SUMMARY
Parallel processing circuits are thought to dramatically expand the network capabilities of the nervous system. Magnocellular and parvocellular oxytocin neurons have been proposed to subserve two parallel streams of social information processing, which allow a single molecule to encode a diverse array of ethologically distinct behaviors. Here we provide the first comprehensive characterization of magnocellular and parvocellular oxytocin neurons in male mice, validated across anatomical, projection target, electrophysiological, and transcriptional criteria. We next used novel multiple feature selection tools in Fmr1 KO mice to provide direct evidence that normal functioning of the parvocellular but not magnocellular oxytocin pathway is required for autism-relevant social reward behavior. Finally, we demonstrate that autism risk genes are enriched in parvocellular compared to magnocellular oxytocin neurons. Taken together these results provide the first evidence that oxytocin pathway specific pathogenic mechanisms account for social impairments across a broad range of autism etiologies.
INTRODUCTION
Parallel processing pathways have been identified in both sensory (Dijkerman and de Haan, 2007; Haberly, 2001; Nassi and Callaway, 2009; Recanzone and Cohen, 2010; Roper, 2009; Zwergal et al., 2009) and striatal (Cui et al., 2013; Kravitz et al., 2012; Lobo and Nestler, 2011) circuits, and are thought to dramatically expand the network capabilities of the nervous system. Interestingly, oxytocin (OT) neurons are comprised of magnocellular and parvocellular subtypes (Gurdjian, 1927) raising the possibility that this organizational principle can be extended to social information processing. Indeed, magnocellular and parvocellular OT neuronal subtypes can be differentiated by a number of characteristic features (van den Pol, 2012), which suggest that they are specialized to subserve distinct behaviors. For example, because magnocellular OT neurons are specialized to release large quantities of OT in both the central and peripheral nervous system, this adaptation may enable coordination of central and systemic responses during lactation and parturition (Ludwig and Leng, 2006). In contrast, parvocellular OT neurons release smaller quantities of OT that are restricted to the central brain, an adaptation that may subserve the measured reward associated with social cognition and peer-peer attachments (Dölen and Malenka, 2014). At this time this view is largely supported by indirect evidence from comparative studies across species (Dölen, 2015). Nevertheless, the existence of parallel circuits for social information processing will have significant implications for understanding the pathogenesis of autism spectrum disorder (ASD), a neurodevelopmental disease characterized by domain specific impairments in social interactions (Vivanti and Nuske, 2017).
Selective vulnerability of neuronal subtypes to brain injury is called ‘pathoclisis’ and has been proposed to explain how despite exposure to a common insult, disease pathogenesis may disproportionately reflect disrupted function of a subset of brain cells (Klatzo, 2003). For example, in Huntington’s disease, even though the genetic lesion exists throughout the brain, characteristic motor impairments and striatal degeneration have been proposed to be the consequence of selective affinity of mutant Huntingtin (HTT) for Rhes, the Ras homologue enriched in the striatum (Paul et al., 2014). The etiology of ASD is also primarily genetic (Sullivan et al., 2012), with over 1000 genes implicated to date. Nevertheless, at this time little is known about the pathogenic mechanisms underlying social behavioral abnormalities in ASD, and whether these are the consequence of selective vulnerability of specific cell types to genetic injury.
In 1943, Julia Bell and James Martin first described the genetic disorder now known as Fragile X (Martin and Bell, 1943), the same year that Leo Kanner published his seminal psychiatric description of Autism (Kanner, 1943). Using narrowly defined diagnostic criteria, early studies revealed the exceptionally high heritability of Autism (Bailey et al., 1995; Folstein and Rutter, 1977; Freitag, 2007; Steffenburg et al., 1989), and identified Fragile X as the first known genetic cause of the disorder (August, 1983; Brown et al., 1982, 1986). Broader modern clinical definitions have given rise to somewhat reduced heritability estimates (Sullivan et al., 2012); nevertheless, 50% of males, and 20% of females with Fragile X meet full diagnostic criteria for ASD (Kaufmann et al., 2017), making monogenic mutation of FMR1, the gene disrupted in Fragile X (Verkerk et al., 1991), still the most common identified cause of the disease (Suhl and Warren, 2015). Moreover, FMRP, the protein product of FMR1, regulates roughly 20% of ASD susceptibility genes (Darnell et al., 2011; Driesche et al., 2019), thus this gene is thought to constitute a “node” of ASD etiology that can be studied to understand how mutations in a diverse array of genes produces the complex behavioral phenotypes that clinically unify the disorder (Dölen and Sahin, 2016).
Unlike the strong evidence implicating FMR1 mutations in the etiology of ASD, evidence supporting a role for the genes encoding OT (OXT) and the OT receptor (OXTR) has been more controversial (Allen-Brady et al., 2009; Ebstein et al., 2009; Hovey et al., 2014; Satterstrom et al., 2020; Willsey et al., 2013; Zhang et al., 2017). At the same time, we and others have demonstrated that the nucleus accumbens (NAc), dorsal raphe nucleus (dRph), and ventral tegmental area (VTA) receive direct or indirect projections from hypothalamic OT neurons (Beier et al., 2015; Dölen et al., 2013; Hung et al., 2017; Nardou et al., 2019), which together comprise a neural circuit encoding social reward behaviors that may be especially relevant to autism (Chevallier et al., 2013). Interestingly, of these brain regions, only the hypothalamus, where OT neurons reside, exhibits the exaggerated protein synthesis phenotype seen in Fmr1 KO mice (Qin et al., 2005). Furthermore, hypothalamic disruptions of OT neurons have been reported in human patients with ASD related disorders such as Prader Willi syndrome (Swaab et al., 1995), as well as in knockout mice for ASD risk genes including Fmr1 (Francis et al., 2014), Cntnap2 (Peñagarikano et al., 2015), and Pten (Page et al., 2009). Nevertheless, social behavioral abnormalities seen in constitutive knockout mice are not recapitulated following deletion of Pten in OT neurons (Clipperton-Allen et al., 2016), underscoring an important gap in our understanding of how pathognomonic features of the disease arise. Here we sought to bridge this gap by examining the interplay between genetic disruption and molecularly defined circuits which subserve social behaviors that are disrupted in ASD.
RESULTS
Distribution of OT neuronal subtypes across the rostral-caudal axis of the PVN
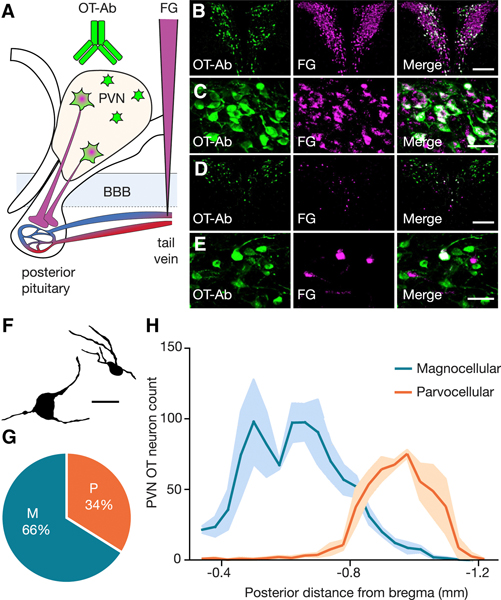
Magnocellular and parvocellular subtypes of OT neurons were first observed nearly a century ago (Gurdjian, 1927; Krieg, 1932; van den Pol, 2012), and early reports suggested that these subtypes can be segregated by their anatomical distribution (Armstrong et al., 1980; Hosoya and Matsushita, 1979; Sawchenko and Swanson, 1982; Swanson and Kuypers, 1980). Nevertheless, to date a quantitative analysis of the proportion and distribution of these OT neuron subtypes within the PVN in mice has not been carried out. Here we used a well-established approach for differentiating between magnocellular and parvocellular OT neuronal subtypes, which is based on the observation that only magnocellular OT neurons send axonal projections outside of the blood-brain barrier (BBB) to the posterior pituitary (van den Pol, 2012). Specifically, intravenous (i.v.) injections of the BBB impermeable retrograde tracer FluoroGold (FG) into the lateral tail vein enables selective labeling of magnocellular but not parvocellular OT neurons (Merchenthaler, 1991). Combining this retrograde tracing approach with OT antibody labeling (Ben-Barak et al., 1985; Whitnall et al., 1985) allowed us to conduct comprehensive quantification of the anatomical distribution and proportion of subtypes of OT neuron in the paraventricular nucleus (PVN) of the hypothalamus in male mice (postnatal day 30–40). OT-Ab labeled neurons were classified as magnocellular if they were FG positive, and parvocellular if they were FG negative (Figures 1A-F; Figure S1). Quantification of these subtypes (n = 3 mice, every other section quantified) revealed that in the PVN, 66% (952 ± 69 neurons) of OTergic neurons are magnocellular, while 34% (488 ± 25 neurons) of OTergic neurons are parvocellular (Figure 1G). Next we examined the distribution of all OT neurons according to their location along the rostral caudal axis of the PVN. As shown in Figure 1H magnocellular OT neurons are predominantly localized to the rostral division of the PVN, while parvocellular OT neurons are concentrated in the caudal division of the PVN. These data provide the first comprehensive quantification of the proportion and rostral-caudal distribution of magnocellular and parvocellular OT neurons, and highlight the importance of examining the entire rostral-caudal axis of the PVN when assessing the contribution of OT neuronal subtypes to functional circuitry.
Figure 1. Distribution of OT neuronal subtypes across the rostral caudal axis of the PVN.
(A) Diagram illustrating labeling strategy. (B-E) Example images of rostral (B,C) and caudal (D,E) sections of the PVN at low magnification (B,D, scale bar 200 μm) and high magnification (C,E, scale bar 50 μm) showing OT-Ab (green, left), FG (magenta, center) and merged (right) images. (F) Drawing of a magnocellular (left) and parvocellular (right) OT neuron identified by i.v. FG labeling. Scale bar indicates 20 μm. (G) Magnocellular OT neurons account for 65.9 ± 2.6% and parvocellular OT neurons account for 34.1 ± 2.6% of PVN OT neurons (n = 3 animals; 1439.3 ± 48.3 OT neurons per animal). (H) Magnocellular OT neurons are predominantly located in the rostral PVN, while parvocellular OT neurons are predominantly located in the caudal PVN. Data represented as mean ± sem. See also Figure S1.
Electrophysiological signatures of magnocellular and parvocellular OT neurons
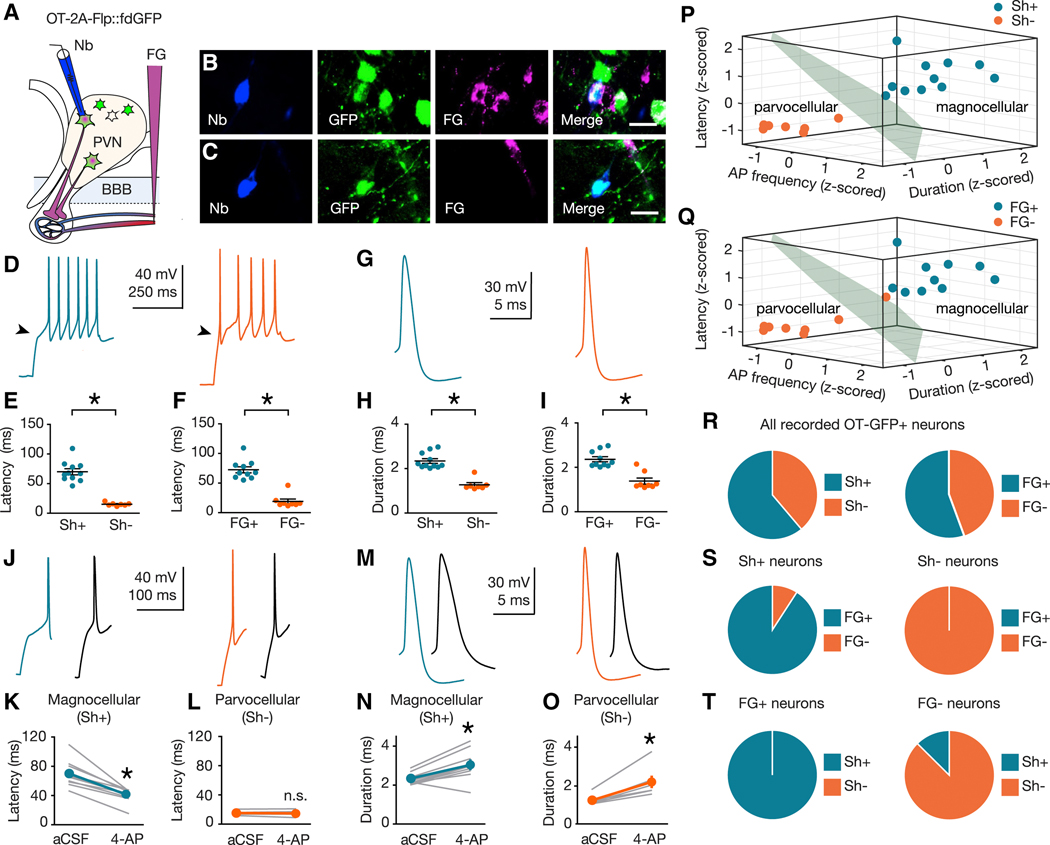
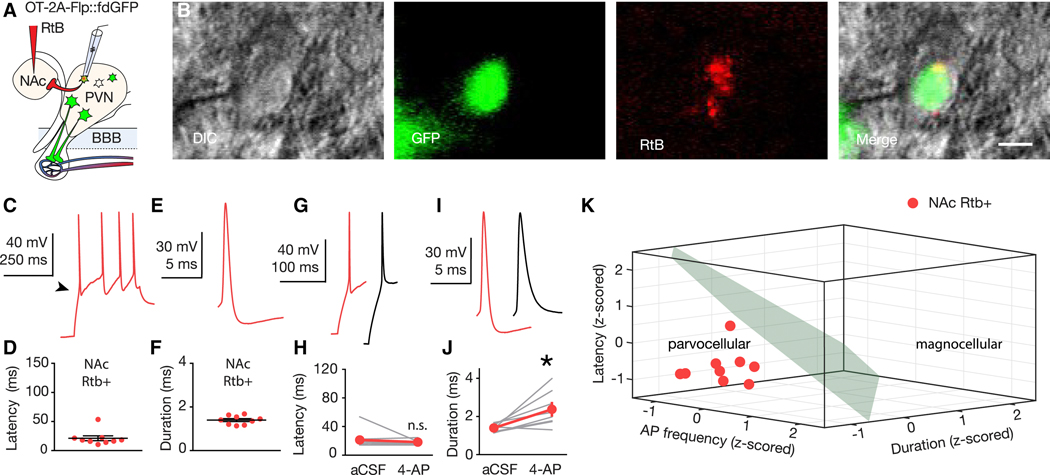
Within the PVN, magnocellular and parvocellular OT cells are intermixed with a wide variety of non-OTergic magnocellular and parvocellular neurons (Biag et al., 2012). Recent studies using electrophysiological circuit mapping of OTergic projections (Eliava et al., 2016; Xiao et al., 2017) relied on the implicit assumption that OT neurons can be classified as magnocellular or parvocellular using criteria established for PVN neurons of unspecified peptidergic identity (Hoffman et al., 1991; Luther et al., 2000, 2002; Tasker and Dudek, 1991). In order to explicitly test this assumption, here we examined magnocellular and parvocellular electrophysiological signatures (Luther and Tasker, 2000) in identified OT neurons using our recently generated OT-Flp recombinase driver line (Nardou et al., 2019), crossed to a Flp-dependent GFP reporter line (fdGFP; Figure S1; Miyoshi et al., 2010). OT-2A-Flp::fdGFP mice (n = 9) were injected with i.v. FG and, blind to FG labeling status, GFP+ neurons were targeted for whole-cell current clamp recording in acute slices of the PVN (Figure 2A). During recording, neurons were labeled with neurobiotin (Nb) to enable post-hoc classification of cells as FG positive (FG+, Figure 2B) or FG negative (FG-, Figure 2C). Neurons were depolarized from a hyperpolarized membrane potential (approximately −100 mV; Figure S2) and the shape of the membrane depolarization preceding action potential (AP) initiation was classified as shoulder positive (Sh+, Figure 2D; blue) or shoulder negative (Sh-, Figure 2D; orange). OT neurons were examined for latency to first AP (Figures 2E,F), AP duration (Figures 2H,I), and sensitivity to the voltage-gated K+ antagonist 4-aminopyridine (4-AP, Figures 2J-O). These studies confirm that latency to first AP is longer in Sh+ compared to Sh- OT neurons (Figure 2E), as well in FG+ compared to FG- OT neurons when neurons are grouped by FG labeling (Figure 2F). Furthermore, AP duration (Figure 2G) is longer in Sh+ compared to Sh- OT neurons (Figure 2H) as well as longer in FG+ versus FG- OT neurons (Figure 2I). Application of the voltage-gated K+ channel blocker 4-AP (5 mM; Figures 2J,M) reduced the latency to first AP and increased AP duration in magnocellular (Sh+) OT neurons, but had no effect on latency in parvocellular (Sh-) OT neurons (Figures 2K,L,N,O).
