Abstract
Nucleic acid‐based therapeutics that regulate gene expression have been developed towards clinical use at a steady pace for several decades, but in recent years the field has been accelerating. To date, there are 11 marketed products based on antisense oligonucleotides, aptamers and small interfering RNAs, and many others are in the pipeline for both academia and industry. A major technology trigger for this development has been progress in oligonucleotide chemistry to improve the drug properties and reduce cost of goods, but the main hurdle for the application to a wider range of disorders is delivery to target tissues. The adoption of delivery technologies, such as conjugates or nanoparticles, has been a game changer for many therapeutic indications, but many others are still awaiting their eureka moment. Here, we cover the variety of methods developed to deliver nucleic acid‐based therapeutics across biological barriers and the model systems used to test them. We discuss important safety considerations and regulatory requirements for synthetic oligonucleotide chemistries and the hurdles for translating laboratory breakthroughs to the clinic. Recent advances in the delivery of nucleic acid‐based therapeutics and in the development of model systems, as well as safety considerations and regulatory requirements for synthetic oligonucleotide chemistries are discussed in this review on oligonucleotide‐based therapeutics.
Keywords: delivery, oligonucleotides, preclinical models, RNA therapeutics, safety
Subject Categories: Synthetic Biology & Biotechnology, Chemical Biology, RNA Biology
Recent advances in the delivery of nucleic acid‐based therapeutics and in the development of model systems, as well as safety considerations and regulatory requirements for synthetic oligonucleotide chemistries are discussed in this review on oligonucleotide‐based therapeutics.
Glossary
- Anti‐drug antibodies (ADAs)
Antibody‐mediated immunogenicity elicited in vivo to a given drug. Drug‐specific antibodies can reduce the efficacy of the treatment and even fully inactivate the drug, and/or they can induce adverse effects.
- Antisense oligonucleotides (ASOs)
Single‐stranded oligonucleotides complementary to RNA target sequences.
- Aptamers
Single‐stranded oligonucleotides (20‐100 nucleotides) which adopt three‐dimensional structures that allow them to bind very specifically to protein target sites.
- Blood–brain barrier (BBB) and blood–spinal cord barrier (BSCB)
Selectively permeable membranes of the central nervous system (CNS) vasculature. Only small molecules (molecular weight below 400‐500 Da) and high lipid solubility (logP value of approximately 2.1) can cross these vascular barriers. Generally, oligonucleotides display a molecular weight of approximately 10 kDa and are hydrophilic; hence, they are too large and hydrophilic to cross biological barriers by passive diffusion.
- Cell‐penetrating peptides (CPPs)
Short cationic and/or amphipathic peptides (usually less than 30 amino acids) capable of translocating different types of cargoes across biological barriers and cell membranes. CPPs can be directly conjugated to oligonucleotides (ONs) or used to encapsulate ONs into nanoparticles.
- European Medicines Agency (EMA)
Agency of the European Union in charge of the evaluation and supervision of medicinal products. The EMA facilitates development and access to medicines, evaluates applications for marketing authorisation and monitors the safety of human and veterinary medicines.
- Food and Drug Administration (FDA)
The federal agency of the United States Department of Health and Human Services, responsible for protecting public health by ensuring the safety, efficacy and security of human and veterinary drugs.
- Gapmer
Chimeric antisense oligonucleotides (ASOs) that contain a central block of DNA nucleotides, flanked by modified sequences, usually containing 2′‐O‐modified or locked nucleic acid (LNA) chemistries. Gapmers are used for gene silencing by stimulating RNA cleavage through the recruitment of RNase H.
- Lipid nanoparticles (LNPs)
Delivery systems based on LNPs are composed of one or several lipid components, often an ionisable cationic lipid used for complexation of polyanionic DNA/RNA and stabilising helper lipids such as distearoylphosphatidylcholine (DSPC) and cholesterol. In addition, LNPs may be stabilised sterically by surface coating with polyethylene glycol (PEG). LNPs have a complex internal lipid architecture that is well suited for stable and efficient encapsulation of DNA/RNA cargoes.
- MicroRNAs (miRNAs)
Small noncoding RNAs (∼22 nt), which regulate gene expression at the post‐transcriptional level by degrading target mRNAs, when complementary to the sequence, or inhibiting their translation when not fully complementary. Each miRNA can influence the expression of hundreds of mRNAs.
- Pharmacodynamics (PD)
The relationship between the drug concentration at the site of action and the observed biochemical response and its efficacy.
- Pharmacokinetics (PK)
The time course of drug absorption, distribution, metabolism, excretion and toxicity (ADMET), as well as the liberation of a drug from its formulation.
- Phosphorodiamidate morpholino oligonucleotides (PMOs)
Oligonucleotides containing uncharged chemistry. The nucleic acid backbone has been replaced with 6‐membered morpholino rings and phosphorodiamidate linkages, while retaining standard nucleobases.
- Peptide nucleic acid (PNA)
Uncharged oligonucleotide chemistry with amide bond linkages between the nucleobases. PNAs are manufactured by peptide synthesis.
- RNAse H cleavage
RNAse H hydrolyses the phosphodiester bonds of RNA when hybridised to DNA.
- Small interfering RNA (siRNA)
Double‐stranded RNA (~21 nt) composed of a guide strand complementary to the target mRNA and a passenger strand. siRNAs act within the endogenous RNA‐induced silencing complex (RISC) to degrade mRNA.
- Toll‐like receptors (TLRs)
Pattern‐recognition receptors usually found on the plasma or endosomal membranes of sentinel cells such as macrophages and dendritic cells (DCs). Activation of TLRs can promote an inflammatory response. For example, TLR9 is activated by unmethylated cytidine‐phosphate‐guanosine (CpG) dinucleotides present in bacterial and viral DNA.
Introduction
Synthetic oligonucleotides (ONs) are small, single‐ or double‐stranded pieces of modified nucleic acids that have been exploited as therapeutic modalities in different ways (Table 1). The unique characteristic of ONs is that they bind to their target via Watson–Crick base pairing, enabling intervention at a genetic level by targeting RNA in a specific manner (Zamecnik & Stephenson, 1978). ONs encompass many types of nucleic acid‐based therapeutics, including antisense oligonucleotides (ASOs), small interfering RNA (siRNA), anti‐miRNA (antagomirs), miRNA mimics (agomirs), aptamers and unmethylated CpG‐containing ONs. Depending on their mechanism of action, treatment with therapeutic nucleic acids may cause decreased, increased or restored protein expression. Currently, 11 ON‐based drugs across many disease areas have received regulatory approval by the US Food and Drug Administration (FDA), the European Medicines Agency (EMA) and/or the Japanese Ministry of Health, Labour and Welfare. However, further therapeutic development is challenged by unfavourable absorption, distribution, metabolism, excretion and toxicity (ADMET) properties for most clinical applications (Godfrey et al, 2017). This review mainly focuses on the development of single‐stranded ONs and covers (i) the numerous methods developed to date to deliver ONs across biological barriers, (ii) the model systems used to test ONs and (iii) the hurdles existing for translating laboratory breakthroughs to the clinic. The content represents the joint efforts of members of the EU Cooperation of Science and Technology (COST) network Delivery of RNA Therapeutics (DARTER, COST Action 17103, www.antisenserna.eu), which aims to facilitate RNA‐targeting nucleic acid‐based drugs to reach their full potential.
Table 1.
Mechanisms of action of therapeutic oligonucleotides.
| Modality | Mechanism | Example(s) |
|---|---|---|
| RNase H | RNase H‐mediated cleavage of target transcript | Gapmers |
| Steric Blockage | Interference with post‐transcriptional RNA‐binding elements, e.g. splicing modulation and blocking endogenous miRNA | 2nd and 3rd generation ASOs and antagomirs |
| Protein Binding | Bind target proteins in a structure‐specific manner | Aptamer |
| Innate Immunity | Inhibits protein expression via target‐specific mRNA degradation | Unmethylated CpG‐containing ONs |
| RNAi | Inhibition of gene expression via target‐specific mRNA degradation | siRNAs, microRNAs |
Chemistry dictates the drug properties of oligonucleotides
Therapeutic nucleic acids are chemically modified in several ways to endow them with properties such as increased resistance to nucleases and improved target binding affinity (Jarver et al, 2014) (Fig 1). Each modification confers the ON different properties, and some may be combined, but other modifications are not compatible or may modify the ON in ways that complicate their synthesis or interfere with the mechanisms by which they exert their effect. First‐generation chemistries include the widely used phosphate backbone modifications, e.g. phosphorothioate (PS), which imparts resistance to endonucleases and improves bioavailability by reducing renal clearance due to increased affinity for serum proteins (Eckstein, 2014). However, this modification also reduces the affinity for the target RNA. Second‐generation chemistries include ribose modifications at the 2′‐O position of RNA and 2′ position of DNA, of which the 2ʹ‐O‐methyl (2ʹ‐OMe), 2ʹ‐O‐methoxy‐ethyl (2ʹ‐MOE) and 2ʹ‐fluoro (2ʹ‐F) modifications are the most commonly used types. These modifications increase the binding affinity to RNA and further improve the nuclease resistance. An even greater binding affinity chemistry is the conformationally constrained DNA analogues locked nucleic acid (LNA) and tricyclo‐DNA (tcDNA). LNA contains a methyl bridge between the 2′‐O and 4′ position of the ribose ring (Koshkin et al, 1998; Obika et al, 1998). The backbone considerably changed for tcDNA via introduction of an ethylene bridge with a cyclopropane ring between the ribose 3' and 5' carbon positions (Renneberg & Leumann, 2002). The bridge imposes a locked conformation on the ribose ring, which is ideal for binding to RNA. All first‐ and second‐generation chemistries are compatible with nucleic acid synthesis and can easily be mixed with DNA and RNA in ON chimeras. Third‐generation chemistries include changes in the nucleobase, e.g. phosphorodiamidate morpholino oligomers (PMO) (Summerton & Weller, 1997) and peptide nucleic acid (PNA) (Nielsen et al, 1991; Hanvey et al, 1992). For PMOs, the nucleic acid backbone has been replaced with a 6‐membered morpholino ring and phosphorodiamidate linkages, while retaining standard nucleobases. The nucleobases of PNAs are linked by amide bonds, which are synthesised similarly to peptides. Both PMO and PNA are uncharged, very resistant to nucleases, and display variable affinity for the target RNA (Smulevitch et al, 1996; Summerton & Weller, 1997). The choice of chemical modifications is largely dictated by the modality and the target tissue.
Figure 1. Oligonucleotide chemistries.
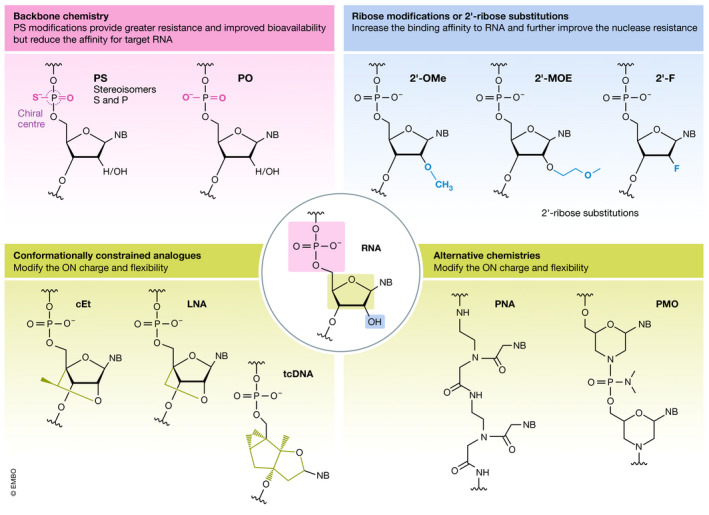
Commonly used nucleic acid chemistries. The often used phophorothioate (PS) backbone replaces the natural phosphodiester (PO). Modifications to the ribose at the 2ʹ‐O position of RNA and 2ʹ‐position of DNA include the 2ʹ‐O‐methyl (2ʹ‐OMe), 2ʹ‐O‐methoxy‐ethyl (2ʹ‐MOE) and 2ʹ‐fluoro (2ʹ‐F) are the most commonly used. Conformationally constrained DNA analogues, locked nucleic acid (LNA), constrained 2′‐O‐ethyl (cEt) and tricyclo‐DNA (tcDNA), provide greater binding affinity. LNA and cEt are constrained by a methyl bridged from the 2′‐O and 4′ position of the ribose. tcDNA introduces of an ethylene bridge with a cyclopropane ring between the 3′ and 5′ carbon positions of ribose. Alternative chemistries include changes in the nucleobase, e.g. phosphorodiamidate morpholino oligomers (PMO) and peptide nucleic acid (PNA).
Single‐stranded ASOs complementary to target RNA were first utilised therapeutically by exploiting RNase H cleavage of DNA/RNA hybrids (Stein & Hausen, 1969; Wu et al, 2004) (Fig 2). RNase H‐inducible ASOs are designed as gapmers, where central DNA nucleotides are flanked by RNase H‐resistant modified nucleotides (Wahlestedt et al, 2000). The modified sequences improve target affinity while the central DNA sequence forms the DNA/RNA hybrid for RNase H recognition and cleavage (Monia et al, 1993). Fully modified second‐ and third‐generation ASO chemistries act through RNase H‐independent mechanisms (Fig 2) (Jarver et al, 2014). Steric blocking ASOs can inhibit or activate translation through the binding to regulatory elements, e.g. upstream open reading frames (Liang et al, 2016b; Liang et al, 2017). A common therapeutic modality is the modulation of pre‐mRNA splicing (Arechavala‐Gomeza et al, 2014), which is used to induce or suppress exon inclusion. In Duchenne muscular dystrophy (DMD) patients, ASO‐induced exon skipping of mutated dystrophin pre‐mRNA restores the reading frame and allows for the production of partially functional, rather than non‐functional, dystrophin protein (Mitrpant et al, 2009). In contrast, for spinal muscular atrophy (SMA) patients, ASOs increase the level of exon 7 inclusion in survival motor neuron 2 (SMN2) mRNA, leading to increased levels of SMN protein (Singh et al, 2006). Similarly, ASOs can also induce the skipping of pseudoexons (Collin et al, 2012) or block RNA‐splicing factors from recognising cryptic splice sites (Rivera‐Barahona et al, 2015). ASOs can also sterically block the union of RNA‐binding factors to repeat expansion regions of pathogenic mRNAs (Fig 2). In myotonic dystrophy 1, expanded microsatellite repeats sequester RNA‐binding factors within nuclear expansion RNA foci (Miller et al, 2000). ASOs targeting the CUG repeat expansion mRNA release the sequestered RNA‐binding factors and reverse the phenotype (Klein et al, 2019). RNA interference (RNAi)‐based therapies, i.e. double‐stranded siRNA and single‐stranded microRNA (miRNA), exploit the endogenous RNAi pathway in the cytosol (Fire et al, 1998) to silence or modulate the expression of specific proteins (Fig 2). Commonly used chemical modifications for siRNA, including 2ʹ‐OMe and 2ʹ‐F modifications, decrease RNase recognition and are well tolerated throughout the entire siRNA duplex (Watts et al, 2008). In addition, these modifications are widely used to decrease immune stimulation (Judge et al, 2006). ASOs can influence miRNA function, either by sequestering a miRNA (antagomir) or by generating a miRNA mimic (agomir). Notably, a single miRNA generally regulates the expression of multiple genes in a given pathway; hence, antagomirs and agomirs have the potential to mediate increased or decreased expression of multiple genes, respectively (Friedman et al, 2009). Finally, two types of ONs which do not work through Watson–Crick base pairing are aptamers and unmethylated CpG‐containing ONs. Aptamers are single‐stranded ONs (20–100 nucleotides) selected from randomised libraries based on their high‐avidity binding to specific targets (Ellington & Szostak, 1990; Tuerk & Gold, 1990). They adopt three‐dimensional structures that bind to protein target sites through attractive electrostatic interactions and pocket‐like structures (Ellington & Szostak, 1990), and they display binding affinities to their receptor targets which are comparable to those of monoclonal antibodies (Jayasena, 1999). Unmethylated CpG‐containing ONs include a cytosine‐guanine motif coupled with a phosphodiester (PO) or PS backbone. Unmethylated CpG motifs are commonly found in bacterial DNA and activate the immune system through Toll‐like receptor 9 (TLR9). Unmethylated CpG‐containing ONs have been tested clinically as vaccine adjuvants and for cancer immunotherapy (Krieg & Davis, 2001; Krieg, 2006, 2007).
Figure 2. Mechanisms and location of action for oligonucleotides.
