Abstract
Introduction
The study evaluated if blood markers reflecting diverse biological pathways differentiate clinical diagnostic groups among Hispanic and non‐Hispanic White adults.
Methods
Within Hispanic (n = 1193) and non‐Hispanic White (n = 650) participants, serum total tau (t‐tau), neurofilament light (NfL), ubiquitin carboxyl‐terminal hydrolase LI, glial fibrillary acidic protein (GFAP), soluble cluster of differentiation‐14, and chitinase‐3‐like protein 1 (YKL‐40) were quantified. Mixed‐effects partial proportional odds ordinal logistic regression and linear mixed‐effects models were used to evaluate the association of biomarkers with diagnostic group and cognition, adjusting for age, sex, ethnicity, apolipoprotein E ε4, education, and site.
Results
T‐tau, NfL, GFAP, and YKL‐40 discriminated between diagnostic groups (receiver operating curve: 0.647–0.873). Higher t‐tau (odds ratio [OR] = 1.671, 95% confidence interval [CI] = 1.457–1.917, P < .001), NfL (OR = 2.150, 95% CI = 1.819–2.542, P < .001), GFAP (OR = 2.283, 95% CI = 1.915–2.722, P < .001), and YKL‐40 (OR = 1.288, 95% CI = 1.125–1.475, P < .001) were associated with increased likelihood of dementia relative to cognitively unimpaired and mild cognitive impairment groups. Higher NfL was associated with poorer global cognition (β = –0.455, standard error [SE] = 0.083, P < .001), semantic fluency (β = –0.410, SE = 0.133, P = .002), attention/processing speed (β = 2.880, SE = 0.801, P < .001), and executive function (β = 5.965, SE = 2.037, P = .003). Higher GFAP was associated with poorer global cognition (β = –0.345, SE = 0.092, P = .001), learning (β = –1.426, SE = 0.359, P < .001), and memory (β = –0.890, SE = 0.266, P < .001). Higher YKL‐40 (β = –0.537, SE = 0.186, P = .004) was associated with lower memory scores. Interactions with ethnicity were observed for learning (NfL, GFAP, YKL‐40), memory (NfL, GFAP), and semantic fluency (NfL; interaction terms P < .008), which were generally no longer significant in a demographically matched subset of Hispanic and non‐Hispanic White participants.
Discussion
Blood biomarkers of neuronal/axonal and glial injury differentiated between clinical diagnostic groups in a bi‐ethnic cohort of Hispanic and non‐Hispanic Whites. Our results add to the growing literature indicating that blood biomarkers may be viable tools for detecting neurodegenerative conditions and highlight the importance of validation in diverse cohorts.
Keywords: Alzheimer's disease, blood biomarkers, chitinase‐3‐like protein 1, ethnicity, glial fibrillary acidic protein, Hispanic, mild cognitive impairment, neurofilament light, tau
1. INTRODUCTION
A significant limiting factor for both timely Alzheimer's disease (AD) treatment and research participation is the vast underdiagnosis of dementia. Diagnostic delays more severely impact Hispanics and other diverse groups, who are more likely to be misdiagnosed or diagnosed at later stages than non‐Hispanic Whites within the United States. 1 The establishment of sensitive blood‐based biomarkers for dementia could facilitate broadly accessible diagnostic tools and may advance our understanding of the biological pathways implicated in AD and related dementias (ADRDs). 2 In the past, establishment of blood‐based biomarkers for ADRDs was hindered by relatively low protein concentrations in the blood relative to the cerebrospinal fluid (CSF). 2 However, single molecule array methods (Simoa) have been developed, improving analytic sensitivity up to 1000‐fold compared to traditional enzyme‐linked immunosorbent assays (ELISAs). 3 , 4 With this technology, prior studies have reported that blood‐based biomarkers, including total tau (t‐tau) and neurofilament light (NfL), have diagnostic accuracy for dementia. 4 , 5 , 6 , 7 However, these studies have been conducted in primarily non‐Hispanic White populations, limiting the generalizability of the findings. As racial/ethnic differences have been reported in dementia presentation and disease biology including genetic predisposition, burden of modifiable risk factors, age of onset, and disease progression, 8 , 9 the associations between biomarkers and cognitive status may vary across racial/ethnic groups. A recent white paper by the Alzheimer's Association International Society to Advance Alzheimer's Research and Treatment (ISTAART) highlighted validation of biofluid markers in diverse ethnoracial groups as a critical need in ADRD research. 10
The goal of the present study was to evaluate the diagnostic accuracy of blood‐based biomarkers for mild cognitive impairment (MCI) and dementia in a bi‐ethnic cohort of Hispanic and non‐Hispanic White older adults. Leveraging findings from genetic and neuropathological studies, 11 , 12 , 13 , 14 , 15 , 16 we assessed serum biomarkers associated with neuronal/axonal injury (t‐tau, NfL), ubiquitin protease system clearance (ubiquitin carboxyl‐terminal hydrolase L1 [UCHL1]), glial injury (soluble cluster of differentiation 14 [sCD14]), chitinase‐3‐like protein 1 (YKL‐40), and glial fibrillary acidic protein (GFAP). In addition to exploring associations across the whole sample, we evaluated interactions with ethnicity and apolipoprotein E (APOE) ε4 status. We hypothesized that the serum biomarkers would distinguish between cognitively unimpaired, MCI, and dementia groups.
2. METHODS
2.1. Participants
Samples were obtained from the baseline visit of participants in the Texas Alzheimer's Research and Care Consortium (TARCC). As previously described, 17 participants were recruited from nine Texas academic medical centers. Inclusion criteria included age 50 years or older at enrollment. A total of 3670 participants completed the baseline visit. As pilot award funds were used to conduct the assays for this project, we were able to process 1879 samples with the available budget. Given our interest in examining ethnicity, we over‐selected samples from individuals of Hispanic ethnicity and randomly selected a total of 1226 samples. The remainder of our funds were allocated to process samples from 653 non‐Hispanic White participants, selected across disease stages as defined by the Clinical Dementia Rating (CDR) global. Of the 1879 participants with available serum data for assay, 36 were excluded for missing data for at least one serum biomarker (missing NfL n = 35, t‐tau n = 34, UCLH1 n = 34, sCD14 n = 32, YKL‐40 = 30). Differences between participants with available serum data for the analyses and those without are presented in Table S1 in supporting information. The final study sample with available data included 1193 Hispanic (98% Mexican American) and 650 non‐Hispanic White adults aged 50 years and older. Analyses examining cognitive outcomes included a smaller subset of participants with available data for the specified measure. The study was approved by the institutional review board at each institution and was conducted in adherence with the Code of Ethics of the World Medical Association. All participants provided written informed consent prior to enrollment with appropriate legal representation for individuals lacking capacity to consent. Local institutional review board approval was obtained to process and analyze de‐identified samples and clinical/demographic data.
2.2. Neuropsychological evaluation
The neuropsychological evaluation was comprised of measures of global cognition (Mini‐Mental State Examination [MMSE]), 18 learning and memory (Weschler Memory Scale [WMS]‐3 Logical Memory [LM] I and II 19 ), attention/processing speed (Trail Making Test, Part A 20 ), executive function (Trail Making Test, Part B 20 ), and language (Animal Fluency 21 ).
