Abstract
Background
Pancreatic ductal adenocarcinoma (PDAC) is the most lethal malignancy: it has a 5-year survival rate of less than 9%. Although surgical resection is an effective treatment for PDAC, only a small number of patients can have their tumors surgically removed. Thus, an urgent need to find new therapeutic targets for PDAC exists. Understanding the molecular mechanism of PDAC development is essential for the treatment of this malignancy. This research aimed to study the mechanisms of pancreatic stellate cells (PSCs), which regulate branched-chain amino acid (BCAA) metabolism in PDAC.
Methods
Differentially expressed proteins were detected via nanoliquid chromatography coupled to mass spectrometry (nano-LC-MS/MS). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment methods were used to find the valine-leucine-isoleucine (BCAA) degradation pathway. The levels of BCAAs in the sera and tissues of patients with PDAC were measured by using nuclear magnetic resonance (NMR). The functions of BCAA concentrations and the effects of activated pancreatic stellate cells (aPSCs) were also evaluated by performing Cell Counting Kit-8, colony formation, and wound healing assays.
Results
A total of 1,519 proteins with significantly differential expression were discovered in PDAC and adjacent tissues by using nano-LC-MS/MS. KEGG pathway enrichment analysis identified the BCAA degradation pathway. The content of BCAA in PDAC clinical samples was up-regulated. However, the addition of different concentrations of BCAA to PDAC cell culture medium failed to promote the proliferation and migration of PDAC cells. Given that analysis based on The Cancer Genome Atlas database showed that the number of aPSCs gradually increased with the progression of PDAC, the effects of aPSCs on PDAC cells were explored. After coculture with aPSCs, PDAC cell proliferation showed a significant increase, and the proteins involved in the BCAA degradation pathway in PDAC cells had also changed.
Conclusions
aPSCs could regulate BCAA metabolism to enhance the progression of PDAC, indicating that the regulation of BCAA metabolism may serve as a new therapeutic direction for PDAC.
Keywords: Pancreatic stellate cells (PSCs), branched-chain amino acid metabolism, pancreatic cancer
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive tumor that is associated with a poor prognosis (1) and is the most deadly major cancer (2). PDAC is the most lethal malignancy and has a 5-year survival rate of less than 9% (3). Although surgical resection is an effective treatment for PDAC (4), surgical tumor removal is suitable only for 20% of patients due to locally advanced or metastatic disease (5). Thus, finding new therapeutic targets for pancreatic cancer is urgently needed.
PDAC is characterized by metabolic deregulation (6). Tumor metabolism is closely related to tumorigenesis and tumor progression, and metabolic alterations must accommodate the cellular processes needed to support transformation and cancer progression. Changes in amino acid metabolism are a key way through which this situation occurs. The stability of amino acid metabolism is essential for maintaining homeostasis in the body (7). Branched-chain amino acids (BCAAs), including leucine, isoleucine, and valine, are a class of essential amino acids that are not synthesized in humans, and their metabolism has been associated with specific cancer phenotypes (8). Changes in BCAA metabolism not only affect the intrinsic cancer characteristics of a cell but also reflect systematic metabolic changes related to some cancers (9). The targeting of the massive difference in amino acid metabolism between cancer cells and normal cells is expected to become an ideal strategy for cancer treatment (4).
Pancreatic stellate cells (PSCs) are the major constitutive component of the pancreatic tumor microenvironment and may be responsible for the aggressive, metastatic, and resilient nature of PDAC (10). PSCs play a crucial role in metabolic reprogramming in PDAC. Normally, PSCs are quiescent; however, with the development of PDAC, quiescent PSCs become activated via various underlying mechanisms. Activated PSCs (aPSCs) can secrete metabolism-altering factors in PDAC (11). The metabolic crosstalk between PSCs and PDA cells can facilitate PDAC progression and invasiveness under nutrient-limiting conditions (12). However, how aPSCs regulate BCAA metabolism in the promotion of PDAC development is unclear.
This study aims to explore how aPSCs regulate BCAA metabolism in PDAC to understand its molecular mechanism for treatment in patients with this malignancy. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-21-761).
