Abstract
Study Objectives
Sleep spindles, a defining feature of stage N2 sleep, are maximal at central electrodes and are found in the frequency range of the electroencephalogram (EEG) (sigma 11–16 Hz) that is known to be heritable. However, relatively little is known about the heritability of spindles. Two recent studies investigating the heritability of spindles reported moderate heritability, but with conflicting results depending on scalp location and spindle type. The present study aimed to definitively assess the heritability of sleep spindle characteristics.
Methods
We utilized the polysomnography data of 58 monozygotic and 40 dizygotic same-sex twin pairs to identify heritable characteristics of spindles at C3/C4 in stage N2 sleep including density, duration, peak-to-peak amplitude, and oscillation frequency. We implemented and tested a variety of spindle detection algorithms and used two complementary methods of estimating trait heritability.
Results
We found robust evidence to support strong heritability of spindles regardless of detector method (h2 > 0.8). However not all spindle characteristics were equally heritable, and each spindle detection method produced a different pattern of results.
Conclusions
The sleep spindle in stage N2 sleep is highly heritable, but the heritability differs for individual spindle characteristics and depends on the spindle detector used for analysis.
Keywords: spindle, heritability, spindle detection, genetics
Statement of Significance.
The sleep spindle, a defining feature of stage N2 sleep, is associated with the consolidation of learning and memory. Sleep spindle dysfunction has also been implicated in several psychiatric and neurological disorders including schizophrenia, autism, and dementia. Utilizing eight distinct automated spindle detector algorithms and two methods to quantify heritability in monozygotic and dizygotic twin pairs, we provide robust evidence that spindles are highly heritable, regardless of the detector used, but that not all spindle characteristics are equally heritable. As spindles could represent an important potential biomarker for impaired cognitive functioning and certain psychiatric disorders, the present results highlight the value of the assessment of sleep spindle characteristics.
Introduction
In the last 15 years, research has demonstrated that the regulation of sleep is genetically controlled and that several sleep disorders, including narcolepsy and restless leg syndrome, have a genetic basis [1]. Similarly, work on the genetic heritability of the sleep electroencephalogram (EEG) has revealed that some characteristics of the individual sleep EEG are also under genetic control. For example, both rodent [2] and twin studies in humans [3] have demonstrated that the delta, theta, alpha, and beta EEG frequencies generated from power spectral analysis are relatively stable and consistent, in addition to being highly heritable. Within these frequencies, the 1–14 Hz range has consistently been shown to have significant genetic variance [3], suggesting that certain NREM EEG events occurring within this range, including delta waves and sleep spindles, may be highly heritable.
Sleep spindles, a hallmark feature of stage N2 sleep, are characterized as transient EEG events with an amplitude that progressively increases and subsequently decreases. This activity, best quantified at central electrodes, occurs within the sigma frequency range (11–16 Hz). Research has shown that the density and duration of sleep spindles increase following a period of learning during waking [4], suggesting that spindles are associated with the consolidation of learning and memory. Moreover, spindle dysfunction has been implicated in several psychiatric and neurological disorders including schizophrenia, [5] autism [6], and dementia [7], suggesting that the presence of spindles is important for healthy brain function.
Power in the 8–16 Hz frequency range, within which spindle frequency falls, is known to be one of the most heritable characteristics of the sleep EEG [8]. Because of this, studies have now begun to examine the heritability of specific spindle characteristics, including spindle density (spindles per minute), duration, amplitude, and oscillation frequency. Utilizing several approaches to estimate heritability, Purcell et al. [9] found consistent evidence of the heritability of spindle density, spindle amplitude, and sigma power. They also showed that both fast spindles, defined as >13 Hz with a centroparietal focus, and slow spindles, defined as <13 Hz with a frontal focus, demonstrated strong heritability. In contrast, Adamczyk et al. [10] determined that although there was a strong genetic basis for all parameters of slow spindles, there was little to no evidence of a genetic basis for parameters of fast spindles using a novel method to automatically detect spindles within the sleep EEG, called the continuous wavelet transform. More recently, Rusterholz et al. [11] also investigated the heritability of the sleep spindle in adolescents. Using high-density EEG and two methods to estimate heritability, they determined that both fast and slow spindles in posterior regions of the brain had a genetic basis, while spindles generated in the anterior regions had an environmental basis. Taken together, although there does seem to be some evidence of the heritability of spindles, the characteristics of spindles that are most heritable remain to be established due to the significant variability in the methodological approaches used.
Due to the interest in spindles as a biological phenomenon, numerous methodological strategies have been developed for the automated detection of sleep spindles in EEG data. It has been shown, however, that there are important differences between the results of different spindle detection algorithms [12, 13]. While all algorithms are efficient and reproducible, the agreement between the different detectors is varied, and therefore produces different results, particularly for basic spindle metrics as spindle density and average spindle duration.
Similarly, methods used to estimate heritability are also varied, which can likewise lead to conflicting results. One of the most common methods to determine heritability is the analysis of twin zygosity differences in interclass correlations, however other methods have also been described. Our previous work has demonstrated that using alternative methods concomitantly provides a more comprehensive approach to estimating heritability than any one approach alone [14]. Therefore, the present study used complementary methods to estimate trait heritability of spindles, in addition to using a variety of spindle detection algorithms with two specific aims: (1) to assess the heritability of specific characteristics of spindles, and (2) to determine the robustness of spindle heritability, by using different but commonly used spindle detection methods, which have previously been shown to give different spindle detection results. Given that the 8–16 Hz range of the sleep EEG has shown strong heritability, we hypothesized that sleep spindles will show moderate to strong heritability. However, given that the varied spindle detection methods detect different subsets of spindles with slightly different characteristics including amplitude, oscillation frequency, and duration, and that these individual characteristics may be more or less heritable, we hypothesized that individual spindle characteristics are differentially heritable.
Methods
We performed a secondary analysis of data from a sleep deprivation research study of monozygotic (MZ) and dizygotic (DZ) same-sex adult twin pairs designed to determine the heritability of sleep homeostasis [14]. Methods of participant recruitment and the sleep deprivation protocol have been previously described [14, 15]. The original study was approved by the Institutional Review Boards at the University of Pennsylvania and the subcontracted sites, the University of Chicago and Virginia Commonwealth University. The subcontracted sites were the source of the twin registries used for recruitment. Written informed consent was obtained from each participant.
