Abstract
Biological systems are composed of heterogeneous populations of cells that intercommunicate to form a functional living tissue. Biological function varies greatly across populations of cells, as each single cell has a unique transcriptome, proteome, and metabolome that translates to functional differences within single species and across kingdoms. Over the past decade, substantial advancements in our ability to characterize omic profiles on a single cell level have occurred, including in multiple spectroscopic and mass spectrometry (MS)-based techniques. Of these technologies, spatially resolved mass spectrometry approaches, including mass spectrometry imaging (MSI), have shown the most progress for single cell proteomics and metabolomics. For example, reporter-based methods using heavy metal tags have allowed for targeted MS investigation of the proteome at the subcellular level, and development of technologies such as laser ablation electrospray ionization mass spectrometry (LAESI-MS) now mean that dynamic metabolomics can be performed in situ. In this Perspective, we showcase advancements in single cell spatial metabolomics and proteomics over the past decade and highlight important aspects related to high-throughput screening, data analysis, and more which are vital to the success of achieving proteomic and metabolomic profiling at the single cell scale. Finally, using this broad literature summary, we provide a perspective on how the next decade may unfold in the area of single cell MS-based proteomics and metabolomics.
Keywords: mass spectrometry imaging, MALDI, SIMS, LAESI, DESI, NanoDESI, LDI, high-throughput omics
I. Introduction
Omics measurements involve the identification and quantification of biomolecules with the overall aim of inferring the physiological state of an organism based on molecular type, location, and any change in abundance.1 Molecular-based omics can be subcategorized into genomics, transcriptomics, proteomics, and metabolomics, all of which provide valuable information toward understanding the complete biotic state of biological systems. In the fields of proteomics and metabolomics, mass spectrometry (MS) has been invaluable as these methods are ubiquitously employed for bulk characterization of proteins and metabolites extracted from homogenized tissue and cell lysates.2−5 Innovations in MS have resulted in an explosion of discoveries and technological advancements within these omic fields over the last two decades6−8 and have allowed the scientific community to provide answers to complex biological questions, including how protein expression regulates the circadian clock and cell signal transduction,9,10 as well as enable the development of tools to artificially evolve proteins or target specific gene sequences for modification.11,12
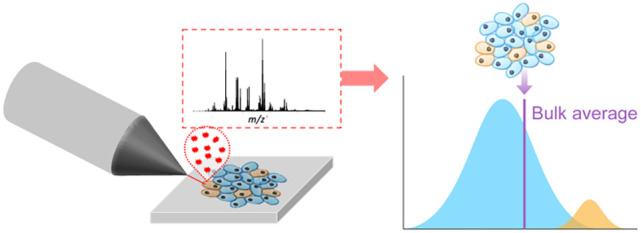
The biological and bioanalytical chemistry research communities have increasingly set their sights on adapting MS-based proteomics and metabolomics to the single cell scale. Single cell omics differs from bulk omics in that analysis and/or extraction of nucleic acids, proteins, and metabolites is constrained and targeted to singular cells within a tissue or a culture, the benefit of which is that intercellular differences can be measured, allowing for the visualization of discrete populations of cells based on their physiological states (Figure 1). Single cell scales are also necessary to elucidate changes in cell biochemistry that occur in the early stages of a disease, for example, which might otherwise be unresolvable by conventional bulk-based measurements.13−16 For this reason, single cell omics is increasingly applied to decipher the degree of biochemical differences between cells within a tissue to understand phenomena such as immune cell plasticity,17 microbial resistance,18 and cellular dysregulation.19
Figure 1.
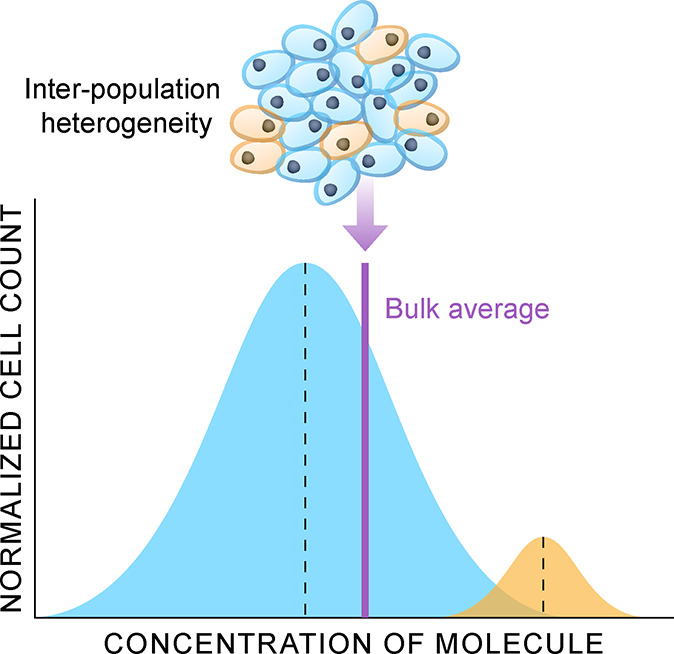
Illustration of how physiological state heterogeneity occurring across a cell population within a sample may not be resolved by bulk omics measurements (purple line). In this example, two discrete subpopulations of cells can be resolved by measuring each cell, which will resolve the predominate cell population (blue) from a minor cell population (yellow).
There are substantial challenges in moving from bulk and multicell analyses to single cell analyses. To begin, eukaryotic cells, for example, exhibit a significant range in size (∼500 nm to >1 cm; Figure 2) across kingdoms (e.g., planta, animalia, etc.).20,21 This means that measurements of biomolecules using probes such as lasers or ion beams must be able to sample across these size scales (Figure 2). Another challenge is that cells contain a wide variety of molecules, all with different abundances ranging from millimolar to subattomolar concentrations.22−24 These attributes matter particularly at small size scales, where math begins to dominate in that extraction and ionization probabilities of each molecule clash with the reduction in the available molecules that can be measured.25 It is also worth noting that eukaryotes present an easier analytical challenge when compared to prokaryotes and archaea, whose smaller sizes (<100 nm to <20 μm) further compound sensitivity issues. This, in part, is the reason minimal molecular-based single cell proteomics and metabolomics research has been performed for these kingdoms in comparison to eukaryotes. Finally, the speed of analysis remains another challenge for single cell omics. Cellular level sampling undoubtedly multiplies the number of data points in an experiment, as multiple points (e.g., pixels) in a sample are analyzed,26 and furthermore, the speed of any extraction processes, digestion steps, and molecular identification further amplifies this issue.27
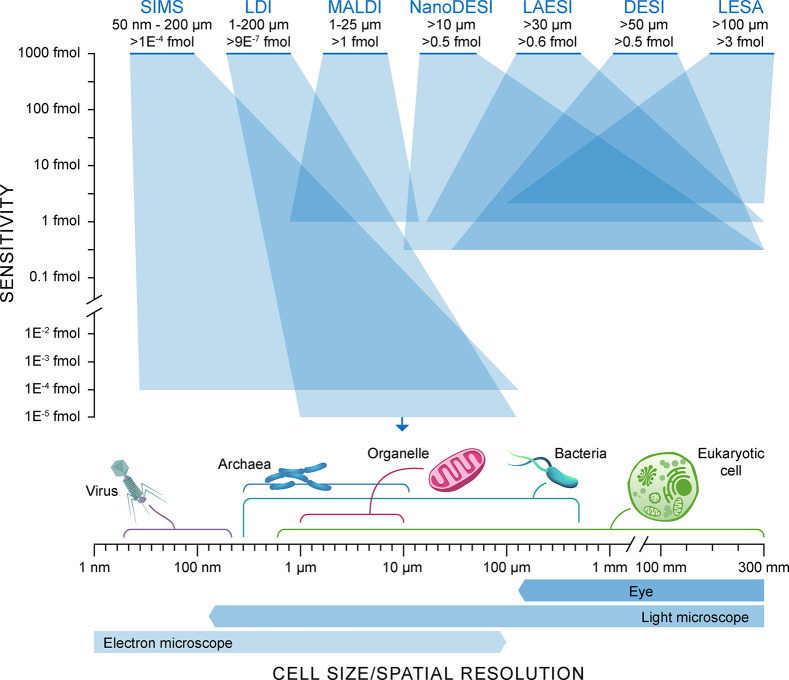
Figure 2.
Representation of the techniques available for MS-based spatial metabolomics and proteomics. The range of sensitivities in femtomoles (y-axis) is compared against the spatial resolution range (x-axis) for these spatial-MS approaches. The spatial dynamic range is illustrated by the transparent blue boxes. Cell size dimensions and the lateral resolution of other structural imaging techniques are displayed along the x-axis for comparison.
Mass spectrometry imaging (MSI) has been an invaluable technique for single cell biochemical analysis.28,29 There are numerous MSI techniques (Figure 2), each with their own unique benefits and limitations for single cell analysis. MSI methods essentially employ an analytical probe (e.g., ion beam, laser, or solvent junction) capable of in situ endogenous chemical desorption and/or ionization.30 There are numerous historical examples of single cell MSI studies involving analysis of tissue mounted substrates by focused ion beam techniques (e.g., secondary ion mass spectrometry; SIMS).31−34 However, these examples are limited in their analytical throughput and molecular coverage, and to complicate the matter, primary ion beams tend to produce high degrees of molecular fragmentation that make data analysis nontrivial.29 Ideally, single cell omics methods allow for intact, sensitive, rapid molecular characterization, in combination with a thorough understanding of how measurement techniques affect molecular ionization yields and fragmentation.
Significant advancements have occurred in MSI and spatially resolved MS-analysis, as a whole, over the past decade, and many of these have focused on enabling spatial proteomics and metabolomics on a single cell scale.29 Advances in single cell metabolomics have allowed for the interrogation and classification of bioactive small molecules (e.g., sugars, fatty acids, amino acids, etc.), drugs, and lipids through improvements in sensitivity and spatial resolution limits,35 whereas innovative approaches have been used to push the spatial limits and automate single cell proteomics.36−38 Increasingly, new detectors and separation methods are being coupled to study the metabolome and proteome for high spatial resolution analysis at the single cell scale, and the benefit of which is improvements in mass resolution, tandem mass spectrometry (MS2), or orthogonal measurements of analytes, such as their collisional cross sections.35 In addition, efforts toward rapid analyte assignment through fragmentation prediction, and hyperspectral data analysis have coalesced with new detector technologies and sampling methods that has improved the ability to probe metabolite and protein classes at the single cell scale.35,39
In this Perspective, we review the recent advancements in MS-based single cell omics over roughly the past decade—focusing on the proteome and metabolome—to address the challenges yet unmet, illustrate the state of the field as a whole, and attempt to present an outlook on the future. In short, we will aim to answer the question: how far can we go toward rapid, automated, sensitive, single cell scale sampling?
Applications for Single Cell Proteomics and Metabolomics
The development of probe-based MS for biological molecular imaging has led to a wealth of knowledge and insight into the spatial distribution of various molecules for a better understanding of physiological processes.40,41 The high chemical specificity and sensitivity, along with the critical spatial information obtained within these experiments, makes spatially resolved MS a technique of increasing interest for single cell omics.42,43 In single cell omics, one must first consider the sensitivity limitations related to the nature of the sample itself, wherein a single cell contains a diverse population of molecules (e.g., carbohydrates, lipids, proteins) that vary greatly in their natural abundances. The abundance of a protein molecule can range from a single molecule to a few million copies per cell, and this consequently directly affects what can be measured in a single cell.44,45 For example, the calculations from McDonnell et al. suggested a peptide molecule would require a concentration in the micromolar (μM) range in order to be detected in a single cell, assuming an attomole limit of detection.46 This effectively restricts the detection of the majority of the molecules present in a single cell to only the most abundant ones due to ion competition during ionization and detection. This is one reason why metabolites and lipids are often measured in untargeted single cell experiments. The second major challenge regarding sensitivity in high-spatial resolution MS experiments is associated with the fact that increasing lateral resolution reduces the analysis area (volume) or pixel (voxel, respectively) size, thus reducing the amount of material available for analysis.47 In spite of these challenges, single cell MS analysis has been achieved in a variety of organisms for numerous applications, which include measuring dynamic cellular biosynthesis, activation or inhibition of signaling pathways, metabolic processes in diseased states, new natural products, chemical defense mechanisms, and toxicological risk assessments.48
Various metabolomic and proteomic MS approaches have demonstrated the potential of obtaining a vast knowledge about mammalian cell processes,44 but with the addition of a spatial dimension due to the increasing interest in developing single cell profiling tools this knowledge base will likely continue to grow.49 The reason metabolomic and proteomic profile characterization is of such great interest is they help predict differing cell types that could be associated with molecular changes due to different perturbations of interest.50−52 One example comes from Jansson et al., who used a microscopy-guided single cell matrix-assisted laser desorption/ionization (MALDI)-MS approach to determine the single cell heterogeneity of islets of Langerhans.53 Additionally with the use of MALDI-2, Bowman et al. demonstrated rich lipid spectra from mouse and human brain tissue containing multiple sclerosis lesions at approximately 6 μm spatial resolution.54 These studies are some examples that demonstrate the utility of single cell MSI for the investigation of mammalian cells, and further examples will be discussed in detail later.
Single cell MS techniques have also been adopted by plant biologists in effort to help elucidate primary metabolism, natural products, and plant defense pathways, for example.55 The vast molecular differences between the leaf, stem, and root are even more apparent through the lens of single cell analyses. Individual plant cells can range from ∼10–100 μm in size, which in part makes single cell analysis more readily attainable than in mammalian cells.56 However, special consideration with regard to the cell wall must be considered, as this can introduce unique challenges into the analysis of plant cells. Specifically, the outside of the plant plasma membrane is completely coated with arranged layers of cellulose microfibers embedded in a matrix that composes a plant cell wall, which can be up to 0.2 μm thick.57 However, this also contributes to the rigidity of the plant, which ultimately makes plant cells often easier to handle for single cell MS experiments.58 As an example, Korte et al. used a modified MALDI-Orbitrap-MS instrument for single plant cell analysis, imaging juvenile maize leaves at a 5 μm spatial resolution.59 This permitted the mapping of a variety of metabolites ranging from amino acids, glycerolipids, and defense-related compounds at below single cell resolution.59 Stopka et al. also utilized an optical fiber-based laser ablation electrospray ionization mass spectrometry (f-LAESI-MS) technique to target specific cell types within Egeria densa leaf blade cells.60 Primary metabolites such as malate, asparate, and ascorbate were found at higher levels in epidermal cells, while lipids and triterpene saponins were found in idioblast cells. These two techniques only scrape the surface of how single cell MS has been applied to plant cells. More examples of single cell plant analysis can be found elsewhere55,61,62 and will be discussed further in this review.
Understanding the function of microbial communities is of major interest, yet they remain a challenge largely due to their enormous diversity and the complexity of interactions between community members.63,64 As single cell MS technologies have advanced, high lateral resolution molecular analysis of bacteria and fungi is now within reach.64,65 However, single cells of bacteria and fungi are inherently smaller in size than mammalian and plant cells, and they contain certain properties that can make high spatial resolution molecular-based MS measurements difficult. Specifically, bacteria typically range in size from approximately 0.2 to 5 μm, whereas fungi can range from 2 to 10 μm with fungal hyphal-cell bundles reaching 5–50 μm (Figure 2).20,21 Other challenges persist with these types of organisms, where fungal cells, for example, are mostly made up of chitin that is difficult to breakdown to access intercellular metabolites and proteins.58 Additionally, diatoms, a single celled alga, contain a cell wall consisting of silica, which makes their analysis extremely difficult for the same reason.66,67 Nonetheless, high-lateral resolution secondary ion mass spectrometry (NanoSIMS) has provided insights into these microorganisms. NanoSIMS can achieve spatial resolutions down to 30 nm allowing for microbes to be imaged.68 Schoffelen et al. utilized NanoSIMS for single cell imaging of phosphorus uptake within algae.69 Additionally, Zimmerman and co-workers used a combination of flow cell sorting and NanoSIMS to determine phenotypic heterogeneity of chlorobium phaeobacteroides.70
Single cell MS as a field has grown immensely over the past decade. Technologies such as MALDI-2, NanoSIMS, multiplexed ion beam imaging by time-of-flight MS (MIBI-TOF), and many more have allowed for a continual growth in research to be conducted on a wide variety of organisms and cell types. As researchers continue to push the boundaries, single cell MS is likely to become a mainstay and an essential piece of research as we continue to strive to elucidate metabolic functions in vivo.
II. Advances in Single-Cell MS Analyses
Single Cell Proteomics
Elucidating the spatial distribution of proteins at the cellular and the subcellular level, as well as having the ability to capture ever-changing protein dynamics within a cell, are vital for a complete understanding of the underlying biology of a system from immune response to cellular stress, for example.71 Genome amplification and sequencing techniques commonly perform single cell analyses and have been shown to resolve rare cell populations and interrogate specific cells and substructures of interest within heterogeneous clinical tissues.72 However, genomic and transcriptomic technologies only provide indirect measurements of cellular states, as these are constantly changing in response to cellular stress.73 The relationship between protein abundance and transcript abundance is complex and dependent on the experimental context, which challenges biological interpretation.16,74 In-depth proteomic measurements provide a more direct characterization of phenotypes and are therefore crucial for understanding cellular functions and regulatory networks.75 Furthermore, MS-based proteomics measurements have the potential to identify post-translational modifications (PTMs), providing insights not available through genomics measurements.76 Single cell proteomics by MS, therefore, promises to revolutionize our understanding of cellular biology.
Single cell proteomics approaches that utilize immunohistochemistry (IHC) methods—bedrocks of clinical diagnostics and basic research77—have thus far demonstrated the highest lateral resolution. Fluorescently labeled antibodies used for the simultaneous detection of multiple targets with optical microscopy and spectrometric tools are ubiquitous, but these techniques have their limitations. In particular, the lateral resolution of conventional optical microscopy is diffraction limited, which constrains the lateral resolution of these measurements.77 Another key challenge in IHC is the availability of high-quality, thoroughly validated antibodies.78,79
Multiplexing fluorescent labeled IHC tags is possible, but there is a need for primary antibodies to be highly selective to the targeted protein (i.e., have very minimal nonspecific interactions) and have nonoverlapping reporter emission spectra.80 Carefully matching fluorescent reporters is essential along with utilizing dichroic mirrors and band-pass filters to limit the overlapping emission spectra.80 Alternatively, metal-based IHC, like those used in mass cytometry approaches, readily enable the targeted detection of up to tens of protein markers from a sample by utilizing heavy metal antibody-bound reporter species.80 Furthermore, imaging mass cytometry and metal-based IHC using MSI has come to the forefront of spatially resolved single cell proteomics measurements. Fluidigm utilizes a laser ablation-inductively coupled plasma (LA-ICP)-MS approach for the detection of antibodies within single cells (i.e., imaging mass cytometry).81 Additionally, MIBI, developed by the Angelo lab (Figure 3), employs a primary ion probing technique with MS to image metal-tagged antibodies at subcellular resolution within clinical tissue sections.71,82 Both approaches have the capability of analyzing up to 100 targets simultaneously over a five-log dynamic range. Imaging mass cytometry and MIBI have frequently been utilized for the analysis of formalin-fixed, paraffin-embedded (FFPE) tissue.83 FFPE preservation has been a common practice for clinical pathological analysis because of its ability to preserve tissue morphologies long-term. Because of this, large tissue repositories containing patient samples exist waiting for analysis.83 MIBI-TOF was recently used to simultaneously quantify 36 proteins at subcellular resolution in triple-negative breast cancer samples.84 In this study, spatial enrichment analysis showed immune mixed and compartmentalized tumors coinciding with the expression of proteins PD1, PD-L1, and IDO, which can be indicative of metastasis and poor prognosis, in a cell type and location specific manner.84 Wang et al. utilized the Fluidigm system to analyze human pancreas islets during the progression of type 1 diabetes.85 Using this system they were able to achieve 1 μm resolution to visualize an altered islets architecture and changes in protein expression levels during disease progression.85 However, utilization of this type of technology for single cell proteomics ultimately is inherently limited by the availability of high-quality antibody reagents—requiring a priori knowledge of the proteins being targeted for analysis—and a finite multiplexing capacity.