Figure. 2. Electrophysiological signatures of magnocellular and parvocellular OT neurons.
(A) Validation strategy using i.v. FG and Nb-filled recording pipette. (B,C) FG+ (B) and FG- (C) neurons recovered after electrophysiological recording. Nb (blue, left), GFP (green, center-left), FG (magenta center-right), merge (right). Scale bars 20 μm. (D,G) Representative traces from magnocellular (blue), and parvocellular (orange) OT neurons. (E,F,H,I) Latency to first AP and AP duration are shorter in Sh- (n = 7) compared to Sh+ (n = 11) OT neurons (U = 0, p > 0.001 (latency); U = 0, p > 0.001 (duration)) (E,H) and FG- (n = 8) compared to FG+ (n = 10) OT neurons (U = 0, p < 0.001 (latency); U = 5, p > 0.001 (duration)) (F,I). (J,M) Representative traces (blue, magnocellular (Sh+) OT; orange, parvocellular (Sh-) OT; black, following 5 mM 4-AP). (K,L) 4-AP reduces latency to first AP in magnocellular (Sh+, n = 10, Sum of signed ranks = 55, p = 0.002), (K) but not parvocellular (Sh-, n = 7, Sum of signed ranks = 10, p = 0.469) (L) OT neurons. (N,O) 4-AP increases AP duration in magnocellular (Sh+; n = 10, Sum of signed ranks = −51, p = 0.006) (N) and parvocellular (Sh-, n = 7, Sum of signed ranks = −28; p = 0.016) (O) OT neurons. (P,Q) K-means cluster analysis using z-scored values for AP frequency, duration, and latency to first AP for each neuron. (P) Clusters coincided completely (100%, 18 of 18 neurons) with qualitative magnocellular or parvocellular categorization based on shoulder. (Q) Cluster assignment matched magnocellular or parvocellular identity of 94% (17 of 18) neurons defined by FG labeling. (R) Left; 61% (n = 11) of sampled neurons were Sh+, and 39% (n = 7) were Sh-. Right; 56% (n = 10) sampled neurons were FG+, and 44% (n = 8) were FG-. (S) Left; 91% (n = 10 of 11) Sh+ neurons were FG+. Right; 100% (n = 7 of 7) Sh- neurons were FG-. (T) Left; 100% (n = 10 of 10) FG+ neurons were also Sh+. Right; 88% (n = 7 of 8) FG- neurons were Sh-. Data represented as mean ± sem. * indicates p < 0.05; Mann-Whitney U Test for unpaired (E,F,H,I) or Wilcoxon Rank Sum Test for paired (K,L,N,O) comparisons (both two-tailed). See also Figure S1, S2.
To quantify whether an individual OT neuron’s magnocellular or parvocellular identity can be predicted from its electrophysiological features, we used unsupervised k-means clustering analysis on the z-scored values for AP frequency, duration, and latency to first AP for each neuron. The resulting best fit to this data are two clusters (Figure S2), which corresponded completely (100%, 18 of 18 OT neurons) with the qualitative magnocellular or parvocellular categorization based on the presence or absence of a shoulder in the electrophysiological trace (Figure 2P) and in 94% (17 of 18 OT neurons) when defined by FG labeling, (Figure 2Q). Consistent with the proportions determined in Figure 1, 61% of sampled neurons were Sh+, and 39% were Sh-. Similarly, 56% of sampled neurons were FG+ and 44% were FG- (Figure 2R). Further supporting our cross-validation, among Sh+ OT neurons 91% were FG+, and 100% of Sh- neurons were FG- (Figure 2S). Similarly, 100% of FG+ neurons were Sh+ and 88% of FG- neurons were Sh- (Figure 2T). These results demonstrate that the electrophysiological signature of magnocellular and parvocellular neurons applies specifically to the subset of these cells that are oxytocinergic (Figure 2D-O). Further, these electrophysiological parameters can be used to build a mathematical model to predict magnocellular and parvocellular OT cell identity (Figure 2P-Q). Since the proportion of electrophysiologically defined subtypes (Figure 2R-T) is consistent with anatomical estimates (Figure 1G), these observations provide cross-validation of magnocellular and parvocellular OT neuronal subtype identities by two independently derived measures and demonstrate near perfect agreement of electrophysiological and anatomical classification systems (94%; 17 of 18 OT neurons).
scRNASeq of OT expressing neurons in the PVN
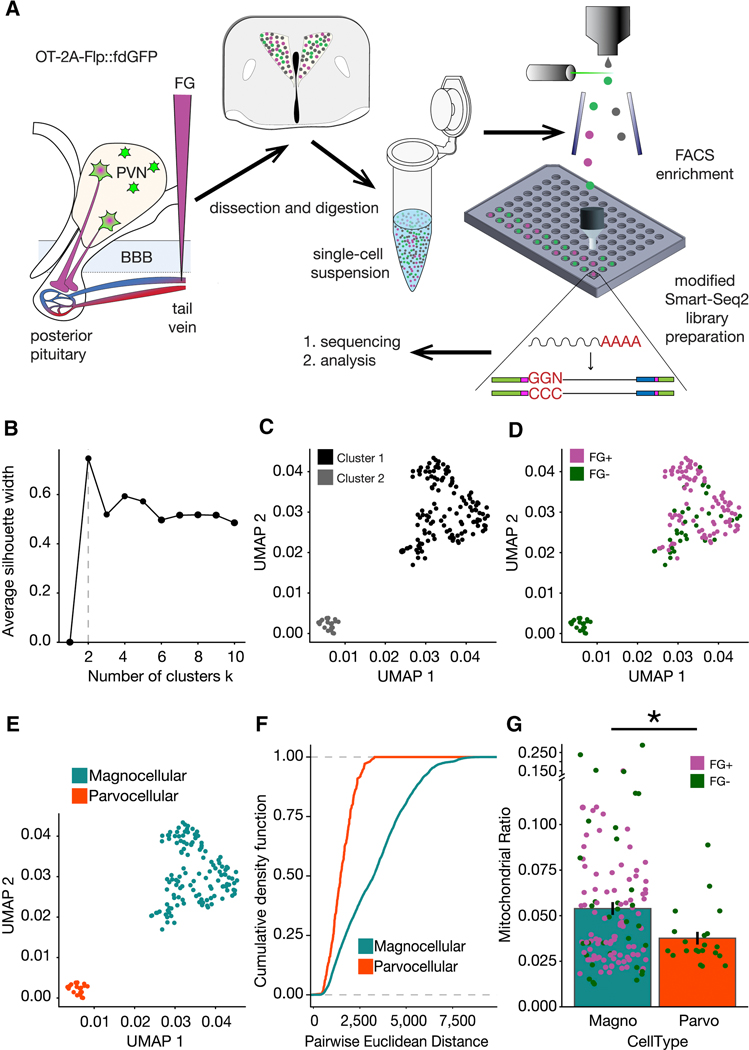
Next we sought to determine whether magnocellular and parvocellular OT neurons exhibit distinct transcriptional profiles. While previous studies have performed single cell RNA sequencing (scRNAseq) analysis of the entire hypothalamus (Chen et al., 2017; Romanov et al., 2017), low recovery rates of OT neurons in these studies limit their power for identifying magnocellular and parvocellular subtypes by transcriptional profiling, leading to confusion about existing classification systems (Althammer and Grinevich, 2017). Others have employed modern molecular strategies to selectively enrich samples for OT neurons, however these studies used relatively low-throughput qRT-PCR to focus on a handful of target genes, and bulk RNA-Seq analyses that can mask important sources of heterogeneity across cell subtypes and states (Maynard et al., 2018). To overcome these technical hurdles, here we combined full-length cDNA scRNA-seq (SMART-Seq2) with molecular targeting of OT neuron subtypes in OT-2A-Flp::fdGFP mice, and additionally utilized projection target mapping (Chevée et al., 2018) to enable cross validation of transcriptional profiles against OT neuronal subtype features with established ground truth (Merchenthaler, 1991) and (Figures 1 and 2). In total, 194 OT neurons were collected by fluorescence assisted cell sorting (FACS) and used as input for a modified SMART-Seq2 scRNA-Seq library preparation (Figure 3A). We next used the Uniform Manifold Approximation and Projection (UMAP) embedding method (Becht et al., 2019) to visualize the transcriptional landscape. We performed a silhouette analysis to quantitatively determine the optimal number of clusters in the UMAP embedding (Figure 3B). This calculation yielded a best fit of 2 clusters, which are plotted in Figure 3C.
Figure 3. scRNASeq of OT expressing neurons in the PVN.
(A) Schematic overview of experiment. (B) Plot of the average silhouette width versus number of clusters for scRNAseq data indicating that the optimal number of clusters is two. (C) UMAP of scRNASeq data with the two clusters indicated by the silhouette analysis. (D) Cluster 1 is majority FG+ (91 of 125 cells; 72.8% FG+) and cluster 2 is devoid of fluorogold positive cells, (21 of 21 cells; 100% FG-).(E) Magnocellular (blue) and parvocellular (orange) neuron identity assigned to cluster 1 and cluster 2, respectively. (F) Empirical Cumulative Distribution Functions for all expressed genes in the two cell types indicated greater intra-cell type heterogeneity within magnocellular neurons (p = 1.665e−15). K-S test for statistical comparison. (G) Comparison of the density of mitochondrial RNA in the two clusters as calculated via the distributions of the per cell ratio of reads mapped to mitochondrial RNA vs total number of mapped reads. One-tailed t-test for statistical comparison (p = 0.0007972). See also Figure S1, S3.
To determine whether these two cell type clusters map onto magnocellular and parvocellular OT neuronal subtypes, we then assessed the distribution of FG labeling across the two clusters (Figure 3D). Since 72.8% of cells in cluster 1 were FG positive (FG+), while cluster 2 contained only cells that were FG negative (FG-; 100%) across all three replicates (Figure S3), we deduce that cluster 1 cells are magnocellular, while cluster 2 cells are parvocellular OT neurons (Figure 3E-F). The FG tail vein injection labeling technique, like all labeling techniques, has a non-zero false negative rate. In our assay, these false negative magnocellular neurons would be represented in the FG- population of cluster 1. Additionally, since the emission wavelength of FG is known to be altered at acidic pH (Schmued and Fallon, 1986), the absence of FG labeling in a subset of cells in the magnocellular cluster may represent acidification of the intracellular environment during dissociation and FACS sorting. Nevertheless, since the FG- cells in the magnocellular cluster have the same characteristics (e.g. mRNA levels, number of genes expressed) as the FG+ cells in that cluster (Figure S3), and since all cells had to pass strict quality metrics to be included in the analysis, it is unlikely that diminished cell health or quality accounts for this finding. Taken together with our observation that only one of the two transcriptionally defined OT populations demonstrated any FG+ staining, these results enabled us to use FG labeling as a supervisory metric to discriminate between the two populations.
To cross validate this observation, we next quantified the density of mitochondrial RNA in the two clusters. Previous reports indicate that magnocellular neurons have increased mitochondrial density (Dayanithi et al., 2012; van den Pol, 1982), a specialization thought to support the increased metabolic demands of magnocellular neurons that synthesize and release significantly larger quantities of OT peptide compared to parvocellular neurons (Dreifuss, 1975; Ludwig and Leng, 2006; Nordmann and Morris, 1984; van den Pol, 1982, 2012; Pow and Morris, 1989). Consistent with these reports, magnocellular neurons had a greater ratio of mitochondrial reads to genomic reads than that of parvocellular neurons (Figure 3G). Moreover, within the magnocellular cluster, where FG+ neurons (purple circles) dominate, the FG- neurons (green circles) are evenly distributed across mitochondrial density measurements (Figure 3G), consistent with our interpretation that FG- cells in this cluster represent a technical artifact, rather than a biologically significant subcategory of magnocellular neuron. Together, these studies identify two distinct transcriptional subtypes of OT neurons using high-resolution full-length sequencing techniques, in samples enriched for OT neurons by molecular techniques. By cross validating these transcriptional profiles across two independent distinguishing features (projection target and mitochondrial density), these studies are the first to establish a transcriptional signature for classifying magnocellular and parvocellular OT neuronal subtypes.
Differential expression analysis of clusters determines unique molecular markers of magnocellular and parvocellular OT neurons
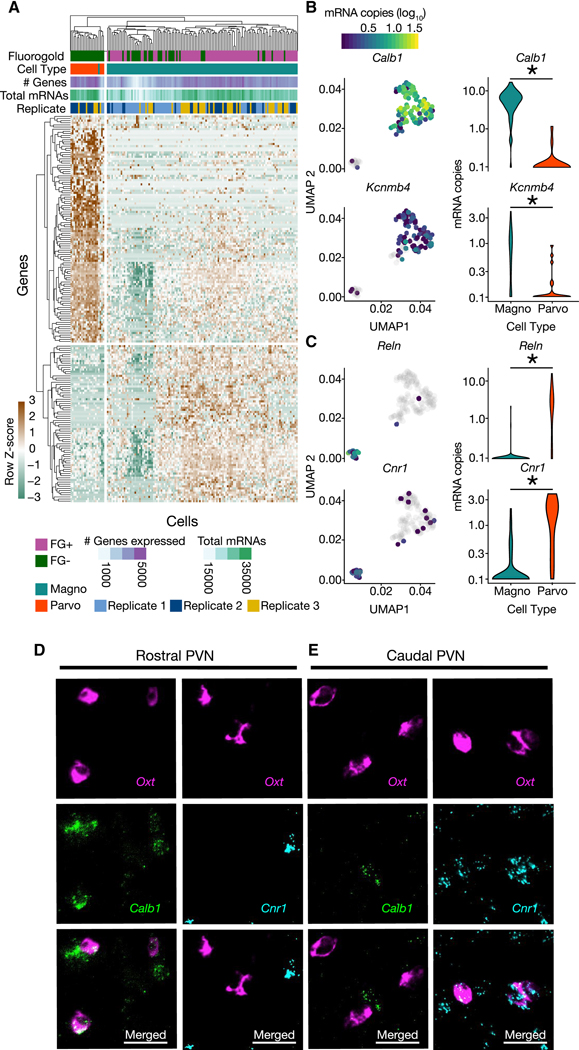
Having identified anatomical, circuit, electrophysiological and transcriptional characteristics that differentiate magnocellular and parvocellular OT neuronal subtypes, next we made use of our high-resolution transcriptional data to identify differentially expressed genes across magnocellular and parvocellular OTergic neurons. Using the monocle2 likelihood ratio test (Trapnell et al., 2014), we identified 181 genes (Table S1) with significant (0.1% FDR; Monocle2; BH-corrected) differential expression (DE) between magnocellular and parvocellular OT cells (Figure 4A), representing novel discriminating marker genes for these two subtypes. Amongst the most significantly differentially expressed genes was the calcium-binding protein Calbindin (Calb1) and a large conductance calcium-activated potassium channel subunit (Kcnmb4), both enriched for expression in the magnocellular OT neuronal population (Figure 4B), as well as the extracellular matrix serine protease Reelin (Reln) and the cannabinoid receptor 1 (Cnr1) genes, both significantly enriched in parvocellular OT neurons (Figure 4C). These results were validated with fluorescent in situ hybridization chain reaction version 3.0 (HCR 3.0; Table S2) (Choi et al., 2018) combined with anatomical criteria (Figure 1), which demonstrated that Oxt+ neurons in the rostral portion of the PVN were specifically colocalized with Calb1 and not Cnr1 mRNA (Figure 4D), while in more caudal sections Oxt+ neurons colocalized with Cnr1 and not Calb1 (Figure 4E). In addition, 18 differentially expressed genes were identified as potentially contributing to the electrophysiological signature of magnocellular and parvocellular OT neurons (Figure S3). These results are the first to establish a set of discriminating molecular identifiers which can distinguish between magnocellular and parvocellular OT neuronal subtypes. While many other differentially expressed genes likely also exist, this conservative list of 181 significantly differentially expressed genes provides a high-value candidate gene list for future studies examining the molecular mechanisms contributing to the functional, physiological, and connectivity differences between these two OT neuronal subtypes. Moreover, as described below, identification of differentially expressed genes paves the way for functional genomics in magnocellular and parvocellular OT neurons.