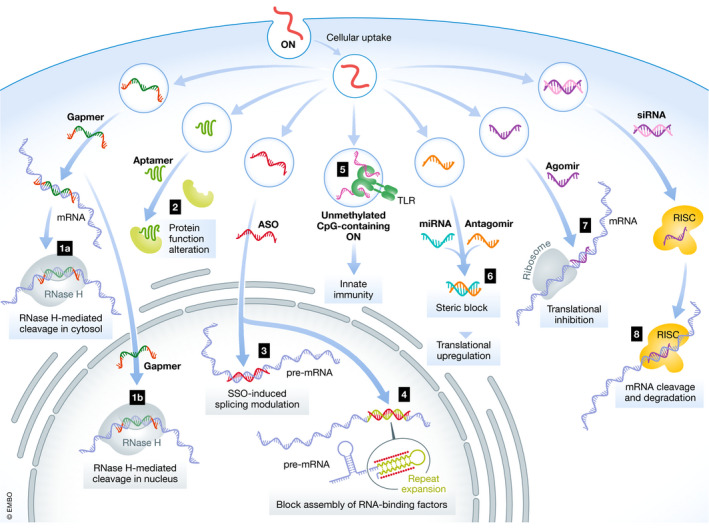
Representative mechanisms of action and intracellular localisation for (1) gapmer and mRNA degradation, (2) aptamer, (3) nuclear steric blockage for splice switching, (4) blockage the assembly of RNA‐binding factors, (5) TLR activation of innate immunity, (6) miRNA and antagomir, steric block, translational upregulation, (7) agomir, translational inhibition, and (8) siRNA, RISC, RNAi silencing ONs.
Delivery systems for oligonucleotides
The sites of action for ONs lay within the intracellular space. Consequently, they need to overcome several biological barriers to reach their pharmacological targets in vivo. PS‐modified ONs bind reversibly to plasma proteins, e.g. albumin, which increases their plasma half‐life and facilitates their distribution and accumulation in the liver, kidneys, spleen, lymph nodes and bone marrow (Geary, 2009). Targeting tissues beyond these organs has had clinical success for local delivery to the eye, brain and spinal cord via intravitreal (IVT) and intrathecal (IT) administration, respectively (Hache et al, 2016; Cideciyan et al, 2019) (Fig 3). Both routes bypass renal clearance and maintain high ON exposure to the cellular microenvironment for efficient uptake. Additionally, significant advances for pulmonary delivery of RNA therapies have been extensively reviewed elsewhere (Chow et al, 2020; Shaffer, 2020). However, systemic administration of ONs has been less successful due to poor tissue uptake. Cellular uptake of ONs occurs predominantly via different types of endocytosis. ONs are subsequently trafficked into the endolysosomal system, from where they need to escape to avoid degradation in the lysosomal environment (Crooke et al, 2017). Only a very small ON dose fraction escapes the endosomes and becomes available at the site of action (Gilleron et al, 2013). Single‐stranded ONs, such as PS ASOs, which are relatively small, uncharged and/or hydrophobic, can productively enter cells and escape the endosomes into the cytoplasm and nucleus without the need for a delivery agent (Liang et al, 2016a) in a process referred to as gymnosis (Stein et al, 2010), but relatively high ON doses are required for this process to take place. However, most of RNA‐based therapeutics, e.g. double‐stranded siRNA, are too large and charged to enter cells unassisted and require a delivery agent. Accelerating the rate of cellular uptake, intracellular trafficking and endosomal escape has been a driving force behind advances in many chemical modifications and delivery agents (Juliano et al, 2018; Biscans et al, 2020). A wide variety of delivery approaches improve the transport and bioavailability of ONs (Fig 4) (Roberts et al, 2020). These include (i) direct conjugation to carriers and (ii) incorporation into nanoparticulate carriers, both with the aim of improving the ADMET properties.
Figure 3. Delivery of oligonucleotides to the brain and eye.
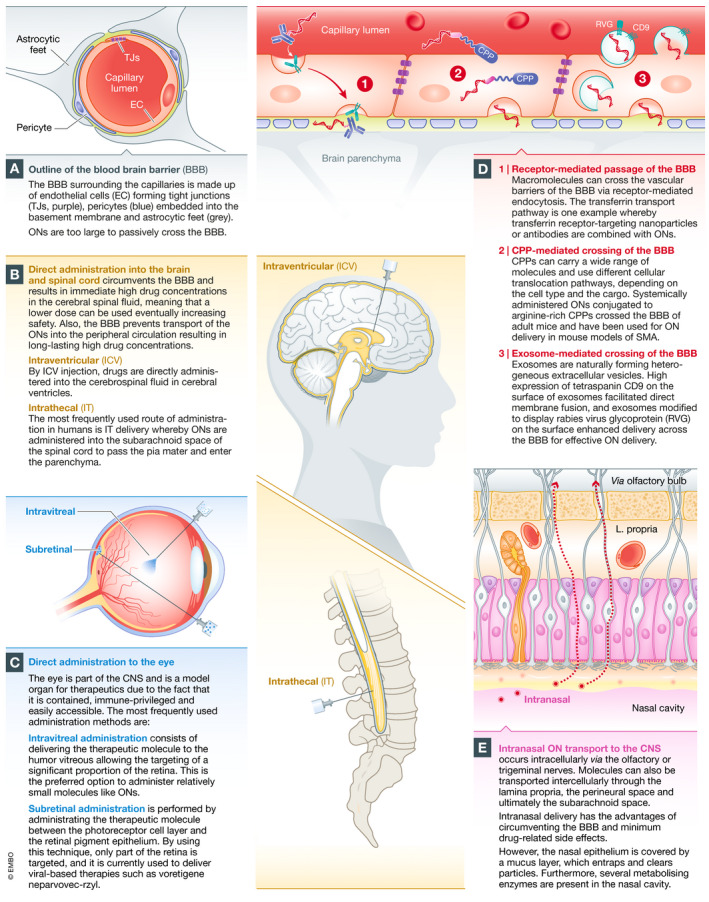
(A) ONs are prevented from passive diffusion into the central nervous system (CNS) by the vascular BBB. (B) ONs without a delivery reagent require direct administration into the brain or spinal cord. The most frequently used CNS administration route in humans is intrathecal (IT) administration, where ONs are administered into the subarachnoid space of the spinal cord to pass the pia mater and enter the parenchyma. This results in an immediate high ON concentration in the cerebral spinal fluid, meaning that a lower dose can be used, which reduces side effects. Also, the BBB prevents transport of ONs into the peripheral circulation resulting in long‐lasting high ON concentrations. (C) The eye is a contained and immune‐privileged organ of the CNS that allows local delivery. ONs are effective and well tolerated when administered directly by intravitreal injection. Subretinal delivery is also possible, but the treated area will be reduced. (D) Certain macromolecules can cross the vascular barriers via receptor‐mediated endocytosis after systemic administration (Pardridge, 2007). The transferrin transport pathway has been exploited in several rodent studies to carry ONs into the brain parenchyma (Lee et al, 2002; Kozlu et al, 2014). Systemically delivered ONs covalently conjugated to arginine‐rich CPPs have been shown to cross the BBB in mice (Du et al, 2011) and have been used for ON delivery in mouse models of SMA (Hammond et al, 2016). Several studies have shown exosome‐mediated delivery of small RNAs across the vascular barriers into the CNS (Alvarez‐Erviti et al, 2011; Yang et al, 2017). (E) Drugs dosed by intranasal administration can be transported into the brain along the olfactory, trigeminal nerve and rostral migratory stream (Curtis et al, 2007).
Figure 4. Delivery technologies for oligonucleotides.
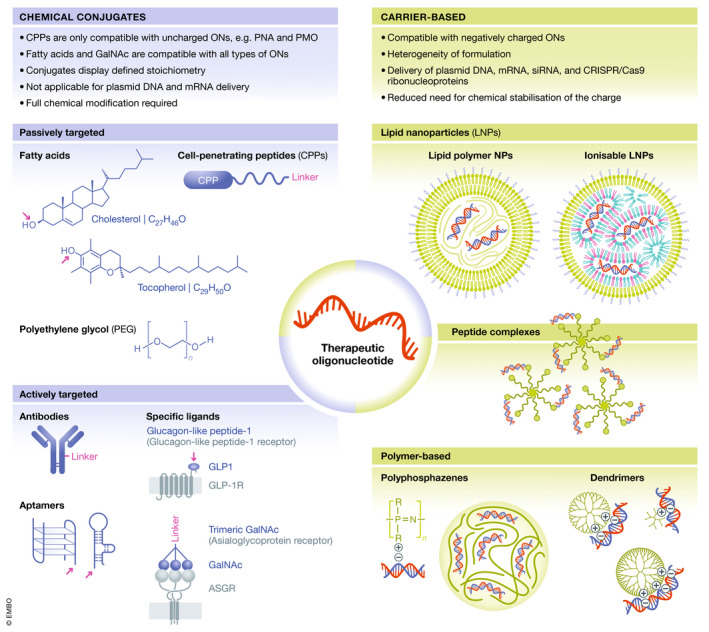
Delivery technologies used to improve the ADMET properties of ONs, including chemical conjugates (left) and nanoparticulate carriers (right). Polymers, cell‐penetrating peptides (CPPs) and lipids represent examples of molecules used for covalent conjugation to ONs for passive targeting, whereas covalent conjugation of ONs to antibodies, receptor ligands and aptamers are applied for active targeting. Drug conjugates display a defined stoichiometry. CPP conjugation is only compatible with uncharged ONs, e.g. PMOs and PNAs, whereas lipids and GalNAc are compatible with all types of ONs. Nanoparticulate carriers can be used to encapsulate negatively charged ONs and can be based on lipids, e.g. lipid nanoparticles (LNPs) and exosomes, polymers, e.g. dendrimers, poly(lactide‐co‐glycolic acid) (PLGA) and polyphosphazenes, and peptides, or on hybrid systems composed of several different types of compounds. The complexity of these systems poses new challenges in the development with respect to cost, manufacturability, safety, quality assurance and quality control.
Chemical conjugates
Chemical conjugation of molecules to therapeutic ONs is an attractive strategy for improving ADMET properties. As chemical conjugates, ONs are exposed to serum, and therefore, full chemical modification of ONs is needed to protect them from degradation. Polymers, peptides, lipids, receptor ligands and aptamers represent examples of molecules used for conjugation (Fig 4).
Polymers
Covalent conjugation of polyethylene glycol (PEG) improves the ADMET properties of drugs. PEGylation has been applied mainly for therapeutic proteins, but more recently also for ONs, e.g. the marketed aptamer‐PEG conjugate pegaptanib directed against vascular endothelial growth factor (VEGF) (Ng et al, 2006). PEG is a highly flexible, non‐charged and hydrophilic polymer with end groups available for functionalisation. PEG shields the conjugated drug cargo via formation of a hydration shell, which sterically blocks other biomacromolecules from binding to the drug. Also, PEGylation prolongs the circulation time by reducing renal excretion and increasing ON stability. The ADMET properties of PEGylated ONs are dependent on the physicochemical properties of the PEG moiety, including the molecular weight, the type of end group modification and the PEG architecture (linear or branched). For example, pegaptanib contains a 40 kDa Y‐shaped PEG, which causes the aptamer binding affinity to decrease fourfold compared with the parent aptamer, whereas the antiangiogenic efficacy is increased, which is attributed to prolonged tissue residence time due to increased half‐life (Ng et al, 2006).
Peptides
Cell‐penetrating peptides (CPPs) are short cationic and/or amphipathic peptides, usually less than 30 amino acids long, capable of translocating different types of cargoes across biological barriers and cell membranes (Foged & Nielsen, 2008; Pooga & Langel, 2015; Lehto et al, 2016). CPPs can be used as direct conjugates or to encapsulate oligonucleotides into nanoparticles, which is discussed further in the next section. Once inside the cells, CPPs may also improve endosomal escape (Cleal et al, 2013). However, the cationic charge often restricts their covalent conjugation to charge‐neutral ON chemistries (PNAs and PMOs) due to electrostatic interactions between anionic ONs and cationic CPPs that result in aggregation. For systemic diseases, CPP‐ONs circumvent cell‐specific receptors, allowing for pharmacological activity across multiple tissues, and they have been developed for uptake into particularly impervious tissues, e.g. skeletal muscle, heart and CNS (Hammond et al, 2016; Betts et al, 2019), as well as targeting viral and bacterial infections (Burrer et al, 2007; Geller et al, 2013; Geller et al, 2018). At the time of this review, a phase I clinical trial for safety and tolerability of an arginine‐rich CPP‐ASO conjugate for DMD (SRP‐5051) has been completed and a phase II is recruiting to determine the optimal dose.
Lipids
Conjugation of hydrophobic compounds such as cholesterol to ONs can improve delivery in vitro by promoting endosomal release (Wang et al, 2019) and results in longer plasma half‐life and accumulation in the liver upon systemic administration (Osborn et al, 2019). Such modifications may enhance delivery, mainly to the liver, but also to peripheral tissues such as muscle (Prakash et al, 2019), via passive targeting by increasing the binding affinity of ONs to plasma proteins and/or via active targeting by hijacking endogenous lipid transport pathways (Osborn et al, 2019).
Receptor ligands
Tissue‐specific active targeting may be achieved through conjugation of ONs to receptor ligands that facilitate specific binding to receptors on the target cells and mediate tissue‐specific delivery. A wide variety of receptor ligands have been investigated, including carbohydrates, peptides/proteins, aptamers, antibodies/antibody fragments and small molecules), and several feasible receptor‐ligand systems have been identified.
Perhaps the most successful tissue targeting ligand is trimeric N‐acetyl galactosamine (GalNAc) (Lee et al, 1984). GalNAc binds to the asialoglycoprotein receptor (AGPR), which is abundantly expressed in the liver (Schwartz et al, 1980). This high affinity‐binding ligand has been directly conjugated to ONs and siRNA and provides highly specific and effective delivery to hepatocytes (Matsuda et al, 2015; Janas et al, 2018; Debacker et al, 2020). Another striking example is the glucagon‐like peptide‐1 (GLP1) receptor (GLPR1) system for specific targeting of pancreatic β cells (Muller et al, 2019). Recent studies showed that GLP1‐ON conjugates are specifically taken up by GLPR1‐expressing cells in the pancreas, including isolated pancreatic islets, and induce strong accumulation and activity in pancreatic β cells in a ligand‐dependent manner upon systemic delivery in mice (Ammala et al, 2018).
Antibodies
A promising recent development in chemical conjugates is antibody–RNA conjugates (ARCs). ARCs typically include monoclonal antibodies, or antibody fragments, with functional ONs, and they have been used for imaging and protein detection. However, antibodies can also be used as a delivery agent for therapeutic ONs. An antibody fragment specific for the transferrin receptor, which is involved in intracellular transport of iron‐laden transferrin, has been used to target siRNA towards skeletal and cardiac muscle tissues (Sugo et al, 2016). Companies are taking this technology forward for diseases such as myotonic dystrophy and Duchenne muscular disease.
Aptamers
Aptamers have been shown to mediate delivery of therapeutic ONs as aptamer‐ON conjugates, or within nanoparticle formulations (Catuogno et al, 2016; Soldevilla et al, 2018). The first aptamer‐siRNA chimeras targeted prostate‐specific membrane antigen‐expressing cancer cells to deliver apoptosis‐inducing siRNAs (McNamara et al, 2006). Further development of aptamer‐ONs involved chemical modifications to protect the ONs from nuclease degradation and increase their plasma half‐life. Aptamer‐ONs have since shown effective in vivo delivery of miRNAs, antagomirs, ASOs and bi‐modular miRNA‐antagomirs within preclinical cancer models (Catuogno et al, 2015; Esposito et al, 2016; Soldevilla et al, 2018).
Carrier‐based delivery systems
The pharmacological properties of carrier‐based delivery systems are largely independent of the physicochemical properties of the ON cargo, and instead depend on the properties of the delivery system. Therefore, the desired properties can be built into them via formulation design, resulting in multifunctional advanced drug delivery systems. These delivery systems may serve (often simultaneously) many different purposes, including (i) protecting the ON cargo from premature degradation, (ii) increasing the effect duration and (iii) enhancing the targeting. This improved targeting can either occur via passive or active targeting. Passive targeting exploits the microanatomical features of tissues, for instance, tissues with enhanced permeability and retention, or tissues with discontinuous/fenestrated epithelium. For active targeting, delivery systems are decorated with active targeting ligands. Particulate carrier‐based delivery systems also facilitate intracellular delivery by enhancing cellular uptake, intracellular trafficking and endosomal escape. In that way, the dose reaching non‐target tissues and/or the toxicological targets may be reduced, whereas the dose reaching the pharmacological target may be increased. The net result is an improved drug therapeutic index. The complexity of these systems leads to new challenges in the development, for example with respect to cost, manufacturability, safety, quality assurance and quality control.
Reflecting the immense interest in delivery of therapeutic ONs, a plethora of nanocarrier types have been investigated for delivery purposes, such as gold nanoparticles (Ding et al, 2014; Morgan et al, 2019), mesoporous silica (Steinbacher & Landry, 2014; Cha et al, 2017) and other inorganic nanocarriers (Malmsten, 2013). Yet, the current focus seems to be on lipid‐, polymer‐ and peptide‐based delivery systems and hybrids of these, which are described further below (Fig 4).
Lipid‐based delivery systems
The recent approval of patisiran (Table 3) (Suhr et al, 2015), together with improvements in manufacturability brought about by the introduction of microfluidics, has reinforced interest in lipid‐based delivery systems to the scientific community and pharmaceutical industry. The term lipid nanoparticles (LNPs) is used generically below to describe ON‐loaded lipid‐based delivery systems, because the structural complexity of most lipid‐based nanocarriers complicates their further classification into, for example, liposomes and solid lipid nanoparticles.
Table 3.
