic pathogen Streptococcus suis, the linkage between erm(B) and tet(O) was more frequently detected in different countries (Martel et al., 2005; Gurung et al., 2015; Huang et al., 2015; Bojarska et al., 2016; Pan et al., 2019), suggesting that MGEs responsible for macrolide and tetracycline resistance might be different from other streptococci (Huang et al., 2016b,c). S. suis is a key antibiotic resistance gene reservoir and a major zoonotic pathogen responsible for severe economic loss to the swine industry. This bacterium causes specific diseases in humans after contact with infected animals or derived food products. It caused human infection outbreaks in China in 1998 and 2005, respectively, and sporadic cases of S. suis infections in humans have occurred occasionally worldwide (Hui et al., 2005; Mazokopakis et al., 2005; Yu et al., 2006; Mai et al., 2008; CDC, 2013; Huang et al., 2019). Recent studies have demonstrated that the erm(B) and tet(O) genes co-existed on different ICEs in S. suis isolates of both pig and human origins (Holden et al., 2009; Zhang et al., 2011; Huang et al., 2016a,c). Previous results from our laboratory and other investigators have confirmed the intra-species transfer of the erm(B)- and tet(O)-carrying ICEs by conjugation (Huang et al., 2016a,c; Zhou et al., 2017; Pan et al., 2019). However, knowledge about types of erm(B) elements responsible for widespread macrolide resistance remains rare. In the present study, we identified three erm(B)-carrying transferable elements, including two erm(B)- and tet(O)-harboring putative ICEs, belonging to the ICESa2603 family, and a novel erm(B)-carrying GI, which can be horizontally transferred among S. suis strains with different serotypes.
Materials and Methods
Bacterial Strains and Culture Condition
In this study, a total of 320 S. suis isolates obtained from humans and pigs in China from 2005 to 2018 were included. All S. suis strains were routinely cultivated on Todd–Hewitt broth (THB) or Todd–Hewitt agar (THA) plates supplemented with 5% calf serum at 37°C.
Genomic DNA Extraction and PCR Amplification
The crude genomic DNA was prepared using boiling extraction. The bacterial cultures were centrifuged (6,000g for 5 min at room temperature), and the pellets were harvested and resuspended in TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH = 8.0). The mixtures were boiled for 10 min and incubated with ice for 10 min, then the mixtures were centrifuged, and the supernatants were collected. The extracted DNA was used as the template for PCR. All S. suis isolates were subjected to screen for the resistance genes of erm(B) and tet(O) in PCR analysis. The ICESa2603 family conserved genes of Inttyr and virB4 were characterized by a PCR mapping assay. To investigate the presence of circular/integrate forms of ICE and GI, two specific primer pairs (P1–P4 for ICESsuYSB17_rplL and P5–P8 for GISsuJHJ17_rpsI) were designed and used in PCR experiments. All the PCR primers were listed in Supplementary Table S1. Amplification reactions were performed in a total volume of 25 μl containing 12.5 μl 2 × Taq Plus Master Mix II (Vazyme, China), 1 μl of each primer (10 μM), 1 μl genomic DNA, and 9.5 μl water. The PCR assay was carried out in a thermocycler, comprising 5 min of pre-incubation at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 50–60°C (determined by primers), and 1 min at 72°C. The final extension was performed for 10 min at 72°C.
Antibiotic Susceptibility Testing
Antimicrobial susceptibility testing was performed for determining the minimum inhibitory concentrations (MICs) to the corresponding antimicrobial agents according to the CLSI M100-ED28 guideline (CLSI, 2018). Staphylococcus aureus ATCC 29213 was used for quality control.
Transfer and Retransfer Experiments
We randomly selected six non-serotype 2 S. suis strains carrying the erm(B), tet(O), virB4, and Inttyr genes that were used as donors (rifampicin and fusidic acid susceptibility and erythromycin resistance) (Supplementary Table S2). S. suis P1/7RF (rifampicin and fusidic acid resistance and erythromycin susceptibility) described in a previous study (Huang et al., 2016a) was utilized as recipients, which was considered to be not competent until the comRS system was activated. For a long time, S. suis was thought to be a bacterium unable to transformation. However, recently, the natural competence of S. suis under laboratory conditions was demonstrated with the addition of a comX-inducing peptide (Zaccaria et al., 2014). S. suis SH28CIP and NP4CIP (ciprofloxacin resistance and erythromycin susceptibility) were used as recipients in retransfer experiments. Transfer and retransfer experiments were performed by filter mating as described previously (Li et al., 2011; Huang et al., 2016b), with minor modifications. In brief, donor and recipient strains were grown separately at 37°C. The bacterial cultures were centrifuged to harvest at the end of the exponential growth phase and then mixed at a ratio of 1:10 (donor to recipient). The mixtures were placed on sterile nitrocellulose filters on THA plates and incubated at 37°C for 4 h. Bacteria were removed from the filters by washing with 2 ml THB medium. Transconjugants were selected by THA plates containing appropriate antibiotics (50 mg/l erythromycin with 100 mg/l rifampicin and 100 mg/l fusidic acid in transfer assays or 100 mg/l ciprofloxacin in retransfer assays) and further confirmed the presence of the erm(B), tet(O), and type IV secretion system (T4SS) core genes by PCR. To rule out spontaneous mutation and the contribution of transformation to the genetic exchange during transfer, filter mating experiments were carried out in the presence of 10 μg/ml DNase I in transfer and retransfer assays, with donor and recipient control plates included. The residual DNA with the treatment of DNase I was quantified by quantitative PCR (qPCR) using primers targeting the virB4 gene in wash buffer. The conjugation experiments were done in triplicate. The transfer frequency was calculated based on the number of observed transconjugants divided by the donors’ initial number.
PFGE and DNA Hybridization
To determine the location of the erm(B) or tet(O) genes, genomic DNA from each of the donor strains, the recipient strains, and the corresponding transconjugants was digested with SmaI endonuclease and subjected to pulsed-field gel electrophoresis (PFGE) as previously described (Vela et al., 2003; Huang et al., 2015), followed by southern blotting and DNA hybridization analysis using erm(B)- or tet(O) probes with specific primers (Supplementary Table S1).
