Abstract
Genetic screens provide a mechanism to identify genes involved with different cellular and organismal processes. Using a Flp/FRT screen in the Drosophila eye we identified mutations that result in alterations and de-regulation of cell growth and division. From this screen a group of undergraduate researchers part of the Fly-CURE consortium mapped and characterized a new allele of the gene Hippo, HpoN.1.2.
Figure 1. The Dark82, HpoN.1.2 mosaic eye results in dramatic tissue overgrowth and pupal lethality.
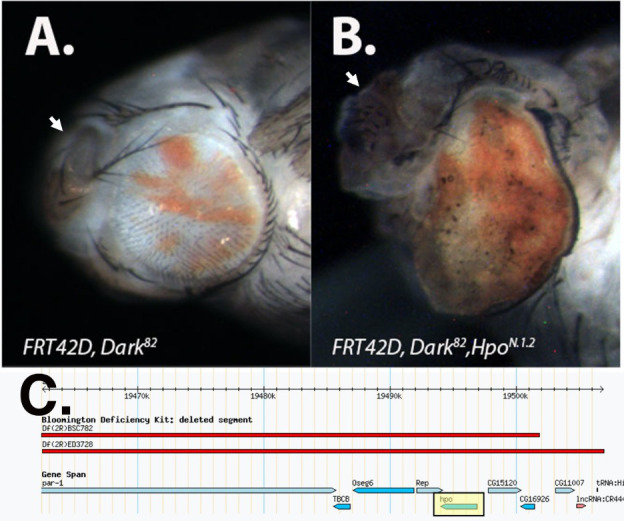
A.) Lateral view of FRT42D, Dark82 mosaic control eye and B.) FRT42D, Dark82,HpoN.1.2 mosaic mutant pupal eyes at the same magnification (40x). In both genotypes, mutant tissue displays red pigmentation (w+mC). Arrows on A and B point to antennae, eyes are oriented with anterior to the left and dorsal to the top. C.) Genomic region of chromosome 2R in which mutant N.1.2 failed to complement (2R:19,462,450..19,506,861). The N.1.2 mutation mapped to the hpo gene locus found in this region (yellow highlight). Image adapted from flybase.org (Gramates 2017).
Description
In order to identify conditional regulators of cell growth and tumorigenesis, an EMS-based genetic screen was conducted in Drosophila melanogaster utilizing the Flp/FRT system in an apoptotic null background (Akdemir et al., 2006). A fly line possessing the FRT42D, Dark82 chromosome and containing a mini-white cassette (w+mC) was used for EMS mutagenesis. Subsequent matings to FRT42D; Ey-Flp flies facilitated phenotypic screening in eye tissue. Phenotypes observed in the screen included alterations in mosaicism (red > white pigmentation pattern), eye and/or antennal overgrowth, defects in patterning, and pupal lethality (Kagey et al. 2012). One of the pupal lethal mutants identified in this screen, N.1.2, is discussed here. Genetic crosses between FRT42D, Dark82, N.1.2 X FRT42D; Ey-Flp resulted in near complete pupal lethality (~ 90%) due to dramatic tissue overgrowth of the eye, antennae, and inter-ocular space (Figure 1B compared to 1A). Due to the pupal lethality associated with the eye tissue overgrowth, flies were dissected from late pupal stages in order to visualize the phenotype. Imaging following dissection shows that the mutant eyes are comprised of mostly mutant (pigmented) tissue that protrudes from the eye cavity creating tissue folding (Figure 1B). Control FRT42D, Dark82 flies exhibited a balance in the red:white ratio and no eye overgrowth (Figure 1A). For direct comparison to N.1.2, the FRT42D, Dark82 mosaic eye was also imaged at the late pupal stage. Eyes were imaged under 70% ethanol on an AM Scope digital camera at 40x using a LED light ring.