Figure 4. Differential expression analysis of clusters determines unique molecular markers of magnocellular and parvocellular OT neurons.
(A) Heatmap of significantly differentially expressed genes (Monocle Likelihood Ratio Test, FDR 0.1%) between magnocellular and parvocellular OT neurons. (B) UMAPs (left) and Violin plots (right) for genes expressed more highly in magnocellular OT neurons: Calb1 and Kcnmb4 (identified using FDR 0.00001%; p = 1.28e−30 and p = 5.46e−7, respectively). (C) UMAPs (left) and Violin plots (right) for genes expressed more highly in parvocellular OT neurons: Reln and Cnr1 (identified using FDR 0.00001%; p = 1.48e−15 and p = 6.67e−11, respectively). (D,E) Validation of differential expression via fluorescent in situ hybridization chain reaction version 3.0 (HCR 3.0) within the PVN. Calb1 but not Cnr1, is colocalized with Oxt within the rostral PVN (D). Cnr1, but not Calb1 is colocalized with Oxt in the caudal PVN (E). Scale bars 20 μm. Distribution of Calb1 (rostral) and Cnr1 (caudal) expression in OT neurons recapitulates differential localization of magnocellular (rostrally enriched) and parvocellular (caudally enriched) neurons respectively. * indicates p < 0.05; Monocle2 likelihood ratio test for statistical comparisons. See also Figure S3, and Tables S1, S2.
Fmr1 KO mice exhibit social domain-specific impairment in reward learning
For over 20 years, ASD researchers have taken advantage of the high heritability of the disease to generate transgenic mice with construct validity for ASD (Nestler and Hyman, 2010), and used these model mice to examine how ASD risk genes might converge at the level of biochemical pathways and synapses (Dölen and Bear, 2009; Dölen and Sahin, 2016). This ‘bottom-up’ approach has led to the discovery of some very interesting fundamental mechanisms, but today we are arguably no closer to understanding how disruption of these synaptic and biochemical functions gives rise to the behavioral phenotypes that unify the disease (Antoine et al., 2019). Because ASD remains a diagnosis that is defined by behavioral abnormalities, taking instead the ‘top-down’ approach, offers a significant opportunity to reconcile the etiological diversity of ASD with the convergence of the clinical presentation (Diester et al., 2015; Dölen et al., 2015). Combining this approach with the Fmr1 KO mouse model, which has established construct validity for the disease (Bakker et al., 1994), here we focused on behaviors with face validity for social impairments in ASD (Nestler and Hyman, 2010).
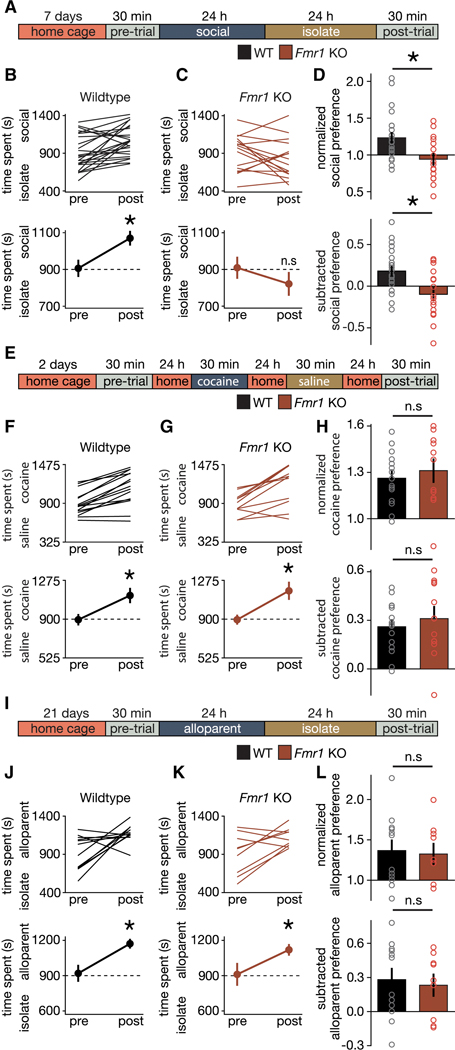
The social motivation theory of autism proposes that a primary deficit in peer-peer social reward learning could account for all of the characteristic behavioral impairments that define the clinical picture in ASD (Chevallier et al., 2013). To test this theory, we began by characterizing three forms of reward learning in the constitutive Fmr1 knockout (KO) using peer-peer social, cocaine, and alloparent social conditioned place preference (CPP) assays (Bardo and Bevins, 2000; Fang and Wang, 2017; Panksepp and Lahvis, 2007). These results demonstrate a significant impairment in peer-peer social CPP (sCPP) in Fmr1 KO mice compared to wildtype (WT; Figures 5A-D, Figure S4), while both WT and Fmr1 KO mice exhibit significant cocaine CPP (cCPP) (Figures 5E-H), as well alloparent CPP (aCPP) (Figures 5I-L, S4). Our finding that Fmr1 KO mice exhibit impairments in social reward but not drug reward (Figures 5A-H) is consistent with human studies that have identified social (Demurie et al., 2011; Stavropoulos and Carver, 2014), but not food or patient salient object (Assaf et al., 2013; Cascio et al., 2012) reward deficits in ASD [but see, (Damiano et al., 2012; Smith et al., 2014)]. Furthermore, our finding that Fmr1 KO mice exhibit no significant impairment in filial attachments (Figures 5I-L) is consistent with the human literature (Baron et al., 2020; Naber et al., 2007, 2008; Rutgers et al., 2004; Stokes et al., 2007; Vivanti and Nuske, 2017). Previously, we have shown that changes in locomotor activity are unlikely to account for differences in sCPP across a variety of experimental manipulations (Dölen et al., 2013; Nardou et al., 2019). Although previous reports raise the possibility that Fmr1 KO mice exhibit altered locomotor behavior, these have been inconsistent across labs (Bernardet and Crusio, 2006); furthermore, since aCPP and cCPP were not different (Figures 5E-L), it is unlikely that changes in locomotor activity could account for the observed differences in sCPP (Figures 5A-D). Taken together, these findings support the view that social reward learning impairments in Fmr1 KO mice are domain specific and restricted to peer-peer social attachments. Furthermore, they provide improved face validity for social impairments in the Fmr1 KO mouse model of ASD, as well as additional support for the social motivation theory of autism.
Figure 5. Fmr1 KO mice exhibit social domain-specific impairment in reward learning.
(A) Timeline for sCPP. (B,C) Individual (top) and average (bottom) data indicate that WT mice (B; t(23) = 3.586; p = 0.002) and not Fmr1 KO mice (C; t(15) = 1.324; p = 0.205) exhibit sCPP. (D) Normalized (t(38) = 2.917; p = 0.006) and subtracted (t(38) = 3.23; p = 0.003) comparisons of social preference reveal a difference in the magnitude of sCPP between WT and Fmr1 KO mice. (E) Protocol and timeline for cocaine CPP. (F,G) Individual (top) and average (bottom) data indicate that WT (F; t(14) = 6.454; p < 0.0001) and Fmr1 KO mice (G; t(10) = 4.297; p = 0.002) exhibit cocaine CPP. (H) Normalized (t(24) = 0.609; p = 0.548) and subtracted (t(24) = 0.644; p = 0.525) comparisons of cocaine preference reveal a similar magnitude of cocaine CPP in WT and Fmr1 KO mice. (I) Protocol and timeline for alloparent CPP. (J,K) Individual (top) and average (bottom) data indicate that WT (J; t(11) = 2.931; p = 0.014) and Fmr1 KO mice (K; t(8) = 2.488; p = 0.038) exhibit alloparent CPP. (L) Normalized (t(19) = 0.234; p = 0.817) and subtracted (t(19) = 0.354; p = 0.727) comparisons of alloparent preference reveal a similar magnitude of alloparent CPP in WT and Fmr1 KO mice. Data represented as mean ± sem. * indicates p < 0.05; Student’s t-test (two-tailed; paired (B,C,F,G,J,K) or unpaired (D,H,L)) for statistical comparisons.
Parvocellular OT neurons innervate NAc
The impairment in peer-peer sCPP in the Fmr1 KO mouse (Figure 5A-D) recapitulates previously reported consequences of OT receptor blockade or ablation in the Nucleus Accumbens (NAc) (Dölen et al., 2013), raising the possibility that these phenotypes share overlapping mechanisms. To test this possibility, we began by taking an anatomical approach that we have used previously to qualitatively identify both magnocellular and parvocellular OT neuronal projections from the PVN to the VTA (Hung et al., 2017). Example images of immunohistochemically identified OT neurons from a wild-type mouse that received an i.v. FG injection, and NAc injection of retrogradely transported fluorescent microspheres (retrobeads; RtB (Katz et al., 1984)), indicated that unlike the VTA, the NAc receives an exclusively parvocellular OTergic projection (Figure S5). To quantify this finding, we next injected RtBs into the NAc (Figure S6) of OT-2A-Flp::fdGFP mice (n = 6 mice), and performed current clamp recordings from RtB/GFP double-positive neurons in acute slices of the PVN (Figures 6A,B). All RtB labeled, OT positive neurons recorded (9 of 9) exhibited a parvocellular electrophysiological phenotype by latency to first AP, AP duration, 4-AP sensitivity (Figures 6C-J), providing quantitative electrophysiological cross validation of our immunohistochemical observations.
Figure 6. Parvocellular OT neurons innervate NAc.
(A) RtB labeling and recording strategy in OT-2A-Flp::fdGFP mice. (B) Example of a GFP/RtB double-positive OT neuron targeted for electrophysiological recording. Left to right; DIC (gray), GFP (green), RtB (red), merge. Scale bar 10 μm. (C,E) Traces recorded from a NAc-projecting OT neuron. (D) Latency to first AP of NAc-projecting OT neurons (n = 9; 21.2 ± 4.2 ms). (F) AP duration of NAc-projecting OT neurons (n = 9; 1.4 ± 0.07 ms). (G,I) Traces (red, RtB+; black, following 5 mM 4-AP). (H,J) 4-AP does not change latency to first AP (n = 8; Sum of signed ranks = 6; p = 0.742) (H), but increases AP duration (n = 8; Sum of signed ranks = −34 p = 0.016) (J) in NAc-projecting OT neurons. (K) Using the k-means clustering algorithm trained using data from Figure 2, 100% (9 of 9) sampled RtB/GFP double-positive cells were assigned to the parvocellular cluster. Data represented as mean ± sem. * indicates p < 0.05; Wilcoxon Rank Sum Test (two-tailed; paired (H,J). See also Figure S1, S2, S5, S6.
Next, we used unsupervised K-means clustering, trained with electrophysiological data from Figure 2, to provide unbiased quantitative cross validation of our electrophysiological findings (Figure 6K). We further used the soft K-means method to calculate the probability that a cell belongs to a given cluster based on the Euclidean distance between that cell and the centers of each cluster in feature space. The probability that a cell belongs to an individual cluster is given by the following softmax equation:
where xi is a cell’s coordinates, cj is the coordinates of the center of the cluster j, p(xi, cj) is the probability that xi belongs to cluster j, and N is the total number of clusters. Using this analysis, we determined that NAc-projecting OT neurons belong to the parvocellular cluster with a mean probability of 92.8 ± 2.7%. This is similar (H(2) = 0.762, p = 0.6833; Kruskal Wallis test) to the probabilities for FG- neurons and Sh- neurons (87.2 ± 8.9%; Figure 2Q, and 96 ± 1.9%; Figure 2P) respectively. Although we and others have identified a number of brain regions involved in the regulation of social behaviors that receive both magnocellular and parvocellular OTergic projections (Hung et al., 2017; Knobloch et al., 2012; Xiao et al., 2017), until technological advancements enable functional genomic interrogation of magnocellular and parvocellular OT neurons separately, identification of the NAc as a brain region that receives an exclusively parvocellular OTergic input in male mice provides a unique opportunity to selectively interrogate the functional role of Fmr1 in parvocellular OT neurons for social reward learning.
Parvocellular OT neurons require Fmr1 to mediate social reward learning
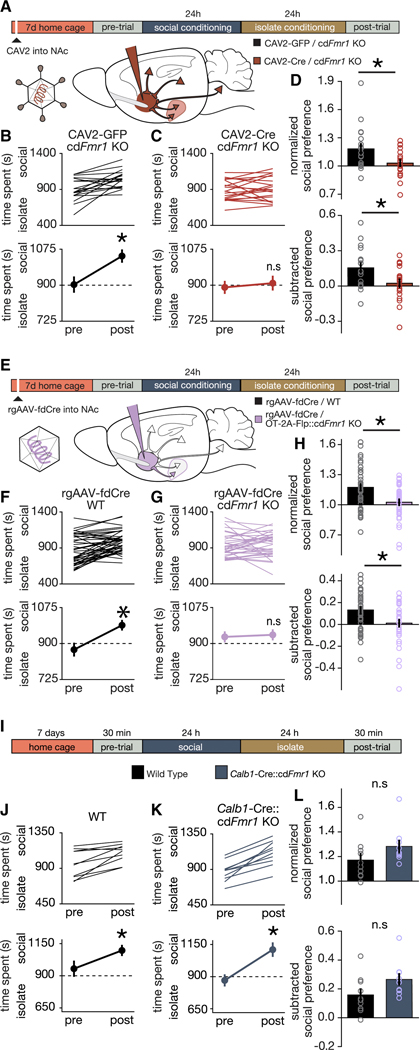
To directly examine the behavioral consequence of disrupting the parvocellular OT input to the NAc, we began by testing whether ongoing Fmr1 function is required for peer-peer social reward learning. We utilized conditional (Cre dependent) Fmr1 KO mice (Mientjes et al., 2006) (cdFmr1 KO) receiving injections of Canine Adenovirus expressing Cre (CAV2-Cre-GFP) or GFP (CAV2-GFP) (Hung et al., 2017; Kremer et al., 2000; Uematsu et al., 2017) bilaterally into the NAc. Since we have previously used the CAV2-Cre-GFP to manipulate gene expression for examining the interplay between genetic and circuit mechanisms of sCPP (Hung et al., 2017), here we controlled for genetic and rearing effects in cdFmr1 KO mice and examined sCPP behaviors following injection of control virus (CAV2-GFP). Injection sites and viral function were confirmed as shown in Figure S6. Seven days later, sCPP was measured, and we observed impaired sCPP in cdFmr1 KO mice injected with CAV2-Cre-GFP, but not CAV2-GFP (Figures 7A-D). Importantly, since acute knockdown of Fmr1 in adolescence is sufficient to recapitulate peer-peer sCPP impairments seen in the constitutive Fmr1 KO, these results demonstrate an ongoing requirement of Fmr1 in regulating these behaviors, and suggest that previous reports that hysteresis plays an important role in the pathogenesis of social impairments in ASD (Orefice et al., 2019), do not generalize across etiologies.
