Abstract

For the first time, it is shown that severe COVID-19 associated genes (CCR2, DPP9, HSPA1L, TYK2, OAS1, ACE2, and TMPRSS2) were also upregulated in the brain tissue of chronic alcoholics. It is proposed that chronic alcohol abuse should be considered as an important factor in COVID-19-associated neurological complications.
Keywords: SARS-CoV-2, COVID-19, alcoholism, neurological manifestations
The clinical manifestations of SARS-CoV-2 infection are highly variable. Whereas the majority of infected individuals remain asymptomatic, many patients show mild to moderate symptoms associated with coronavirus disease 2019 (COVID-19). However, others also develop severe pneumonia and demonstrate the involvement of the central nervous system leading to neurological complications. Among underlying risk factors, ethnicity, age, underlying diseases, obesity, and the microbiome have been discussed. More recently, a history of excessive and chronic alcohol consumption has been suggested to correlate with an increased risk of severe COVID-19;1 however, there is no evidence that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes COVID-19, has severe effects on the brain of alcoholics. For the first time, using in silico analysis, we present a genetic predisposition of SARS-CoV-2-induced brain pathology in chronic alcohol drinkers.
A genome-wide association study identified five genes (CCR2, DPP9, IFNAR2, OAS1, and TYK2) linked to immune system response to SARS-CoV-2 infection in hospitalized COVID-19 patients exhibiting disease severity.2 Additionally, the HSPA1L gene was of interest as it is involved in SARS-CoV-2 modulation of host epigenetic regulation, leading to increased viral replication in the host cells.3 Along with these, we included in our candidate gene list genes related to SARS-CoV-2 host cell entry (ACE2 and TMPRSS2). Next, transcriptomic data sets available from the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) were explored. First, we selected a data set (i.e., GSE150316), which included the spectrum of viral load and host response seen in autopsies of SARS-CoV-2 infected lungs. In this data set, at least two tissue sections from areas devoid of acute inflammation were used per sample, and 11 different lung samples from autopsies of severe COVID-19 patients were utilized. Second, we selected the GSE53808 data set, which consisted of autopsy samples of brain white matter from individuals with a history of chronic alcohol consumption (lifelong intake > 80 g/day) without a pathological diagnosis of hepatic encephalopathy versus controls (lifelong intake < 20 g/day). For both the data sets, the gene expression profiles were downloaded, and the raw data were processed using R statistical software (version 3.5.1). According to the expression profiling data, relative mRNA expression in infected/alcoholics versus healthy controls was identified using the Limma package (available at http://www.bioconductor.org/packages/release/bioc/html/limma.html) in Bioconductor (package version 1.0.2). A (log 2)-fold-change (log2FC) was calculated to present differentially expressed genes, and an adjusted p-value < 0.05 using classical t-test was applied.
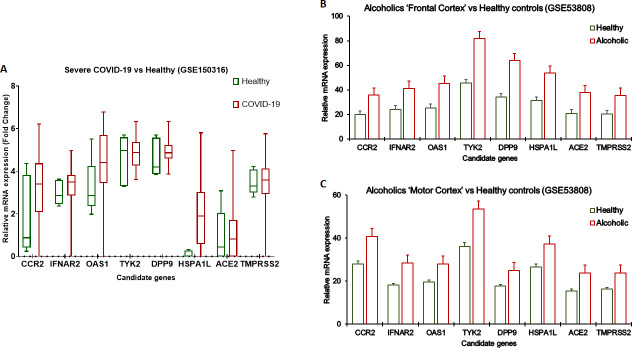
Our analysis showed a higher expression of all eight candidate genes in severe COVID-19 cases [Figure 1A]. Furthermore, it was found that the aforementioned genes were significantly upregulated in the white matter brain tissues of chronic alcoholics compared to nonalcoholics. For the first time, it is shown that severe COVID-19 associated genes (CCR2, DPP9, HSPA1L, TYK2, OAS1, ACE2, and TMPRSS2) were also upregulated in the brain tissue of chronic alcoholics [Figure 1B,C]. CCR2 and TYK2 are key drivers of inflammation, and OAS1 and HSPA1L are associated with increased susceptibility to viral infection. The function of DPP9 in SARS-CoV-2 is yet to be characterized. In contrast, we found a higher expression of IFNAR2, which encodes interferon receptors, in severe COVID as well in alcoholics, but a lower expression of this gene was reported to be linked with COVID-19 severity.2
Figure 1.
Transcriptomic analysis using publicly available databases from NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). (A) Boxplot showing analysis of GSE150316 data set containing RNaseq data obtained from autopsy samples from severe COVID-19 patients (n = 11) compared to healthy controls (n = 5). (B and C) Bar graphs showing analysis of GSE53808 data set containing expression array data obtained from brain white matter tissues from alcoholics without hepatic encephalopathy (n = 4) and neurologically normal controls (n = 3). Data is presented as mean ± SE in error bars.
On the basis of these genetic correlations, we believe that chronic alcoholics are at very high risk of developing severe neurological manifestations if exposed to SARS-CoV-2. These findings suggest that targeting these molecules could prevent the development of severe symptoms; however, extensive research is needed to realize these expectations. Regardless, there is a need for increased public awareness about the use of excessive alcoholic beverages and having a higher probability of developing severe brain damage due to COVID-19.
Author Contributions
R.S. and N.A.K. envisioned the concept amid critical discussions with J.S.M. J.S.M. reviewed the literature and prepared the first draft of the article and the figures under the supervision of N.A.K. All authors contributed to the manuscript and will act as guarantors.
The authors declare no competing financial interest.
References
- Testino G. (2020) Are patients with alcohol use disorders at increased risk for COVID-19 infection?. Alcohol Alcohol. 55 (4), 344–346. 10.1093/alcalc/agaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E.; Clohisey S.; Klaric L.; Bretherick A. D.; Rawlik K.; Pasko D.; Walker S.; Parkinson N.; Fourman M. H.; Russell C. D. (2021) Genetic mechanisms of critical illness in COVID-19. Nature 591, 92–98. 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- Muhammad J. S.; Saheb Sharif-Askari N.; Cui Z.-G.; Hamad M.; Halwani R. (2021) SARS-CoV-2 infection-induced promoter hypomethylation as an epigenetic modulator of heat shock protein A1L (HSPA1L) gene. Front. Genetics 12 (129), 622271. 10.3389/fgene.2021.622271. [DOI] [PMC free article] [PubMed] [Google Scholar]