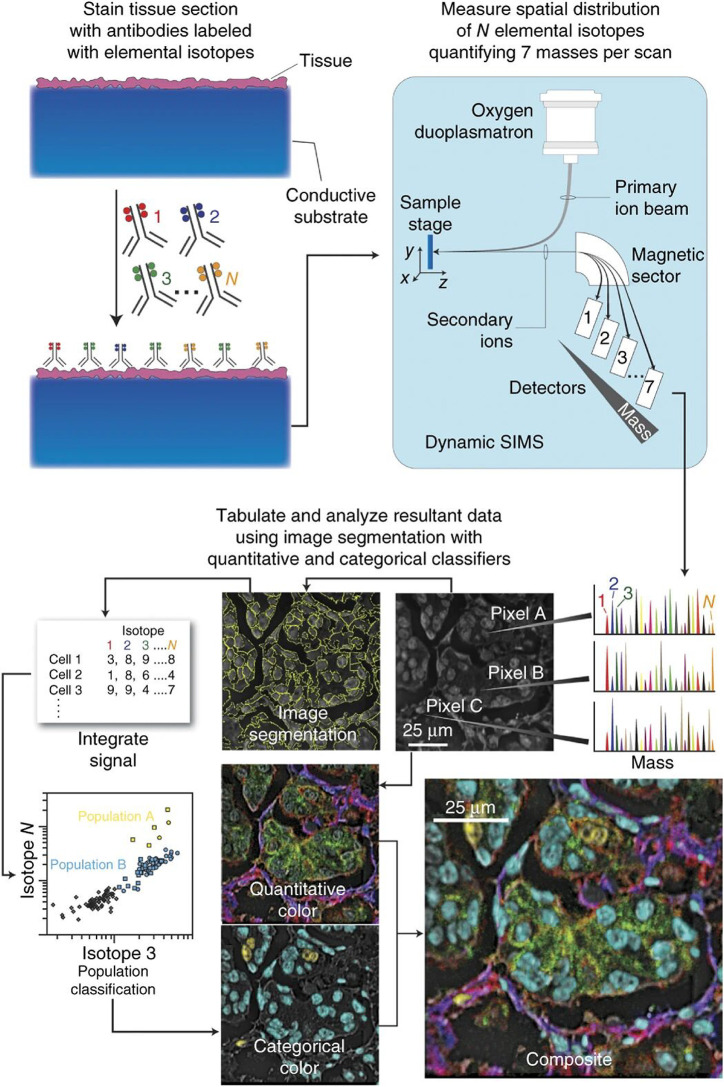
Figure 3.
Multiplexed ion beam imaging workflow for high-resolution spatial proteomics. Here, preserved tissue sections are mounted on conductive substrates and incubated with unique isotopic transition metal-tagged antibody reporters. An oxygen primary ion beam rasters the sample surface, ejecting and ionizing the isotope reporters, and their masses are subsequently measured via a mass analyzer. In this example, MIBI analysis of human breast tissue displaying multichannel overlays, where each color represents a separate protein specific reporter. Adapted with permission from ref (82). 2014 Nature Research.
With the challenges of using IHC-based single cell MS in mind, untargeted, direct single cell measurements are also a common technique used for proteomics analysis. Historically, MALDI-MS was a common method of protein analysis, and only in the recent decade has it found broad applicability in metabolomics.43 The detection of peptides within individual cells was reported over two decades ago (in the year 2000).86 MALDI-MS offers many advantages for single cell proteomics, including good tolerance for salts, simple sample preparation, and attomole detection limits with little sample consumption.87,88 A few early MALDI-MS-based studies revealed neuropeptide profiles in single neurons of invertebrates,89 as well as the discovery of many novel neurohormones in single invertebrate neurons.90 Imaging proteins with MALDI-MS presents a challenge when it comes to balancing throughput, sensitivity at high spatial resolution, molecular specificity, and identification. Spraggins et al. worked to address these issues by improving acquisition rates and the spatial resolution down to a single cell level, and they were able to spatially resolve and identify proteins directly associated with host immune response within specific lung tissue structures.91 Laser ablation electrospray ionization (LAESI) has also been used for the measurement of intact proteins, where Kiss et al. used a LAESI source combined with a hybrid ion-trap Fourier transform ion cyclotron resonance (FTICR)-MS for the detection of intact proteins directly from a tissue using a top-down approach.92 These laser ablation-based spatially resolved proteomics techniques, however, provide limited molecular coverage of only a handful of proteins within an analysis.
While MALDI-MS analysis of proteins presents its own set of challenges, untargeted molecular separation-based MS techniques, using liquid chromatography (LC) and nanoelectrospray ionization (ESI), have excelled in multi- and near single cell proteomics, in part, by providing broad molecular coverage. Currently, most LC–MS-based proteomic approaches require samples comprising of thousands of cells in order to provide in-depth protein profiling.38 However, through the use of laser capture microdissection (LCM), which permits targeting areas of interest within a tissue, or flow cytometry cell sorting of disrupted tissue, specific cellular populations can be isolated to provide comprehensive molecular coverage of their proteomes.93 The recent development of nanodroplet processing in one pot for trace samples (nanoPOTS) provides the ability to probe small to single cell populations in depth.94 NanoPOTS effectively enhances the efficiency and recovery of sample processing by downscaling preparation volumes to less than 200 nL to minimize surface losses.94 Using nanoPOTS combined with a differential tandem mass tag (TMT) labeling approach,95 over 1400 proteins were able to be quantified from 152 individual single cells.38 This finding demonstrates how this technology can be used for spatially resolved proteomic measurements, with broad molecular coverage, within clinical tissues of interest.38
Even though LCM and flow cytometry provide reliable dissociative methods for single cell analysis, there remains a need for nondisruptive single cell analysis tools. The ability to carry out microsampling single cell proteomics experiments opens the door for studies that can directly probe the developmental processes in complex tissues or whole organisms. Within this arena, Xenopus laevis has been extensively used as a model system for microsampling single cell proteomic studies due to the large cell size and protein concentrations in the early stages of their development.96 Sun and co-workers were able to quantify 1400 protein groups in a single run using LC–MS2 from a single blastomere obtained from a 16-cell X. laevis embryo.94 Blastomere-to-blastomere heterogeneity was observed in 8-, 16-, 32-, and 50-cell embryos, indicating that comprehensive quantitative proteomics of this model organism can lead to valuable insights into cellular differentiation and organ development.97 Lombard-Banek and co-workers pushed further and isolated a single D11 cell from X. laevis from the 32-, 64-, and 128-cell stage of development. Using optically guided in situ subcellular capillary microsampling and one-pot extraction and digestion method, they were able to identify between 750 and 800 protein groups in each cell stage by analyzing 5 ng of protein by capillary zone electrophoresis MS.98 As technologies continue to push the boundaries of single cell MS, more in-depth proteomic measurements will be able to aid in elucidating the critical roles of cellular functions.
Single Cell Metabolomics
Individual cells will exhibit a wide degree of molecular diversity across their population, therefore screening and identifying biomolecules over this expansive range is a significant challenge. In the case of metabolites (e.g., lipids, sugars, and organic acids), the number of identified molecules orders in the hundreds of thousands in mammals (according to the human metabolome database),99 and over 200,000 in the plant kingdom (determined from early calculations).100 Metabolite heterogeneity is a multifaceted problem for numerous reasons. To begin, lipids and polysaccharides, for example, act as membrane and cell wall supports in mammalian and plant cells, respectively, but the overall membrane composition can vary across cells of the same genus.101,102 This compounds the analytical challenge as the dynamic nature of metabolism can result in metabolic asymmetry.103 For these reasons, establishing powerful spatial MS tools to sample the metabolome on a cell-to-cell basis may provide the ability to answer critical questions such as the speed and degree of metabolite exchange that occurs in a cell population and, consequently, how this impacts metabolic asymmetry.104
To date, MSI has been the most impactful tool for MS-based single cell metabolomics.6−8,42,43,55 MSI methods have primarily involved probes including ion beams, lasers, or solvent junctions for in situ analysis of metabolites.105 These techniques, summarized in Figure 2, do not require molecular labels and are considered label-free imaging techniques. Of all the techniques, MALDI-MS is utilized the most for spatial metabolomics,43 and has been used for tissue level studies of bacterial,65,106 fungal,107−109 invertebrate,110,111 plant,112,113 and mammalian systems,114,115 for example.
Reflective geometry MALDI-MS, where the laser directly impacts the sample surface, is the most common configuration. As this is usually employed as a microprobe technique, lateral resolution is considered laser spot size limited, and therefore, single cell analysis typically requires subcellular laser spot sizes.116−118 Reflective geometry MALDI-MS involving spot sizes greater than a single cell have been explored; however, this has involved either dispersed cells in a culture or cell microarrays rather than analyzing single cells in a tissue.119−124 Over the past decade, several research groups have improved the source design for MALDI-MS to enable single cell imaging in tissues. The Caprioli and Lee groups have used small diameter pinholes to filter the beam down to spot sizes of ∼5 μm.59,125 However, oversampling, where individual pixels are partially overlaid, was necessary to compensate for low ion yields in these studies.59 Recently, Feenstra et al. developed a beam expander able to vary spot sizes between 4 and 50 μm.126 They visualized lipids, sodiated sugars, and glucosides within maize roots at 5 μm, but significant decreases in ion yield were observed for all species, demonstrating the limitation of a pinhole approach.126 There have also been many new instrumental designs developed to increase the imaging speed of MALDI-MSI since imaging overall large (tissue level) areas at single cell resolution increases the number of data points (pixels) needed to be obtained.127,128 Recently, Potocnik et al. used continuous laser acquisition to image lipids at a rate of 50 pixel/s and a lateral resolution of 10 μm in sections of mouse brain using a Bruker rapifleX MALDI Tissuetyper TOF-MS instrument.127
Over the past decade, the Spengler group has developed and improved upon their so-called scanning microprobe MALDI (SMALDI) source. SMALDI uses a coaxial ion source geometry with a novel objective for laser focusing, which permits spot sizes down to 0.25 μm (as observed by the ablation crater), making it readily applicable for analyzing single cells.129 Recently, SMALDI has been adapted for ambient pressure analysis (AP-SMALDI).130 Khalil et al. coupled the AP-SMALDI to a Thermo Q Exactive to assign 67 lipids at 12 μm resolution in mosquito sections.130 Kompauer et al. decreased pixel sizes to 1.5 μm using a custom nine lens objective, imaging lipids, metabolites, and peptides at subcellular resolution in mouse brain sections.131 Most recently, Garikapati et al. optimized sample preparation for 10 μm lipid imaging in sections of mouse fetus.132
An alternative optical configuration for MALDI is using transmission geometry (TG-MALDI), where the laser impacts the sample through the backside of an optically transparent substrate.133 This delivers smaller spot sizes than reflective geometry via high-resolution focusing lenses with focal lengths typically too small to be used in reflective geometry.133 Another benefit of TG is that ions can be desorbed directly into the MS inlet without ion losses that occur due to orientation of the optics and ion extraction in reflective geometry mode.134 Recently, Zavalin et al. achieved 1 μm for lipids in a HEK-293 culture and mouse cerebellum tissues using TG-MALDI.133
It is worth reiterating that the reduction in probe diameter size will decrease sensitivity.118 However, postionization can compensate for sensitivity reductions.135 Soltwisch et al. first successfully reported laser post ionization (MALDI-2) in the reflective geometry configuration.136 Recently, Bowman et al. mapped 147 unique lipids in sclerotic mammalian lesions at 6 μm,54 and separately, McMillen et al. and Heijs et al. used the timsTOF flex for neutral lipid and N-glycan profiling, respectively.137,138 In transmission geometry, Spivey et al. analyzed lipid standards, showing that UV postionization was able to compensate for sensitivity losses at <1 μm spot sizes.139 Niehaus et al. took this approach further, visualizing neutral phospho- and glycolipid species in the cerebellum and kidneys of a mouse model at a submicron resolution (Figure 4).140
Figure 4.
Example single cell imaging results from TG-MALDI-2 in the analysis of Vero-B4 cell culture: (a) Bright-field microscopy image of Vero B cells with deposited DHB matrix; (b) background (m/z = 633.042) and (c–e) single ion images of PE (36:2), PC (34:1), and PC (34:1). (f) High resolution bright-field microscopy image of highlighted red region in a, and (g) overlay of ion images in b, c, and e. (h) in-line bright-field microscopy image from the TG-MALDI-2 source outlined region in f. Adapted with permission from ref (140). 2019 Nature Research.
While microprobe analysis is the most common approach for MALDI-MS, microscope mode is an alternate method.117 Here, 2D ion detectors provide ion positions and register ion packets while maintaining their spatial dimensions, which permits subcellular lateral resolution imaging using spot sizes larger than the diameter of the analyzed cell.141 Furthermore, this reduces throughput time in an imaging experiment, as fewer sample positions are required for an image.141 Many of these notable experiments have involved the triple focusing TRIFT II mass spectrometer to achieve lateral resolutions of below 10 μm in the detection of lipid and peptides.142−145
A common aim of MSI is absolute analyte quantification with spatial analysis.146−148 The ability to quantify molecules in a tissue section opens up the possibility of quantitative molecular histology, which would permit MSI to be more readily applied as a diagnostic tool for precise monitoring of disease states based on spatial changes in molecular concentrations.149 However, analyte quantification is complex for a number of reasons. Using MALDI-MS as an example, several factors inhibit quantitative MSI. First, tissue-specific ion suppression may occur in the analytes surrounding environment, so-called “matrix effects,” which can change measured analyte yields.150 Second, molecules have specific ionization energies, meaning that the ionization potential of a molecule must first be known to convert relative abundance to absolute concentration.151 Finally, in the case of MSI techniques involving artificial matrices, analyte yields are dependent upon the matrix composition, relative amount, and uniformity across a tissue section.152 Currently, quantification is achieved with the aid of calibration standards;153−155 however, this is far from scalable for metabolite analysis as the number of known and unknown metabolites orders in the hundreds of thousands.
To overcome challenges related to MALDI-MSI analysis,152,156 matrix-free LDI methods have been explored.157 Mid-IR lasers (i.e., λ = 2940 nm) capable of vibrationally exciting endogenous water molecules within cells have been of particular interest.158 In this approach, probing water-rich samples enables the ablation of molecules from the sample.100 However, since the vast majority of these molecules are neutral, a postionization step is necessary.159 An orthogonal electrospray that intercepts the ablation plume and funnels the ions to the mass analyzer is used in LAESI160 and infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI).161,162 Recent examples by Kulkami et al. have explored metabolite distribution in the roots of native and invasive plant species at 100 μm lateral resolution.163 Agtuca et al. explored the metabolic asymmetry between infected and noninfected soybean nodules with LAESI where elevated fatty acid, purine, and lipid levels occurred as a result of Bradyrhizobium japonicum infection.164 The Muddiman group used ice matrices to increase lipid yields,165 explored dynamic neurotransmitter imaging,166 used IR-MALDESI for imaging metabolomics in plant and mammalian samples,167,168 and coupled a drift tube for ion mobility direct collision cross section (CCS) measurements from several tissue types.169 Most recently, cell-to-cell lipid heterogeneity in a dispersed HeLa cell culture was demonstrated by Xing et al. with IR-MALDESI.170
Given that spot size increases with wavelength,171 single cell analysis can be more challenging with methods using IR lasers in comparison to those employing UV lasers (e.g., MALDI). Nevertheless, there have been efforts in this arena. The Vertes lab have pioneered f-LAESI, which uses an optical fiber to transmit the IR laser to the sample surface, rather than a series of mirrors and objectives for focusing.60 This configuration can produce spot sizes <30 μm, and therefore can be used to ablate single cells.172 Stopka et al. used f-LAESI to elucidate the metabolic noise within Egeria densa and infected soybean root nodule cells.60 Recently, Samarah et al. used f-LAESI in combination with a 21-T FTICR-MS to study the fine isotopic structure (IFS) of endogenous metabolites (Figure 5).159 It should also be noted that transmission geometry has also been explored by the Vertes group,173 but most recent efforts have been directed at f-LAESI.
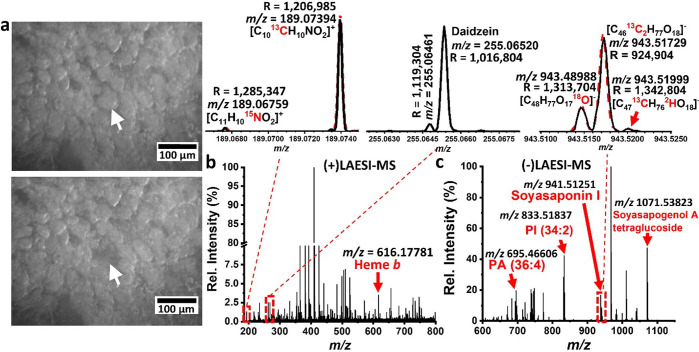
Figure 5.
Example of f-LAESI analysis using a 21T-FTICR-MS. Here, single soybean root nodule cells infected with B. japonicum were analyzed. (a) Identification and ablation of an infected soybean cell and (b, c) the resulting mass spectra data from positive- and negative-ion modes, respectively. The top left inset shows the captured IFS (black) for N-acetylglutamic acid overlaid on top of its simulated mass spectrum (red dashed line). The middle inset shows the resolving of two peaks that correspond to two different metabolites. The top right inset shows the IFS of dehydrosoyasaponin I (black) captured at a longer transient length. The peaks drawn with a red dashed line correspond to the simulated mass spectrum of the same ion. Adapted from ref (159).
An alternative ionization approach for laser desorption is using structured array supports. Here, lithography and ion etching techniques are used to fabricate nanopost arrays, which act as a chemical ionization reagent through surface plasmon wave excitation when tissues mounted on the array are irradiated.174−176 Korte et al. demonstrated that silicon nanopost array (NAPA) devices produced better metabolite coverage in LDI-MS than MALDI-MS by detecting 88 unique low-molecular weight metabolites.177 NAPA studies have shown that this method can also expand to other metabolite classes including amino acids, nucleotides, carbohydrates, xenobiotics, and lipids.178 Experiments by Fincher et al. have shown that the sensitivity of NAPA-based LDI is in the subfemtomole range, which is significantly more sensitive than techniques involving modification of tissues after mounting on a substrate.179 Additionally, it is theorized that understanding the effect of array variation on ion yield quantitation is far easier using the NAPA platform as rapid prototyping techniques typically can be optimized to minimize array variation.180 Nanostructure-initiator mass spectrometry (NIMS) is another technique that utilizes nanostructured surfaces, which use porous silicon to facilitate analyte desorption by laser irradiation.181−183 However, this technology is in its infancy, and limited examples of cellular resolution imaging with NIMS have been shown.184,185
Desorption electrospray ionization (DESI) is a desirable technique for single cell MS, as molecule desorption via a solvent stream can be performed under ambient analysis conditions and with minimal sample preparation.186 However, the limited spatial resolution of this approach has relegated it to primarily tissue-level imaging.187−189 Over the past decade, Laskin and co-workers have developed nanospray-DESI (or nanoDESI) utilizing a microliquid junction between two capillaries to desorb analytes which has resulted in improvements in sensitivity and lateral resolution.190 Sub-10 μm lateral resolution nanoDESI-MS has been applied for lipid imaging in lung, brain, and pancreatic tissues.191,192 The addition of shear force probes to standardize capillary-to-sample distance has allowed for combined topography/molecular imaging, permitting the ability to assess ion yield variation effects with topographic changes, for example.193
Of all the MSI techniques, SIMS maintains the highest spatial resolution for MSI.194 The ability to obtain molecular images of intact species depends upon ion beam characteristics such as the composition, the configuration of the ion optics, and the energy spread of the beam.195,196 Ion beams composed of majority monatomic species readily provide subcellular resolution imaging capability, as they can be focused to below 50 nm.194,197,198 However, the limitation for analysis of biological samples is that they generate significant fragmentation, therefore molecules are rarely desorbed (and consequently measured) intact.196 Gas cluster ion beams (GCIBs), which can be focused to <3 μm and are used on instruments including the Ionoptika J105,199,200 have improved intact lipid imaging due to softer desorption processes that generate less fragmentation than the liquid metal ion guns (LMIG) typically used in SIMS.201−203 Subcellular imaging of phosphocholine (PC) lipids in the brain, lipid visualization in 3D within bacterial envelopes, and de novo purine biosynthesis have been demonstrated at a single cell scale with (CO2)n/Arn gas clusters.204−206 Recently, high energy water clusters have been shown to improve ion yields for single cell imaging with SIMS.207 Due to SIMS introducing significant fragmentation, instruments with MS2 capabilities are desired as they aid assignment of fragment ions (produced by molecule fragmentation) to their corresponding parent ions in a fragment rich SIMS spectra.208,209 However, many instruments only employ a simple TOF-multichannel plate detector, which limits the mass resolving power to <10000, making structural assignments nontrivial.210 The PHI nanoTOF II has MS2 capabilities and operates a triple focusing electrostatic analyzer, where precursor ions can be selected and directed into a collision induced dissociation cell for MS2.211 Single cell imaging with the nanoTOF II has included the imaging endoplasmic reticulum stains in single cells,212 among other examples.213 Recently, IONTOF have developed MS2 capabilities for the TOF-SIMS V by the addition of a Thermo Q Exactive Orbitrap (FTMS) to the instrument (termed “the hybrid orbiSIMS”).214 Passarelli et al. have used this novel instrument for imaging lipids and isotopically labeled amiodarone in 3D within single cells at either high lateral or high mass resolution (Figure 6).215
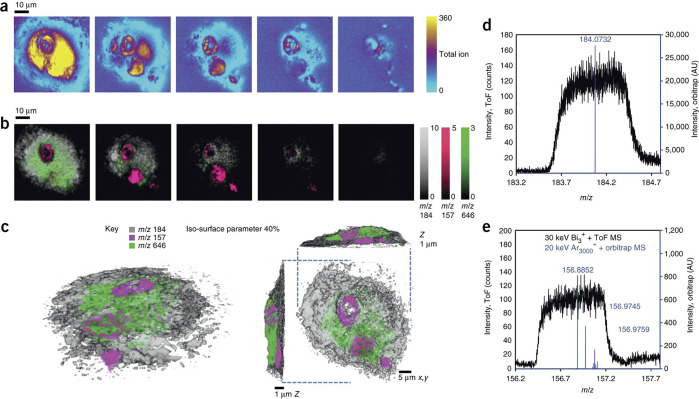
Figure 6.