Whole-Genome Sequencing and Analysis
Bacterial cells were centrifuged, and the pellets were harvested and resuspended in TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8.0). Total genomic DNA was extracted using an Omega Bacteria DNA Kit (OMEGA, China) according to the manufacturer’s instructions. Purified genomic DNAs were submitted for 150 bp paired-end whole-genome sequencing (WGS) on the Illumina Hiseq 2000 platform (Biozeron, Shanghai, China). ABySS v2.0.2 was used for genome assembly with multiple-Kmer parameters (Jackman et al., 2017). The genomes were annotated using the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) annotation server2 (Overbeek et al., 2014), and the genetic elements were predicted using the ICEfinder3. ICEs and GI were identified by comparison with other MGEs from GenBank and were visualized using Mauve and Easyfig 2.2.2 (Sullivan et al., 2011).
Growth Curve and Fitness Measurements
The fitness difference between transconjugants and the recipient strains was calculated by in vitro growth and competition assays as described previously (Gagneux et al., 2006; Kodio et al., 2019). In in vitro growth assay, a single colony of each strain was picked from the agar plate and incubated overnight at 37°C Cultures were adjusted into the same optical density (OD), diluted 1:100 in fresh THB medium, and aliquoted to 1 ml at an interval of every hour, and the OD600 of bacterial cultures was measured for 24 h.
In in vitro competition assay, cultures of each competitor were adjusted to OD600 = 0.1, mixed in a 1:1 ratio, and diluted to 1:100 in 10 ml at 37°C for 24 h. The mixtures at both startpoint (0 h) and endpoint (24 h) were plated on THA plates without or with 50 mg/l erythromycin and incubated at 37°C for 48 h. The relative competitive fitness W was calculated using the formula W = ln(Rf/Ri)/ln(Sf/Si). Ri and Si indicate the number of transconjugant and recipient cells at 0 h, respectively, and Rf and Sf indicate the number of transconjugant and recipient cells at 24 h, respectively.
GenBank Accession Numbers
The complete nucleotide sequences of ICESsuYSB17_rplL and GISsuJHJ17_rpsI have been deposited in the GenBank database under accession numbers MN876247 and MN876248, respectively.
Results
Co-transfer of erm(B) With Other Antimicrobial Resistance Genes
There is a strong association between erm(B) and tet(O) in S. suis isolated from China and worldwide (Huang et al., 2015, 2016c). In this study, 221 S. suis strains (86.33%, 221/320) were co-existed of erm(B) and tet(O). In order to test the co-transfer frequency of erm(B) with tet(O), we randomly selected six S. suis strains as donors for conjugative transfer, which were all co-harboring the erm(B), tet(O), and T4SS core genes (Supplementary Table S2). Transconjugants were observed from strains YSB17, YSJ15, and JHJ17 under erythromycin selection with or without DNase I treatment. The residual DNA with the treatment of DNase I in mating experiments was detected by qPCR using primers targeting the virB4 gene in wash buffer but with a negative result. For each strain, about 30–50 transconjugant clones were picked and detected to be positive for the erm(B), tet(O), and T4SS core genes by PCR. Retransfer assays using S. suis SH28CIP and NP4CIP as recipients were performed, but no transconjugant was obtained.
Two strains, YSB17 and YSJ15, successfully transferred the erythromycin and tetracycline resistance to recipient S. suis P1/7RF, with a calculated transfer frequency of (5.75 ± 1.18) × 10–8 and (3.84 ± 1.29) × 10–8. The two transconjugants, designated as SScYSB17 and SScYSJ15, respectively, exhibited macrolide and tetracycline resistance phenotypes and were tested positive for erm(B) and tet(O) (Table 1). The transfer frequency of the third transconjugant SScJHJ17 was (4.31 ± 1.53) × 10–8, and SScJHJ17 showed erythromycin, streptomycin, and spectinomycin resistance but tetracycline-sensitive phenotype. It acquired not only erm(B) and aadE from donor strain JHJ17, which is responsible for erythromycin and high-level streptomycin resistance, respectively, but also spw-like, which exhibited 96.58% identity to spw in E. faecalis strain E211 (MK784777) (Wendlandt et al., 2013), and might mediate resistance to spectinomycin in S. suis SScJHJ17. However, tet(O) carried by donor strain JHJ17 was not detected in this conjugant strain.
TABLE 1.
Characteristics of strains included in the filter mating conjugation experiments performed in this study.
| Strains | Conjugation frequencya | MIC (mg/l) | |||||
| RIF | FUS | ERY | TET | STR | SPC | ||
| P1/7RF | 256 | 256 | 0.125 | 0.25 | 1,024 | 32 | |
| YSB17 | ≤0.0625 | 32 | >256 | 64 | >2,048 | 8 | |
| SScYSB17 | (5.75 ± 1.18) × 10–8 | 256 | 256 | >256 | 64 | 1,024 | 32 |
| YSJ15 | ≤0.0625 | 32 | >256 | 256 | >2,048 | 8 | |
| SScYSJ15 | (3.84 ± 1.29) × 10–8 | 256 | 256 | >256 | 128 | 1,024 | 32 |
| JHJ17 | ≤0.0625 | 32 | >256 | 32 | >2,048 | 256 | |
| SScJHJ17 | (4.31 ± 1.53) × 10–8 | 256 | 256 | >256 | 0.25 | >2,048 | 256 |
RIF, rifampin; FUS, fusidic acid; ERY, erythromycin; TET, tetracycline; STR, streptomycin; SPC, spectinomycin. aThe frequency is calculated by CFUs of transconjugants/donors. Resistance-related phenotypes related to transfer in conjugation assays were shown in bold.
Following PFGE separation, southern blotting, and hybridization with the erm(B) or tet(O) probes, the sizes of the transferable DNA fragments were deduced by comparing the profiles of the donor strains, the recipient strains, and the transconjugants. SmaI-PFGE analysis of YSB17 and JHJ17 conjugation pairs showed differences in three bands between the recipients and transconjugants (Supplementary Figure S1). An ∼460 kb fragment existed in the recipient P1/7RF but could not be detected in transconjugant SScYSB17. Instead, two fragments, with the sizes of ∼390 and ∼140 kb, were present in SScYSB17. These results suggested the successfully transferred element with an estimated size of approximately 70 kb, most probably ICE that carried erm(B) and tet(O), into the recipient’s genome. Subsequent DNA hybridization revealed that the genes erm(B) and tet(O) were located on the different fragments, indicating the presence of SmaI restriction sites within this element (Supplementary Figure S1A). Similarly, the maximal fragment of recipient P1/7RF was replaced with two smaller fragments of transconjugant SScJHJ17, and the erm(B) gene was located in one of the fragments that differed from the recipient P1/7RF (Supplementary Figure S1B).