Clinically approved oligonucleotides.
| Approval date | Drug name | Disease | Target | ASO sequence 5′–3′ | Administration route/target tissues | |
|---|---|---|---|---|---|---|
| FDA | EMA | |||||
| RNaseH | ||||||
| 26 August 1998 | 29 July 1999 | Fomivirsen (Vitravene) | Cytomegalovirus retinitis in immunocompromised patients | CMV major immediate‐early gene mRNA | dGs‐dCs‐dGs‐dTs‐dTs‐dTs‐dGs‐dCs‐dTs‐dCs‐dTs‐dTs‐dCs‐dTs‐dTs‐dCs‐dTs‐dTs‐dGs‐dCs‐dG | IVT / eye |
| 29 January 2013 | refused authorisation | Mipomersen (Kynamro) | Homozygous familial hypercholesterolemia | Apolipoprotein B‐100 | Gs*‐mCs*‐mCs*‐Ts*‐mCs*‐dAs‐dGs‐dTs‐dmCs‐dTs‐dGs‐dmCs‐dTs‐dTs‐dmCs‐Gs*‐mCs*‐As*‐mCs*‐mC* | SC / liver |
| 05 October 2018 | 05 July 2018 | Inotersen (Tegsedi) | Hereditary transthyretin amyloidosis | Transthyretin | mTs*‐mCs*‐mTs*‐mTs*‐Gs*‐dGs‐dTs‐dTs‐dAs‐dmCs‐dAs‐dTs‐dGs‐dAs‐dAs‐dAs‐mTs*‐mCs*‐mCs*‐mC*‐3′ | SC / liver |
| Under review | 03 May 2019 | Volanesorsen (Waylivra) | Familial chylomicronemia syndrome, hypertriglyceridemia and familial partial lipodystrophy | Apolipoprotein CIII | As*‐Gs*‐mCs*‐Ts*‐Ts*‐dmCs‐dTs‐dTs‐dGs‐dTs‐dmCs‐dmCs‐dAs‐dGs‐dmCs‐Ts*‐Ts*‐Ts*‐As*‐T* | SC / liver |
| Splice modulation | ||||||
| 01 June 2019‡ | N/A | Milasen | Neuronal ceroid lipofuscinosis 7, Batten's disease | MFSD8 exon 6 | As*‐As*‐Ts*‐Gs*‐Ts*‐Ts*‐As*‐Gs*‐Ts*‐Gs*‐mCs*‐Ts*‐Ts*‐Gs*‐Ts*‐Ts*‐Gs*‐As*‐Gs*‐Gs*‐Gs*‐mC* | IT / CNS |
| 19 September 2016 | refused authorisation | Eteplirsen (Exondys51) | Duchenne muscular dystrophy | Dystrophin exon 51 | CTCCAACATCAAGGAAGATGGCATTTCTAG | IV / skeletal muscle |
| 12 December 2019 | Under review | Golodirsen (Vyondys 53) | Duchenne muscular dystrophy | Dystrophin exon 53 | GTTGCCTCCGGTTCTGAAGGTGTTC | IV / skeletal muscle |
| August 2020 | Under review | Viltolarsen† (Viltepso) | Duchenne muscular dystrophy | Dystrophin exon 53 | CCTCCGGTTCTGAAGGTGTTC | IV / skeletal muscle |
| 23 December 2016 | 30 May 2017 | Nusinersen (Spinraza) | Spinal muscular dystrophy | Survival motor neuron 2 exon 7 | mTs*‐mCs*‐As*‐mCs*‐mTs*‐mTs*‐mTs*‐mCs*‐As*‐mTs*‐As*‐As*‐mTs*‐Gs*‐mCs*‐mTs*‐Gs*‐G* | IT / CNS |
| Aptamer | ||||||
| 17 December 2004 | 31 January 2006 | Pegaptanib (Macugen) | Age‐related macular degeneration | Vascular endothelial growth factor | 40 kDa PEG‐5′‐CF‐G^‐G^‐A‐A‐UF‐CF‐A^‐G^‐UF‐G^‐A^‐A^‐UF‐G^‐CF‐UF‐UF‐A^‐UF‐A^‐CF‐A^‐UF‐CF‐CF‐G^‐3′‐3′‐dT‐5′ | IVT / eye |
| RNAi | ||||||
| 10 August 2018 | 27 August 2018 | Patisiran (Onpattro) | Hereditary transthyretin amyloidosis | Transthyretin |
5′‐G‐U^‐A‐A‐C^‐C^‐A‐A‐G‐A‐G‐U^‐A‐U^‐U^‐C^‐C^‐A‐U^‐dT‐dT‐3′ 3′‐dT‐dT‐C‐A‐U^‐U‐G‐G‐U‐U‐C‐U‐C‐A‐U^‐A‐A‐G‐G‐U‐A‐5′ LNP formulated |
IV / liver |
| 20 November 2019 | 02 March 2020 | Givosiran (Givlaari) | Acute hepatic porphyria | Aminolevulinate synthase 1 |
5′‐Cs^‐As^‐G^‐A^‐A^‐A^‐GF‐A^‐GF‐U^‐GF‐U^‐CF‐U^‐CF‐A^‐U^‐C^‐U^‐U^‐A^‐3′ 3′‐Us^‐Gs^‐G^‐UF‐C^‐UF‐U^‐UF‐C^‐UF‐C^‐AF‐C^‐AF‐G^‐AF‐G^‐UF‐A^‐GF‐AsF‐AsF‐U^‐5′ GalNAc Conjugate |
SC / liver |
| 16 October 2020¥ | Lumasiran (Oxlumo) | Primary hyperoxaluria type 1 | Hydroxiacid oxidase 1 |
5′‐As^‐Cs^‐C^‐U^‐G^‐A^‐A^‐AF‐G^‐UF‐A^‐G^‐G^‐A^‐CF‐CF‐U^‐UF‐U^‐A^‐Us^‐AsF‐U^‐3′ 3′‐Gs^‐As^‐Cs^‐U^‐U^‐U^‐CF‐A^‐UF‐CF‐CF‐U^‐G^‐G^‐A^‐A^‐A^‐U^‐A^‐U^‐A^‐5′ GalNAc Conjugate |
SC / liver | |
| 16 October 2020¥ | Inclisiran | Atherosclerotic cardiovascular disease | Proprotein convertase subtilisin‐kexin type 9 |
5′‐As^‐CFs‐As^‐AF‐AF‐AF‐G^‐CF‐A^‐AF‐A^‐A^‐C^‐AF‐G^‐GF‐U^‐CF‐U^‐A^‐Gs^‐As^‐A^‐3′ 3′‐U^‐G^‐U^‐U^‐U^‐U^‐C^‐G^‐U^‐U^‐dT‐U^‐GF‐U^‐CF‐C^‐A^‐G^‐As^‐Us^‐C^‐5′ GalNAc Conjugate |
SC / liver | |
s, phosphorothioate linkage; *, 2′‐MOE; d, 2′‐deoxy; m, 5‐methyl; F, 2′‐F; ^, 2′‐OMe; italicised, PMO; † Viltolarsen approval by Japanese Ministry of Health Labour and Welfare, 25 March 2020; ‡ Milasen approved by FDA for clinical testing only, ¥, lumasiran and inclisiran received positive opinion for marketing by the CHMP.
Cationic lipids entrap ONs via attractive electrostatic interactions (Felgner et al, 1987), and highly efficient commercial in vitro transfection reagents are based on cationic lipids. However, as systemic toxicity of cationic lipids is often dose‐limiting for in vivo application, ionisable lipids that are positively charged at low pH, e.g. during LNP manufacture, and typically neutral at physiological pH, are favoured (Semple et al, 2001). Today, a vast number of ionisable lipids have been developed covering a wide range of different structures. These include, among others, lipidoids (Akinc et al, 2008; Dong et al, 2014) and the ionisable lipid DLin‐MC3‐DMA (Jayaraman et al, 2012), which is considered the gold standard of ionisable cationic lipids. In general, they display headgroups containing tertiary amines, which are protonated under acidic conditions and uncharged at neutral pH. The hydrophobic lipid tails stabilise the LNP structure during formation and in formulation via hydrophobic interactions.
Clinically approved patisiran contains DLin‐MC3‐DMA, helper lipids (Kulkarni et al, 2019) and PEG‐lipid encapsulating siRNA directed against transthyretin (TTR) mRNA (Adams et al, 2018). The PEG lipid stabilises the LNPs during manufacturing and storage, and it increases the circulation half‐life. However, PEG lipids inhibit cellular transfection; hence, they are designed to rapidly diffuse from the LNPs after IV administration (Chen et al, 2016). The LNPs are passively targeted to the liver (Shi et al, 2011), and the size of the LNPs permits delivery through the fenestrated endothelium in the liver to the underlying hepatocytes (Chen et al, 2016). In addition, active hepatocyte targeting has been shown to occur via surface adsorption of apolipoprotein E, which targets the LNPs to the internalising low‐density lipoprotein receptor expressed on hepatocytes (Akinc et al, 2010; Chen et al, 2016). After cellular uptake, endosomal escape of siRNA into the cytosol may be facilitated via interactions between the re‐protonated ionisable cationic lipid in the acidic endosomal environment and anionic endogenous lipids in the endosomal membrane (Habrant et al, 2016).
Exosomes are particular lipid‐based nanocarriers (Barile & Vassalli, 2017). These nanosized vesicles are shed from the cells, encapsulating part of the cellular cytoplasm in the process (Pathan et al, 2019). They are remarkable in their biocompatibility and potential for highly specific active targeting through surface display of endogenous cellular ligands. The main challenges for using exosomes as delivery systems are (i) reproducible, large‐scale production and (ii) effective loading of drugs. Additionally, exosome heterogeneity is caused by their natural content of proteins and nucleic acids derived from the host cell (Willms et al, 2018; Jeppesen et al, 2019). This complicates their use as therapeutic delivery agents. The therapeutic promise of exosomes has been extensively reviewed elsewhere (Wiklander et al, 2019).
Polymer‐based delivery systems
Although less clinically advanced, polymer‐based systems are also interesting carriers for ON delivery, largely due to the chemical flexibility of polymers, in particular synthetic polymers (Fig 4) (Freitag & Wagner, 2020). Both monomer sequence and side/end group functionalities can be engineered. Additionally, polymeric nanocarriers exhibit high structural integrity and stability during storage.
One polymer with high biocompatibility that has been studied and used extensively is the copolymer poly(lactic‐co‐glycolic acid) (PLGA) (Rezvantalab et al, 2018). For small‐molecule drugs, highly efficient encapsulation in polymeric nanoparticles can be achieved, e.g. through miniemulsion‐based synthesis, followed by in situ polymerisation (Fusser et al, 2019). However, due to their negative charge, anionic ONs cannot be encapsulated using this approach. Instead, encapsulation can be achieved through attractive electrostatic interactions between the anionic ONs and polycationic polymers. Dendrimers are hyperbranched polymers, which are well suited for this purpose because they can complex many ON molecules. Several cationic polymers have been used, including poly(amidoamine), poly(propyleneimine) and poly(L‐lysine) [reviewed by (Mignani et al, 2019)].
Among the synthetic polymers, polyphosphazenes are notable in their high biocompatibility and chemical flexibility, and they have successfully been used to deliver therapeutic ONs (Peng et al, 2016; Hsu et al, 2020). Polyphosphazenes can be tailored to exhibit responsivity to external (bio)chemical stimuli (Teasdale, 2019), e.g. local pH. This allows for a targeted release of the cargo at the desired site of action. Complementing the use of synthetic polymers, there is long‐standing interest in the use of naturally occurring biopolymers for ON encapsulation; the most notable example is the use of the polycation chitosan, often in complex with another, anionic polymer, e.g. PLGA (Taetz et al, 2009) or alginate (Lee & Mooney, 2012).
Recently, there has been significant interest in lipid–polymer hybrid nanoparticles (Thanki et al, 2017). These hybrids combine desirable properties from both nanoparticle types, i.e. the serum stability of PLGA‐based matrix system with the biocompatibility and high loading capacity of ONs in delivery systems based on cationic lipids.
Peptide‐based delivery systems
CPPs represent another group of compounds that have been also successfully used as a carrier‐based drug delivery system (Lehto et al, 2016). In this context, formation of CPP/ON nanoparticles is driven by electrostatic and hydrophobic interactions between cationic CPPs and anionic ONs. Compared with directly conjugated CPP‐ONs, peptide‐based vectors are more amphipathic and usually carry additional chemical modifications that make them compatible with encapsulating ONs. Commonly, such modifications include incorporation of various hydrophobic modifications, such as fatty acid derivatives, to the CPP sequences, which increase the stability of the formulation and enhance their cellular uptake and endosomal escape. Various types of CPPs have demonstrated considerable potential for ON delivery in a nanoparticle‐based format, including MPG and PepFect peptide derivatives [reviewed in (Boisguerin et al, 2015; Lehto et al, 2016)].
Antibody complexation delivery systems
Antibodies are another promising form of carrier delivery system used both as direct conjugates or unconjugated carriers. As unconjugated carriers, antibodies or antibody fragments have been fused with either avidin or protamine peptide. Taking advantage of the natural avidin–biotin complexation system, antibody–avidin fusion molecules bind to biotinylated ONs (Penichet et al, 1999). The peptide protamine is a positively charged RNA‐binding peptide, which binds to siRNA and condenses it into antibody–siRNA complex (Song et al, 2005). This system has been used to link cytotoxic siRNAs with Her2‐positive cancer cell‐targeted antibodies (Yao et al, 2012). Like all complexation systems, these two systems have the advantage of an established target‐specific antibody carrier, which can easily be complexed with any siRNA.
Model systems for oligonucleotide development
Successful development of ON‐based drugs depends on detailed knowledge about pharmacokinetic (PK) and pharmacodynamic (PD) properties. PK/PD analyses describe the relationship between PK (drug concentration in the organism) and PD (the organism's biological response to the drugs) in a time‐dependent manner (Negus & Banks, 2018). PK/PD modelling and simulations are used to rapidly characterise the efficacy and safety of drugs, and PK/PD simulation models containing in vitro and in vivo preclinical studies can anticipate potential risks in humans (Li et al, 2016). The use of predictive model systems for PK/PD analyses save time, costs and minimise the need for in vivo studies, facilitating the translation from bench to bedside.
Methodologies for in vitro testing of oligonucleotides
In vitro models can be implemented to test pharmacological activity, transfection efficiency, hepatotoxicity and intracellular half‐life. However, it is usually difficult to correlate in vitro findings to preclinical and clinical in vivo findings (Table 2). Novel technologies, such as reprogramming patient‐derived cells into induced pluripotent stem cells (iPSCs) (Takahashi & Yamanaka, 2006; Takahashi et al, 2007) and genome editing techniques to make isogenic cell lines, have revolutionised the field (Ran et al, 2013). Two‐dimensional (2D) and three‐dimensional (3D) cell cultures, including organoids, are used to improve the understanding of pathological disease mechanisms, as well as ON efficacy studies. One example of successful translation from a 3D‐model to a clinical trial is sepofarsen for the treatment of the inherited retinal disease Leber congenital amaurosis (LCA) (Collin et al, 2012; den Hollander et al, 2006). Combining patient‐derived retinal organoids with toxicity studies in non‐human primates (NHPs) was sufficient to initiate a phase I/II clinical trial (NCT03140969, NCT03913143) (Cideciyan et al, 2019). The eye is an exceptional target organ, given its isolated and immune‐privileged status, which allows for translation of results from organoids in culture to the human eye. However, for other (multi‐)organ diseases, establishing predictive cellular models to mimic the functions of entire organs remains a challenge.
Table 2.
Comparison of different disease models.
| In vivo | In vitro | 3D Organoids | Organs‐on‐chips | |
|---|---|---|---|---|
| Human‐derived tissue | No | Yes | Yes | Yes |
| Personalised medicine | No | Yes | Yes | Yes |
| Realistic microenvironment | Yes | No | Yes | Yes |
| Organ‐level function | Yes | Limited | Potentially/Limited | Potentially |
| Real‐time readouts | No | Limited | Limited | Yes |
| High‐throughput testing | No | Yes | Limited | Possibly |
| Pharmacodynamics / ‐kinetics | Yes | No | No/Limited | Potentially |
An interesting alternative to 2D and 3D tissue culture techniques is the microfluidics‐based organ‐on‐chip technology (van der Meer & van den Berg, 2012), which consists of micro‐engineered iPSC‐derived models that combine the advantages of current in vitro and in vivo models. The technology breaks down organs into the most essential components, including biological barriers, for drug delivery, efficacy, toxicity and PK/PD studies. Organ‐on‐chips reproduce the interaction between cultures of multiple tissue types using microfluidic channels and chambers (Huh et al, 2010; Kim et al, 2012; Westein et al, 2013). This interaction can be monitored in real time to study the PK/PD of a specific drug as well as drug–drug interactions (Lee et al, 2017; Shinha et al, 2020). For instance, the PK/PD evaluation of terfenadine (a type of antihistamine) has been assessed by using a cellular model combining heart and liver cells in two interconnected chambers. This model, combined with microelectrode arrays, also contributed to predict the potential cardiotoxicity of the drug (McAleer et al, 2019). Interestingly, recent drug permeability studies in blood–brain barrier (BBB)‐on‐chip models were found to be more predictive compared with existing in vitro models (van der Helm et al, 2016). Other cellular models under development include retina‐on‐chip (Achberger et al, 2019; Seo et al, 2019) and lung‐on‐chip (Huh et al, 2010) models. Mimicking the function of entire organ(s) in a dish by combining several cell types in a single device may have valuable potential for drug screening and development, as well as PK/PD and toxicity studies. In the future, organ‐on‐chip models might, to some extent, replace experimental animal models.