Figure 7. Parvocellular OT neurons require Fmr1 to mediate social reward learning.
(A) Protocol for sCPP following acute knockdown of Fmr1 in neurons projecting to NAc. (B,C) Individual (top) and average (bottom) data indicating that cdFmr1 KO mice injected with CAV2-GFP (B; t(18) = 3.95; p = 0.001) but not CAV2-Cre-GFP (C; t(19) = 0.69; p = 0.499) exhibit sCPP. (D) Normalized (t(37) = 2.453; p = 0.019) and subtracted (t(37) = 2.566; p = 0.015) comparisons reveal reduced magnitude sCPP in cdFmr1 KO mice injected with CAV2-Cre-GFP compared to CAV2-GFP. (E) Protocol for sCPP following acute knockdown of Fmr1 in OT neurons projecting to NAc. (F,G) Individual (top) and average (bottom) data indicating that WT (F; t(43) = 4.947; p > 0.0001) but not OT-2A-Flp::cdFmr1 KO (G; t(35) = 0.349; p = 0.729) mice injected with rgAAV-fdCre exhibit sCPP. (H) Normalized (t(78) = 3.295; p = 0.002) and subtracted (t(78) = 2.937; p = 0.004) comparisons reveal reduced magnitude sCPP in OT-2A-Flp::cdFmr1 KO compared to WT mice injected with rgAAV-fdCre. (I) Protocol and timeline for sCPP. (J,K) Individual (top) and average (bottom) data indicate that WT (J; t(9) = 3.901; p = 0.004) and Calb1-IRES-Cre::cdFmr1 KO mice (K; t(9) = 7.146; p > 0.0001) exhibit social CPP. (L) Normalized (t(17.941) = 1.642; p = 0.118) and subtracted (t(17.874) = −1.965; p = 0.065) comparisons of social preference reveal that WT and Calb1-IRES-Cre::cdFmr1 KO mice exhibit similar magnitude sCPP. Data represented as mean ± sem. * indicates p < 0.05; Student’s t-test (two-tailed; paired (B,C,F,G,J,K) two-tailed; unpaired (D,H,L)) for statistical comparisons. Welch’s correction was used to account for unequal variance and small sample size in (L). See also Figure S6.
To further circumscribe the OT cell type specific role of Fmr1 in regulating social reward learning, we next crossed cdFmr1 KO mice to OT-2A-Flp mice, and generated retrograde Adeno-Associated Virus expressing Flp-dependent-Cre (rgAAV-fdCre) (Penzo et al., 2015; Schneeberger et al., 2019; Tervo et al., 2016) (Figure 7E). Since our experiments above indicated that control cdFmr1 mice injected with CAV-GFP (Figure 7B) exhibit sCPP that is comparable to that seen in WT mice (Figure 5B), here we decided to control for potential off-target effects of rgAAV-fdCre, which although well validated for selectivity and efficiency, had not been previously tested in this behavioral and injection target context. As shown in Figures 7F-H, sCPP was absent in OT-2A-Flp::cdFmr1 KO, while WT animals injected with rgAAV-fdCre exhibited normal social reward learning. Injection sites and viral function were confirmed as shown in Figure S6. Taken together with the studies above identifying the source of these OTergic inputs to the NAc as parvocellular (Figure 6), these results demonstrate that Fmr1 in OT neurons projecting to the NAc specifically, is required for peer-peer social reward learning. Furthermore, since previously parvocellular OT neurons had only been directly implicated in non-social functions (e.g. feeding, yawning, erection, cardiovascular function, pain (Althammer and Grinevich, 2017; Eliava et al., 2016; Melis et al., 1986; Petersson, 2002; Valassi et al., 2008)), these studies identify the first social function directly encoded by this subtype of OT neuron.
While the studies above provide direct evidence that normal function of OT neurons is necessary for social reward learning, in order to test the specificity of this function, we next sought to determine the consequences of Fmr1 deletion in magnocellular OT neurons. To date we have yet to identify a brain region that receives an exclusively magnocellular OTergic projection (Hung et al., 2017), thus the projection targeting method used above could not be utilized to selectively knockdown Fmr1 in magnocellular OT neurons. Instead, we capitalized on our discovery that Calb1 is a discriminatory marker of magnocellular compared to parvocellular OT neurons. Crossing Calb1-IRES-Cre mice (Daigle et al., 2018; Nigro et al., 2018) with cdFmr1 KO mice allowed us to selectively knock down Fmr1 in magnocellular OT neurons expressing Calb1. Although this manipulation also knocked down Fmr1 in other, non-OTergic Calb1-expressing neuronal populations elsewhere in the brain (see also Figures 4D-E), remarkably, sCPP was not impaired in Calb1-IRES-Cre::cdFmr1 KO mice (Figures 7I-L), consistent with the selective requirement of Fmr1 in parvocellular OT neurons for peer-peer social reward learning. Furthermore, since Calb1 is also expressed in the Purkinje cells of the cerebellum, these results suggest that previous reports implicating the cerebellum in the pathogenesis of social impairments in the Tsc1 KO model of ASD (Kelly et al., 2020; Tsai et al., 2012) do not generalize across ASD etiologies. Taken together, these anatomical, electrophysiological, and functional genomic data demonstrate that social reward learning deficits seen in constitutive Fmr1 KO mice are not the consequence of hysteresis, but rather the ongoing requirement of Fmr1 in parvocellular, but not magnocellular OT neurons, and directly implicate these neurons in the pathogenesis of a peer-peer social reward learning behavior that has clear face validity for social impairments that define ASD.
ASD risk and FMRP target genes are enriched in parvocellular OT neurons
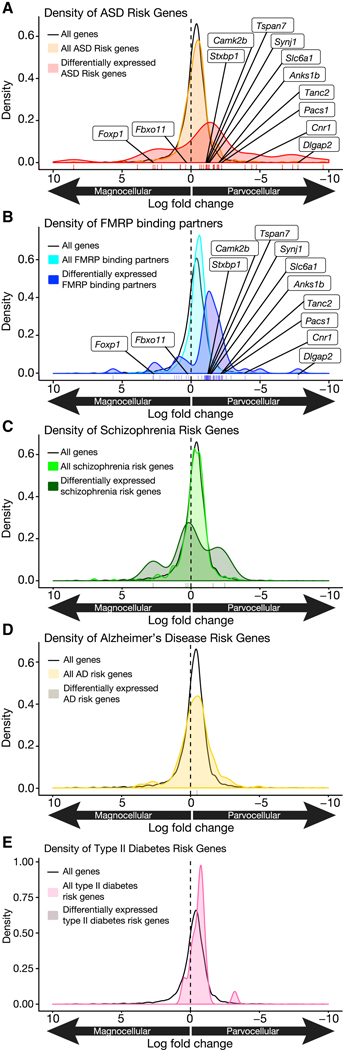
Having identified a role for Fmr1 in parvocellular OT neuronal regulation of domain specific social reward learning, we next sought to determine whether this pathogenic mechanism in Fragile X may generalize to other ASD etiologies. Given the diversity of ASD risk gene functions, we hypothesized that the pathogenic overlap might occur at the level of selective vulnerability of parvocellular OT neurons to genetic injury, even if cellular phenotypes are not shared across all ASD etiologies. If so, we predict that this overlap would be evident in the transcriptional profile of these cells. In order to test this hypothesis, we performed gene set enrichment analysis for ASD risk genes (Abrahams et al., 2013) and FMRP binding partners (Driesche et al., 2019). In both cases, genes were significantly enriched in parvocellular relative to magnocellular neurons (Figures 8A,B; Tables S3 and S4). Additionally, 10 of the 12 differentially expressed genes in the intersection of the 2 groups (Dlgap2, Cnr1, Pacs1, Tanc2, Anks1b, Slc6a1, Synj1, Tspan7, Camk2b, Stxbp1, Fbxo11, Foxp1; Figures 8A,B, individually labeled genes; Figure S7) were also enriched in parvocellular cells. Since OT neurons in both the rostral and caudal PVN (Figure S7) express FMRP, these results indicate that subtype specific expression of FMRP itself is unlikely to account for selective enrichment of ASD risk genes and FMRP binding partners in parvocellular OT neurons.
Figure 8. ASD risk and FMRP target genes are enriched in parvocellular OT neurons.
(A) Density of the log2 fold change for (black) all expressed genes, (orange) all ASD risk genes, and (red) all significantly differentially expressed (FDR = 0.1%) ASD risk genes. ASD risk genes (orange vs, black; p = 0.0009279; one-sided Wilcoxon Rank Sum Test) and differentially expressed ASD risk genes (red vs black; p = 8.589e−9) are enriched in parvocellular OT neurons. Individually labeled genes are the union of ASD risk genes and FMRP binding partners that are differentially expressed between neuron types. (B) Density of the log2 fold change for (black) all expressed genes, (light blue) all FMRP binding partners, and (dark blue) all significantly differentially expressed (FDR 0.1%) FMRP binding partners. FMRP binding partners (light blue vs black; p = 6.83e−16; one-sided Wilcoxon Rank Sum Test) and differentially expressed FMRP binding partners (dark blue vs black; p = 1.751e−9) are enriched in parvocellular OT neurons. Individually labeled genes are the union of ASD risk genes and FMRP binding partners that are differentially expressed between neuron types (FDR 0.1%). (C) Density of the log2 fold change for (black) all expressed genes, (light green) all schizophrenia risk genes, and (dark green) all significantly differentially expressed (FDR 0.1%) schizophrenia risk genes. Schizophrenia risk genes are not enriched in magnocellular or parvocellular OT neurons (light green vs black; p = 0.98) and differentially expressed schizophrenia risk genes (dark green vs black; p = 0.21; one-sided Wilcoxon Rank Sum Test) are not enriched in parvocellular OT neurons. (D) Density of the log2 fold change for (black) all expressed genes, (light yellow) all Alzheimer’s disease (AD) risk genes, and (dark yellow) all significantly differentially expressed (FDR 0.1%) AD risk genes. AD risk genes are not enriched in magnocellular or parvocellular OT neurons (p = 0.99). (E) Density of the log2 fold change for (black) all expressed genes, (light pink) all type II diabetes risk genes, and (dark magenta) all significantly differentially expressed (FDR 0.1%) type II diabetes risk genes. Type II diabetes risk genes are not enriched in magnocellular or parvocellular OT neurons (p = 0.33). Vertical tick marks below density plots indicate the individual differentially expressed ASD risk genes (A), FMRP binding partners (B), schizophrenia risk genes (C), and the single differentially expressed AD risk gene (D). Note the lack of differentially expressed type II diabetes risk genes (E). See also Figure S7, and Tables S1, S3, S4.
Next we repeated this analysis on risk genes for Schizophrenia, Alzheimer’s Disease, and Type II Diabetes (Kanehisa and Goto, 2000; Scelsi et al., 2018; Sullivan et al., 2012). While a handful of Schizophrenia risk genes were differentially expressed in OT neuronal subtypes, these were not significantly enriched in either magnocellular or parvocellular OT neurons (p = 0.98, Figure 8C). Moreover, Alzheimer’s Disease had only a single differentially expressed gene (Lhx5), and Type II Diabetes risk genes were completely absent amongst differentially expressed genes in OT cells (Figure 8D,E). These results support the view that cell type specific vulnerability of parvocellular OT neurons to genetic injury generalizes across multiple ASD etiologies, but is not apparent in unrelated genetic disorders that are not defined by impairments in social behavior.
DISCUSSION
In the current studies, we took a comprehensive approach to provide validation of 8 of the 14 characteristics identified as distinguishing between magnocellular and parvocellular OT neuron, having discovered 2 of these characteristic features here (Figure S8, Center; Figures 1–4, 8). To date, OT’s role as a regulator of social behaviors has largely focused on reproductive behaviors such as pair bonding and parental behaviors. Here we provide evidence that these filial social behaviors (Figure S8, Left) are governed by mechanisms distinct from those governing peer-peer social interactions (Figure S8, Right). Specifically, peer-peer, but not filial social attachments are disrupted in the Fmr1 KO (Figure 5). Furthermore, in parvocellular OT neurons (Figure 6), but not magnocellular OT neurons (Figure 4) Fmr1 is required for peer-peer social reward learning (Figure 7). Taken together these results delineate a novel example of parallel processing, which enables a single peptide to encode a wide diversity of social behaviors, significantly expanding the network capabilities of the social brain (Figure S8).
For more than a decade, research into the pathogenesis of ASD has largely focused on understanding unifying biochemical mechanisms based on the hypothesis that ASD is a ‘synaptopathy’ (Bear et al., 2008; Dölen and Bear, 2009; Grabrucker et al., 2011). One implicit assumption of this hypothesis is that identification of synaptic phenotypes anywhere in the brain provides insight into disease pathogenesis (Dölen et al., 2007). Our current findings provide evidence that the pathogenesis of social impairments in ASD can be better understood using the framework of ‘pathoclisis’ (Klatzo, 2003), whereby subtype-specific differences render a specific neuronal population vulnerable to genetic lesion. Furthermore, by examining mechanisms at the intersection of both construct and face validity (Dölen and Sahin, 2016; Dölen et al., 2015), the current studies have identified a critical role of parvocellular OT in the pathogenesis of ASD. Importantly, this mechanistic link has been overlooked by genetic association studies, underscoring the importance of taking a neurobiological approach to the disease that does not forfeit etiological considerations to genetics alone.
Clinical trials examining core symptoms of ASD have failed to consistently demonstrate efficacy of intranasal OT treatment (Anagnostou et al., 2012; Dadds et al., 2014; Guastella et al., 2015; Hollander et al., 2007), likely because intranasal OT targets magnocellular but not parvocellular OT neurons (Dölen, 2015; Leng and Ludwig, 2016). Furthermore, despite the fact that much of the understanding of OT as a regulator of social behaviors is based on the importance of magnocellular OT neurons for pair bonding, we found no evidence to support the view that the magnocellular subtype of OT neuron is involved in the pathogenesis of ASD. Rather, our findings implicate the parvocellular OT neurons as the appropriate therapeutic target for correcting social impairments in ASD. Moreover, the only study to date that has examined the therapeutic potential of intranasal OT specifically in the Fragile X population (Hall et al., 2012), found a correlation between improvements in a measure of social anxiety (a syndromic symptom that is seen more frequently in Fragile X than ASD (Thurman et al., 2014)) and decreased serum cortisol levels (Hall et al., 2012). Since intranasally delivered OT also reaches systemic OT receptors in the adrenal cortex (Hammock and Levitt, 2013), whose activation suppresses cortisol release (Chiodera and Legros, 1981; Legros et al., 1984; de Oliveira et al., 2007; Stachowiak et al., 1995), these anxiolytic effects likely reflect indirect activity of OT in the periphery.
Future studies dissecting input and output connections, defining developmental programs, and computational modeling of this circuitry will be enabled by the technical achievements of this work, not the least of which is the identification of novel molecular markers, which can be used in combination with our OT-Flp driver line to specify (by multiple feature selection) circuit elements for functional genomics (including optogenetics and calcium imaging). Multiple feature selection-based approaches will be particularly useful in the study of OTergic projections to brain regions such as the VTA, which are innervated by mixed populations of magnocellular and parvocellular neurons, as well as to enable studies to strengthen the mechanistic link between cellular and behavioral consequences of Fmr1 deletion. Ultimately, such studies will lay the foundation for understanding the universal motifs governing the brain circuit organization of sociality.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Gül Dölen (gul@jhu.edu).
Materials Availability
OT-2A-Flp mice will be made available with a materials transfer agreement.