Characterization of Two erm(B)- and tet(O)-Carrying ICEs and an erm(B)-Carrying GI
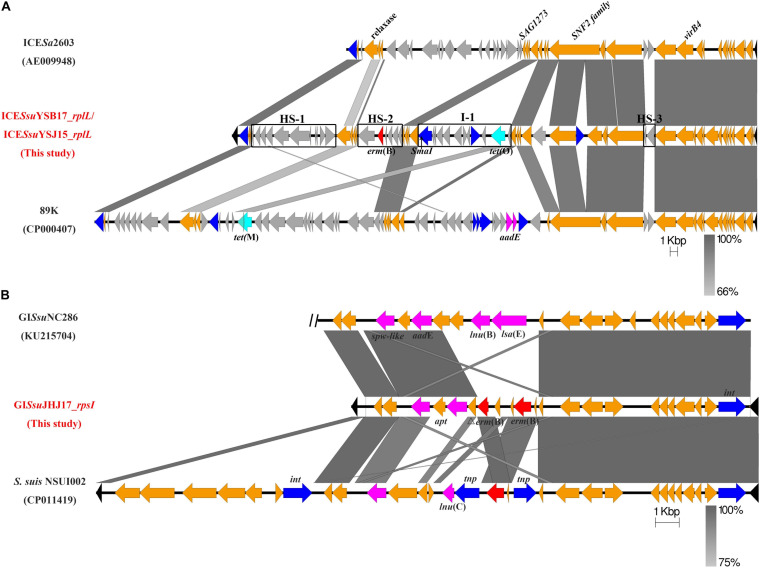
To better understand the genetic context of the erm(B)-carrying elements, we determined the whole genomes of the donors YSB17, YSJ15, and JHJ17 and their respective transconjugants by WGS. In both YSB17 and its transconjugant SScYSB17, a single putative ICE carrying erm(B) and tet(O) in the chromosome was identified using the ICEfinder and designated as ICESsuYSB17_rplL. The hyd and rplL (Figure 1A, black color) are located at the terminals of ICESsuYSB17_rplL, which encoded a predicted hydrolase and 50S ribosomal protein L7/L12, respectively. ICESsuYSB17_rplL is 69,442 bp in length, with an average G + C content of 38%, and consists of 68 putative open reading frames (ORFs). A SmaI site existed at 4,843 bp downstream of the erm(B) gene and 9,665 bp upstream of the tet(O) gene, which is consistent with the SmaI-PFGE and hybridization results (Figure 1A). A 15-bp conserved sequence (5’-TTATTTAAGAGTAAC-3’) was presented at both the left (L) and right (R) ends of the integrated ICESsuYSB17_rplL element. ICESsuYSB17_rplL was integrated into the 3’-end of the rplL gene and contained all 30 conserved core genes compared with ICESa2603. In addition, three intergenic hotspots (HS-1, HS-2, and HS-3) and three additional insertion sites were presented (Figure 1A). Among the three insertions, one was inserted in a previously identified site I-1, and one reverse transcriptase gene and one integrase gene were integrated within the SNF2 protein gene sequence, whereas erm(B) and tet(O) are located in HS-2 and I-1 variable regions, respectively. To trace the derivation of the resistance genes, comparative genome analyses were performed for HS-2 and I-1. The 5,869 bp HS-2 segment shared higher similarity with the corresponding sequences of the S. suis 9401240 (LR738724), ICESsu32457 (FR823304), ICESsuYS108 (MK211815), and S. suis (MN437484) (Supplementary Figure S2A). The content of 10,906-bp I-1 segment showed identical nucleotide sequence with the S. suis NSUI060 (CP012911), Blautia hansenii DSM 20583 (CP022413), Enterocloster clostridioformis FDAARGOS_739 (CP050964), Enterococcus cecorum NCTC12421 (LS483306), Streptococcus pyogenes NCTC12057 (LS483331), and Eubacterium hallii EH1 (LT907978). The only difference is that the I-1 region contained an additional ORF (1,503 bp) encoding IS4 family transposase (Supplementary Figure S2B). Comparison of ICESsuYSB17_rplL with some other erm(B)- and tet(O)-carrying ICEs revealed highly conserved core genes but differed greatly in non-conserved regions (Supplementary Figure S3). In YSJ15 and its transconjugant SScYSJ15, we also detected a putative ICE, designated as ICESsuYSJ15_rplL, neatly identical to ICESsuYSB17_rplL with only five nucleotide differences. In JHJ17 but not the transconjugant SScJHJ17, a tet(O)-carrying putative ICE with all conserved modules was integrated into the 3’-end of the rplL gene (data not shown).
FIGURE 1.
Genetic features of the mobile genetic elements ICESsuYSB17_rplL and GISsuJHJ17_rpsI. The direction of the arrow indicates the direction of transcription. Homologous regions are shaded in gray. Genes are shown in different colors: the ICE and GI flanking chromosomal genes were shown in black, the 30 conserved core genes of the ICESa2603 family backbone and GISsuJHJ17_rpsI backbone are in orange, and variable genes are in light gray. Integrase/transposase/recombinase genes were highlighted in blue, erm(B) is marked in red, tet-resistant genes are in pale blue, and other resistant genes are labeled in pink. (A) Comparison of ICESsuYSB17_rplL/ICESsuYSJ15_rplL with ICESa2603 and 89K. Intergenic hotspots HS-1, HS-2, and HS-3 and insertion site I-2 were indicated. SmaI restriction site is marked by a black arrow. (B) GISsuJHJ17_rpsI from S. suis JHJ17 and linear DNA comparison against part of ICESsuNC286 and fragments of S. suis NSUI002. The two vertical black diagonal lines on the left of NC286 indicate that there is still a part of the sequence that is not shown.