Example of TOF-SIMS subcellular imaging, where the distribution of a drug is visualized in a single cell using the hybrid OrbiSIMS. (a) A sequence of total ion images captured every ∼400 nm of depth, as the cell was sputtered away. (b) Overlaid ion images of the PC headgroup in gray, m/z 157 (a nuclear marker) in magenta, and the drug (amiodarone) in green at each of the respective spatial locations in a, and (c) a 3D rendering of the cell that tomographically illustrates the location of PC, nucleus, and drug markers. Mass spectrum obtained from the ToF-MS (black) and the Orbitrap-MS (blue) of the (d) PC headgroup and (e) nuclear marker. Adapted with permission from ref (215). 2017 Nature Research.
III. Correlative Imaging and Multimodal Spatial Omics
Single cell MS experiments can provide an abundance of informative data about molecular processes related to a cell’s phenotype. Nonetheless, linking data of this technology with data from other analytical approaches can increase the amount of knowledge obtained from a sample. Combining two or more imaging modalities (i.e., “multimodal imaging”) provides advantages that include (i) improved sensitivity and specificity of chemical classes that cannot be easily identified or analyzed by a single imaging modality alone and (ii) enhanced data mining capabilities.216 Moreover, the utilization of multiple spatially resolved MS tools in conjunction with one another can provide a more effective means to probe the biomolecular complexity of biological systems by providing higher confidence in molecular annotations and localizations.217,218 Beyond MS-based omics, there are other major technologies for proteomics and metabolomics, such as NMR,219−221 arrays,222 and other reporter-based methods that can provide spatially resolved biochemical information.195,223,224
The addition of labels can be used to correlate data from multitechnique analyses and enhance the interpretation in single cell biology. For example, the use of hybrid tracers enables a direct comparison between fluorescence confocal microscopy and LA-ICP-MS imaging techniques, further showing how multimodal techniques can be complementary to one another.81 Recently, breast cancer cell lines stained using receptor specific hybrid tracers, which contained both a fluorophore and a DTPA single lanthanide chelate, permitted fluorescence confocal microscopy to guide subsequent LA-ICP-MS analysis.81 Laser ablation isotope ratio mass spectrometry (LA-IRMS) has the ability to trace 13C through a variety of samples and can be used to inform, as well as in conjunction with, higher spatial resolution imaging techniques like SIMS to confirm labeled molecule distributions across tissues and down to each cell.225,226
Raman spectroscopy is an advancing cell imaging method that, unlike MS methods, is nondestructive. Single cell Raman microspectroscopy, for example, can obtain biochemical fingerprints of individual microbial cells.227 A key advantage of Raman spectroscopy is its ability to determine the biochemical makeup of a cell, wherein proteins, lipids, and DNA can be visualized according to their unique vibrational spectra.228 Using the unique vibrational spectra, functional groups and chemical bonding in molecules can be identified. The weak Raman signal of water affords the ability to image cells within aqueous environments more readily, which means live cell imaging is feasible under normal physiological conditions.229 Stable isotope probing (SIP)-Raman spectroscopy can also help reveal metabolic activity within a cell. A number of bands within a single cell Raman spectrum can shift upon metabolic labeling with 13C, 15N, and 2H.230 Furthermore, the results from SIP-Raman spectroscopy can be combined with fluorescence in situ hybridization (FISH) specific bands shifts to link metabolic activity to cell identity.231 SIP can also be used to bridge NanoSIMS and Raman microspectroscopy imaging modalities, by tracking isotopically labeled substrates (i.e., 13C, 2H, 15N) incorporation into metabolic pathways at single cell and subcellular spatial resolution. Numerous reviews detail the application areas of SIP-NanoSIMS232 and SIP-Raman microspectroscopy.231 Recently, SIP-NanoSIMS has been used to identify the presence of cholesterol-rich sphingolipid patches in plasma membranes of kidney cells,233 image the root cell-soil interface in permafrost,234 image carbon transfer between the root-rhizosphere,235 identify the dependence of Bacillus subtilis sporulation on the presence of mycelia in nutrient starved regions of soil,236 and image the accumulation of drugs in multilamellar lysozymes at nanometer scale resolution.237
Microscopy is no doubt the most utilized tool for single cell imaging, and it is highly versatile in the types of measurements it can make. These capabilities include live-cell fluorescence imaging for the understanding of signaling dynamics,238 FISH for measuring gene expression,239 immunofluorescence,238 and other antibody-based methods for measuring protein expression230 (as detailed in the proteomics section). In addition, recent studies have combined other modalities such as RNA-sequencing240 and microfluidics239 with live-cell imaging to expand the possible measurements and knowledge gained from a single cell. The use of microfluidics in MS has also grown in recent years due to the advent of “biology-on-a-chip” devices. These devices have the ability to monitor dynamic chemical processes in a controlled environment, and having the ability to spatially map these chemical signals within the devices may allow for new insights to be gained.241 For example, Cahill et al. recently developed a porous membrane sealed microfluidic device that can be used in conjunction with liquid microjunction surface sampling probe MS.241 In this study, they were able to demonstrate in situ MS chemical analysis of fluid within a microfluidic flow cell device by forming a liquid junction between the porous membrane and the sampling probe.241 Further examples of how microfluidic technologies have been used in single cell metabolomics and proteomics can be found in other recent reviews.242−244 Combining atomic force microscopy with single cell force spectroscopy is also useful for single cell analysis, as this can be used to study the adhesion of living cells in near-physiological conditions,245 while scanning electron microscopy246 and transmission electron microscopy (TEM) can be used to create a high resolution image of the single cell.247 For example, NanoSIMS and TEM were used together to visualize the uptake of cisplatin, a common chemotherapeutic, into resistant and nonresistant ovarian cancer cells.248
All of these techniques described here can provide a wealth of knowledge individually, but when combined with additional analytical approaches, they can provide a more comprehensive picture of complex biochemical processes. Moreover, since each technique has its own unique advantage(s) for single cell analyses, when they are used in a multimodal fashion, one can enhance their results through the combination of data as more biologically relevant signals can be produced. MSI experiments can give a wealth of chemical meaning, while other techniques such as Raman spectroscopy can give meaningful chemical bonding and structural information. However, there can be technical challenges when coregistering images acquired with different modalities. One major issue can be the differences in spatial resolution between the images that are being coregistered. Recent work has been done in an effort to help mitigate coregistration issues. For example, Patterson et al. have developed experimental and computational pipelines to help bridge data sets between high spatial resolution optical microscopy and MALDI-MSI studies.249,250 Multimodal imaging analysis is therefore an important aspect to contemplate and consider when attempting to elucidate the complexity of a single cell.
IV. Data Acquisition and High-Throughput Screening
The commercial viability of single cell screening with MS requires technologies able to rapidly and efficiently extract and assign biomolecules from multicellular samples. Introducing high throughput screening (HTS) methods into MS workflows is attractive, as this method of experimentation involves analysis of thousands of samples on a rapid time scale and is usually reliant on automation and robotics that provide reproducibility and robustness.251 Historically, HTS of biological samples has involved fluorometric or calorimetric assays to provide molecular information, but these techniques require analytes to be chromophoric probing methods that may limit molecular specificity, decrease throughput from the extra labeling steps, and are blind to the majority of biomolecules present in a cell.252 The aim of MS-based HTS is for biomolecule extraction, ionization, separation, and molecular assignment all to be automated. However, many of the methods we have referenced illustrate that proteome or metabolome MS measurements at the single cell often require lengthy sample preparation steps and/or long data acquisition and analysis times, which brings to question, can certain types of MS analyses be used in HTS?
The microarray format has been useful for rapid screening in drug discovery for decades.253 HTS has been adapted from drug discovery to single cell MS-metabolic analysis through manufacturing single cells arrays. Ellis et al. combined liquid extraction surface analysis with bioprinting to measure the lipid profile of mammalian single cell lines, where they showed impressive spectral quality.124 However, acquisition times were nonideal, and only 37% of the droplets on the array contained single cells.124 Yang et al. used microcontact printing to deposit poly-l-lysine and electrostatically adhere single mammalian lung cancer cells with a capture efficiency of 40% and demonstrated a linear relationship between PC lipid abundance and cell number using MALDI-MS.122
Ambient sampling methods have been developed to diminish the need for extensive sample preparation. LAESI, for example, allows for in situ metabolomics of native tissue.60 However, HTS single cells with LAESI is still quite complex, as this often relies on fiber-based LAESI (f-LAESI), where the etched optical fiber is placed in contact with the cell surface, requiring manual positioning.60 This also presents a challenge in measuring dynamic systems with ongoing metabolic processes, where tissues and cells are not fixed. Metabolic quenching can be compensated for by freeze-fixed analysis, but the speed of how rapidly activity is quenched also remains a question.254
Direct infusion MS instruments are attractive for HTS, in part because sample processing can be readily automated. Examples of direct infusion commercial instruments include the TriVERSA NanoMate nanoelectrospray/LESA-NanoMate,255 the Agilent RapidFire,256 and the Labcyte Echo acoustic droplet ejection system,257 which are all robotically driven. However, these approaches have had little application on the single cell scale. The invention of the nanoPOTS system (Figure 7) shows promise for MS-HTS for single cell proteomics, in part because sample processing can be automated. In this technique, nanoliter cell-containing droplets are dispensed into a multiwall chip, followed by extraction, alkylation, protein digestion, and peptide collection. Zhu et al. demonstrated the utility of the nanoPOTS platforms by quantifying over 3000 proteins in as little as 10 cells.75 Recently, Williams et al. developed a nanoPOTS autosampler compatible for automated LC–MS analysis, removing labor intensive steps such as manual sample loading.38 256 individual proteins were able to be identified in single MCF10A cells at a rate of 24 cells per day.38
Figure 7.
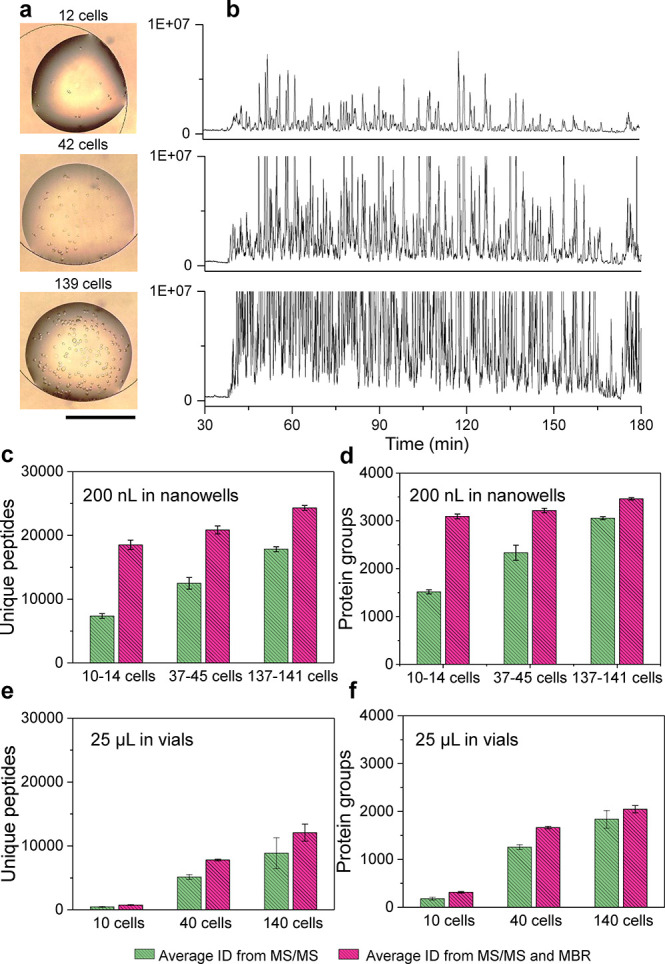
Example of near-single cell untargeted proteomic analysis using nanoPOTS, which is adaptable method for high-throughput single cell screening applications. (a) Bright-field images of nanowells with dispersed HeLa cells in droplets. (b) Ion chromatograms corresponding to analysis of 12, 42, and 139 cells and (c–f) the unique peptides and protein group coverage from nanoPOTS processing of different liquid volumes. Data are expressed as means ± SD for experimental triplicates, and the scale bar in (a) is 500 μm. Adapted with permission from ref (75). 2018 Nature Research.
There are limited examples of HTS-MS imaging workflows in tissue sections, in part due to the challenges of sample variation, image correlation, and automatically assigning molecular structures to multidimensional imaging data sets.258 Many of the efforts have been aimed toward increasing sample analysis through source and detector improvements. Imaging experiments are frequently performed with sample replicates to assess the biological reproducibility.259−261 To this end, fusion of microscopy and MSI data sets have been explored by the Caprioli group as a method of training ion distributions for rapid parallel sample imaging, theoretically allowing for microscopy images to inform analyte distributions in sample replicates. The idea is that once the software is trained sufficiently to recognize ion distributions optical images can be used to predict MS images allowing for rapid sample screening. Van de Plas et al. demonstrated this technique for lipid profiling to accurately predict the distribution of PE(16:0/22:6), and built on the work of Tarolli and co-workers, who first used pan-sharpening to improve lateral resolution of MS images,262 to visualize lipids at 10 μm lateral resolution in mammalian brain tissue using a 100 μm resolution MS image.263
V. Challenges and Future Perspectives
A core challenge for single cell proteomic and metabolomic analysis is moving from the tissue to the cell level, which requires greater detection sensitivity and will ultimately require faster analysis times.264,265 Per sensitivity, development of mass analyzers able to measure the potential attomolar concentrations of metabolites and proteins present within individual cells are necessary.141,266,267 Ideally, these detectors are also capable of high mass resolution and mass accuracy analyses to provide increased confidence in molecular formula annotations. The FTMS-based instruments from Bruker (magnetic field-based, FTICR) and Thermo (electric field-based, Orbitrap) continue to advance in this direction.159,268 As detailed throughout this Perspective, these detectors are regularly being coupled to single and near single cell proteomic and metabolomic methods. Nevertheless, it is important to consider analysis time scales, since inferring biological meaning from single cell measurements will require measurement of hundreds to thousands of single cells.159 To this end, the advent and increased implementation of new FT-based mass analyzers capable of detection at multiples of the cyclotron frequency promise an increase FT performance (e.g., duty cycle, mass resolution, etc.) and are already being implemented into new commercial MSI instrumentation.269 Alternatively, new high speed extended path TOF mass analyzers, utilizing spectral multiplexing techniques,270 may also become more widespread.
As a reminder, orthogonal measurements beyond MS1 are required for obtaining higher confidence molecular identifications.271 Tandem MS methods (MS2, MS3, etc.) can provide more confident structural information, but obtaining more than one spectrum per pixel in a spatially resolved MS measurement remains a challenge. Premass analysis chromatographic separations, like using LC, are a key orthogonal technique in bulk omic analyses, but these methods are limited in their throughput. The past decade has seen the integration of postionization gas phase ion mobility separation MS as a method of circumventing analyte retention time differences in column chromatography, where ion separation is based differences in electrophoretic mobility in a carrier gas.272 This approach is rapid and has enabled measurement of the physical size of ions (i.e., CCS) and can be used to separate and identify structural isomers.272 Waters and Bruker have incorporated ion mobility mass spectrometry into the SYNAPT and timsTOF (trapped ion mobility separation, TIMS) for LDI analysis, respectively,273−275 whereas the integration of field asymmetric-waveform ion-mobility spectrometry (FAIMS) into Thermo systems has the potential to increase proteomics sensitivity and throughput.276 As such, it is expected to see the use of ion mobility approaches expand within single cell proteomics and metabolomics over the next decade.
Any omics scientist will attest that obtaining raw MS data can be challenging, but a significant obstacle still remains in data analysis. The first issue is data handling. Spatial MS data can be “Big Data” due to the spatial dimension beyond that of bulk MS methods. Furthermore, this data can become even larger with another dimension like ion mobility.276 A typical imaging experiment can be represented as
where FLOPS is the number of floating-point operations, k is the number of pixels, and N is the number of variables, or separate mass channels.26 For example, a TOF-MS imaging data set with 200000 pixels equates to a data set with 200000 separate spectra and 70000 mass channels in turn would mean the total number of floating-point operations to be 1 × 1016.277 Moreover, this example is with a theoretical TOF-MS data set. Data handling can be significantly more problematic with FT data that can contain millions of mass channels. Efficient peak picking algorithms and subsampling have been explored for hyperspectral data sets to address this problem,278,279 along with software developments for image analysis.280 However, data set sizes will remain a noteworthy issue in MSI as higher resolution imaging methods are developed. The universal datafile format developed for the MSI community, imzML,281 allows for data export into multiple image processing software, but it has been shown to be 3–4 times slower in write speed when compared to the HDF5 format.282 There are also several other research groups attempting to address this issue by exploring file format optimization for MS files.283,284 However, these alternative formats have yet to gain traction with the broader community. Computational power continues to improve on basic lab computer workspaces, and cloud-based imaging processing (e.g., through Amazon Web Services) are becoming increasingly utilized for processing imaging data sets,285,286 and as such, we anticipate these will be the key to handling such complicated data over the next decade.
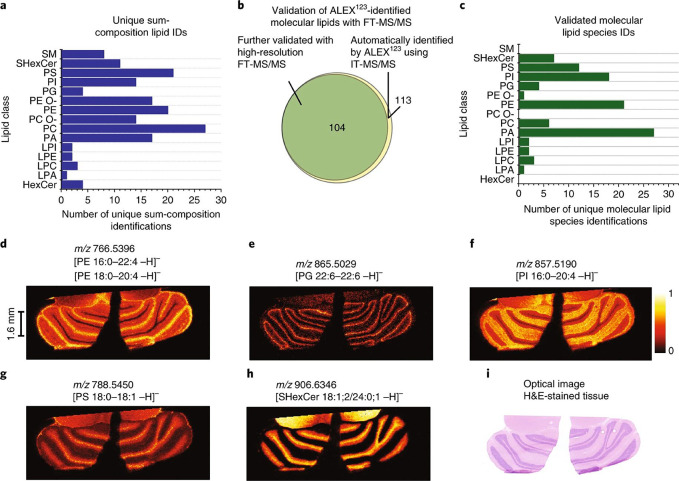
The next issue is turning raw spectra into biological meaning. Molecular assignment in MS-based proteomics and metabolomics for single cells should be performed through reference libraries to assign identities using the molecular ions and fragment species.271,287,288 There are numerous examples of databases built on spectral data from standards.289,290 However, obtaining accurate standards to build these databases for the vast number of known and unknown species is far from practical. As an alternative, in silico approaches are promising for structural identification and streamlining spatially resolved MS workflows. Incorporation of MS in silico fragmentation MS2 modeling into MS studies has been explored by multiple research groups.291−295 Also, in silico methods are being developed for annotating known and unknown molecules detected in ion mobility-based MS measurements.296,297 In the near future, we anticipate these spectral annotation tools will become fully incorporated in spatial MS and MSI workflows. One example that suggests this might be the case was recently reported by Ellis et al., where ALEX, an in silico lipid fragmentation calculator, was incorporated into their MALDI-MSI workflow.298 This automated capability allowed them to structurally assign up to 430,000 lipid parent and fragment ions (Figure 8), and MALDI-MS imaging with MS2 capability permitted rapid assignment of 104 unique lipids in rat cerebellum.298 As molecular databases continue to expand and automated in silico MS2 and CCS prediction becomes more ubiquitous, these will become key tools in single cell proteomics and metabolomics, expanding the types of inter- and intracellular biological mechanisms and biochemical pathways we can resolve.
Figure 8.
Automated parallel mass spectrometry imaging approach that provides high-confidence molecular structural identification. Here, (a) lipid classes and unique lipids were identified within the cerebellum of a rat brain. (b) Number of automatically assigned lipids using the ALEX123 framework and high-resolution FT-MS/MS in comparison to the number validated assignments from parallel MS/MS data and the (c) validated IDs across the different lipid classes. (d–h) Example single MS ion images of the different validated lipid species detected in (i) comparison to the H&E-stained tissue section. Adapted with permission from ref (298). 2018 Nature Research.