Participants
A total of 58 MZ same-sex twin pairs and 40 DZ same-sex twin pairs were recruited as part of the larger study [14]. Two twin pairs were excluded due to irregularity in recording parameters or poor recording quality. Twin zygosity was established by a DNA-based polymerase chain reaction (PCR) analysis of peripheral blood using 12 highly polymorphic short-tandem repeat loci and Amelogenin [16]. All participants were between the ages of 18 and 55 years and had a same-sex twin who was also willing and eligible to participate. Exclusion criteria included the following: an apnea-hypopnea index or periodic limb movement index ≥5 events/h on a full night diagnostic polysomnogram (PSG) that occurred at least 2 weeks prior to start of the study; the presence of depression as indicated by a score >16 on the Center for Epidemiological Studies Depression Scale [17] (CES-D); irregular work hours or shift work; consumption of two or more alcoholic drinks per day as assessed with the CAGE questionnaire [18, 19]; regular use of sedative/hypnotic medications; initiation of any new medication in the previous 3 months; a medical or neurologic condition that would prevent travel; previous diagnosis of obstructive sleep apnea or any other sleep disorder; travel across a different time zone in the previous 6 weeks; abnormal blood tests, i.e. blood hemoglobin <11.3 g/dL, serum bilirubin >2.0 mg/dL, serum creatinine >3 mg/dL; or a positive urine toxicology screening. Individuals were excluded if they were taking the following medications: methylphenidate, modafinil, antidepressants, or beta-blockers. They were also excluded if they were taking medications for pain, including aspirin, nonsteroidal anti-inflammatory drugs, and COX-2 inhibitors, more than three times per week.
Procedures
At least 2 weeks prior to the start of the protocol, participants performed an overnight polysomnogram (diagnostic PSG). For 1 week prior to the study, participants were instructed to maintain a regular sleep/wake schedule which was verified by sleep diary and wrist actigraphy. The actigraph recordings were manually scored with the aid of computer software. Non-compliance with maintaining a regular sleep/wake schedule was considered exclusionary. Participants were also instructed to refrain from smoking, drinking alcohol, or caffeinated beverages for 24 h prior to, and during the entirety of the laboratory stay.
Participants spent 4.5 consecutive days in the sleep laboratory, with electrodes for PSG recording applied on the first night. For the present study, the focus of analyses was restricted to the PSG on day 2 (baseline PSG). The first night served as an adaptation night, while the second night served as the baseline night. Participants were allowed to sleep ad libitum on days 1 and 2. During the days in the sleep laboratory, participants were not permitted to drink caffeinated beverages, use the telephone, read newspapers, or watch TV in order to avoid any extraneous environmental influences on their results. Individuals in each twin pair were studied on different days to prevent their social interaction. The twin who was studied first was instructed not to share his/her experience with the other twin until that person had completed testing. Female participants were studied during the non-menstruating phase of their menstrual cycle, determined by self-report.
Sleep EEG
The diagnostic PSG recorded the following signals: EEG (C3M2, C4M1, O1M2, O2M1), bilateral electrooculograms (EOG), chin muscle electromyogram (EMG), airflow (nasal pressure and oronasal thermistor), rib cage and abdominal movement, bilateral anterior tibialis electromyogram, electrocardiogram, pulse oximetry, and body position. For each successive night (adaptation, baseline), the montage included EEG, EOG, and EMG. The PSGs were scored for conventional sleep variables by the automated scoring system (YRT Limited, Winnipeg, Canada) that was validated in an independent multicenter study [20]. The automated scoring followed the 2012 AASM guidelines for scoring sleep and arousals [21]. A summary of baseline polysomnographic variables can be found in Supplementary Table 1.
Spindle detection and characterization
To identify the heritable characteristics of spindles, we utilized automated spindle detection algorithms to detect spindles in stage N2 sleep. To determine whether methodological differences in spindle detection would influence the heritability results, we tested and compared seven previously published and one proprietary spindle detectors. We used the established nomenclature [12, 13] of these previously validated spindle detectors: a2 [5], a3 [22], a4 [23], a5 [24], a7 [12], a8 [25], a9 [26], and a10 [27]. A summary of these algorithms can be found in the Supplementary Text.
First, spindle density was calculated at the individual level and was defined as the average number of spindles per minute. Next, three spindle characteristics were computed from each detected spindle event: spindle duration, spindle amplitude, and spindle frequency. Spindle duration is the average length (in s) of each spindle event per subject. For all spindle detectors, spindles were limited to 0.3 to 2.5 s in duration. Detected spindles outside this duration range were discarded from the analysis. Spindle amplitude is the average maximum peak-to-peak amplitude (µV) of each spindle per subject. The oscillation frequency is the average of the peak frequencies in the sigma range for each spindle event per subject [12]. In all cases, spindles are taken as the average across both C3/A2 and C4/A1 when artifact-free.
In addition to the spindle detectors, we also compared average total power between 0.5 and 30 Hz (S0), average power in sigma (S1), and relative power in sigma (S2) from power spectral analysis (PSA). S2 is the ratio of S1 over the average power in the broad band (4.5 to 30 Hz). The PSA was calculated using a window length of 2 s and a step of 1 s across the average of C3/A2 and C4/A1 in stage N2 for each subject. S0 was used as a non-specific measure of EEG power. S1 and S2 are estimates of spindling in the sigma range (11–16 Hz) and, although they are not based on individual spindles, they are simpler to compute than the spindle algorithms, and the sigma range was previously shown to be heritable [8].
Data standardization and transformation
Due to non-normal distributions of the spindle density results, all data were transformed using a Box–Cox transformation prior to the heritability analysis. The Box–Cox transformation, a non-linear parametric power transformation used to improve normality in non-negative data [28, 29], was applied to all data with the MZ and DZ twin pairs pooled together to keep measurements on the same scale when comparing between zygosities. The Box–Cox transformation selects the most appropriate transformation to normalize the data. The optimal tuning parameter was allowed to vary across spindle parameters to maximize normality on a case-by-case basis. The ICC and heritability calculations were then performed on the Box–Cox transformed results. Confirmation that normality was improved after the Box–Cox transformation was performed with a Shapiro–Wilk’s test.
Estimation of heritability
Two complementary methods were used to evaluate heritability: the classic approach based on zygosity differences in ICC [30], and the maximum likelihood estimation of model-specific covariance matrices [31].
Descriptive heritability estimates were obtained as h2 = 2(ICCMZ – ICCDZ) [30]. Initial descriptive assessments of shared common environment variance were determined using the formula: C2 = 2 × ICCDZ – ICCMZ. Analyses of covariance with fixed effects of age, gender, and age by gender interaction were used to obtain age and gender-adjusted differences in ICC.
Maximum likelihood estimation was used to determine variance components from mixed-effect multilevel models (MEMA-VCM) [31]. This approach produces the same covariance expectations as path analysis and structural equation modeling with the exception that variances are not restricted to be positive; thus providing an opportunity to identify poor model fit [32]. Age, gender, and age by gender interaction were included as fixed covariates.
Using these methods, we assessed the different models to discriminate between different patterns of genetic transmission. One model examined was the ACE model, which includes additive genetic effects (A), common environmental effects (C), and unique individual effects (E). Another model—the ADE model—includes additive genetic effects (A), dominance genetic effects (D), and unique individual effects (E). Alternative models included only C and E effects.