In both JHJ17 and its transconjugant SScJHJ17, a 16,195-bp sequence with 34% GC content was considered a putative GI and designated as GISsuJHJ17_rpsI. GISsuJHJ17_rpsI carried the erm(B) gene and was found to be integrated into a locus rpsI, the 3’-end of the gene encoding the ribosomal protein S9. Apart from a gene coding an integrase, no other putative conjugative elements, such as coupling proteins or elements participating in T4SS, were observed in GISsuJHJ17_rpsI. GISsuJHJ17_rpsI encodes 22 putative ORFs, 19 of them with the same direction of transcription as that of erm(B). An 8-bp conserved direct repeat sequence (5’-CCTGGTTT-3’) was detected at both flanking of the GISsuJHJ17. BLAST analysis of GISsuJHJ17_rpsI showed that it had the highest similarity to GISsuNC286 (KU215704) and the genomic sequence of S. suis NSUI002 (CP011419) (Figure 1B). In addition to the erm(B) gene (two copies), GISsuJHJ17_rpsI also contained the high-level streptomycin resistance gene aadE and the spectinomycin resistance gene spw-like. These genes are in agreement with the resistance profile of JHJ17 (Table 1).
Detection of the Extrachromosomal Circular Intermediate Forms of ICESsuYSB17_rplL and GISsuJHJ17_rpsI
ICEs and GIs can be excised from the chromosome with the aid of the integrase to generate the extrachromosomal circular form, which is the first step of its transfer lifecycle. In this study, two specific primer pairs (P1–P4 for ICESsuYSB17_rplL and P5–P8 for GISsuJHJ17_rpsI, the location of the primers were shown in Supplementary Figures S4A,B, respectively), were designed to detect the integrated and the extrachromosomal circular forms of ICESsuYSB17_rplL and GISsuJHJ17_rpsI (Supplementary Table S1). More specifically, P1/P2 and P3/P4 amplify the integrated form of ICESsuYSB17_rplL left and right terminals, respectively. P2/P3 detects whether there is a circular form of ICESsuYSB17_rplL. After ICESsuYSB17_rplL excision, P1/P4 detects an empty att site. For GISsuJHJ17_rpsI identification, the pairs used for P5–P8 are analogous to P1–P4. The results confirmed the presence of both the integrated and the extrachromosomal circular forms of ICESsuYSB17_rplL and GISsuJHJ17_rpsI in the original donors and the transconjugants, but absent in the recipient strain S. suis P1/7RF (Supplementary Figures S4C,D). The relatively low probability of occurrence of an excised form of ICESsuYSB17_rplL and GISsuJHJ17_rpsI, as reflected by the shallow bands of P2/P3 and P6/P7 PCR amplification, might be one of the causes of low frequency for transfer of these genetic elements. Analysis of the attICE/attB and attL/attR amplicon sequences identified the 15-bp identical sequence (5’-TTATTTAAGAGTAAC-3’). Both the circular and excised forms of GISsuJHJ17_rpsI contained a copy of the 8-bp conserved sequence corresponding to the direct repeat sequence (5’-CCTGGTTT-3’) site (data not shown).
Fitness of SScYSB17 and SScJHJ17
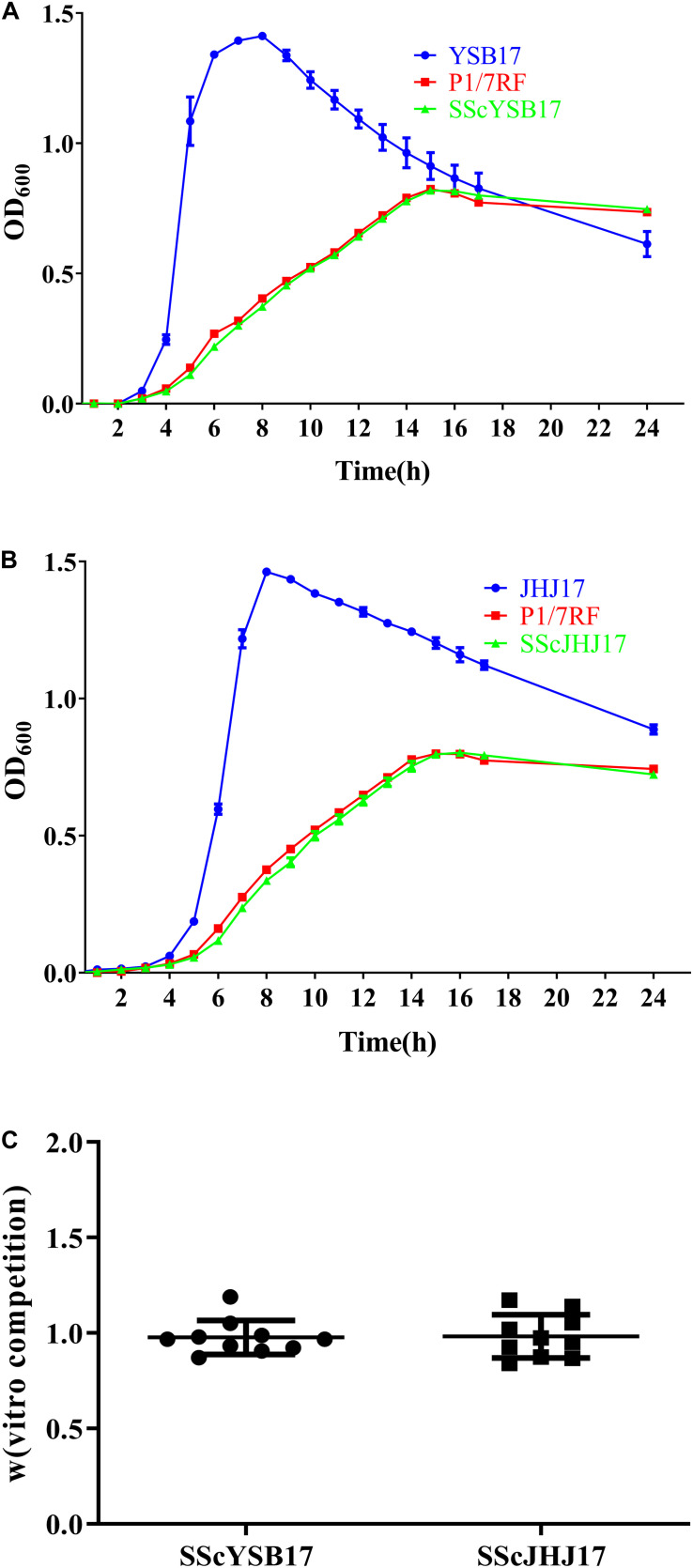
The biological cost of the horizontal acquisition of ICESsuYSB17_rplL or GISsuJHJ17_rpsI was investigated by in vitro growth and competition assays. During the in vitro growth assays, no significant differences were observed between the recipient S. suis P1/7RF and the two transconjugants SScYSB17 and SScJHJ17, suggesting that the acquisition of ICESsuYSB17_rplL (Figure 2A) and GISsuJHJ17_rpsI (Figure 2B) did not affect bacterial growth in THB medium, although both S. suis P1/7RF and the transconjugants showed growth delay compared with the donors and the original S. suis P1/7 strain.