Participants and their study partners also completed the CDR. 22
2.3. Consensus reviews
Clinical diagnoses were assigned based on review of clinical exams and neuropsychological assessments by a consensus committee at each site comprised of at least one physician, neuropsychologist, and research coordinator. Possible or probable diagnoses for AD were assigned based on National Institute of Neurological and Communicative Disorders and Stroke—Alzheimer's Disease and Related Disorders Association criteria. 23 Given the absence of biological confirmation of disease etiology, 24 the terms “dementia” and “ADRD” are used in place of specific diagnostic labeling. MCI subtypes (amnestic versus non‐amnestic) were assigned using established criteria defined by Petersen. 25
2.4. Blood draw and storage
The TARCC collected and processed blood in accordance with established guidelines. 26 Briefly, non‐fasting blood was collected in the morning using a 21g needle. Serum tubes were allowed to clot for 30 minutes. Plasma tubes were inverted 5 to 10 times and centrifuged for 10 minutes at 2000 x g within 1 hour of collection. 500uL aliquots were transferred to polypropylene tubes and samples were placed into –80° freezer within 2 hours of collection. APOE genotyping was performed with polymerase chain reaction as previously described. 17 Samples were shipped to the Laboratory for Clinical Biochemistry Research at the University of Vermont, which has a strong quality assurance program for assays and is equipped with Simoa HD‐1 Analyzer (Quanterix).
2.5. Quantification of serum biomarker levels
Serum levels of t‐tau, NfL, UCHL1, and GFAP were analyzed using the Simoa Neurology 4‐Plex Kit on a Simoa HD‐1 Analyzer (Quanterix). Serum levels of sCD14 and YKL‐40 were quantified using commercial ELISAs (R&D Systems). Analytical ranges and inter‐assay coefficients of variance are presented in Table S2 in supporting information. A certified laboratory technician, blinded to diagnostic and ethnic groups, performed all assays between November and December 2019 using a single batch of reagents.
2.6. Statistical analysis
GFAP, t‐tau, NfL, UCHL1, and YKL‐40 displayed left‐skewed, non‐normal distributions and were natural log transformed, and subsequently winsorized to four standard deviations within the mean. Winsorization was applied to <1% of the biomarker values (UCHL1 = 10, NfL = 7, t‐tau = 4). In effort to limit data loss, t‐tau values below the detection limit were set equal to the detection limit prior to log transformation because values were known to be at this level or below. 27 Differences in demographics and clinical characteristics across the diagnostic groups and by ethnicity were assessed with the chi‐squared statistic for categorical variables or with independent t‐tests or Mann‐Whitney U‐tests for continuous variables.
HIGHLIGHTS
Serum total tau, neurofilament light (NfL), glial fibrillary acidic protein (GFAP), and chitinase‐3‐like protein 1 (YKL‐40) differed across control, mild cognitive impairment, and dementia groups .
Elevated serum NfL, GFAP, and YKL‐40 levels were associated with poorer cognition.
Findings were consistent in demographically matched Hispanics and non‐Hispanic Whites.
RESEARCH IN CONTEXT
Systematic review: Blood‐based biomarkers for dementia hold potential to improve diagnostic accuracy and provide insight into underlying pathophysiological pathways. To date, the majority of biomarker studies have been conducted primarily in non‐Hispanic White populations.
Interpretation: Blood markers of neuronal/axonal injury (total tau [t‐tau], neurofilament light [NfL]) and glial injury (glial fibrillary acidic protein [GFAP], chitinase‐3‐like protein 1 [YKL‐40]) differentiated clinical diagnostic groups in a bi‐ethnic Hispanic and non‐Hispanic White cohort. Higher serum NfL, GFAP, and YKL‐40 levels were associated with poorer cognition. Interactions between biomarkers and ethnicity for some cognitive outcomes were observed, which were generally no longer significant in a demographically matched subset of Hispanic and non‐Hispanic White participants.
Future directions: Future research is needed to determine the interplay of multidimensional sociocultural, environmental, and biological factors that may contribute to ethnic differences in Alzheimer's disease and related disorders serum biomarker values.
The associations between blood‐based biomarkers and clinical diagnostic group were evaluated with separate mixed‐effects partial proportional odds ordinal logistic regression models adjusting for age, sex, ethnicity, APOE ε4 status, and education as fixed effects, and site as a random effect. All covariates were selected a priori and were tested for the assumption of proportional odds using likelihood ratio tests. Serum biomarkers, ethnicity, and APOE violated the assumption of proportional odds (P < .05) and were fitted with unequal slopes. All six biomarkers were also included in a single mixed‐effects partial proportional odds ordinal logistic regression with adjustment for age, sex, ethnicity, APOE ε4 status, education, site (as a random effect), diabetes, systolic blood pressure, and body mass index (BMI). As supplementary analyses, biomarker values within the amnestic (n = 325) and non‐amnestic MCI (n = 115) subtypes were compared to the cognitively unimpaired group, as well as to one another, using multinominal logistic regression with adjustment for age, sex, ethnicity, APOE ε4 status, education, and site.
Diagnostic accuracies between pairs of outcomes were calculated using area under the receiver operating characteristic curve (AUROC) from pairwise mixed‐effects binary logistic regression models, and volume under the ROC surface (VUS) for the three‐category outcome was calculated from mixed‐effects partial proportional odds ordinal logistic regression models, with covariates for age, sex, ethnicity, APOE ε4 status, education, and site (as a random effect). For the purpose of calculating sensitivity and specificity, optimal cut‐points were defined as those maximizing the sum of sensitivity and specificity, and were found using the cutpointr package in R.
The associations between blood‐based biomarkers and cognitive outcomes were evaluated with separate linear mixed‐effects models adjusting for age, sex, ethnicity, APOE ε4 status, education, site (as a random effect), and clinical diagnostic group. In addition, models were assessed that also included interaction terms between biomarkers with ethnicity, and separate models were fit that included interaction terms between biomarkers and APOE ε4 status.
Finally, in exploratory analyses, the associations between biomarkers with clinical diagnostic group, as well as interactions between ethnicity with clinical diagnostics and cognition, were evaluated in a subsample of more closely matched Hispanic and non‐Hispanic White participants using separate mixed‐effects partial proportional odds ordinal logistic regression and linear mixed‐effects models adjusting for age, sex, ethnicity, APOE ε4 status, education, and site as a random effect. One‐to‐one participant matching across the two ethnic groups for age, sex, APOE status, education, and diagnostic group was performed using the optmatch package in R.
For ordinal logistic regression and linear mixed‐effects models, biomarker values were standardized to z‐scores to facilitate comparisons across measures. Statistical tests were two‐sided and statistical significance was set at P < .05. For all models assessing the effect of biomarkers, P‐values were Bonferroni‐corrected for the number of biomarkers, which placed the raw P‐value for statistical significance at P < .008. Statistical analyses were performed using R version 3.6.2.
3. RESULTS
3.1. Participant characteristics
As seen in Table 1, the Hispanic and non‐Hispanic White groups across the entire sample and within clinical diagnostic groups differed across most demographic and clinical variables, as well as blood biomarker levels. Table S3 in supporting information presents the demographic and clinical variables for a subgroup of Hispanics (N = 321) and non‐Hispanic White (N = 321) participants matched for age, sex, APOE status, education, and diagnostic group.
TABLE 1.