Data and Code Availability
All data are archived in the Gene Expression Omnibus (GEO Accession number GSE147092). Raw and processed scRNA-Seq data and code to reproduce the complete analysis are also publicly available at https://github.com/gofflab/OT_neuron_study_2020.
EXPERIMENTAL MODEL AND SUBJECTS DETAILS
Animals
Experiments were conducted in 3 – 7 week-old male mice. Wild-type C57BL6/J (Stock # 000664; 3 – 4 weeks old) and Calb1-IRES-Cre (Stock # 028532) mice were obtained from Jackson Laboratories. Flp-dependent GFP reporter mice (Stock # 32038) were obtained from the Mutant Mouse Resource Center. Transgenic mice were bred in-house and weaned at 3 weeks of age. OT-2A-Flp-optimized (OT-2A-Flp) knockin mice were generated by Cyagen Biosciences (Santa Clara, CA) (Figure S1) (Nardou et al., 2019). Similar to the strategy employed by the OT-IRES-Cre knockin mouse (Wu et al., 2012) the stop codon in exon 3 of the endogenous OT gene was replaced with a 2A-Flp-optimized (FlpO) construct. The co-translational cleavage 2A peptide strategy (known to have a more robust expression of its downstream gene compared to IRES (Furler et al., 2001)) has been used to separate Flp and OT, since Flp is directed to the nucleus, while OT is directed to the cytoplasm. OT-2A-Flp mice were kept on a C57BL/6 background. Despite these designed-in safeguards against disrupting native OT function, mice homozygous for the OT-2A-Flp knockin, like the OT-IRES-Cre mice, exhibit an impaired lactation phenotype. The mice are otherwise phenotypically normal. Therefore, in order to control for possible effects of knockin on Oxt gene function, all OT-Flp mice used in these experiments were heterozygous, leaving one WT copy of the Oxt gene.
To generate hemizygous Fmr1 KO males for behavior, homozygous Fmr1 KO females were bred to hemizygous Fmr1 KO males. The same strategy was used to generate Cre-dependent conditional Fmr1 KO (cdFmr1 KO) males. To generate hemizygous Fmr1 KO males for cocaine CPP, homozygous Fmr1 KO females were bred to OT-2A-Flp::fdGFP homozygous males. To generate OT-2A-Flp::Ai9 and OT-2A-Flp::cdFmr1 KO mice, homozygous OT-2A-Flp males were crossed with homozygous Ai9 or cdFmr1 KO females respectively. To generate Calb1-Cre::cdFmr1 KO males for behavior, homozygous Calb1-IRES-Cre males were crossed with homozygous cdFmr1 KO females. Homozygous or heterozygous OT-2A-Flp and fdGFP mice were crossed to generate OT-2A-Flp::fdGFP mice. All mice were maintained on a on a 12:12h natural light dark cycle, starting at 7:30am with food and water ad libitum. All procedures were conducted in accordance with protocols approved by the Johns Hopkins Animal Care and Use Committee
Genotyping
OT-2A-Flp knockin mice, fdGFP, Fmr1 KO, and cdFmr1 KO mice were genotyped using polymerase chain reaction (PCR) analysis of DNA isolated from tail snips taken before weaning. For OT-2A-Flp mice, the wild-type allele was identified by a 622-bp PCR product and the mutant allele by a 308-bp PCR product using the primers mOxtR1: TCCGACAATTAGACACCAGTCAA; mOxtF1: CTACCTGAGCAGCTACATCAACAG; mOxtF2: AGGGCTTTGGGAAGTGTTAGGCT. The reactions were run under the following conditions: 94°C × 3 min, (94°C × 30 s, 60°C × 35 s, 72°C × 35 s) × 38 cycles, 72°C × 5 min. For fdGFP mice, the wild-type allele was identified by a 603-bp PCR product, and the mutant allele by a 320-bp PCR product using the following primers; mutant: CCA GGC GGG CCA TTT ACC GTA AG; common: AAA GTC GCT CTG AGT TGT TAT; wild-type: GGA GCG GGA GAA ATG GAT ATG. The reactions were run using the Gt(ROSA)26Sortm(CAG) protocol published by Jackson Laboratories. For the Fmr1 KO mice, the wild-type allele was identified by a 131-bp PCR product, and the mutant allele by a 400-bp PCR product using the following primers; wild-type: TGT GAT AGA ATA TGC AGC ATG TGA; common: CTT CTG GCA CCT CCA GCT T; mutant: CAC GAG ACT AGT GAG ACG TG. The reactions were run using the Fmr1tm1Cgr protocol published by Jackson Laboratories. For cdFmr1 KO mice, the wild-type allele was identified by a 120-bp PCR product, and the mutant allele was detected by a 220-bp PCR product using the following primers: CCC ACT GGG AGA GGA TTA TTT GGG and GTT GAG CGG CCG AGT TTG TGA G. Reactions were run under the following conditions: 95°C × 3 min, (95°C × 30 s, 55°C × 30 s, 72°C × 60 s) × 35 cycles, 72°C × 7 min.
METHOD DETAILS
Social Conditioned Place Preference
The sCPP assay (Figure S4) was conducted as previously described (Nardou et al., 2019). Briefly, animals were weaned and socially housed (3–5 same sex cage mates based on unpublished observations that group number does not impact sCPP magnitude within this range) in a cage containing corncob bedding (Anderson Cob, 1/4” cob or 1/8” cob; Animal Specialties and Provisions). At approximately 6 weeks of age, animals were placed individually in one of 5open field activity chambers (ENV-510, Med Associates) employing infrared beams and a software interface (Activity Monitor, Med Associates) to monitor mouse position. The chamber was partitioned into two equally sized zones using a clear Plexiglas wall with a hole at floor level to allow the mouse to pass freely between zones. Each zone contained one type of novel bedding (Alpha-Dri; Animal Specialties and Provisions, Kaytee Soft Granule; Petco, Anderson Cob, 1/4” cob; Animal Specialties and Provisions, or Aspen Chip; Northeastern Products). To establish each animal’s baseline preference for the bedding cues, the amount of time each mouse spent exploring each zone was recorded during a 30 min pre-conditioning trial. Immediately after this trial, mice received social conditioning with cage mates for 24 hr on one type of bedding used in the pre-conditioning trial, followed by 24 hr isolation conditioning on the other bedding used in the pre-conditioning trial. Immediately following isolation conditioning, a 30 min post-conditioning trial was conducted to establish preference for the two conditioned cues. Note that clean bedding was used for each phase of testing. Chamber assignments were counterbalanced for side and bedding cues. Exclusion criteria for this behavior were defined as a pre-conditioning preference score of > 1.5 or < 0.5. Experimental conditions were compared using normalized social preference scores , and subtracted social preference scores (Dölen et al., 2013; Nardou et al., 2019).
Cocaine Conditioned Place Preference
The protocol for cocaine CPP was conducted as previously described (Nardou et al., 2019). Experiments were performed in the same open field activity chambers as sCPP using an identical configuration. After 3 days of habituation to i.p. saline injections in the home cage, the pre-conditioning trial was performed as stated above. After 24 h, mice received an i.p. injection of cocaine (20 mg/kg) immediately followed by 30 min conditioning on one bedding (Soft Granule or Alpha Dri), which was randomly assigned in a counterbalanced fashion. A second 30 min conditioning session was conducted 24 h later on the other bedding after an i.p. injection of saline (equal volume to cocaine). A 30 min post-conditioning test session was conducted 24 h later to determine each mouse’s preference for the cocaine versus saline associated beddings.
Alloparent Conditioned Place Preference
The alloparent CPP assay was adapted from a previously used assay (Fang and Wang, 2017), and experiments were conducted in the same open field activity chambers using an identical configuration to the sCPP assay. To generate familiar alloparent stimulus animals, adult (> 9 weeks old) virgin wild type C57BL6/J females were pair housed with pregnant WT or Fmr1 KO females. This housing configuration remained intact following the birth of WT or Fmr1 KO pups until alloparent CPP testing began. Alloparent conditioning was conducted as follows. Beginning at postnatal day 19–20, and continuing until each male littermate was tested, male mice were individually weaned from their home cage and placed directly in an activity chamber with two novel beddings. To establish each animal’s baseline preference for the novel bedding cues, the amount spent exploring each zone of the activity chamber was recorded during a 30 min pre-conditioning trial. Immediately after this trial, mice (postnatal day 19–24) individually received alloparent conditioning with their virgin female alloparent for 24 hr on one type of bedding, followed by 24 hr isolation conditioning on the other bedding. Immediately following the isolation conditioning, a 30 min post-conditioning trial was conducted to establish preference for the two conditioned cues. This process was repeated individually until each male littermate had been tested. Identical to the methods used for sCPP, chamber assignments were counterbalanced for side and bedding cues. Exclusion criteria for this behavior were defined as a pre-conditioning preference score of > 1.5 or < 0.5. Experimental conditions were compared using normalized social preference scores , and subtracted social preference scores (Dölen et al., 2013; Nardou et al., 2019).
Stereotaxic and tail-vein injections
All stereotaxic injections into the nucleus accumbens (NAc; distance from bregma: anterior +1.54 mm; lateral ±1.065 mm; ventral - 4.1 mm)) were performed under general ketamine-medetomidine anesthesia using a stereotaxic instrument (David Kopf). For electrophysiological analysis of NAc-projecting OT neurons, a small volume (~;30 nl) of diluted Rtb solution (1:4; Lumafluor; Red Retrobeads) was injected unilaterally or bilaterally into NAc core at a slow rate (20 nl/min) using a syringe pump (Harvard Apparatus, MA). For conditional knockdown of Fmr1, 1 μl viral suspension (0.1 μl/min; CAV2-GFP, CAV2-Cre-GFP, rgAAV-fDIO-Cre-GFP, or rgAAV-fDIO-Cre-HA) was injected bilaterally into NAc 7 days before behavioral testing. CAV2-GFP and CAV2-Cre-GFP viruses were obtained from Plateforme de Vectorologie de Montpellier. The fDIO-Cre-GFP plasmid was a gift of Dr. Bo Li and Dr. Linda Van Aelst and viral packaging was conducted by Stanford University’s Neuroscience Gene Vector and Virus Core. rgAAV-fDIO-Cre-HA was obtained from Addgene (Catalog # 121675-AAVrg). To confirm viral-mediated Cre function, 1 μl CAV2-Cre-GFP or 1.5 μl rgAAV-fDIO-Cre-HA was injected into NAc of Ai9 (tdTomato Cre-reporter) or OT-2A-Flp::Ai9 mice respectively. After all injections, the injection pipette was left in place for at least 5 min prior to removal from the brain. Injection sites were confirmed post-hoc by preparing sections (20–50 μm) containing the NAc. For i.v. labeling of magnocellular OT neurons with FG, 1 or 2 lateral tail-vein injections of 15 μl 4% FG (Fluorochrome) diluted in ~;100 μl sterile saline were performed using a 1 ml syringe and 25-gauge needle at least 24 hours prior to sacrifice. To facilitate i.v. injections, mice were placed under a warm lamp for 3–5 min and were briefly restrained using a tail vein restrainer (TV-150; Braintree Scientific). Prior to injection, the tail was rubbed firmly with 70% ethanol to clean the injection area and enhance visibility of the tail vein.
Electrophysiology
Using a Leica VT 1200s vibrating microtome, coronal sections (250 μm) containing the paraventricular nucleus were cut in an ice-cold sucrose solution containing the following (in mM); Sucrose 228; NaHCO3 26; Glucose 11; KCl 2.5; NaH2PO4 1; MgSO4 7; CaCl2 0.5. Immediately after cutting, slices were transferred into 32˚C aCSF containing (in mM); NaCl 119; KCl 2.5; NaH2PO4 1; NaHCO3 26.2; Glucose 11; MgCl2 1.3; CaCl2 2.5. Slices were allowed to recover at 29˚C for at least 1 hour prior to recording. GFP+ and RtB+ neurons were visualized using 470 nm and 535 nm LEDs respectively (Cool LED). LED excitation was delivered through a 40x microscope objective, (Olympus) and fluorescence was detected using a camera and visualized using SliceScope Pro software (Scientifica). Recordings were conducted in current clamp using patch pipettes (2–4 MΩ) filled with internal solution containing the following (in mM: 130 K-Gluconate, 10 HEPES, 1 NaCl, 1 CaCl2, 10 EGTA, 1 MgCl, 2 Mg-ATP, 0.5 Na-GTP, and 0.125 % Neurobiotin) and the liquid junction potential (13 mV) was corrected after recording. Recording was discontinued for neurons that failed to maintain a hyperpolarized resting membrane potential in the absence of current injection, and neurons that failed to produce APs > 50 mV were excluded from further analysis. Data was acquired and analyzed using the Recording Artist plugin in Igor Pro and custom software written in MATLAB (Mathworks). AP initiation was determined as the first point where the membrane potential accelerated past 40 ms/s2. This threshold was confirmed with comparison to manual determination of AP initiation. AP peaks were identified prior to correction of the liquid junction potential by finding local maxima with a minimum peak of 0 mV and a minimum separation of 5 ms and absolute membrane potential values were subsequently corrected. Nine mice, with a maximum of 4 neurons per animal were used to cross-validate FG and electrophysiological differentiation of magnocellular and parvocellular neurons. Neurons were targeted and recorded blind to FG labeling status. Six mice, with a maximum of 2 neurons per animal were used for electrophysiological characterization of OTergic projections to NAc.
Cell Isolation, Enrichment, and cDNA Library Preparation
OT-2A-Flp::fdGFP mice were weaned at P21 and given i.v. injections of FluoroGold on that and the following day. At P23 mice were sacrificed. Brains were rapidly removed and 250 μm thick coronal slices (n = 7 mice) containing the paraventricular nucleus of the hypothalamus were sectioned using a Leica VT-1200s vibrating microtome in ice-cold ACSF solution containing the following (in mM): 124 NaCl; 2.5 KCl; 1.2 NaH2PO4; 24 NaHCO3; 5 HEPES; 13 glucose; 2 MgSO4’; 2 CaCl2, oxygenated with carbogen gas (95% O2 and 5% CO2) to pH 7.3 – 7.4. To microdissect the PVN, slices were placed in a petri-dish containing ice-cold, oxygenated ACSF solution and the PVN was identified using the 3rd ventricle and other structural markers. Following dissection, tissue was placed into 2.6 mL equilibrated Papain DNase-I solution. The dissociation protocol was similar to Chevée et al., 2018, itself an adaptation from the trehalose-enhanced neuronal isolation protocol (Saxena et al., 2012) using the Worthington Papain Dissociation System (Worthington Biochemical Corporation). The following modifications from Chevée et al., 2018 were made: a single low speed centrifugation (300xg for 5 min) was performed after dissociation, and the pellet was resuspended in 250 μL media (DMEM, 5% trehalose (w/v), 25 μM AP-V, 0.4 mM kynurenic acid, 6 μL of 40 U/μl RNase inhibitor) and 250 uL of EBSS#2 (EBSS, 25 mM AP-V, 100 mM Kynurenic acid, ovomucoid protease inhibitor with BSA, DNase-I, 5% w/v Trehalose, 40 U/μl RNase inhibitor) at room temperature. Following resuspension, single-cell suspensions were introduced into a FACS machine (Beckman Coulter MoFlo Cell Sorter).
Neurons were sorted based on fluorescence (GFP+/FG- or GFP+/FG+) directly into individual wells of a 96-well plate containing 2 μL Smart-Seq2 lysis buffer + RNAase inhibitor, 1μL oligo-dT primer, and 1 μL dNTPs(Picelli et al., 2014). Three plates were collected across 2 separate dates (Plate 1: n = 1 mouse, 32 GFP+/FG- neurons and 47 GFP+/FG+ neurons; Plate 2: n = 1 mouse, 32 GFP+/FG- neurons and 33 GFP+/FG+ neurons; Plate 3: n = 5 mice, 25 GFP+/FG-neurons and 48 GFP+/FG+ neurons). Following the sort, plates were briefly spun-down in a table-top microcentrifuge and immediately placed on dry ice. Single-cell lysates were kept at - 80°C until cDNA library preparation. Library preparation and amplification of single-cell samples was performed using a modified version of the Smart-Seq2 protocol (Chevée et al., 2018).