FIGURE 2.
The biological cost of the horizontal acquisition of ICESsuYSB17_rplL or GISsuJHJ17_rpsI. (A) Growth curves of the YSB17, P1/7RF, and SScYSB17 under the same conditions in vitro. (B) Growth curves of the JHJ17, P1/7RF, and SScJHJ17 under the same conditions in vitro. (C) The relative competitive fitness W of recipient P1/7RF and two transconjugants SScYSB17 and SScJHJ17 strain. The values are represented as mean ± SD of 10 independent experiments.
In vitro competition assays showed that the transconjugants SScYSB17 and SScJHJ17 had relative fitness values W of 0.977 ± 0.085 and 0.982 ± 0.108, respectively, when compared with the recipient strain P1/7RF (Figure 2C). These results further suggest that there was no visible fitness cost when recipient strain acquired ICESsuYSB17_rplL or GISsuJHJ17_rpsI.
Discussion
Macrolide-resistant Streptococcus pneumoniae, S. pyogenes, and Streptococcus agalactiae are 3 of the top 18 drug-resistant threats as declared by the Centers for Disease Control and Prevention (CDC) in the United States in 2013 (CDC, 2013). Previous studies have suggested that S. suis is a reservoir of antimicrobial resistance (AMR) genes for other streptococcal pathogens (Palmieri et al., 2011; Huang et al., 2016a). The erm(B) gene is the most prevalent determinant conferring resistance to macrolide in streptococci clinical isolates (Chu et al., 2009; Haenni et al., 2018; Ichikawa et al., 2020). However, knowledge about the transfer of erm(B) as well as the related MGEs in S. suis remains unclear. In this study, we reported the co-transfer of erm(B) with other AMR genes among S. suis strains mediated by ICEs or GI, which could reveal the reason for the fast spread of macrolide-resistant S. suis in recent years in China.
Co-transfer of erm(B) and tet(O) was confirmed in two strains of S. suis serotype 21, which is co-located on ICEs of the ICESa2603 family. ICESa2603 family is highly prevalent in major Streptococcus species (Davies et al., 2009; Ambroset et al., 2015; Huang et al., 2016b). A variety of resistance genes responsible for resistance to tetracyclines, macrolides, or phenicols have been shown to be transferred inter-strains or inter-species by this family of ICEs (Chen et al., 2007; Palmieri et al., 2012; Marini et al., 2015; Huang et al., 2016b,c; Libante et al., 2019; Pan et al., 2019). Since erm(B) and tet(O) are located on two different variable regions, namely, HS-2 and I-1, and these two segments showed nearly identical sequence similarity to the corresponding sequences in S. suis and other Gram-positive cocci (Supplementary Figure S2), it is reasonable to speculate that ICESsuYSB17_rplL was evolved from acquisition of erm(B)-carrying HS-2 and tet(O)-carrying I-1 elements through a multi-step process. These results revealed the important role of the acquisition of AMR genes in ICEs diversity and evolution.
Co-transfer of erm(B) and aadE-spw-like elements was mediated by a novel GI, GISsuJHJ17_rpsI, which is integrated at the rpsI site, a conserved hotspot in Streptococcus species that was commonly integrated by IMEs and ICEs (Ambroset et al., 2015; Coluzzi et al., 2017; Libante et al., 2019). GIs are usually detected integrated into the 3’-end of the tRNA gene. However, two GIs were found in the rpsI gene, one carrying the ant(9)–lnu(C)–erm(B) genes (Libante et al., 2019) and another carrying the aadE-lnu(B)–lsa(E)–spw-like genes (Huang et al., 2016c). Moreover, GIs integrated into rpsI could be mobilized by subverting the relaxase and mating apparatus of a co-resident ICE (Libante et al., 2019). In this study, we also confirmed that GISsuJHJ17_rpsI was able to transfer from a S. suis serotype 29 isolate to serotype 2. We speculated that the transfer of GISsuJHJ17_rpsI was mobilized by a tet(O)-carrying ICE that harbored a fully functional mobilization module. It needs to be further proven by the inactivation of the tet(O)-carrying ICE. Studies have shown that some GIs not only need conjugative elements to promote their own transfer but also influence the transfer or stability of the helper co-resident elements (Guedon et al., 2017).
ICEs could integrate into the chromosome of bacteria and are capable to transfer to a new host uponconjugative transfer (Johnson and Grossman, 2015; Santoro et al., 2018). Functional ICEs were shown to excise from chromosome by site-specific recombination between attL and attR recombination sites, thus producing a covalently closed circular form of the ICE and a chromosomal excised attB site (Puymege et al., 2013). Under normal growth conditions, ICESsuYSB17_rplL and GISsuJHJ17_rpsI are mainly integrated into the chromosome. To check this integration state, we used primer pairs P1 + P2 and P3 + P4 to detect ICESsuYSB17_rplL in the chromosome and primer pairs P5 + P6 and P7 + P8 for GISsuJHJ17_rpsI. However, both MGEs could be excised from the bacterial genome and generated the extrachromosomal circular forms of ICESsuYSB17_rplL and GISsuJHJ17_rpsI, which identified the product by primers of P2 + P3 and P6 + P7, respectively. Furthermore, the empty rplL attB or rpsI attB’ sites were detected by primers P1 + P4 and P5 + P8, respectively (Supplementary Table S4). This suggests that ICESsuYSB17_rplL and GISsuJHJ17_rpsI are functional and thus have the potential to be transferred. Previous studies have shown that excision of ICEs could be induced under environmental stress, including antimicrobials, such as ciprofloxacin and tetracycline (Beaber et al., 2004; Liu et al., 2017; Scornec et al., 2017). Considering the extensive use of antimicrobials in livestock and poultry, it is of great significance to evaluate the selection stress, especially antimicrobials, which are involved in inducing the excision and thereafter the transfer of the ICEs/GIs.
