Abstract
DNA damage activates the checkpoint protein CHK1 to arrest cell cycle progression, providing time for repair and recovery. Consequently, inhibitors of CHK1 (CHK1i) enhance damage-induced cell death. Additionally, CHK1i elicits single agent cytotoxicity in some cell lines. We compared three CHK1i that have undergone clinical trials and exhibited different toxicities. Each CHK1i inhibits other targets at higher concentrations, and whether these contribute to the toxicity is unknown. We compared their sensitivity in a panel of cell lines, their efficacy at inhibiting CHK1 and CHK2, and their ability to induce DNA damage and abrogate damage-induced S phase arrest. Published in vitro kinase analyses were a poor predictor of selectivity and potency in cells. LY2606368 was far more potent at inhibiting CHK1 and inducing growth arrest, while all three CHK1i inhibited CHK2 at concentrations 10- (MK-8776 and SRA737) to 100- (LY2606368) fold higher. MK-8776 and SRA737 exhibited similar off-target effects: higher concentrations demonstrated transient protection from growth inhibition, circumvented DNA damage, and prevented checkpoint abrogation, possibly due to inhibition of CDK2. Acquired resistance to LY2606368 resulted in limited cross-resistance to other CHK1i. LY2606368-resistant cells still abrogated DNA damage-induced S phase arrest, which requires low CDK2 activity, whereas inappropriately high CDK2 activity is responsible for sensitivity to CHK1i alone. All three CHK1i inhibited protein synthesis in a sensitive cell line correlating with cell death, whereas resistant cells failed to inhibit protein synthesis and underwent transient cytostasis. LY2606368 appears to be the most selective CHK1i, suggesting that further clinical development of this drug is warranted.
Keywords: cancer, DNA damage response, CHK1, CHK2, CDK2, protein translation
Checkpoint kinase 1 (CHK1) is a major intermediary protein in the DNA damage response pathway.1 Single-strand regions in DNA activate ATR (ataxia telangiectasia and RAD3-related), which in turn phosphorylates and activates CHK1. Alternately, DNA double-strand breaks activate ATM (ataxia telangiectasia mutated), leading to resection of DNA at the break, creating single-strand DNA that also activates ATR/CHK1. CHK1 is responsible for arresting cell cycle progression permitting time for DNA repair and cell survival, thereby preventing the DNA damage from becoming lethal. A second checkpoint kinase, CHK2, is also activated directly by ATM, and may play a role in signaling to the p53 tumor suppressor.2,3
Many anticancer drugs function by inducing DNA damage, either by alkylating or cross-linking DNA, or indirectly by starving cells for deoxyribonucleotides thereby stalling replication. The checkpoint pathways protect the cells until they can repair the damage and thereby impede the desired therapeutic action. Consequently, inhibitors of CHK1 (CHK1i) have been developed because they can sensitize cells to DNA damaging drugs by inducing replication and mitosis before DNA repair is complete.1 While this CHK1i-mediated abrogation of cell cycle arrest occurs in most if not all cell lines, we have demonstrated hypersensitivity to CHK1i as a single agent in about 15% of cell lines.4 Our goal is to define the mechanisms of the single agent activity so that clinical trials can be targeted to appropriate patients.
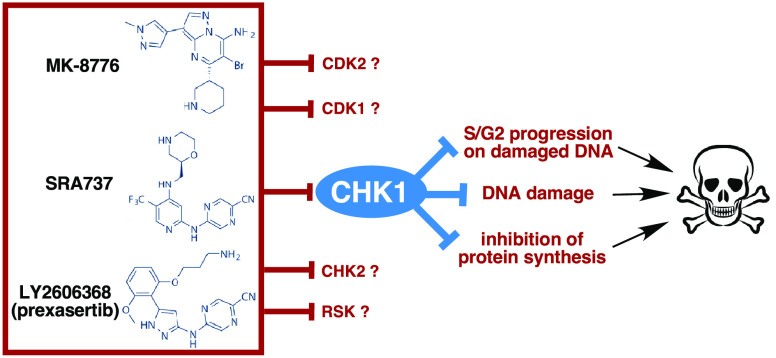
Our recent research used the CHK1i MK-8776,4−7 but Merck discontinued its development. Furthermore, it may have limited in vivo stability due to demethylation.8 Hence, we sought a different CHK1i that might still have clinical potential. Clinical trials with most CHK1i have been terminated for toxicity, lack of efficacy, or for business reasons.9 LY2606368 (prexasertib; Lilly) was the most advanced with 14 clinical trials listed on www.clinicaltrials.gov either as a single agent or in various drug combinations, but its development has recently been terminated due to neutropenia. SRA737 (Sierra Oncology), previously known as CCT245737,10 has completed Phase I trials as a monotherapy and in combination with gemcitabine, and limited neutropenia was observed.11,12 Clinical activity of this combination was observed in tumor types such as anal, cervical, and rectal.12 Furthermore, SRA737 is orally bioavailable, which should permit more continuous treatment schedules than LY2606368 that requires intravenous administration.
Each CHK1i has been reported to inhibit additional targets, albeit usually at considerably higher concentrations. These targets were identified from in vitro kinome-wide assays and whether they inhibit these targets in cells has generally not been reported. Furthermore, kinome-wide assays do not assess the potential for nonkinase targets. These off-target effects could also impact efficacy in a patient because it is difficult to limit the drug at the tumor to the concentration that inhibits only a single target. Here, we compared MK-8776, LY2606368, and SRA737 in a variety of cell-based assays. We conclude that MK-8776 and SRA737 have very similar activities, but both exhibit an off-target effect at concentrations a little above those that inhibit CHK1. This off-target effect appears to protect cells from inhibition of CHK1, at least over a short time frame, and therefore might provide protection from neutropenia in patients. In contrast, LY2606368 appears much more selective for CHK1, thereby suggesting that the observed neutropenia in patients may be due to on-target activity. The impact of these observations on further clinical development is discussed.
Results
Comparison of Growth Inhibition by CHK1i
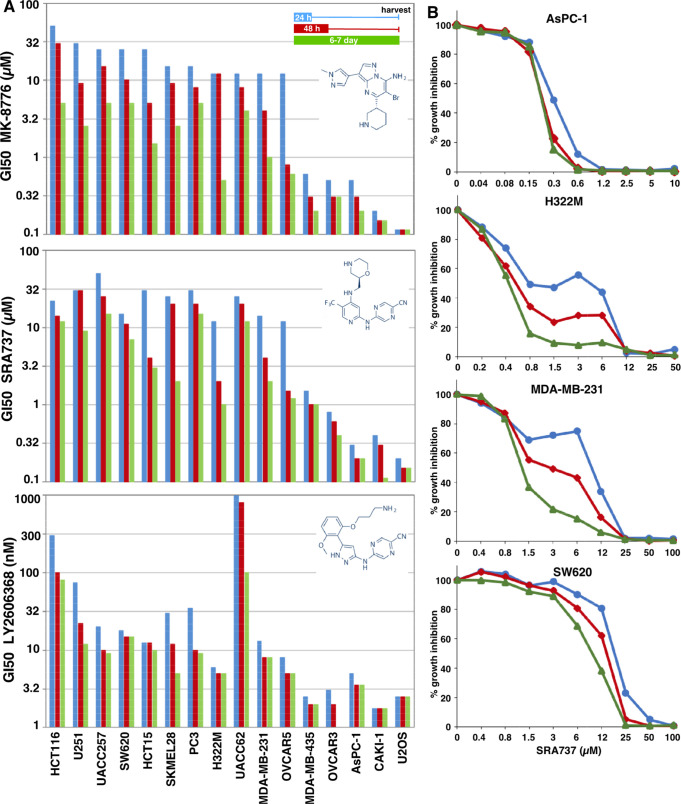
The primary goal of these experiments was to compare and contrast the effects of three CHK1i as an explanation for the different experiences observed in patients. Initially, the growth inhibitory activity of the three CHK1i as single agents was tested in a panel of cell lines selected from a larger panel of known sensitivity to MK-8776.4 Cells were incubated with drug for either 24 or 48 h, then the drug was removed and incubation continued for another 5 or 6 days. Alternately, cells were incubated with drug continuously for 6 or 7 days. At the time of harvest, the untreated cells were usually close to confluence. Wells were then scored for the amount of DNA as a surrogate for cell number, and results are expressed as the concentration that inhibits growth by 50% (GI50). This provides data on both the time and concentration required to inhibit growth.
There is a significant difference in the potency of the three CHK1i. While the cells most sensitive to MK-8776 and SRA737 exhibited a GI50 of 0.1–1 μM, LY2606368 was about 100-fold more potent (Figure 1A). Another major difference between the CHK1i is the range of GI50 concentrations that discriminate the most sensitive from the most resistant cell lines, with MK-8776 and SRA737 exhibiting ∼500-fold difference in response, as opposed to only 50-fold for LY2606368 (with the exception of UACC62 that are very resistant to LY2606368).