Following McArdle and Prescott [31], the ACE and ADE models were formulated by including additive genetic variance (σ2A), variance arising from dominance effects (σ2D), family-specific variance (σ2C), and unique variance (σ2A) into the model as random effects. These random effects are either shared or not shared between twin members and were weighted according to zygosity to produce the desired variance components of the MEMA-VCM models [31] using SAS Proc Mixed. Then, these parameters were re-estimated using the general-purpose nonlinear mixed model algorithm in Proc NLMIXED to obtain p values and standard errors for the desired functions of the various components. Although the parameter estimates are identical, Proc NLMIXED provides asymptotic standard errors for the proportions of phenotypic variance explained by A + D, A alone, and D alone as well as p values for testing null hypotheses that each proportion is equal to zero assuming the ADE model is true.
Factor analysis of spindle detector algorithms
Multivariable analyses were performed to enhance the interpretability of the heritability results. The aim of these analyses was to evaluate the degree of redundancy in the set of spindle detector algorithms and the degree to which heritability findings relate to the underlying dimensionality of the datasets. To this end, multivariate factor analysis [33] was applied to each phenotype (density, duration, amplitude, and frequency). First, eigenvalues were extracted from each of the four covariance matrices pooling monozygotic and dizygotic twin pairs using principal components analysis. The cumulative percentage of total explained variance as a function of the number of eigenvalues was evaluated in a Scree plot [34] and the number of underlying dimensions was identified at the point where the slope of the Scree plot approached zero. The number of factors was set to this number of dimensions. The factor loading matrix was obtained through principal factor analysis and rotated using the oblique Promax rotation to obtain a “simple” solution, one in which each observed phenotype loads highly on only one latent factor. Consequently, the latent factors were correlated. Analyses were performed using SAS Proc Factor.
Results
A total of 58 MZ and 40 DZ same-sex twin pairs completed the protocol (Table 1). Forty-four (75.9%) of the MZ twin pairs and 26 (65.0%) of the DZ twin pairs were female.
Table 1.
Mean ± SD of participant characteristics
| Measure | Total (N = 196) |
Monozygotic twins (MZ) (N = 116) |
Dizygotic twins (DZ) (N = 80) |
Mixed model* p-value |
|---|---|---|---|---|
| Male gender pairs | 28.6% (28/98) | 24.1% (14/58) | 35.0% (14/40) | 0.242† |
| Age (years) | 28.1 ± 7.2 | 29.1 ± 6.8 | 26.6 ± 7.6 | 0.100‡ |
| Height (cm) | 167.5 ± 11.3 | 165.9 ± 10.2 | 169.9 ± 12.4 | 0.096 |
| Weight (kg) | 69.3 ± 16.7 | 68.4 ± 14.8 | 70.6 ± 19.1 | 0.459 |
| BMI (kg/m2) | 24.2 ± 4.2 | 24.4 ± 4.5 | 23.9 ± 3.8 | 0.483 |
| CES-D total score | 14.9 ± 4.0 | 14.5 ± 3.7 | 15.4 ± 4.3 | 0.147 |
| ESS total score | 5.6 ± 3.2 | 5.3 ± 2.8 | 6.0 ± 3.6 | 0.237 |
| Global PSQI score | 3.7 ± 2.2 | 3.5 ± 2.0 | 3.8 ± 2.4 | 0.517 |
| AHI (events/h) | 1.8 ± 1.4 | 1.6 ± 1.4 | 2.0 ± 1.4 | 0.128 |
| PLMI with arousal (events/h) | 1.8 ± 5.2 | 2.3 ± 6.2 | 1.1 ± 3.2 | 0.105 |
Abbreviations: MZ, monozygotic; DZ, dizygotic; BMI, body mass index; CES-D, Center for Epidemiologic Studies Depression Scale; ESS, Epworth Sleepiness Scale; PSQI, Pittsburgh Sleep Quality Index; AHI, apnea-hypopnea index; PLMI, periodic limb movement index.
*Mixed model analyses of variance for continuous measures that accounted for correlations within family pairs but which allowed for variances and co-variances to differ between MZ and DZ were used to compare groups (MZ vs DZ). Covariance parameters were estimated using restricted maximum likelihood (REML) to reduce bias.
†Chi-square test based on the numbers of male and female pairs.
‡Student’s t-test based on pair specific ages.
Spindle characteristics vary by the detector
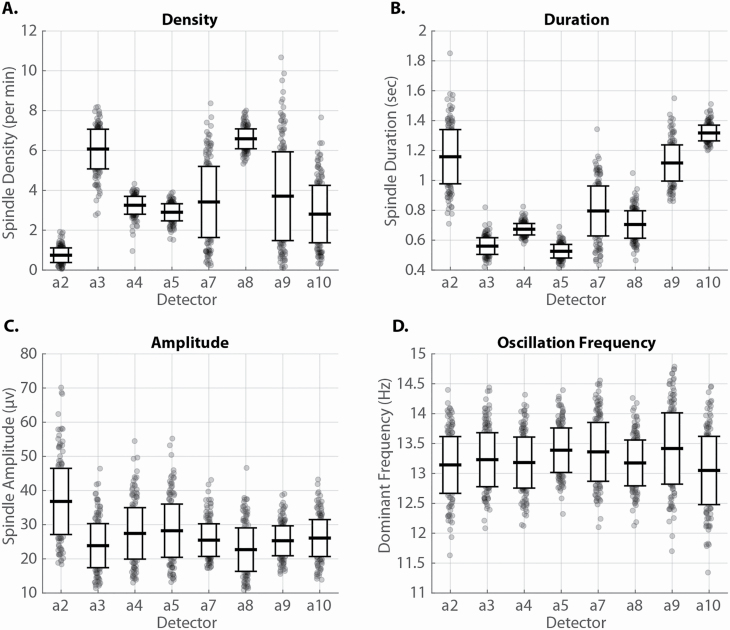
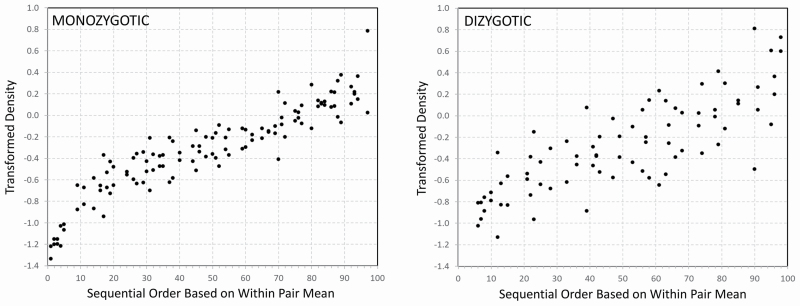
The spindle characteristic results from the eight spindle detection algorithms in stage N2 are presented in Figure 1. Overall, the estimates of spindle density in all subjects ranged from 0.74 to 6.59 per minute depending on the detection method used. Considerable variability in individual sleep density was present for any given spindle detector. For example, using the analysis of spindle detector a2, individual spindle density in stage N2 sleep for each MZ (upper panel) and DZ (lower panel) twin pair is shown in Figure 2. The panels reveal a substantial range of responses. A greater similarity of results within MZ twin pairs than within DZ twin pairs is visually apparent.