Demographic and clinical characteristics of Hispanic and non‐Hispanic White groups by clinical diagnostic group (N = 1843)
| Overall | Cognitively unimpaired | MCI | Dementia | |||||
|---|---|---|---|---|---|---|---|---|
| Hispanic N = 1193 | Non‐Hispanic White N = 650 | Hispanic N = 711 | Non‐Hispanic White N = 184 | Hispanic N = 325 | Non‐Hispanic White N = 115 | Hispanic N = 157 | Non‐Hispanic White N = 351 | |
| Age, years | 67 ± 9* | 74 ± 8 | 63 ± 8* | 72 ± 8 | 70 ± 9* | 73 ± 9 | 75 ± 8 | 75 ± 9 |
| Female, N (%) | 840 (70%)* | 358 (55%) | 515 (72%)* | 117 (64%) | 223 (69%)* | 52 (45%) | 102 (65%)* | 189 (54%) |
| Education, years | 10 ± 5* | 15 ± 3 | 10 ± 5* | 16 ± 2 | 10 ± 4* | 15 ± 3 | 10 ± 5* | 15 ± 3 |
| BMI, m/kg2 | 31 ± 6* | 27 ± 4 | 31 ± 7* | 27 ± 4 | 31 ± 6* | 28 ± 4 | 29 ± 5* | 26 ± 4 |
| Blood pressure, mmHg | ||||||||
| Systolic | 139 ± 21* | 134 ± 18 | 137 ± 21* | 130 ± 27 | 139 ± 19 | 135 ± 18 | 144 ± 21* | 135 ± 18 |
| Diastolic | 78 ± 12* | 76 ± 11 | 79 ± 12 | 74 ± 10 | 76 ± 11 | 76 ± 11 | 76 ± 11 | 76 ± 11 |
| Diabetes, N (%) | 398 (33%)* | 64 (10%) | 230 (32%)* | 14 (8%) | 112 (34%)* | 17 (15%) | 56 (36%)* | 33(9%) |
| Presence of APOE ε4 allele, N (%) | 247 (21%)* | 309 (48%) | 130 (18%) | 38 (21%) | 58 (18%)* | 45 (39%) | 59 (38%)* | 226 (64%) |
| CDR Global, N (%) | ||||||||
| 0 | 737(62%) | 194 (30%) | 711(100%) | 183 (99%) | 26 (8%) | 11 (10%) | 0 (0%) | 0 (0%) |
| 0.5 | 377 (32%) | 225 (35%) | 0 (0%) | 1 (1%) | 298 (92%) | 103 (90%) | 79 (50%) | 121 (34%) |
| ≥1 | 79 (7%) | 231 (36%) | 0 (0%) | 0 (0%) | 1 (< 1%) | 1 (< 1%) | 78 (50%) | 230 (66%) |
| Serum t‐tau, median (quartile 1, quartile 3) | 0.3 (0.1, 0.4)* | 0.3 (0.2, 0.6) | 0.3 (0.1, 0.4) | 0.2 (0.1, 0.4) | 0.3 (0.1, 0.4)* | 0.2 (0.1, 0.4) | 0.4 (0.2, 0.6) | 0.4 (0.3, 0.6) |
| Serum NfL median (quartile 1, quartile 3) | 17 (12, 26)* | 28 (20, 39) | 15 (11, 21)* | 22 (17, 31) | 20 (14, 29)* | 24 (17, 35) | 31 (22, 50) | 33 (24, 45) |
| Serum GFAP median (quartile 1, quartile 3) | 155 (110, 224)* | 344 (203, 488) | 134 (98, 186)* | 206 (145, 367) | 174 (120, 249)* | 253 (175, 380) | 279 (182, 432)* | 429 (308, 591) |
| Serum UCHL1 median (quartile 1, quartile 3) | 28 (22, 38)* | 36 (28, 51) | 27 (22, 37)* | 33 (24, 44) | 29 (22, 38)* | 35 (26, 50) | 34 (26, 44)* | 39 (30, 56) |
| Serum YKL‐40 median (quartile 1, quartile 3) | 60,645 (37,681, 109,885)* | 49,005 (31,140, 82,929) | 55,126 (35,181, 101,850)* | 41,825 (27,811, 74,587) | 64,210 (38,685, 111,221)* | 45,589 (29,075, 67,417) | 83,807 (55,101, 150,856)* | 55,437 (33,577, 91,645) |
| Serum sCD14 median (quartile 1, quartile 3) | 1306 (1163, 1506)* | 1422 (1221, 1636) | 1299 (1154, 1466)* | 1422 (1226, 1591) | 1324 (1167, 1553) | 1349 (1186, 1524) | 1347 (1182, 1623)* | 1452 (1247, 1676) |
| MMSE | 26 ± 4* | 25 ± 4 | 28 ± 2* | 29 ± 0.9 | 26 ± 3* | 28 ± 2 | 21 ± 5* | 22 ± 4 |
| WMS LM I | 30 ± 11* | 32 ± 16 | 34 ± 9* | 44 ± 9 | 27 ± 10* | 30 ± 11 | 16 ± 11 | 14 ± 9 |
| WMS LM 2 | 17 ± 9 | 17 ± 12 | 20 ± 7* | 27 ± 7 | 15 ± 8 | 16 ± 9 | 6 ± 7* | 3 ± 5 |
| Animal Fluency | 15 ± 5* | 14 ± 6 | 16 ± 4* | 18 ± 4 | 14 ± 4* | 16 ± 6 | 10 ± 4* | 11 ± 5 |
| Trails A, seconds to complete | 57 ± 32* | 51 ± 30 | 49 ± 24* | 36 ± 11 | 62 ± 33* | 40 ± 14 | 91 ± 41* | 63 ± 35 |
| Trails B, seconds to complete | 148 ± 80 | 144 ± 87 | 121 ± 61* | 79 ± 29 | 178 ± 82* | 113 ± 59 | 242 ± 75* | 199 ± 86 |
Note: *P < .05; Group differences were assessed with independent t‐tests or Mann‐Whitney U tests for continuous variables and the chi‐squared statistic for categorical variables. Fisher's exact test was applied to the CDR Global variable due to low cell counts. All values represent mean ± standard deviation unless otherwise noted.
Abbreviations: APOE, apolipoprotein E; BMI, body mass index; CDR, Clinical Dementia Rating scale; GFAP, glial fibrillary acidic protein; MMSE, Mini‐Mental State Examination; NfL, neurofilament light; sCD14, soluble cluster of differentiation 14; Trails, Trail Making Test; t‐tau, total tau; UCHL1, ubiquitin carboxyl‐terminal hydrolase L1; YKL‐40, chitinase‐3‐like protein 1; WMS LM, Weschler Memory Scale Logical Memory.
3.2. Associations between serum biomarkers and diagnostic group
As seen in Table 2 and Figure S1 in supporting information, higher levels of t‐tau, NfL, and GFAP were associated with increased likelihood of MCI or dementia relative to the cognitively unimpaired group. Increased levels of t‐tau, NfL, GFAP, and YKL‐40 were associated with greater likelihood of dementia relative to the cognitively unimpaired and MCI groups. The same pattern of findings was observed in a subset of demographically matched Hispanic and non‐Hispanic White participants (Table S4 in supporting information).
TABLE 2.
Associations of serum biomarkers with cognitive diagnostics group based on mixed‐effects partial proportional odds ordinal logistic regression (N = 1843)
| MCI or dementia relative to the cognitively unimpaired group | Dementia relative to the cognitively unimpaired and MCI groups | |
|---|---|---|
| T‐tau | OR = 1.320, 95% CI = 1.171–1.489, P < .001* | OR = 1.671, 95% CI = 1.457–1.917, P < .001* |
| NfL | OR = 1.466, 95% CI = 1.269–1.694, P < .001* | OR = 2.150, 95%CI = 1.819–2.542, P < 0.001* |
| GFAP | OR = 1.548, 95% CI = 1.321–1.813, P < 0.001* | OR = 2.283, 95% CI = 1.915–2.722, P < 0.001* |
| UCHL1 | OR = 1.013, 95% CI = 0.892–1.152, P = .837 | OR = 1.179, 95% CI = 1.027–1.354, P = .019 |
| sCD14 | OR = 1.007, 95% CI = 0.894–1.133, P = .914 | OR = 1.144, 95% CI = 1.009–1.299, P = .036 |
| YKL‐40 | OR = 1.157, 95% CI = 1.031–1.299, P = .013 | OR = 1.288, 95% CI = 1.125–1.475, P < .001* |
Note: Separate mixed‐effects partial proportional odds ordinal logistic regression models on clinical diagnostic group were conducted with covariates for age, sex, ethnicity, APOE ε4 status, education, site, and serum biomarkers.