Methods
Serum and tissue samples
Samples of PDAC tissues were collected from Tianjin Medical University Cancer Institute and Hospital between May 2020 and September 2020. The study included 121 serum samples, including 67 samples from patients with PDAC and 54 samples from healthy controls (HCs). HC blood samples were obtained from healthy individuals who visited the hospital for medical check-ups and agreed to participate in the study. The diagnosis of PDAC was based on clinical evaluation and imaging studies and histologically confirmed by surgery or imaging-guided biopsy. Serum samples were collected from all patients before treatment. The samples were centrifuged at 3,000 ×g for 10 min. After centrifugation, the supernatants were collected and stored at −80 °C. The training set comprised 38 samples from HCs and 50 samples from patients with PDAC. Validation was performed with 16 samples from HCs and 17 samples from patients with PDAC. PDAC tissues and corresponding adjacent normal tissues were obtained from 6 patients during surgical procedures. Tissue fragments were promptly frozen in liquid nitrogen at the time of surgery and stored at −80 °C. The tumor tissues were histologically confirmed as PDAC. The usage of serum samples in this study was reviewed and approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (Scientific Ethical Approval No. bc2020101), and informed consent was obtained from all the patients. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture and reagents
The human PDAC cell lines SW1990 and BxPC-3and aPSCs were acquired from the American Type Culture Collection and tested regularly for mycoplasma contamination. SW1990 and BxPC-3 cells were cultured in Roswell Park Memorial Institute-1640 medium (Gibco, Rockville, MD, USA). aPSC cells were cultured in Dulbecco’s modified Eagle medium (Gibco, Rockville, MD, USA) supplemented with 10% fetal bovine serum (BI) and penicillin/streptomycin and maintained at 37 °C in a humidified atmosphere with 5% CO2. BCAA powders (Sigma-Aldrich) were added to the cell cultures.
Nuclear magnetic resonance
Plasma was harvested through the centrifugation of the collected blood samples. The plasma was mixed thoroughly with 340 µL of plasma buffer (Bruker, H170046) for 1 min. A total of 50 µL of tissue samples (~5 mg) were cut into small pieces, placed into 600 µL of extraction buffer (MeOH:H2O 2:1), and maintained at –40 °C overnight. Then, the samples were sonicated (2 s on and 3 s off) on ice for 10 min. The supernatants were collected via centrifugation for 10 min at 12,000 ×g and 4 °C. A total of 600 µL of extraction buffer was added to the sediment and sonicated. This step was done twice. The supernatants from the three replicates were combined, freeze dried, and further dissolved in 600 µL of loading buffer (0.15 M Na/K buffer, TSP 0.001%, NaN3, 0.01% pH =7.4). After this step, 550 µL of samples were transferred to 5 mm nuclear magnetic resonance (NMR) tubes and loaded into an NMR machine (Bruker 600 MHz). The acquired data were uploaded and analyzed by using Bruker Topspin 3.6.0 software on NMRPC.
Protein extraction and tryptic digestion
Formalin-fixed and paraffin-embedded samples were pre-prepared through dewaxing with xylene and gradient ethanol reduction. The tissue samples were ground with liquid nitrogen before lysis. The cell samples were added directly into the lysis buffer. Different samples were lysed in lysis buffer (0.1 M Tris-hydrochloride, pH 8.0, 4% sodium dodecyl sulfate, 1% NP-40, 2 mM ethylenediaminetetraacetic acid, and 1% protease inhibitor cocktail) for 15 min on ice. This step was followed by 10 min of sonication (2 s on and 3 s off) on ice. The samples were further decrosslinked at 100 °C for 60 min. The supernatants were collected by centrifugation at 20,000 ×g for 10 min, and protein concentration was measured through BCA protein assay. The reductive alkylation of extracted proteins was performed by adding 5 mM dithiothreitol at 56 °C for 1 h followed by incubation with 50 mM iodoacetamide at room temperature for 30 min in the dark. Protein purification and digestion were performed by using SP3 magnetic beads (1).
Nanoliquid chromatography coupled to mass spectrometry
Data were acquired from lysed peptide samples by using Orbitrap Q Exactive HF mass spectrometry (Thermo Fisher Scientific, Waltham, MA) with a Thermo Scientific UltiMate 3000 UHPLC system. Peptides were redissolved in loading buffer (2% ACN) with indexed retention time standards (Biognosys, Schlieren, Switzerland). The peptides were separated by using a 150 min gradient (0–3 min, 3%–9% of buffer B; 3–127 min, 9%–63% of buffer B; 127–131 min, 63% of buffer B; and 131–149 min, 63% of buffer B). The digested peptides were ionized at 2 kV and introduced into the mass spectrometer under data-independent acquisition mode. For the MS1 full scan, ions with m/z ranging from 300 to 1,400 were acquired with an Orbitrap mass analyzer at the high resolution of 120,000. The maximal ion injection time was 50 ms. MS2 acquisition was performed in top-speed mode, and the duty cycle time was 3 s. Precursor ions were selected and fragmented with higher energy collision dissociation with a normalized collision energy of 32%. The maximal ion injection time of MS2 was 35 ms, and the dynamic exclusion was 60 s.