VI. Conclusion
Is sensitive single cell scale MS sampling realistic for the future, or have we set our sights too high? The recent advancements in MS and MSI over the past decade have dramatically addressed issues related to sensitivity, spatial resolution, and throughput challenges for molecular characterization. There are multiple instrument configurations available for spatial MS and MSI studies, and many of the recent developments in this area have been in effort to transition from the tissue scale to the single cell level. SIMS has historically been a valuable tool in single cell analysis, and the advancement of new cluster sources and adaptation of higher performing mass analyzers position it well for future single cell studies. Perhaps MALDI of all the spatial probing MS methods has seen the most growth over the past decade. There are now a number of commercial instruments that can achieve ∼10 μm or less lateral resolution, and the intermediate vacuum pressure it can operate under has been key to its ability to successfully employ postionization; permitting MALDI-2 the ability to detect and image a broader number of molecules and molecular classes.136 Advancements in ambient ionization sources, like nanoDESI190 and LAESI,159 have also shown promise for single cell metabolomics and may play a larger role in HTS approaches because limited sample preparation is needed to employ these methods. Untargeted proteomics utilizing MSI remains a significant challenge for single cells, yet the recent development of metal-based IHC with MSI and imaging cyTOF holds significant promise for multiplexed subcellular proteomic imaging.71,81,82 New advancements in sample handling have started to transform untargeted proteomics, like those utilized in nanoPOTS, and we are now at the precipice of being able to readily perform untargeted single cell proteomics with broad molecular coverage.94
To address the question posed at the beginning of this section: yes, single cell MS-based proteomics and metabolomics is very realistic in the near future. If the past decade of advancements in this arena detailed in this Perspective are an indicator, then we should continue to see enhancements in single cell MS probing approaches, increased sensitivity and throughput of mass analyzers, further incorporation of orthogonal methods like ion mobility, and improvements in data computation and annotations over the next decade. Moreover, we are starting to see advancements in many of these methods that are directly applicable to HTS approaches, meaning that rapid single cell screening of thousands of cells from culture or a tissue can provide greater biological understanding from resolving the cellular heterogeneity across and within phenotypes. As such, these methods will complement single cell transcriptomics and structural imaging techniques and provide a full phenome of single cells.
Acknowledgments
We thank Paul Piehowski for providing edits and useful feedback. This work was supported by the NIH-NIDDK through Grant No. 1UG3DK114920-01. Much of this research was performed using the Environmental Molecular Sciences Laboratory, a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory (PNNL). PNNL is operated for the DOE by Battelle Memorial Institute under Contract No. DE-AC05-76RLO1830.
The authors declare no competing financial interest.
References
- Zhu C.; Preissl S.; Ren B. Single-Cell Multimodal Omics: The Power of Many. Nat. Methods 2020, 17 (1), 11–14. 10.1038/s41592-019-0691-5. [DOI] [PubMed] [Google Scholar]
- Gemperline E.; Keller C.; Li L. Mass Spectrometry in Plant-Omics. Anal. Chem. 2016, 88 (7), 3422–3434. 10.1021/acs.analchem.5b02938. [DOI] [PubMed] [Google Scholar]
- Boughton B. A.; Thinagaran D.; Sarabia D.; Bacic A.; Roessner U. Mass Spectrometry Imaging for Plant Biology: A Review. Phytochem. Rev. 2016, 15 (3), 445–488. 10.1007/s11101-015-9440-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber S. A.; Rush J.; Stemman O.; Kirschner M. W.; Gygi S. P. Absolute Quantification of Proteins and Phosphoproteins from Cell Lysates by Tandem MS. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (12), 6940–6945. 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankrah N. Y. D.; May A. L.; Middleton J. L.; Jones D. R.; Hadden M. K.; Gooding J. R.; LeCleir G. R.; Wilhelm S. W.; Campagna S. R.; Buchan A. Phage Infection of an Environmentally Relevant Marine Bacterium Alters Host Metabolism and Lysate Composition. ISME J. 2014, 8 (5), 1089–1100. 10.1038/ismej.2013.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez A. J.; Fagerer S. R.; Schmidt A. M.; Urban P. L.; Jefimovs K.; Geiger P.; Dechant R.; Heinemann M.; Zenobi R. Mass Spectrometry-Based Metabolomics of Single Yeast Cells. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (22), 8790–8794. 10.1073/pnas.1209302110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svatoš A. Single-Cell Metabolomics Comes of Age: New Developments in Mass Spectrometry Profiling and Imaging. Anal. Chem. 2011, 83 (13), 5037–5044. 10.1021/ac2003592. [DOI] [PubMed] [Google Scholar]
- Amantonico A.; Urban P. L.; Zenobi R. Analytical Techniques for Single-Cell Metabolomics: State of the Art and Trends. Anal. Bioanal. Chem. 2010, 398 (6), 2493–2504. 10.1007/s00216-010-3850-1. [DOI] [PubMed] [Google Scholar]
- Dong L.; Zhang B.; Wu L.; Shang Z.; Liu S.; Jiang X.; Wang H.; Fan L.; Zhang Y.; Xiao H. Proteomics Analysis of Cellular BRS3 Receptor Activation Reveals Potential Mechanism for Signal Transduction and Cell Proliferation. J. Proteome Res. 2020, 19 (4), 1513–1521. 10.1021/acs.jproteome.9b00760. [DOI] [PubMed] [Google Scholar]
- Zhang R.; Lahens N. F.; Ballance H. I.; Hughes M. E.; Hogenesch J. B. A Circadian Gene Expression Atlas in Mammals: Implications for Biology and Medicine. Proc. Natl. Acad. Sci. U. S. A. 2014, 111 (45), 16219–16224. 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M.; Chylinski K.; Fonfara I.; Hauer M.; Doudna J. A.; Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science (Washington, DC, U. S.) 2012, 337 (6096), 816–821. 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold F. H. Design by Directed Evolution. Acc. Chem. Res. 1998, 31 (3), 125–131. 10.1021/ar960017f. [DOI] [Google Scholar]
- Muhl L.; Genové G.; Leptidis S.; Liu J.; He L.; Mocci G.; Sun Y.; Gustafsson S.; Buyandelger B.; Chivukula I. V.; Segerstolpe Å.; Raschperger E.; Hansson E. M.; Björkegren J. L. M.; Peng X.-R.; Vanlandewijck M.; Lendahl U.; Betsholtz C. Single-Cell Analysis Uncovers Fibroblast Heterogeneity and Criteria for Fibroblast and Mural Cell Identification and Discrimination. Nat. Commun. 2020, 11 (1), 3953. 10.1038/s41467-020-17740-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschuler S. J.; Wu L. F. Cellular Heterogeneity: Do Differences Make a Difference?. Cell 2010, 141 (4), 559–563. 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman S. L.; MacKay M.; Afshinnekoo E.; Melnick A. M.; Wu S.; Mason C. E. The Impact of Heterogeneity on Single-Cell Sequencing. Front. Genet. 2019, 10, 8. 10.3389/fgene.2019.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasdekis A. E.; Singh A. Microbial Metabolic Noise. Wiley Interdiscip. Rev. Syst. Biol. Med. 2020, e1512 10.1002/wsbm.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirahara K.; Poholek A.; Vahedi G.; Laurence A.; Kanno Y.; Milner J. D.; O’Shea J. J. Mechanisms Underlying Helper T-Cell Plasticity: Implications for Immune-Mediated Disease. J. Allergy Clin. Immunol. 2013, 131 (5), 1276–1287. 10.1016/j.jaci.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B.; Li H.; Peng X. Proteomics Approach to Understand Bacterial Antibiotic Resistance Strategies. Expert Rev. Proteomics 2019, 16 (10), 829–839. 10.1080/14789450.2019.1681978. [DOI] [PubMed] [Google Scholar]
- Lee H.-J.; Jedrychowski M. P.; Vinayagam A.; Wu N.; Shyh-Chang N.; Hu Y.; Min-Wen C.; Moore J. K.; Asara J. M.; Lyssiotis C. A.; Perrimon N.; Gygi S. P.; Cantley L. C.; Kirschner M. W. Proteomic and Metabolomic Characterization of a Mammalian Cellular Transition from Quiescence to Proliferation. Cell Rep. 2017, 20 (3), 721–736. 10.1016/j.celrep.2017.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Lin X. Morphogenesis in Fungal Pathogenicity: Shape, Size, and Surface. PLoS Pathog. 2012, 8 (12), e1003027. 10.1371/journal.ppat.1003027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin P. A.; Angert E. R. Small but Mighty: Cell Size and Bacteria. Cold Spring Harbor Perspect. Biol. 2015, 7 (7), a019216 10.1101/cshperspect.a019216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera V.; Hsu S.-C. J.; Rahim M. K.; Chen C.; Nguyen L.; Liu W. F.; Haun J. B. Pushing the Limits of Detection for Proteins Secreted from Single Cells Using Quantum Dots. Analyst 2019, 144 (3), 980–989. 10.1039/C8AN01083H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler S. A.; Rottner K.; Lai F.; Block J.; Vinzenz M.; Small J. V. F- and G-Actin Concentrations in Lamellipodia of Moving Cells. PLoS One 2009, 4 (3), e4810 10.1371/journal.pone.0004810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong Q.; Koch K.; Yoon J. M.; Everard J. D.; Shanks J. V. Influence of Carbon to Nitrogen Ratios on Soybean Somatic Embryo (Cv. Jack) Growth and Composition. J. Exp. Bot. 2013, 64 (10), 2985–2995. 10.1093/jxb/ert138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther S.; Koestler M.; Schulz O.; Spengler B. Laser Spot Size and Laser Power Dependence of Ion Formation in High Resolution MALDI Imaging. Int. J. Mass Spectrom. 2010, 294 (1), 7–15. 10.1016/j.ijms.2010.03.014. [DOI] [Google Scholar]
- Lippens J. L.; Nshanian M.; Spahr C.; Egea P. F.; Loo J. A.; Campuzano I. D. G. Fourier Transform-Ion Cyclotron Resonance Mass Spectrometry as a Platform for Characterizing Multimeric Membrane Protein Complexes. J. Am. Soc. Mass Spectrom. 2018, 29 (1), 183–193. 10.1007/s13361-017-1799-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanka D.-T.; Konzer A.; Edelmann D.; Berghoff B. A. High-Throughput Proteomics Identifies Proteins With Importance to Postantibiotic Recovery in Depolarized Persister Cells. Front. Microbiol. 2019, 10 (MAR), 378. 10.3389/fmicb.2019.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A.; Abouleila Y.; Shimizu Y.; Hiyama E.; Watanabe T. M.; Yanagida T.; Germond A. Single-Cell Screening of Tamoxifen Abundance and Effect Using Mass Spectrometry and Raman-Spectroscopy. Anal. Chem. 2019, 91 (4), 2710–2718. 10.1021/acs.analchem.8b04393. [DOI] [PubMed] [Google Scholar]
- Gilmore I. S.; Heiles S.; Pieterse C. L. Metabolic Imaging at the Single-Cell Scale: Recent Advances in Mass Spectrometry Imaging. Annu. Rev. Anal. Chem. 2019, 12 (1), 201–224. 10.1146/annurev-anchem-061318-115516. [DOI] [PubMed] [Google Scholar]
- Porta Siegel T.; Hamm G.; Bunch J.; Cappell J.; Fletcher J. S.; Schwamborn K. Mass Spectrometry Imaging and Integration with Other Imaging Modalities for Greater Molecular Understanding of Biological Tissues. Mol. Imaging Biol. 2018, 20 (6), 888–901. 10.1007/s11307-018-1267-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan N. T. N.; Munem M.; Ewing A. G.; Fletcher J. S. MS/MS Analysis and Imaging of Lipids across Drosophila Brain Using Secondary Ion Mass Spectrometry. Anal. Bioanal. Chem. 2017, 409 (16), 3923–3932. 10.1007/s00216-017-0336-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarelli M. K.; Winograd N. Lipid Imaging with Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS). Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2011, 1811 (11), 976–990. 10.1016/j.bbalip.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama Y.; Aoyagi S.; Fujii M.; Matsuo J.; Fletcher J. S.; Lockyer N. P.; Vickerman J. C.; Passarelli M. K.; Havelund R.; Seah M. P. Peptide Fragmentation and Surface Structural Analysis by Means of ToF-SIMS Using Large Cluster Ion Sources. Anal. Chem. 2016, 88 (7), 3592–3597. 10.1021/acs.analchem.5b04133. [DOI] [PubMed] [Google Scholar]
- Angerer T. B.; Velickovic D.; Nicora C. D.; Kyle J. E.; Graham D. J.; Anderton C.; Gamble L. J. Exploiting the Semidestructive Nature of Gas Cluster Ion Beam Time-of-Flight Secondary Ion Mass Spectrometry Imaging for Simultaneous Localization and Confident Lipid Annotations. Anal. Chem. 2019, 91 (23), 15073–15080. 10.1021/acs.analchem.9b03763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K. D.; Fyrestam J.; Lanekoff I. Advances in Mass Spectrometry Based Single-Cell Metabolomics. Analyst 2019, 144 (3), 782–793. 10.1039/C8AN01581C. [DOI] [PubMed] [Google Scholar]
- Lundberg E.; Borner G. H. H. Spatial Proteomics: A Powerful Discovery Tool for Cell Biology. Nat. Rev. Mol. Cell Biol. 2019, 20 (5), 285–302. 10.1038/s41580-018-0094-y. [DOI] [PubMed] [Google Scholar]
- Dou M.; Clair G.; Tsai C.-F.; Xu K.; Chrisler W. B.; Sontag R. L.; Zhao R.; Moore R. J.; Liu T.; Pasa-Tolic L.; Smith R. D.; Shi T.; Adkins J. N.; Qian W.-J.; Kelly R. T.; Ansong C.; Zhu Y. High-Throughput Single Cell Proteomics Enabled by Multiplex Isobaric Labeling in a Nanodroplet Sample Preparation Platform. Anal. Chem. 2019, 91 (20), 13119–13127. 10.1021/acs.analchem.9b03349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. M.; Liyu A. V.; Tsai C.-F.; Moore R. J.; Orton D. J.; Chrisler W. B.; Gaffrey M. J.; Liu T.; Smith R. D.; Kelly R. T.; Pasa-Tolic L.; Zhu Y. Automated Coupling of Nanodroplet Sample Preparation with Liquid Chromatography-Mass Spectrometry for High-Throughput Single-Cell Proteomics. Anal. Chem. 2020, 92, 10588–10596. 10.1021/acs.analchem.0c01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonville J. M.; Carter C. L.; Pizarro L.; Steven R. T.; Palmer A. D.; Griffiths R. L.; Lalor P. F.; Lindon J. C.; Nicholson J. K.; Holmes E.; Bunch J. Hyperspectral Visualization of Mass Spectrometry Imaging Data. Anal. Chem. 2013, 85 (3), 1415–1423. 10.1021/ac302330a. [DOI] [PubMed] [Google Scholar]
- Baker T. C.; Han J.; Borchers C. H. Recent Advancements in Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging. Curr. Opin. Biotechnol. 2017, 43, 62–69. 10.1016/j.copbio.2016.09.003. [DOI] [PubMed] [Google Scholar]
- Hansen R. L.; Lee Y. J. High-Spatial Resolution Mass Spectrometry Imaging: Toward Single Cell Metabolomics in Plant Tissues. Chem. Rec. 2018, 18 (1), 65–77. 10.1002/tcr.201700027. [DOI] [PubMed] [Google Scholar]
- Sturtevant D.; Lee Y.-J.; Chapman K. D. Matrix Assisted Laser Desorption/Ionization-Mass Spectrometry Imaging (MALDI-MSI) for Direct Visualization of Plant Metabolites in Situ. Curr. Opin. Biotechnol. 2016, 37, 53–60. 10.1016/j.copbio.2015.10.004. [DOI] [PubMed] [Google Scholar]
- Palmer A.; Trede D.; Alexandrov T. Where Imaging Mass Spectrometry Stands: Here Are the Numbers. Metabolomics 2016, 12 (6), 1–3. 10.1007/s11306-016-1047-0. [DOI] [Google Scholar]
- Labib M.; Kelley S. O. Single-Cell Analysis Targeting the Proteome. Nat. Rev. Chem. 2020, 4 (3), 143–158. 10.1038/s41570-020-0162-7. [DOI] [PubMed] [Google Scholar]
- Levy E.; Slavov N. Single Cell Protein Analysis for Systems Biology. Essays Biochem. 2018, 62 (4), 595–605. 10.1042/EBC20180014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell L. A.; Corthals G. L.; Willems S. M.; van Remoortere A.; van Zeijl R. J. M.; Deelder A. M. Peptide and Protein Imaging Mass Spectrometry in Cancer Research. J. Proteomics 2010, 73 (10), 1921–1944. 10.1016/j.jprot.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Yin L.; Zhang Z.; Liu Y.; Gao Y.; Gu J. Recent Advances in Single-Cell Analysis by Mass Spectrometry. Analyst 2019, 144 (3), 824–845. 10.1039/C8AN01190G. [DOI] [PubMed] [Google Scholar]
- Hu Y.; An Q.; Sheu K.; Trejo B.; Fan S.; Guo Y. Single Cell Multi-Omics Technology: Methodology and Application. Front. Cell Dev. Biol. 2018, 6 (APR), 28. 10.3389/fcell.2018.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzberg M. B.; Kafri R.; Kirschner M. On Being the Right (Cell) Size. Science (Washington, DC, U. S.) 2015, 348 (6236), 1245075–1245075. 10.1126/science.1245075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov T. Spatial Metabolomics and Imaging Mass Spectrometry in the Age of Artificial Intelligence. Annu. Rev. Biomed. Data Sci. 2020, 3 (1), 61–87. 10.1146/annurev-biodatasci-011420-031537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann E. K.; Comi T. J.; Rubakhin S. S.; Sweedler J. V. Lipid Heterogeneity between Astrocytes and Neurons Revealed by Single-Cell MALDI-MS Combined with Immunocytochemical Classification. Angew. Chem., Int. Ed. 2019, 58 (18), 5910–5914. 10.1002/anie.201812892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do T. D.; Comi T. J.; Dunham S. J. B.; Rubakhin S. S.; Sweedler J. V. Single Cell Profiling Using Ionic Liquid Matrix-Enhanced Secondary Ion Mass Spectrometry for Neuronal Cell Type Differentiation. Anal. Chem. 2017, 89 (5), 3078–3086. 10.1021/acs.analchem.6b04819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson E. T.; Comi T. J.; Rubakhin S. S.; Sweedler J. V. Single Cell Peptide Heterogeneity of Rat Islets of Langerhans. ACS Chem. Biol. 2016, 11 (9), 2588–2595. 10.1021/acschembio.6b00602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman A. P.; Bogie J. F. J.; Hendriks J. J. A.; Haidar M.; Belov M.; Heeren R. M. A.; Ellis S. R. Evaluation of Lipid Coverage and High Spatial Resolution MALDI-Imaging Capabilities of Oversampling Combined with Laser Post-Ionisation. Anal. Bioanal. Chem. 2020, 412 (10), 2277–2289. 10.1007/s00216-019-02290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueñas M. E.; Larson E. A.; Lee Y. J. Toward Mass Spectrometry Imaging in the Metabolomics Scale: Increasing Metabolic Coverage Through Multiple On-Tissue Chemical Modifications. Front. Plant Sci. 2019, 10, 860. 10.3389/fpls.2019.00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K. H.; Zwieniecki M. A. Physical Limits to Leaf Size in Tall Trees. Phys. Rev. Lett. 2013, 110 (1), 018104. 10.1103/PhysRevLett.110.018104. [DOI] [PubMed] [Google Scholar]