Abbreviations: APOE, apolipoprotein E; CI, confidence interval; GFAP, glial fibrillary acidic protein; MCI, mild cognitive impairment; NfL, neurofilament light; OR, odds ratio; sCD14, soluble cluster of differentiation 14; t‐tau, total tau; UCHL1, ubiquitin carboxyl‐terminal hydrolase L1; YKL‐40, chitinase‐3‐like protein 1.
P < .008 level of significance after Bonferroni correction for six biomarkers.
In a model that included all six biomarkers with additional covariates for diabetes, systolic blood pressure, and BMI (Table S5 in supporting information), higher levels of t‐tau and GFAP were associated with increased likelihood of MCI or dementia. In addition, elevated levels of t‐tau, NfL, and GFAP were associated with higher likelihood of dementia relative to the cognitively unimpaired and MCI groups. Across biomarkers, there were no significant differences between amnestic and non‐amnestic MCI subtypes compared to one another and compared to the cognitively unimpaired group (Table S6 in supporting information).
3.3. Diagnostic accuracy of serum biomarkers for MCI and AD
As displayed in Figure 1, serum t‐tau, NfL, GFAP, and YKL‐40 differentiated between diagnostic groups. With inclusion of all six biomarkers and covariates, the AUROC was 0.876 (0.857–0.894, sensitivity = 0.823, specificity = 0.784) for cognitively unimpaired and dementia groups, 0.643 (0.612–0.674, sensitivity = 0.632, specificity = 0.617) for cognitively unimpaired and MCI groups, and 0.803 (0.775–0.831, sensitivity = 0.809, specificity = 0.664) for MCI and dementia groups. Multicategory classification analyses indicated that the performance for all biomarkers was above chance (VUS: 0.428–0.461, chance: 0.167, Figure S2 in supporting information).
FIGURE 1.
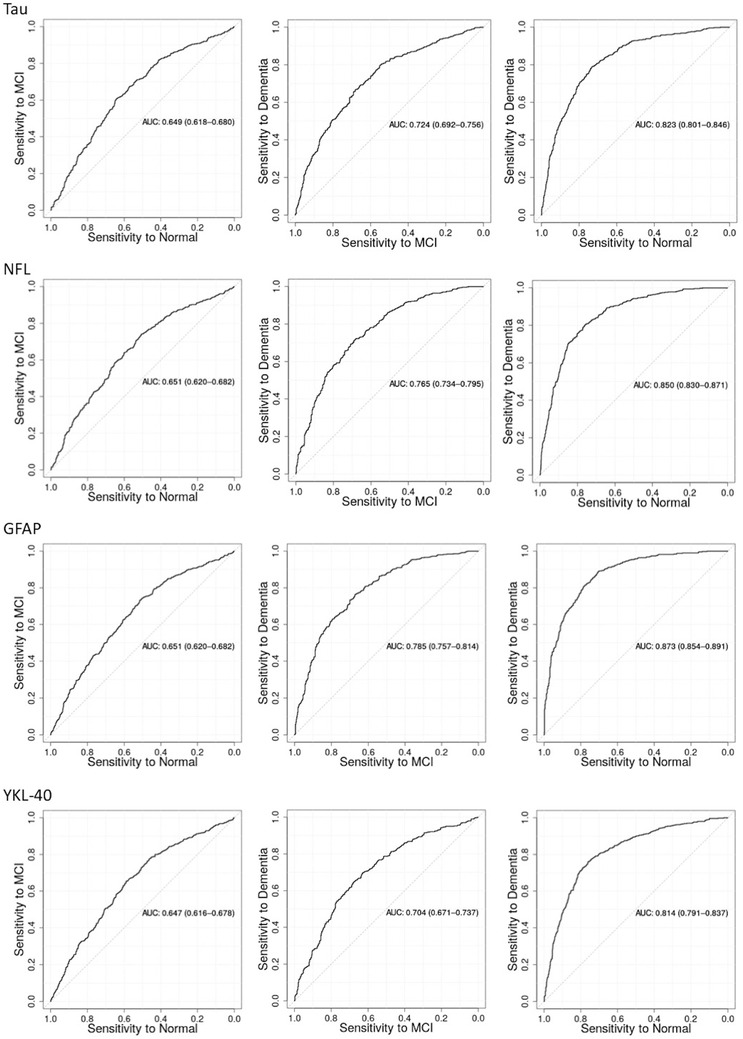
Area under the receiver operating characteristic curve (AUROC) for total tau, neurofilament light (NfL), glial fibrillary acidic protein (GFAP), and chitinase‐3‐like protein 1 (YLK‐40) with diagnostic group
3.4. Interactions between serum biomarkers with ethnicity and APOE ε4 status for diagnostic group
As displayed in Table 3, GFAP and NfL had a significant interaction with APOE ε4 status for odds of MCI and dementia relative to the cognitively unimpaired group. For these interactions, stronger associations were observed in APOE ε4 carriers. There was also a strong trend toward an interaction between t‐tau and ethnicity for odds of dementia relative to the cognitively unimpaired and MCI groups. No significant interactions with ethnicity were observed in the subset of matched Hispanic and non‐Hispanic White participants (Table S7 in supporting information).
TABLE 3.
Interactions between serum biomarkers with ethnicity and APOE ε4 status for clinical diagnostic group based on mixed‐effects partial proportional odds ordinal logistic regression models (N = 1843)
| Ethnicity x biomarkers | APOE ε4 x biomarkers | |||
|---|---|---|---|---|
| MCI or dementia relative to cognitively unimpaired | Dementia relative to MCI or cognitively unimpaired | MCI or dementia relative to cognitively unimpaired | Dementia relative to MCI or cognitively unimpaired | |
| T‐tau | Interaction P = .040 | Interaction P = .008 | Interaction P = .815 | Interaction P = .608 |
| Hispanic OR: 1.216, 95% CI = 1.050–1.408 | Hispanic OR: 1.401, 95% CI = 1.149–1.709 | APOE ε4 OR: 1.291, 95% CI = 1.030–1.618 | APOE ε4 OR: 1.601, 95% CI = 1.291–1.985 | |
| Non‐Hispanic OR: 1.592, 95% CI = 1.289–1.968 | Non‐Hispanic OR: 2.052, 95% CI = 1.675–2.154 | Non‐APOE ε4 OR: 1.332, 95% CI = 1.159–1.530 | APOE ε4 OR: 1.720, 95% CI = 1.441–2.054 | |
| NfL | Interaction P = .549 | Interaction P = .647 | Interaction P = .002* | Interaction P = .656 |
| Hispanic OR: 1.434, 95% CI = 1.219–1.688 | Hispanic OR: 2.104, 95% CI = 1.701–2.603 | APOE ε4 OR: 2.203, 95% CI = 1.630–2.979 | APOE ε4 OR: 2.340, 95% CI = 1.739–3.149 | |
| Non‐Hispanic OR: 1.574, 95% CI = 1.198–2.067 | Non‐Hispanic OR: 2.273, 95% CI = 1.738–2.975 | Non‐APOE ε4 OR: 1.319, 95% CI = 1.129–1.540 | Non‐APOE ε4 OR: 2.166, 95% CI = 1.788–2.625 | |
| GFAP | Interaction P = .099 | Interaction P = .082 | Interaction P < .001* | Interaction P = .958 |
| Hispanic OR: 1.423, 95% CI = 1.179–1.717 | Hispanic OR: 2.008, 95% CI = 1.582–2.548 | APOE ε4 OR: 2.528, 95% CI = 1.865–3.425 | APOE ε4 OR: 2.489, 95% CI = 1.870–3.314 | |
| Non‐Hispanic OR: 1.828, 95% CI = 1.419–2.355 | Non‐Hispanic OR: 2.700, 95% CI = 2.096–3.478 | Non‐APOE ε4 OR: 1.307, 95% CI = 1.100–1.554 | Non‐APOE ε4 OR: 2.466, 95% CI = 1.972–3.085 | |
| UCHL1 | Interaction P = .101 | Interaction P = .152 | Interaction P = .193 | Interaction P = .862 |
| Hispanic OR: 0.935, 95% CI = 0.794–1.101 | Hispanic OR: 1.076, 95% CI = 0.872–1.326 | APOE ε4 OR: 1.173, 95% CI = 0.903–1.524 | APOE ε4 OR: 1.219, 95% CI = 0.962–1.546 | |
| Non‐Hispanic OR: 1.171, 95% CI = 0.944–1.453 | Non‐Hispanic OR: 1.329, 95% CI = 1.086–1.627 | Non‐APOE ε4 OR: 0.964, 95% CI = 0.831–1.118 | Non‐APOE ε4 OR: 1.188, 95% CI = 0.996–1.417 | |