Individual libraries were quality controlled, pooled, and sequenced on two lanes of an Illumina HiSeq 2500 sequencer to an average depth of 1,424,499 ±77,731 paired-end 50bp reads per neuron. Reads were aligned to the reference mouse genome (mm10) using the Hisat 2 aligner and transcript abundances were estimated using CuffNorm (Qiu et al., 2017) against the mouse GENCODE reference transcriptome (vM8).(Qiu et al., 2017) Relative abundances were converted to absolute estimates of gene expression using the Monocle2 CENSUS approach and used as input for the dpFeature workflow (Qiu et al., 2017). 146 neurons passed our quality assessment, and we identified a total of 10,172 genes with detectable expression in at least 5 neurons. For those cells that passed QC, the mean mRNA-copies per cell was 27,551 and mean number of genes detected across all cells was 3,713. A Uniform Manifold Approximation and Projection (UMAP)(Becht et al., 2019) embedding was obtained by collapsing the top 50 principal components learned on the top 1000 genes with highest residuals to the Monocle model fit. All data are archived in the Gene Expression Omnibus (GEO Accession number GSE147092). Raw and processed scRNA-Seq data and code to reproduce the complete analysis are also publicly available at https://github.com/gofflab/OT_neuron_study_2020.
In situ hybridization chain reaction
Experiments were performed on 3 week-old FG injected male wild-type C57BL6/J mice obtained from Jackson Laboratories. Brains were extracted and immediately frozen in OCT with liquid nitrogen. Brains were sectioned on a cryostat at 7 μm. Serial sections were collected onto charged slides, briefly washed in PBS and then fixed in 4% PFA for 10 mins. Slides were then washed twice in PBS, transferred to 70% ethanol, and kept at 4˚C. In situ HCR v3.0 was performed using the protocols previously detailed (Choi et al., 2018). Probe hybridization and amplification steps were both performed overnight (12–16 hours). In situ probes (Table S2) were design using publicly available software (Raj et al., 2008).
Immunohistochemistry
Prior to immunostaining, sections were mounted on slides and rinsed 4 × 10 min in PBS followed by 1 hr in blocking solution (0.5% Triton X-100, 10% horse serum, 0.2% bovine serum albumin in PBS). Antibodies were diluted in PBS containing the following: 0.5% Triton X-100, 1% horse serum, 0.2% bovine serum albumin. Primary antibodies were applied overnight at room temperature (RT), and after rinsing slides in PBS, secondary antibodies were applied for 2 hr at RT. With the exception of FMRP immunohistochemistry, OT neurons were labeled using the antibody PS38 (gift of Dr. Harold Gainer; 1:150). PS38 was detected using Alexa 488, (magnocellular and parvocellular distribution; Life Technologies; donkey anti-mouse; 1:1000) Alexa 647 (GFP or tdTomato colocalization; Life Technologies; goat-anti mouse or donkey-anti mouse respectively; 1:1000), or Alexa 350 (RtB colocalization; Life Technologies; goat-anti mouse; 1:200). FluoroGold was labeled with an anti-fluorogold antibody, (Fluorochrome; 1:100), and detected using Alexa 647 (Life Technologies; donkey anti-rabbit; 1:1000). This approach was chosen instead of visualizing the native FG fluorescence to maximize the detectable signal with our microscope configuration. GFP signal was amplified using the anti-GFP antibody, ab13970, (ABCAM; 1:1000) and visualized using Alexa 488 (Life Technologies; goat-anti chicken; 1:1000). tdTomato signal was amplified using the anti-mCherry antibody AB0040 (SICGEN; 1:4000) and visualized using Alexa 555 (Life Technologies; donkey-anti goat; 1:1000). For post-hoc identification of electrophysiologically recorded neurons, Neurobiotin was detected using streptavidin-conjugated Alexa 350 (Life Technologies; 1:800).
FMRP co-labeling experiments were conducted as described above with the following exceptions. After rinsing in PBS, slides were transferred to a hot (90°C) sodium citrate (10 mM Sodium Citrate, 0.05% Tween 20, pH 6.0) bath for antigen retrieval (30 min) followed by 1hr in blocking solution at RT. Sections were labeled with anti-FMRP 2F5–1 ((Gabel et al., 2004) Developmental Studies Hybridoma Bank; 1:1) which was detected using Alexa 647 (Jackson ImmunoResearch; goat anti-mouse IgG2b; 1:500). OT neurons were labeled with the anti-OT antibody VA10 (Altstein and Gainer, 1988) (gift of Dr. Harold Gainer; 1:1000) and detected using Alexa 488 (Life Technologies; donkey anti-rabbit; 1:1000).
Histology
Following transcardial perfusion with 1M PBS and 10% formalin, brains were kept in formalin at 4˚C overnight and then transferred to PBS. To prepare them for re-sectioning, acute brain slices used for electrophysiology were placed in 10% formalin immediately after recording, and then transferred to PBS. For all experiments in which fluorescent colocalization was analyzed, serial brain sections (20 μm thickness) were cut using a cryostat following cryoprotection of tissue with a 30% sucrose solution containing 0.01% azide. Following immunostaining, images were acquired using an EVOS or Olympus BX41 microscope with 4x, 10x, and 40x objectives and analyzed using ImageJ.
QUANTIFICATION AND STATISTICAL ANALYSIS
All behavioral and electrophysiological analyses were performed with MATLAB (Mathworks), Prism (GraphPad), or R (The R Project). Electrophysiological comparisons were made using a two-tailed Mann-Whitney U Test or Wilcoxon Signed-Rank Test for unpaired and paired comparisons, respectively. p < 0.05 was considered significant. Behavioral comparisons were made using a two-tailed, Students t-test (paired or unpaired and with or without Welch’s correction as appropriate per the Central Limit Theorem), with p < 0.05 considered significant. Statistical parameters can be found in figure legends. All data were subjected to the Shapiro-Wilk normality test and F test to compare variances. For k-means clustering analysis, two clusters were specified, as this number of clusters was indicated by the elbow method (Figure S2). Electrophysiological feature values used for the clustering analysis were z-scored across all neurons, while the clustering algorithm was trained only on the values from neurons from Fig. 1. Calb1-Cre::cdFmr1KO experiments were done blind, as were a subset of OT-Flp::cdFmr1KO experiments. For electrophysiological analysis, neurons were targeted for recording blind to FG labeling status.
All scRNAseq analysis was performed in R/Bioconductor following alignment with Hisat2. Log2 expression estimates (with a pseudocount of 1) of high-variance genes were used as input for PCA analysis and UMAP clustering of individual cells. After cluster assignment, differential expression testing was performed using the Monocle2 VGAM model comparison test(Trapnell et al., 2014) (FDR 0.1%, Monocle2 test, Benjamini-Hochberg corrected). Gene set enrichment was computed using the hypergeometric test unless stated otherwise. Using the full dataset with a q value of 0.001 (0.1 % FDR), Post hoc power calculations made for observing 181 differentially expressed genes. Based on these calculations, there is 99.84% power (probability of true positives) to observe 181 genes with a significant two-fold change (log2 Magno/Parvo ≥1 or log2 Magno/Parvo ≤−1) in gene expression. Given that the observed log2-fold changes (magnocellular vs. parvocellular) for the mean expression of Calb1, Kcnmb4, Reln, Cnr1 are 6.59, 2.91, −7.38, and −3.92 respectively, it was determined that there was more than sufficient (greater than 99.84%) power for this analysis. Limiting the power calculation to a balanced design by using only enough magnocellular cells to match the reduced number of parvocellular cells (thereby disregarding the observed ratio of cell types), results in 96.94% power for and FDR of 0.001 to observe 181 genes with a significant two-fold change (log2 Magno/Parvo ≥1 or log2 Magno/Parvo ≤−1). Code to reproduce the complete analysis is publicly available at https://github.com/gofflab/OT_neuron_study_2020.
Supplementary Material
Differentially expressed ASD risk genes. Related to Figure 8. (Excel File)
Differentially expressed FMRP binding targets. Related to Figure 8. (Excel File)
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-OT neurophysin | Whitnall et al., 1985 Ben-Barak et al., 1985; gift from Harold Gainer, PhD | PS38 |
| Alexa 488 donkey anti-mouse | Life Technologies | A-21202 |
| Alexa 647 donkey anti-mouse | Life Technologies | A-31571 |
| Alexa 647 goat anti-mouse | Life Technologies | A-21235 |
| Alexa 350 goat anti-mouse | Life Technologies | A-11045 |
| Rabbit anti-FluoroGold | Fluorochrome | NA |
| Alexa 647 donkey anti-rabbit | Life Technologies | A-31573 |
| Chicken anti-GFP | ABCAM | Ab13970 |
| Alexa 488 goat anti-chicken | Life Technologies | A-11039 |
| Goat anti-mCherry | SICGEN | AB0040 |
| Alexa 555 donkey anti-goat | Life Technologies | A-21432 |
| Mouse anti-FMRP | Developmental Studies Hybridoma Bank | 2F5-1 |
| Alexa 647 goat anti-mouse IgG2b | Jackson ImmunoResearch | 115-607-187; RRID: AB 2632546 |
| Rabbit anti-OT | Altstein and Gainer, 1988; gift from Harold Gainer, PhD | VA10 |
| Alexa 488 donkey anti-rabbit | Life Technologies | A-21206 |
| Bacterial and Virus Strains | ||
| rgAAV-EF1 a-fDIO-Cre | Schneeberger et al., 2019 | Addgene 121675- AAVrg |
| rgAAV-CAG-fDIO-GFP-Cre | Stanford University Neuroscience Gene Vector and Virus Core | N/A |
| CAV-2-GFP | Plateforme de Vectorologie de Montpellier | N/A |
| CAV-2-Cre-GFP | Plateforme de Vectorologie de Montpellier | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Neurobiotin | Vector Laboratories | SP-1120 |
| Alexa 350 streptavidin conjugate | Life Technologies | S11249 |
| FluoroGold | Fluorochrome | N/A |
| Red Retrobeads | Lumafluor | N/A |
| 4-Aminopyridine | Sigma-Aldrich | 275875 |
| Cocaine hydrochloride | Sigma-Aldrich | C5776 |
| Critical Commercial Assays | ||
| Worthington Papain Dissociation System | Worthington Biochemical Corporation | Cat#LK003182 |
| Illumina TruSeq kit | Illumina | Cat#RS-122-200 |
| Deposited Data | ||
| Accession number GSE147092 | GEO | |
| Experimental Models: Organisms/Strains | ||
| C57BL6/J | Jackson Laboratories | Stock # 000664 |
| Calbl-IRES-Cre | Jackson Laboratories | Stock # 028532 |
| Flp-dependent GFP reporter mice | Mutant Mouse Resource Center | Stock # 32038 |
| Ai9 Cre-dependent TdTomato reporter mice | Jackson Laboratories | Stock # 007909 |
| Constitutive Fmr1 knockout | Jackson Laboratories | Stock # 003025 |
| Conditional Fmr1 knockout | Mientjes et al., 2006 | N/A |
| Oxytocin-2A-Flp-optimized | Nardou et al., 2019 | N/A |
| Oligonucleotides | ||
| see supplemental tables (HCR) | ||
| Recombinant DNA | ||
| fDIO-Cre-GFP | Penzo et al., 2015 | Dr. Bo Li and Dr. Linda Van Aelst |
| Software and Algorithms | ||
| R version 3.5 | The R project | https://www.r-project.org/ |
| MATLAB | Mathworks | https://www.mathworks.com/ |
| Prism 5 | GraphPad | https://www.graphpad.com/ |
| Igor Pro | WaveMetrics | https://www.wavemetrics.com/ |
| Recording Artist (plugin for Igor Pro) | Richard C Gerkin, PhD | https://github.com/rgerkin/recording-artist |
ACKNOWLEDGMENTS
We thank members of the Dölen and Goff labs for comments on the manuscript, Elizabeth Vincent for histological consultation, Ben Emmert and Arjun Raj (UPenn) for assistance with the HCR 3.0 protocol. PS38 and VA10 antibodies were kind gifts from Dr. Harold Gainer. Funding: This work was supported by funds to G.D. (NIMH R56MH115177, NIMH R01MH117127) and L.A.G. (the Chan-Zuckerberg Initiative DAF 2018–183445 and NSF IOS-1656592).