Figure 1.
Sensitivity of cell lines to CHK1 inhibitors. (A) Each cell line was incubated with MK-8776 (top), SRA737 (middle), or LY2606368 (bottom) for either 24 h (blue) or 48 h (red); then, drug was removed, and cells were incubated in fresh media for an additional 5–6 days. Alternately, cells were incubated in drug continuously for 6–7 days (green). Cells were lysed, stained for DNA, and the concentration that inhibited growth by 50% was recorded. The cell lines are ordered based on the 24-h drug exposure to MK-8776, then 48-h exposure, and finally continuous exposure. The same order of cell lines is then retained for SRA737 and LY2606368. Values for many of these lines reflect 2–4 determinations; all values are tabulated in Table S1. Values for MK-8776 are similar to those previously reported in an independent analysis but whose highest concentration was 10 μM.4 (B) Representative growth curves are shown for four cell lines incubated with SRA-737. The biphasic curves rarely resulted in two possible GI50 values, but when this did occur, the higher value was used in panel A.
Intriguingly, some cells lines exhibited a biphasic growth curve when incubated with MK-8776 or SRA737 but not LY2606368. This was most evident for cell lines with intermediate sensitivity; several examples are shown in Figure 1B. This only occurred after short drug exposure. The biphasic response was not previously observed as the maximum concentration of MK-8776 tested was 10 μM.4 As discussed below, this appears to be due to an off-target effect of MK-8776 and SRA737 that limits damage after a short drug exposure. Even after continuous exposure to CHK1i, most cell lines were still 20–50-fold less sensitive to MK-8776 and SRA737 than the hypersensitive subset of cells (U2OS, CAKI-1, AsPC-1, and OVCAR3), whereas LY2606368 elicited a much smaller differential. These results suggest that while LY2606368 may be the more potent drug, the other two CHK1i exhibit a greater differential between cell lines.
Potential reasons why LY2606368 did not discriminate a hypersensitive subset of cells include drug instability or serum binding, either of which might have greater impact at low concentrations of drug. To test these possibilities, we incubated AsPC-1 cells with LY2606368 for 24 h and then removed the drug and added it to a second set of cells. The sensitivity of the cells remained the same, as shown in Figure 1. Cells were also incubated with LY2606368 for 6 h, and then the media was added to other cells and assessed for γH2AX (as discussed below). Again, the levels of γH2AX did not change. These observations rule out inactivation of the drug by metabolism or serum protein binding as an explanation for the limited discrimination of the hypersensitive population.
Inhibition of CHK1 in Cells
In cell-free protein kinase assays, the three CHK1i all compete for ATP binding. Published inhibitory concentrations for MK-8776,13 LY2606368,14 and SRA73710,15 are presented in Table 1. The inhibition of CHK1 occurs in the low nanomolar range for all three inhibitors, but they each inhibit other kinases at different concentrations. MK-8776 was compared against a limited panel of kinases, and IC50 values were compared only for CHK1, CHK2, and CDK2 (Table 1). For LY2606368, 6 additional kinases were found to have an IC50 < 100 nM, but only CHK2 and the RSK family kinases had IC50s of less than 10 nM.14 SRA737 was at least 93-fold selective for CHK1 compared to 124 kinases tested.10 One limitation of comparing these values is that they were performed in different laboratories with different assay conditions, yet these are the published values commonly referred to in many investigations and by commercial suppliers. Other limitations of these kinase assays are that they were only tested in a partial kinase panel, the kinase activities are not in a biological context (e.g., necessary binding partners or activation status), and there are many potential targets that are not kinases. Consequently, we investigated the effects of these three CHK1i on various targets in cells.
Table 1. Summary of the Efficacy of Each CHK1i in Various Assays.
| MK-8776 | SRA737 | LY2606368 | |
|---|---|---|---|
| in vitro kinase inhibition (IC50; as published) | |||
| CHK1 | 3 nM | 1.4 nM | <1 nM |
| CHK2 | 1.5 μM | 2.4 μM | 8 nM |
| CDK1 | 9 μM | >10 μM | |
| CDK2 | 160 nM | 3.85 μM | ≥10 μM |
| RSK1 | 361 nM | 9 nM | |
| kinase inhibition in cells (IC50; Figures 2 and 3) | |||
| CHK1 (AsPC-1) | 0.3 μM | 1 μM | 3 nM |
| CHK1 (SW620) | 0.3 μM | 3 μM | 3 nM |
| CHK2 (H322M) | 5 μM | 10 μM | 250 nM |
| CHK2 after SN38 (AsPC-1) | 10 μM | 5 μM | 40 nM |
| GI50 in AsPC-1 cells (Figure 1) | |||
| 24 h | 0.5 μM | 0.3 μM | 5 nM |
| 48 h | 0.3 μM | 0.2 μM | 3.5 nM |
| 7 day | 0.2 μM | 0.2 μM | 3.5 nM |
| GI50 in MDA-MB-231 cells (Figure 1) | |||
| 24 h | 12 μM | 14 μM | 13 nM |
| 48 h | 4 μM | 4 μM | 8 nM |
| 7 day | 1 μM | 2 μM | 8 nM |
| lowest concentration giving maximum γH2AX (Figure 4) | |||
| AsPC-1 | 1 μM | 3 μM | 30 nM |
| abrogation of S phase arrest (minimum cells in S; Figure 5) | |||
| MDA-MB-231 | 0.3 μM | 0.3 μM | 3 nM |
| MDA-MB-231-LY-R | 0.3 μM | 1 μM | 10 nM |
| AsPC-1 | 0.03 μM | 0.1 μM | 1 nM |
| AsPC-776-R | 0.3 μM | 1 μM | 10 nM |
| AsPC-LY-R | 0.3 μM | 1 μM | 10 nM |
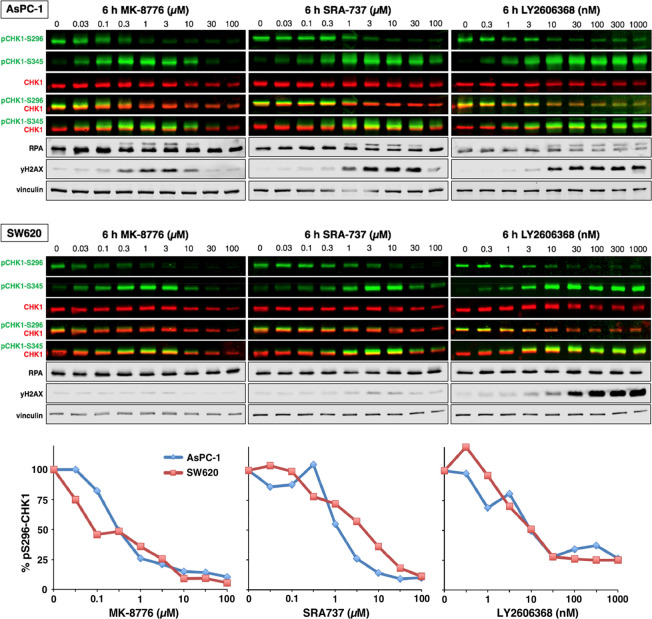
AsPC-1 cells exhibit constitutive activation of CHK1 as reflected in autophosphorylation on serine 296. Following a 6 h incubation, 50% inhibition of pS296-CHK1 was observed at 0.3 μM MK-8776, 1 μM SRA737, and 3 nM LY2606368 (Figure 2A). These values are close to the concentrations that inhibit growth of these cells. Phosphorylation of CHK1 by ATR on serine-345 is observed, either because of the concurrent appearance of DNA damage (as indicated by γH2AX), or because of a feedback loop in which CHK1 phosphorylates and activates PP2A thereby limiting pS345-CHK1.16 These data demonstrate that LY2606368 is almost as potent in cells as in in vitro kinase assays, whereas MK-8776 and SRA737 require about 100-fold higher concentrations to inhibit CHK1 in cells.
Figure 2.
Inhibition of CHK1 and induction of DNA damage markers by CHK1i. ASPC-1 and SW620 cells were incubated with the indicated concentrations of each CHK1i for 6 h and then analyzed by Western blotting using the indicated primary antibodies and fluorescent secondary antibodies. All images were generated using a fluorescent scanner. Dual-color images represent multiplexed total and phospho-proteins on the same membrane. Single color images represent individual channels split from the multiplexed images. The lower panel shows the quantitation of pS296-CHK1.
At higher concentrations of MK-8776 and SRA737 but not LY2606368, both pS345-CHK1 and γH2AX decrease. These data are consistent with pS345-CHK1 being a consequence of DNA damage activating ATR that then phosphorylates CHK1, and with the self-limiting, off-target effect of MK-8776 and SRA737 that limits the DNA damage.