In order to map the genetic location of the N.1.2 mutation, we conducted complementation tests with deficiency strains, and looked for lethality, consistent with the phenotype of homozygous mutant flies of the N.1.2 stock. Deficiency crosses were conducted by mating FRT42D, Dark82, N.1.2/CyO virgin females to males from each of 86 deficiency stocks with deletions distal to the FRT42D site on the right arm of chromosome two. All stocks used for mapping were part of the Bloomington Deficiency 2R Kit (Cook et. al., 2012). The genetic mapping was conducted by undergraduate research students from Nevada State College, Ohio Northern University, Ohio Wesleyan University, and the University of Detroit Mercy as part of the Fly-CURE consortium (Vrailas-Mortimer et al. 2021; Bieser et al. 2019; Stamm et al. 2019). The N.1.2 mutant failed to complement two of these deficiencies, Df(2R)BSC782 (2R: 19,451,027..19,501,804) and Df(2R)ED3728 (2R:19,462,450..19,726,747). These two deficiencies overlapped from cytological bands 56D10-56D14 (2R:19,462,450..19,506,861) and included the hippo (hpo) gene locus and nine other protein coding genes (2R:19,492,996..19,496,856) (Figure 1C). Subsequent crosses to individual lethal alleles of hpo: hpoMGH1and hpoKS240, failed to complement N.1.2, confirming that the N.1.2 mutant is a newly isolated hpo allele, hpoN.1.2 (Harvey et al., 2003; Udan et al., 2003).
The hpo gene functions as a negative regulator of cell growth and is a part of a highly conserved signaling pathway first characterized in Drosophila (reviewed in Harvey and Hariharan 2012). Overall, research has shown that the hippo pathway is critical for regulating organ size through regulation of apoptosis, cell survival, cell polarity, and cell proliferation (Harvey et.al., 2003; Jia et.al. 2003; Pantalacci et. al., 2003; Udan et. al., 2003; reviewed in Yu et. al, 2015). Consistent with the eye phenotypes observed in the N.1.2 mutation, mutations in hpo are known to result in striking overgrowth phenotypes in a variety of tissue types from flies to humans (Pan 2010, Pluoffe et al., 2015; Yu et. al., 2015). Identification of this novel hpoN.1.2 allele will support further research into the molecular mechanisms by which multicellularity is regulated and restricted by this critical signaling pathway.
Reagents
FRT42D, Dark82/CyO (Akdemir et al., 2006)
FRT42D, Dark82, hpoN.1.2/CyO (this manuscript)
FRT42D; Ey-Flp (BDSC 8211)
Bloomington Drosophila Stock Center 2R Deficiency Kit(Cook et al., 2012)
w1118; Df(2R)BSC782/SM6a (BDSC 27354)
w1118; Df(2R)ED3728/SM6a (BDSC 9067)
yw; FRT42D, hpoKS240/CyO (BDSC 25085)
hpoMGH1 (Harvey et al. 2003)
Acknowledgments
Acknowledgments
Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Funding
Fly-CURE (K. Bieser, J. Kagey) National Science Foundation IUSE Award (NSF 2021146). A. Murry is funded by the NIH ReBUILDetroit (TL4GM118983)
References
- Akdemir F, Farkas R, Chen P, Juhasz G, Medved'ová L, Sass M, Wang L, Wang X, Chittaranjan S, Gorski SM, Rodriguez A, Abrams JM. Autophagy occurs upstream or parallel to the apoptosome during histolytic cell death. Development. 2006 Mar 15;133(8):1457–1465. doi: 10.1242/dev.02332. [DOI] [PubMed] [Google Scholar]