MS database searching
DIA data were searched against the human UniProt database (20 365 sequences) by using Spectronaut (v14.5.200813.47784). Default settings were applied for library generation with a trypsin/P digest rule, a high protein and peptide confidence level, and a false discovery rate of 0.01. Output quantified protein intensities were normalized by using Spectronaut with median correction. Proteins appearing in at least 30% of all samples were selected for further analysis. Absent values were replaced with the half-minimum value of each protein intensity.
Pathway enrichment analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional gene enrichment analyses were carried out by using R package (clusterProfiler, v3.16.1). The annotation database used was org.Hs.eg.db (v3.11.4). Background proteins were set with all the quantified proteins. Differentially expressed proteins were input to generate the enrichment pathway list and figures.
Cell proliferation assay
SW1990 and BxPC-3 cells (3,000 cells per well) were seeded into 96-well plates. After the cells had attached to the walls of the wells, the cell culture was replaced with a new cell culture medium with different BCAA concentrations (normal, 2, 4, or 6 mM) or aPSCs supernatant, and culture was conducted for 24 and 48 h. At the corresponding time points, Cell Counting Kit (CCK)-8 reagents were added, and the incubation was continued for 3 h. The optical density value was measured by using the µQuant Universal Microplate Spectrophotometer (Bio-Tek Instruments, Winooski, VT, USA). Data were processed to generate cell proliferation curves in Graphpad Prism 6.0 software.
Colony formation assay
SW1990 and BxPC-3 cells were seeded into 6-well plates at the density of 500 cells per well. After the cells had adhered, the initial cell culture was discarded and replaced with culture medium containing different concentrations of BCAAs (normal, 2, 4, or 6 mM). The medium was replaced every 3 days. The cell colonies were observed under a microscope (Olympus, Tokyo, Japan) when most had amassed >50 cells. This step was followed by the use of paraformaldehyde (Solarbio) to fix the colonies, which were subsequently stained with crystal violet (Solarbio) and counted.
Wound healing assay
SW1990 and BxPC-3 cells were seeded into 6-well plates (density: 6×105 cells/well). After the cells had adhered, the initial cell culture medium was discarded and replaced with different concentrations of BCAAs (normal, 2, 4, or 6 mM). When approximately 95–100% of the dish was covered by cells, a 100 µL pipette tip was used to create scratches. The cells were incubated in the medium for 24 h. 0H and 24H images were acquired. Six fields of view were captured for each well under ×100 magnification with a microscope (Olympus) to prevent false negative or false positive results.
Cocultivation with aPSCs
SW1990 and BxPC-3 cell suspensions were placed into 6-well plates at the density of 6×105 cells/mL, and aPSCs were seeded into chambers (density: 2×105 cells/well). After the cells had adhered to the walls, the medium was removed from each well, and nonadherent cells were washed off. The same culture system was used for the two cell types.
Animal experiment
BALB/c nude mice aged 4–6 weeks old were purchased from Vital River Company. All mice were maintained under specific pathogen-free conditions. All animal experiment procedures were approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital and were performed in compliance with the principles and procedures of the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Statistical analysis
SPSS software version 21.0 was used to perform statistical analyses. Differentially expressed proteins with P<0.05 were considered statistically significant. All statistical analyses and data plotting were performed by using R software (http:///www.r-project.org).
Results
BCAA degradation pathway was enriched in PDAC
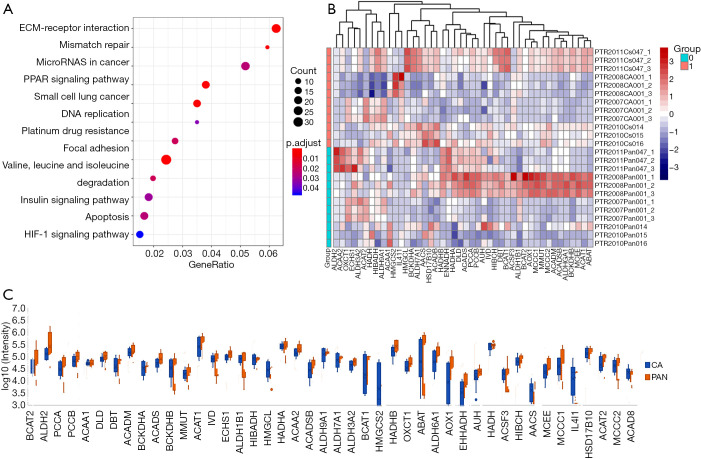
The levels of protein expression in six sets of PDAC and adjacent tissues were detected by using nano-LC-MS/MS. Figure 1A shows that differential proteins were enriched in some pathways, including the BCAA degradation pathway. Figure 1B shows the heatmap of protein expression involved in the BCAA degradation pathway in PDAC and adjacent tissues. The changes in these proteins are illustrated in Figure 1C. Together, these results might highlight the importance of BCAAs in PDAC.