The acquisition of MGEs was thought to impose an immediate biological cost (Leon-Sampedro et al., 2016). However, the acquisition of ICESsuYSB17_rplL or GISsuJHJ17_rpsI in this study showed negligible fitness cost (Figure 2), which is consistent with our previous study (Huang et al., 2016b). In addition, the AMR-carrying ICEs or GIs enhance their survival under the corresponding antimicrobials. Those may explain the observation that the AMR-carrying ICESsuYSB17_rplL, GISsuJHJ17_rpsI, and similar ICEs are widely distributed in streptococci (Ambroset et al., 2015; Libante et al., 2019).
In summary, we identified three erm(B)-carrying transferable elements, including two erm(B)- and tet(O)-harboring ICEs of the ICESa2603 family and a novel erm(B)-carrying GI, which can be transferred between S. suis of different serotypes. The intraspecific transfer of erm(B)-carrying MGEs among different serotypes of S. suis strains might have contributed to the worldwide spread of macrolide resistance. This reinforces the need for strategies that inhibit the horizontal gene transfer of AMR-carrying MGEs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: (Repository: Genbank) (Accessions: BankIt2297772 Seq1 MN876247; BankIt2297772 Seq2 MN876248).
Author Contributions
LC, JH, and LW developed the concept and designed the experiments. LC, JS, XD, and XW performed the experiments and collected the data. LC, XH, and YH conducted all bioinformatics analyses. LC, JH, MS, and LW prepared the manuscript. All authors have contributed to, seen, and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported by the National Natural Science Foundation of China (Nos. 31872517, 31702292, 31572567), the Fundamental Research Funds for the Central Universities (KJQN201827), the Natural Science Foundation of Shanghai City (19ZR1417400), the National Key R&D Program of China (2018YFD0500300), the Innovation Project for Postgraduate Training in Jiangsu Province (KYCX18_0714), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.628740/full#supplementary-material
References
- Ambroset C., Coluzzi C., Guedon G., Devignes M. D., Loux V., Lacroix T., et al. (2015). New insights into the classification and integration specificity of Streptococcus integrative conjugative elements through extensive genome exploration. Front. Microbiol. 6:1483. 10.3389/fmicb.2015.01483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaber J. W., Hochhut B., Waldor M. K. (2004). SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427 72–74. 10.1038/nature02241 [DOI] [PubMed] [Google Scholar]
- Bellanger X., Payot S., Leblond-Bourget N., Guedon G. (2014). Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol. Rev. 38 720–760. 10.1111/1574-6976.12058 [DOI] [PubMed] [Google Scholar]
- Bojarska A., Molska E., Janas K., Skoczynska A., Stefaniuk E., Hryniewicz W., et al. (2016). Streptococcus suis in invasive human infections in Poland: clonality and determinants of virulence and antimicrobial resistance. Eur. J. Clin. Microbiol. Infect. Dis. 35 917–925. 10.1007/s10096-016-2616-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenciani A., Bacciaglia A., Vecchi M., Vitali L. A., Varaldo P. E., Giovanetti E. (2007). Genetic elements carrying erm(B) in Streptococcus pyogenes and association with tet(M) tetracycline resistance gene. Antimicrob. Agents Chemother. 51 1209–1216. 10.1128/AAC.01484-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (2013). Antibiotic resistance threats in the United States, 2013 [Online]. Available: http://www.cdc.gov/drugresistance/threat-report-2013 (accessed January 12, 2016). [Google Scholar]
- Chen C., Tang J., Dong W., Wang C., Feng Y., Wang J., et al. (2007). A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS One 2:e315. 10.1371/journal.pone.0000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y. W., Cheung T. K., Chu M. Y., Tsang V. Y., Fung J. T., Kam K. M., et al. (2009). Resistance to tetracycline, erythromycin and clindamycin in Streptococcus suis serotype 2 in Hong Kong. Int. J. Antimicrob. Agents 34 181–182. 10.1016/j.ijantimicag.2009.01.007 [DOI] [PubMed] [Google Scholar]
- CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Cochetti I., Tili E., Mingoia M., Varaldo P. E., Montanari M. P. (2008). erm(B)-carrying elements in tetracycline-resistant pneumococci and correspondence between Tn1545 and Tn6003. Antimicrob. Agents Chemother. 52 1285–1290. 10.1128/AAC.01457-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluzzi C., Guedon G., Devignes M. D., Ambroset C., Loux V., Lacroix T., et al. (2017). A glimpse into the world of integrative and mobilizable elements in streptococci reveals an unexpected diversity and novel families of mobilization proteins. Front. Microbiol. 8:443. 10.3389/fmicb.2017.00443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daccord A., Ceccarelli D., Burrus V. (2010). Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol. Microbiol. 78 576–588. 10.1111/j.1365-2958.2010.07364.x [DOI] [PubMed] [Google Scholar]
- Davies M. R., Shera J., Van Domselaar G. H., Sriprakash K. S., McMillan D. J. (2009). A novel integrative conjugative element mediates genetic transfer from group G Streptococcus to other β-hemolytic Streptococci. J. Bacteriol. 191 2257–2265. 10.1128/JB.01624-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feßler A. T., Wang Y., Wu C., Schwarz S. (2018). Mobile macrolide resistance genes in staphylococci. Plasmid 99 2–10. 10.1016/j.plasmid.2018.05.001 [DOI] [PubMed] [Google Scholar]
- Fyfe C., Grossman T. H., Kerstein K., Sutcliffe J. (2016). Resistance to macrolide antibiotics in public health pathogens. Cold Spring Harb. Perspect. Med. 6:a25395. 10.1101/cshperspect.a025395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux S., Long C. D., Small P. M., Van T., Schoolnik G. K., Bohannan B. J. (2006). The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312 1944–1946. 10.1126/science.1124410 [DOI] [PubMed] [Google Scholar]