| sCD14 | Interaction P = .281 | Interaction P = .889 | Interaction P = .507 | Interaction P = .237 |
| Hispanic OR: 1.061, 95% CI = 0.913–1.232 | Hispanic OR: 1.131, 95% CI = 0.925–1.383 | APOE ε4 OR: 1.074, 95% CI = 0.852–1.353 | APOE ε4 OR: 1.061, 95% CI = 0.868–1.296 | |
| Non‐Hispanic OR: 0.932, 95% CI = 0.776–1.121 | Non‐Hispanic OR: 1.110, 95% CI = 0.938–1.315 | Non‐APOE ε4 OR: 0.981, 95% CI = 0.854–1.127 | Non‐APOE ε4 OR: 1.244, 95% CI = 1.053–1.470 | |
| YKL‐40 | Interaction P = .804 | Interaction P = .027 | Interaction P = .731 | Interaction P = .151 |
| Hispanic OR: 1.163, 95% CI = 1.016–1.331 | Hispanic OR: 1.537, 95% CI = 1.260–1.876 | APOE ε4 OR: 1.117, 95% CI = 0.886–1.408 | APOE ε4 OR: 1.150, 95% CI = 0.927–1.425 | |
| Non‐Hispanic OR: 1.126, 95% CI = 0.907–1.398 | Non‐Hispanic OR: 1.127, 95% CI = 0.927–1.371 | Non‐APOE ε4 OR: 1.169, 95% CI = 1.025–1.332 | Non‐APOE ε4 OR: 1.399, 95% CI = 1.177–1.663 | |
Note: Interaction terms displayed for separate mixed‐effects partial proportional odds ordinal logistic regression models on clinical diagnostic group with additional covariates for age, sex, ethnicity, APOE ε4 status, education, site, and serum biomarkers.
Abbreviations: APOE, apolipoprotein E; CI, confidence interval; GFAP, glial fibrillary acidic protein; MCI, mild cognitive impairment; NfL, neurofilament light; OR, odds ratio; sCD14, soluble cluster of differentiation 14; t‐tau, total tau; UCHL1, ubiquitin carboxyl‐terminal hydrolase L1; YKL‐40, chitinase‐3‐like protein 1.
P < .008 level of significance after Bonferroni correction for six biomarkers.
3.5. Associations between serum biomarkers and cognition
As seen in Table 4, higher levels of NfL were associated with poorer global cognition, semantic fluency, attention/processing speed, and executive function scores. Higher levels of GFAP were associated with poorer global cognition, learning, and memory. Additionally, higher YKL‐40 levels were associated with lower memory scores.
TABLE 4.
Results of linear mixed‐effects models displaying associations between serum biomarkers and cognitive outcomes
| T‐tau | NfL | GFAP | UCHL1 | YKL‐40 | sCD14 | |
|---|---|---|---|---|---|---|
| MMSE N = 1842 | β = –0.176, SE = 0.069, P =.011 | β = –0.455, SE = 0.083, P<.001* | β = –0.345, SE = 0.092, P =.001* | β = –0.140, SE = 0.073, P =.056 | β = –0.114, SE = 0.068, P =.092 | β = –0.150, SE = 0.066, P =.024 |
| WMS LM 1 N = 1406 | β = –0.307 SE = 0.265, P =.248 | β = –0.542, SE = 0.315, P =.085 | β = –1.426, SE = 0.359, P<.001* | β = –0.192, SE = 0.297, P =.517 | β = –0.658 SE = 0.252, P =.009 | β = 0.104, SE = 0.266, P =.696 |
| WMS LM II N = 1403 | β = –0.183, SE = 0.197, P =.353 | β = –0.224, SE = 0.233, P =.335 | β = –0.890, 0.266, P<.001* | β = –0.085, SE = 0.220 P =.698 | β = –0.537, SE = 0.186, P =.004* | β = –0.025, SE = 0.197, P =.899 |
| Animal Fluency N = 1583 | β = –0.190, SE = 0.113, P =.092 | β = –0.410, SE = 0.133, P =.002* | β = –0.346, SE = 0.151, P =.022 | β = –0.064, SE = 0.121, P =.596 | β = –0.072, SE = 0.109, P =.506 | β = –0.229, SE = 0.109, P =.037 |
| Trails A N = 1783 | β = 0.816 SE = 0.668, P =.215 | β = 2.880, SE = 0.801, P<.001* | β = –0.466, SE = 0.882, P =.600 | β = 1.281, SE = 0.699, P =.067 | β = 0.400, SE = 0.645, P =.535 | β = −‐1.257, SE = 0.639, P =.049 |
| Trails B N = 1608 | β = 3.265, SE = 1.644, P =.047 | β = 5.965, SE = 2.037, P =.003* | β = –1.621, SE = 2.216, P =.464 | β = –1.318, SE = 1.802, P =.465 | β = 1.556, SE = 1.631, P =.340 | β = –1.321, SE = 1.613, P =.413 |
Note: Linear mixed‐effects models with cognitive data regressed on age, sex, ethnicity, APOE ε4 status, education, site, clinical diagnostic group, and serum biomarkers.
Abbreviations: APOE, apolipoprotein E; BMI, body mass index; CDR, Clinical Dementia Rating scale; GFAP, glial fibrillary acidic protein; MMSE, Mini‐Mental State Examination; NfL, neurofilament light; sCD14, soluble cluster of differentiation 14; SE, standard error; Trails, Trail Making Test; t‐tau, total tau; UCHL1, ubiquitin carboxyl‐terminal hydrolase L1; YKL‐40, chitinase‐3‐like protein 1; WMS LM, Weschler Memory Scale Logical Memory.
P<.008 level of significance after Bonferroni correction for six biomarkers
3.6. Interactions between serum biomarkers with ethnicity and APOE ε4 status for cognition
As displayed in Table 5, the associations between NfL and GFAP with learning and memory differed by ethnicity. Additionally, NfL had an interaction with ethnicity for semantic fluency and YKL‐40 had an interaction with ethnicity for learning. For these cognitive outcomes, Hispanics displayed smaller changes in serum biomarker levels relative to non‐Hispanic Whites. The associations between serum levels of GFAP with learning and memory also differed by APOE ε4 status with presence of the APOE ε4 allele associated with larger effect sizes.
TABLE 5.