Figure 1.
The BCAA degradation pathway was enriched in PDAC. (A) BCAA degradation pathway was enriched in PDAC through KEGG enrichment analysis; (B) a heatmap of protein expression involved in the BCAA degradation pathway; (C) the changes of these proteins. BCAA, branched-chain amino acid; PDAC, pancreatic ductal adenocarcinoma; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Clinicopathological data of study participants
The clinicopathological data of the PDAC patients (n=67) and HCs (n=54) are provided in Tables 1,2. Patients with PDAC over 70 years of age were excluded to match the HCs. The patients with PDAC had a mean age of 60 years, and 42 were male and 25 were female. Classification according to the American Joint Committee on Cancer (AJCC) 8th edition revealed that 12, 19, 16, and 20 tumors were stage I, II, III, and IV tumors, respectively. The HCs had a mean age of 46.5 years, and 30 were male and 24 were female.
Table 1. Characteristics of samples from patients with PDAC.
| Characteristic | Training set | Validation set | Total |
|---|---|---|---|
| Sex, male/female | 33/17 | 9/8 | 42/25 |
| Median age (years) | 59.5 | 61 | 60 |
| Clinical stage | 50 | 17 | 67 |
| Stage IA | 1 | 0 | 1 |
| Stage IB | 9 | 2 | 11 |
| Stage II | 15 | 4 | 19 |
| Stage III | 10 | 6 | 16 |
| Stage IV | 15 | 5 | 20 |
| PDAC CA19-9+ | 38 | 15 | 43 |
| PDAC CEA+ | 25 | 9 | 34 |
| PDAC CA242+ | 25 | 8 | 33 |
PDAC, pancreatic ductal adenocarcinoma.
Table 2. Characteristics of samples from healthy controls (HC).
| Characteristic | Training set | Validation set | Total |
|---|---|---|---|
| Healthy controls | 38 | 16 | 54 |
| Sex, male/female | 21/17 | 9/7 | 30/24 |
| Median age (years) | 45.5 | 47.5 | 46.5 |
Up-regulation of BCAAs in PDAC clinical samples
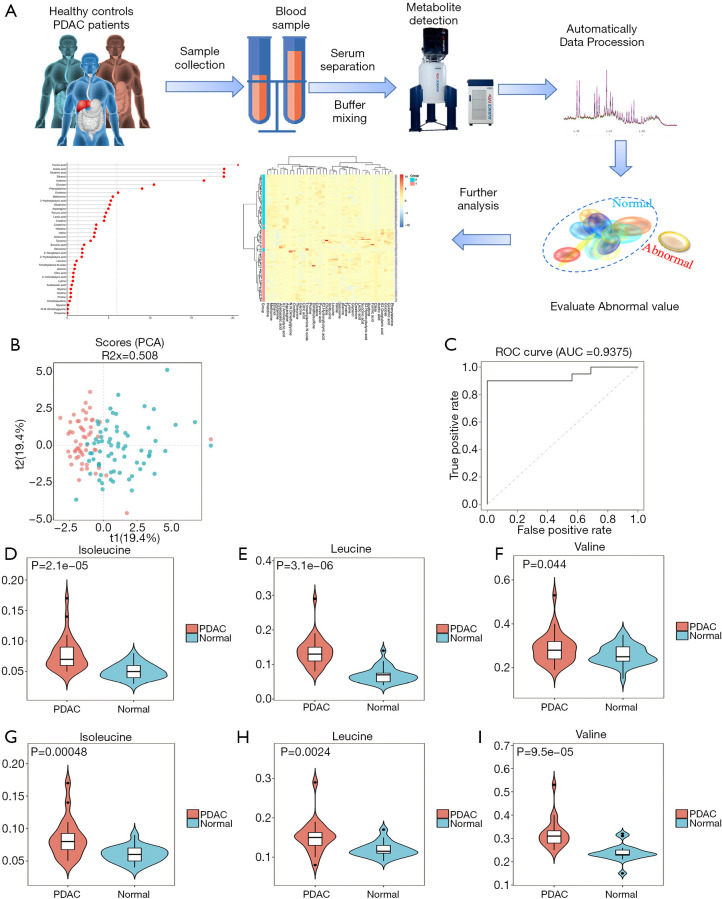
An NMR-based metabolomics approach that mainly targeted water-soluble metabolites was used to detect the content of BCAAs in PDAC and HC. Figure 2A shows the workflow of the NMR process.