Figure 1.
Spindle characteristics in stage N2 sleep for each spindle detector: (A) spindle density, (B) spindle duration, (C) maximum peak to peak amplitude, and (D) peak oscillation frequency. Mean (middle horizontal line) and standard deviation (upper and lower horizontal lines) are shown. Each dot is one subject. Density is spindles/min per subject. Duration (s), amplitude (µV), and oscillation frequency (Hz) are averages for each subject.
Figure 2.
Individual spindle density (spindles/min) in stage N2 sleep from detector a2 for each monozygotic (MZ, upper panel) and dizygotic (DZ, lower panel) twin pair is plotted. For every twin pair, there are two values, with each dot representing one subject. In each panel, the pairs are ordered by the magnitude of spindle density (averaged over each pair) on the X-axis, with the twin pair with the lowest spindle density on the left and the twin pair with the highest spindle density on the right. The transformed ICC values, unadjusted, are plotted on the Y-axis. The panels reveal substantial differences in individual results. As is visually apparent, the MZ twin pairs cluster more closely than the DZ twin pairs, as the ICC reveals greater similarity within MZ twin pairs than within DZ twin pairs. 80.4% of the total variance in the MZ twins was due to variance between pairs whereas only 36.9% of the total variance in DZ twins was due to variance between pairs. ICC, intraclass correlation.
The greatest differences between detectors were found for estimates of spindle density and duration. In contrast, there was much less variability in the spindle oscillation frequency and peak-to-peak amplitude. The average of each characteristic is found in Supplementary Table 2. The distribution of oscillation frequency was found to be primarily unimodal (Supplementary Figure 1), suggesting that no clear distinction exists between faster and slower spindles at the C3/4 scalp position.
Analysis of genetic effects on spindle density
In preliminary descriptive analyses based on ICCs, the heritability estimates (h2) for spindle density ranged from 0.08 to 1.18 depending on the spindle detection algorithm used, with a mean heritability estimate of 0.74 ± 0.30 (Table 2; see Supplementary Table 3 for unadjusted ICC). Only one spindle detection algorithm, a4, showed a heritability estimate of less than 0.65, indicating that the heritability of spindle density is high. Values larger than 1 can occur using the classical approach and can sometimes suggest a dominant genetic transmission. Negative values of C2 = 2 × ICCDZ – ICCMZ suggest that shared common environment is not an important factor in explaining within-pair correlation, and, in those situations, the ADE model, that includes a dominant genetic factor rather than the ACE model which includes a shared common environment, is the preferred model of genetic transmission [35]. These issues were examined in more detail using the maximum likelihood approach described below.
Table 2.
Intraclass correlation coefficients (ICC*), classic heritability estimates (h2 †) and common environmental variance (CEV‡) for sigma power and spindle density from eight spindle detector algorithms in monozygotic (MZ) and dizygotic (DZ) twin pairs, adjusted§
| Characteristic | Variable | ICC_MZ | ICC_DZ | h2 | CEV |
|---|---|---|---|---|---|
| Total power | S0 | 0.59 | 0.51 | 0.16 | 0.43 |
| Average sigma | S1 | 0.76 | 0.58 | 0.37 | 0.39 |
| Relative sigma | S2 | 0.81 | 0.43 | 0.77 | 0.04 |
| Detector | |||||
| Density | a2 | 0.78 | 0.37 | 0.83 | -0.05 |
| a3 | 0.81 | 0.44 | 0.72 | 0.08 | |
| a4 | 0.57 | 0.53 | 0.08 | 0.49 | |
| a5 | 0.66 | 0.30 | 0.71 | -0.05 | |
| a7 | 0.77 | 0.38 | 0.78 | -0.01 | |
| a8 | 0.75 | 0.16 | 1.18 | -0.43 | |
| a9 | 0.77 | 0.40 | 0.75 | 0.02 | |
| a10 | 0.76 | 0.34 | 0.83 | -0.08 |
Heritability is calculated using Falconer’s method; values closer to one indicate a pattern expected where variance in a trait is genetically controlled. Negative CEV values suggest that common environmental effects are likely not very important and that genetic dominance effects may be present.
*ICC = σ 2B/[σ 2W + σ 2B].
† h 2, heritability = 2(ICCMZ – ICCDZ).
‡ CEV = (2 × ICCDZ) – ICCMZ.
§Adjusted for age, sex, and age by sex interaction.
The proportion of variance explained by common (shared) environment factors can be estimated using the equation: 2 (ICCDZ) – ICCMZ. For spindle density, this average value among all detection algorithms was equal to –0.004 ± 0.25 and was negative for five of the eight algorithms. A negative value supports a genetic transmission model that does not include common environmental effects [32].
Primary analyses were derived from the evaluation of alternative genetic transmission models based on maximum likelihood estimation [31] (Table 3) controlling for age, gender, and age by gender interaction. For spindle density, with the exception of detector a4, these analyses demonstrated no evidence that common environmental factors contributed significantly to the phenotypic variance. In the AE models for spindle density, seven of eight detectors emerged with a significant additive genetic transmission. The mean percentage of phenotypic variance explained by additive genetic variance (A) was 77.1%. For all but a4, the p-value was <0.0001, which provides substantial evidence that an important fraction of phenotypic variance is explained by additive genetic transmission based on the twin model. For a summary of the performance of the detector algorithms for spindle density, please see Supplementary Text.
Table 3.
Maximum likelihood estimation of variance components from adjusted * mixed-effects multilevel models for spindle density
| Characteristic | Variable | Optimal model | Genetic variance | Common variance |
|---|---|---|---|---|
| Total power | S0 | AE | 0.637 (0.496, 0.779)** | |
| Average sigma | S1 | AE | 0.817 (0.738, 0.895)** | |
| Relative sigma | S2 | AE | 0.811 (0.729, 0.893)** | |
| Detector | ||||
| Density | a2 | AE | 0.800 (0.712, 0.888)** | |
| a3 | AE | 0.805 (0.722, 0.888)** | ||
| a4 | CE | 0.538 (0.395, 0.68) | ||
| a5 | AE | 0.648 (0.507, 0.789)** | ||
| a7 | AE | 0.816 (0.733, 0.899)** | ||
| a8 | AE | 0.737 (0.622, 0.852)** | ||
| a9 | AE | 0.791 (0.700, 0.882)** | ||
| a10 | AE | 0.801 (0.711, 0.891)** |
Variances with confidence intervals. Mean for the automated detectors with SD.
*Adjusted for age, sex, and age by sex interaction.