Investigation of PK/PD properties in vivo
In vivo models have been extensively used for dose‐finding studies. PK properties are largely comparable across multiple species including mouse, rat, NHP and human (Yu et al, 2009; Geary et al, 2015). Hence, cross‐species PK/PD relationships are very valuable for the prediction of human dosing. Animal models have been vital for determining in vivo efficacy of ONs, tissue‐specific delivery, and optimising the route of administration for systemic and neurological diseases (Schoch & Miller, 2017; Buijsen et al, 2019). Preclinical in vivo testing in a transgenic mouse model for SMA predicted the enhanced benefit of treating pre‐symptomatic stages of the disease, which was later validated in the clinic.
However, detailed knowledge of the disease model is vital for interpreting data: A study in the mdx mouse model for DMD of the PK/PD of 2ʹ‐OMe ONs for DMD revealed higher ON levels in dystrophin‐deficient muscle fibres than in healthy fibres, as well as an enhanced exon skipping efficiency (Heemskerk et al, 2010). However, ON efficiency was lower in clinical trials in DMD patients, potentially due to a better regenerative capacity in mice. Also, animal models may not always reciprocate the human condition due to the different genomic context of the mutations, even when using humanised animal models. This is evident for pre‐mRNA splicing, which is differentially regulated between tissues, organs and species (Rivera‐Barahona et al, 2015). Between tissues, DNA variants have been observed to affect pre‐mRNA splicing, complicating the interpretation of in vitro studies. An example is the aforementioned deep‐intronic change underlying LCA: while lymphoblastoid and fibroblast cells derived from patients suggested a hypomorphic effect (Garanto et al, 2016), reprogrammed patient‐derived iPSCs differentiated towards a retinal fate revealed that the percentage of aberrantly spliced mRNA was highly increased in photoreceptor cells, explaining the retinal phenotype observed in LCA patients (Parfitt et al, 2016). Follow‐up studies revealed that a pseudoexon present in humans was differentially recognised in cell lines derived from other species (Garanto et al, 2015). Thus, care is warranted when selecting a model system for assessing the effects of a certain genetic variant, as well as for the development of splice‐modulation therapies.
Safety assessment of oligonucleotide‐based therapeutics
While new chemistries and delivery technologies might lead to higher efficacy, it is important to screen for potential side effects in early phases of preclinical development to avoid subsequent failure. Toxicological aspects of therapeutic ONs have been comprehensively summarised previously (Andersson & den Besten, 2019). The Oligonucleotide Safety Working Group (OSWG) has also published extensive guidelines for assessing the various aspects of ON safety. Our understanding of ON‐mediated toxicity increases as more preclinical and clinical data become available. While the concept of class toxicity appears nuanced in the light of the expanding knowledge on various chemistries, ON‐related side effects still falls under two main categories: (i) hybridisation‐dependent effects, including on‐ and off‐target effects, and (ii) hybridisation‐independent effects, mostly caused by protein‐binding properties (Fig 5).
Figure 5. ASO mediated toxicities.
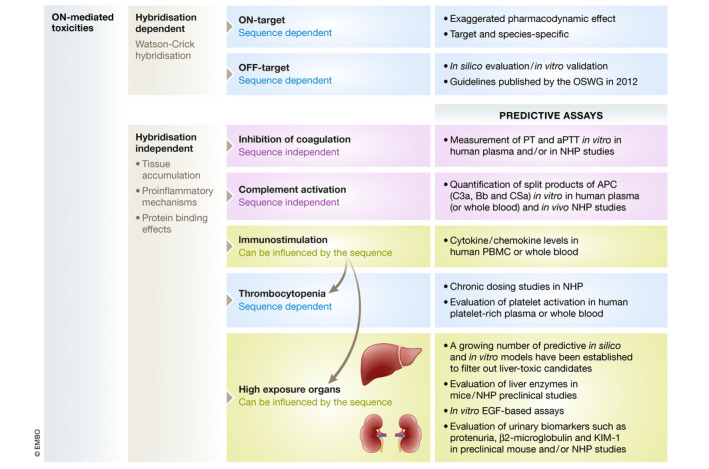
Schematic representation of the most common ON‐mediated toxicities, which are mainly classified as hybridisation‐dependent (Watson–Crick hybridisation) or hybridisation‐independent effects (tissue accumulation, proinflammatory mechanisms and/ or protein binding effects). Some of them are strictly class specific (sequence independent), while others can be influenced by the sequence (sequence specific).
Hybridisation‐dependent effects
On‐target safety, also referred to as exaggerated pharmacology, relates to the possible toxicities induced by excessive or prolonged activity of the ON in target or non‐target organs. These effects are considered rare and are generally discovered in preclinical studies. However, due to the sequence‐specific action of ON‐based drugs, target sequences may not be conserved across species. Therefore, human sequences might not display efficacy in rodents or NHPs; hence, species‐specific surrogates are needed for on‐target risk assessment (Levin & Henry, 2008).
Off‐target effects correspond to the potential toxicities associated with ON hybridisation to unintended RNA targets (complete or partial complementarity). They have increased with the development of high‐affinity chemistries, e.g. LNA, tcDNA and constrained ethyl (cEt), which allow the use of much shorter sequences. Off‐target effects are of particular concern for gapmer ONs and siRNA, which aim at downregulating their targets, as they could downregulate the expression of unintended ones (Fedorov et al, 2006; Burel et al, 2016). Several studies have characterised off‐target effect‐associated mechanisms and described elegant ways to reduce risks and improve the design of specific gapmers and siRNAs (Hagedorn et al, 2017; Janas et al, 2018). In contrast, splice‐switching ASOs must bind specific splicing regulatory elements to be efficacious, and they are therefore less likely to induce off‐target effects. With the development of more stable ONs and efficient delivery systems, systemic administrations might distribute to target but also non‐target tissues; hence, off‐target effects should be carefully evaluated during preclinical development. The guidelines published by the OSWG for assessing off‐target effects recommend (i) in silico evaluation, (ii) interpretation of in silico hits using auxiliary data (e.g. time‐ and spatiotemporal‐dependent expression of off‐target RNA) and (iii) in vivo evaluation of ON drugs (Lindow et al, 2012).
Hybridisation‐independent effects
Most ON‐mediated toxicities are not caused by Watson–Crick base pairing to RNA, but are rather a result of ON–protein interactions and therefore depend on the chemistry and/or the delivery system. Single‐stranded PS‐modified ON display particularly high protein binding affinities, and the majority of the hybridisation‐independent effects have thus been reported for this class of ON, as opposed to siRNA containing less PS‐modified residues.
Inhibition of blood coagulation
Inhibition of the intrinsic blood coagulation pathway is a well‐documented side effect of the PS chemistry (Henry et al, 1997b; Echevarria et al, 2019). It is considered a class effect, modulated by interactions of the ON with plasma proteins in a sequence‐independent way. The PS modification selectively prolongs the partial thromboplastin time at low plasma concentrations by inhibiting the tenase complex. However, at high plasma concentrations, both the intrinsic and extrinsic pathways are affected, suggesting additional inhibitory effects (Sheehan & Lan, 1998). Prolongation of clotting times is correlated with the maximal plasma concentration (C max) of circulating ONs, and it has not been associated to relevant clinical signs, as it can be controlled by dose reduction or by extending infusion times. Nevertheless, it should be included in screening studies, which can be performed both in vivo and in vitro in mouse, NHP and human serum, respectively, since results can be extrapolated across species (Andersson & den Besten, 2019).
Complement activation
Systemic administration of PS‐modified ONs has been reported to activate the alternative complement pathway as a consequence of plasma protein binding (Henry et al, 2002). Although this hybridisation‐independent effect is mainly related to the ON chemistry (class effect), unexpected complement activation has been observed with some sequence specificity, as in the case of tcDNA (Aupy et al, 2020). Activation of the alternative complement pathway has been thoroughly studied in NHP models, which are particularly sensitive (Henry et al, 2016). The effect is dependent on the plasma concentration and can be controlled by increasing the IV infusion time to reduce the Cmax. PS‐modified ONs have been shown to interact directly with plasma factor H, which is a negative regulator of the complement cascade that reduces the free levels of inhibitor, permitting uncontrolled amplification of the cascade and release of split products such as Bb and anaphylotoxins C3a and C5a (Henry et al, 1997a). Complement can be activated similarly at every dose; hence, chronic administration of toxic ONs can result in C3 depletion, eventually leading to altered complement function, secondary inflammation and vasculitis (Engelhardt et al, 2015; Shen et al, 2016; Andersson & den Besten, 2019). Although humans appear less sensitive to complement activation, it is recommended to routinely evaluate complement activation in preclinical safety studies of new ON‐drug candidates in NHPs.
Complement activation can be assessed in vitro in NHP or human serum, or whole blood, to measure split products of the alternative complement pathway (Bb, C3a and C5a). Nevertheless, one should be cautious when interpreting the results, as it is difficult to extrapolate and predict dose–response relationships (Andersson & den Besten, 2019).
Immunostimulation
ON‐induced immunostimulation is a complex side effect that depends on several aspects, including chemistry and nucleotide sequence (Krieg, 1998; Agrawal & Kandimalla, 2004). ONs can activate the innate immune system through binding to pattern‐recognition receptors (PRRs) such as the Toll‐like receptors (TLRs). Activation of the innate immune system by CpG‐containing ONs is comparable to that observed for bacterial DNA and CpG‐containing ONs are used for cancer and autoimmune disease therapies as well as vaccine adjuvants (Krieg & Davis, 2001; Krieg, 2006; Kline & Krieg, 2008). However, the immunostimulatory activity of ONs designed for antisense purposes constitutes a potential side effect. In this regard, modified ONs with 2ʹ‐ribose modifications, 5‐methyl cytosine residues, or without CpG motifs, have been designed to avoid TLR9 activation. Additional studies have demonstrated that CpG‐free, PS‐modified ONs can also elicit proinflammatory responses, although the molecular mechanism is still debated (Vollmer et al, 2004; Senn et al, 2005; Younis et al, 2006). Of note, immunostimulatory effects have never been reported for ONs with neutral backbones, e.g. PMOs (Zhang et al, 2015). Rodents are particularly sensitive to immune stimulation. Mice treated with high doses of PS‐ONs display increased levels of circulating cytokines (IL‐1b, IL‐6, interferon, tumour necrosis factor‐α) and chemokines, as well as proliferation of B‐lymphocytes (Monteith et al, 1997). Although generally less critical, some significant inflammatory responses, e.g. vasculitis, related to complement activation mediated by PS‐ONs, have been described in NHP studies (Levin & Henry, 2008; Engelhardt et al, 2015; Frazier, 2015; EMA, 2016). Differences in immune response between species have been attributed to the differential sequence, expression and function of the germline‐encoded PRRs (Barchet et al, 2008).
In clinical trials, inflammatory adverse effects may manifest as flu‐like symptoms and injection site reactions following subcutaneous (SC) administration (Rudin et al, 2001; Thomas et al, 2013; Voit et al, 2014). Understanding the underlying mechanisms of therapeutic ON‐mediated induction of proinflammatory adverse effects has facilitated the design of safer and more potent sequences that are efficacious at lower doses. Nevertheless, some sequences still display unexpected toxicity, and specific screening for immunostimulatory adverse effects is recommended. In addition to in vivo studies in rodents and NHPs, proinflammatory evaluation is usually performed in vitro using human peripheral blood mononuclear cells or whole blood (Apter et al, 1990; Lankveld et al, 2010).
The potential immunogenicity of ONs is poorly documented but recent data show that anti‐drug antibodies (ADAs) are present in NHPs and humans (Andersson & den Besten, 2019). More than 30 and 70% of patients treated with drisapersen and mipomersen, respectively, were found positive for ADA after 24 weeks of treatment. Recently, ADA plasma levels were shown to increase both in monkeys and humans, while no impact on efficacy and safety was reported (Bosgra et al, 2019; Yu et al, 2020).
Formulations based on nanoparticles administered IV may also induce infusion‐related reactions (IRRs), e.g. hypersensitivity, evident as flu‐like symptoms and even cardiac anaphylaxis (Szebeni, 2018). Hence, before IV infusion of patisiran, patients are required to be premedicated with IV antihistamines (H1/H2 blockers), IV corticosteroid and oral acetaminophen or paracetamol to suppress IRRs. Mild to moderate IRRs were observed in a phase III trial of patisiran in approximately 20% of the patients, who were all premedicated, the incidence of which decreased over time (Adams et al, 2018). In contrast, premedication is not required before administration of ONs and GalNAc–siRNA conjugates.
Thrombocytopenia
ON‐associated thrombocytopenia is an occasional event that has been observed in rodents and NHPs, as well as in three recent clinical trials with unrelated PS‐ONs [volanesorsen (FDA, 2018), inotersen (Benson et al, 2018; Mathew & Wang, 2019) and drisapersen (EMA, 2016; Goemans et al, 2016)]. The exact underlying mechanisms of thrombocytopenia is still debated, and several immune and non‐immune mediated mechanisms have been proposed. Direct activation of platelets by PS‐ONs through the binding to platelet receptors has been demonstrated (Flierl et al, 2015; Sewing et al, 2017). In addition, a heparin‐induced thrombocytopenia‐like mechanism through the induction of anti‐platelet factor 4 IgG antibodies has also been proposed, based on the binding of nucleic acids to platelet factor 4 (Jaax et al, 2013), although contradictory results have been reported. A recent study suggests that sequestration of platelets in the liver and spleen occurs through the activation of monocytes, but not platelets, and is accompanied by increased serum IgM levels (Narayanan et al, 2018). In most cases, thrombocytopenia after treatment with ONs is mild to moderate and reversible. The number of platelets does not drop below the normal limit during treatment and normalises after withdrawal from treatment. However, a concerning and severe decline (< 50,000 platelets/µl) has been observed in NHP studies after repeated dosing (Henry et al, 2017). To date, severe thrombocytopenia has not been reported for siRNA drugs, neither in preclinical studies nor in clinical trials, but encapsulation of siRNA to LNPs has been shown to cause thrombocytopenia in rats, presumably induced by the cationic lipid molecules themselves (Chi et al, 2017).
High‐exposure organs
Following IV administration and independently of the chemistry, the highest concentrations of ONs are found in the liver and the kidneys, which are considered high‐exposure organs (Fig 5). The toxicities observed in these organs are not necessarily associated with the accumulation of ONs per se but can also be due to sequence‐specific effects. Accumulated ONs are often apparent as basophilic granules (ONs in lysosomal compartments) in tissue sections. However, these effects are regarded as non‐adverse because of their reversible nature upon termination of treatment. In contrast, acute toxicities characterised by large areas of necrosis, pronounced elevation of liver enzyme levels, morbidity and mortality have been reported for some high‐affinity gapmers after a single or few doses in mice (Hagedorn et al, 2013; Burdick et al, 2014). The mechanisms underlying these sequence‐specific acute toxicities may be accumulation of RNase H‐cleaved mRNA products and/or protein interactions (Burel et al, 2016; Sewing et al, 2016; Shen et al, 2019). While the screening for these acute toxicities previously relied on in vivo studies assessing levels of liver enzymes following IV administration in rodents, a growing number of predictive in silico (Hagedorn et al, 2013; Burdick et al, 2014) and in vitro models (Sewing et al, 2016; Dieckmann et al, 2018) have been established.
Renal lesions are generally restricted to the proximal tubules and appear only in animals treated with much higher ON doses than the clinically relevant doses. No clinically significant renal dysfunction was reported in a large retrospective study of 2ʹ‐MOE gapmer trials (Crooke et al, 2018). Renal toxicity was mostly regarded as accumulation‐related toxicity and primarily sequence unspecific until more acute tubular lesions were reported with high‐affinity ONs, e.g. LNAs (Engelhardt et al, 2015). Beyond the classical biomarkers for renal injury, e.g. increased excretion of β2‐microglobulin and kidney injury molecule‐1, a predictive epidermal growth factor‐based assay has recently been developed to exclude this type of nephrotoxic candidates (Moisan et al, 2017).
Approved oligonucleotide‐based therapeutics
Advances in therapeutic ON technology in recent decades provide a unique opportunity for addressing previously inaccessible drug targets (Bennett et al, 2017). Since the approval of fomivirsen in 1998 by the FDA for treating cytomegalovirus (CMV) retinitis (Marwick, 1998), 11 ON‐based drugs have received marketing authorisation to be used in humans, and two additional ON drugs have received positive opinion for marketing by the EMA (Table 3). Here, we discuss approved ON therapeutics according to their functional modalities.
Fomivirsen (Vitravene) is a 21‐mer PS DNA‐based ON developed for treating CMV retinitis patients, especially those with acquired immunodeficiency syndrome (AIDS) (Vitravene Study G, 2002). This first‐generation ASO targets the human CMV major immediate‐early gene mRNA for RNAse H degradation (Geary et al, 2002). Fomivirsen is delivered locally by IVT administration and hence does not require a delivery agent. While a second‐generation 2ʹ‐MOE‐based gapmer sequence (ISS 13312) was in clinical development (Henry et al, 2001), Novartis discontinued development and withdrew marketing (Wathion, 2002). The number of CMV retinitis cases had decreased dramatically due to the development of highly active antiretroviral therapy. Nevertheless, fomivirsen was a success and established ON therapies as viable for clinical development.