- Bacete L.; Mélida H.; Miedes E.; Molina A. Plant Cell Wall-Mediated Immunity: Cell Wall Changes Trigger Disease Resistance Responses. Plant J. 2018, 93 (4), 614–636. 10.1111/tpj.13807. [DOI] [PubMed] [Google Scholar]
- Dong Y.; Li B.; Malitsky S.; Rogachev I.; Aharoni A.; Kaftan F.; Svatoš A.; Franceschi P. Sample Preparation for Mass Spectrometry Imaging of Plant Tissues: A Review. Front. Plant Sci. 2016, 7, 60. 10.3389/fpls.2016.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte A. R.; Yandeau-Nelson M. D.; Nikolau B. J.; Lee Y. J. Subcellular-Level Resolution MALDI-MS Imaging of Maize Leaf Metabolites by MALDI-Linear Ion Trap-Orbitrap Mass Spectrometer. Anal. Bioanal. Chem. 2015, 407 (8), 2301–2309. 10.1007/s00216-015-8460-5. [DOI] [PubMed] [Google Scholar]
- Stopka S. A.; Khattar R.; Agtuca B. J.; Anderton C. R.; Paša-Tolić L.; Stacey G.; Vertes A. Metabolic Noise and Distinct Subpopulations Observed by Single Cell LAESI Mass Spectrometry of Plant Cells in Situ. Front. Plant Sci. 2018, 9, 1646. 10.3389/fpls.2018.01646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez C. J.; Bagga A. K.; Prova S. S.; Yousefi Taemeh M.; Ifa D. R. Review and Perspectives on the Applications of Mass Spectrometry Imaging under Ambient Conditions. Rapid Commun. Mass Spectrom. 2019, 33 (S3), 27–53. 10.1002/rcm.8145. [DOI] [PubMed] [Google Scholar]
- Pozebon D.; Scheffler G. L.; Dressler V. L. Recent Applications of Laser Ablation Inductively Coupled Plasma Mass Spectrometry (LA-ICP-MS) for Biological Sample Analysis: A Follow-up Review. J. Anal. At. Spectrom. 2017, 32 (5), 890–919. 10.1039/C7JA00026J. [DOI] [Google Scholar]
- Saleem M.; Hu J.; Jousset A. More Than the Sum of Its Parts: Microbiome Biodiversity as a Driver of Plant Growth and Soil Health. Annu. Rev. Ecol. Evol. Syst. 2019, 50 (1), 145–168. 10.1146/annurev-ecolsys-110617-062605. [DOI] [Google Scholar]
- Herrmann A. M.; Ritz K.; Nunan N.; Clode P. L.; Pett-Ridge J.; Kilburn M. R.; Murphy D. V.; O’Donnell A. G.; Stockdale E. A. Nano-Scale Secondary Ion Mass Spectrometry — A New Analytical Tool in Biogeochemistry and Soil Ecology: A Review Article. Soil Biol. Biochem. 2007, 39 (8), 1835–1850. 10.1016/j.soilbio.2007.03.011. [DOI] [Google Scholar]
- Dunham S. J. B.; Ellis J. F.; Li B.; Sweedler J. V. Mass Spectrometry Imaging of Complex Microbial Communities. Acc. Chem. Res. 2017, 50 (1), 96–104. 10.1021/acs.accounts.6b00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand M.; Lerch S. J. L.; Shrestha R. P. Understanding Diatom Cell Wall Silicification—Moving Forward. Front. Mar. Sci. 2018, 5 (APR), 125. 10.3389/fmars.2018.00125. [DOI] [Google Scholar]
- Leefmann T.; Heim C.; Kryvenda A.; Siljeström S.; Sjövall P.; Thiel V. Biomarker Imaging of Single Diatom Cells in a Microbial Mat Using Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS). Org. Geochem. 2013, 57, 23–33. 10.1016/j.orggeochem.2013.01.005. [DOI] [Google Scholar]
- Schoffelen N. J.; Mohr W.; Ferdelman T. G.; Littmann S.; Duerschlag J.; Zubkov M. V.; Ploug H.; Kuypers M. M. M. Single-Cell Imaging of Phosphorus Uptake Shows That Key Harmful Algae Rely on Different Phosphorus Sources for Growth. Sci. Rep. 2018, 8 (1), 17182. 10.1038/s41598-018-35310-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musat N.; Musat F.; Weber P. K.; Pett-Ridge J. Tracking Microbial Interactions with NanoSIMS. Curr. Opin. Biotechnol. 2016, 41, 114–121. 10.1016/j.copbio.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Zimmermann M.; Escrig S.; Hübschmann T.; Kirf M. K.; Brand A.; Inglis R. F.; Musat N.; Müller S.; Meibom A.; Ackermann M.; Schreiber F. Phenotypic Heterogeneity in Metabolic Traits among Single Cells of a Rare Bacterial Species in Its Natural Environment Quantified with a Combination of Flow Cell Sorting and NanoSIMS. Front. Microbiol. 2015, 06 (MAR), 243. 10.3389/fmicb.2015.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren L.; Bosse M.; Thompson S.; Risom T.; Vijayaragavan K.; McCaffrey E.; Marquez D.; Angoshtari R.; Greenwald N. F.; Fienberg H.; Wang J.; Kambham N.; Kirkwood D.; Nolan G.; Montine T. J.; Galli S. J.; West R.; Bendall S. C.; Angelo M. MIBI-TOF: A Multiplexed Imaging Platform Relates Cellular Phenotypes and Tissue Structure. Sci. Adv. 2019, 5 (10), eaax5851. 10.1126/sciadv.aax5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achim K.; Pettit J.-B.; Saraiva L. R.; Gavriouchkina D.; Larsson T.; Arendt D.; Marioni J. C. High-Throughput Spatial Mapping of Single-Cell RNA-Seq Data to Tissue of Origin. Nat. Biotechnol. 2015, 33 (5), 503–509. 10.1038/nbt.3209. [DOI] [PubMed] [Google Scholar]
- Shapiro E.; Biezuner T.; Linnarsson S. Single-Cell Sequencing-Based Technologies Will Revolutionize Whole-Organism Science. Nat. Rev. Genet. 2013, 14 (9), 618–630. 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Beyer A.; Aebersold R. On the Dependency of Cellular Protein Levels on MRNA Abundance. Cell 2016, 165 (3), 535–550. 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- Zhu Y.; Piehowski P. D.; Zhao R.; Chen J.; Shen Y.; Moore R. J.; Shukla A. K.; Petyuk V. A.; Campbell-Thompson M.; Mathews C. E.; Smith R. D.; Qian W.-J.; Kelly R. T. Nanodroplet Processing Platform for Deep and Quantitative Proteome Profiling of 10–100 Mammalian Cells. Nat. Commun. 2018, 9 (1), 882. 10.1038/s41467-018-03367-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L.; Tsai C.-F.; Dirice E.; Swensen A. C.; Chen J.; Shi T.; Gritsenko M. A.; Chu R. K.; Piehowski P. D.; Smith R. D.; Rodland K. D.; Atkinson M. A.; Mathews C. E.; Kulkarni R. N.; Liu T.; Qian W.-J. Boosting to Amplify Signal with Isobaric Labeling (BASIL) Strategy for Comprehensive Quantitative Phosphoproteomic Characterization of Small Populations of Cells. Anal. Chem. 2019, 91 (9), 5794–5801. 10.1021/acs.analchem.9b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson R. M.; Borowsky A. D.; Angelo M. Immunohistochemistry and Mass Spectrometry for Highly Multiplexed Cellular Molecular Imaging. Lab. Invest. 2015, 95 (4), 397–405. 10.1038/labinvest.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M.; Bandrowski A.; Carr S.; Edwards A.; Ellenberg J.; Lundberg E.; Rimm D. L.; Rodriguez H.; Hiltke T.; Snyder M.; Yamamoto T. A Proposal for Validation of Antibodies. Nat. Methods 2016, 13 (10), 823–827. 10.1038/nmeth.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. Antibody Anarchy: A Call to Order. Nature 2015, 527 (7579), 545–551. 10.1038/527545a. [DOI] [PubMed] [Google Scholar]
- Bendall S. C.; Simonds E. F.; Qiu P.; Amir E. A. D.; Krutzik P. O.; Finck R.; Bruggner R. V.; Melamed R.; Trejo A.; Ornatsky O. I.; Balderas R. S.; Plevritis S. K.; Sachs K.; Pe’er D.; Tanner S. D.; Nolan G. P. Single-Cell Mass Cytometry of Differential Immune and Drug Responses across a Human Hematopoietic Continuum. Science (Washington, DC, U. S.) 2011, 332 (6030), 687–696. 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker T.; Buckle T.; Van Malderen S. J. M.; van Willigen D. M.; van Unen V.; van Leeuwen F. W. B.; Vanhaecke F. High-Resolution Imaging and Single-Cell Analysis via Laser Ablation-Inductively Coupled Plasma-Mass Spectrometry for the Determination of Membranous Receptor Expression Levels in Breast Cancer Cell Lines Using Receptor-Specific Hybrid Tracers. Anal. Chim. Acta 2019, 1074, 43–53. 10.1016/j.aca.2019.04.064. [DOI] [PubMed] [Google Scholar]
- Angelo M.; Bendall S. C.; Finck R.; Hale M. B.; Hitzman C.; Borowsky A. D.; Levenson R. M.; Lowe J. B.; Liu S. D.; Zhao S.; Natkunam Y.; Nolan G. P. Multiplexed Ion Beam Imaging of Human Breast Tumors. Nat. Med. 2014, 20 (4), 436–442. 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scurrah C. R.; Simmons A. J.; Lau K. S.. Single-Cell Mass Cytometry of Archived Human Epithelial Tissue for Decoding Cancer Signaling Pathways. In Methods Mol. Biol.; Humana Press, Inc., 2019; Vol. 1884, pp 215–229. 10.1007/978-1-4939-8885-3_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren L.; Bosse M.; Marquez D.; Angoshtari R.; Jain S.; Varma S.; Yang S.-R.; Kurian A.; Van Valen D.; West R.; Bendall S. C.; Angelo M. A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 2018, 174 (6), 1373–1387. 10.1016/j.cell.2018.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. J.; Traum D.; Schug J.; Gao L.; Liu C.; Atkinson M. A.; Powers A. C.; Feldman M. D.; Naji A.; Chang K. M.; Kaestner K. H. Multiplexed In Situ Imaging Mass Cytometry Analysis of the Human Endocrine Pancreas and Immune System in Type 1 Diabetes. Cell Metab. 2019, 29 (3), 769–783. 10.1016/j.cmet.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Garden R. W.; Sweedler J. V. Single-Cell MALDI: A New Tool for Direct Peptide Profiling. Trends Biotechnol. 2000, 18 (4), 151–160. 10.1016/S0167-7799(00)01427-X. [DOI] [PubMed] [Google Scholar]
- Yang N.; Irving S. J.; Romanova E. V.; Mitchell J. W.; Gillette M. U.; Sweedler J. V. Neuropeptidomics. In Molecular Neuroendocrinology 2016, 155–169. 10.1002/9781118760369.ch8. [DOI] [Google Scholar]
- Clark A. E.; Kaleta E. J.; Arora A.; Wolk D. M. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry: A Fundamental Shift in the Routine Practice of Clinical Microbiology. Clin. Microbiol. Rev. 2013, 26 (3), 547–603. 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummon A. B.; Amare A.; Sweedler J. V. Discovering New Invertebrate Neuropeptides Using Mass Spectrometry. Mass Spectrom. Rev. 2006, 25 (1), 77–98. 10.1002/mas.20055. [DOI] [PubMed] [Google Scholar]
- Neupert S.; Predel R. Mass Spectrometric Analysis of Single Identified Neurons of an Insect. Biochem. Biophys. Res. Commun. 2005, 327 (3), 640–645. 10.1016/j.bbrc.2004.12.086. [DOI] [PubMed] [Google Scholar]
- Spraggins J. M.; Rizzo D. G.; Moore J. L.; Noto M. J.; Skaar E. P.; Caprioli R. M. Next-Generation Technologies for Spatial Proteomics: Integrating Ultra-High Speed MALDI-TOF and High Mass Resolution MALDI FTICR Imaging Mass Spectrometry for Protein Analysis. Proteomics 2016, 16 (11–12), 1678–1689. 10.1002/pmic.201600003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss A.; Smith D. F.; Reschke B. R.; Powell M. J.; Heeren R. M. A. Top-down Mass Spectrometry Imaging of Intact Proteins by Laser Ablation ESI FT-ICR MS. Proteomics 2014, 14 (10), 1283–1289. 10.1002/pmic.201300306. [DOI] [PubMed] [Google Scholar]
- Gutstein H. B.; Morris J. S. Laser Capture Sampling and Analytical Issues in Proteomics. Expert Rev. Proteomics 2007, 4 (5), 627–637. 10.1586/14789450.4.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R.; Zhu Y.; Liang Y.; Cong Y.; Piehowski P.; Dou M.; Zhao R.; Qian W.-J.; Burnum-Johnson K.; Ansong C. Single Cell Proteome Mapping of Tissue Heterogeneity Using Microfluidic Nanodroplet Sample Processing and Ultrasensitive LC–MS. J. Biomol. Technol. 2019, 30 (Suppl), S61. [Google Scholar]
- Budnik B.; Levy E.; Harmange G.; Slavov N. SCoPE-MS: Mass Spectrometry of Single Mammalian Cells Quantifies Proteome Heterogeneity during Cell Differentiation. Genome Biol. 2018, 19 (1), 161. 10.1186/s13059-018-1547-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.; Bertke M. M.; Champion M. M.; Zhu G.; Huber P. W.; Dovichi N. J. Quantitative Proteomics of Xenopus Laevis Embryos: Expression Kinetics of Nearly 4000 Proteins during Early Development. Sci. Rep. 2015, 4, 4365. 10.1038/srep04365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.; Dubiak K. M.; Peuchen E. H.; Zhang Z.; Zhu G.; Huber P. W.; Dovichi N. J. Single Cell Proteomics Using Frog (Xenopus Laevis) Blastomeres Isolated from Early Stage Embryos, Which Form a Geometric Progression in Protein Content. Anal. Chem. 2016, 88 (13), 6653–6657. 10.1021/acs.analchem.6b01921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard-Banek C.; Moody S. A.; Manzini M. C.; Nemes P. Microsampling Capillary Electrophoresis Mass Spectrometry Enables Single-Cell Proteomics in Complex Tissues: Developing Cell Clones in Live Xenopus Laevis and Zebrafish Embryos. Anal. Chem. 2019, 91 (7), 4797–4805. 10.1021/acs.analchem.9b00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S.; Tzur D.; Knox C.; Eisner R.; Guo A. C.; Young N.; Cheng D.; Jewell K.; Arndt D.; Sawhney S.; Fung C.; Nikolai L.; Lewis M.; Coutouly M.-A.; Forsythe I.; Tang P.; Shrivastava S.; Jeroncic K.; Stothard P.; Amegbey G.; Block D.; Hau D. D.; Wagner J.; Miniaci J.; Clements M.; Gebremedhin M.; Guo N.; Zhang Y.; Duggan G. E.; MacInnis G. D.; Weljie A. M.; Dowlatabadi R.; Bamforth F.; Clive D.; Greiner R.; Li L.; Marrie T.; Sykes B. D.; Vogel H. J.; Querengesser L. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka S. A.; Agtuca B. J.; Koppenaal D. W.; Paša-Tolić L.; Stacey G.; Vertes A.; Anderton C. R. Laser-Ablation Electrospray Ionization Mass Spectrometry with Ion Mobility Separation Reveals Metabolites in the Symbiotic Interactions of Soybean Roots and Rhizobia. Plant J. 2017, 91 (2), 340–354. 10.1111/tpj.13569. [DOI] [PubMed] [Google Scholar]
- van Meer G.; Voelker D. R.; Feigenson G. W. Membrane Lipids: Where They Are and How They Behave. Nat. Rev. Mol. Cell Biol. 2008, 9 (2), 112–124. 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez-Lopez D.; Laurens F.; Quéméner B.; Lahaye M. Variability of Cell Wall Polysaccharides Composition and Hemicellulose Enzymatic Profile in an Apple Progeny. Int. J. Biol. Macromol. 2011, 49 (5), 1104–1109. 10.1016/j.ijbiomac.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Van Norman J. M.Asymmetry and Cell Polarity in Root Development. Dev. Biol.; Academic Press, Inc., 2016; pp 165–174. 10.1016/j.ydbio.2016.07.009. [DOI] [PubMed] [Google Scholar]
- Veličković D.; Agtuca B. J.; Stopka S. A.; Vertes A.; Koppenaal D. W.; Paša-Tolić L.; Stacey G.; Anderton C. R. Observed Metabolic Asymmetry within Soybean Root Nodules Reflects Unexpected Complexity in Rhizobacteria-Legume Metabolite Exchange. ISME J. 2018, 12 (9), 2335–2338. 10.1038/s41396-018-0188-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y.-N.; Shu L.-J.; Yang Y.-L. Imaging Mass Spectrometry for Metabolites: Technical Progress, Multimodal Imaging, and Biological Interactions. Wiley Interdiscip. Rev. Syst. Biol. Med. 2017, 9 (5), e1387 10.1002/wsbm.1387. [DOI] [PubMed] [Google Scholar]
- Anderton C. R.; Chu R. K.; Tolić N.; Creissen A.; Paša-Tolić L. Utilizing a Robotic Sprayer for High Lateral and Mass Resolution MALDI FT-ICR MSI of Microbial Cultures. J. Am. Soc. Mass Spectrom. 2016, 27 (3), 556–559. 10.1007/s13361-015-1324-6. [DOI] [PubMed] [Google Scholar]
- Boya P. C. A.; Fernández-Marín H.; Mejiá L. C.; Spadafora C.; Dorrestein P. C.; Gutiérrez M. Imaging Mass Spectrometry and MS/MS Molecular Networking Reveals Chemical Interactions among Cuticular Bacteria and Pathogenic Fungi Associated with Fungus-Growing Ants. Sci. Rep. 2017, 7 (1), 1–13. 10.1038/s41598-017-05515-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P. C. A.; Christian M. H.; Fernandez-Marin H.; Gutierrez M. Fungus-Growing Ant’s Microbial Interaction of Streptomyces Sp. and Escovopsis Sp. through Molecular Networking and MALDI Imaging. Nat. Prod. Commun. 2019, 14 (1), 63–66. 10.1177/1934578X1901400117. [DOI] [Google Scholar]
- Sica V. P.; Raja H. A.; El-Elimat T.; Oberlies N. H. Mass Spectrometry Imaging of Secondary Metabolites Directly on Fungal Cultures. RSC Adv. 2014, 4 (108), 63221–63227. 10.1039/C4RA11564C. [DOI] [Google Scholar]
- Ly A.; Ragionieri L.; Liessem S.; Becker M.; Deininger S. O.; Neupert S.; Predel R. Enhanced Coverage of Insect Neuropeptides in Tissue Sections by an Optimized Mass-Spectrometry-Imaging Protocol. Anal. Chem. 2019, 91 (3), 1980–1988. 10.1021/acs.analchem.8b04304. [DOI] [PubMed] [Google Scholar]
- Yang F.; Chen J.; Ruan Q.; Saqib H. S. A.; He W.; You M. Mass Spectrometry Imaging: An Emerging Technology for the Analysis of Metabolites in Insects. Arch. Insect Biochem. Physiol. 2020, 103 (4), e21643. 10.1002/arch.21643. [DOI] [PubMed] [Google Scholar]
- Gorzolka K.; Kölling J.; Nattkemper T. W.; Niehaus K. Spatio-Temporal Metabolite Profiling of the Barley Germination Process by MALDI MS Imaging. PLoS One 2016, 11 (3), e0150208 10.1371/journal.pone.0150208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi R.; Hashimoto K.; Toyooka K.; Saito K. Keeping the Shape of Plant Tissue for Visualizing Metabolite Features in Segmentation and Correlation Analysis of Imaging Mass Spectrometry in Asparagus Officinalis. Metabolomics 2019, 15 (2), 24. 10.1007/s11306-019-1486-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C.; Xie P.; Yong T.; Wang H.; Chung A. C. K.; Cai Z. MALDI-MS Imaging Reveals Asymmetric Spatial Distribution of Lipid Metabolites from Bisphenol S-Induced Nephrotoxicity. Anal. Chem. 2018, 90 (5), 3196–3204. 10.1021/acs.analchem.7b04540. [DOI] [PubMed] [Google Scholar]
- Dilillo M.; Ait-Belkacem R.; Esteve C.; Pellegrini D.; Nicolardi S.; Costa M.; Vannini E.; De Graaf E. L.; Caleo M.; McDonnell L. A. Ultra-High Mass Resolution MALDI Imaging Mass Spectrometry of Proteins and Metabolites in a Mouse Model of Glioblastoma. Sci. Rep. 2017, 7 (1), 1–11. 10.1038/s41598-017-00703-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehaus M.; Soltwisch J. New Insights into Mechanisms of Material Ejection in MALDI Mass Spectrometry for a Wide Range of Spot Sizes. Sci. Rep. 2018, 8 (1), 7755. 10.1038/s41598-018-25946-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurchen J. C.; Rubakhin S. S.; Sweedler J. V. MALDI-MS Imaging of Features Smaller than the Size of the Laser Beam. J. Am. Soc. Mass Spectrom. 2005, 16 (10), 1654–1659. 10.1016/j.jasms.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Wiegelmann M.; Dreisewerd K.; Soltwisch J. Influence of the Laser Spot Size, Focal Beam Profile, and Tissue Type on the Lipid Signals Obtained by MALDI-MS Imaging in Oversampling Mode. J. Am. Soc. Mass Spectrom. 2016, 27 (12), 1952–1964. 10.1007/s13361-016-1477-y. [DOI] [PubMed] [Google Scholar]