** p < 0.0001.
Analysis of genetic effects on spindle amplitude, duration, and frequency
The ICCs of spindle duration, amplitude, and frequency measures from the eight spindle detection algorithms in stage N2 of the MZ and DZ twins are shown in Table 4 (see Supplementary Table 4 for unadjusted ICC). The heritability estimates (h2) for spindle duration ranged from 0.05 to 1.34 with a mean heritability estimate of 0.89 ± 0.47, indicating that heritability is high for spindle duration. In contrast, the estimates (h2) for spindle amplitude ranged from 0.19 to 0.56 with a mean heritability estimate of 0.35 ± 0.11, and spindle frequency from 0.23 to 0.57, with a mean heritability estimate of 0.39 ± 0.10.
Table 4.
Intraclass correlation coefficients (ICC*), classic heritability estimates (h2 †), and common environmental variance (CEV‡) for spindle characteristics in monozygotic (MZ) and dizygotic (DZ) twin pairs, adjusted§
| Characteristic | Detector | ICC_MZ | ICC_DZ | h2 | CEV |
|---|---|---|---|---|---|
| Duration | a2 | 0.73 | 0.10 | 1.25 | –0.52 |
| a3 | 0.79 | 0.12 | 1.34 | –0.55 | |
| a4 | 0.64 | 0.17 | 0.94 | –0.30 | |
| a5 | 0.74 | 0.18 | 1.13 | –0.39 | |
| a7 | 0.63 | 0.41 | 0.45 | 0.19 | |
| a8 | 0.79 | 0.13 | 1.30 | –0.52 | |
| a9 | 0.73 | 0.41 | 0.64 | 0.09 | |
| a10 | 0.66 | 0.64 | 0.05 | 0.61 | |
| Amplitude | a2 | 0.74 | 0.58 | 0.34 | 0.41 |
| a3 | 0.70 | 0.51 | 0.39 | 0.31 | |
| a4 | 0.67 | 0.48 | 0.38 | 0.30 | |
| a5 | 0.69 | 0.50 | 0.39 | 0.30 | |
| a7 | 0.64 | 0.53 | 0.24 | 0.41 | |
| a8 | 0.73 | 0.45 | 0.56 | 0.17 | |
| a9 | 0.60 | 0.51 | 0.19 | 0.41 | |
| a10 | 0.66 | 0.51 | 0.30 | 0.35 | |
| Frequency | a2 | 0.70 | 0.50 | 0.41 | 0.29 |
| a3 | 0.80 | 0.58 | 0.43 | 0.37 | |
| a4 | 0.75 | 0.57 | 0.36 | 0.39 | |
| a5 | 0.78 | 0.64 | 0.29 | 0.49 | |
| a7 | 0.80 | 0.69 | 0.23 | 0.57 | |
| a8 | 0.79 | 0.51 | 0.57 | 0.22 | |
| a9 | 0.85 | 0.68 | 0.35 | 0.50 | |
| a10 | 0.75 | 0.53 | 0.44 | 0.31 |
Heritability is calculated using Falconer’s method; values closer to one indicate a pattern expected where variance in a trait is genetically controlled. Negative CEV values suggest that common environmental effects are likely not very important and that genetic dominance effects may be present.
*ICC = σ 2B/[σ 2W + σ 2B].
† h 2, heritability = 2(ICCMZ – ICCDZ).
‡ CEV = (2 × ICCDZ) – ICCMZ.
§ Adjusted for age, sex, and age by sex interaction.
The average estimated proportion of variance explained by common (shared) environment factors for spindle duration, amplitude and frequency among all detection algorithms was –0.17 ± 0.42, 0.3 ± 0.08, and 0.39 ± 0.12, respectively. As previously stated, a negative value supports a genetic transmission model that does not include common environmental effects, supporting the finding that spindle duration is highly heritable.
Using the maximum likelihood approach and controlling for age, sex, and age by sex interaction, AE was the best fitting model for spindle duration in six of eight detection algorithms (Table 5). For the a2 detector, a dominant genetic factor was observed. The total genetic variance (A + D) for a2 was 74.8%. For detector a10, no significant genetic variance for spindle duration was observed with 63.2% of phenotypic variance explained by shared environment (p < 0.001). For amplitude, all eight detectors exhibited large and highly statistically significant additive genetic variance (AE, p < 0.001), with a minimum value of 71.6%. For frequency, a significant additive genetic variance was observed in two of the eight algorithms. The optimal models for detectors a3 and a9 included both additive and shared environment variance (ACE), but the additive genetic variances were only marginally significant (p = 0.08). Significant additive variance (p < 0.001) was observed for detectors a8 and a10 (AE). Only shared common variance was observed for a2, a4, a5, and a7 (p < 0.001). For a summary of the performance of the detector algorithms for spindle duration, amplitude, and frequency, please see Supplementary Text.
Table 5.
Maximum likelihood estimation of variance components from adjusted* mixed-effects multilevel models for spindle amplitude and duration variances with confidence intervals
| Characteristic | Detector | Optimal model | Genetic variance | Common variance |
|---|---|---|---|---|
| Duration | a2 | ADE | 0.748 (0.643, 0.854)** | |
| a3 | AE | 0.794 (0.701, 0.888)** | ||
| a4 | AE | 0.637 (0.485, 0.790)** | ||
| a5 | AE | 0.742 (0.627, 0.857)** | ||
| a7 | AE | 0.719 (0.598, 0.840)** | ||
| a8 | AE | 0.777 (0.677, 0.877)** | ||
| a9 | AE | 0.774 (0.675, 0.873)** | ||
| a10 | CE | 0.632 (0.508, 0.755)** | ||
| Amplitude | a2 | AE | 0.800 (0.715, 0.884)** | |
| a3 | AE | 0.781 (0.685, 0.877)** | ||
| a4 | AE | 0.775 (0.674, 0.875)** | ||
| a5 | AE | 0.783 (0.687, 0.879)** | ||
| a7 | AE | 0.735 (0.616, 0.855)** | ||
| a8 | AE | 0.798 (0.707, 0.889)** | ||
| a9 | AE | 0.716 (0.593, 0.838)** | ||
| a10 | AE | 0.752 (0.645, 0.860)** | ||
| Frequency | a2 | CE | 0.649 (0.533, 0.765) | |
| a3 | ACE | 0.311 (–0.045, 0.666) | 0.485 (0.145, 0.825) | |
| a4 | CE | 0.699 (0.596, 0.802) | ||
| a5 | CE | 0.740 (0.649, 0.831) | ||
| a7 | CE | 0.776 (0.696, 0.856) | ||
| a8 | AE | 0.807 (0.728, 0.886)** | ||
| a9 | ACE | 0.242 (–0.032, 0.516) | 0.604 (0.339, 0.869)* | |
| a10 | AE | 0.759 (0.664, 0.855)** |
* Adjusted for age, sex, and age by sex interaction.