This experiment was repeated in SW620 cells that are relatively resistant to CHK1i (Figure 1), yet all three CHK1i were as potent at inhibiting pS296-CHK1 (Figure 2B). Hence, the ability to inhibit CHK1 is not the sole determinant for growth inhibition and DNA damage. Other events downstream of CHK1 inhibition such as activation of CDK2 determine whether a cell will die.4 The SW620 cells also exhibited biphasic phosphorylation and then inhibition of pS345-CHK1 at higher concentrations of MK-8776 and SRA737, although they induced little γH2AX, suggesting the initial increase in pS345-CHK1 is more likely a result of the feedback regulation of PP2A in these cells. The decrease in pS345 at higher concentrations appears to be an off-target effect. In contrast, high γH2AX occurs in response to LY2606368 and correlates with the persistence of pS345-CHK1; whether this is due to DNA damage or the feedback loop (or both) is unknown.
Impact of CHK1i on CHK2 in Cells
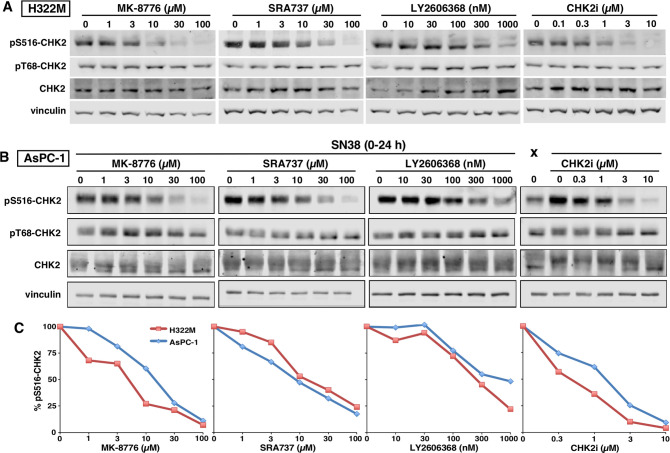
LY2606368 is well-recognized as a CHK2 inhibitor from in vitro kinase assays, while MK-8776 and SRA737 are ∼1000-fold less potent against CHK2 than CHK1 (Table 1). To assess the inhibition of CHK2 in cells, we used H322M cells that exhibit constitutive CHK2 activation as reflected in autophosphorylation on S516. Surprisingly, MK-8776 and SRA737 inhibited CHK2 at 10 and 30 μM respectively, whereas LY2606368 inhibited CHK2 at 300 nM (Figure 3A). Importantly, there was no change in basal pT68-CHK2, a site phosphorylated by ATM, suggesting the effects of CHK1i were directly on CHK2. As a positive control, we also demonstrated the efficacy of a CHK2i at selectively inhibiting pS516-CHK2.
Figure 3.
Inhibition of CHK2 by CHK1i. (A) H322M cells were incubated with the indicated concentrations of each CHK1i or a CHK2 inhibitor (CHK2i) for 3 h. Cell lysates were analyzed by Western blotting using the indicated primary antibodies and fluorescent secondary antibodies. (B) AsPC-1 cells were incubated with 30 ng/mL (75 nM) SN38 for 24 h and then with each CHK1i or CHK2i for 30 min and analyzed as in panel A. (C) Quantitation of pS516-CHK2 from panels A and B.
Given this unexpected observation regarding inhibition of CHK2, we repeated the analysis in AsPC-1 cells following induction of DNA damage with the topoisomerase I inhibitor SN38 (Figure 3B; “X” shows an undamaged sample). Selective inhibition of pS516-CHK2 was again seen with all three CHK1i, at similar concentrations to those observed in H322M cells. Hence in cells, MK-8776 and SRA737 exhibit only 10–20-fold selectivity for CHK1 over CHK2, whereas LY2606368 demonstrates 100-fold selectivity (Table 1).
CHK1i-Mediated Induction of DNA Damage
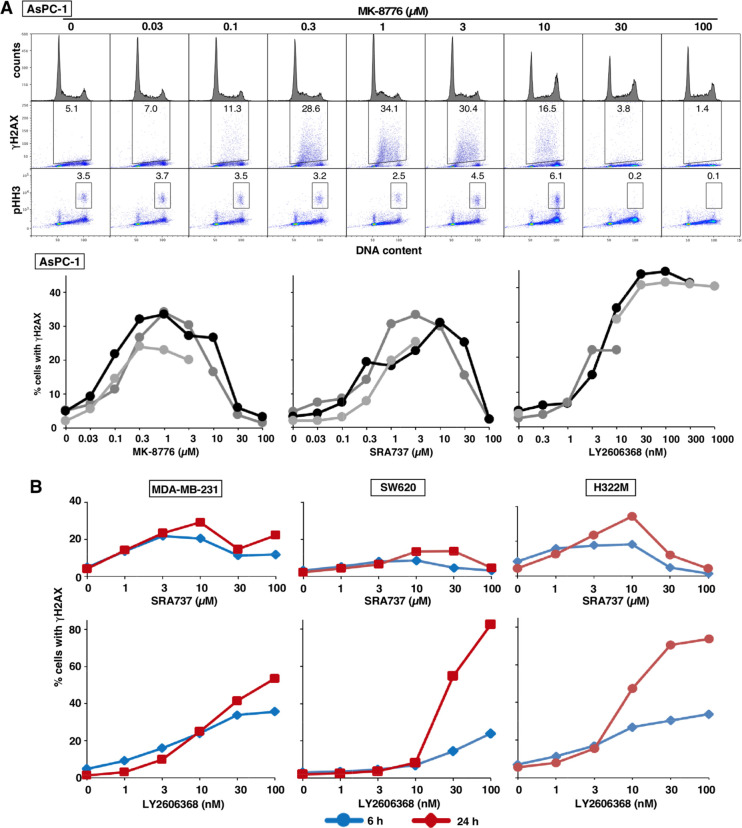
DNA damage, reflected in γH2AX, occurs rapidly in cell lines hypersensitive to CHK1i.4 Prior experiments using the neutral comet assay have demonstrated that this damage is consistent with DSB.4 AsPC-1 cells were incubated with each CHK1i for 6 h and then analyzed by flow cytometry. γH2AX was only induced in S phase cells and comparable levels (30–40%) occurred at 0.3 μM MK-8776, 1 μM SRA737, and 10 nM LY2606368 (Figures 4A, S1). These concentrations were comparable to the concentrations of each drug that inhibited both cell growth and pS296-CHK1.
Figure 4.
Induction of γH2AX by CHK1i. (A) ASPC-1 cells were incubated with the indicated concentrations of MK-8776 for 6 h and then analyzed by flow cytometry for DNA content, γH2AX, and pHH3. The percentage of cells positive for γH2AX or pHH3 is indicated. The percentage of γH2AX-positive cells from replicate experiments is presented. (B) MDA-MB-231, SW620, and H322M cells were incubated with the indicated concentrations of each CHK1i for 6 or 24 h and analyzed as in panel A. The flow cytometry data for these analyses are presented in Figure S1.
At higher concentrations of MK-8776 and SRA737 (30–100 μM) but not LY2606368, there was a clear decrease in γH2AX. A plausible explanation for this observation is that MK-8776 and SRA737 also inhibit CDK2. We have previously demonstrated that inhibition of γH2AX can be elicited by inhibition of CDK2,4 which is a reported off-target substrate of MK-8776 at 50-fold higher concentrations than inhibit CHK1 (Table 1). The potential inhibition of CDK2 by SRA737 was unexpected as in vitro kinase data suggested a 1000-fold higher concentration would be required.
It has been suggested that sensitivity to CHK1i is due to premature mitosis of S phase cells,17,18 but we detected minimal phosphorylation of histone H3 (pHH3; a common marker of mitosis), except in M phase, and not in the γH2AX-positive cells (Figures 4, S1). However, we noted an increase of pHH3 in G2/M cells at 10 μM MK-8776 and 30 μM SRA737. This increase has previously been seen with the CDK1/2 inhibitor Ro33064 and may occur when CDK1 is partially inhibited such that cells can enter but not complete mitosis. At higher concentrations of MK-8776 and SRA737, pHH3 was dramatically reduced. These results are consistent with inhibition of CDK1 and/or CDK2 at the higher concentrations of these two CHK1i.
We next assessed γH2AX induced by SRA737 and LY2606368 in relatively resistant cells. Higher concentrations of both CHK1i were required to induce γH2AX (Figures 4B, S1). For SRA737, there was little increase between 6 and 24 h, and inhibition was again observed at the highest concentrations. The decrease in γH2AX was associated with S phase progression and accumulation in G2/M, the latter of which might be attributable to inhibition of CDK1. In contrast, upon incubation with LY2606368, γH2AX continued to increase up to 24 h as more cells entered S phase.