- LaBonia G. J.; Lockwood S. Y.; Heller A. A.; Spence D. M.; Hummon A. B. Drug Penetration and Metabolism in 3D Cell Cultures Treated in a 3D Printed Fluidic Device: Assessment of Irinotecan via MALDI Imaging Mass Spectrometry. Proteomics 2016, 16 (11–12), 1814–1821. 10.1002/pmic.201500524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappez L.; Stadler M.; Triana Sierra S. H.; Phapale P.; Heikenwalder M.; Alexandrov T. Spatial Single-Cell Profiling of Intracellular Metabolomes in Situ: Supplementary Data S1. bioRxiv 2019, 510222. 10.1101/510222. [DOI] [Google Scholar]
- Papagiannopoulou C.; Parchen R.; Rubbens P.; Waegeman W. Fast Pathogen Identification Using Single-Cell Matrix-Assisted Laser Desorption/Ionization-Aerosol Time-of-Flight Mass Spectrometry Data and Deep Learning Methods. Anal. Chem. 2020, 92 (11), 7523–7531. 10.1021/acs.analchem.9b05806. [DOI] [PubMed] [Google Scholar]
- Yang T.; Gao D.; Jin F.; Jiang Y.; Liu H. Surface-Printed Microdot Array Chips Coupled with Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry for High-Throughput Single-Cell Patterning and Phospholipid Analysis. Rapid Commun. Mass Spectrom. 2016, 30, 73–79. 10.1002/rcm.7628. [DOI] [PubMed] [Google Scholar]
- Krismer J.; Sobek J.; Steinhoff R. F.; Fagerer S. R.; Pabst M.; Zenobi R. Screening of Chlamydomonas Reinhardtii Populations with Single-Cell Resolution by Using a High-Throughput Microscale Sample Preparation for Matrix-Assisted Laser Desorption Ionization Mass Spectrometry. Appl. Environ. Microbiol. 2015, 81 (16), 5546–5551. 10.1128/AEM.01201-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis S. R.; Ferris C. J.; Gilmore K. J.; Mitchell T. W.; Blanksby S. J.; In het Panhuis M. Direct Lipid Profiling of Single Cells from Inkjet Printed Microarrays. Anal. Chem. 2012, 84 (22), 9679–9683. 10.1021/ac302634u. [DOI] [PubMed] [Google Scholar]
- Zavalin A.; Yang J.; Caprioli R. Laser Beam Filtration for High Spatial Resolution MALDI Imaging Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2013, 24 (7), 1153–1156. 10.1007/s13361-013-0638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feenstra A. D.; Dueñas M. E.; Lee Y. J. Five Micron High Resolution MALDI Mass Spectrometry Imaging with Simple, Interchangeable, Multi-Resolution Optical System. J. Am. Soc. Mass Spectrom. 2017, 28 (3), 434–442. 10.1007/s13361-016-1577-8. [DOI] [PubMed] [Google Scholar]
- Ogrinc Potočnik N.; Porta T.; Becker M.; Heeren R. M. A.; Ellis S. R. Use of Advantageous, Volatile Matrices Enabled by next-Generation High-Speed Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Imaging Employing a Scanning Laser Beam. Rapid Commun. Mass Spectrom. 2015, 29 (23), 2195–2203. 10.1002/rcm.7379. [DOI] [PubMed] [Google Scholar]
- Spraggins J. M.; Caprioli R. M. High-Speed MALDI-TOF Imaging Mass Spectrometry: Rapid Ion Image Acquisition and Considerations for Next Generation Instrumentation. J. Am. Soc. Mass Spectrom. 2011, 22 (6), 1022–1031. 10.1007/s13361-011-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler B.; Hubert M. Scanning Microprobe Matrix-Assisted Laser Desorption Ionization (SMALDI) Mass Spectrometry: Instrumentation for Sub-Micrometer Resolved LDI and MALDI Surface Analysis. J. Am. Soc. Mass Spectrom. 2002, 13 (6), 735–748. 10.1016/S1044-0305(02)00376-8. [DOI] [PubMed] [Google Scholar]
- Khalil S. M.; Römpp A.; Pretzel J.; Becker K.; Spengler B. Phospholipid Topography of Whole-Body Sections of the Anopheles Stephensi Mosquito, Characterized by High-Resolution Atmospheric-Pressure Scanning Microprobe Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging. Anal. Chem. 2015, 87 (22), 11309–11316. 10.1021/acs.analchem.5b02781. [DOI] [PubMed] [Google Scholar]
- Kompauer M.; Heiles S.; Spengler B. Atmospheric Pressure MALDI Mass Spectrometry Imaging of Tissues and Cells at 1.4-Mm Lateral Resolution. Nat. Methods 2017, 14 (1), 90–96. 10.1038/nmeth.4071. [DOI] [PubMed] [Google Scholar]
- Garikapati V.; Karnati S.; Bhandari D. R.; Baumgart-Vogt E.; Spengler B. High-Resolution Atmospheric-Pressure MALDI Mass Spectrometry Imaging Workflow for Lipidomic Analysis of Late Fetal Mouse Lungs. Sci. Rep. 2019, 9 (1), 1–14. 10.1038/s41598-019-39452-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavalin A.; Todd E. M.; Rawhouser P. D.; Yang J.; Norris J. L.; Caprioli R. M. Direct Imaging of Single Cells and Tissue at Sub-Cellular Spatial Resolution Using Transmission Geometry MALDI MS. J. Mass Spectrom. 2012, 47 (11), 1473–1481. 10.1002/jms.3108. [DOI] [PubMed] [Google Scholar]
- Richards A. L.; Lietz C. B.; Wager-Miller J. B.; Mackie K.; Trimpin S. Imaging Mass Spectrometry in Transmission Geometry. Rapid Commun. Mass Spectrom. 2011, 25 (6), 815–820. 10.1002/rcm.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff A.; Dreisewerd K.; Soltwisch J. Detailed Characterization of the Postionization Efficiencies in MALDI-2 as a Function of Relevant Input Parameters. J. Am. Soc. Mass Spectrom. 2020, 31 (9), 1844–1853. 10.1021/jasms.0c00072. [DOI] [PubMed] [Google Scholar]
- Soltwisch J.; Kettling H.; Vens-Cappell S.; Wiegelmann M.; Muthing J.; Dreisewerd K. Mass Spectrometry Imaging with Laser-Induced Postionization. Science (Washington, DC, U. S.) 2015, 348 (6231), 211–215. 10.1126/science.aaa1051. [DOI] [PubMed] [Google Scholar]
- McMillen J.; Fincher J.; Klein D.; Spraggins J.; Caprioli R.. Effect of MALDI Matrices on Lipid Analyses of Biological Tissues Using MALDI-2 Post-Ionization Mass Spectrometry. J. Mass Spectrom. 2020. 10.26434/chemrxiv.12494705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijs B.; Potthoff A.; Soltwisch J.; Dreisewerd K. MALDI-2 for the Enhanced Analysis of N -Linked Glycans by Mass Spectrometry Imaging. Anal. Chem. 2020, 92, 13904. 10.1021/acs.analchem.0c02732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivey E. C.; McMillen J. C.; Ryan D. J.; Spraggins J. M.; Caprioli R. M. Combining MALDI-2 and Transmission Geometry Laser Optics to Achieve High Sensitivity for Ultra-High Spatial Resolution Surface Analysis. J. Mass Spectrom. 2019, 54 (4), 366–370. 10.1002/jms.4335. [DOI] [PubMed] [Google Scholar]
- Niehaus M.; Soltwisch J.; Belov M. E.; Dreisewerd K. Transmission-Mode MALDI-2 Mass Spectrometry Imaging of Cells and Tissues at Subcellular Resolution. Nat. Methods 2019, 16 (9), 925–931. 10.1038/s41592-019-0536-2. [DOI] [PubMed] [Google Scholar]
- Jungmann J. H.; MacAleese L.; Buijs R.; Giskes F.; de Snaijer A.; Visser J.; Visschers J.; Vrakking M. J. J.; Heeren R. M. A. Fast, High Resolution Mass Spectrometry Imaging Using a Medipix Pixelated Detector. J. Am. Soc. Mass Spectrom. 2010, 21 (12), 2023–2030. 10.1016/j.jasms.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Klerk L. A.; Altelaar A. F. M.; Froesch M.; McDonnell L. A.; Heeren R. M. A. Fast and Automated Large-Area Imaging MALDI Mass Spectrometry in Microprobe and Microscope Mode. Int. J. Mass Spectrom. 2009, 285 (1–2), 19–25. 10.1016/j.ijms.2009.02.032. [DOI] [Google Scholar]
- Luxembourg S. L.; Mize T. H.; McDonnell L. A.; Heeren R. M. A. High-Spatial Resolution Mass Spectrometric Imaging of Peptide and Protein Distributions on a Surface. Anal. Chem. 2004, 76 (18), 5339–5344. 10.1021/ac049692q. [DOI] [PubMed] [Google Scholar]
- Jungmann J. H.; Smith D. F.; MacAleese L.; Klinkert I.; Visser J.; Heeren R. M. A. Biological Tissue Imaging with a Position and Time Sensitive Pixelated Detector. J. Am. Soc. Mass Spectrom. 2012, 23 (10), 1679–1688. 10.1007/s13361-012-0444-5. [DOI] [PubMed] [Google Scholar]
- Soltwisch J.; Göritz G.; Jungmann J. H.; Kiss A.; Smith D. F.; Ellis S. R.; Heeren R. M. A. MALDI Mass Spectrometry Imaging in Microscope Mode with Infrared Lasers: Bypassing the Diffraction Limits. Anal. Chem. 2014, 86 (1), 321–325. 10.1021/ac403421v. [DOI] [PubMed] [Google Scholar]
- Tobias F.; Hummon A. B. Considerations for MALDI-Based Quantitative Mass Spectrometry Imaging Studies. Journal of Proteome Research. 2020, 19, 3620–3630. 10.1021/acs.jproteome.0c00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta T.; Lesur A.; Varesio E.; Hopfgartner G. Quantification in MALDI-MS Imaging: What Can We Learn from MALDI-Selected Reaction Monitoring and What Can We Expect for Imaging?. Anal. Bioanal. Chem. 2015, 407 (8), 2177–2187. 10.1007/s00216-014-8315-5. [DOI] [PubMed] [Google Scholar]
- Abu Sammour D.; Marsching C.; Geisel A.; Erich K.; Schulz S.; Ramallo Guevara C.; Rabe J.-H.; Marx A.; Findeisen P.; Hohenberger P.; Hopf C. Quantitative Mass Spectrometry Imaging Reveals Mutation Status-Independent Lack of Imatinib in Liver Metastases of Gastrointestinal Stromal Tumors. Sci. Rep. 2019, 9 (1), 10698. 10.1038/s41598-019-47089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N.; Walch A. Qualitative and Quantitative Mass Spectrometry Imaging of Drugs and Metabolites in Tissue at Therapeutic Levels. Histochem. Cell Biol. 2013, 140 (2), 93–104. 10.1007/s00418-013-1127-4. [DOI] [PubMed] [Google Scholar]
- Perry W. J.; Patterson N. H.; Prentice B. M.; Neumann E. K.; Caprioli R. M.; Spraggins J. M. Uncovering Matrix Effects on Lipid Analyses in MALDI Imaging Mass Spectrometry Experiments. J. Mass Spectrom. 2020, 55 (4), e4491 10.1002/jms.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj C.; Hanley L. Ion Sources for Mass Spectrometric Identification and Imaging of Molecular Species. Nat. Prod. Rep. 2014, 31 (6), 756–767. 10.1039/C3NP70094A. [DOI] [PubMed] [Google Scholar]
- Hankin J. A.; Barkley R. M.; Murphy R. C. Sublimation as a Method of Matrix Application for Mass Spectrometric Imaging. J. Am. Soc. Mass Spectrom. 2007, 18 (9), 1646–1652. 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu Sammour D.; Marsching C.; Geisel A.; Erich K.; Schulz S.; Ramallo Guevara C.; Rabe J.-H.; Marx A.; Findeisen P.; Hohenberger P.; Hopf C. Quantitative Mass Spectrometry Imaging Reveals Mutation Status-Independent Lack of Imatinib in Liver Metastases of Gastrointestinal Stromal Tumors. Sci. Rep. 2019, 9 (1), 10698. 10.1038/s41598-019-47089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncelet L.; Ait-Belkacem R.; Bonnel D.; Stauber J. Advantages and Limitations of Quantitative Mass Spectrometry Imaging in Pharmaco-Toxicology Area. Toxicol. Anal. Clin. 2019, 31 (2), S51–S52. 10.1016/j.toxac.2019.03.073. [DOI] [Google Scholar]
- Koeniger S. L.; Talaty N.; Luo Y.; Ready D.; Voorbach M.; Seifert T.; Cepa S.; Fagerland J. A.; Bouska J.; Buck W.; Johnson R. W.; Spanton S. A Quantitation Method for Mass Spectrometry Imaging. Rapid Commun. Mass Spectrom. 2011, 25 (4), 503–510. 10.1002/rcm.4891. [DOI] [PubMed] [Google Scholar]
- Li S.; Zhang Y.; Liu J.; Han J.; Guan M.; Yang H.; Lin Y.; Xiong S.; Zhao Z. Electrospray Deposition Device Used to Precisely Control the Matrix Crystal to Improve the Performance of MALDI MSI. Sci. Rep. 2016, 6 (1), 37903. 10.1038/srep37903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud G.; Barry J.; Garrard K.; Muddiman D. Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI) Imaging Source Coupled to a FT-ICR Mass Spectrometer. J. Am. Soc. Mass Spectrom. 2013, 24 (1), 92. 10.1007/s13361-012-0505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka S. A.; Samarah L. Z.; Shaw J. B.; Liyu A. V.; Veličković D.; Agtuca B. J.; Kukolj C.; Koppenaal D. W.; Stacey G.; Paša-Tolić L.; Anderton C. R.; Vertes A. Ambient Metabolic Profiling and Imaging of Biological Samples with Ultrahigh Molecular Resolution Using Laser Ablation Electrospray Ionization 21 T FTICR Mass Spectrometry. Anal. Chem. 2019, 91 (8), 5028–5035. 10.1021/acs.analchem.8b05084. [DOI] [PubMed] [Google Scholar]
- Samarah L. Z.; Khattar R.; Tran T. H.; Stopka S. A.; Brantner C. A.; Parlanti P.; Veličković D.; Shaw J. B.; Agtuca B. J.; Stacey G.; Paša-Tolić L.; Tolić N.; Anderton C. R.; Vertes A. Single-Cell Metabolic Profiling: Metabolite Formulas from Isotopic Fine Structures in Heterogeneous Plant Cell Populations. Anal. Chem. 2020, 92 (10), 7289–7298. 10.1021/acs.analchem.0c00936. [DOI] [PubMed] [Google Scholar]
- Shrestha B.; Vertes A. In Situ Metabolic Profiling of Single Cells by Laser Ablation Electrospray Ionization Mass Spectrometry. Anal. Chem. 2009, 81 (20), 8265–8271. 10.1021/ac901525g. [DOI] [PubMed] [Google Scholar]
- Bokhart M. T.; Muddiman D. C. Infrared Matrix-Assisted Laser Desorption Electrospray Ionization Mass Spectrometry Imaging Analysis of Biospecimens. Analyst 2016, 141 (18), 5236–5245. 10.1039/C6AN01189F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha B.; Sripadi P.; Reschke B. R.; Henderson H. D.; Powell M. J.; Moody S. A.; Vertes A. Subcellular Metabolite and Lipid Analysis of Xenopus Laevis Eggs by LAESI Mass Spectrometry. PLoS One 2014, 9 (12), e115173 10.1371/journal.pone.0115173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni P.; Wilschut R. A.; Verhoeven K. J. F.; van der Putten W. H.; Garbeva P. LAESI Mass Spectrometry Imaging as a Tool to Differentiate the Root Metabolome of Native and Range-Expanding Plant Species. Planta 2018, 248 (6), 1515–1523. 10.1007/s00425-018-2989-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agtuca B. J.; Stopka S. A.; Evans S.; Samarah L.; Liu Y.; Xu D.; Stacey M. G.; Koppenaal D. W.; Paša-Tolić L.; Anderton C. R.; Vertes A.; Stacey G. Metabolomic Profiling of Wild-type and Mutant Soybean Root Nodules Using Laser-ablation Electrospray Ionization Mass Spectrometry Reveals Altered Metabolism. Plant J. 2020, 103, 1937. 10.1111/tpj.14815. [DOI] [PubMed] [Google Scholar]
- Robichaud G.; Barry J. A.; Muddiman D. C. IR-MALDESI Mass Spectrometry Imaging of Biological Tissue Sections Using Ice as a Matrix. J. Am. Soc. Mass Spectrom. 2014, 25 (3), 319–328. 10.1007/s13361-013-0787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley M. C.; Ekelöf M.; Rock K.; Patisaul H.; Muddiman D. C. IR-MALDESI Mass Spectrometry Imaging of Underivatized Neurotransmitters in Brain Tissue of Rats Exposed to Tetrabromobisphenol A. Anal. Bioanal. Chem. 2018, 410 (30), 7979–7986. 10.1007/s00216-018-1420-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley M. C.; Pace C. L.; Ekelöf M.; Muddiman D. C. Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI) Mass Spectrometry Imaging Analysis of Endogenous Metabolites in Cherry Tomatoes. Analyst 2020, 145 (16), 5516–5523. 10.1039/D0AN00818D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokhart M. T.; Manni J.; Garrard K. P.; Ekelöf M.; Nazari M.; Muddiman D. C. IR-MALDESI Mass Spectrometry Imaging at 50 Micron Spatial Resolution. J. Am. Soc. Mass Spectrom. 2017, 28 (10), 2099–2107. 10.1007/s13361-017-1740-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekelöf M.; Dodds J.; Khodjaniyazova S.; Garrard K. P.; Baker E. S.; Muddiman D. C. Coupling IR-MALDESI with Drift Tube Ion Mobility-Mass Spectrometry for High-Throughput Screening and Imaging Applications. J. Am. Soc. Mass Spectrom. 2020, 31 (3), 642–650. 10.1021/jasms.9b00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y.; Tu A.; Muddiman D. C. Lipidomic Profiling of Single Mammalian Cells by Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI). Anal. Bioanal. Chem. 2020, 412 (29), 8211–8222. 10.1007/s00216-020-02961-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehaus M.; Schnapp A.; Koch A.; Soltwisch J.; Dreisewerd K. New Insights into the Wavelength Dependence of MALDI Mass Spectrometry. Anal. Chem. 2017, 89 (14), 7734–7741. 10.1021/acs.analchem.7b01744. [DOI] [PubMed] [Google Scholar]
- Stolee J. A.; Vertes A. Toward Single-Cell Analysis by Plume Collimation in Laser Ablation Electrospray Ionization Mass Spectrometry. Anal. Chem. 2013, 85 (7), 3592–3598. 10.1021/ac303347n. [DOI] [PubMed] [Google Scholar]
- Jacobson R. S.; Thurston R. L.; Shrestha B.; Vertes A. In Situ Analysis of Small Populations of Adherent Mammalian Cells Using Laser Ablation Electrospray Ionization Mass Spectrometry in Transmission Geometry. Anal. Chem. 2015, 87 (24), 12130–12136. 10.1021/acs.analchem.5b02971. [DOI] [PubMed] [Google Scholar]
- Morris N. J.; Anderson H.; Thibeault B.; Vertes A.; Powell M. J.; Razunguzwa T. T. Laser Desorption Ionization (LDI) Silicon Nanopost Array Chips Fabricated Using Deep UV Projection Lithography and Deep Reactive Ion Etching. RSC Adv. 2015, 5 (88), 72051–72057. 10.1039/C5RA11875A. [DOI] [Google Scholar]
- Muthu M.; Gopal J.; Chun S. Nanopost Array Laser Desorption Ionization Mass Spectrometry (NAPA-LDI MS): Gathering Moss?. TrAC, Trends Anal. Chem. 2017, 97, 96–103. 10.1016/j.trac.2017.08.016. [DOI] [Google Scholar]
- Stopka S. A.; Holmes X. A.; Korte A. R.; Compton L. R.; Retterer S. T.; Vertes A. Trace Analysis and Reaction Monitoring by Nanophotonic Ionization Mass Spectrometry from Elevated Bowtie and Silicon Nanopost Arrays. Adv. Funct. Mater. 2018, 28 (29), 1801730. 10.1002/adfm.201801730. [DOI] [Google Scholar]
- Korte A. R.; Morris N. J.; Vertes A. High Throughput Complementary Analysis and Quantitation of Metabolites by Maldi- A Nd Silicon Nanopost Array-Laser Desorption/Ionization-Mass Spectrometry. Anal. Chem. 2019, 91 (6), 3951–3958. 10.1021/acs.analchem.8b05074. [DOI] [PubMed] [Google Scholar]
- Korte A. R.; Stopka S. A.; Morris N.; Razunguzwa T.; Vertes A. Large-Scale Metabolite Analysis of Standards and Human Serum by Laser Desorption Ionization Mass Spectrometry from Silicon Nanopost Arrays. Anal. Chem. 2016, 88 (18), 8989–8996. 10.1021/acs.analchem.6b01186. [DOI] [PubMed] [Google Scholar]