** p < 0.0001.
Analysis of genetic effects on sigma power
With regard to sigma power, ICCs of all measures are shown in Table 2 (see Supplementary Table 3 for unadjusted ICC). The heritability estimates (h2) ranged from 0.16 to 0.77, demonstrating differential heritability based on the measure used. Using the maximum likelihood approach, AE was the best fitting model for all measures of sigma power (Table 3). All measures exhibited large and highly statistically significant additive genetic variance (AE, p < 0.001), with a minimum value of 63.7%
Factor analysis of spindle detector algorithms
For spindle density, the cumulative percentages were 66.8%, 83.0%, 91.0%, 95.9%, 98.6%, 99.2%, 99.7%, and 100% for one through eight dimensions. Had the various spindle density algorithms been entirely “redundant,” then the first eigenvalue would have been associated with 100% of the latent variance. Instead, it took five latent dimensions to explain about 99% of the variance and little additional explained variance is obtained when a sixth latent factor is considered. In contrast, removing the fifth factor reduces explained variance by nearly 5%. Therefore, a five-factor model was interpreted. A “simple” and interpretable rotated factor pattern matrix was obtained. The data from the Scree plots, rotated factor pattern matrices, and the reference axis correlation matrices are provided for each phenotype in Supplementary material. To summarize, detectors a9 (0.971), a10 (0.943), and a7 (0.915) all loaded highly on factor 1. The factor loadings on the remaining detectors were all small in absolute value (|all| < 0.183). Detectors a5 (0.954) and a3 (0.787) loaded high on factor 2 with all remaining factors small (|all| < 0.104). Only unique detectors had large loadings on Factor 3 (a8 = 0.984, |remaining| < 0.075), factor 4 (a2 = 0.922, |remaining| < 0.195), and factor 5 (a4 = 0.832, |remaining| < 0.086). These results demonstrate that, as a set, the spindle density detectors are non-redundant, spanning a five-dimensional space. These results help to understand why a4 was found to lack heritability (Table 3). The latent trait behind detector a4 is not shared with any other detector. The three detectors sharing factor 1, a9, a10, and a7 have nearly identical heritability, 0.79, 0.80, 0.82, respectively. The detector with the lowest, but still significant heritability, a5, also uniquely loads on its own factor.
The same analysis was applied to duration. Similar to density, the cumulative percentages were 69.0, 85.2, 91.6, 96.2, 97.6, 98.9, 99.5, and 100% for one through eight dimensions. Considerations of the Scree plot and the cumulative percentages of variance explained first suggested five latent factors explaining 98% of the variance, although a sixth dimension adds 1.2% more variance explained and eliminating the fifth factor reduces explained variance by only 1.4%. However, when the rotated factor pattern matrix was examined, detector a2 had a larger loading on factor 1 (0.789) than on factor 5 (0.418) and a2 was the only detector with a loading larger than 0.106 in absolute value on factor 5. Therefore, the four-factor model was interpreted, which captured 96.2% of the latent variance. Factor 1 was characterized by detectors a8, a2, a5, and a3. Factor 2 was characterized by a9 and a7. Factors 3 and 4 were characterized by a10, and a4, respectively. Interestingly, the underlying factor structure appeared different between density and duration. Detector a10 did not exhibit significant heritability for the duration phenotype. Consistent with this finding, a10 appeared as its own latent trait reflected in factor 3. The detector with the smallest, but significant genetic variance was a4. This detector also was reflected by its own latent trait as factor 4. Detectors a8, a2, a5, and a3 (factor 1) had relatively similar genetic variance (0.78, 0.75, 0.74, and 0.79, respectively).
In contrast, to density and duration, amplitude exhibited only one underlying dimension explaining 97.8% of variance. This is consistent with all of the detectors exhibiting significant genetic variance. Although the first dimension for frequency was dominant, explaining 94% of variance, a second factor explained an additional 3.1% of variance and so was retained. The only two frequency detectors with statistically significant genetic variance, a8 (0.72) and a10 (0.89) loaded on the first dominant factor. Detector a3 had weak evidence suggesting genetic variance and loaded on this factor but with a much lower factor loading of 0.54. The only association displaying inconsistency between the factor analysis and the analysis of genetic variance involved a4 frequency. This phenotype also loaded in factor 1 (0.80) but significant genetic variance was not observed.
Discussion
The present study demonstrated that the sleep spindle is highly heritable. This finding is consistent with the previous literature which showed that one of the most heritable characteristics of the sleep EEG is power in the 8–16 Hz frequency range [8]. However, it is important to note that while our results indicate that spindles are a heritable characteristic of the sleep EEG, not all spindle characteristics are equally heritable. The present results revealed that while spindle density, duration, and amplitude showed significant heritability using both classic heritability assessment and the MLE method across most detector types, spindle frequency did not. Similarly, it is equally crucial to recognize that each of the automated spindle detector algorithms used in this study yielded a different pattern of results. For example, although all spindle detectors analyzed identical data, the spindle density reported by the different detectors varied, ranging from 0.74 to 6.59 spindles per minute. In the future, it will be essential for researchers looking to take advantage of these detector methods to understand the strengths and weaknesses of the detector(s) used in their analysis.
A strength of the present study was the use of eight distinct automated spindle detector algorithms. Our results demonstrated that across most detectors used, estimates of heritability were high. The study also employed two methods of measuring heritability, the classic heritability estimate using ICC and Falconer’s h2, and the MLE method. This multimodal approach provides an opportunity to substantiate and enhance the findings of any one method alone. As expected, both the classic and MLE models suggest that the heritability of spindles is high with spindle density demonstrating a mean h2 of 0.74 ± 0.30, in addition to 77.1% of the phenotypic variance explained by additive genetic variance, lending additional support to our conclusions. Other studies have previously examined the heritability of spindles and have generally found evidence of moderate heritability. The present results suggest that the estimates of moderate heritability (0.3 to 0.5) may be a result of the limitations of using one detection algorithm. For example, due to the fact that each spindle detection algorithm identifies different spindles, and that these subsets of spindles display distinctive characteristics which can be more (density) or less (oscillation frequency) heritable, if only one spindle detection algorithm is used, overall estimates of heritability will be skewed towards the heritability estimate of the spindle characteristic best detected.
Our results are in line with other studies demonstrating the heritability of the sleep EEG and its associated components, including but not limited to sigma power [8], K complexes [36], and arousal intensity [15]. As the sleep EEG has been suggested to be one of the most heritable human traits [11]. our findings establishing that the sleep spindle is highly heritable is consistent with this notion. As previously mentioned, previous studies have generally reported moderate heritability of the sleep spindle, although with conflicting patterns of results, especially among slow and fast spindles, and different topographical regions. While our analyses did not differentiate between fast and slow spindles or anterior and posterior regions, our results indicate that the sleep spindle generally shows strong evidence of heritability. It is interesting to note, however, that all 8 spindle detection algorithms showed an average spindle oscillation frequency of between 13 and 13.5 Hz. This would seem to indicate that at C3/C4, the site at which our analyses were focused, fast spindles, defined as >13 Hz, predominate. Because our analyses were limited to the central electrode location, the present data could not determine the heritability of slow vs fast spindles based on topography (anterior vs posterior). We found a bell-shaped unimodal distribution of spindle oscillation frequency at C3/C4, which made finding a meaningful cut point dividing slow vs fast spindles at a single electrode location difficult and arbitrary. However, our results may suggest that the previous mixed findings with regard to spindle heritability may be due to methodological limitations, and not the variability in the heritability of the sleep spindle.