Replication protein A (RPA) is recruited to single-strand DNA and phosphorylated on several sites, including S4/S8, resulting in a significant electrophoretic band shift. In AsPC-1 cells, pRPA was observed at concentrations of CHK1i that correlated with the appearance of γH2AX (Figure 2). pRPA was inhibited at higher concentrations of MK-8776 and SRA737, but not LY2606368, correlating with the inhibition of γH2AX. However, little if any pRPA was observed in SW620 cells, even following incubation with LY2606368, despite the high level of γH2AX (Figure 2). This may be attributed to the observation that γH2AX in these cells occurs at late S or G2 phase, rather than in early to mid-S phase, and suggests single-strand DNA is not essential for γH2AX (Figure S1C).
Abrogation of DNA Damage-Induced Cell Cycle Arrest
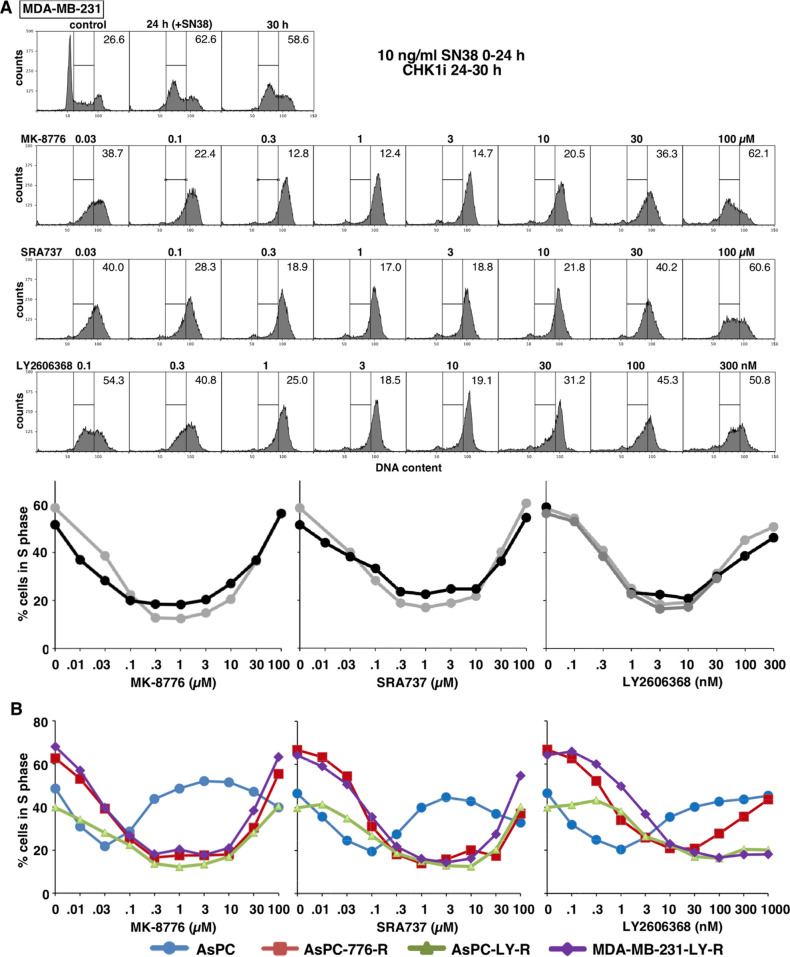
MDA-MB-231 cells were incubated with SN38 for 24 h to arrest the cells in mid-S phase. Upon removal of SN38, cells were incubated with each CHK1i for 6 h. All three CHK1i abrogated arrest, driving the cells into G2 (Figure 5A). Maximum abrogation occurred at approximately 0.3 μM MK-8776, 0.3 μM SRA737, and 3 nM LY2606368. These values are similar to those required to inhibit CHK1 in cells (Figure 2, Table 1).
Figure 5.
Abrogation of DNA damage-induced cell cycle arrest by CHK1i in constitutive and acquired-resistant cell lines. (A) MDA-MB-231 cells were incubated with the 10 ng/mL SN38 (25 nM) for 24 h; then, SN38 was removed, and cells were incubated with the indicated concentrations of CHK1i from 24 to 30 h. Cells were fixed and analyzed by flow cytometry for DNA content. The percentage of cells in S phase are presented, and replicate experiments are graphed. (B) ASPC-1 cells and their resistant derivatives, as well as MDA-MB-231 cells selected for resistance to LY2606368 were analyzed as in panel A. The data for these analyses are presented in Figure S2.
At the higher concentrations of all three CHK1i, checkpoint abrogation was completely inhibited. In the case of MK-8776 and SRA737, the failure to abrogate arrest is likely due to their off-target effect and is consistent with the possible inhibition of CDK2 that is required for S phase progression. However, for LY2606368, failure to abrogate arrest is likely due to the high level of γH2AX induced as a single agent, which then impedes S phase progression.
To further dissect the inability to abrogate S phase arrest at higher drug concentrations, we repeated the experiment in cells with different sensitivity to CHK1i. In CHK1i-sensitive AsPC-1 cells, low concentrations of CHK1i abrogated S phase arrest, but at slightly higher concentrations, the cells remained in S phase (Figures 5B, S2A). These concentrations correlated with the induction of γH2AX by single agent CHK1i, suggesting this contributed to the inhibition of checkpoint abrogation. As discussed below, we generated various cell lines with resistance to each CHK1i. AsPC-1 cells selected for resistance to MK-8776 were able to abrogate S phase arrest up to all but the highest concentrations (Figures 5B, S2B). Similar results were observed in AsPC-1 cells selected for resistance to LY2606368, with high concentrations of MK-8776 and SRA737 inhibiting abrogation. Importantly, abrogation still occurred at high concentrations of LY2606368 (Figures 5B, S2C). Similarly, in MDA-MB-231 cells selected for resistance to LY2606368, S phase abrogation was still prevented at high concentrations of MK-8776 and SRA737, but not at high concentrations of LY2606368 (Figures 5B, S2D). These observations are consistent with resistance to the monotherapy activity of LY2606368 occurring at these high concentrations, while response to the other two CHK1i is impeded by the off-target effect.
The Off-Target Effect of MK-8776 and SRA737 Involves Inhibition of CDK2
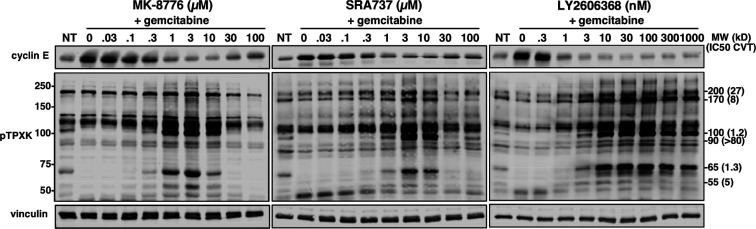
CDK1/2 can phosphorylate threonine when followed by proline. Some of these target proteins can therefore be detected using a phospho-specific antibody to the sequence pTPXK. Using this approach, we have previously demonstrated that target proteins can be variably sensitive to CVT-313, which inhibits CDK2 at low concentrations, and CDK1 at higher concentrations.19,20 The current experiment used MDA-MB-231 cells as in our prior studies. Cells were initially incubated with gemcitabine which activates CHK1 and reduces the existing phosphorylation of these target proteins. The addition of CHK1i to gemcitabine-arrested cells does not induce S phase progression due to the lack of dNTPs, nor does it induce premature mitosis.20 The cells were then incubated with each of the three CHK1i for 6 h (Figure 6). There was a clear induction of at least 6 bands at concentrations that activate CHK1, consistent with activation of CDK2. Importantly, the induction of these bands was inhibited at high concentrations of MK-8776 and SRA737 but not LY2606368. Previously, when using CVT-313 to inhibit these bands, we noted the effective concentration ranged from 1.2 to >80 μM;20 the lower values were consistent with inhibition of CDK2. The fact that all of the bands are inhibited at the same concentrations of MK-8776 and SRA737 suggests that the off-target effects are likely due to inhibition of both CDK1 and CDK2.
Figure 6.
Impact of CHK1i on CDK1/2 substrates. MDA-MB-231 cells were incubated with 50 nM gemcitabine for 18 h, removed, and CHK1i added for another 6 h. Cells were lysed and analyzed by Western blotting for the indicated antigens. For the pTPXK blot, the size of molecular weight markers are shown on the left. The molecular weights on the right refer to the target proteins previously observed, together with the concentration of CVT-313 that inhibited their appearance by 50%.20
One target of CDK2 is cyclin E; active CDK2 phosphorylates cyclin E leading to its degradation.21 We also probed the above lysates for cyclin E (Figure 6). Incubation with gemcitabine resulted in accumulation of cyclin E which was then repressed at effective concentrations of each CHK1i. At high concentrations of MK-8776 and SRA737 but not LY2606368, partial recovery of cyclin E occurred, consistent with inhibition of CDK2. Overall, these results strongly suggest that inhibition of CDK2 (and probably CDK1) is the cause of the off-target effects.