- Fincher J. A.; Jones D. R.; Korte A. R.; Dyer J. E.; Parlanti P.; Popratiloff A.; Brantner C. A.; Morris N. J.; Pirlo R. K.; Shanmugam V. K.; Vertes A. Mass Spectrometry Imaging of Lipids in Human Skin Disease Model Hidradenitis Suppurativa by Laser Desorption Ionization from Silicon Nanopost Arrays. Sci. Rep. 2019, 9 (1), 17508. 10.1038/s41598-019-53938-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch-Månson N.; Spangenberg A.; Gomez L. P. C.; Malval J.-P.; Soppera O.; Martinez K. L. Rapid Prototyping of Polymeric Nanopillars by 3D Direct Laser Writing for Controlling Cell Behavior. Sci. Rep. 2017, 7 (1), 9247. 10.1038/s41598-017-09208-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanes O.; Woo H.-K.; Northen T. R.; Oppenheimer S. R.; Shriver L.; Apon J.; Estrada M. N.; Potchoiba M. J.; Steenwyk R.; Manchester M.; Siuzdak G. Nanostructure Initiator Mass Spectrometry: Tissue Imaging and Direct Biofluid Analysis. Anal. Chem. 2009, 81 (8), 2969–2975. 10.1021/ac802576q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H. K.; Northen T. R.; Yanes O.; Siuzdak G. Nanostructure-Initiator Mass Spectrometry: A Protocol for Preparing and Applying NIMS Surfaces for High-Sensitivity Mass Analysis. Nat. Protoc. 2008, 3 (8), 1341–1349. 10.1038/nprot.2008.110. [DOI] [PubMed] [Google Scholar]
- Lee D. Y.; Platt V.; Bowen B.; Louie K.; Canaria C. A.; McMurray C. T.; Northen T. Resolving Brain Regions Using Nanostructure Initiator Mass Spectrometry Imaging of Phospholipids. Integr. Biol. 2012, 4 (6), 693. 10.1039/c2ib20043k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northen T. R.; Yanes O.; Northen M. T.; Marrinucci D.; Uritboonthai W.; Apon J.; Golledge S. L.; Nordström A.; Siuzdak G. Clathrate Nanostructures for Mass Spectrometry. Nature 2007, 449 (7165), 1033–1036. 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- O’Brien P. J.; Lee M.; Spilker M. E.; Zhang C. C.; Yan Z.; Nichols T. C.; Li W.; Johnson C. H.; Patti G. J.; Siuzdak G. Monitoring Metabolic Responses to Chemotherapy in Single Cells and Tumors Using Nanostructure-Initiator Mass Spectrometry (NIMS) Imaging. Cancer Metab. 2013, 1 (1), 4. 10.1186/2049-3002-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman H. M.; Lindfors L.; Palm F.; Kihlberg J.; Lanekoff I. Metabolite Aberrations in Early Diabetes Detected in Rat Kidney Using Mass Spectrometry Imaging. Anal. Bioanal. Chem. 2019, 411 (13), 2809–2816. 10.1007/s00216-019-01721-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman J. M.; Ifa D. R.; Zhu Y.; Kissinger C. B.; Manicke N. E.; Kissinger P. T.; Cooks R. G. Desorption Electrospray Ionization Mass Spectrometry: Imaging Drugs and Metabolites in Tissues. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (47), 18120–18125. 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous J.; Roach P.; Heath B.; Alexandrov T.; Laskin J.; Dorrestein P. C. Metabolic Profiling Directly from the Petri Dish Using Nanospray Desorption Electrospray Ionization Imaging Mass Spectrometry. Anal. Chem. 2013, 85 (21), 10385–10391. 10.1021/ac4023154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen S. N.; Kyle J. E.; Dautel S. E.; Sontag R.; Luders T.; Corley R.; Ansong C.; Carson J.; Laskin J. Lipid Coverage in Nanospray Desorption Electrospray Ionization Mass Spectrometry Imaging of Mouse Lung Tissues. Anal. Chem. 2019, 91 (18), 11629–11635. 10.1021/acs.analchem.9b02045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach P. J.; Laskin J.; Laskin A. Nanospray Desorption Electrospray Ionization: An Ambient Method for Liquid-Extraction Surface Sampling in Mass Spectrometry. Analyst 2010, 135 (9), 2233. 10.1039/c0an00312c. [DOI] [PubMed] [Google Scholar]
- Nguyen S. N.; Sontag R. L.; Carson J. P.; Corley R. A.; Ansong C.; Laskin J. Towards High-Resolution Tissue Imaging Using Nanospray Desorption Electrospray Ionization Mass Spectrometry Coupled to Shear Force Microscopy. J. Am. Soc. Mass Spectrom. 2018, 29 (2), 316–322. 10.1007/s13361-017-1750-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R.; Burnum-Johnson K. E.; Sun X.; Dey S. K.; Laskin J. High Spatial Resolution Imaging of Biological Tissues Using Nanospray Desorption Electrospray Ionization Mass Spectrometry. Nat. Protoc. 2019, 14 (12), 3445–3470. 10.1038/s41596-019-0237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen S. N.; Liyu A. V.; Chu R. K.; Anderton C. R.; Laskin J. Constant-Distance Mode Nanospray Desorption Electrospray Ionization Mass Spectrometry Imaging of Biological Samples with Complex Topography. Anal. Chem. 2017, 89 (2), 1131–1137. 10.1021/acs.analchem.6b03293. [DOI] [PubMed] [Google Scholar]
- Kollmer F.; Paul W.; Krehl M.; Niehuis E. Ultra High Spatial Resolution SIMS with Cluster Ions - Approaching the Physical Limits. Surf. Interface Anal. 2013, 45 (1), 312–314. 10.1002/sia.5093. [DOI] [Google Scholar]
- Sadler N. C.; Wright A. T. Activity-Based Protein Profiling of Microbes. Curr. Opin. Chem. Biol. 2015, 24, 139–144. 10.1016/j.cbpa.2014.10.022. [DOI] [PubMed] [Google Scholar]
- Anderton C. R.; Gamble L. J. Secondary Ion Mass Spectrometry Imaging of Tissues, Cells, and Microbial Systems. Microsc. Today 2016, 24 (2), 24–31. 10.1017/S1551929516000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski S. G. Mass Spectrometric Imaging of Highly Curved Membranes During Tetrahymena Mating. Science (Washington, DC, U. S.) 2004, 305 (5680), 71–73. 10.1126/science.1099791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurczy M. E.; Piehowski P. D.; Van Bell C. T.; Heien M. L.; Winograd N.; Ewing A. G. Mass Spectrometry Imaging of Mating Tetrahymena Show That Changes in Cell Morphology Regulate Lipid Domain Formation. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (7), 2751–2756. 10.1073/pnas.0908101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer T. B.; Chakravarty N.; Taylor M. J.; Nicora C. D.; Graham D. J.; Anderton C. R.; Chudler E. H.; Gamble L. J. Insights into the Histology of Planarian Flatworm Phagocata Gracilis Based on Location Specific, Intact Lipid Information Provided by GCIB-ToF-SIMS Imaging. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2019, 1864 (5), 733–743. 10.1016/j.bbalip.2019.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H.; Maciażek D.; Postawa Z.; Garrison B. J.; Winograd N. CO2 Cluster Ion Beam, an Alternative Projectile for Secondary Ion Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2016, 27 (9), 1476–1482. 10.1007/s13361-016-1423-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer T. B.; Magnusson Y.; Landberg G.; Fletcher J. S. Lipid Heterogeneity Resulting from Fatty Acid Processing in the Human Breast Cancer Microenvironment Identified by GCIB-ToF-SIMS Imaging. Anal. Chem. 2016, 88 (23), 11946–11954. 10.1021/acs.analchem.6b03884. [DOI] [PubMed] [Google Scholar]
- Sämfors S.; Ståhlman M.; Klevstig M.; Borén J.; Fletcher J. S. Localised Lipid Accumulation Detected in Infarcted Mouse Heart Tissue Using ToF-SIMS. Int. J. Mass Spectrom. 2019, 437, 77–86. 10.1016/j.ijms.2017.09.012. [DOI] [Google Scholar]
- Sämfors S.; Fletcher J. S. Lipid Diversity in Cells and Tissue Using Imaging SIMS. Annu. Rev. Anal. Chem. 2020, 13 (1), 249–271. 10.1146/annurev-anchem-091619-103512. [DOI] [PubMed] [Google Scholar]
- Tian H.; Sparvero L. J.; Blenkinsopp P.; Amoscato A. A.; Watkins S. C.; Bayır H.; Kagan V. E.; Winograd N. Secondary-Ion Mass Spectrometry Images Cardiolipins and Phosphatidylethanolamines at the Subcellular Level. Angew. Chem. 2019, 131 (10), 3188–3193. 10.1002/ange.201814256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimovska Nilsson K.; Palm M.; Hood J.; Sheriff J.; Farewell A.; Fletcher J. S. Chemical Changes On, and Through, the Bacterial Envelope in Escherichia Coli Mutants Exhibiting Impaired Plasmid Transfer Identified Using Time-of-Flight Secondary Ion Mass Spectrometry. Anal. Chem. 2019, 91 (17), 11355–11361. 10.1021/acs.analchem.9b02533. [DOI] [PubMed] [Google Scholar]
- Pareek V.; Tian H.; Winograd N.; Benkovic S. J. Metabolomics and Mass Spectrometry Imaging Reveal Channeled de Novo Purine Synthesis in Cells. Science (Washington, DC, U. S.) 2020, 368 (6488), 283–290. 10.1126/science.aaz6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheraz S.; Tian H.; Vickerman J. C.; Blenkinsopp P.; Winograd N.; Cumpson P. Enhanced Ion Yields Using High Energy Water Cluster Beams for Secondary Ion Mass Spectrometry Analysis and Imaging. Anal. Chem. 2019, 91 (14), 9058–9068. 10.1021/acs.analchem.9b01390. [DOI] [PubMed] [Google Scholar]
- Bruinen A. L.; Fisher G. L.; Heeren R. M. A.. ToF-SIMS Parallel Imaging MS/MS of Lipid Species in Thin Tissue Sections. In Methods Mol. Biol.; Humana Press, Inc., 2017; Vol. 1618, pp 165–173. 10.1007/978-1-4939-7051-3_14. [DOI] [PubMed] [Google Scholar]
- Lanni E. J.; Dunham S. J. B.; Nemes P.; Rubakhin S. S.; Sweedler J. V. Biomolecular Imaging with a C60-SIMS/MALDI Dual Ion Source Hybrid Mass Spectrometer: Instrumentation, Matrix Enhancement, and Single Cell Analysis. J. Am. Soc. Mass Spectrom. 2014, 25 (11), 1897–1907. 10.1007/s13361-014-0978-9. [DOI] [PubMed] [Google Scholar]
- Vanbellingen Q. P.; Elie N.; Eller M. J.; Della-Negra S.; Touboul D.; Brunelle A. Time-of-flight Secondary Ion Mass Spectrometry Imaging of Biological Samples with Delayed Extraction for High Mass and High Spatial Resolutions. Rapid Commun. Mass Spectrom. 2015, 29 (13), 1187–1195. 10.1002/rcm.7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher G. L.; Bruinen A. L.; Ogrinc Potočnik N.; Hammond J. S.; Bryan S. R.; Larson P. E.; Heeren R. M. A. A New Method and Mass Spectrometer Design for TOF-SIMS Parallel Imaging MS/MS. Anal. Chem. 2016, 88 (12), 6433–6440. 10.1021/acs.analchem.6b01022. [DOI] [PubMed] [Google Scholar]
- Chini C. E.; Fisher G. L.; Johnson B.; Tamkun M. M.; Kraft M. L. Observation of Endoplasmic Reticulum Tubules via TOF-SIMS Tandem Mass Spectrometry Imaging of Transfected Cells. Biointerphases 2018, 13 (3), 03B409. 10.1116/1.5019736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agüi-Gonzalez P.; Jähne S.; Phan N. T. N. SIMS Imaging in Neurobiology and Cell Biology. J. Anal. At. Spectrom. 2019, 34 (7), 1355–1368. 10.1039/C9JA00118B. [DOI] [Google Scholar]
- Kotowska A. M.; Trindade G. F.; Mendes P. M.; Williams P. M.; Aylott J. W.; Shard A. G.; Alexander M. R.; Scurr D. J. Protein Identification by 3D OrbiSIMS to Facilitate in Situ Imaging and Depth Profiling. Nat. Commun. 2020, 11 (1), 5832. 10.1038/s41467-020-19445-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarelli M. K.; Pirkl A.; Moellers R.; Grinfeld D.; Kollmer F.; Havelund R.; Newman C. F.; Marshall P. S.; Arlinghaus H.; Alexander M. R.; West A.; Horning S.; Niehuis E.; Makarov A.; Dollery C. T.; Gilmore I. S. The 3D OrbiSIMS - Label-Free Metabolic Imaging with Subcellular Lateral Resolution and High Mass-Resolving Power. Nat. Methods 2017, 14 (12), 1175–1183. 10.1038/nmeth.4504. [DOI] [PubMed] [Google Scholar]
- Wang D.; Bodovitz S. Single Cell Analysis: The New Frontier in ‘Omics.’. Trends Biotechnol. 2010, 28 (6), 281–290. 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy G.; Veličković D.; Chu R. K.; Carrell A. A.; Weston D. J.; Ibrahim Y. M.; Anderton C. R.; Smith R. D. Towards Resolving the Spatial Metabolome with Unambiguous Molecular Annotations in Complex Biological Systems by Coupling Mass Spectrometry Imaging with Structures for Lossless Ion Manipulations. Chem. Commun. 2019, 55 (3), 306–309. 10.1039/C8CC07482H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veličković D.; Chu R. K.; Carrell A. A.; Thomas M.; Paša-Tolić L.; Weston D. J.; Anderton C. R. Multimodal MSI in Conjunction with Broad Coverage Spatially Resolved MS2 Increases Confidence in Both Molecular Identification and Localization. Anal. Chem. 2018, 90 (1), 702–707. 10.1021/acs.analchem.7b04319. [DOI] [PubMed] [Google Scholar]
- Qi M.; Philip M. C.; Yang N.; Sweedler J. V. Single Cell Neurometabolomics. ACS Chem. Neurosci. 2018, 9 (1), 40–50. 10.1021/acschemneuro.7b00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchinat E.; Banci L. In-Cell NMR: A Topical Review. IUCrJ 2017, 4 (2), 108–118. 10.1107/S2052252516020625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markley J. L.; Brüschweiler R.; Edison A. S.; Eghbalnia H. R.; Powers R.; Raftery D.; Wishart D. S. The Future of NMR-Based Metabolomics. Curr. Opin. Biotechnol. 2017, 43, 34–40. 10.1016/j.copbio.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.; George J.; Wang J. Deep Profiling of Cellular Heterogeneity by Emerging Single-Cell Proteomic Technologies. Proteomics 2020, 20 (13), 1900226. 10.1002/pmic.201900226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T.; Stadtfeld M. Heterogeneity of Embryonic and Adult Stem Cells. Cell Stem Cell 2008, 3 (5), 480–483. 10.1016/j.stem.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Irish J. M.; Kotecha N.; Nolan G. P. Mapping Normal and Cancer Cell Signalling Networks: Towards Single-Cell Proteomics. Nat. Rev. Cancer 2006, 6 (2), 146–155. 10.1038/nrc1804. [DOI] [PubMed] [Google Scholar]
- Denis E. H.; Ilhardt P. D.; Tucker A. E.; Huggett N. L.; Rosnow J. J.; Moran J. J. Spatially Tracking Carbon through the Root-Rhizosphere-Soil System Using Laser Ablation-IRMS. J. Plant Nutr. Soil Sci. 2019, 182 (3), 401–410. 10.1002/jpln.201800301. [DOI] [Google Scholar]
- Moran J. J.; Newburn M. K.; Alexander M. L.; Sams R. L.; Kelly J. F.; Kreuzer H. W. Laser Ablation Isotope Ratio Mass Spectrometry for Enhanced Sensitivity and Spatial Resolution in Stable Isotope Analysis. Rapid Commun. Mass Spectrom. 2011, 25 (9), 1282–1290. 10.1002/rcm.4985. [DOI] [PubMed] [Google Scholar]
- Ember K. J. I.; Hoeve M. A.; McAughtrie S. L.; Bergholt M. S.; Dwyer B. J.; Stevens M. M.; Faulds K.; Forbes S. J.; Campbell C. J. Raman Spectroscopy and Regenerative Medicine: A Review. npj Regen. Med. 2017, 2 (1), 12. 10.1038/s41536-017-0014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.; Wright K. L.; Ashton L. Raman Spectroscopy: An Evolving Technique for Live Cell Studies. Analyst 2016, 141 (12), 3590–3600. 10.1039/C6AN00152A. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Pakoulev A.; Pang Y.; Dlott D. D. Vibrational Substructure in the OH Stretching Transition of Water and HOD. J. Phys. Chem. A 2004, 108 (42), 9054–9063. 10.1021/jp048545t. [DOI] [Google Scholar]
- Cui L.; Yang K.; Zhu Y. G. Stable Isotope-Labeled Single-Cell Raman Spectroscopy Revealing Function and Activity of Environmental Microbes. Methods Mol. Biol. 2019, 2046, 95–107. 10.1007/978-1-4939-9721-3_8. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Huang W. E.; Cui L.; Wagner M. Single Cell Stable Isotope Probing in Microbiology Using Raman Microspectroscopy. Curr. Opin. Biotechnol. 2016, 41, 34–42. 10.1016/j.copbio.2016.04.018. [DOI] [PubMed] [Google Scholar]
- Nuñez J.; Renslow R.; Cliff J. B.; Anderton C. R. NanoSIMS for Biological Applications: Current Practices and Analyses. Biointerphases 2018, 13 (3), 03B301. 10.1116/1.4993628. [DOI] [PubMed] [Google Scholar]
- Yeager A. N.; Weber P. K.; Kraft M. L. Cholesterol Is Enriched in the Sphingolipid Patches on the Substrate near Nonpolarized MDCK Cells, but Not in the Sphingolipid Domains in Their Plasma Membranes. Biochim. Biophys. Acta, Biomembr. 2018, 1860 (10), 2004–2011. 10.1016/j.bbamem.2018.04.008. [DOI] [PubMed] [Google Scholar]
- Mueller C. W.; Hoeschen C.; Steffens M.; Buddenbaum H.; Hinkel K.; Bockheim J. G.; Kao-Kniffin J. Microscale Soil Structures Foster Organic Matter Stabilization in Permafrost Soils. Geoderma 2017, 293, 44–53. 10.1016/j.geoderma.2017.01.028. [DOI] [Google Scholar]
- Vidal A.; Hirte J.; Bender S. F.; Mayer J.; Gattinger A.; Höschen C.; Schädler S.; Iqbal T. M.; Mueller C. W. Linking 3D Soil Structure and Plant-Microbe-Soil Carbon Transfer in the Rhizosphere. Front. Environ. Sci. 2018, 6 (FEB), 9. 10.3389/fenvs.2018.00009. [DOI] [Google Scholar]
- Worrich A.; Stryhanyuk H.; Musat N.; König S.; Banitz T.; Centler F.; Frank K.; Thullner M.; Harms H.; Richnow H. H.; Miltner A.; Kästner M.; Wick L. Y. Mycelium-Mediated Transfer of Water and Nutrients Stimulates Bacterial Activity in Dry and Oligotrophic Environments. Nat. Commun. 2017, 8 (1), 1–9. 10.1038/ncomms15472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H.; Passarelli M. K.; Munro P. M. G.; Kilburn M. R.; West A.; Dollery C. T.; Gilmore I. S.; Rakowska P. D. High-Resolution Sub-Cellular Imaging by Correlative NanoSIMS and Electron Microscopy of Amiodarone Internalisation by Lung Macrophages as Evidence for Drug-Induced Phospholipidosis. Chem. Commun. 2017, 53 (9), 1506–1509. 10.1039/C6CC08549K. [DOI] [PubMed] [Google Scholar]
- Grimm J. B.; English B. P.; Choi H.; Muthusamy A. K.; Mehl B. P.; Dong P.; Brown T. A.; Lippincott-Schwartz J.; Liu Z.; Lionnet T.; Lavis L. D. Bright Photoactivatable Fluorophores for Single-Molecule Imaging. Nat. Methods 2016, 13 (12), 985–988. 10.1038/nmeth.4034. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Mateos P.; Azevedo N. F.; Almeida C.; Pamme N. FISH and Chips: A Review of Microfluidic Platforms for FISH Analysis. Med. Microbiol. Immunol. 2020, 209 (3), 373–391. 10.1007/s00430-019-00654-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J.; Sheng J.; Sims P. A. SCOPE-Seq: A Scalable Technology for Linking Live Cell Imaging and Single-Cell RNA Sequencing. Genome Biol. 2018, 19 (1), 1–5. 10.1186/s13059-018-1607-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A.; Sved Skottvoll F.; Rayner S.; Pedersen-Bjergaard S.; Sullivan G.; Krauss S.; Ray Wilson S.; Harrison S. 3D Cell Culture Models and Organ-on-a-chip: Meet Separation Science and Mass Spectrometry. Electrophoresis 2020, 41 (1–2), 56–64. 10.1002/elps.201900170. [DOI] [PubMed] [Google Scholar]