RNase‐dependent second‐generation ASOs targeting the liver have been approved for polyneuropathy of hereditary transthyretin‐mediated amyloidosis (hATTR) (inotersen) as well as familial chylomicronemia syndrome (FCS), hypertriglyceridemia and familial partial lipodystrophy (volanesorsen), and familial hypercholesterolemia (mipomersen). The rare disease hATTR is linked to missense mutations in the TTR gene, which result in TTR protein misfolding. The TTR protein is secreted into the blood and cerebral spinal fluid, and accumulation of amyloid deposits (both wild type and mutant) in tissues causes polyneuropathy, multiorgan dysfunction and cardiomyopathy. Inotersen targets the hepatic expression of both wild‐type and mutant TTR mRNA. Patients treated with inotersen display a reduction in serum TTR protein levels and enhanced quality of life (Benson et al, 2018). Volanesorsen, although still awaiting FDA approval at the time of writing this review, was awarded EMA approval in May 2019. By targeting the 3ʹ UTR of apolipoprotein C3 mRNA, volanesorsen reduces the levels of triglycerides and apolipoprotein C3, which represent two known risk factors for cardiovascular disease, while increasing the levels of low‐density and high‐density lipoprotein cholesterol and apolipoprotein B in patients with FCS and hypertriglyceridemia (Graham et al, 2013; EMA, 2019). Mipomersen also targets apolipoprotein B‐100 to reduce circulating low‐density lipoprotein cholesterol, which constitutes another major risk factor for cardiovascular disease (Wong & Goldberg, 2014). In contrast to inotersen and volanesorsen, mipomersen was given FDA approval, but EMA authorisation was denied due to safety concerns related to liver toxicity and severe cardiovascular events (EMA, 2012). It has since been discontinued by the FDA and is only available through a restricted risk evaluation and mitigation strategy. All three ON therapies are dosed by SC administration without a delivery agent due to the natural uptake of ONs by the liver. In 2004, the aptamer pegaptanib (Macugen) was approved by the FDA for the prevention of the eye‐related disorder age‐related macular degeneration (Ng et al, 2006). Pegaptanib is a covalent conjugate of a highly modified single‐stranded aptamer and two 20‐kDa PEG units. It binds with high specificity and affinity to the extracellular VEGF isoform 165 and blocks its neo‐angiogenic activity (Ruckman et al, 1998). Patients dosed with pegaptanib demonstrated reduced vision loss compared with placebo controls (Gragoudas et al, 2004). Common to degenerative diseases, early treatment results in improved therapeutic outcome (Gonzales CR & Group VISiONCT, 2005).
Several splice modifying ON‐based drugs are approved to treat the paediatric disorders DMD and SMA, focussing on splice modification and targets tissues beyond liver. The first approved drug, i.e. eteplirsen (Exondys51), is a PMO‐based splice‐switching ASO that interacts specifically with DMD exon 51, and is used in DMD patients with dystrophin deletions amenable to exon 51 skipping (~14% of patients) (Cirak et al, 2011). Dystrophin expression is limited mainly to skeletal and cardiac muscles, and eteplirsen is expected to be most efficacious in skeletal muscle. However, conducive with all PMOs, high accumulation in the kidneys and rapid urine excretion is also expected (Heemskerk et al, 2009). The approval of eteplirsen by the FDA was accompanied by controversy due to the trial design and difficulties in quantifying increased expression of dystrophin, which leaves doubt on the efficacy of eteplirsen (Aartsma‐Rus & Arechavala‐Gomeza, 2018). As a result, it was not approved by the EMA. Strengthened by improved clinical trial designs, two additional PMO‐based ASOs have recently been approved for DMD patients amenable for dystrophin exon 53 skipping, i.e. golodirsen and viltolarsen, by the FDA, and both the FDA and Japanese Ministry of Health Labour and Welfare, respectively (Dhillon, 2020; Heo, 2020).
The only ON‐based therapeutic approved for a neurological disease is nusinersen, used for the treatment of SMA (Aartsma‐Rus, 2017; Finkel et al, 2017; Mercuri et al, 2018). Nusinersen targets the alternatively spliced exon 7 of SMN2 pre‐mRNA, increasing exon inclusion and producing a functional SMN protein. It is administered directly to the cerebral spinal fluid surrounding the spinal cord by IT injection (Hache et al, 2016). IT administration directs uptake into the CNS, allowing low doses and circumvention of liver metabolism and kidney excretion. Patients, especially young pre‐symptomatic patients, report extended survival and reaching motor milestones over the expected natural history of the disease. Controversy related to nusinersen is not over efficacy but rather the exceedingly high cost, which has delayed approval and prevented marketing in countries with national health services (Starner & Gleason, 2019).
The success of nusinersen has led to the use of ONs as personalised medicines, exemplified in the development of milasen, which targets a mutation specific to a single patient with a form of Batten’s disease (Kim et al, 2019). In this case, the insertion of an SVA (SINE‐VNTR‐Alu) retrotransposon altered the splicing of the major facilitator superfamily domain containing 8 (MFSD8) exon 6 into a cryptic splice‐acceptor site. Clinicians followed the preclinical studies and trial designs from the nusinersen studies to accelerate the FDA approval of the clinical study: milasen dosing was initiated 14 months after clinical diagnosis and just 4.5 months after identification of a therapeutic ASO. The patient’s rate of deterioration meant that dosing had to be initiated as soon as possible; hence, the patient was dosed in parallel to toxicology studies in animals. Although therapeutic efficacy in a single patient cannot be defined, milasen reduced the frequency and duration of seizures and potentially diminished the neurodegenerative decline.
Two ON therapeutics based on RNAi, i.e. patisiran and givosiran, were approved by the FDA in 2018 and 2019, respectively. Patisiran represents an important milestone, because it is the first marketed drug based on siRNA, launched only 20 years after the discovery of the RNAi mechanism (Fire et al, 1998). Like inotersen, patisiran inhibits hepatocyte expression of TTR in patients with hATTR (Adams et al, 2018). Patisiran consists of siRNA directed against TTR mRNA formulated as LNPs, which are administered systemically by IV infusion. The latest breakthrough is givosiran, which represents the first approved GalNAc‐siRNA conjugate. Givosiran inhibits hepatic synthesis of delta aminolevulinate synthase 1 (ALAS1) in patients with acute hepatic porphyria (AHP), which is a rare inherited disease of haem biosynthesis (Sardh et al, 2019). Monthly subcutaneous administration of givosiran results in hepatocyte‐specific distribution and downregulation of elevated ALAS1 mRNA in the liver.
Recently, two new liver‐targeting GalNAc‐siRNA drugs received positive opinion for marketing in Europe, i.e. lumasiran and inclisiran (Fitzgerald et al, 2017; McGregor et al, 2020). Lumasiran targets hydroxyacid oxidase 1 (HAO1) for the treatment of primary hyperoxaluria type 1 (PH1), which is a rare inherited disorder characterised by the overproduction of oxalate. Targeting HAO1 reduces the substrate needed for oxalate production in the liver (McGregor et al, 2020). Inclisiran targets the proprotein convertase subtilisin–kexin type 9 (PCSK9) to reduce low‐density lipoprotein (LDL) cholesterol. PCSK9 is a serine protease, which binds to the LDL receptors to induce their lysosomal degradation. Therefore, silencing PCSK9 enhances the half‐life of LDL receptors responsible for cholesterol clearance (Fitzgerald et al, 2017). Inclisiran reduces more than 50% LDL cholesterol levels in treated patients with minimal side effects (Khvorova, 2017; Ray et al, 2020). Approval of Inclisiran will expand the indications for ONs to not only include rare but also common diseases.
Concluding comments and future perspectives
In 1978, it was demonstrated that a 13‐mer DNA‐based ON binding to Rous sarcoma virus RNA could inhibit protein expression in cell culture (Zamecnik & Stephenson, 1978), but it was not until 20 years later (1998) that the first ON‐based therapeutic drug fomiversen was approved. By 2016, only two additional drugs (pegaptanib and mipomersen) had been approved (Table 3) but since then the development pace of ON‐based drugs accelerated with 11 ON‐based drugs currently approved (Aartsma‐Rus & Corey, 2020). Yet, many of these drugs display limited efficacy (eteplirsen, golodirsen, viltolersen), and the more efficacious drugs take advantage of local administration (nusinersen). However, GalNAc conjugation and the LNP technology represent delivery breakthroughs that have completely changed the perspective for therapies targeting hepatocytes: in one stroke, this tissue is now accessible for treatment with ONs. These examples of how delivery technologies can be used to overcome delivery hurdles has provided the whole field with a new impetus that will accelerate discoveries for targeting of tissues beyond the liver.
Design and manufacturing of efficient delivery systems is not the only hurdle: the safety of these and their combination with ONs is also paramount. Testing ON safety has not been easy, primarily because many of these drugs have been developed to treat rare diseases. This implies an abundance of preclinical models but limited clinical data. Being a whole new class of drugs, this makes stakeholders wary of missing any step of the development. A striking exception to this is the recent n‐of‐one clinical studies: the development of milasen (Kim et al, 2019) was achieved in record time, but it took a high risk/high reward gamble, relying on the safety of IT administration of a chemistry already approved for nusinersen.
A likely leapfrog in the clinical application of therapeutic ONs may come from the results of current clinical trial (NCT04023552; testing APO(a)‐LRx, a GalNAc3‐conjugated ASO) for the lowering of lipoprotein (a) in cardiovascular disease. This trial includes 7,680 patients and the large data set that will be generated is due to change in the landscape for these drugs. By then, many new delivery technologies may have successfully been developed for other targets making this decade the era of ON therapeutics.
Pending issues
A vast array of delivery systems could be used to deliver ONs; however, the majority target the liver or deliver throughout the body without specificity. Further advances are needed to enhance tissue‐specific delivery.
Our understanding of ON‐mediated toxicities has improved, and many predictive in vitro tests have been developed to exclude toxic candidates early in development. Since some toxicities are sequence dependent, it will be important to implement toxicity screening early in the preclinical development of ONs.
Following the example of milasen, it is anticipated that bespoke ON therapies will be developed for additional brain diseases. A process to guide this development is required.
Very limited clinical data have been available for many therapeutic ONs, as most target rare disorders and have been tested in dozens or, at best, a few hundred patients. However, this may be about to change thanks to new ON drugs targeting common disorders, such as hyperlipidaemia, which could produce clinical data from thousands of patients and further accelerate development of future therapeutic ONs.
Conflict of interest
A.A‐R discloses being employed by LUMC which has patents on exon skipping technology. As co‐inventor of some of these patents, AAR is entitled to a share of royalties. AAR further discloses being ad hoc consultant for PTC Therapeutics, Sarepta Therapeutics, Eisa Pharmaceuticals, WaVe Life Sciences, Alpha Anomeric, CRISPR Therapeutics, BioMarin Pharmaceuticals Inc., Global Guidepoint and GLG consultancy, Grunenthal and BioClinica, being a member of the Duchenne Network Steering Committee (BioMarin) and of the scientific advisory boards of ProQR and Philae Pharmaceuticals. Remuneration for these activities is paid to LUMC. LUMC also received speaker honoraria from PTC Therapeutics and BioMarin Pharmaceuticals. A.G and R.W.J.C are inventors of several patents describing the use of antisense oligonucleotides for the treatment of inherited retinal diseases. C.F is ad hoc consultant for Lundbeck Pharma A/S, Valby, DK. W.vR.M discloses being employed by LUMC which has patents on exon skipping technology for brain disorders. As co‐inventor of some of these patents WvRM is entitled to a share of royalties. T.L is a consultant for and has equity interests in Evox Therapeutics Ltd., Oxford, UK. L.E is an employee of SQY therapeutics developing tcDNA antisense oligonucleotides. M.A.D and G.C are co‐inventors of patent WO2016/151523 (RNA interference mediated therapy for neurodegenerative diseases) filed by the University of Trento and are entitled to a share of royalties. S.M.H is an inventor on a patent describing cell‐penetrating peptides and is employed by Oxford Biomedica Plc, Oxford UK. Co‐authors, R.A.M.B, G.G, S.A, L.R.D, S.K, V.A‐G, S.E.B and A.T.G declare no conflict of interests.
For more information
Acknowledgements
This work was supported by funding from Cooperation of Science and Technology (COST) Action CA17103 (networking grant to V.A‐G). V.A‐G holds a Miguel Servet Fellowship from the ISCIII [grant reference CPII17/00004] that is part‐funded by the European Regional Development Fund (ERDF/FEDER) and also acknowledges funding from Ikerbasque (Basque Foundation for Science). S.M.H is funded by the Medical Research Council and Muscular Dystrophy UK. A.A‐R receives funding from amongst others the Duchenne Parent Project, Spieren voor Spieren, the Prinses Beatrix Spierfonds, Duchenne UK and through Horizon2020 project BIND. A.G and R.W.J.C are supported by several foundations including the Algemene Nederlandse Vereniging ter Voorkoming van Blindheid, Stichting Blinden‐Penning, Landelijke Stichting voor Blinden en Slechtzienden, Stichting Oogfonds Nederland, Stichting Macula Degeneratie Fonds, and Stichting Retina Nederland Fonds (who contributed through UitZicht 2015‐31 and 2018‐21), together with the Rotterdamse Stichting Blindenbelangen, Stichting Blindenhulp, Stichting tot Verbetering van het Lot der Blinden, Stichting voor Ooglijders, and Stichting Dowilvo; as well as the Foundation Fighting Blindness USA, grant no. PPA‐0517‐0717‐RAD. R.A.M.B is supported by Hersenstichting Nederland Grant DR‐2018‐00253. G.G. is supported by Ministry of Research and Innovation in Romania/National Program 31N/2016/PN 16.22.02.05. S.A is supported by Project PTDC/BBB‐BMD/6301/2014 (Fundação para a Ciência e a Tecnologia—MCTES, Portugal). L.R.D. is supported by Fundación Ramón Areces Grant XVII CN and Spanish Ministry of Science and Innovation (MICINN, grant PID2019‐105344RB‐I00). T.L is supported by Estonian Research Council grant PSG226. S.K is supported by the Friedrich‐Baur‐Stiftung. C.F is funded by The Danish Council for Independent Research, Technology and Production Sciences (grant number DFF‐4184‐00422). W.vRM is supported by ZonMw Programme Translational Research 2 [Project number 446002002], Campaign Team Huntington and AFM Telethon [Project number 20577]. S.E.B is supported by the H2020 projects B‐SMART, Grant number 721058, and REFINE, Grant number 761104. A.T.G is supported by the Institut National de la santé et la recherche médicale (INSERM) and the Association Monegasque contre les myopathies (AMM). L.E. is founded by the Association Monegasque contre les myopathies (AMM).
EMBO Mol Med (2021) 13: e13243.
See the Glossary for abbreviations used in this article.