Development of Resistance to CHK1i
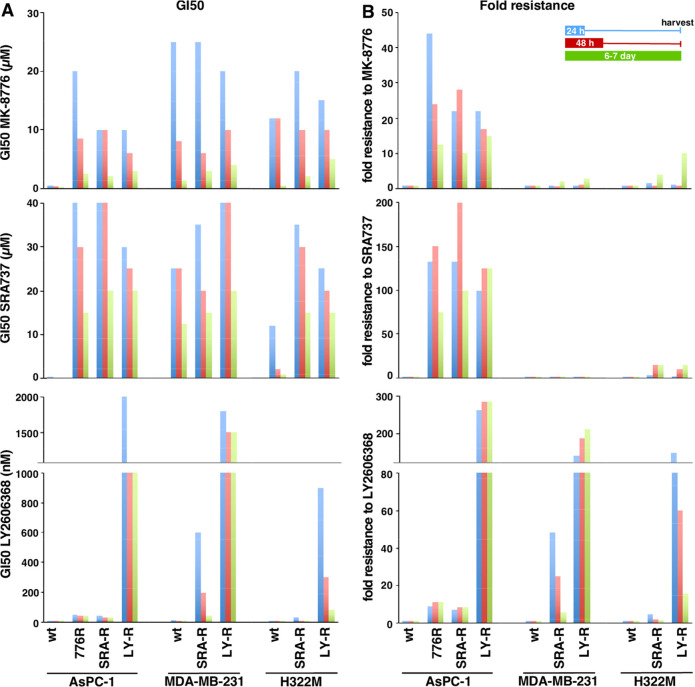
To further compare the three CHK1i, we developed resistance by slowly increasing the drug concentration over 3–6 months (“acquired resistance”). ASPC-1 cells initially exhibited about 10-fold resistance to the drug of selection and this remained stable for several months after removal of drug. Further selection resulted in an increase in resistance to only LY2606368 (Figure 7). The primary question was whether these cells are cross-resistant to the other CHK1i, thereby suggesting a similar target is impacted. Cross-resistance was observed for all three CHK1i. However, AsPC-1 cells selected for resistance to LY2606368, while being cross-resistant to MK-8776 and SRA737, exhibited much greater resistance to LY2606368.
Figure 7.
Sensitivity of cell lines with acquired resistance to CHK1i. AsPC-1 cells were selected for resistance to MK-8776, SRA737, and LY2606368. Similarly, MDA-MB-231 and H322M cells were selected for resistance to SRA737 and LY2606368. Panel A represents the GI50 values; Panel B represents fold resistance. The numerical values for these data are presented in Table S2.
We subsequently attempted to raise resistance to SRA737 and LY2606368 in MDA-MB-231 and H322M cells. Interestingly, selection with SRA737 for 6 months failed to increase resistance. These cell lines were relatively resistant to SRA737 at the outset, and we suspect the cells have difficulty circumventing off-target mediated toxicity. In contrast, MDA-MB-231 and H322M cells selected on LY2606368 developed very high levels of resistance with negligible cross-resistance to the other two CHK1i. This suggests that LY2606368 is selective for CHK1, but higher resistance to the other CHK1i is difficult as concurrent resistance to two critical targets would have a low probability of developing.
To determine whether resistance might be mediated by drug uptake, efflux or inactivation we determined whether each CHK1i can still abrogate SN38-mediated S phase arrest. There was little change in the concentration of each CHK1i required to abrogate arrest in the resistant cells, confirming that the drugs still inhibit CHK1 at low concentrations (Figure 5B). These results demonstrate that acquired resistance is due to inhibition of the single agent activity of each CHK1i, but has no impact on their efficacy in combination with a DNA damaging agent.
Inhibition of Protein Synthesis
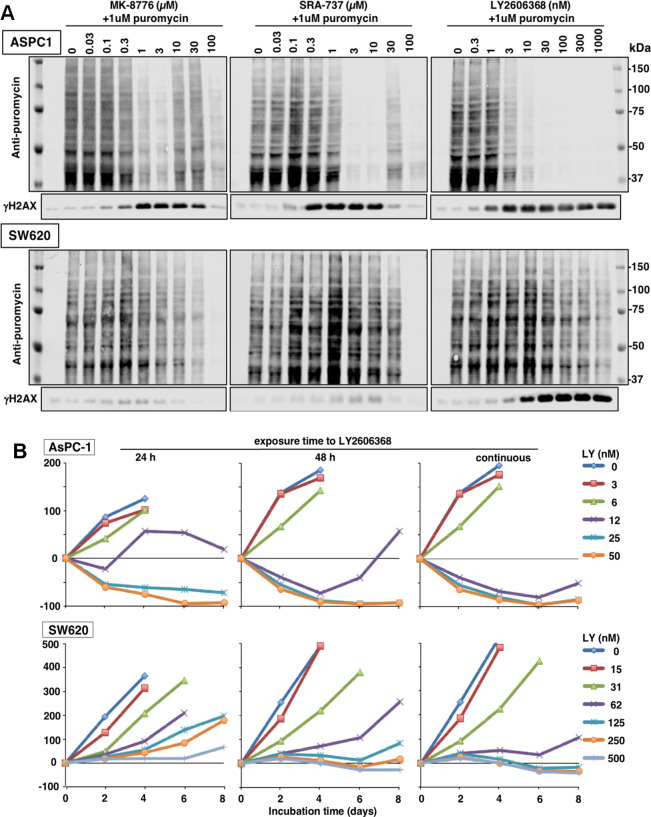
LY2606368 has also been reported to inhibit RSK1 in an in vitro kinase assay (Table 1), but this was ruled out as a target in cells as no inhibition of the RSK1 substrate pS6 (S235/S236) was observed.14 However, S6 is not a unique target of RSK and thus may not adequately report on its inhibition.22 Since the consequence of RSK inhibition is decreased protein synthesis, we assessed the potential inhibition of protein synthesis by adding puromycin 1 h before harvest of cells. The incorporated puromycin was detected by Western blotting.23 AsPC-1 cells revealed a dramatic inhibition of protein synthesis 24 h after addition of all three CHK1i that correlated with the concentrations that inhibited growth and induced γH2AX (Figure 8A). There was no overall change in protein levels during this drug treatment (Figure S3A). Furthermore, the decrease in protein synthesis occurred ∼12 h after the appearance of γH2AX (Figure S3B). Partial protection from inhibition of protein synthesis occurred at intermediate concentrations of MK-8776 and SRA737 consistent with the self-limiting off-target effect of these two drugs. That inhibition of translation was observed with all three compounds at concentrations that inhibited CHK1 suggests this is likely a consequence of the on-target inhibition of CHK1, albeit a late event.
Figure 8.
Inhibition of protein synthesis by CHK1i. (A) ASPC-1 and SW620 cells were incubated with each CHK1i for 24 h. During the final hour, 1 μM puromycin was added to label proteins being synthesized. Cells were analyzed by Western blotting using an antibody to puromycin, followed by fluorescent secondary antibody. Images were generated using a fluorescent scanner. A Coomassie blue stained membrane of the same lysates is shown in Figure S3. (B) AsPC-1 and SW620 cells were incubated with the indicated concentrations of LY2606368 for either 24 h, 48 h, or 8 days. Cells were harvested every 2 days and stained for DNA. A decrease below the starting inoculum reflects cell death.
This experiment was repeated in SW620 cells that are more resistant to CHK1i. Protein synthesis was only inhibited at high concentrations of each drug, although MK-8776 and SRA737 exhibited little γH2AX at any concentration (Figure 8A). In contrast, the more pronounced appearance of γH2AX elicited by LY2606368 correlated with growth inhibition.
Growth inhibition and cell death are not synonymous end points and may explain the poor correlation between γH2AX and inhibition of protein synthesis. While GI50 values only reflect growth inhibition (Figure 1), we have previously demonstrated that AsPC-1 cells can be killed after incubation with ∼1 μM MK-8776.21 This analysis requires that experiments start with ten times more cells such that a decrease from starting inoculum can be assessed.22 This strategy was used here to compare the impact of LY2606368 in AsPC-1 and SW620 cells.
Cells were exposed to LY2606368 for either 24 h, 48 h, or continuously and harvested every 2 days (Figure 8B). Following a 24 h drug exposure, the AsPC-1 cells were dying by day 2 at concentrations in excess of 12 nM. This concentration is about 4-fold higher than the GI50 (Figure 1). In contrast, the SW620 cells, which had a GI50 of 18 nM (Figure 1), appeared to remain viable throughout, even at high concentrations. Hence, it appears that γH2AX may correlate with growth inhibition in SW620 cells but does not predict that the cells will die.