Figure 2.
BCAAs were upregulated in clinical samples of PDAC. (A) workflow for the NMR process to detect metabolites in serum; (B) principal component analysis score plots showing differences in metabolic molecules between the PDAC patients and HCs; (C) the AUC was 0.9375, reflecting the accuracy of the results. (D,E,F) BCAA levels were significantly higher in PDAC patients compared to HCs in serum. (G,H,I) BCAA levels were significantly higher in PDAC patients compared to HCs in tissues. BCAAs, branched-chain amino acids; PDAC, pancreatic ductal adenocarcinoma; HC, healthy controls.
We compared the plasma metabolomic profiles of 38 samples from the HCs and 50 samples from the PDAC patients in the training set to identify a metabolic signature of PDAC. The score plots of principal component analysis revealed differences in metabolic molecules between the HC and PDAC groups (Figure 2B). The remaining 16 samples from HCs and 17 samples from PDAC were used to verify the accuracy of the results from the training set. The resulting AUC was 0.9375 (Figure 2C). We found that serum BCAA levels were significantly higher in PDAC patients compared to HCs (Figure 2D,E,F). Next, we determined whether the levels of BCAAs also increased in patients with PDAC at the organizational level. Compared with those in adjacent normal tissues, the levels of isoleucine, leucine and valine in PDAC tissues were increased (Figure 2G,H,I). Therefore, BCAA showed significant up-regulation in the clinical samples of PDAC.
BCAAs cannot promote the proliferation or migration of PDAC cells
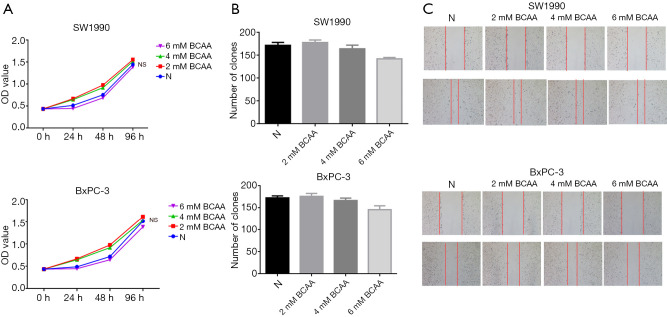
Given that the role of BCAAs in tumor growth is complex and depends on tumor type, we next explored the effects of BCAAs on PDAC growth by using SW1990 and BxPC-3 cells. We conducted CCK-8 and colony formation assays to explore the effects of BCAAs on PDAC cell proliferation. As shown in Figure 3A and B, BCAAs could not promote PDAC cell proliferation. Next, we conducted a wound healing assay (Figure 3C) to verify the effect of BCAAs on PDAC cell migration. Together, these data indicated that BCAAs had no effect on the proliferation or migration of PDAC cells in vitro.
Figure 3.
BCAAs can’t promote the proliferation and migration of PDAC cells. (A,B) The addition of different concentrations of BCAAs to the cell culture medium failed to promote PDAC cell proliferation; (C) the addition of different concentrations of BCAAs failed to promote PDAC cell migration. BCAAs, branched-chain amino acids; PDAC, pancreatic ductal adenocarcinoma.
Importance of aPSCs in the development of PDAC
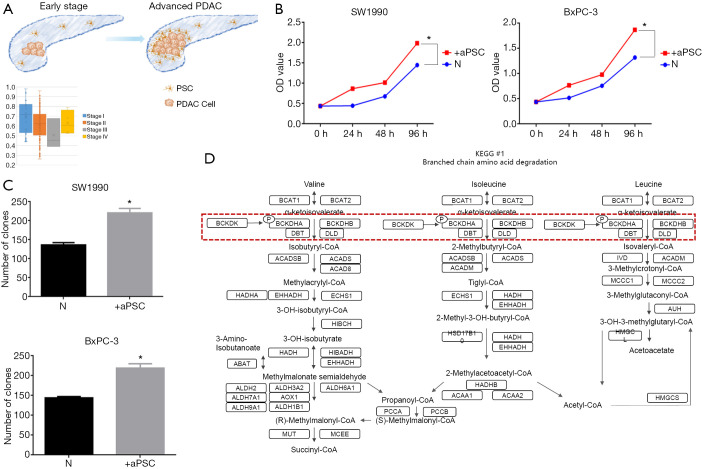
We analyzed the correlation between the number of aPSCs and the progression of PDAC to investigate the role of aPSCs in PDAC. The number of tumor cells was inversely related to the number of aPSCs. The purity of PDAC cells was calculated on the basis of the RNA sequencing data of TCGA, and the different clinical stages of PDAC were compared. Given the small number of stage III and IV cases in the study, only the results for stage I and stage II PDAC were reported. We found that the number of PDAC cells gradually decreased with the progression of PDAC, whereas the number of aPSCs gradually increased (Figure 4A). Next, added aPSCs supernatant into PDAC cell culture medium to perform CCK-8. And co-cultured aPSCs and PDAC cells to perform clone formation assay. The CCK-8 and clone formation assay results demonstrated that the addition of aPSCs could promote cell proliferation (Figure 4B,C). We also used nano-LC-MS/MS to detect changes in intracellular protein levels. Comparison with the HC group revealed that cancer cells cocultured with aPSC cells showed changes in the BCAA degradation pathway. The altered proteins are labeled in Figure 4D.