- Bieser K, Sanford J, Saville K, Arreola K, Ayres Z, Basulto D, Benito S, Breen C, Brix J, Brown N, Burton K, Chadwick T, Chen M, Chu K, Corbett B, Dill Z, Faughender M, Hickey A, Julia J, Kelty S, Jobs B, Krason B, Lam B, McCullough C, McEwen B, McKenzie J, McQuinn K, Moritz C, Myers K, Naugle E, Nutter A, O'Conke D, O'Grondik M, Patel K, Rudowski S, Sberna E, Stall G, Steiner T, Tanriverdi E, Torres Patarroyo N, Traster V, Tsai L, Valenti A, Villegas M, Voors S, Watson K, Wright M, Kagey J. Genetic mapping of shnE.3.2 in Drosophila melanogaster. MicroPubl Biol. 2019 Jun 01;2019 doi: 10.17912/micropub.biology.000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook RK, Christensen SJ, Deal JA, Coburn RA, Deal ME, Gresens JM, Kaufman TC, Cook KR. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 2012;13(3):R21–R21. doi: 10.1186/gb-2012-13-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates LS, Marygold SJ, Santos GD, Urbano JM, Antonazzo G, Matthews BB, Rey AJ, Tabone CJ, Crosby MA, Emmert DB, Falls K, Goodman JL, Hu Y, Ponting L, Schroeder AJ, Strelets VB, Thurmond J, Zhou P, the FlyBase Consortium. FlyBase at 25: looking to the future. Nucleic Acids Res. 2016 Oct 30;45(D1):D663–D671. doi: 10.1093/nar/gkw1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003 Aug 22;114(4):457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Hariharan IK. The hippo pathway. Cold Spring Harb Perspect Biol. 2012 Aug 01;4(8):a011288–a011288. doi: 10.1101/cshperspect.a011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003 Oct 15;17(20):2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey JD, Brown JA, Moberg KH. Regulation of Yorkie activity in Drosophila imaginal discs by the Hedgehog receptor gene patched. Mech Dev. 2012 Jun 15;129(9-12):339–349. doi: 10.1016/j.mod.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010 Oct 19;19(4):491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Léopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003 Sep 21;5(10):921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- Plouffe SW, Hong AW, Guan KL. Disease implications of the Hippo/YAP pathway. Trends Mol Med. 2015 Feb 18;21(4):212–222. doi: 10.1016/j.molmed.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm J, Joshi G, Anderson MA, Bussing K, Houchin C, Elinsky A, Flyte J, Husseini N, Jarosz D, Johnson C, Johnson A, Jones C, Kooner T, Myhre D, Rafaill T, Sayed S, Swan K, Toma J, Kagey J. Genetic mapping of EgfrL.3.1 in Drosophila melanogaster. MicroPubl Biol. 2019 Apr 26;2019 doi: 10.17912/micropub.biology.000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003 Sep 21;5(10):914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Vrailas-Mortimer AD, Aggarwal N, Ahmed NN, Alberts IM, Alhawasli M, Aljerdi IA, Allen BM, Alnajar AM, Anderson MA, Armstong R, Avery CC, Avila EJ, Baker TN, Basardeh S, Bates NA, Beidas FN, Bosler AC, Brewer DM, Buenaventura RS, Burrell NJ, Cabrera-Lopez AP, Cervantes-Gonzalez AB, Cezar RP, Coronel J, Croslyn C, Damery KR, Diaz-Alavez L, Dixit NP, Duarte DL, Emke AR, English K, Eshun AA, Esterly SR, Estrada AJ, Feng M, Freund MM, Garcia N, Ghotra CS, Ghyasi H, Hale CS, Hulsman L, Jamerson L, Jones AK, Kuczynski M, Lacey-Kennedy TN, Lee MJ, Mahjoub T, Mersinger MC, Muckerheide AD, Myers DW, Nielsen K, Nosowicz PJ, Nunez JA, Ortiz AC, Patel TT, Perry NN, Poser WSA, Puga DM, Quam C, Quintana-Lopez P, Rennerfeldt P, Reyes NM, Rines IG, Roberts C, Robinson DB, Rossa KM, Ruhlmann GJ, Schmidt J, Sherwood JR, Shonoda DH, Soellner H, Torrez JC, Velide M, Weinzapfel Z, Ward AC, Bieser KL, Merkle JA, Stamm JC, Tillett RL, Kagey JD. B.2.16 is a non-lethal modifier of the Dark82 mosaic eye phenotype in Drosophila melanogaster. MicroPubl Biol. 2021 Jan 18;2021 doi: 10.17912/micropub.biology.000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Guan KL. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell. 2015 Nov 01;163(4):811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]