Discussion
CHK1 inhibitors were initially developed to sensitize cancer cells to DNA damaging agents. While the combination of gemcitabine plus CHK1i became the predominant combination tested clinically, the discovery of a large range of sensitivity to single agent CHK1i stimulated research to define the underlying mechanism so patients could be appropriately stratified for clinical trials. This goal has still not been realized with many mechanisms of sensitivity having been proposed.9 We noted in our recent review that the proportion of cells considered sensitive to CHK1i varied with the drug as well as the threshold defining sensitivity in each publication.9 In addition, different toxicities have been observed in patients, and whether this is due to on-target or off-target effects remains unknown. Here, we compared three CHK1i selected because of their clinical experience.
MK-8776 and SRA737 had very similar activities in all the assays used, and both appeared to have an off-target effect at 30–100-fold higher concentrations than those that inhibited CHK1. While very sensitive cells were not impacted by this off-target effect, it became important for more resistant cells as it appeared to protect them from CHK1 inhibition, at least for a limited time period. This was evident in the biphasic growth curves, and the increased growth inhibition observed following longer drug exposures. However, the short exposures are more relevant to the administration schedule in patients (see ref (24) for critique).
Cells sensitive to all three CHK1i were insensitive to the time of exposure to drug, with extensive γH2AX detected in S phase cells within 6 h. However, cells that were slightly more resistant to LY2606368 were significantly more resistant to MK-8776 and SRA737. We surmise that this difference relates to the off-target effect that protects cells from MK-8776 and SRA737. Whether the protective effect elicited by these drugs could provide an advantage in limiting toxicity to a patient while still killing a very sensitive tumor remains to be established.
Inhibition of CHK1 results in active CDK2 that is required for progression through S phase, both to initiate firing of replication forks, and to restart replication forks that have stalled on damaged DNA. However, this mechanism does not appear critical for the single agent activity of CHK1i. For example, cells resistant to CHK1i (both constitutive and acquired resistance) still abrogated S phase arrest. The sensitivity to single agent CHK1i requires high CDK2 activity that is erroneously activated in S phase. The resulting γH2AX can be inhibited by low concentrations of the CDK2 inhibitor CVT-313 and by suppression of cyclin A.4,19 In contrast, S phase progression on damaged DNA is only inhibited at high concentrations of CVT-313. Hence, the different response to single agent versus combination activity depends on different activity thresholds of CDK2 and hence different CDK2 substrates.9 These different thresholds likely depend on the different cyclin (A or E) to which CDK2 is complexed. Resistant cells prevent the hyperactivity of CDK2 while in S phase but permit low activity CDK2, presumably complexed to cyclin E, that is required for ongoing replication.
Considering that higher concentrations of both MK-8776 and SRA737 inhibited γH2AX, it is possible that the off-target effect observed is through inhibition of CDK2. MK-8776 reportedly inhibits CDK2 at 50-fold higher concentrations than inhibit CHK1,13 and this is consistent with the concentration difference that inhibits γH2AX. These higher concentrations also inhibited S phase abrogation which can also be attributed to inhibition of CDK2. Concurrent inhibition of pHH3 also suggests inhibition of CDK1. Most CDK1/2 inhibitors elicit little selectivity for CDK1 or CDK2 so it is possible that MK-8776 inhibits both at these higher concentrations.
In contrast, SRA737 was reported to require 2000-fold higher concentrations to inhibit CDK2 and CDK1 compared to CHK1,15 yet in all the assays presented here, it performed like MK-8776, suggesting that it also inhibits CDK2 at only 50-fold higher concentrations. This conclusion is clearly supported by the observed inhibition of phosphorylation at the CDK2 consensus site TPXK (Figure 6). Furthermore, CDK1/2 phosphorylates S286/S301-CHK1 facilitating phosphorylation at 345-CHK1.25 This could explain the decrease in p345-CHK1 observed at high concentrations of MK-8776 and SRA737. There may be a second off-target effect at the highest concentrations as inhibition of CDK1/2 can no longer protect the cells, albeit inhibitors of CDK1/2 also kill cells;26 perhaps the difference between protection and killing relates to the same target but different drug exposure times.
The development of acquired resistance provided additional information on the off-target effects. We were able to develop low levels of resistance to MK-8776 and SRA737 in the sensitive AsPC-1 cells but not in the constitutively resistance MDA-MB-231 and H322M cells. In contrast, high levels of resistance to LY2606368 were obtained in all three cell lines. Importantly, this resistance was only to single agent CHK1i, and did not impede sensitivity to the drug combination, demonstrating that CHK1i still accessed its target. The relatively facile development of resistance to LY2606368 suggests only a single target is involved, whereas it is presumably more difficult to generate resistance to multiple targets, as seems to be the case with MK-8776 and SRA737.
Two recent papers have also addressed resistance to LY2606368. Nair et al.27 suggested that resistance occurs through decreased expression of cyclin B and thereby decreased CDK1 activity in G2, preventing mitotic catastrophe. Unfortunately, this explanation does not explain the dramatic accumulation of cells they observe in S phase upon incubation with LY2606368 (as we show in Figures 4 and S1), nor how this is circumvented in their resistant cells. Li et al.28 performed a CRISPR screen that identified loss of FAM122A as a mediator of resistance to LY2606368. FAM122A is an inhibitor of protein phosphatase 2A which, when phosphorylated by CHK1, leads to active PP2A, dephosphorylation and stabilization of WEE1, thereby preventing CDK1 activity. While they did not address the impact of this pathway on CDK2, it may be relevant to our observations on the critical role of CDK2/cyclin A in eliciting sensitivity to CHK1i.
One important lesson from this study is that the published in vitro kinase assays are poor predictors of concentrations that inhibit a kinase in cells. For example, while all three CHK1i exhibited similar potency in vitro, when added to cells, MK-8776 and SRA737 required 100-fold higher concentrations to inhibit CHK1, while LY2606368 is almost as potent in cells as in vitro; this could be due to different cell permeability for each drug or differences in the vitro kinase assays performed in different laboratories. In addition, only MK-8776 was predicted to inhibit CDK2, yet our data suggest that SRA737 may also inhibit CDK2. Kinase assays also suggested that LY2606368 had the least selectivity for CHK1 over CHK2, yet all three drugs inhibited CHK2 in cells, and LY2606368 exhibited the greater selectivity for CHK1. The fact that LY2606368 also inhibits CHK2 but does not impede sensitivity to the CHK1i effect suggests CHK2 is not the source of the off-target effects observed.
Intriguingly, all three CHK1i inhibited protein synthesis in a sensitive cell line at concentrations that induced γH2AX and killed cells. However, in a constitutively resistant cell line (SW620), γH2AX was still induced by LY2606368 but with little inhibition of protein synthesis, and the cells underwent cytostasis rather than cell death. Perhaps continuing protein synthesis facilitates DNA repair and survival. Hence for a cell to die, perhaps protein synthesis needs to be inhibited in addition to the induction of DNA damage. Inhibition of protein synthesis does not appear to be simply a consequence of DNA damage but whether it is a cause or consequence of cell death will be the subject of future studies.
One important observation here and in prior studies6,7 is that the ability to enhance the cytotoxicity induced by DNA damaging agents is independent of the sensitivity to the single agent activity of CHK1i. A recent report has demonstrated that gemcitabine plus SRA737 has clinical efficacy in some tumors.12 Whether the off-target effect of SRA737 limits the toxicity of the combination may be worth considering.
When administered to patients, all three compounds are reported to achieve a plasma concentration that exceeds that predicted to be effective from preclinical models (5 μM MK-8776;29 6 μM SRA737;11 1–2 μM LY260636830). However, these concentrations decrease rapidly to 0.5 μM MK-8776 by 6 h, 1 μM SRA737 by 24 h, and 20 nM LY2606368 by 24 h. More important are the concentrations in the tumor. This was assessed in one murine model with SRA737 where 10–25-fold higher concentrations were observed.10 Considering the lipophilic nature of all three CHK1i, it is likely that the tissue concentrations are much higher than the plasma concentrations. Hence, it is possible that both the on-target and off-target effects reported here can be engaged in patient tumors.
Preclinical experiments in xenografts have shown impressive single agent activity in some tumors.31,32 The fact that the development of LY2606368 was terminated due to toxicity is unfortunate because it suggests the recommended phase II dose was too high. Additionally, further analysis of the schedule may improve outcome. The best xenograft regression resulted from LY2606368 administered twice daily for 3 days. Such scheduling may be difficult unless an oral CHK1i is available. SRA737 has the advantage of being oral and may still have therapeutic activity if administered in repeated low dose to patients with sensitive tumors. Unfortunately, further development of SRA737 is currently on hold. We have noted recent clinical development of another oral CHK1i, LY2880070 (www.clinicaltrials.gov), although no preclinical data are available. It will be important to ensure this novel drug, and any future CHK1i, have undergone extensive analyses similar to that presented here to define on-target and off-target effects.