Figure 4.
The importance of aPSCs in the development of PDAC. (A) With the development of PDAC, the number of aPSCs gradually increased; (B) added aPSCs supernatant into PDAC cell culture medium can promote PDAC proliferation; (C) co-culture with aPSCs significantly enhanced PDAC cell proliferation; (D) the BCAA degradation pathway proteins with alterations following co-cultivation with aPSCs. *, statistically significant (P<0.05). aPSCs, activated pancreatic stellate cells; BCAAs, branched-chain amino acids; PDAC, pancreatic ductal adenocarcinoma.
Tumor growth was reduced by a branched-chain ketoacid dehydrogenase kinase inhibitor
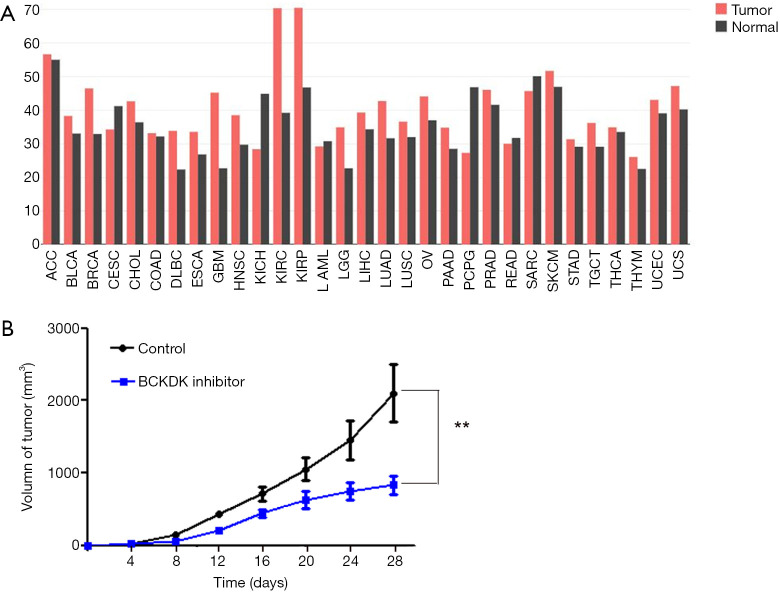
Considering that branched-chain ketoacid dehydrogenase kinase (BCKDK) is a key negative regulatory enzyme in BCAA catabolism that regulates the expression of the branched-chain α-keto acid dehydrogenase complex and plays an important role in many human diseases, we analyzed the expression levels of BCKDK in PDAC and normal tissues in the TCGA database (Figure 5A). The expression level of BCKDK in PDAC tissue was higher than that in normal tissue. Next, we subcutaneously injected PDAC cells into the right flanks of male athymic BALB/c nude mice. On day 7 after inoculation, the tumor-bearing mice were randomly divided into the treatment and control groups. We injected BCKDK inhibitor (BT2) into one of these groups. Tumor growth was significantly inhibited by BT2 therapy. The tumor growth curve is given in Figure 5B. Together, these data demonstrated that blocking BCKDK expression significantly reduced the tumorigenic properties of PDAC cells in vivo.
Figure 5.
BCKDK inhibitor (BT2) reduces the tumorigenic properties of PDAC cells. (A) The expression levels of BCKDK in various cancer tissues in the TCGA database in the GEPIA http://gepia.cancer-pku.cn/detail.php; (B) the final average tumor growth curve of mice injected with control or BCKDK inhibitor. **, statistically significant (P<0.01). BCKDK, branched-chain ketoacid dehydrogenase kinase; PDAC, pancreatic ductal adenocarcinoma; TCGA, The Cancer Genome Atlas.