Interactions between serum biomarkers, ethnicity, and APOE ε4 status for cognition derived from linear mixed‐effects models
| Outcome variable | Interaction term | T‐tau | NfL | GFAP | UCHL1 | YKL‐40 | sCD14 |
|---|---|---|---|---|---|---|---|
| MMSE M = 1842 | Ethnicity x Biomarker | β = 0.039, SE = 0.139, P =.776 | β = 0.187, SE = 0.151, P =.216 | β = 0.096, SE = 0.153, P =.528 | β = 0.204, SE = 0.144, P =.155 | β = –0.007, SE = 0.137, P =.957 | β = –0.020, SE = 0.129, P =.875 |
| APOE x Biomarker | β = ‐0.250, SE = 0.140, P =.075 | β = 0.071, SE = 0.141, P =.618 | β = ‐0.233, SE = 0.143, P =.105 | β = ‐0.197, SE = 0.147, P =.181 | β = 0.146, SE = 0.141, P =.299 | β = 0.071, SE = 0.134, P = 0.597 | |
| WMS LM 1 N = 1406 | Ethnicity x Biomarker | β = 1.023, SE = 0.624, P =.102 | β = 3.441, SE = 0.680, P<.001 * | β = 3.708, SE = 0.670, P<.001* | β = 1.614, SE = 0.679, P =.018 | β = 1.717, SE = 0.608, P =.005* | β = –0.111, SE = 0.614, P =.857 |
| APOE x Biomarker | β = –0.803, SE = 0.580, P =.167 | β = –0.537, SE = 0.571, P =.348 | β = –1.843, SE = 0.623, P =.003* | β = –0.493, SE = 0.696, P = .479 | β = 0.086, SE = 0.582, P =.883 | β = 0.152, SE = 0.590, P =.796 | |
| WMS LM II N = 1403 | Ethnicity x Biomarker | β = 0.549, SE = 0.462, P =.235 | β = 1.595, SE = 0.506, P =.002* | β = 1.849, SE = 0.500, P<.001* | β = 0.979, SE = 0.502, P =.051 | β = 0.516, SE = 0.451, P =.252 | β = 0.054, SE = 0.456, P =.905 |
| APOE x Biomarker | β = –0.506, SE = 0.429, P =.239 | β = –0.761, SE = 0.424, P =.073 | β = –1.819, SE = 0.460, P<.001* | β = –0.580, SE = 0.514, P =.260 | β = –0.150, SE = 0.432, P =.729 | β = 0.085, SE = 0.437, P =.846 | |
| Animal Fluency N = 1583 | Ethnicity x Biomarker | β = 0.570, SE = 0.248, P =.022 | β = 0.767, SE = 0.278, P =.006* | β = 0.630, SE = 0.287, P =.028 | β = 0.455, SE = 0.256, P =.076 | β = 0.373, SE = 0.247, P =.130 | β = 0.296, SE = 0.225, P =.188 |
| APOE x Biomarker | β = –0.340, SE = 0.232, P =.142 | β = –0.170, SE = 0.231, P =.461 | β = –0.268, SE = 0.238, P =.260 | β = –0.052, SE = 0.252, P =.835 | β = 0.183, SE = 0.235, P =.436 | β = 0.212, SE = 0.227, P =.352 | |
| Trails A N = 1783 | Ethnicity x Biomarker | β = ‐0.577, SE = 1.332, P =.665 | β = 1.906, SE = 1.478, P =.197 | β = 0.460, SE = 1.481, P =.756 | β = 0.200, SE = 1.388, P =.885 | β = 2.826, SE = 1.320, P =.032 | β = 0.806, SE = 1.251, P =.520 |
| APOE x Biomarker | β = 1.351, SE = 1.339, P =.313 | β = ‐2.310, SE = 1.396, P = .098 | β = –0.853, SE = 1.390, P =.540 | β = 1.090, SE = 1.414, P =.441 | β = –0.411, SE = 1.358, P =.762 | β = –0.862, SE = 1.296, P =.506 | |
| Trails B N = 1608 | Ethnicity x Biomarker | β = –5.858, SE = 3.314, P =.077 | β = –6.978, SE = 3.727, P =.061 | β = –8.613, SE = 3.710, P =.020 | β = –3.370, SE = 3.585, P =.347 | β = 0.521, SE = 3.330, P =.876 | β = –3.832, SE = 3.166, P =.226 |
| APOE x Biomarker | β = 3.380, SE = 3.371, P =.316 | β = 5.166, SE = 3.570, P =.148 | β = 3.321, SE = 3.508, P =.344 | β = 3.030, SE = 3.683, P =.411 | β = –3.882, SE = 3.435, P =.259 | β = 4.317, SE = 3.269,P =.187 |
Note: Interaction terms displayed for separate linear mixed‐effects models on cognitive tests with additional covariates for age, sex, ethnicity, APOE ε4 status, education, site, clinical diagnostic group, and serum biomarkers.
Abbreviations: APOE, apolipoprotein E; BMI, body mass index; CDR, Clinical Dementia Rating scale; GFAP, glial fibrillary acidic protein; MMSE, Mini‐Mental State Examination; NfL, neurofilament light; sCD14, soluble cluster of differentiation 14; SE, standard error; Trails, Trail Making Test; t‐tau, total tau; UCHL1, ubiquitin carboxyl‐terminal hydrolase L1; YKL‐40, chitinase‐3‐like protein 1; WMS LM, Weschler Memory Scale Logical Memory.
P<.008 level of significance after Bonferroni correction for six biomarkers
Within the matched Hispanic and non‐Hispanic White sample, NfL had an interaction with ethnicity for learning (Table S8 in supporting information).
4. DISCUSSION
The present study examined six blood‐based biomarkers that reflect diverse biological pathways implicated in ADRDs, within a bi‐ethnic cohort of Hispanic and non‐Hispanic White older adults. Our results validate the findings from previous studies reporting that blood‐derived levels of t‐tau and NfL have discriminability for dementia 4 , 5 , 6 , 7 and further extend the results to a bi‐ethnic cohort of Hispanics and non‐Hispanic Whites. Moreover, we identified less established blood‐based biomarkers, GFAP and YKL‐40, that differentiated between clinical diagnostic groups. With inclusion of all biomarkers in a single model, t‐tau, NfL, and GFAP remained significant, suggesting unique contributions from each biomarker. Higher levels of serum NfL, GFAP, and YKL‐40 were associated with poorer cognitive performance. 28 For learning, memory, and semantic fluency, ethnicity had significant interactions with biomarker values with weaker associations in Hispanics relative to non‐Hispanic Whites. Ethnic differences were generally no longer significant within a demographically matched subset of Hispanic and non‐Hispanic White participants.
In our study, t‐tau was significantly associated with diagnostic group. T‐tau levels in the blood are presumed to be derived from brain clearance and are considered to reflect neuronal injury. 4 , 29 In a prior meta‐analysis, plasma t‐tau had a large effect size for discriminating between AD and cognitively unimpaired groups. 7 While the correlation between plasma and CSF t‐tau levels are low, data from the Framingham Heart and Memento Studies indicated that plasma t‐tau is equally robust as CSF for predicting conversion to dementia. 4 Our findings add to the growing literature demonstrating the efficacy of blood‐derived t‐tau for dementia detection and further broaden the findings to a bi‐ethnic cohort.
Serum NfL levels in our study were associated with diagnostic group and poorer cognition on measures of global cognition, semantic fluency, attention/processing speed, and executive function. NfL is an intermediate filament protein found in the axons of myelinated neurons and is considered a marker of axonal/neuronal damage. 30 In contrast to t‐tau, CSF and blood‐derived levels of NfL are significantly correlated. 5 , 6 , 30 Elevations in NfL within the CSF and blood have been associated with clinical diagnosis, amyloid positron emission tomography (PET) positivity, cerebral atrophy, and cognitive decline. 5 , 6 , 30 Prior research has indicated that individuals with MCI who displayed more pronounced increases in NFL experienced accelerated cognitive decline, suggesting that NfL may dynamically change across disease stages. 6
Serum GFAP levels were strongly associated with diagnostic group, global cognition, learning, and memory. GFAP is an intermediate filament that forms the cytoskeleton of mature astrocytes. 31 In the presence of plaques and neurofibrillary tangles, astrocytes undergo morphological changes, transforming into a reactive state with greater expression of GFAP. 32 Elevations in CSF GFAP levels have been reported in individuals with ADRD relative to cognitively unimpaired groups. 33 In addition, a small study found that serum GFAP had high sensitivity and specificity for distinguishing between AD and cognitively unimpaired groups. 34 Enhanced knowledge of this pathophysiological process, as well as the ability to monitor and track levels in vivo, has the potential to enhance the development of dementia treatments targeting neuroinflammatory mechanisms.