Methods
Cell Culture
Most cell lines were obtained from the Developmental Therapeutics Program, National Cancer Institute, as part of the NCI60 cell line panel. Other cell lines were obtained from the American Type Culture Collection. Cells were expanded and stored at low passage number and cultures were replaced approximately every 3 months. Cells were maintained in RPMI1640 (Corning/Mediatech), 10% fetal bovine serum (Hyclone), and 1% antibiotic/antimycotic (Gibco). Cell lines were tested for mycoplasma using the MycoAlert Mycoplasma Detection Kit (Lonza).
Chemicals
MK-8776 (Merck) and SRA737 (Sierra Oncology) were stored at 10 and 100 mM solutions in DMSO. LY2606368 (Selleckchem) was stored at 0.1 and 1 mM in DMSO. CHK2 inhibitor II was obtained from Sigma. When added to cell cultures, there was no indication of drug precipitation at the high concentrations used.
Growth Inhibition
Cells (100 μL) were plated at 500–2000/cells per well of a 96-well plate (depending on growth rate). The following day, drugs were added at 2-fold dilutions (8 wells per concentration). After 24 or 48 h, drug was removed; wells were washed with phosphate buffered saline, and fresh media was added. Alternately, drugs were left on the cells until harvest. Cells were harvested on day 6 or 7 when close to confluence. Plates were washed in phosphate buffered saline and stored at −80 °C. Cells were lysed and DNA was stained with Hoechst 33258 as previously described.5,33 Fluorescence was read on a microplate spectrofluorometer. Values for each concentration were averaged (technical SD ∼ 10% of the average) and graphed in Excel. Results were expressed as the concentration that inhibited growth by 50% (GI50), derived from the values that bisect the 50% point of each curve. For MK-8776, this screen replicated a prior study but with concentrations extended to 100 μM.4 SRA737 was also tested up to 100 μM, while LY2606368 was tested up to 1 μM. As the starting cell number in this assay is low, an observation of no growth can not distinguish cytostasis from cytotoxicity. To assess true cytotoxicity, cells were plated at 10 000 cells/well and followed for 8 days. This facilitated assessment of the decrease in cell number below the starting inoculum and therefore scores cell death.24 In this modified assay, cells incubated at low concentrations of drug will saturate each well limiting or even decreasing the apparent growth.
Cell Cycle
Cell cycle analysis was conducted by flow cytometry using propidium iodide.6 As required, cells were labeled concurrently with Alexa 488-conjugated anti-γH2AX and Alexa 647-conjugated anti-pHH3 (Cell Signaling Technology). Cells were analyzed on a Becton Dickinson Gallios flow cytometer.
Acquired Resistance to Chk1 Inhibitors
Cells were incubated in T25 flasks with a range of concentrations of each CHK1i above the GI50. After 72 h, the drugs were removed and the cells recovered in fresh media. This incubation time was selected to ensure that the surviving cells were not simply those that had not entered S phase during the incubation time. After cells recovered (3–14 days), they were replated and incubated at the same or higher drug concentrations. This process was repeated over 3–4 months with increasing concentrations as the cells became more resistant.
Western Blotting
Cells were rinsed in PBS, lysed in Laemmli lysis buffer, and boiled for 5 min. Proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Western blotting was performed with the following primary and secondary antibodies: Cell Signaling Technology: pS296-CHK1 (2349S), pS345-CHK1 (2348S), pT68-CHK2 (2197S), pS516-CHK2 (2669S), γH2AX (9718S), pTPXK (14371S), cyclin E (20808S), mouse IgG-DyLight 800 (5257), rabbit IgG-DyLight 800 (5151), mouse IgG-DyLight 680 (5470). Santa Cruz Biotechnology: CHK1 (sc8408), vinculin (sc073541). EMD Millipore: CHK2 (05-649), puromycin (MABE343). Abcam: RPA (ab2175). Images were generated using a Licor Odyssey imager and processed using Image Studio Lite.
Acknowledgments
We thank W. Davis for help with H322M experiments and the NCCC Immunology and Flow Cytometry Shared Resource for facilitating the acquisition of flow cytometry data.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.0c00201.
Author Contributions
J.P.D. and A.E. designed and performed all of the experiments. J.P.D. performed protein analyses. A.E. performed flow cytometry. J.P.D. and A.E. wrote the manuscript.
This work was supported by grant CA117874 from the National Cancer Institute and a Cancer Center Support Grant to the Norris Cotton Cancer Center (CA23108). Research funds were also provided by Sierra Oncology.
The authors declare the following competing financial interest(s): Partial financial support for the research was provided by Sierra Oncology.
Supplementary Material
References
- Thompson R.; Eastman A. (2013) The cancer chemotherapeutic potential of Chk1 inhibitors: how mechanistic studies impact clinical trial design. Br. J. Clin. Pharmacol. 76, 358–369. 10.1111/bcp.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H.; Naka K.; Okada Y.; Watanabe M.; Harada N.; Saito S.; Anderson C. W.; Appella E.; Nakanishi M.; Suzuki H.; Nagashima K.; Sawa H.; Ikeda K.; Motoyama N. (2002) Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 21, 5195–5205. 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squatrito M.; Brennan C. W.; Helmy K.; Huse J. T.; Petrini J. H.; Holland E. C. (2010) Loss of ATM/Chk2/p53 pathway components accelerates tumor development and contributes to radiation resistance in gliomas. Cancer Cell 18, 619–629. 10.1016/j.ccr.2010.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurikar N.; Thompson R.; Montano R.; Eastman A. (2016) A subset of cancer cell lines is acutely sensitive to the Chk1 inhibitor MK8776 as monotherapy due to CDK2 activation in S phase. Oncotarget 7, 1380–1394. 10.18632/oncotarget.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montano R.; Chung I.; Garner K. M.; Parry D.; Eastman A. (2012) Preclinical development of the novel Chk1 inhibitor SCH900776 in combination with DNA damaging agents and antimetabolites. Mol. Cancer Ther. 11, 427–438. 10.1158/1535-7163.MCT-11-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montano R.; Thompson R.; Chung I.; Hou H.; Khan N.; Eastman A. (2013) Sensitization of human cancer cells to gemcitabine by the Chk1 inhibitor MK-8776: cell cycle perturbation and impact of administration schedule in vitro and in vivo. BMC Cancer 13, 604. 10.1186/1471-2407-13-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montano R.; Khan N.; Hou H.; Seigne J.; Ernstoff M. S.; Lewis L. D.; Eastman A. (2017) Cell cycle perturbation induced by gemcitabine in human tumor cell culture, xenografts and bladder cancer patients: implications for clinical trial designs combining gemcitabine with a Chk1 inhibitor. Oncotarget 8, 67754–67768. 10.18632/oncotarget.18834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samadder P.; Suchankova T.; Hylse O.; Khirsariya P.; Nikulenkov F.; Drapela S.; Strakova N.; Vanahara P.; Vaickova K.; Kolarova H.; Bino L.; Bittova M.; Ovesna P.; Kollar P.; Fedr R.; Esner M.; Jaros J.; Hampl A.; Krejci L.; Paruch K.; Soucek K. (2017) Synthesis and profiling of a novel potent selective inhibitor of CHK1 kinase possessing unusual N-trifluoromethylpyrazole pharmacophore resistant to metabolic N-dealkylation. Mol. Cancer Ther. 16, 1831–1842. 10.1158/1535-7163.MCT-17-0018. [DOI] [PubMed] [Google Scholar]