Since spindle frequency was the only spindle characteristic to not show evidence of high heritability, we were interested in examining the ICC, specifically. These data demonstrated moderate to high ICC values for both the MZ and DZ twin pairs which may suggest that spindle frequency is influenced more by environmental factors than by genetic factors. For example, spindle frequency may vary with age, as the sigma frequency range has been shown to decrease in power as age increases [37]. Likewise, spindle frequency may also be related to general cognitive or health factors that can covary with similar experience or environment. For example, higher spindle frequency from centroparietal leads has been shown to be associated with better verbal learning [38], possibly suggesting that educational experiences, commonly similar among siblings, can influence spindles. As previously mentioned, health status has been shown to be related to sleep spindle characteristics [5–7] and may represent another factor contributing to the high degree of consistency in spindle frequency between both MZ and DZ twins.
Broadly, these data demonstrate the high heritability of most features of spindles, despite the conclusion that spindle characteristics are differentially heritable. None of the detector algorithms utilized in the present study demonstrated low heritability across all features, while at the same time no detector consistently showed high heritability across all features. This point is worth noting as there is presently no gold standard detection algorithm. The selection of detector algorithm, therefore, should be based on the characteristics of that algorithm. For example, if the research question is focused on spindle duration, a3 or a8, the detector algorithms that showed both high and consistent estimates of both methods of heritability, may be an appropriate choice. In order to examine the issue of redundancy among spindle detector algorithms, we also conducted a factor analysis on each of the spindle characteristics: density, duration, amplitude, and frequency. With regard to amplitude, results showed only one underlying dimension which was in line with the heritability results demonstrating high genetic variance among all detector algorithms. Frequency demonstrated two significant factors, and both detector algorithms that showed significant genetics variance loaded onto this factor. In contrast, with regard to density and duration, our results indicated that a five-factor model and four-factor model, respectively, best accounted for the patterns in the data. This demonstrates that these detector algorithms are not entirely redundant and instead represent a multi-dimensional space that parallels the heritability findings. Details regarding these factor models, including the cumulative proportions of phenotypic variance explained, rotated factor pattern matrices and inter-factor correlations, are provided in the supplement.
As an alternative to using spindle detection algorithms, one common method for quantifying spindle activity within the EEG is to use sigma power (11 to 16 Hz) as a proxy metric, since spindle activity generates power within this frequency range. Absolute average sigma power is generated from the raw PSA, while relative sigma power is a metric which takes into account individual differences by dividing sigma power by total power by individual. The present results suggest that relative sigma power (s2) is highly heritable, similar to results found using the spindle detection algorithms. In contrast, total power generated from the EEG, in addition to average absolute measures of sigma power (s0, s1) were not heritable. This may indicate that sigma power and spindles, specifically, play a special role in sleep biology, and are not simply markers of overall brain activity. Furthermore, these findings suggest that when quantifying spindle activity, in certain circumstances (e.g. relatively few arousals or no evidence of alpha intrusion), relative sigma power seems to provide adequate information.
The present results should be interpreted in light of study limitations. First, our analyses were focused on data from the average of the C3 and C4 electrode sites. This limited our ability to examine differences between spindles generated from different scalp locations. As fast and slow spindles have been suggested to be associated with distinct brain regions, this study could not disentangle the differences in heritability between fast and slow spindles or spindles generated in anterior and posterior brain regions. It is true that combining all individual results in an aggregate plot may have obscured the purported double peak. However, we did inspect the frequency distribution of all individual participants and found no evidence for a clear separation between fast and slow spindles. We would argue that the relevancy of two spectral peaks at C3/C4 is an open debate, and worthy of further investigation. Future studies could consider replicating the present study with high-density EEG. Second, our analyses were also based on N2 sleep, the stage in which spindles are most likely to be present and one of the stages’s defining features. Spindles, however, are not restricted to this sleep stage, and may occur during N3 sleep, where they may go undetected due to the presence of large-amplitude slow oscillations [39]. In future analyses, it will be useful to assess the heritability of spindles across the sleep period to confirm the present results. Furthermore, our data demonstrated a significant difference in the time in N2 sleep between monozygotic and dizygotic twins (see Supplementary Table 1). Although spindle metrics were calculated per unit time (spindles per minute) which accounts for most differences in duration, it is important to acknowledge that this effect could have influenced our results. Our method of calculating relative sigma power is also somewhat different from previous methods of calculating this metric in that we used total power between 4.5 and 35 Hz as the denominator, rather that total power in the entire EEG spectrum. We chose to calculate relative sigma power this way as we have found that using the total power including delta, which is very dependent on sleep history, has a large impact on the normalization. Our method has been shown to yield a more stable measurement [12]. Lastly, we must acknowledge that our analyses were conducted on data that underwent a Box–Cox transformation due to the non-normal distribution of our spindle density results. This transformation was chosen on the basis that it selects the most appropriate transformation to normalize the data at a case-by-case level. Despite these limitations, we are confident of our results due to our comprehensive methodology including the use of eight distinct, previously validated spindle detection algorithms and two heritability assessment methods in a large sample of twin pairs.
In summary, the present study demonstrated that sleep spindles are highly heritable, regardless of the method of detection or heritability method used. These results extend previous literature showing evidence of heritability of sleep spindles, and recent research demonstrating the heritability of the K complex, another EEG graphoelement. Our results also add to the mounting evidence that suggests that the sleep EEG is one of the most heritable human traits. The generally high level of consistency among detector algorithms also illustrates that spindle detection algorithms and sigma power can be reliably used to assess spindles in the sleep EEG, with the important caveat that while sleep spindles are highly heritable, not all spindle characteristics are equally heritable. Lastly, it is important to note that each detector algorithm produces a different pattern of results.
Supplementary Material
Acknowledgments
Funding for this work was provided by the Chaire Pfizer, Bristol-Myers Squibb, SmithKline Beecham, Eli Lilly en psychopharmacologie de l’ Université de Montréal, the Centre de Recherche Hôpital du Sacré-Coeur de Montréal, the Canadian Institutes of Health Research (CIHR), NIH K23 MH118580, NIH P50 HL060287 and NIH P01 HL094307.
Disclosure Statement
Financial Disclosure: M.Y. developed the a10 algorithm which is included within a commercial sleep scoring system (Michele Sleep Scoring; MSS). M.Y. receives royalties and consultation fees from Cerebra Health which licensed MSS.
Non-financial Disclosure: none.