Discussion
PDAC is a lethal malignancy, and its incidence is increasing (13). Given the difficulty in detecting PDAC, as well as the atypical early symptoms of this malignancy, the majority of PDAC cases are diagnosed at a late stage of the disease (14). Consequently, most patients with PDAC are unsuitable for surgery and, with limited therapeutic options, their 5-year survival rate is low. Given the poor prognosis of PDAC, further research to identify new therapeutic targets is desperately needed.
BCAAs are a group of amino acids that include valine, leucine, and isoleucine, which are essential amino acids and cannot be synthesized by the human body. BCAAs have unique properties and perform various physiological and metabolic roles. The best-characterized metabolic phenotype observed in tumor cells is the Warburg effect. Recently, disorders of BCAA metabolism have been reported in various malignancies, including liver cancer (15), pancreatic cancer (16), and leukemia (17). Compared with adjacent tissues, PDAC tissues showed increased BCAA uptake (4). BCAAs are strongly associated with the progression of PDAC, although the mechanisms underlying this progression remain poorly understood. Here, we detected differentially expressed proteins between cancerous and adjacent tissues from patients with PDAC and found them to be enriched in the BCAA degradation pathway. Mayers (18) found that the plasma levels of BCAAs are elevated in samples collected 2–5 years before diagnosis. These elevations are associated with a greater than 2-fold increase in the risk of a future diagnosis of pancreatic cancer. Therefore, we measured the levels of BCAAs in the serum of patients with PDAC and HCs. We found that BCAA concentrations in the serum of patients with PDAC were significantly higher than those in HCs. We also measured the BCAA content in cancerous and adjacent tissues and obtained similar results. Ericksen (15) found that a decrease in the expression of BCAA catabolic enzymes can lead to an increase in BCAAs, and the degree of the inhibition of these enzymes is correlated with tumor growth and aggressiveness. In other words, the loss of BCAA catabolism can induce the mTORC1 pathway to promote tumor development and growth. The inhibition of mTORC1 can be beneficial to Kras-driven pancreatic cancers (19); however, the specific mechanism of this benefit is unclear. Most tissues can oxidize BCAAs rapidly into the tricarboxylic acid cycle to provide a carbon source for cancer cells (20). Therefore, we speculated that the proliferation and migration of PDAC cells would be promoted by BCAAs. However, after adding different concentrations of BCAAs to the cell culture medium, we found that cell proliferation was not promoted by high levels of BCAAs. Given that the levels of BCAAs in a typical cell culture medium may be far above the physiological level, we will explore the effect of low BCAA levels in future studies. We investigated other initiating factors that regulate BCAA metabolism to promote tumor progression.
PSCs are located at the basolateral aspect of acinar cells or in the surrounding perivascular and periductal regions in the healthy pancreas. PSCs are star-shaped stromal cells and constitute most of the PDAC matrix. Quiescent PSCs are involved in the storage of vitamin A-rich lipid droplets, normal exocrine and endocrine secretion, phagocytosis, immunity, and the maintenance of normal stromal composition. When disease occurs, quiescent PSCs are activated through various underlying mechanisms. aPSCs play an important role in the aggressive, metastatic, and resilient nature of PDAC. aPSCs can secrete some cytokines such as IL-6, IL-10 and some growth factors, including insulin-like growth factor 1 (IGF1), platelet-derived growth factor (PDGF), fibroblast growth factor (FGF), CTGF. These cytokines and growth factors can promote cancer cells proliferation, migration, and invasion, which leads cancer cells metastasis. Sousa (6) found that aPSCs are critical for PDAC metabolism through the secretion of nonessential amino acids. We intended to explore the novel metabolic interaction between aPSCs and PDAC cells. Thus, we cocultured aPSCs and PDAC cells to detect changes in the BCAA metabolism pathway in PDAC. We found that the expression of proteins related to the BCAA degradation pathway was decreased. This decrement led to the accumulation of BCAAs. Further research is needed to determine whether an increase in BCAAs can result in the development of PDAC.
In conclusion, this study is the first to explore the effect of aPSCs on the BCAA degradation pathway in PDAC cells. Our results indicated that aPSCs could regulate BCAA metabolism in PDAC. This capability might become a meaningful research direction. Although aPSCs have previously been shown to promote PDAC cell proliferation through the secretion of alanine, the underlying mechanism by which aPSCs regulate the BCAA degradation pathway remains unclear. Additional experiments are necessary to explore the molecular mechanism of the interaction between aPSCs and PDAC cells.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by the Natural Science Foundation of Tianjin (grant No. 18JCZDJC32600), the National Natural Science Foundation of China (grant No. 81802432), and the Key Laboratory of Immune Microenvironment and Disease (Tianjin Medical University), Ministry of Education (grant No. 20200201).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The usage of serum samples in this study was reviewed and approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (Scientific Ethical Approval No. bc2020101), and informed consent was obtained from all the patients. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All animal experiment procedures were approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital and were performed in compliance with the principles and procedures of the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-761
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-761
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-761). The authors have no conflicts of interest to declare.