- Warren N. J. H.; Eastman A. (2020) Comparison of the different mechanisms of cytotoxicity induced by checkpoint kinase 1 inhibitors when used as single agents or in combination with DNA damage. Oncogene 39, 1389–1401. 10.1038/s41388-019-1079-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton M. I.; Eve P. D.; Hayes A.; Henley A. T.; Valenti M. R.; De Haven Brandon A. K.; Box G.; Boxall K. J.; Tall M.; Swales K.; Matthews T. P.; McHardy T.; Lainchbury M.; Osborne J.; Hunter J. E.; Perkins N. D.; Aherne G. W.; Reader J. C.; Raynaud F. I.; Eccles S. A.; Collins I.; Garrett M. D. (2016) The clinical development candidates CCT245737 is an orally active CHK1 inhibitor with preclinical activity in RAS mutant NSCLC and Eμ-MYC driven B-cell lymphoma. Oncotarget 7, 2329–2342. 10.18632/oncotarget.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer E. R.; Kristeleit R. S.; Cojocaru E.; Haris N. M.; Carter L.; Jones R. H.; Blagden S. P.; Evans T.R. J.; Arkenau H.-T.; Sarker D.; Danson S.; Symeonides S. N.; Walter H.; Ocen J.; Randhawa M.; Kowalski M. M.; Verdon I.; Dye A.; Banerji U. (2019) A first-in-human phase I/II trial of SRA737 (a Chk1 inhibitor) in subjects with advanced cancer. J. Clin. Oncol. 37, 3094. 10.1200/JCO.2019.37.15_suppl.3094. [DOI] [Google Scholar]
- Banerji U.; Plummer E. R.; Moreno V.; Ang J. E.; Quinton A.; Drew Y.; Hernandez T.; Roda D.; Carter L.; Navarro A.; Kristeleit R. S.; Arkenau H. T.; Sarker D.; Castellano D. E.; Walter H.; Roxburgh P.; Blagden S. P.; Anthoney A.; Verdon I.; Jones R. H. (2019) A phase I/II first--in-human trial of oral SRA737 (a Chk1 inhibitor) given in combination with low-dose gemcitabine in subjects with advanced cancer. J. Clin. Oncol. 37, 3095. 10.1200/JCO.2019.37.15_suppl.3095. [DOI] [Google Scholar]
- Guzi T.; Paruch K.; Dwyer M. P.; Labroli M.; Shanahan F.; Davis N.; Taricani L.; Wiswell D.; Seghezzi W.; Penaflor E.; Bhagwat B.; Wang W.; Gu D.; Hsieh Y.; Lee S.; Liu M.; Parry D. (2011) Targeting the replication checkpoint using SCH 900776, a potent and functionally selective CHK1 inhibitor identified via high content functional screening. Mol. Cancer Ther. 10, 591–602. 10.1158/1535-7163.MCT-10-0928. [DOI] [PubMed] [Google Scholar]
- King C.; Diaz H..B.; McNeely S.; Barnard D.; Dempsey J.; Blosser W.; Beckmann R.; Barda D.; Marshall M. S. (2015) LY2606368 causes replication catastrophe and antitumor effects through CHK1-dependent mechanisms. Mol. Cancer Ther. 14, 2004–2013. 10.1158/1535-7163.MCT-14-1037. [DOI] [PubMed] [Google Scholar]
- Collins I., and Garrett M. D. (2018) Preclinical profiles and contexts for CHK1 and CHK2 inhibitors. In Targeting the DNA Damage Response for Anti-Cancer Therapy; Cancer Drug Discovery and Development (Pollard J., and Curtin N.,, Eds.) pp 241–276, Humana Press, Totowa, NJ. [Google Scholar]
- Leung-Pineda V.; Ryan C. E.; Piwnica-Worms H. (2006) Phosphorylation of Chk1 by ATR is antagonized by a Chk1-regulated protein phosphatase 2A circuit. Mol. Cell. Biol. 26, 7529–7538. 10.1128/MCB.00447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarts M.; Sharpe R.; Garcia-Murillas I.; Gevensleben H.; Hurd M. S.; Shumway S. D.; Toniatti C.; Ashworth A.; Turner N. C. (2012) Forced mitotic entry of S phase cells as a therapeutic strategy induced by inhibition of WEE1. Cancer Discovery 2, 524–539. 10.1158/2159-8290.CD-11-0320. [DOI] [PubMed] [Google Scholar]
- Ruiz S.; Mayor-Ruiz C.; Lafarga V.; Murga M.; Vega-Sendino M.; Ortega S.; Fernandez-Capetillo O. (2016) A genome-wide CRISPR screen identifies CDC25A as a determinant of sensitivity to ATR inhibitors. Mol. Cell 62, 307–313. 10.1016/j.molcel.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren N. J. H.; Donahue K.; Eastman A. (2019) Differential sensitivity to CDK2 inhibition discriminates the molecular mechanisms of CHK1 inhibitors as monotherapy or in combination with the topoisomerase I inhibitor SN38. ACS Pharmacol. Transl. Sci. 2, 168–182. 10.1021/acsptsci.9b00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren N. J. H.; Eastman A. (2019) Inhibition of checkpoint kinase 1 following gemcitabine-mediated S phase arrest results in CDC7- and CDK2-dependent replication catastrophe. J. Biol. Chem. 294, 1763–1778. 10.1074/jbc.RA118.005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurikar N.; Eastman A. (2016) Critical reanalysis of the methods that discriminate the activity of CDK2 from CDK1. Cell Cycle 15, 1184–1188. 10.1080/15384101.2016.1160983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roffe M.; Lupinacci F. C.; Soares L. C.; Hajj G. N.; Martins V. R. (2015) Two widely used RSK inhibitors, BI-D1870 and SL101, alter mTORC1 signaling in a RSK-independent manner. Cell. Signalling 27, 1630–1642. 10.1016/j.cellsig.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Schmidt E. K.; Clavarino G.; Ceppi M.; Pierre P. (2009) SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277. 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- Payton M.; Chung G.; Yakowec P.; Wong A.; Powers D.; Xiong L.; Zhang N.; Leal J.; Bush T. L.; Santora V.; Askew B.; Tasker A.; Radinsky R.; Kendall R.; Coats S. (2006) Discovery and evaluation of dual CDK1 and CDK2 inhibitors. Cancer Res. 66, 4299–4308. 10.1158/0008-5472.CAN-05-2507. [DOI] [PubMed] [Google Scholar]
- Nair J.; Huang T.-T.; Murai J.; Haynes B.; Steeg P. S.; Pommier Y.; Lee J.-M. (2020) Resistance to the CHK1 inhibitor prexasertib involves functionally distinct activities in BRCA wild-type ovarian cancer. Oncogene 39, 5520–5535. 10.1038/s41388-020-1383-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.; Kozono D.; Deraska P.; Branigan T.; Dunn C.; Zheng X. F.; Parmar K.; Nguyen H.; DeCaprio J.; Shapiro G. I.; Chowdhury D.; D’Andrea A. D. (2020) Chk1 inhibitor blocks phosphorylation of FAM122A and promotes replication stress. Mol. Cell 80, 410–422. 10.1016/j.molcel.2020.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp J. E.; Thomas B. M.; Greer J. M.; Sorge C.; Gore S. D.; Pratz K. W.; Smith B. D.; Flatten K. S.; Peterson K.; Schneider P.; Mackey K.; Freshwater T.; Levis M. J.; McDevitt M. A.; Carraway H. E.; Gladstone D. E.; Showel M. M.; Loechner S.; Parry D. A.; Horowitz J. A.; Isaacs R.; Kaufmann S. H. (2012) Phase I and pharmacologic trial of cytosine arabinoside with the selective checkpoint 1 inhibitor Sch 900776 in refractory acute leukemias. Clin. Cancer Res. 18, 6723–6731. 10.1158/1078-0432.CCR-12-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D.; Infante J.; Janku F.; Jones S.; Nguyen L. M.; Burris H.; Naing A.; Bauer T. M.; Piha-Paul S.; Johnson F. M.; Kurzrock R.; Golden L.; Hynes S.; Lin J.; Lin A. B.; Bendell J. (2016) Phase I study of LY2606368, a checkpoint kinase 1 inhibitor, in patients with advanced cancer. J. Clin. Oncol. 34, 1764–1771. 10.1200/JCO.2015.64.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery C. D.; VanWye A. B.; Dowless M.; Blosser W.; Falcon B. L.; Stewart J.; Stephens J.; Beckmann R. P.; Bence Lin A.; Stancato L. F. (2017) The checkpoint kinase 1 inhibitor prexasertib induces regression of preclinical models of human neuroblastoma. Clin. Cancer Res. 23, 4354–4363. 10.1158/1078-0432.CCR-16-2876. [DOI] [PubMed] [Google Scholar]
- Lowery C. D.; Dowless M.; Renschler M.; Blosser W.; VanWye A. B.; Stephens J. R.; Iversen P. W.; Lin A. B.; Beckmann R. P.; Krytska K.; Cole K. A.; Maris J. M.; Hawkins D. S.; Rubin B. P.; Kurmasheva R. T.; Houghton P. J.; Gorlick R.; Kolb E. A.; Kang M. H.; Reynolds C. P.; Erickson S. W.; Teicher B. A.; Smith M. A.; Stancato L. F. (2019) Broad spectrum activity of the checkpoint kinase 1 inhibitor prexasertib as a single agent or chemopotentiator across a range of preclinical pediatric tumor models. Clin. Cancer Res. 25, 2278–2289. 10.1158/1078-0432.CCR-18-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao J.; Otto W. R. (1992) Fluorometric DNA assay for cell growth estimation. Anal. Biochem. 207, 186–192. 10.1016/0003-2697(92)90521-8. [DOI] [PubMed] [Google Scholar]
- Eastman A. (2017) Improving anticancer drug development begins with cell culture: misinformation perpetrated by the misuse of cytotoxicity assays. Oncotarget 8, 8854–8866. 10.18632/oncotarget.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N.; Libertini S.; Black E. J.; Lao Y.; Hegarat N.; Walker M.; Gillespie D. A. (2012) Cdk-mediated phosphorylation of Chk1 is required for efficient activation and full checkpoint proficiency in response to DNA damage. Oncogene 31, 1086–1094. 10.1038/onc.2011.310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.