- Guedon G., Libante V., Coluzzi C., Payot S., Leblond-Bourget N. (2017). The obscure world of integrative and mobilizable elements, highly widespread elements that pirate bacterial conjugative systems. Genes 8:337. 10.3390/genes8110337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung M., Tamang M. D., Moon D. C., Kim S. R., Jeong J. H., Jang G. C., et al. (2015). Molecular basis of resistance to selected antimicrobial agents in the emerging zoonotic pathogen Streptococcus suis. J. Clin. Microbiol. 53 2332–2336. 10.1128/JCM.00123-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni M., Lupo A., Madec J. Y. (2018). Antimicrobial resistance in Streptococcus spp. Microbiol. Spectr. 6 1–25. 10.1128/microbiolspec.ARBA-0008-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden M. T., Hauser H., Sanders M., Ngo T. H., Cherevach I., Cronin A., et al. (2009). Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS One 4:e6072. 10.1371/journal.pone.0006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horaud T., Bouguenec C. L., Pepper K. (1985). Molecular genetics of resistance to macrolides, lincosamides and streptogramin B (MLS) in streptococci. J. Antimicrob. Chemother. 16 111–135. 10.1093/jac/16.suppl_a.111 [DOI] [PubMed] [Google Scholar]
- Huang J., Liang Y., Guo D., Shang K., Ge L., Kashif J., et al. (2016a). Comparative genomic analysis of the ICESa2603 family ICEs and spread of erm(B)- and tet(O)-carrying transferable 89K-subtype ICEs in swine and bovine isolates in China. Front. Microbiol. 7:55. 10.3389/fmicb.2016.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Ma J., Shang K., Hu X., Liang Y., Li D., et al. (2016b). Evolution and diversity of the antimicrobial resistance associated mobilome in Streptococcus suis: a probable mobile genetic elements reservoir for other streptococci. Front. Cell Infect. Microbiol. 6:118. 10.3389/fcimb.2016.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Shang K., Kashif J., Wang L. (2015). Genetic diversity of Streptococcus suis isolated from three pig farms of China obtained by acquiring antibiotic resistance genes. J. Sci. Food Agric. 95 1454–1460. 10.1002/jsfa.6841 [DOI] [PubMed] [Google Scholar]
- Huang K., Zhang Q., Song Y., Zhang Z., Zhang A., Xiao J., et al. (2016c). Characterization of spectinomycin resistance in Streptococcus suis leads to two novel insights into drug resistance formation and dissemination mechanism. Antimicrob. Agents Chemother. 60 6390–6392. 10.1128/AAC.01157-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Wang M., Hao H., Yang R., Xie J., Su J., et al. (2019). Genomic epidemiological investigation of a Streptococcus suis outbreak in Guangxi, China, 2016. Infect. Genet. Evol. 68 249–252. 10.1016/j.meegid.2018.12.023 [DOI] [PubMed] [Google Scholar]
- Hui A. C., Ng K. C., Tong P. Y., Mok V., Chow K. M., Wu A., et al. (2005). Bacterial meningitis in Hong Kong: 10-years’ experience. Clin. Neurol. Neurosurg. 107 366–370. 10.1016/j.clineuro.2004.10.006 [DOI] [PubMed] [Google Scholar]
- Ichikawa T., Oshima M., Yamagishi J., Muramatsu C., Asai T. (2020). Changes in antimicrobial resistance phenotypes and genotypes in Streptococcus suis strains isolated from pigs in the Tokai area of Japan. J. Vet. Med. Sci. 82 9–13. 10.1292/jvms.19-0449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman S. D., Vandervalk B. P., Mohamadi H., Chu J., Yeo S., Hammond S. A., et al. (2017). ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 27 768–777. 10.1101/gr.214346.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. M., Grossman A. D. (2015). Integrative and conjugative elements (ICEs): what they do and how they work. Annu. Rev. Genet. 49 577–601. 10.1146/annurev-genet-112414-055018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodio O., Georges Togo A. C., Sadio Sarro Y. D., Fane B., Diallo F., Somboro A., et al. (2019). Competitive fitness of Mycobacterium tuberculosis in vitro. Int. J. Mycobacteriol. 8 287–291. 10.4103/ijmy.ijmy_97_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Sampedro R., Novais C., Peixe L., Baquero F., Coque T. M. (2016). Diversity and evolution of the Tn5801-tet(M)-like integrative and conjugative elements among Enterococcus, Streptococcus, and Staphylococcus. Antimicrob. Agents Chemother. 60 1736–1746. 10.1128/AAC.01864-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Shen X., Yan J., Han H., Zheng B., Liu D., et al. (2011). GI-type T4SS-mediated horizontal transfer of the 89K pathogenicity island in epidemic Streptococcus suis serotype 2. Mol. Microbiol. 79 1670–1683. 10.1111/j.1365-2958.2011.07553.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libante V., Nombre Y., Coluzzi C., Staub J., Guedon G., Gottschalk M., et al. (2019). Chromosomal conjugative and mobilizable elements in Streptococcus suis: major actors in the spreading of antimicrobial resistance and bacteriocin synthesis genes. Pathogens 9 1–23. 10.3390/pathogens9010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Wu Z., Xue H., Zhao X. (2017). Antibiotics trigger initiation of SCCmec transfer by inducing SOS responses. Nucleic Acids Res. 45 3944–3952. 10.1093/nar/gkx153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai N. T., Hoa N. T., Nga T. V., Linh le D., Chau T. T., Sinh D. X., et al. (2008). Streptococcus suis meningitis in adults in Vietnam. Clin. Infect. Dis. 46 659–667. 10.1086/527385 [DOI] [PubMed] [Google Scholar]
- Marini E., Palmieri C., Magi G., Facinelli B. (2015). Recombination between Streptococcus suis ICESsu32457 and Streptococcus agalactiae ICESa2603 yields a hybrid ICE transferable to Streptococcus pyogenes. Vet. Microbiol. 178 99–104. 10.1016/j.vetmic.2015.04.013 [DOI] [PubMed] [Google Scholar]
- Marosevic D., Kaevska M., Jaglic Z. (2017). Resistance to the tetracyclines and macrolide-lincosamide-streptogramin group of antibiotics and its genetic linkage - a review. Ann. Agric. Environ. Med. AAEM 24 338–344. 10.26444/aaem/74718 [DOI] [PubMed] [Google Scholar]
- Martel A., Decostere A., Leener E. D., Marien M., Graef E. D., Heyndrickx M., et al. (2005). Comparison and transferability of the erm(B) genes between human and farm animal streptococci. Microb. Drug Resist. 11 295–302. 10.1089/mdr.2005.11.295 [DOI] [PubMed] [Google Scholar]