References
- Aartsma‐Rus A (2017) FDA approval of nusinersen for spinal muscular atrophy makes 2016 the year of splice modulating oligonucleotides. Nucleic Acid Ther 27: 67–69 [DOI] [PubMed] [Google Scholar]
- Aartsma‐Rus A, Arechavala‐Gomeza V (2018) Why dystrophin quantification is key in the eteplirsen saga. Nat Rev Neurol 14: 454–456 [DOI] [PubMed] [Google Scholar]
- Aartsma‐Rus A, Corey DR (2020) The 10th oligonucleotide therapy approved: golodirsen for duchenne muscular dystrophy. Nucleic Acid Ther 30: 67–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achberger K, Probst C, Haderspeck J, Bolz S, Rogal J, Chuchuy J, Nikolova M, Cora V, Antkowiak L, Haq W et al (2019) Merging organoid and organ‐on‐a‐chip technology to generate complex multi‐layer tissue models in a human retina‐on‐a‐chip platform. Elife 8: e46188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D, Gonzalez‐Duarte A, O'Riordan WD, Yang CC, Ueda M, Kristen AV, Tournev I, Schmidt HH, Coelho T, Berk JL et al (2018) Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med 379: 11–21 [DOI] [PubMed] [Google Scholar]
- Agrawal S, Kandimalla ER (2004) Role of Toll‐like receptors in antisense and siRNA [corrected]. Nat Biotechnol 22: 1533–1537 [DOI] [PubMed] [Google Scholar]
- Akinc A, Querbes W, De S, Qin J, Frank‐Kamenetsky M, Jayaprakash KN, Jayaraman M, Rajeev KG, Cantley WL, Dorkin JR et al (2010) Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand‐based mechanisms. Mol Ther 18: 1357–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R et al (2008) A combinatorial library of lipid‐like materials for delivery of RNAi therapeutics. Nat Biotechnol 26: 561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez‐Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ (2011) Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 29: 341–345 [DOI] [PubMed] [Google Scholar]
- Ammala C, Drury WJ 3rd, Knerr L, Ahlstedt I, Stillemark‐Billton P, Wennberg‐Huldt C, Andersson EM, Valeur E, Jansson‐Lofmark R, Janzen D et al (2018) Targeted delivery of antisense oligonucleotides to pancreatic beta‐cells. Science Adv 4: eaat3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson P, den Besten C (2019) CHAPTER 20 preclinical and clinical drug‐metabolism, pharmacokinetics and safety of therapeutic oligonucleotides. In Advances in nucleic acid therapeutics, Agrawal S, Gait MJ (eds), pp 474–531. London: The Royal Society of Chemistry; [Google Scholar]
- Apter S, Hertz M, Rubinstein ZJ, Zissin R (1990) Gossypiboma in the early post‐operative period: a diagnostic problem. Clin Radiol 42: 128–129 [DOI] [PubMed] [Google Scholar]
- Arechavala‐Gomeza V, Khoo B, Aartsma‐Rus A (2014) Splicing modulation therapy in the treatment of genetic diseases. Appl Clin Genet 7: 245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aupy P, Echevarria L, Relizani K, Zarrouki F, Haeberli A, Komisarski M, Tensorer T, Jouvion G, Svinartchouk F, Garcia L et al (2020) Identifying and avoiding tcDNA‐ASO sequence‐specific toxicity for the development of DMD exon 51 skipping therapy. Mol Ther Nucleic Acids 19: 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchet W, Wimmenauer V, Schlee M, Hartmann G (2008) Accessing the therapeutic potential of immunostimulatory nucleic acids. Curr Opin Immunol 20: 389–395 [DOI] [PubMed] [Google Scholar]
- Barile L, Vassalli G (2017) Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol Ther 174: 63–78 [DOI] [PubMed] [Google Scholar]
- Bennett CF, Baker BF, Pham N, Swayze E, Geary RS (2017) Pharmacology of antisense drugs. Annu Rev Pharmacol Toxicol 57: 81–105 [DOI] [PubMed] [Google Scholar]
- Benson MD, Waddington‐Cruz M, Berk JL, Polydefkis M, Dyck PJ, Wang AK, Plante‐Bordeneuve V, Barroso FA, Merlini G, Obici L et al (2018) Inotersen treatment for patients with hereditary transthyretin amyloidosis. N Engl J Med 379: 22–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts CA, McClorey G, Healicon R, Hammond SM, Manzano R, Muses S, Ball V, Godfrey C, Merritt TM, van Westering T et al (2019) Cmah‐dystrophin deficient mdx mice display an accelerated cardiac phenotype that is improved following peptide‐PMO exon skipping treatment. Hum Mol Genet 28: 396–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biscans A, Caiazzi J, Davis S, McHugh N, Sousa J, Khvorova A (2020) The chemical structure and phosphorothioate content of hydrophobically modified siRNAs impact extrahepatic distribution and efficacy. Nucleic Acids Res 48: 7665–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisguerin P, Deshayes S, Gait MJ, O'Donovan L, Godfrey C, Betts CA, Wood MJ, Lebleu B (2015) Delivery of therapeutic oligonucleotides with cell penetrating peptides. Adv Drug Deliv Rev 87: 52–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosgra S, Sipkens J, de Kimpe S, den Besten C, Datson N, van Deutekom J (2019) The pharmacokinetics of 2'‐O‐methyl phosphorothioate antisense oligonucleotides: experiences from developing exon Skipping therapies for duchenne muscular dystrophy. Nucleic Acid Ther 29: 305–322 [DOI] [PubMed] [Google Scholar]
- Buijsen RAM, Toonen LJA, Gardiner SL, van Roon‐Mom WMC (2019) Genetics, mechanisms, and therapeutic progress in polyglutamine spinocerebellar ataxias. Neurotherapeutics 16: 263–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick AD, Sciabola S, Mantena SR, Hollingshead BD, Stanton R, Warneke JA, Zeng M, Martsen E, Medvedev A, Makarov SS et al (2014) Sequence motifs associated with hepatotoxicity of locked nucleic acid–modified antisense oligonucleotides. Nucleic Acids Res 42: 4882–4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burel SA, Hart CE, Cauntay P, Hsiao J, Machemer T, Katz M, Watt A, Bui HH, Younis H, Sabripour M et al (2016) Hepatotoxicity of high affinity gapmer antisense oligonucleotides is mediated by RNase H1 dependent promiscuous reduction of very long pre‐mRNA transcripts. Nucleic Acids Res 44: 2093–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrer R, Neuman BW, Ting JP, Stein DA, Moulton HM, Iversen PL, Kuhn P, Buchmeier MJ (2007) Antiviral effects of antisense morpholino oligomers in murine coronavirus infection models. J Virol 81: 5637–5648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catuogno S, Esposito CL, De Franciscis V (2016) Aptamer‐mediated targeted delivery of therapeutics: an update. Pharmaceuticals 9: 69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catuogno S, Rienzo A, Di Vito A, Esposito CL, de Franciscis V (2015) Selective delivery of therapeutic single strand antimiRs by aptamer‐based conjugates. J Control Release 210: 147–159 [DOI] [PubMed] [Google Scholar]
- Cha W, Fan R, Miao Y, Zhou Y, Qin C, Shan X, Wan X, Li J (2017) Mesoporous silica nanoparticles as carriers for intracellular delivery of nucleic acids and subsequent therapeutic applications. Molecules 22: 782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Tam YYC, Lin PJC, Sung MMH, Tam YK, Cullis PR (2016) Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J Control Release 235: 236–244 [DOI] [PubMed] [Google Scholar]
- Chi X, Gatti P, Papoian T (2017) Safety of antisense oligonucleotide and siRNA‐based therapeutics. Drug Discov Today 22: 823–833 [DOI] [PubMed] [Google Scholar]
- Chow MYT, Qiu Y, Lam JKW (2020) Inhaled RNA therapy: from promise to reality. Trends Pharmacol Sci 41: 715–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cideciyan AV, Jacobson SG, Drack AV, Ho AC, Charng J, Garafalo AV, Roman AJ, Sumaroka A, Han IC, Hochstedler MD et al (2019) Effect of an intravitreal antisense oligonucleotide on vision in Leber congenital amaurosis due to a photoreceptor cilium defect. Nat Med 25: 225–228 [DOI] [PubMed] [Google Scholar]
- Cirak S, Arechavala‐Gomeza V, Guglieri M, Feng L, Torelli S, Anthony K, Abbs S, Garralda ME, Bourke J, Wells DJ et al (2011) Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open‐label, phase 2, dose‐escalation study. Lancet 378: 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleal K, He L, Watson PD, Jones AT (2013) Endocytosis, intracellular traffic and fate of cell penetrating peptide based conjugates and nanoparticles. Curr Pharm Des 19: 2878–2894 [DOI] [PubMed] [Google Scholar]
- Collin RW, den Hollander AI, van der Velde‐Visser SD, Bennicelli J, Bennett J, Cremers FP (2012) Antisense oligonucleotide (AON)‐based therapy for leber congenital amaurosis caused by a frequent mutation in CEP290. Mol Ther Nucleic Acids 1: e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke ST, Baker BF, Pham NC, Hughes SG, Kwoh TJ, Cai D, Tsimikas S, Geary RS, Bhanot S (2018) The effects of 2'‐O‐methoxyethyl oligonucleotides on renal function in humans. Nucleic Acid Ther 28: 10–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke ST, Wang S, Vickers TA, Shen W, Liang XH (2017) Cellular uptake and trafficking of antisense oligonucleotides. Nat Biotechnol 35: 230–237 [DOI] [PubMed] [Google Scholar]
- Curtis MA, Kam M, Nannmark U, Anderson MF, Axell MZ, Wikkelso C, Holtas S, van Roon‐Mom WM, Bjork‐Eriksson T, Nordborg C et al (2007) Human neuroblasts migrate to the olfactory bulb via a lateral ventricular extension. Science 315: 1243–1249 [DOI] [PubMed] [Google Scholar]
- Debacker AJ, Voutila J, Catley M, Blakey D, Habib N (2020) Delivery of oligonucleotides to the liver with GalNAc: from research to registered therapeutic drug. Mol Ther 28: 1759–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon S (2020) Viltolarsen: first approval. Drugs 80: 1027–1031 [DOI] [PubMed] [Google Scholar]
- Dieckmann A, Hagedorn PH, Burki Y, Brugmann C, Berrera M, Ebeling M, Singer T, Schuler F (2018) A sensitive in vitro approach to assess the hybridization‐dependent toxic potential of high affinity gapmer oligonucleotides. Mol Ther Nucleic Acids 10: 45–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Jiang Z, Saha K, Kim CS, Kim ST, Landis RF, Rotello VM (2014) Gold nanoparticles for nucleic acid delivery. Mol Ther 22: 1075–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Love KT, Dorkin JR, Sirirungruang S, Zhang Y, Chen D, Bogorad RL, Yin H, Chen Y, Vegas AJ et al (2014) Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc Natl Acad Sci USA 111: 3955–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Kayali R, Bertoni C, Fike F, Hu H, Iversen PL, Gatti RA (2011) Arginine‐rich cell‐penetrating peptide dramatically enhances AMO‐mediated ATM aberrant splicing correction and enables delivery to brain and cerebellum. Hum Mol Genet 20: 3151–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echevarria L, Aupy P, Relizani K, Bestetti T, Griffith G, Blandel F, Komisarski M, Haeberli A, Svinartchouk F, Garcia L et al (2019) Evaluating the impact of variable phosphorothioate content in tricyclo‐DNA antisense oligonucleotides in a duchenne muscular dystrophy mouse model. Nucleic Acid Ther 29: 148–160 [DOI] [PubMed] [Google Scholar]
- Eckstein F (2014) Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther 24: 374–387 [DOI] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822 [DOI] [PubMed] [Google Scholar]
- EMA (2012) Refusal of the marketing authorisation for Kynamro (mipomersen). https://www.ema.europa.eu/en/medicines/human/EPAR/kynamro
- EMA (2016) Assessment Report: Kyndrisa EMA/439369/2016. https://www.ema.europa.eu/en/medicines/human/withdrawn-applications/kyndrisa
- EMA (2019) Volanesorsen: EU summary of product characteristics. https://www.ema.europa.eu/en/medicines/human/EPAR/waylivra
- Engelhardt JA, Fant P, Guionaud S, Henry SP, Leach MW, Louden C, Scicchitano MS, Weaver JL, Zabka TS, Frazier KS et al (2015) Scientific and regulatory policy committee points‐to‐consider paper*: drug‐induced vascular injury associated with nonsmall molecule therapeutics in preclinical development: part 2. Antisense oligonucleotides. Toxicol Pathol 43: 935–944 [DOI] [PubMed] [Google Scholar]
- Esposito CL, Nuzzo S, Kumar SA, Rienzo A, Lawrence CL, Pallini R, Shaw L, Alder JE, Ricci‐Vitiani L, Catuogno S et al (2016) A combined microRNA‐based targeted therapeutic approach to eradicate glioblastoma stem‐like cells. J Control Release 238: 43–57 [DOI] [PubMed] [Google Scholar]
- FDA (2018) Final Summary Minutes of the Endocrinologic and Metabolic Drugs Advisory Committee Meeting. https://www.fda.gov/media/113979/download
- Fedorov Y, Anderson EM, Birmingham A, Reynolds A, Karpilow J, Robinson K, Leake D, Marshall WS, Khvorova A (2006) Off‐target effects by siRNA can induce toxic phenotype. RNA 12: 1188–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, Northrop JP, Ringold GM, Danielsen M (1987) Lipofection: a highly efficient, lipid‐mediated DNA‐transfection procedure. Proc Natl Acad Sci USA 84: 7413–7417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel RS, Mercuri E, Darras BT, Connolly AM, Kuntz NL, Kirschner J, Chiriboga CA, Saito K, Servais L, Tizzano E et al (2017) Nusinersen versus sham control in infantile‐onset spinal muscular atrophy. N Engl J Med 377: 1723–1732 [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double‐stranded RNA in Caenorhabditis elegans . Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Fitzgerald K, White S, Borodovsky A, Bettencourt BR, Strahs A, Clausen V, Wijngaard P, Horton JD, Taubel J, Brooks A et al (2017) A Highly durable RNAi therapeutic inhibitor of PCSK9. N Engl J Med 376: 41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flierl U, Nero TL, Lim B, Arthur JF, Yao Y, Jung SM, Gitz E, Pollitt AY, Zaldivia MT, Jandrot‐Perrus M et al (2015) Phosphorothioate backbone modifications of nucleotide‐based drugs are potent platelet activators. J Exp Med 212: 129–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foged C, Nielsen HM (2008) Cell‐penetrating peptides for drug delivery across membrane barriers. Exp Opin Drug Deliv 5: 105–117 [DOI] [PubMed] [Google Scholar]
- Frazier KS (2015) Antisense oligonucleotide therapies: the promise and the challenges from a toxicologic pathologist's perspective. Toxicol Pathol 43: 78–89 [DOI] [PubMed] [Google Scholar]
- Freitag F, Wagner E (2020) Optimizing synthetic nucleic acid and protein nanocarriers: The chemical evolution approach. Adv Drug Deliv Rev 168: 30–54 [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusser M, Overbye A, Pandya AD, Morch Y, Borgos SE, Kildal W, Snipstad S, Sulheim E, Fleten KG, Askautrud HA et al (2019) Cabazitaxel‐loaded Poly(2‐ethylbutyl cyanoacrylate) nanoparticles improve treatment efficacy in a patient derived breast cancer xenograft. J Control Release 293: 183–192 [DOI] [PubMed] [Google Scholar]
- Garanto A, Chung DC, Duijkers L, Corral‐Serrano JC, Messchaert M, Xiao R, Bennett J, Vandenberghe LH, Collin RW (2016) In vitro and in vivo rescue of aberrant splicing in CEP290‐associated LCA by antisense oligonucleotide delivery. Hum Mol Genet 25: 2552–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garanto A, Duijkers L, Collin RW (2015) Species‐dependent splice recognition of a cryptic exon resulting from a recurrent intronic CEP290 mutation that causes congenital blindness. Int J Mol Sci 16: 5285–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary RS (2009) Antisense oligonucleotide pharmacokinetics and metabolism. Expert Opin Drug Metab Toxicol 5: 381–391 [DOI] [PubMed] [Google Scholar]
- Geary RS, Baker BF, Crooke ST (2015) Clinical and preclinical pharmacokinetics and pharmacodynamics of mipomersen (kynamro((R))): a second‐generation antisense oligonucleotide inhibitor of apolipoprotein B. Clin Pharmacokinet 54: 133–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary RS, Henry SP, Grillone LR (2002) Fomivirsen: clinical pharmacology and potential drug interactions. Clin Pharmacokinet 41: 255–260 [DOI] [PubMed] [Google Scholar]
- Geller BL, Li L, Martinez F, Sully E, Sturge CR, Daly SM, Pybus C, Greenberg DE (2018) Morpholino oligomers tested in vitro, in biofilm and in vivo against multidrug‐resistant Klebsiella pneumoniae. J Antimicrob Chemother 73: 1611–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller BL, Marshall‐Batty K, Schnell FJ, McKnight MM, Iversen PL, Greenberg DE (2013) Gene‐silencing antisense oligomers inhibit acinetobacter growth in vitro and in vivo. J Infect Dis 208: 1553–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stoter M et al (2013) Image‐based analysis of lipid nanoparticle‐mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol 31: 638–646 [DOI] [PubMed] [Google Scholar]