- Yu J.; Zhou J.; Sutherland A.; Wei W.; Shin Y. S.; Xue M.; Heath J. R. Microfluidics-Based Single-Cell Functional Proteomics for Fundamental and Applied Biomedical Applications. Annu. Rev. Anal. Chem. 2014, 7 (1), 275–295. 10.1146/annurev-anchem-071213-020323. [DOI] [PubMed] [Google Scholar]
- Yin H.; Marshall D. Microfluidics for Single Cell Analysis. Curr. Opin. Biotechnol. 2012, 23 (1), 110–119. 10.1016/j.copbio.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Chen X.; Zhang Y.; Liu J. Advancing Single-Cell Proteomics and Metabolomics with Microfluidic Technologies. Analyst 2019, 144 (3), 846–858. 10.1039/C8AN01503A. [DOI] [PubMed] [Google Scholar]
- Helenius J.; Heisenberg C.-P.; Gaub H. E.; Muller D. J. Single-Cell Force Spectroscopy. J. Cell Sci. 2008, 121 (11), 1785–1791. 10.1242/jcs.030999. [DOI] [PubMed] [Google Scholar]
- Ahmad M. R.; Nakajima M.; Kojima S.; Homma M.; Fukuda T. In Situ Single Cell Mechanics Characterization of Yeast Cells Using Nanoneedles Inside Environmental SEM. IEEE Trans. Nanotechnol. 2008, 7 (5), 607–616. 10.1109/TNANO.2008.2003185. [DOI] [Google Scholar]
- Penen F.; Malherbe J.; Isaure M. P.; Dobritzsch D.; Bertalan I.; Gontier E.; Le Coustumer P.; Schaumlöffel D. Chemical Bioimaging for the Subcellular Localization of Trace Elements by High Contrast TEM, TEM/X-EDS, and NanoSIMS. J. Trace Elem. Med. Biol. 2016, 37, 62–68. 10.1016/j.jtemb.2016.04.014. [DOI] [PubMed] [Google Scholar]
- Lee R. F. S.; Riedel T.; Escrig S.; Maclachlan C.; Knott G. W.; Davey C. A.; Johnsson K.; Meibom A.; Dyson P. J. Differences in Cisplatin Distribution in Sensitive and Resistant Ovarian Cancer Cells: A TEM/NanoSIMS Study. Metallomics 2017, 9 (10), 1413–1420. 10.1039/C7MT00153C. [DOI] [PubMed] [Google Scholar]
- Patterson N. H.; Tuck M.; Lewis A.; Kaushansky A.; Norris J. L.; Van de Plas R.; Caprioli R. M. Next Generation Histology-Directed Imaging Mass Spectrometry Driven by Autofluorescence Microscopy. Anal. Chem. 2018, 90 (21), 12404–12413. 10.1021/acs.analchem.8b02885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N. H.; Tuck M.; Van de Plas R.; Caprioli R. M. Advanced Registration and Analysis of MALDI Imaging Mass Spectrometry Measurements through Autofluorescence Microscopy. Anal. Chem. 2018, 90 (21), 12395–12403. 10.1021/acs.analchem.8b02884. [DOI] [PubMed] [Google Scholar]
- Rohman M.; Wingfield J. High-Throughput Screening Using Mass Spectrometry within Drug Discovery. Methods Mol. Biol. 2016, 1439, 47–63. 10.1007/978-1-4939-3673-1_3. [DOI] [PubMed] [Google Scholar]
- Inglese J.; Johnson R. L.; Simeonov A.; Xia M.; Zheng W.; Austin C. P.; Auld D. S. High-Throughput Screening Assays for the Identification of Chemical Probes. Nat. Chem. Biol. 2007, 3 (8), 466–479. 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- Mayr L. M.; Bojanic D. Novel Trends in High-Throughput Screening. Curr. Opin. Pharmacol. 2009, 9 (5), 580–588. 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Amato P.; Christner B. C. Energy Metabolism Response to Low-Temperature and Frozen Conditions in Psychrobacter Cryohalolentis. Appl. Environ. Microbiol. 2009, 75 (3), 711–718. 10.1128/AEM.02193-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S. S.; Randall E. C.; Regan M. S.; Lopez B. G. C.; Clark A. R.; Schmitt N. D.; Agar J. N.; Dillon D. A.; Agar N. Y. R. In Vitro Liquid Extraction Surface Analysis Mass Spectrometry (IvLESA-MS) for Direct Metabolic Analysis of Adherent Cells in Culture. Anal. Chem. 2018, 90 (8), 4987–4991. 10.1021/acs.analchem.8b00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer W. S.; Srikumar N.; Carver J.; Chu P. Y.; Shen A.; Xu A.; Williams A. J.; Spiess C.; Wu C.; Liu Y.; Tran J. C. High-Throughput Antibody Screening from Complex Matrices Using Intact Protein Electrospray Mass Spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (18), 9851–9856. 10.1073/pnas.1917383117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts K.; Callis R.; Ikeda T.; Paunovic A.; Simpson C.; Tang E.; Turton N.; Walker G. Implementation and Challenges of Direct Acoustic Dosing into Cell-Based Assays. J. Lab. Autom. 2016, 21 (1), 76–89. 10.1177/2211068215595212. [DOI] [PubMed] [Google Scholar]
- Kempa E. E.; Hollywood K. A.; Smith C. A.; Barran P. E. High Throughput Screening of Complex Biological Samples with Mass Spectrometry-from Bulk Measurements to Single Cell Analysis. Analyst 2019, 144 (3), 872–891. 10.1039/C8AN01448E. [DOI] [PubMed] [Google Scholar]
- O’Rourke M. B.; Djordjevic S. P.; Padula M. P. The Quest for Improved Reproducibility in MALDI Mass Spectrometry. Mass Spectrom. Rev. 2018, 37 (2), 217–228. 10.1002/mas.21515. [DOI] [PubMed] [Google Scholar]
- Morikawa-Ichinose T.; Fujimura Y.; Murayama F.; Yamazaki Y.; Yamamoto T.; Wariishi H.; Miura D. Improvement of Sensitivity and Reproducibility for Imaging of Endogenous Metabolites by Matrix-Assisted Laser Desorption/Ionization-Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2019, 30 (8), 1512–1520. 10.1007/s13361-019-02221-7. [DOI] [PubMed] [Google Scholar]
- Tu A.; Muddiman D. C. Systematic Evaluation of Repeatability of IR-MALDESI-MS and Normalization Strategies for Correcting the Analytical Variation and Improving Image Quality. Anal. Bioanal. Chem. 2019, 411 (22), 5729–5743. 10.1007/s00216-019-01953-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarolli J.; Tian H.; Winograd N. Application of Pan-Sharpening to SIMS Imaging. Surf. Interface Anal. 2014, 46 (S1), 217–220. 10.1002/sia.5540. [DOI] [Google Scholar]
- Van de Plas R.; Yang J.; Spraggins J.; Caprioli R. M. Image Fusion of Mass Spectrometry and Microscopy: A Multimodality Paradigm for Molecular Tissue Mapping. Nat. Methods 2015, 12 (4), 366–372. 10.1038/nmeth.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa A.; Saito K. Metabolite Analyses of Single Cells. Plant J. 2012, 70 (1), 30–38. 10.1111/j.1365-313X.2012.04967.x. [DOI] [PubMed] [Google Scholar]
- Specht H.; Slavov N. Transformative Opportunities for Single-Cell Proteomics. J. Proteome Res. 2018, 17 (8), 2565–2571. 10.1021/acs.jproteome.8b00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman A. P.; Blakney G. T.; Hendrickson C. L.; Ellis S. R.; Heeren R. M. A.; Smith D. F. Ultra-High Mass Resolving Power, Mass Accuracy, and Dynamic Range MALDI Mass Spectrometry Imaging by 21-T FT-ICR MS. Anal. Chem. 2020, 92 (4), 3133–3142. 10.1021/acs.analchem.9b04768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J. H.; Heeren R. M. A. Detection Systems for Mass Spectrometry Imaging: A Perspective on Novel Developments with a Focus on Active Pixel Detectors. Rapid Commun. Mass Spectrom. 2013, 27 (1), 1–23. 10.1002/rcm.6418. [DOI] [PubMed] [Google Scholar]
- Cong Y.; Liang Y.; Motamedchaboki K.; Huguet R.; Truong T.; Zhao R.; Shen Y.; Lopez-Ferrer D.; Zhu Y.; Kelly R. T. Improved Single-Cell Proteome Coverage Using Narrow-Bore Packed NanoLC Columns and Ultrasensitive Mass Spectrometry. Anal. Chem. 2020, 92 (3), 2665–2671. 10.1021/acs.analchem.9b04631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Zhu N.; Tang C.; Duan H.; Wang Y.; Zhao G.; Liu J.; Ye Y. Differential Distribution of Characteristic Constituents in Root, Stem and Leaf Tissues of Salvia Miltiorrhiza Using MALDI Mass Spectrometry Imaging. Fitoterapia 2020, 146, 104679. 10.1016/j.fitote.2020.104679. [DOI] [PubMed] [Google Scholar]
- Willis P.; Jaloszynski J.; Artaev V. Improving Duty Cycle in the Folded Flight Path High-Resolution Time-of-Flight Mass Spectrometer. Int. J. Mass Spectrom. 2021, 459, 116467. 10.1016/j.ijms.2020.116467. [DOI] [Google Scholar]
- Sumner L. W.; Amberg A.; Barrett D.; Beale M. H.; Beger R.; Daykin C. A.; Fan T. W. M.; Fiehn O.; Goodacre R.; Griffin J. L.; Hankemeier T.; Hardy N.; Harnly J.; Higashi R.; Kopka J.; Lane A. N.; Lindon J. C.; Marriott P.; Nicholls A. W.; Reily M. D.; Thaden J. J.; Viant M. R. Proposed Minimum Reporting Standards for Chemical Analysis. Metabolomics 2007, 3 (3), 211–221. 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu T.; Oetjen J.; Chapelle M.; Verdu A.; Szesny M.; Chaumot A.; Degli-Esposti D.; Geffard O.; Clément Y.; Salvador A.; Ayciriex S. In Situ Isobaric Lipid Mapping by MALDI-Ion Mobility Separation-Mass Spectrometry Imaging. J. Mass Spectrom. 2020, 55 (9), e4531 10.1002/jms.4531. [DOI] [PubMed] [Google Scholar]
- Peacock P. M.; Zhang W.-J.; Trimpin S. Advances in Ionization for Mass Spectrometry. Anal. Chem. 2017, 89 (1), 372–388. 10.1021/acs.analchem.6b04348. [DOI] [PubMed] [Google Scholar]
- Taylor A. J.; Dexter A.; Bunch J. Exploring Ion Suppression in Mass Spectrometry Imaging of a Heterogeneous Tissue. Anal. Chem. 2018, 90 (9), 5637–5645. 10.1021/acs.analchem.7b05005. [DOI] [PubMed] [Google Scholar]
- Spraggins J. M.; Djambazova K. V.; Rivera E. S.; Migas L. G.; Neumann E. K.; Fuetterer A.; Suetering J.; Goedecke N.; Ly A.; Van de Plas R.; Caprioli R. M. High-Performance Molecular Imaging with MALDI Trapped Ion-Mobility Time-of-Flight (TimsTOF) Mass Spectrometry. Anal. Chem. 2019, 91 (22), 14552–14560. 10.1021/acs.analchem.9b03612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert A. S.; Prasad S.; Belford M. W.; Bailey D. J.; McAlister G. C.; Abbatiello S. E.; Huguet R.; Wouters E. R.; Dunyach J.-J.; Brademan D. R.; Westphall M. S.; Coon J. J. Comprehensive Single-Shot Proteomics with FAIMS on a Hybrid Orbitrap Mass Spectrometer. Anal. Chem. 2018, 90 (15), 9529–9537. 10.1021/acs.analchem.8b02233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. A.; van Zeijl R. J. M.; Andrén P. E.; Deelder A. M.; Wolters L.; McDonnell L. A. High Speed Data Processing for Imaging MS-Based Molecular Histology Using Graphical Processing Units. J. Am. Soc. Mass Spectrom. 2012, 23 (4), 745–752. 10.1007/s13361-011-0327-1. [DOI] [PubMed] [Google Scholar]
- McDonnell L. A.; van Remoortere A.; de Velde N.; van Zeijl R. J. M.; Deelder A. M. Imaging Mass Spectrometry Data Reduction: Automated Feature Identification and Extraction. J. Am. Soc. Mass Spectrom. 2010, 21 (12), 1969–1978. 10.1016/j.jasms.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Cumpson P. J.; Fletcher I. W.; Sano N.; Barlow A. J. Rapid Multivariate Analysis of 3D ToF-SIMS Data: Graphical Processor Units (GPUs) and Low-Discrepancy Subsampling for Large-Scale Principal Component Analysis. Surf. Interface Anal. 2016, 48 (12), 1328–1336. 10.1002/sia.6042. [DOI] [Google Scholar]
- Tortorella S.; Tiberi P.; Bowman A. P.; Claes B. S. R.; Ščupáková K.; Heeren R. M. A.; Ellis S. R.; Cruciani G. LipostarMSI: Comprehensive, Vendor-Neutral Software for Visualization, Data Analysis, and Automated Molecular Identification in Mass Spectrometry Imaging. J. Am. Soc. Mass Spectrom. 2020, 31 (1), 155–163. 10.1021/jasms.9b00034. [DOI] [PubMed] [Google Scholar]
- Schramm T.; Hester Z.; Klinkert I.; Both J.-P.; Heeren R. M. A.; Brunelle A.; Laprévote O.; Desbenoit N.; Robbe M.-F.; Stoeckli M.; Spengler B.; Römpp A. ImzML — A Common Data Format for the Flexible Exchange and Processing of Mass Spectrometry Imaging Data. J. Proteomics 2012, 75 (16), 5106–5110. 10.1016/j.jprot.2012.07.026. [DOI] [PubMed] [Google Scholar]
- Askenazi M.; Ben Hamidane H.; Graumann J. The Arc of Mass Spectrometry Exchange Formats Is Long, but It Bends toward HDF5. Mass Spectrom. Rev. 2017, 36 (5), 668–673. 10.1002/mas.21522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhamber R.; Jankevics A.; Deutsch E.; Jones A.; Dowsey A.. MzMLb: A Future-Proof Raw Mass Spectrometry Data Format Based on Standards-Compliant MzML and Optimized for Speed and Storage Requirements. bioRxiv 2021, 2020.02.13.947218. 20172. 10.1021/acs.jproteome.0c00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M.; Kirchner M.; Steen J. A. J.; Steen H. Mz5: Space- and Time-Efficient Storage of Mass Spectrometry Data Sets. Mol. Cell. Proteomics 2012, 11 (1), 1–5. 10.1074/mcp.O111.011379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikova K.; Kovalev V.; Stuart L.; Alexandrov T. OffsampleAI: Artificial Intelligence Approach to Recognize off-Sample Mass Spectrometry Images. BMC Bioinf. 2020, 21 (1), 129. 10.1186/s12859-020-3425-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey N. K.; Diwakar M.. A Review on Cloud Based Image Processing Services. In 2020 7th International Conference on Computing for Sustainable Global Development (INDIACom); IEEE, 2020; pp 108–112. 10.23919/INDIACom49435.2020.9083718. [DOI]
- Taylor M. J.; Zhang K. Y.; Graham D. J.; Gamble L. J. Fatty Acid and Lipid Reference Spectra. Surf. Sci. Spectra 2018, 25 (2), 025001. 10.1116/1.5063595. [DOI] [Google Scholar]
- Guijas C.; Montenegro-Burke J. R.; Domingo-Almenara X.; Palermo A.; Warth B.; Hermann G.; Koellensperger G.; Huan T.; Uritboonthai W.; Aisporna A. E.; Wolan D. W.; Spilker M. E.; Benton H. P.; Siuzdak G. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90 (5), 3156–3164. 10.1021/acs.analchem.7b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy E.; Subramaniam S.; Murphy R. C.; Nishijima M.; Raetz C. R. H.; Shimizu T.; Spener F.; Van Meer G.; Wakelam M. J. O.; Dennis E. A.. Update of the LIPID MAPS Comprehensive Classification System for Lipids. J. Lipid Res.; American Society for Biochemistry and Molecular Biology, 2009, p S9. 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Carver J. J.; Phelan V. V.; Sanchez L. M.; Garg N.; Peng Y.; Nguyen D. D.; Watrous J.; Kapono C. A.; Luzzatto-Knaan T.; Porto C.; Bouslimani A.; Melnik A. V.; Meehan M. J.; Liu W.-T.; Crüsemann M.; Boudreau P. D.; Esquenazi E.; Sandoval-Calderón M.; Kersten R. D.; Pace L. A.; Quinn R. A.; Duncan K. R.; Hsu C.-C.; Floros D. J.; Gavilan R. G.; Kleigrewe K.; Northen T.; Dutton R. J.; Parrot D.; Carlson E. E.; Aigle B.; Michelsen C. F.; Jelsbak L.; Sohlenkamp C.; Pevzner P.; Edlund A.; McLean J.; Piel J.; Murphy B. T.; Gerwick L.; Liaw C.-C.; Yang Y.-L.; Humpf H.-U.; Maansson M.; Keyzers R. A.; Sims A. C.; Johnson A. R.; Sidebottom A. M.; Sedio B. E.; Klitgaard A.; Larson C. B.; Boya P C. A.; Torres-Mendoza D.; Gonzalez D. J.; Silva D. B.; Marques L. M.; Demarque D. P.; Pociute E.; O’Neill E. C.; Briand E.; Helfrich E. J. N.; Granatosky E. A.; Glukhov E.; Ryffel F.; Houson H.; Mohimani H.; Kharbush J. J.; Zeng Y.; Vorholt J. A.; Kurita K. L.; Charusanti P.; McPhail K. L.; Nielsen K. F.; Vuong L.; Elfeki M.; Traxler M. F.; Engene N.; Koyama N.; Vining O. B.; Baric R.; Silva R. R.; Mascuch S. J.; Tomasi S.; Jenkins S.; Macherla V.; Hoffman T.; Agarwal V.; Williams P. G.; Dai J.; Neupane R.; Gurr J.; Rodríguez A. M. C.; Lamsa A.; Zhang C.; Dorrestein K.; Duggan B. M.; Almaliti J.; Allard P.-M.; Phapale P.; Nothias L.-F.; Alexandrov T.; Litaudon M.; Wolfender J.-L.; Kyle J. E.; Metz T. O.; Peryea T.; Nguyen D.-T.; VanLeer D.; Shinn P.; Jadhav A.; Müller R.; Waters K. M.; Shi W.; Liu X.; Zhang L.; Knight R.; Jensen P. R.; Palsson B. Ø.; Pogliano K.; Linington R. G.; Gutiérrez M.; Lopes N. P.; Gerwick W. H.; Moore B. S.; Dorrestein P. C.; Bandeira N. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34 (8), 828–837. 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandy J.; Davies V.; Van Der Hooft J. J. J.; Weidt S.; Daly R.; Rogers S. In Silico Optimization of Mass Spectrometry Fragmentation Strategies in Metabolomics. Metabolites 2019, 9 (10), 219. 10.3390/metabo9100219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachran A. D.; Balabin I.; Cathey T.; Transue T. R.; Al-Ghoul H.; Grulke C.; Sobus J. R.; Williams A. J. Linking in Silico MS/MS Spectra with Chemistry Data to Improve Identification of Unknowns. Sci. Data 2019, 6 (1), 1–9. 10.1038/s41597-019-0145-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A.; Al-Ghoul H.; McEachran A. D.; Balabin I.; Transue T.; Cathey T.; Grossman J. N.; Singh R. R.; Ulrich E. M.; Williams A. J.; Sobus J. R. In Silico MS/MS Spectra for Identifying Unknowns: A Critical Examination Using CFM-ID Algorithms and ENTACT Mixture Samples. Anal. Bioanal. Chem. 2020, 412 (6), 1303–1315. 10.1007/s00216-019-02351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaženović I.; Kind T.; Torbašinović H.; Obrenović S.; Mehta S. S.; Tsugawa H.; Wermuth T.; Schauer N.; Jahn M.; Biedendieck R.; Jahn D.; Fiehn O. Comprehensive Comparison of in Silico MS/MS Fragmentation Tools of the CASMI Contest: Database Boosting Is Needed to Achieve 93% Accuracy. J. Cheminf. 2017, 9 (1), 32. 10.1186/s13321-017-0219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S.; Schmidt S.; Müller-Hannemann M.; Neumann S. In Silico Fragmentation for Computer Assisted Identification of Metabolite Mass Spectra. BMC Bioinf. 2010, 11 (1), 148. 10.1186/1471-2105-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby S. M.; Nuñez J. R.; Hodas N. O.; Corley C. D.; Renslow R. R. Deep Learning to Generate in Silico Chemical Property Libraries and Candidate Molecules for Small Molecule Identification in Complex Samples. Anal. Chem. 2020, 92 (2), 1720–1729. 10.1021/acs.analchem.9b02348. [DOI] [PubMed] [Google Scholar]
- Colby S. M.; Thomas D. G.; Nunez J. R.; Baxter D. J.; Glaesemann K. R.; Brown J. M.; Pirrung M. A.; Govind N.; Teeguarden J. G.; Metz T. O.; Renslow R. S. ISiCLE: A Quantum Chemistry Pipeline for Establishing in Silico Collision Cross Section Libraries. Anal. Chem. 2019, 91 (7), 4346–4356. 10.1021/acs.analchem.8b04567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis S. R.; Paine M. R. L.; Eijkel G. B.; Pauling J. K.; Husen P.; Jervelund M. W.; Hermansson M.; Ejsing C. S.; Heeren R. M. A. Automated, Parallel Mass Spectrometry Imaging and Structural Identification of Lipids. Nat. Methods 2018, 15 (7), 515–518. 10.1038/s41592-018-0010-6. [DOI] [PubMed] [Google Scholar]