References
- 1. Sehgal A, et al. Genetics of sleep and sleep disorders. Cell. 2011;146(2):194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Franken P, et al. Genetic variation in EEG activity during sleep in inbred mice. Am J Physiol. 1998;275(4):R1127–R1137. [DOI] [PubMed] [Google Scholar]
- 3. Ambrosius U, et al. Heritability of sleep electroencephalogram. Biol Psychiatry. 2008;64(4):344–348. [DOI] [PubMed] [Google Scholar]
- 4. Fogel SM, et al. The function of the sleep spindle: a physiological index of intelligence and a mechanism for sleep-dependent memory consolidation. Neurosci Biobehav Rev. 2011;35(5):1154–1165. [DOI] [PubMed] [Google Scholar]
- 5. Ferrarelli F, et al. Reduced sleep spindle activity in schizophrenia patients. Am J Psychiatry. 2007;164(3):483–492. [DOI] [PubMed] [Google Scholar]
- 6. Farmer CA, et al. Spindle activity in young children with autism, developmental delay, or typical development. Neurology. 2018;91(2):e112–e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Petit D, et al. Sleep and quantitative EEG in neurodegenerative disorders. J Psychosom Res. 2004;56(5):487–496. [DOI] [PubMed] [Google Scholar]
- 8. De Gennaro L, et al. The electroencephalographic fingerprint of sleep is genetically determined: a twin study. Ann Neurol. 2008;64(4):455–460. [DOI] [PubMed] [Google Scholar]
- 9. Purcell SM, et al. Characterizing sleep spindles in 11,630 individuals from the National Sleep Research Resource. Nat Commun. 2017;8:15930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Adamczyk M, et al. Automatic sleep spindle detection and genetic influence estimation using continuous wavelet transform. Front Hum Neurosci. 2015;9:624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rusterholz T, et al. Nature and nurture: brain region-specific inheritance of sleep neurophysiology in adolescence. J Neurosci. 2018;38(43):9275–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lacourse K, et al. A sleep spindle detection algorithm that emulates human expert spindle scoring. J Neurosci Methods. 2019;316:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Warby SC, et al. Sleep-spindle detection: crowdsourcing and evaluating performance of experts, non-experts and automated methods. Nat Methods. 2014;11(4):385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuna ST, et al. Heritability of performance deficit accumulation during acute sleep deprivation in twins. Sleep. 2012;35(9):1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao X, et al. Heritability of heart rate response to arousals in twins. J Sleep Disord Res. 2017;40(6):zsx055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Becker A, et al. Twin zygosity. Automated determination with microsatellites. J Reprod Med. 1997;42(5):260–266. [PubMed] [Google Scholar]
- 17. Radloff LS. The CES-D scale: a self-report depression scale for research in the general population. Appl Psychol Meas. 1977;1(3):385–401. [Google Scholar]
- 18. Liskow B, et al. Validity of the CAGE questionnaire in screening for alcohol dependence in a walk-in (triage) clinic. J Stud Alcohol. 1995;56(3):277–281. [DOI] [PubMed] [Google Scholar]
- 19. Mayfield D, et al. The CAGE questionnaire: validation of a new alcoholism screening instrument. Am J Psychiatry. 1974;131(10):1121–1123. [DOI] [PubMed] [Google Scholar]
- 20. Malhotra A, et al. Performance of an automated polysomnography scoring system versus computer-assisted manual scoring. Sleep. 2013;36(4):573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Berry RB, et al. The AASM manual for the scoring of sleep and associated events. Rules Terminol Tech Specific Darien Illinois Am Acad Sleep Med. 2012;176:2012. [Google Scholar]
- 22. Mölle M, et al. Grouping of spindle activity during slow oscillations in human non-rapid eye movement sleep. J Neurosci. 2002;22(24):10941–10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin N, et al. Topography of age-related changes in sleep spindles. Neurobiol Aging. 2013;34(2):468–476. [DOI] [PubMed] [Google Scholar]
- 24. Wamsley EJ, et al. Reduced sleep spindles and spindle coherence in schizophrenia: mechanisms of impaired memory consolidation? Biol Psychiatry. 2012;71(2):154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ray LB, et al. Expert and crowd-sourced validation of an individualized sleep spindle detection method employing complex demodulation and individualized normalization. Front Hum Neurosci. 2015;9:507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parekh A, et al. Detection of K-complexes and sleep spindles (DETOKS) using sparse optimization. J Neurosci Methods. 2015;251:37–46. [DOI] [PubMed] [Google Scholar]
- 27. Guadagni V, et al. Association of sleep spindle characteristics with executive functioning in healthy sedentary middle-aged and older adults. J Sleep Res. 2020. doi: 10.1111/jsr.13037. [DOI] [PubMed] [Google Scholar]
- 28. Box GE, et al. An analysis of transformations. J R Stat Soc: Ser B (Methodological). 1964;26(2):211–243. [Google Scholar]
- 29. Sakia RM. The box‐cox transformation technique: a review. J R Stat Soc: Ser D (The Statistician). 1992;41(2):169–178. [Google Scholar]
- 30. Falconer DS, et al. Introduction to Quantitative Genetics. Harlow, Essex, UK: Longmans Green; 1996: 3. [Google Scholar]
- 31. McArdle JJ, et al. Mixed-effects variance components models for biometric family analyses. Behav Genet. 2005;35(5):631–652. [DOI] [PubMed] [Google Scholar]
- 32. Heath AC, et al. Evidence for genetic influences on sleep disturbance and sleep pattern in twins. Sleep. 1990;13(4):318–335. [DOI] [PubMed] [Google Scholar]
- 33. Kim JO, Mueller CW. Factor Analysis: Beverly Hills. CA: Sage; 1978. [Google Scholar]
- 34. Cattell RB. The Scree test for the number of factors. Multivariate Behav Res. 1966;1(2):245–276. [DOI] [PubMed] [Google Scholar]
- 35. Austin MA, et al. Lipoprotein(a) in women twins: heritability and relationship to apolipoprotein(a) phenotypes. Am J Hum Genet. 1992;51(4):829–840. [PMC free article] [PubMed] [Google Scholar]
- 36. Gorgoni M, et al. The heritability of the human K-complex: a twin study. Sleep. 2019;42(6). doi: 10.1093/sleep/zsz053 [DOI] [PubMed] [Google Scholar]
- 37. Carrier J, et al. The effects of age and gender on sleep EEG power spectral density in the middle years of life (ages 20–60 years old). Psychophysiology. 2001;38(2):232–242. [PubMed] [Google Scholar]
- 38. Lafortune M, et al. Sleep spindles and rapid eye movement sleep as predictors of next morning cognitive performance in healthy middle-aged and older participants. J Sleep Res. 2014;23(2):159–167. [DOI] [PubMed] [Google Scholar]
- 39. Cox R, et al. Individual differences in frequency and topography of slow and fast sleep spindles. Front Hum Neurosci. 2017;11:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.