(English Language Editor: J. Reynolds)
References
- 1.Pothuraju R, Rachagani S, Junker WM, et al. Pancreatic cancer associated with obesity and diabetes: an alternative approach for its targeting. J Exp Clin Cancer Res 2018;37:319. 10.1186/s13046-018-0963-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halbrook CJ, Lyssiotis CA. Employing Metabolism to Improve the Diagnosis and Treatment of Pancreatic Cancer. Cancer Cell 2017;31:5-19. 10.1016/j.ccell.2016.12.006 [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 4.Lee JH, Cho YR, Kim JH, et al. Branched-chain amino acids sustain pancreatic cancer growth by regulating lipid metabolism. Exp Mol Med 2019;51:1-11. 10.1038/s12276-019-0299-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morani AC, Taher A, Ramani NS, et al. Pancreatic Cancer Imaging: What the Surgeon Wants to Know? Abdom Radiol (NY) 2020;45:964-81. [DOI] [PubMed] [Google Scholar]
- 6.Sousa CM, Biancur DE, Wang X, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 2016;536:479-83. 10.1038/nature19084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabe Y, Lorenzi PL, Konopleva M. Amino Acid Metabolism in Hematologic Malignancies and the Era of Targeted Therapy. Blood 2019;134:1014-23. 10.1182/blood.2019001034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sivanand S, Heiden MGV. Emerging Roles for Branched-Chain Amino Acid Metabolism in Cancer. Cancer Cell 2020;37:147-56. 10.1016/j.ccell.2019.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lei MZ, Li XX, Zhang Y, et al. Acetylation promotes BCAT2 degradation to suppress BCAA catabolism and pancreatic cancer growth. Signal Transduct Target Ther 2020;5:70. 10.1038/s41392-020-0168-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schnittert J, Bansal R, Prakash J. Targeting Pancreatic Stellate Cells in Cancer. Trends Cancer 2019;5:128-42. 10.1016/j.trecan.2019.01.001 [DOI] [PubMed] [Google Scholar]
- 11.Kuninty PR, Bansal R, Geus SWLD, et al. ITGA5 inhibition in pancreatic stellate cells attenuates desmoplasia and potentiates efficacy of chemotherapy in pancreatic cancer. Sci Adv 2019;5:eaax2770. 10.1126/sciadv.aax2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Y, Liu S, Zeng S, et al. The critical roles of activated stellate cells-mediated paracrine signaling, metabolism and onco-immunology in pancreatic ductal adenocarcinoma. Mol Cancer 2018;17:62. 10.1186/s12943-018-0815-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costello E. A metabolomics-based biomarker signature discriminates pancreatic cancer from chronic pancreatitis. Gut 2018;67:2-3. 10.1136/gutjnl-2016-313665 [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Bamlet WR, Oberg AL, et al. Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19-9 blood markers. Sci Transl Med 2017;9:eaah5583. 10.1126/scitranslmed.aah5583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ericksen RE, Lim SL, McDonnell E, et al. Loss of BCAA Catabolism during Carcinogenesis Enhances mTORC1 Ac-tivity and Promotes Tumor Development and Progression. Cell Metab 2019;29:1151-1165.e6. 10.1016/j.cmet.2018.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li JT, Yin M, Wang D, et al. BCAT2-mediated BCAA catabolism is critical for development of pancreatic ductal adenocarcinoma. Nat Cell Biol 2020;22:167-74. 10.1038/s41556-019-0455-6 [DOI] [PubMed] [Google Scholar]
- 17.Hattori A, Tsunoda M, Konuma T, et al. Cancer progression by reprogrammed BCAA metabolism in myeloid leukaemia. Nature 2017;545:500-4. 10.1038/nature22314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayers JR, Wu C, Clish CB, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med 2014;20:1193-8. 10.1038/nm.3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palm W, Park Y, Wright K, et al. The Utilization of Extracellular Proteins as Nutrients Is Suppressed by mTORC1. Cell 2015;162:259-70. 10.1016/j.cell.2015.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Z, Achreja A, Meurs N, et al. Tumour-reprogrammed stromal BCAT1 fuels branched-chain ketoacid dependency in stromal-rich PDAC tumours. Nat Metab 2020;2:775-92. 10.1038/s42255-020-0226-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as