- Godfrey C, Desviat LR, Smedsrod B, Pietri‐Rouxel F, Denti MA, Disterer P, Lorain S, Nogales‐Gadea G, Sardone V, Anwar R et al (2017) Delivery is key: lessons learnt from developing splice‐switching antisense therapies. EMBO Mol Med 9: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goemans NM, Tulinius M, van den Hauwe M, Kroksmark AK, Buyse G, Wilson RJ, van Deutekom JC, de Kimpe SJ, Lourbakos A, Campion G (2016) Long‐term efficacy, safety, and pharmacokinetics of drisapersen in duchenne muscular dystrophy: results from an open‐label extension study. PLoS One 11: e0161955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales CR, Group VISiONCT (2005) Enhanced efficacy associated with early treatment of neovascular age‐related macular degeneration with pegaptanib sodium: an exploratory analysis. Retina 25: 815–827 [DOI] [PubMed] [Google Scholar]
- Gragoudas ES, Adamis AP, Cunningham ET, Feinsod M, Guyer DR, Group VISiONCT (2004) Pegaptanib for neovascular age‐related macular degeneration. N Engl J Med 351: 2805–2816 [DOI] [PubMed] [Google Scholar]
- Graham MJ, Lee RG, Bell TA 3rd, Fu W, Mullick AE, Alexander VJ, Singleton W, Viney N, Geary R, Su J et al (2013) Antisense oligonucleotide inhibition of apolipoprotein C‐III reduces plasma triglycerides in rodents, nonhuman primates, and humans. Circ Res 112: 1479–1490 [DOI] [PubMed] [Google Scholar]
- Habrant D, Peuziat P, Colombani T, Dallet L, Gehin J, Goudeau E, Evrard B, Lambert O, Haudebourg T, Pitard B (2016) Design of ionizable lipids to overcome the limiting step of endosomal escape: application in the intracellular delivery of mRNA, DNA, and siRNA. J Med Chem 59: 3046–3062 [DOI] [PubMed] [Google Scholar]
- Hache M, Swoboda KJ, Sethna N, Farrow‐Gillespie A, Khandji A, Xia S, Bishop KM (2016) Intrathecal injections in children with spinal muscular atrophy: nusinersen clinical trial experience. J Child Neurol 31: 899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn PH, Hansen BR, Koch T, Lindow M (2017) Managing the sequence‐specificity of antisense oligonucleotides in drug discovery. Nucleic Acids Res 45: 2262–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn PH, Yakimov V, Ottosen S, Kammler S, Nielsen NF, Hog AM, Hedtjarn M, Meldgaard M, Moller MR, Orum H et al (2013) Hepatotoxic potential of therapeutic oligonucleotides can be predicted from their sequence and modification pattern. Nucleic Acid Ther 23: 302–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Hazell G, Shabanpoor F, Saleh AF, Bowerman M, Sleigh JN, Meijboom KE, Zhou H, Muntoni F, Talbot K et al (2016) Systemic peptide‐mediated oligonucleotide therapy improves long‐term survival in spinal muscular atrophy. Proc Natl Acad Sci USA 113: 10962–10967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanvey JC, Peffer NJ, Bisi JE, Thomson SA, Cadilla R, Josey JA, Ricca DJ, Hassman CF, Bonham MA, Au KG et al (1992) Antisense and antigene properties of peptide nucleic acids. Science 258: 1481–1485 [DOI] [PubMed] [Google Scholar]
- Heemskerk HA, de Winter CL, de Kimpe SJ, van Kuik‐Romeijn P, Heuvelmans N, Platenburg GJ, van Ommen GJ, van Deutekom JC, Aartsma‐Rus A (2009) In vivo comparison of 2'‐O‐methyl phosphorothioate and morpholino antisense oligonucleotides for Duchenne muscular dystrophy exon skipping. J Gene Med 11: 257–266 [DOI] [PubMed] [Google Scholar]
- Heemskerk H, de Winter C, van Kuik P, Heuvelmans N, Sabatelli P, Rimessi P, Braghetta P, van Ommen GJ, de Kimpe S, Ferlini A et al (2010) Preclinical PK and PD studies on 2'‐O‐methyl‐phosphorothioate RNA antisense oligonucleotides in the mdx mouse model. Mol Ther 18: 1210–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Helm MW, van der Meer AD, Eijkel JC, van den Berg A, Segerink LI (2016) Microfluidic organ‐on‐chip technology for blood‐brain barrier research. Tissue Barriers 4: e1142493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry SP, Beattie G, Yeh G, Chappel A, Giclas P, Mortari A, Jagels MA, Kornbrust DJ, Levin AA (2002) Complement activation is responsible for acute toxicities in rhesus monkeys treated with a phosphorothioate oligodeoxynucleotide. Int Immunopharmacol 2: 1657–1666 [DOI] [PubMed] [Google Scholar]
- Henry SP, Giclas PC, Leeds J, Pangburn M, Auletta C, Levin AA, Kornbrust DJ (1997a) Activation of the alternative pathway of complement by a phosphorothioate oligonucleotide: potential mechanism of action. J Pharmacol Exp Ther 281: 810–816 [PubMed] [Google Scholar]
- Henry SP, Miner RC, Drew WL, Fitchett J, York‐Defalco C, Rapp LM, Levin AA (2001) Antiviral activity and ocular kinetics of antisense oligonucleotides designed to inhibit CMV replication. Invest Ophthalmol Vis Sci 42: 2646–2651 [PubMed] [Google Scholar]
- Henry SP, Narayanan P, Shen L, Bhanot S, Younis HS, Burel SA (2017) Assessment of the effects of 2'‐methoxyethyl antisense oligonucleotides on platelet count in cynomolgus nonhuman primates. Nucleic Acid Ther 27: 197–208 [DOI] [PubMed] [Google Scholar]
- Henry SP, Novotny W, Leeds J, Auletta C, Kornbrust DJ (1997b) Inhibition of coagulation by a phosphorothioate oligonucleotide. Antisense Nucleic Acid Drug Dev 7: 503–510 [DOI] [PubMed] [Google Scholar]
- Henry SP, Seguin R, Cavagnaro J, Berman C, Tepper J, Kornbrust D (2016) Considerations for the characterization and interpretation of results related to alternative complement activation in monkeys associated with oligonucleotide‐based therapeutics. Nucleic Acid Ther 26: 210–215 [DOI] [PubMed] [Google Scholar]
- Heo YA (2020) Golodirsen: first approval. Drugs 80: 329–333 [DOI] [PubMed] [Google Scholar]
- den Hollander AI, Koenekoop RK, Yzer S, Lopez I, Arends ML, Voesenek KE, Zonneveld MN, Strom TM, Meitinger T, Brunner HG et al (2006) Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet 79: 556–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu WH, Csaba N, Alexander C, Garcia‐Fuentes M (2020) Polyphosphazenes for the delivery of biopharmaceuticals. J Appl Polym Sci 137: 48688 [Google Scholar]
- Huh D, Matthews BD, Mammoto A, Montoya‐Zavala M, Hsin HY, Ingber DE (2010) Reconstituting organ‐level lung functions on a chip. Science 328: 1662–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaax ME, Krauel K, Marschall T, Brandt S, Gansler J, Furll B, Appel B, Fischer S, Block S, Helm CA et al (2013) Complex formation with nucleic acids and aptamers alters the antigenic properties of platelet factor 4. Blood 122: 272–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas MM, Schlegel MK, Harbison CE, Yilmaz VO, Jiang Y, Parmar R, Zlatev I, Castoreno A, Xu H, Shulga‐Morskaya S et al (2018) Selection of GalNAc‐conjugated siRNAs with limited off‐target‐driven rat hepatotoxicity. Nat Commun 9: 723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarver P, O'Donovan L, Gait MJ (2014) A chemical view of oligonucleotides for exon skipping and related drug applications. Nucleic Acid Ther 24: 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman M, Ansell SM, Mui BL, Tam YK, Chen J, Du X, Butler D, Eltepu L, Matsuda S, Narayanannair JK et al (2012) Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew Chem 51: 8529–8533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasena SD (1999) Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem 45: 1628–1650 [PubMed] [Google Scholar]
- Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R et al (2019) Reassessment of exosome composition. Cell 177: 428–445.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge AD, Bola G, Lee AC, MacLachlan I (2006) Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther 13: 494–505 [DOI] [PubMed] [Google Scholar]
- Juliano RL, Wang L, Tavares F, Brown EG, James L, Ariyarathna Y, Ming X, Mao C, Suto M (2018) Structure‐activity relationships and cellular mechanism of action of small molecules that enhance the delivery of oligonucleotides. Nucleic Acids Res 46: 1601–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A (2017) Oligonucleotide therapeutics ‐ a new class of cholesterol‐lowering drugs. N Engl J Med 376: 4–7 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Huh D, Hamilton G, Ingber DE (2012) Human gut‐on‐a‐chip inhabited by microbial flora that experiences intestinal peristalsis‐like motions and flow. Lab Chip 12: 2165–2174 [DOI] [PubMed] [Google Scholar]
- Kim J, Hu C, Moufawad El Achkar C, Black LE, Douville J, Larson A, Pendergast MK, Goldkind SF, Lee EA, Kuniholm A et al (2019) Patient‐customized oligonucleotide therapy for a rare genetic disease. N Engl J Med 381: 1644–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AF, Varela MA, Arandel L, Holland A, Naouar N, Arzumanov A, Seoane D, Revillod L, Bassez G, Ferry A et al (2019) Peptide‐conjugated oligonucleotides evoke long‐lasting myotonic dystrophy correction in patient‐derived cells and mice. J Clin Invest 129: 4739–4744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline JN, Krieg AM (2008) Toll‐like receptor 9 activation with CpG oligodeoxynucleotides for asthma therapy. Drug News Perspect 21: 434–439 [DOI] [PubMed] [Google Scholar]
- Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J (1998) LNA (Locked Nucleic Acids): synthesis of the adenine, cytosine, guanine, 5‐methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron 54: 3607–3630 [Google Scholar]
- Kozlu S, Caban S, Yerlikaya F, Fernandez‐Megia E, Novoa‐Carballal R, Riguera R, Yemisci M, Gursoy‐Ozdemir Y, Dalkara T, Couvreur P et al (2014) An aquaporin 4 antisense oligonucleotide loaded, brain targeted nanoparticulate system design. Pharmazie 69: 340–345 [PubMed] [Google Scholar]
- Krieg AM (1998) The CpG motif: implications for clinical immunology. BioDrugs 10: 341–346 [DOI] [PubMed] [Google Scholar]
- Krieg AM (2006) Therapeutic potential of Toll‐like receptor 9 activation. Nat Rev Drug Discov 5: 471–484 [DOI] [PubMed] [Google Scholar]
- Krieg AM (2007) Antiinfective applications of toll‐like receptor 9 agonists. Proc Am Thorac Soc 4: 289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg AM, Davis HL (2001) Enhancing vaccines with immune stimulatory CpG DNA. Curr Opin Mol Ther 3: 15–24 [PubMed] [Google Scholar]
- Kulkarni JA, Witzigmann D, Leung J, van der Meel R, Zaifman J, Darjuan MM, Grisch‐Chan HM, Thony B, Tam YYC, Cullis PR (2019) Fusion‐dependent formation of lipid nanoparticles containing macromolecular payloads. Nanoscale 11: 9023–9031 [DOI] [PubMed] [Google Scholar]
- Lankveld DP, Van Loveren H, Baken KA, Vandebriel RJ (2010) In vitro testing for direct immunotoxicity: state of the art. Methods Mol Biol 598: 401–423 [DOI] [PubMed] [Google Scholar]
- Lee HJ, Boado RJ, Braasch DA, Corey DR, Pardridge WM (2002) Imaging gene expression in the brain in vivo in a transgenic mouse model of Huntington's disease with an antisense radiopharmaceutical and drug‐targeting technology. J Nucl Med 43: 948–956 [PubMed] [Google Scholar]
- Lee H, Kim DS, Ha SK, Choi I, Lee JM, Sung JH (2017) A pumpless multi‐organ‐on‐a‐chip (MOC) combined with a pharmacokinetic‐pharmacodynamic (PK‐PD) model. Biotechnol Bioeng 114: 432–443 [DOI] [PubMed] [Google Scholar]
- Lee KY, Mooney DJ (2012) Alginate: properties and biomedical applications. Prog Polym Sci 37: 106–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RT, Lin P, Lee YC (1984) New synthetic cluster ligands for galactose/N‐acetylgalactosamine‐specific lectin of mammalian liver. Biochemistry 23: 4255–4261 [DOI] [PubMed] [Google Scholar]
- Lehto T, Ezzat K, Wood MJA, El Andaloussi S (2016) Peptides for nucleic acid delivery. Adv Drug Deliv Rev 106: 172–182 [DOI] [PubMed] [Google Scholar]
- Levin AA, Henry SP (2008) Toxicology of oligonucleotide therapeutics and understanding the relevance of the toxicities. In Preclinical safety evaluation of biopharmaceuticals: a science‐based approach to facilitating clinical trials, Cavagnaro JA (ed), pp 537–574. Hoboken, NJ: John Wiley & Sons Inc; [Google Scholar]
- Li JY, Ren YP, Yuan Y, Ji SM, Zhou SP, Wang LJ, Mou ZZ, Li L, Lu W, Zhou TY (2016) Preclinical PK/PD model for combined administration of erlotinib and sunitinib in the treatment of A549 human NSCLC xenograft mice. Acta Pharmacol Sin 37: 930–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Shen W, Sun H, Kinberger GA, Prakash TP, Nichols JG, Crooke ST (2016a) Hsp90 protein interacts with phosphorothioate oligonucleotides containing hydrophobic 2'‐modifications and enhances antisense activity. Nucleic Acids Res 44: 3892–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Shen W, Sun H, Migawa MT, Vickers TA, Crooke ST (2016b) Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat Biotechnol 34: 875–880 [DOI] [PubMed] [Google Scholar]
- Liang XH, Sun H, Shen W, Wang S, Yao J, Migawa MT, Bui HH, Damle SS, Riney S, Graham MJ et al (2017) Antisense oligonucleotides targeting translation inhibitory elements in 5' UTRs can selectively increase protein levels. Nucleic Acids Res 45: 9528–9546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow M, Vornlocher HP, Riley D, Kornbrust DJ, Burchard J, Whiteley LO, Kamens J, Thompson JD, Nochur S, Younis H et al (2012) Assessing unintended hybridization‐induced biological effects of oligonucleotides. Nat Biotechnol 30: 920–923 [DOI] [PubMed] [Google Scholar]
- Malmsten M (2013) Inorganic nanomaterials as delivery systems for proteins, peptides, DNA, and siRNA. Curr Opin Colloid Interface Sci 18: 468–480 [Google Scholar]
- Marwick C (1998) First "antisense" drug will treat CMV retinitis. JAMA 280: 871 [PubMed] [Google Scholar]
- Mathew V, Wang AK (2019) Inotersen: new promise for the treatment of hereditary transthyretin amyloidosis. Drug Design Dev Ther 13: 1515–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Keiser K, Nair JK, Charisse K, Manoharan RM, Kretschmer P, Peng CG, Kelin AV, Kandasamy P, Willoughby JL et al (2015) siRNA conjugates carrying sequentially assembled trivalent N‐acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chem Biol 10: 1181–1187 [DOI] [PubMed] [Google Scholar]
- McAleer CW, Pointon A, Long CJ, Brighton RL, Wilkin BD, Bridges LR, Narasimhan Sriram N, Fabre K, McDougall R, Muse VP et al (2019) On the potential of in vitro organ‐chip models to define temporal pharmacokinetic‐pharmacodynamic relationships. Sci Rep 9: 9619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor TL, Hunt KA, Yee E, Mason D, Nioi P, Ticau S, Pelosi M, Loken PR, Finer S, Lawlor DA et al (2020) Characterising a healthy adult with a rare HAO1 knockout to support a therapeutic strategy for primary hyperoxaluria. Elife 9: e54363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara JO 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH (2006) Cell type‐specific delivery of siRNAs with aptamer‐siRNA chimeras. Nat Biotechnol 24: 1005–1015 [DOI] [PubMed] [Google Scholar]
- van der Meer AD, van den Berg A (2012) Organs‐on‐chips: breaking the in vitro impasse. Integr Biol 4: 461–470 [DOI] [PubMed] [Google Scholar]
- Mercuri E, Darras BT, Chiriboga CA, Day JW, Campbell C, Connolly AM, Iannaccone ST, Kirschner J, Kuntz NL, Saito K et al (2018) Nusinersen versus sham control in later‐onset spinal muscular atrophy. N Engl J Med 378: 625–635 [DOI] [PubMed] [Google Scholar]
- Mignani S, Shi X, Zablocka M, Majoral JP (2019) Dendrimer‐enabled therapeutic antisense delivery systems as innovation in medicine. Bioconjug Chem 30: 1938–1950 [DOI] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng‐Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS (2000) Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J 19: 4439–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrpant C, Adams AM, Meloni PL, Muntoni F, Fletcher S, Wilton SD (2009) Rational design of antisense oligomers to induce dystrophin exon skipping. Mol Ther 17: 1418–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisan A, Gubler M, Zhang JD, Tessier Y, Dumong Erichsen K, Sewing S, Gerard R, Avignon B, Huber S, Benmansour F et al (2017) Inhibition of EGF uptake by nephrotoxic antisense drugs in vitro and implications for preclinical safety profiling. Mol Ther Nucleic Acids 6: 89–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monia BP, Lesnik EA, Gonzalez C, Lima WF, McGee D, Guinosso CJ, Kawasaki AM, Cook PD, Freier SM (1993) Evaluation of 2'‐modified oligonucleotides containing 2'‐deoxy gaps as antisense inhibitors of gene expression. J Biol Chem 268: 14514–14522 [PubMed] [Google Scholar]
- Monteith DK, Henry SP, Howard RB, Flournoy S, Levin AA, Bennett CF, Crooke ST (1997) Immune stimulation–a class effect of phosphorothioate oligodeoxynucleotides in rodents. Anticancer Drug Des 12: 421–432 [PubMed] [Google Scholar]
- Morgan E, Wupperfeld D, Morales D, Reich N (2019) Shape matters: gold nanoparticle shape impacts the biological activity of siRNA delivery. Bioconjug Chem 30: 853–860 [DOI] [PubMed] [Google Scholar]