Footnotes
DECLARATION OF INTERESTS
Authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, Menashe I, Wadkins T, Banerjee-Basu S, and Packer A. (2013). SFARI Gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 4, 2–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen-Brady K, Miller J, Matsunami N, Stevens J, Block H, Farley M, Krasny L, Pingree C, Lainhart J, Leppert M, et al. (2009). A high-density SNP genome-wide linkage scan in a large autism extended pedigree. Mol. Psychiatry 14, 590–600. [DOI] [PubMed] [Google Scholar]
- Althammer F, and Grinevich V. (2017). Diversity of oxytocin neurons: beyond magno- and parvocellular cell types? J. Neuroendocrinol 140, 874–888. [DOI] [PubMed] [Google Scholar]
- Altstein M, and Gainer H. (1988). Differential biosynthesis and posttranslational processing of vasopressin and oxytocin in rat brain during embryonic and postnatal development. J. Neurosci 8, 3967–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anagnostou E, Soorya L, Chaplin W, Bartz J, Halpern D, Wasserman S, Wang AT, Pepa L, Tanel N, Kushki A, et al. (2012). Intranasal oxytocin versus placebo in the treatment of adults with autism spectrum disorders: a randomized controlled trial. Mol. Autism 3, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine MW, Langberg T, Schnepel P, and Feldman DE (2019). Increased Excitation-Inhibition Ratio Stabilizes Synapse and Circuit Excitability in Four Autism Mouse Models. Neuron 101, 648–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong WE, Warach S, Hatton GI, and Mcneill TH (1980). Subnuclei in the rat hypothalamic paraventricular nucleus: A cytoarchitectural, horseradish peroxidase and immunocytochemical analysis. Neuroscience 5, 1931–1958. [DOI] [PubMed] [Google Scholar]
- Assaf M, Hyatt CJ, Wong CG, Johnson MR, Schultz RT, Hendler T, and Pearlson GD (2013). Mentalizing and motivation neural function during social interactions in autism spectrum disorders. NeuroImage Clin. 3, 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- August GJ (1983). A genetic marker associated with infantile autism. Am. J. Psychiatry 140, 813. [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, and Rutter M. (1995). Autism as a strongly genetic disorder: evidence from a British twin study. Psychol. Med 25, 63. [DOI] [PubMed] [Google Scholar]
- Bakker C, Verheij C, Willemsen R, van der Helm R, Oerlemans F, Vermey M, Bygrave, a, Hoogeveen A, Reyniers E, De Boulle K, et al. (1994). Fmrl Knockout M ice : A Model to Study Fragile X Mental Retardation. Cell 78, 23–33.8033209 [Google Scholar]
- Bardo MT, and Bevins RA (2000). Conditioned place preference: What does it add to our preclinical understanding of drug reward? Psychopharmacology (Berl). 153, 31–43. [DOI] [PubMed] [Google Scholar]
- Baron D, Holland CM, Carlson K, Wolfrum E, and Thompson BL (2020). Adapting social conditioned place preference for use in young children. Neurobiol. Learn. Mem 172, 107235. [DOI] [PubMed] [Google Scholar]
- Bear MF, Dölen G, Osterweil E, and Nagarajan N. (2008). Fragile X: Translation in action. Neuropsychopharmacology 33, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre C-AA, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2019). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol 37, 38–47. [DOI] [PubMed] [Google Scholar]
- Beier KT, Steinberg EE, Deloach KE, Xie S, Miyamichi K, Schwarz L, Gao XJ, Kremer EJ, Malenka RC, and Luo L. (2015). Circuit Architecture of VTA Dopamine Neurons Revealed by Systematic Input-Output Mapping. Cell 162, 622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Barak Y, Russell JT, Whitnall MH, Ozato K, and Gainer H. (1985). Neurophysin in the hypothalamo-neurohypophysial system. I. Production and characterization of monoclonal antibodies. J. Neurosci 5, 81–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardet M, and Crusio WE (2006). Fmr1 KO Mice as a Possible Model of Autistic Features. Sci. World J 6, 1164–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biag J, Huang Y, Gou L, Hintiryan H, Askarinam A, Hahn JD, Toga AW, and Dong HW (2012). Cyto- and chemoarchitecture of the hypothalamic paraventricular nucleus in the C57BL/6J male mouse: A study of immunostaining and multiple fluorescent tract tracing. J. Comp. Neurol 520, 6–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WT, Friedman E, Jenkins EC, Brooks J, Wisniewski K, Raguthu S, and French JH (1982). Association of fragile X syndrome with autism. Lancet (London, England) 1, 100. [DOI] [PubMed] [Google Scholar]
- Brown WT, Jenkins EC, Cohen IL, Fisch GS, Wolf-Schein EG, Gross A, Waterhouse L, Fein D, Mason-Brothers A, and Ritvo E. (1986). Fragile X and autism: a multicenter survey. Am. J. Med. Genet 23, 341–352. [DOI] [PubMed] [Google Scholar]
- Cascio CJ, Foss-Feig JH, Heacock JL, Newsom CR, Cowan RL, Benningfield MM, Rogers BP, and Cao A. (2012). Response of neural reward regions to food cues in autism spectrum disorders. J. Neurodev. Disord 4, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel M, Morris JF, and Road SP (1988). The neurophysin-containing innervation of the forebrain of the mouse. Neuroscience 24, 937–966. [DOI] [PubMed] [Google Scholar]
- Chen R, Wu X, Jiang L, and Zhang Y. (2017). Single-Cell RNA-Seq Reveals Hypothalamic Cell Diversity. Cell Rep. 18, 3227–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallier C, Kohls G, Troiani V, Brodkin ES, and Schultz RT (2013). The social motivation theory of autism. Trends Cogn. Neurosci 16, 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevée M, Robertson JDJ, Cannon GH, Brown SP, and Goff LA (2018). Variation in Activity State, Axonal Projection, and Position Define the Transcriptional Identity of Individual Neocortical Projection Neurons. Cell Rep. 22, 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodera P, and Legros JJ (1981). Intravenous injection of synthetic oxytocin induces a decrease of cortisol plasma level in normal man. C. R. Seances Soc. Biol. Fil 175, 546–549. [PubMed] [Google Scholar]
- Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, and Pierce NA (2018). Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clipperton-Allen AE, Chen Y, and Page DT (2016). Autism-relevant behaviors are minimally impacted by conditional deletion of Pten in oxytocinergic neurons. Autism Res. 9, 1248–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Jun SB, Jin X, Pham MD, Vogel SS, Lovinger DM, and Costa RM (2013). Concurrent activation of striatal direct and indirect pathways during action initiation. Nature 494, 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadds MR, MacDonald E, Cauchi A, Williams K, Levy F, and Brennan J. (2014). Nasal oxytocin for social deficits in childhood autism: a randomized controlled trial. J. Autism Dev. Disord 44, 521–531. [DOI] [PubMed] [Google Scholar]
- Daigle TL, Madisen L, Hage TA, Valley MT, Knoblich U, Larsen RS, Takeno MM, Huang L, Gu H, Larsen R, et al. (2018). A Suite of Transgenic Driver and Reporter Mouse Lines with Enhanced Brain-Cell-Type Targeting and Functionality. Cell 174, 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiano CR, Aloi J, Treadway M, Bodfish JW, and Dichter GS (2012). Adults with autism spectrum disorders exhibit decreased sensitivity to reward parameters when making effort-based decisions. J. Neurodev. Disord 4, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. (2011). FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayanithi G, Forostyak O, Ueta Y, Verkhratsky A, and Toescu EC (2012). Segregation of calcium signalling mechanisms in magnocellular neurones and terminals. Cell Calcium 51, 293–299. [DOI] [PubMed] [Google Scholar]
- Demurie E, Roeyers H, Baeyens D, and Sonuga-Barke E. (2011). Common alterations in sensitivity to type but not amount of reward in ADHD and autism spectrum disorders. J. Child Psychol. Psychiatry 52, 1164–1173. [DOI] [PubMed] [Google Scholar]
- Diester I, Hefti F, Mansuy I, Pascual-Leone A, Robbins TW, Rubin LL, Sawa A, Wernig M, Dölen G, Hyman SE, et al. (2015). Bridging the Gap between Patients and Models. In Translational Neuroscience: Toward New Therapies. Strüngmann Forum Reports, Nikolich S, and K Hyman SE; Lupp J, ed. (MIT Press; ), pp. 209–244. [PubMed] [Google Scholar]
- Dijkerman HC, and de Haan EHF (2007). Somatosensory processes subserving perception and action. Behav. Brain Sci 30, 189–201. [DOI] [PubMed] [Google Scholar]
- Dölen G. (2015). Oxytocin: Parallel Processing in the Social Brain? J. Neuroendocrinol 27, 516–535. [DOI] [PubMed] [Google Scholar]
- Dölen G, and Bear MF (2009). Fragile x syndrome and autism: From disease model to therapeutic targets. J. Neurodev. Disord 1, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölen G, and Malenka RC (2014). The emerging role of nucleus accumbens oxytocin in social cognition. Biol. Psychiatry 76, 354–355. [DOI] [PubMed] [Google Scholar]
- Dölen G, and Sahin M. (2016). Editorial: Essential pathways and circuits of autism pathogenesis. Front. Neurosci 10, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölen G, Osterweil E, Rao BSS, Smith GB, Auerbach BD, Chattarji S, and Bear MF (2007). Correction of Fragile X Syndrome in Mice. Neuron 56, 955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölen G, Darvishzadeh A, Huang KW, and Malenka RC (2013). Social reward requires coordinated activity of nucleus accumbens oxytocin and serotonin. Nature 501, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölen G, Malenka R, Perlumutter J, Brose N, Frackowiak R, Cuthbert B, Diester I, Mansuy I, Kroker K, Boekers T, et al. (2015). Pathophysiological Toolkit: Genes to Circuits. In Translational Neuroscience: Toward New Therapies. Strüngmann Forum Reports, series editor. Nikolich K and Hyman SE; Lupp J, ed. (MIT Press; ), pp. 139–163. [PubMed] [Google Scholar]
- Dreifuss JJ (1975). A review on neurosecretory granules: their contents and mechanisms of release. Ann. N. Y. Acad. Sci 248, 184–201. [DOI] [PubMed] [Google Scholar]
- Driesche SJ Van, Sawicka K, Zhang C, Hung SKY, Park CY, Fak JJ, Yang C, Darnell RB, and Darnell JC (2019). FMRP binding to a ranked subset of long genes is revealed by coupled CLIP and TRAP in specific neuronal cell types. BioRxiv 762500. [Google Scholar]
- Ebstein RP, Israel S, Lerer E, Uzefovsky F, Shalev I, Gritsenko I, Riebold M, Salomon S, and Yirmiya N. (2009). Arginine vasopressin and oxytocin modulate human social behavior. Ann. N. Y. Acad. Sci 1167, 87–102. [DOI] [PubMed] [Google Scholar]
- Edsinger E, and Dölen G. (2018). A Conserved Role for Serotonergic Neurotransmission in Mediating Social Behavior in Octopus. Curr. Biol 28, 3136–3142.e4. [DOI] [PubMed] [Google Scholar]
- Eliava M, Melchior M, Knobloch-Bollmann HS, Wahis J, da Silva Gouveia M, Tang Y, Ciobanu AC, Triana del Rio R, Roth LC, Althammer F, et al. (2016). A New Population of Parvocellular Oxytocin Neurons Controlling Magnocellular Neuron Activity and Inflammatory Pain Processing. Neuron 89, 1291–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Q, and Wang J. (2017). Place preferences associated with pups or cocaine change the expression of D2R, V1aR and OTR in the NAcc and MeA and the levels of plasma AVP, OT, T and E2 in mandarin vole fathers. Psychoneuroendocrinology 80, 147–154. [DOI] [PubMed] [Google Scholar]
- Folstein S, and Rutter M. (1977). Genetic influences and infantile autism. Nature 265, 726–728. [DOI] [PubMed] [Google Scholar]
- Francis SM, Sagar A, Levin-Decanini T, Liu W, Carter CS, and Jacob S. (2014). Oxytocin and vasopressin systems in genetic syndromes and neurodevelopmental disorders. Brain Res. 1580, 199–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag CM (2007). The genetics of autistic disorders and its clinical relevance: a review of the literature. Mol. Psychiatry 12, 2–22. [DOI] [PubMed] [Google Scholar]
- Furler S, Paterna JC, Weibel M, and Büeler H. (2001). Recombinant AAV vectors containing the foot and mouth disease virus 2A sequence confer efficient bicistronic gene expression in cultured cells and rat substantia nigra neurons. Gene Ther. 8, 864–873. [DOI] [PubMed] [Google Scholar]
- Gabel LA, Won S, Kawai H, McKinney M, Tartakoff AM, and Fallon JR (2004). Visual Experience Regulates Transient Expression and Dendritic Localization of Fragile X Mental Retardation Protein. J. Neurosci 24, 10579–10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabrucker AM, Schmeisser MJ, Schoen M, and Boeckers TM (2011). Postsynaptic ProSAP/Shank scaffolds in the cross-hair of synaptopathies. Trends Cell Biol. 21, 594–603. [DOI] [PubMed] [Google Scholar]
- Guastella AJ, Gray KM, Rinehart NJ, Alvares GA, Tonge BJ, Hickie IB, Keating CM, Cacciotti-Saija C, and Einfeld SL (2015). The effects of a course of intranasal oxytocin on social behaviors in youth diagnosed with autism spectrum disorders: a randomized controlled trial. J. Child Psychol. Psychiatry 56, 444–452. [DOI] [PubMed] [Google Scholar]
- Gurdjian ES (1927). The diencephalon of the albino rat: Studies on the brain of the rat. No. 2. J. Comp. Neurol 43, 1–114. [Google Scholar]
- Haberly LB (2001). Parallel-distributed Processing in Olfactory Cortex: New Insights from Morphological and Physiological Analysis of Neuronal Circuitry. Chem. Senses 26, 551–576. [DOI] [PubMed] [Google Scholar]
- Hall SS, Lightbody AA, McCarthy BE, Parker KJ, and Reiss AL (2012). Effects of intranasal oxytocin on social anxiety in males with fragile X syndrome. Psychoneuroendocrinology 37, 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammock EAD, and Levitt P. (2013). Oxytocin receptor ligand binding in embryonic tissue and postnatal brain development of the C57BL/6J mouse. Front. Behav. Neurosci 7, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman NW, Tasker JG, and Dudek FE (1991). Immunohistochemical differentiation of electrophysiologically defined neuronal populations in the region of the rat hypothalamic paraventricular nucleus. J. Comp. Neurol 307, 405–416. [DOI] [PubMed] [Google Scholar]
- Hollander E, Bartz J, Chaplin W, Phillips A, Sumner J, Soorya L, Anagnostou E, and Wasserman S. (2007). Oxytocin increases retention of social cognition in autism. Biol. Psychiatry 61, 498–503. [DOI] [PubMed] [Google Scholar]
- Hosoya Y, and Matsushita M. (1979). Identification and distribution of the spinal and hypophyseal projection neurons in the paraventricular nucleus of the rat. A light and electron microscopic study with the horseradish peroxidase method. Exp. Brain Res 35, 315–331. [DOI] [PubMed] [Google Scholar]
- Hovey D, Zettergren A, Jonsson L, Melke J, Anckarsäter H, Lichtenstein P, and Westberg L. (2014). Associations between oxytocin-related genes and autistic-like traits. Soc. Neurosci 9, 378–386. [DOI] [PubMed] [Google Scholar]
- Hung LW, Neuner S, Polepalli JS, Beier KT, Wright M, Walsh JJ, Lewis EM, Luo L, Deisseroth K, Dölen G, et al. (2017). Gating of social reward by oxytocin in the ventral tegmental area. Science. 357, 1406–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamm O, Aldrich TB, Grote IW, Rowe LW, and Bugbee EP (1928). The active principles of the pituitary gland. J. Am. Chem. Soc 50, 573–598. [Google Scholar]
- Kanehisa M, and Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanner L. (1943). Autistic disturbances of affective contact. Nerv. Child J. Psychopathol. Psychother. Ment. Hyg. Guid. Child 2, 217–250. [Google Scholar]
- Katz LC, Burkhalter A, and Dreyer WJ (1984). Fluorescent latex microspheres as a retrograde neuronal marker for in vivo and in vitro studies of visual cortex. Nature 310, 498–500. [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Kidd SA, Andrews HF, Budimirovic DB, Esler A, Haas-Givler B, Stackhouse T, Riley C, Peacock G, Sherman SL, et al. (2017). Autism Spectrum Disorder in Fragile X Syndrome: Cooccurring Conditions and Current Treatment. Pediatrics 139, S194–S206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly E, Meng F, Fujita H, Morgado F, Kazemi Y, Rice LC, Ren C, Escamilla CO, Gibson JM, Sajadi S, et al. (2020). Regulation of autism-relevant behaviors by cerebellar-prefrontal cortical circuits. Nat. Neurosci 9, 1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatzo I. (2003). Cécile & Oskar Vogt: the significance of their contributions in modern neuroscience. Acta Neurochir. Suppl 86, 29–32. [DOI] [PubMed] [Google Scholar]
- Knobloch HS, Charlet A, Hoffmann LC, Eliava M, Khrulev S, Cetin AH, Osten P, Schwarz MK, Seeburg PH, Stoop R, et al. (2012). Evoked axonal oxytocin release in the central amygdala attenuates fear response. Neuron 73, 553–566. [DOI] [PubMed] [Google Scholar]
- Kravitz AV, Tye LD, and Kreitzer AC (2012). Distinct roles for direct and indirect pathway striatal neurons in reinforcement. Nat. Neurosci 15, 816–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer EJ, Boutin S, Chillon M, and Danos O. (2000). Canine Adenovirus Vectors: an Alternative for Adenovirus-Mediated Gene Transfer. J. Virol 74, 505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg WJS (1932). The hypothalamus of the albino rat. J. Comp. Neurol 55, 19–89. [Google Scholar]