- Mazokopakis E. E., Kofteridis D. P., Papadakis J. A., Gikas A. H., Samonis G. J. (2005). First case report of Streptococcus suis septicaemia and meningitis from Greece. Eur. J. Neurol. 12 487–489. 10.1111/j.1468-1331.2005.00998.x [DOI] [PubMed] [Google Scholar]
- Overbeek R., Olson R., Pusch G. D., Olsen G. J., Davis J. J., Disz T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42 D206–D214. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri C., Magi G., Mingoia M., Bagnarelli P., Ripa S., Varaldo P. E., et al. (2012). Characterization of a Streptococcus suis tet(O/W/32/O)-carrying element transferable to major streptococcal pathogens. Antimicrob. Agents Chemother. 56 4697–4702. 10.1128/AAC.00629-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri C., Varaldo P. E., Facinelli B. (2011). Streptococcus suis, an emerging drug-resistant animal and human pathogen. Front. Microbiol. 2:235. 10.3389/fmicb.2011.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z., Liu J., Zhang Y., Chen S., Ma J., Dong W., et al. (2019). A novel integrative conjugative element mediates transfer of multi-drug resistance between Streptococcus suis strains of different serotypes. Vet. Microbiol. 229 110–116. 10.1016/j.vetmic.2018.11.028 [DOI] [PubMed] [Google Scholar]
- Princivalli M. S., Palmieri C., Magi G., Vignaroli C., Manzin A., Camporese A., et al. (2009). Genetic diversity of Streptococcus Suis clinical isolates from pigs and humans in Italy (2003-2007). Euro Surveill. 14:19310. 10.2807/ese.14.33.19310-en [DOI] [PubMed] [Google Scholar]
- Puymege A., Bertin S., Chuzeville S., Guedon G., Payot S. (2013). Conjugative transfer and cis-mobilization of a genomic island by an integrative and conjugative element of Streptococcus agalactiae. J. Bacteriol. 195 1142–1151. 10.1128/JB.02199-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. C. (2008). Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol. Lett. 282 147–159. 10.1111/j.1574-6968.2008.01145.x [DOI] [PubMed] [Google Scholar]
- Santoro F., Romeo A., Pozzi G., Iannelli F. (2018). Excision and circularization of integrative conjugative element Tn5253 of Streptococcus pneumoniae. Front. Microbiol. 9:1779. 10.3389/fmicb.2018.01779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scornec H., Bellanger X., Guilloteau H., Groshenry G., Merlin C. (2017). Inducibility of Tn916 conjugative transfer in Enterococcus faecalis by subinhibitory concentrations of ribosome-targeting antibiotics. J. Antimicrob. Chemother. 72 2722–2728. 10.1093/jac/dkx202 [DOI] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27 1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomich P. K., An F. Y., Clewell D. B. (1979). A transposon (Tn917) in Streptococcus faecalis that exhibits enhanced transposition during induction of drug resistance. Cold Spring Harb. Symp. Quant. Biol. 43(Pt 2), 1217–1221. 10.1101/sqb.1979.043.01.138 [DOI] [PubMed] [Google Scholar]
- Varaldo P. E., Montanari M. P., Giovanetti E. (2009). Genetic elements responsible for erythromycin resistance in streptococci. Antimicrob. Agents Chemother. 53 343–353. 10.1128/AAC.00781-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela A. I., Goyache J., Tarradas C., Luque I., Mateos A., Moreno M. A., et al. (2003). Analysis of genetic diversity of Streptococcus suis clinical isolates from pigs in Spain by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41 2498–2502. 10.1128/jcm.41.6.2498-2502.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela A. I., Villalon P., Saez-Nieto J. A., Chacon G., Dominguez L., Fernandez-Garayzabal J. F. (2017). Characterization of Streptococcus pyogenes from animal clinical specimens, Spain. Emerg. Infect. Dis. 23 2013–2016. 10.3201/eid2312.151146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendlandt S., Li B., Lozano C., Ma Z., Torres C., Schwarz S. (2013). Identification of the novel spectinomycin resistance gene spw in methicillin-resistant and methicillin-susceptible Staphylococcus aureus of human and animal origin. J. Antimicrob. Chemother. 68 1679–1680. 10.1093/jac/dkt081 [DOI] [PubMed] [Google Scholar]
- Wilson D. N. (2014). Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 12 35–48. 10.1038/nrmicro3155 [DOI] [PubMed] [Google Scholar]
- Woodbury R. L., Klammer K. A., Xiong Y., Bailiff T., Glennen A., Bartkus J. M., et al. (2008). Plasmid-Borne erm(T) from invasive, macrolide-resistant Streptococcus pyogenes strains. Antimicrob. Agents Chemother. 52 1140–1143. 10.1128/AAC.01352-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Cai L., Xiao M., Kong F., Oftadeh S., Zhou F., et al. (2010). Distribution of serotypes, genotypes, and resistance determinants among macrolide-resistant Streptococcus pneumoniae isolates. Antimicrob. Agents Chemother. 54 1152–1159. 10.1128/AAC.01268-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Jing H., Chen Z., Zheng H., Zhu X., Wang H., et al. (2006). Human Streptococcus suis outbreak, Sichuan, China. Emerg. Infect. Dis. 12 914–920. 10.3201/eid1206.051194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccaria E., van Baarlen P., de Greeff A., Morrison D. A., Smith H., Wells J. M. (2014). Control of competence for DNA transformation in Streptococcus suis by genetically transferable pherotypes. PLoS One 9:e99394. 10.1371/journal.pone.0099394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Yang M., Hu P., Wu J., Chen B., Hua Y., et al. (2011). Comparative genomic analysis of Streptococcus suis reveals significant genomic diversity among different serotypes. BMC Genomics 12:523. 10.1186/1471-2164-12-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K., Xie L., Han L., Guo X., Wang Y., Sun J. (2017). ICESag37, a novel integrative and conjugative element carrying antimicrobial resistance genes and potential virulence factors in Streptococcus agalactiae. Front. Microbiol. 8:1921. 10.3389/fmicb.2017.01921 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: (Repository: Genbank) (Accessions: BankIt2297772 Seq1 MN876247; BankIt2297772 Seq2 MN876248).