The potential role of neuroinflammation in dementia was further highlighted by the associations of serum YKL‐40 with clinical diagnostic group and memory performance. YKL‐40 is considered a marker of microglia‐ and astroglia‐derived neuroinflammation. 13 Elevations in CSF and plasma levels of YKL‐40 have been associated with AD and other neurodegenerative conditions in previous studies. 35 , 36 These results, combined with the current findings, suggest that YKL‐40 levels may be a marker of cognitive impairment and further implicate glial injury underlying neurodegenerative processes.
Of the six biomarkers examined, two, sCD14 and UCHL1, were not associated with diagnostic group or cognition. sCD14 is a surface myeloid glycoprotein, expressed in microglial cells in the central nervous system, 37 as well as monocytes and neutrophils in the peripheral nervous system. 38 It remains unclear if sCD14 derived from the central nervous system is detected in blood. UCHL1 is a de‐ubiquitinating enzyme involved in proteasomal degradation including the breakdown of the amyloid precursor protein. 15 Within the CSF, individuals with AD have been found to have higher UCHL1 levels relative to individuals without cognitive impairment. 39 Further research is needed to evaluate if UCHL1 levels in blood correlate with CSF values.
One of the strengths of our study was inclusion of a bi‐ethnic Hispanic and non‐Hispanic White cohort. The majority of the findings were consistent across ethnic groups. However, the associations of some biomarkers with cognitive outcomes displayed significant interactions with weaker effects observed in Hispanics relative to non‐Hispanic Whites. It is important to note that our study lacked validated biomarker confirmation of neurodegenerative disease pathology such as amyloid beta and tau derived from CSF or PET imaging. 24 As clinical diagnoses were assigned from consensus reviews of clinical data, which may be confounded by factors such as culture, education, and literacy, 40 neuropathological burden may have differed across ethnic groups assigned to the same diagnostic category. Moreover, ethnic interactions, with the exception of NfL for learning, were no longer significant when examining a subsample of Hispanics and non‐Hispanic Whites matched across the factors of age, sex, education, APOE ε4 status, and clinical diagnostic group. The results highlight the potential influence of ethnic disparities across environmental and biological factors, including education, access to care, and burden of comorbidities, on ADRD biomarker values. 10 , 41 , 42 Ethnic and/or racial differences in ADRD biomarkers have been reported in other studies. For example, older Black adults have been reported to display lower CSF total and phosphorylated tau levels than non‐Hispanic White adults. 43 Additionally, a prior study using the TARCC cohort reported that Mexican Americans displayed a different multiplex biomarker protein signature for AD than non‐Hispanic Whites with a greater extent of proteins reflecting metabolic perturbations. 44 Therefore, additional research is necessary to determine the interplay of environmental, behavioral, and biological factors that may contribute to variances in ADRD biofluid marker values across diverse ethnic and racial groups. 10 , 41 , 42
While our study had many strengths, including a large bi‐ethnic cohort, several limitations need to be considered. First, the distribution of Hispanic and White non‐Hispanic participants varied across core demographics as well as diagnostic groups. Ethnic group interactions were generally no longer significant when examined in a subset of demographically matched Hispanic and non‐Hispanic White participants. A limitation of our study is the lack of available data to explore contextual and individual factors, such as household income, occupation, exposure to stress, and genetics, which may contribute to variances in biomarker values across and within diverse ethnoracial groups. 10 , 41 , 42 Another limitation is that our study used a retrospective cohort with data collection across several institutions and many years. As such, differences in specimen collection and timing likely exist, which may be contributing to the variability in some markers. 2 Finally, our study does not have established PET or CSF biomarkers for AD, 24 precluding our ability to confirm clinical diagnoses. Importantly, etiological contributions to MCI and dementia may have differed across the ethnic groups, contributing to divergence in some findings.
In summary, we identified four serum biomarkers, t‐tau, NfL, GFAP, and YKL‐40, that discriminated among cognitively unimpaired, MCI, and dementia groups within a bi‐ethnic cohort of Hispanic and non‐Hispanic White older adults. Our results add to the growing literature indicating that blood‐based biomarkers may be viable tools for detecting cognitive impairment and neurodegenerative conditions—at least at the population‐level. 4 , 5 , 7 , 45 Moreover, our study reported interactions between ethnicity and biomarker levels for some cognitive outcomes, which were generally no longer significant within a demographically matched subsample of Hispanic and non‐Hispanic White participants. Future research is needed to determine the interplay of multidimensional sociocultural, environmental, and biological factors that may contribute to ethnic differences in ADRD biofluid marker values. 10 , 41 , 42
Supporting information
Supporting Information
Supporting Information
Supporting Information
ACKNOWLEDGMENTS
This work was made possible by the Texas Alzheimer's Research Consortium (TARCC) funded by the state of Texas through the Texas Council on Alzheimer's Disease and Related Disorders, a TARCC pilot grant (2018‐28‐81‐JI), National Institute of Aging grants AG054076, AG063507, and AG059421, and National Institute of Neurological Disorders and Stroke NS100605.
Gonzales MM, Short MI, Satizabal CL, et al. Blood biomarkers for dementia in Hispanic and non‐Hispanic White adults. Alzheimer's Dement. 2021;7:e12164. 10.1002/trc2.12164
Contributor Information
Mitzi M. Gonzales, Email: GonzalesM20@uthscsa.edu.
Habil Zare, Email: zare@uthscsa.edu.