- Lambert RC, Moos FC, and Richard P. (1993). Action of endogenous oxytocin within the paraventricular or supraoptic nuclei: A powerful link in the regulation of the bursting pattern of oxytocin neurons during the milk-ejection reflex in rats. Neuroscience 57, 1027–1038. [DOI] [PubMed] [Google Scholar]
- Legros JJ, Chiodera P, Geenen V, Smitz S, and von Frenckell R. (1984). Dose-response relationship between plasma oxytocin and cortisol and adrenocorticotropin concentrations during oxytocin infusion in normal men. J. Clin. Endocrinol. Metab 58, 105–109. [DOI] [PubMed] [Google Scholar]
- Leng G, and Ludwig M. (2016). Intranasal Oxytocin: Myths and Delusions. Biol. Psychiatry 79, 243–250. [DOI] [PubMed] [Google Scholar]
- Lobo MK, and Nestler EJ (2011). The striatal balancing act in drug addiction: distinct roles of direct and indirect pathway medium spiny neurons. Front. Neuroanat 5, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M, and Leng G. (2006). Dendritic peptide release and peptide-dependent behaviours. Nat. Rev. Neurosci 7, 126–136. [DOI] [PubMed] [Google Scholar]
- Luther JA, and Tasker JG (2000). Voltage-gated currents distinguish parvocellular from magnocellular neurones in the rat hypothalamic paraventricular nucleus. J. Physiol 523 Pt 1, 193–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther JA, Halmos KC, and Tasker JG (2000). A slow transient potassium current expressed in a subset of neurosecretory neurons of the hypothalamic paraventricular nucleus. J. Neurophysiol 84, 1814–1825. [DOI] [PubMed] [Google Scholar]
- Luther JA, Daftary SS, Boudaba C, Gould GC, Halmos KC, and Tasker JG (2002). Neurosecretory and non-neurosecretory parvocellular neurones of the hypothalamic paraventricular nucleus express distinct electrophysiological properties. J. Neuroendocrinol 14, 929–932. [DOI] [PubMed] [Google Scholar]
- Martin JP, and Bell J. (1943). A Pedigree of Mental Defect Showing Sex-Linkage. J. Neurol. Psychiatry 6, 154–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard KR, Hobbs JW, Phan BN, Gupta A, Rajpurohit S, Williams C, Rajpurohit A, Shin JH, Jaffe AE, and Martinowich K. (2018). BDNF-TrkB signaling in oxytocin neurons contributes to maternal behavior. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis MR, Argiolas A, and Gessa GL (1986). Oxytocin-Induced Penile Erection and Yawning : Site of Action in the Brain. Brain Res. 398, 259–265. [DOI] [PubMed] [Google Scholar]
- Merchenthaler I. (1991). Neurons with access to the general circulation in the central nervous system of the rat: a retrograde tracing study with fluoro-gold. Neuroscience 44, 655–662. [DOI] [PubMed] [Google Scholar]
- Mientjes EJ, Nieuwenhuizen I, Kirkpatrick L, Zu T, Hoogeveen-Westerveld M, Severijnen L, Rifé M, Willemsen R, Nelson DL, and Oostra BA (2006). The generation of a conditional Fmr1 knock out mouse model to study Fmrp function in vivo. Neurobiol. Dis 21, 549–555. [DOI] [PubMed] [Google Scholar]
- Miyoshi G, Hjerling-leffler J, Karayannis T, Sousa VH, Butt JB, Battiste J, Johnson JE, Machold RP, and Fishell G. (2010). Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. J. Neurosci 30, 1582–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naber FBA, Swinkels SHN, Buitelaar JK, Dietz C, van Daalen E, Bakermans-Kranenburg MJ, van Ijzendoorn MH, and van Engeland H. (2007). Joint attention and attachment in toddlers with autism. J. Abnorm. Child Psychol 35, 899–911. [DOI] [PubMed] [Google Scholar]
- Naber FBA, Bakermans-Kranenburg MJ, van Ijzendoorn MH, Swinkels SHN, Buitelaar JK, Dietz C, van Daalen E, and van Engeland H. (2008). Play behavior and attachment in toddlers with autism. J. Autism Dev. Disord 38, 857–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardou R, Lewis EM, Rothhass R, Xu R, Yang A, Boyden E, and Dölen G. (2019). Oxytocin-dependent reopening of a social reward learning critical period with MDMA. Nature 569, 116–120. [DOI] [PubMed] [Google Scholar]
- Nassi JJ, and Callaway EM (2009). Parallel processing strategies of the primate visual system. Nat. Rev. Neurosci 10, 360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, and Hyman SE (2010). Animal models of neuropsychiatric disorders. Nat. Neurosci 13, 1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro MJ, Hashikawa-Yamasaki Y, and Rudy B. (2018). Diversity and connectivity of layer 5 somatostatin-expressing interneurons in the mouse barrel cortex. J. Neurosci 38, 1622–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann JJ, and Morris JF (1984). Method for quantitating the molecular content of a subcellular organelle: hormone and neurophysin content of newly formed and aged neurosecretory granules. Proc. Natl. Acad. Sci. U. S. A 81, 180–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira LF, Camboim C, Diehl F, Consiglio AR, and Quillfeldt JA (2007). Glucocorticoid-mediated effects of systemic oxytocin upon memory retrieval. Neurobiol. Learn. Mem 87, 67–71. [DOI] [PubMed] [Google Scholar]
- Orefice LL, Mosko JR, Morency DT, Wells MF, Tasnim A, Mozeika SM, Ye M, Chirila AM, Emanuel AJ, Rankin G, et al. (2019). Targeting Peripheral Somatosensory Neurons to Improve Tactile-Related Phenotypes in ASD Models. Cell 178, 867–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott I, and Scott JC (1911). The action of infundibulin upon the mammary secretion. Proc. Soc. Exp. Biol. Med 8, 48–49. [Google Scholar]
- Page DT, Kuti OJ, Prestia C, and Sur M. (2009). Haploinsufficiency for Pten and Serotonin transporter cooperatively influences brain size and social behavior. Proc. Natl. Acad. Sci. U. S. A 106, 1989–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panksepp JB, and Lahvis GP (2007). Social reward among juvenile mice. Genes. Brain. Behav 6, 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BD, Sbodio JI, Xu R, Vandiver MS, Cha JY, Snowman AM, and Snyder SH (2014). Cystathionine γ-lyase deficiency mediates neurodegeneration in Huntington’s disease. Nature 508, 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen CA, Ascher JA, Monroe YL, and Prange AJ (1982). Oxytocin induces maternal behavior in virgin female rats. Science 216, 648–650. [DOI] [PubMed] [Google Scholar]
- Peñagarikano O, Lázaro MT, Lu X-H, Gordon A, Dong H, Lam HA, Peles E, Maidment NT, Murphy NP, Yang XW, et al. (2015). Exogenous and evoked oxytocin restores social behavior in the Cntnap2 mouse model of autism. Sci. Transl. Med 7, 271ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzo MA, Robert V, Tucciarone J, De Bundel D, Wang M, Van Aelst L, Darvas M, Parada LF, Palmiter RD, He M, et al. (2015). The paraventricular thalamus controls a central amygdala fear circuit. Nature 519, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson M. (2002). Cardiovascular effects of Oxytocin. Prog. Brain Res 139, 281–288. [DOI] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, and Sandberg R. (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc 9, 171–181. [DOI] [PubMed] [Google Scholar]
- Pietropaolo S, Guilleminot A, Martin B, D’Amato FR, and Crusio WE (2011). Genetic-background modulation of core and variable autistic-like symptoms in Fmr1 knock-out mice. PLoS One 6, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Pol AN (1982). The magnocellular and parvocellular paraventricular nucleus of rat: Intrinsic organization. J. Comp. Neurol 206, 317–345. [DOI] [PubMed] [Google Scholar]
- van den Pol AN (2012). Neuropeptide Transmission in Brain Circuits. Neuron 76, 98–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pow DV, and Morris JF (1989). Dendrites of hypothalamic magnocellular neurons release neurohypophysial peptides by exocytosis. Neuroscience 32, 435–439. [DOI] [PubMed] [Google Scholar]
- Qin M, Kang J, Burlin TV, Jiang C, and Smith CB (2005). Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. J. Neurosci 25, 5087–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma Y-A, and Trapnell C. (2017). Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, and Tyagi S. (2008). Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recanzone GH, and Cohen YE (2010). Serial and parallel processing in the primate auditory cortex revisited. Behav. Brain Res 206, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov RA, Zeisel A, Bakker J, Girach F, Hellysaz A, Tomer R, Alpár A, Mulder J, Clotman F, Keimpema E, et al. (2017). Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat. Neurosci 20, 176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper SD (2009). Parallel processing in mammalian taste buds? Physiol. Behav 97, 604–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutgers AH, Bakermans-Kranenburg MJ, van Ijzendoorn MH, and van Berckelaer-Onnes IA (2004). Autism and attachment: a meta-analytic review. J. Child Psychol. Psychiatry 45, 1123–1134. [DOI] [PubMed] [Google Scholar]
- Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, Peng M, Collins R, Grove J, Klei L, et al. (2020). Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 180, 568–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawchenko PE, and Swanson LW (1982). Immunohistochemical identification of neurons in the paraventricular nucleus of the hypothalamus that project to the medulla or to the spinal cord in the rat. J. Comp. Neurol 205, 260–272. [DOI] [PubMed] [Google Scholar]
- Saxena A, Wagatsuma A, Noro Y, Kuji T, Asaka-Oba A, Watahiki A, Gurnot C, Fagiolini M, Hensch TK, and Carninci P. (2012). Trehalose-enhanced isolation of neuronal sub-types from adult mouse brain. Biotechniques 52, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scelsi MA, Khan RR, Lorenzi M, Christopher L, Greicius MD, Schott JM, Ourselin S, and Altmann A. (2018). Genetic study of multimodal imaging Alzheimer’s disease progression score implicates novel loci. Brain 141, 2167–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmued LC, and Fallon JH (1986). Fluoro-gold: a new fluorescent retrograde axonal tracer with numerous unique properties. Brain Res. 377, 147–154. [DOI] [PubMed] [Google Scholar]
- Schneeberger M, Parolari L, Das Banerjee T, Bhave V, Wang P, Patel B, Topilko T, Wu Z, Choi CHJ, Yu X, et al. (2019). Regulation of Energy Expenditure by Brainstem GABA Neurons. Cell 178, 672–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz S, Opie C, and Atkinson QD (2011). Stepwise evolution of stable sociality in primates. Nature 479, 219–222. [DOI] [PubMed] [Google Scholar]
- Smith LN, Jedynak JP, Fontenot MR, Hale CF, Dietz KC, Taniguchi M, Thomas FS, Zirlin BC, Birnbaum SG, Huber KM, et al. (2014). Fragile X Mental Retardation Protein Regulates Synaptic and Behavioral Plasticity to Repeated Cocaine Administration. Neuron 82, 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder-Mackler N, Burger JR, Gaydosh L, Belsky DW, Noppert GA, Campos FA, Bartolomucci A, Yang YC, Aiello AE, O’Rand A, et al. (2020). Social determinants of health and survival in humans and other animals. Science. 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak A, Macchi C, Nussdorfer GG, and Malendowicz LK (1995). Effects of oxytocin on the function and morphology of the rat adrenal cortex: in vitro and in vivo investigations. Res. Exp. Med. (Berl) 195, 265–274. [DOI] [PubMed] [Google Scholar]
- Stavropoulos KKM, and Carver LJ (2014). Reward anticipation and processing of social versus nonsocial stimuli in children with and without autism spectrum disorders. J. Child Psychol. Psychiatry Allied Discip 55, 1398–1408. [DOI] [PubMed] [Google Scholar]
- Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg IC, Jakobsson G, and Bohman M. (1989). A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J. Child Psychol. Psychiatry 30, 405–416. [DOI] [PubMed] [Google Scholar]
- Stokes M, Newton N, and Kaur A. (2007). Stalking, and social and romantic functioning among adolescents and adults with autism spectrum disorder. J. Autism Dev. Disord 37, 1969–1986. [DOI] [PubMed] [Google Scholar]
- Suhl JA, and Warren ST (2015). Single-Nucleotide mutations in FMR1 reveal novel functions and regulatory mechanisms of the fragile X syndrome protein FMRP. J. Exp. Neurosci 2015, 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, and O’Donovan M. (2012). Genetic architectures of psychiatric disorders: The emerging picture and its implications. Nat. Rev. Genet 13, 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaab D, Purba J, and Hofman M. (1995). Alterations in the hypothalamic paraventricular nucleus and its oxytocin neurons (putative satiety cells) in Prader-Willi syndrome: a study of five cases. J Clin Endocrinol Metab 80, 573–579. [DOI] [PubMed] [Google Scholar]
- Swanson LW, and Kuypers HGJM (1980). The paraventricular nucleus of the hypothalamus: Cytoarchitectonic subdivisions and organization of projections to the pituitary, dorsal vagal complex, and spinal cord as demonstrated by retrograde fluorescence doublelabeling methods. J. Comp. Neurol 194, 555–570. [DOI] [PubMed] [Google Scholar]
- Tasker JG, and Dudek FE (1991). Electrophysiological properties of neurones in the region of the paraventricular nucleus in slices of rat hypothalamus. J. Physiol 434, 271–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tervo DGR, Hwang BY, Viswanathan S, Gaj T, Lavzin M, Ritola KD, Lindo S, Michael S, Kuleshova E, Ojala D, et al. (2016). A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron 92, 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman AJ, McDuffie A, Hagerman R, and Abbeduto L. (2014). Psychiatric symptoms in boys with fragile X syndrome: a comparison with nonsyndromic autism spectrum disorder. Res. Dev. Disabil 35, 1072–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PT, Hull C, Chu Y, Greene-Colozzi E, Sadowski AR, Leech JM, Steinberg J, Crawley JN, Regehr WG, and Sahin M. (2012). Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature 488, 647–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu A, Tan BZ, Ycu EA, Cuevas JS, Koivumaa J, Junyent F, Kremer EJ, Witten IB, Deisseroth K, and Johansen JP (2017). Modular organization of the brainstem noradrenaline system coordinates opposing learning states. Nat. Neurosci 20, 1602–1611. [DOI] [PubMed] [Google Scholar]
- Valassi E, Scacchi M, and Cavagnini F. (2008). Neuroendocrine control of food intake. Nutr. Metab. Cardiovasc. Dis 18, 158–168. [DOI] [PubMed] [Google Scholar]
- Verkerk AJMH, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DPA, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang F, et al. (1991). Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65, 905–914. [DOI] [PubMed] [Google Scholar]
- Vivanti G, and Nuske HJ (2017). Autism, attachment, and social learning: Three challenges and a way forward. Behav. Brain Res 325, 251–259. [DOI] [PubMed] [Google Scholar]
- Whitnall MH, Key S, Ben-Barak Y, Ozato K, and Gainer H. (1985). Neurophysin in the hypothalamo-neurohypophysial system. II. Immunocytochemical studies of the ontogeny of oxytocinergic and vasopressinergic neurons. J. Neurosci 5, 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, Reilly SK, Lin L, Fertuzinhos S, Miller JA, et al. (2013). XCoexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Xu Y, Zhu Y, Sutton AK, Zhao R, Lowell BB, Olson DP, and Tong Q. (2012). An Obligate Role of Oxytocin Neurons in Diet Induced Energy Expenditure. PLoS One 7, 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Priest MF, Nasenbeny J, Lu T, and Kozorovitskiy Y. (2017). Biased Oxytocinergic Modulation of Midbrain Dopamine Systems. Neuron 95, 368–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, and Hernández VS (2013). Synaptic innervation to rat hippocampus by vasopressin-immuno-positive fibres from the hypothalamic supraoptic and paraventricular nuclei. Neuroscience 228, 139–162. [DOI] [PubMed] [Google Scholar]
- Zhang G, Bai H, Zhang H, Dean C, Wu Q, Li J, Guariglia S, Meng Q, and Cai D. (2011). Neuropeptide Exocytosis Involving Synaptotagmin-4 and Oxytocin in Hypothalamic Programming of Body Weight and Energy Balance. Neuron 69, 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Zhang HF, Han JS, and Han SP (2017). Genes Related to Oxytocin and Arginine-Vasopressin Pathways: Associations with Autism Spectrum Disorders. Neurosci. Bull 33, 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Bhalla A, Dean C, Chapman ER, and Jackson MB (2009). Synaptotagmin IV: A multifunctional regulator of peptidergic nerve terminals. Nat. Neurosci 12, 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwergal A, Strupp M, Brandt T, and Büttner-Ennever JA (2009). Parallel ascending vestibular pathways: Anatomical localization and functional specialization. Ann. N. Y. Acad. Sci 1164, 51–59. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially expressed ASD risk genes. Related to Figure 8. (Excel File)
Differentially expressed FMRP binding targets. Related to Figure 8. (Excel File)
Data Availability Statement
All data are archived in the Gene Expression Omnibus (GEO Accession number GSE147092). Raw and processed scRNA-Seq data and code to reproduce the complete analysis are also publicly available at https://github.com/gofflab/OT_neuron_study_2020.