- Muller TD, Finan B, Bloom SR, D'Alessio D, Drucker DJ, Flatt PR, Fritsche A, Gribble F, Grill HJ, Habener JF et al (2019) Glucagon‐like peptide 1 (GLP‐1). Mol Metab 30: 72–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan P, Shen L, Curtis BR, Bourdon MA, Nolan JP, Gupta S, Hoffmaster C, Zhou F, Christian B, Schaubhut JL et al (2018) Investigation into the mechanism(s) that leads to platelet decreases in Cynomolgus monkeys during administration of ISIS 104838, a 2'‐MOE‐modified antisense oligonucleotide. Toxicol Sci 164: 613–626 [DOI] [PubMed] [Google Scholar]
- Negus SS, Banks ML (2018) Pharmacokinetic‐pharmacodynamic (PKPD) analysis with drug discrimination. Curr Top Behav Neurosci 39: 245–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EW, Shima DT, Calias P, Cunningham ET Jr, Guyer DR, Adamis AP (2006) Pegaptanib, a targeted anti‐VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov 5: 123–132 [DOI] [PubMed] [Google Scholar]
- Nielsen PE, Egholm M, Berg RH, Buchardt O (1991) Sequence‐selective recognition of DNA by strand displacement with a thymine‐substituted polyamide. Science 254: 1497–1500 [DOI] [PubMed] [Google Scholar]
- Obika S, Nanbu D, Hari Y, Andoh J‐I, Morio K‐I, Doi T, Imanishi T (1998) Stability and structural features of the duplexes containing nucleoside analogues with a fixed N‐type conformation, 2′‐O, 4′‐C‐methyleneribonucleosides. Tetrahedron Lett 39: 5401–5404 [Google Scholar]
- Osborn MF, Coles AH, Biscans A, Haraszti RA, Roux L, Davis S, Ly S, Echeverria D, Hassler MR, Godinho B et al (2019) Hydrophobicity drives the systemic distribution of lipid‐conjugated siRNAs via lipid transport pathways. Nucleic Acids Res 47: 1070–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardridge WM (2007) Blood‐brain barrier delivery. Drug Discov Today 12: 54–61 [DOI] [PubMed] [Google Scholar]
- Parfitt DA, Lane A, Ramsden CM, Carr AJ, Munro PM, Jovanovic K, Schwarz N, Kanuga N, Muthiah MN, Hull S et al (2016) Identification and correction of mechanisms underlying inherited blindness in human iPSC‐derived optic cups. Cell Stem Cell 18: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan M, Fonseka P, Chitti SV, Kang T, Sanwlani R, Van Deun J, Hendrix A, Mathivanan S (2019) Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res 47: D516–D519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Zhu X, Qiu L (2016) Electroneutral composite polymersomes self‐assembled by amphiphilic polyphosphazenes for effective miR‐200c in vivo delivery to inhibit drug resistant lung cancer. Biomaterials 106: 1–12 [DOI] [PubMed] [Google Scholar]
- Penichet ML, Kang YS, Pardridge WM, Morrison SL, Shin SU (1999) An antibody‐avidin fusion protein specific for the transferrin receptor serves as a delivery vehicle for effective brain targeting: initial applications in anti‐HIV antisense drug delivery to the brain. J Immunol 163: 4421–4426 [PubMed] [Google Scholar]
- Pooga M, Langel U (2015) Classes of Cell‐Penetrating Peptides. Methods Mol Biol 1324: 3–28 [DOI] [PubMed] [Google Scholar]
- Prakash TP, Mullick AE, Lee RG, Yu J, Yeh ST, Low A, Chappell AE, Ostergaard ME, Murray S, Gaus HJ et al (2019) Fatty acid conjugation enhances potency of antisense oligonucleotides in muscle. Nucleic Acids Res 47: 6029–6044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR‐Cas9 system. Nat Protoc 8: 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray KK, Wright RS, Kallend D, Koenig W, Leiter LA, Raal FJ, Bisch JA, Richardson T, Jaros M, Wijngaard PLJ et al (2020) Two phase 3 trials of inclisiran in patients with elevated LDL cholesterol. N Engl J Med 382: 1507–1519 [DOI] [PubMed] [Google Scholar]
- Renneberg D, Leumann CJ (2002) Watson‐Crick base‐pairing properties of tricyclo‐DNA. J Am Chem Soc 124: 5993–6002 [DOI] [PubMed] [Google Scholar]
- Rezvantalab S, Drude NI, Moraveji MK, Guvener N, Koons EK, Shi Y, Lammers T, Kiessling F (2018) PLGA‐based nanoparticles in cancer treatment. Front Pharmacol 9: 1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera‐Barahona A, Sanchez‐Alcudia R, Viecelli HM, Rufenacht V, Perez B, Ugarte M, Haberle J, Thony B, Desviat LR (2015) Functional characterization of the spf/ash splicing variation in OTC deficiency of mice and man. PLoS One 10: e0122966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TC, Langer R, Wood MJA (2020) Advances in oligonucleotide drug delivery. Nat Rev Drug Discov 19: 673–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD, Claesson‐Welsh L, Janjic N (1998) 2'‐Fluoropyrimidine RNA‐based aptamers to the 165‐amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF‐induced vascular permeability through interactions requiring the exon 7‐encoded domain. J Biol Chem 273: 20556–20567 [DOI] [PubMed] [Google Scholar]
- Rudin CM, Holmlund J, Fleming GF, Mani S, Stadler WM, Schumm P, Monia BP, Johnston JF, Geary R, Yu RZ et al (2001) Phase I Trial of ISIS 5132, an antisense oligonucleotide inhibitor of c‐raf‐1, administered by 24‐hour weekly infusion to patients with advanced cancer. Clin Cancer Res 7: 1214–1220 [PubMed] [Google Scholar]
- Sardh E, Harper P, Balwani M, Stein P, Rees D, Bissell DM, Desnick R, Parker C, Phillips J, Bonkovsky HL et al (2019) Phase 1 trial of an RNA interference therapy for acute intermittent porphyria. N Engl J Med 380: 549–558 [DOI] [PubMed] [Google Scholar]
- Schoch KM, Miller TM (2017) Antisense oligonucleotides: translation from mouse models to human neurodegenerative diseases. Neuron 94: 1056–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz AL, Rup D, Lodish HF (1980) Difficulties in the quantification of asialoglycoprotein receptors on the rat hepatocyte. J Biol Chem 255: 9033–9036 [PubMed] [Google Scholar]
- Semple SC, Klimuk SK, Harasym TO, Dos Santos N, Ansell SM, Wong KF, Maurer N, Stark H, Cullis PR, Hope MJ et al (2001) Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: formation of novel small multilamellar vesicle structures. Biochim Biophys Acta 1510: 152–166 [DOI] [PubMed] [Google Scholar]
- Senn JJ, Burel S, Henry SP (2005) Non‐CpG‐containing antisense 2'‐methoxyethyl oligonucleotides activate a proinflammatory response independent of Toll‐like receptor 9 or myeloid differentiation factor 88. J Pharmacol Exp Ther 314: 972–979 [DOI] [PubMed] [Google Scholar]
- Seo J, Byun WY, Alisafaei F, Georgescu A, Yi YS, Massaro‐Giordano M, Shenoy VB, Lee V, Bunya VY, Huh D (2019) Multiscale reverse engineering of the human ocular surface. Nat Med 25: 1310–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewing S, Boess F, Moisan A, Bertinetti‐Lapatki C, Minz T, Hedtjaern M, Tessier Y, Schuler F, Singer T, Roth AB (2016) Establishment of a predictive in vitro assay for assessment of the hepatotoxic potential of oligonucleotide drugs. PLoS One 11: e0159431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewing S, Roth AB, Winter M, Dieckmann A, Bertinetti‐Lapatki C, Tessier Y, McGinnis C, Huber S, Koller E, Ploix C et al (2017) Assessing single‐stranded oligonucleotide drug‐induced effects in vitro reveals key risk factors for thrombocytopenia. PLoS One 12: e0187574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer C (2020) Mist begins to clear for lung delivery of RNA. Nat Biotechnol 38: 1110–1112 [DOI] [PubMed] [Google Scholar]
- Sheehan JP, Lan HC (1998) Phosphorothioate oligonucleotides inhibit the intrinsic tenase complex. Blood 92: 1617–1625 [PubMed] [Google Scholar]
- Shen L, Engelhardt JA, Hung G, Yee J, Kikkawa R, Matson J, Tayefeh B, Machemer T, Giclas PC, Henry SP (2016) Effects of repeated complement activation associated with chronic treatment of cynomolgus monkeys with 2'‐O‐methoxyethyl modified antisense oligonucleotide. Nucleic Acid Ther 26: 236–249 [DOI] [PubMed] [Google Scholar]
- Shen W, De Hoyos CL, Migawa MT, Vickers TA, Sun H, Low A, Bell TA 3rd, Rahdar M, Mukhopadhyay S, Hart CE et al (2019) Chemical modification of PS‐ASO therapeutics reduces cellular protein‐binding and improves the therapeutic index. Nat Biotechnol 37: 640–650 [DOI] [PubMed] [Google Scholar]
- Shi B, Keough E, Matter A, Leander K, Young S, Carlini E, Sachs AB, Tao W, Abrams M, Howell B et al (2011) Biodistribution of small interfering RNA at the organ and cellular levels after lipid nanoparticle‐mediated delivery. J Histochem Cytochem 59: 727–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinha K, Nihei W, Ono T, Nakazato R, Kimura H (2020) A pharmacokinetic–pharmacodynamic model based on multi‐organ‐on‐a‐chip for drug–drug interaction studies. Biomicrofluidics 14: 44108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NK, Singh NN, Androphy EJ, Singh RN (2006) Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol Cell Biol 26: 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smulevitch SV, Simmons CG, Norton JC, Wise TW, Corey DR (1996) Enhancement of strand invasion by oligonucleotides through manipulation of backbone charge. Nat Biotechnol 14: 1700–1704 [DOI] [PubMed] [Google Scholar]
- Soldevilla MM, Meraviglia‐Crivelli de Caso D, Menon AP, Pastor F (2018) Aptamer‐iRNAs as therapeutics for cancer treatment. Pharmaceuticals 11: 108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P et al (2005) Antibody mediated in vivo delivery of small interfering RNAs via cell‐surface receptors. Nat Biotechnol 23: 709–717 [DOI] [PubMed] [Google Scholar]
- Starner CI, Gleason PP (2019) Spinal muscular atrophy therapies: ICER grounds the price to value conversation in facts. J Manage Care Special Pharm 25: 1306–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CA, Hansen JB, Lai J, Wu S, Voskresenskiy A, Hog A, Worm J, Hedtjarn M, Souleimanian N, Miller P et al (2010) Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res 38: e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein H, Hausen P (1969) Enzyme from calf thymus degrading the RNA moiety of DNA‐RNA Hybrids: effect on DNA‐dependent RNA polymerase. Science 166: 393–395 [DOI] [PubMed] [Google Scholar]
- Steinbacher JL, Landry CC (2014) Adsorption and release of siRNA from porous silica. Langmuir 30: 4396–4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugo T, Terada M, Oikawa T, Miyata K, Nishimura S, Kenjo E, Ogasawara‐Shimizu M, Makita Y, Imaichi S, Murata S et al (2016) Development of antibody‐siRNA conjugate targeted to cardiac and skeletal muscles. J Control Release 237: 1–13 [DOI] [PubMed] [Google Scholar]
- Suhr OB, Coelho T, Buades J, Pouget J, Conceicao I, Berk J, Schmidt H, Waddington‐Cruz M, Campistol JM, Bettencourt BR et al (2015) Efficacy and safety of patisiran for familial amyloidotic polyneuropathy: a phase II multi‐dose study. Orphanet J Rare Dis 10: 109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerton J, Weller D (1997) Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev 7: 187–195 [DOI] [PubMed] [Google Scholar]
- Szebeni J (2018) Mechanism of nanoparticle‐induced hypersensitivity in pigs: complement or not complement? Drug Discov Today 23: 487–492 [DOI] [PubMed] [Google Scholar]
- Taetz S, Nafee N, Beisner J, Piotrowska K, Baldes C, Murdter TE, Huwer H, Schneider M, Schaefer UF, Klotz U et al (2009) The influence of chitosan content in cationic chitosan/PLGA nanoparticles on the delivery efficiency of antisense 2'‐O‐methyl‐RNA directed against telomerase in lung cancer cells. Eur J Pharma Biopharm 72: 358–369 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Teasdale I (2019) Stimuli‐responsive phosphorus‐based polymers. Eur J Inorg Chem 2019: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanki K, Zeng X, Justesen S, Tejlmann S, Falkenberg E, Van Driessche E, Morck Nielsen H, Franzyk H, Foged C (2017) Engineering of small interfering RNA‐loaded lipidoid‐poly(DL‐lactic‐co‐glycolic acid) hybrid nanoparticles for highly efficient and safe gene silencing: A quality by design‐based approach. Eur J Pharma Biopharma 120: 22–33 [DOI] [PubMed] [Google Scholar]
- Thomas GS, Cromwell WC, Ali S, Chin W, Flaim JD, Davidson M (2013) Mipomersen, an apolipoprotein B synthesis inhibitor, reduces atherogenic lipoproteins in patients with severe hypercholesterolemia at high cardiovascular risk: a randomized, double‐blind, placebo‐controlled trial. J Am Coll Cardiol 62: 2178–2184 [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510 [DOI] [PubMed] [Google Scholar]
- Vitravene Study G (2002) A randomized controlled clinical trial of intravitreous fomivirsen for treatment of newly diagnosed peripheral cytomegalovirus retinitis in patients with AIDS. Am J Ophthalmol 133: 467–474 [DOI] [PubMed] [Google Scholar]
- Voit T, Topaloglu H, Straub V, Muntoni F, Deconinck N, Campion G, De Kimpe SJ, Eagle M, Guglieri M, Hood S et al (2014) Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): an exploratory, randomised, placebo‐controlled phase 2 study. Lancet Neurol 13: 987–996 [DOI] [PubMed] [Google Scholar]
- Vollmer J, Weeratna RD, Jurk M, Samulowitz U, McCluskie MJ, Payette P, Davis HL, Schetter C, Krieg AM (2004) Oligodeoxynucleotides lacking CpG dinucleotides mediate Toll‐like receptor 9 dependent T helper type 2 biased immune stimulation. Immunology 113: 212–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlestedt C, Salmi P, Good L, Kela J, Johnsson T, Hokfelt T, Broberger C, Porreca F, Lai J, Ren K et al (2000) Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci USA 97: 5633–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Allen N, Prakash TP, Liang XH, Crooke ST (2019) Lipid conjugates enhance endosomal release of antisense oligonucleotides into cells. Nucleic Acid Ther 29: 245–255 [DOI] [PubMed] [Google Scholar]
- Wathion N (2002) Public Statement on Vitravene (fomivirsen). https://www.ema.europa.eu/en/documents/public-statement/public-statement-vitravene-fomivirsen-withdrawal-marketing-authorisation-european-union_en.pdf
- Watts JK, Deleavey GF, Damha MJ (2008) Chemically modified siRNA: tools and applications. Drug Discov Today 13: 842–855 [DOI] [PubMed] [Google Scholar]
- Westein E, van der Meer AD, Kuijpers MJ, Frimat JP, van den Berg A, Heemskerk JW (2013) Atherosclerotic geometries exacerbate pathological thrombus formation poststenosis in a von Willebrand factor‐dependent manner. Proc Natl Acad Sci USA 110: 1357–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiklander OPB, Brennan MA, Lotvall J, Breakefield XO, El Andaloussi S (2019) Advances in therapeutic applications of extracellular vesicles. Sci Transl Med 11: eaav8521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willms E, Cabanas C, Mager I, Wood MJA, Vader P (2018) Extracellular vesicle heterogeneity: subpopulations, isolation techniques, and diverse functions in cancer progression. Front Immunol 9: 738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Goldberg T (2014) Mipomersen (kynamro): a novel antisense oligonucleotide inhibitor for the management of homozygous familial hypercholesterolemia. P T 39: 119–122 [PMC free article] [PubMed] [Google Scholar]
- Wu H, Lima WF, Zhang H, Fan A, Sun H, Crooke ST (2004) Determination of the role of the human RNase H1 in the pharmacology of DNA‐like antisense drugs. J Biol Chem 279: 17181–17189 [DOI] [PubMed] [Google Scholar]
- Yang T, Fogarty B, LaForge B, Aziz S, Pham T, Lai L, Bai S (2017) Delivery of small interfering RNA to inhibit vascular endothelial growth factor in zebrafish using natural brain endothelia cell‐secreted exosome nanovesicles for the treatment of brain cancer. AAPS J 19: 475–486 [DOI] [PubMed] [Google Scholar]
- Yao YD, Sun TM, Huang SY, Dou S, Lin L, Chen JN, Ruan JB, Mao CQ, Yu FY, Zeng MS et al (2012) Targeted delivery of PLK1‐siRNA by ScFv suppresses Her2+ breast cancer growth and metastasis. Sci Transl Med 4: 130ra148 [DOI] [PubMed] [Google Scholar]
- Younis HS, Vickers T, Levin AA, Henry SP (2006) CpG and Non‐CpG oligodeoxynucleotides induce differential proinflammatory gene expression profiles in liver and peripheral blood leukocytes in mice. J Immunotoxicol 3: 57–68 [DOI] [PubMed] [Google Scholar]
- Yu AM, Choi YH, Tu MJ (2020) RNA drugs and RNA targets for small molecules: principles, progress, and challenges. Pharmacol Rev 72: 862–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu RZ, Lemonidis KM, Graham MJ, Matson JE, Crooke RM, Tribble DL, Wedel MK, Levin AA, Geary RS (2009) Cross‐species comparison of in vivo PK/PD relationships for second‐generation antisense oligonucleotides targeting apolipoprotein B‐100. Biochem Pharmacol 77: 910–919 [DOI] [PubMed] [Google Scholar]
- Zamecnik PC, Stephenson ML (1978) Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc Natl Acad Sci USA 75: 280–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Uaesoontrachoon K, Shaughnessy C, Das JR, Rayavarapu S, Brown KJ, Ray PE, Nagaraju K, van den Anker JN, Hoffman EP et al (2015) The use of urinary and kidney SILAM proteomics to monitor kidney response to high dose morpholino oligonucleotides in the mdx mouse. Toxicol Rep 2: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]