REFERENCES
- 1. Amjad H, Roth DL, Sheehan OC, Lyketsos CG, Wolff JL, Samus QM, Underdiagnosis of dementia: an observational study of patterns in diagnosis and awareness in US older adults. J Gen Intern Med. 2018;33:1131‐1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hampel H, O'Bryant SE, Molinuevo JL, et al. Blood‐based biomarkers for Alzheimer disease: mapping the road to the clinic. Nature Rev Neurol. 2018:14:639‐652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rissin DM, Kan CW, Campbell TG, et al. Single‐molecule enzyme‐linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat biotechnol. 2010;28:595‐599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pase MP, Beiser AS, Himali JJ, et al. Assessment of plasma total tau level as a predictive biomarker for dementia and related endophenotypes. JAMA Neurol. 2019;76:598‐606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mattsson N, Andreasson U, Zetterberg H, Blennow K, Association of plasma neurofilament light with neurodegeneration in patients with Alzheimer disease. JAMA Neurol. 2017;74:557‐566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mattsson N, Cullen NC, Andreasson U, Zetterberg H, Blennow K, Association between longitudinal plasma neurofilament light and neurodegeneration in patients with Alzheimer disease. JAMA Neurol. 2019;76:791‐799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Olsson B, Lautner R, Andreasson U, et al. CSF and blood biomarkers for the diagnosis of Alzheimer's disease: a systematic review and meta‐analysis. Lancet Neurol. 2016;15:673‐684. [DOI] [PubMed] [Google Scholar]
- 8. Reitz C, Mayeux R, Genetics of Alzheimer's disease in caribbean hispanic and african american populations. Biol Psychiatry. 2014;75:534‐541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fernández LE, Johnson NJ, In: Adames HY, Tazeau YN (Eds). Demographics and the Epidemiological Risk Factors for Dementia in Hispanic/Latino Populations Caring for Latinxs with Dementia in a Globalized World. New York: Springer; 2020:3‐16. [Google Scholar]
- 10. Babulal GM, Quiroz YT, Albensi BC, et al. Perspectives on ethnic and racial disparities in Alzheimer's disease and related dementias: update and areas of immediate need. Alzheimers Dement. 2019;15:292‐312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kunkle BW, Grenier‐Boley B, Sims R, et al. Genetic meta‐analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet. 2019;51:414‐430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao Y, Xin Y, Meng S, He Z, Hu W, Neurofilament light chain protein in neurodegenerative dementia: a systematic review and network meta‐analysis. Neurosci Biobehav Rev. 2019;102:123‐138. [DOI] [PubMed] [Google Scholar]
- 13. Groblewska M, Mroczko B, YKL‐40 as a potential biomarker and a possible target in therapeutic strategies of Alzheimer's disease. Curr Neuropharmacol. 2017;15:906‐917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Y, Walter S, Stagi M, et al. LPS receptor (CD14): a receptor for phagocytosis of Alzheimer's amyloid peptide. Brain. 2005;128:1778‐1789. [DOI] [PubMed] [Google Scholar]
- 15. Zhang M, Cai F, Zhang S, Zhang S, Song W, Overexpression of ubiquitin carboxyl‐terminal hydrolase L1 (UCHL1) delays Alzheimer's progression in vivo. Sci Rep. 2014;4:7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Korolainen MA, Auriola S, Nyman TA, Alafuzoff I, Pirttilä T, Proteomic analysis of glial fibrillary acidic protein in Alzheimer's disease and aging brain. Neurobiol Dis. 2005;20:858‐870. [DOI] [PubMed] [Google Scholar]
- 17. Waring S, O'Bryant SE, Reisch JS, Diaz‐Arrastia R, Knebl J, Doody R, The Texas Alzheimer's Research Consortium longitudinal research cohort: study design and baseline characteristics. Texas Public Health J. 2008;60:9‐13. [Google Scholar]
- 18. Folstein MF, Folstein SE, McHugh PR, “Mini‐mental state”: a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189‐198. [DOI] [PubMed] [Google Scholar]
- 19. Tulsky D, Zhu J, Ledbetter M, WAIS‐III and WMS‐III Technical Manual. New York: Harcourt Brace & Company; 1997. [Google Scholar]
- 20. Reitan RM, Validity of the trail making test as an indicator of organic brain damage. Percept Mot Ski. 1958;8:271‐276. [Google Scholar]
- 21. Tombaugh TN, Kozak J, Rees L, Normative data stratified by age and education for two measures of verbal fluency: fAS and animal naming. Arch Clin Neuropsy. 1999;14:167‐177. [PubMed] [Google Scholar]
- 22. Doody RS, Dunn JK, Huang E, Azher S, Kataki M, A method for estimating duration of illness in Alzheimer's disease. Dement Geriatr Cogn Disord. 2004;17:1‐4. [DOI] [PubMed] [Google Scholar]
- 23. McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM, Clinical diagnosis of Alzheimer's disease: report of the NINCDS‐ADRDA Work Group* under the auspices of Department of Health and Human Services Task Force on Alzheimer's disease. Neurology. 1984;34:939‐944. [DOI] [PubMed] [Google Scholar]
- 24. Jack CR Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, et al. NIA‐AA research framework: toward a biological definition of Alzheimer's disease. Alzheimers Dement. 2018;14:535‐562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Petersen RC, Mild Cognitive Impairment: Aging to Alzheimer's Disease: Oxford University Press; 2003. [Google Scholar]
- 26. O'Bryant SE, Gupta V, Henriksen K, et al. Guidelines for the standardization of preanalytic variables for blood‐based biomarker studies in Alzheimer's disease research. Alzheimers Dement. 2015;11:549‐560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ahmad S, del Campo Milan M, Hansson O, et al. CDH6 and HAGH protein levels in plasma associate with Alzheimer's disease in APOE ε4 carriers. Sci Rep. 2020;10:1‐13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Papp KV, Rentz DM, Orlovsky I, Sperling RA, Mormino EC, Optimizing the preclinical Alzheimer's cognitive composite with semantic processing: the PACC5. Alzheimers Dement. 2017;3:668‐677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Randall J, Mörtberg E, Provuncher GK, et al. Tau proteins in serum predict neurological outcome after hypoxic brain injury from cardiac arrest: results of a pilot study. Resuscitation. 2013;84:351‐356. [DOI] [PubMed] [Google Scholar]
- 30. Jin M, Cao L, Dai Y‐p, Role of neurofilament light chain as a potential biomarker for Alzheimer's disease: a correlative meta‐analysis. Front Aging Neurosci. 2019;11:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eng LF, Ghirnikar RS, Lee YL, Glial fibrillary acidic protein: gFAP‐thirty‐one years (1969‐2000). Neurochem Res. 2000;25:1439‐1451. [DOI] [PubMed] [Google Scholar]
- 32. Pekny M, Nilsson M, Astrocyte activation and reactive gliosis. Glia. 2005;50:427‐434. [DOI] [PubMed] [Google Scholar]
- 33. Ishiki A, Kamada M, Kawamura Y, et al. Glial fibrillar acidic protein in the cerebrospinal fluid of Alzheimer's disease, dementia with Lewy bodies, and frontotemporal lobar degeneration. J Neurochem. 2016;136:258‐261. [DOI] [PubMed] [Google Scholar]
- 34. Oeckl P, Halbgebauer S, Anderl‐Straub S, et al. Glial fibrillary acidic protein in serum is increased in Alzheimer's disease and correlates with cognitive impairment. J Alzheimers Dis. 2019;67:481‐488. [DOI] [PubMed] [Google Scholar]
- 35. Janelidze S, Hertze J, Zetterberg H, et al. Cerebrospinal fluid neurogranin and YKL‐40 as biomarkers of Alzheimer's disease. Ann Clin Transl Neurol. 2016;3:12‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baldacci F, Toschi N, Lista S, et al. Two‐level diagnostic classification using cerebrospinal fluid YKL‐40 in Alzheimer's disease. Alzheimers Dement. 2017;13:993‐1003. [DOI] [PubMed] [Google Scholar]
- 37. Heneka MT, Carson MJ, El Khoury J, et al. Neuroinflammation in Alzheimer's disease. Lancet Neurol. 2015;14:388‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pujol BF, Lucibello FC, Gehling UM, et al. Endothelial‐like cells derived from human CD14 positive monocytes. Differentiation. 2000;65:287‐300. [DOI] [PubMed] [Google Scholar]
- 39. Öhrfelt A, Johansson P, Wallin A, et al. Increased cerebrospinal fluid levels of ubiquitin carboxyl‐terminal hydrolase L1 in patients with Alzheimer's disease. Dement Geriatr Cogn Disord Extra. 2016;6:283‐294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Manly JM, Mayeux R, in: Anderson NB, Bulatao RA, Cohen B (Eds). Ethnic Differences in Dementia and Alzheimer's Disease, on Race, Critical Perspectives on Racial and Ethnic Differences in Health in Late Life. Washington DC: National Academies Press; 2004:95‐142. [PubMed] [Google Scholar]
- 41. Quiñones AR, Kaye J, Allore HG, Botoseneanu A, Thielke SM, An agenda for addressing multimorbidity and racial and ethnic disparities in Alzheimer's disease and related dementia. Am J Alzheimers Dis Other Dement. 2020;35:1533317520960874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barnes LL, Biomarkers for Alzheimer dementia in diverse racial and ethnic minorities—a public health priority. JAMA Neurol. 2019;76:251‐253. [DOI] [PubMed] [Google Scholar]
- 43. Howell JC, Watts KD, Parker MW, et al. Race modifies the relationship between cognition and Alzheimer's disease cerebrospinal fluid biomarkers. Alzheimers Res Ther. 2017;9:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O'Bryant SE, Xiao G, Edwards M, et al. Biomarkers of Alzheimer's disease among Mexican Americans. J Azheimers Dis. 2013;34:841‐849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blennow K, A review of fluid biomarkers for Alzheimer's disease: moving from CSF to blood. Neurol Ther. 2017;6:15‐124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information


