Abstract
Introduction
Standard testing for infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is based on RT-PCR tests, but detection of viral genetic material alone does not indicate ongoing infectious potential. The ability to isolate whole virus represents a better proxy for infectivity.
Aim
The objective of this study was to gain an understanding of the current literature and compare the reported periods of positive SARS-CoV-2 detection from studies that conducted RT-PCR testing in addition to experiments isolating whole virus.
Methods
Using a rapid review approach, studies reporting empirical data on the duration of positive RT-PCR results and/or successful viral isolation following SARS-CoV-2 infection in humans were identified through searches of peer-reviewed and pre-print health sciences literature. Articles were screened for relevance, then data were extracted, analysed, and synthesised.
Results
Of the 160 studies included for qualitative analysis, 84% (n = 135) investigated duration of positive RT-PCR tests only, 5% (n = 8) investigated duration of successful viral isolations, while 11% (n = 17) included measurements on both. There was significant heterogeneity in reported data. There was a prolonged time to viral clearance when deduced from RT-PCR tests compared with viral isolations (median: 26 vs 9 days).
Discussion
Findings from this review support a minimum 10-day period of isolation but certain cases where virus was isolated after 10 days were identified. Given the extended time to viral clearance from RT-PCR tests, future research should ensure standard reporting of RT-PCR protocols and results to help inform testing policies aimed at clearance from isolation.
Keywords: COVID-19, SARS-CoV-2, transmission, communicable, duration, rapid review
Introduction
Understanding how long individuals may continue to transmit virus after infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is important to inform testing policies and isolation procedures required to prevent nosocomial and community spread. Numerous studies have been conducted to date to address this question; however, the literature has been predominantly represented by studies using methods based on reverse transcription-PCR (RT-PCR) tests. As RT-PCR tests detect the presence of viral genetic material and thus do not differentiate between live (or viable) and non-infective virus [1], inferences from these results as to potential infectious periods are limited.
A more accurate proxy of infectious potential is based on the ability of whole virus to be successfully isolated and cultured in laboratory settings, with clinical confirmation of transmission where possible. While fewer studies have been conducted that have successfully isolated and cultured live virus, results from this literature have contributed to a number of reviews [2-7]. The findings from these reviews have drawn consistent conclusions that align with previously recommended isolation strategies [8]: overall, infectious potential appears to be greatly reduced by day 10 following symptom onset of coronavirus disease (COVID-19) [2,7,9].
However, a growing concern relates to individuals who continue to test positive by RT-PCR over extended periods of time [10-16], including those who re-test positive after an initial negative test result [10]. As RT-PCR tests of respiratory samples have been and will remain the de facto method of confirming initial and ongoing infection with SARS-CoV-2, it is important to be able to interpret positive test results that are obtained throughout the disease course, including during convalescence. Against a backdrop of dramatically rising cases internationally, limited testing resources and public health capacity for ongoing case management, and ongoing restrictions on mobility, there is a need for as much evidence as possible to help understand the likely implication of ongoing positive tests or re-tests after cessation of the recommended period of isolation, on potential disease spread.
The purpose of this review was to conduct a rapid review of existing literature in order to directly compare the duration of potential infectivity of SARS-CoV-2 from studies that obtained measurements of the duration of infectious potential using both RT-PCR and viral isolation methods. An additional, and related, objective was to understand and be able to provide an overview of the literature at the time the review was conducted (until end of September 2020), especially given the evolving nature of the pandemic.
Methods
We conducted a rapid review of the literature using the methods outlined in the National Collaborating Centre for Methods and Tools Guidebook [17]. We used the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines to report our rapid review [18]. We did not publish or pre-register a protocol for our review given time pressures for this information. From the available data, duration (maximums, medians or means values) was retrieved or calculated. Ethics approval was not required.
Search strategy
Databases of peer-reviewed and pre-print health sciences literature (Ovid MEDLINE, Embase, Google Scholar, medRxiv and arXiv) and the grey literature for reports or guidelines on discontinuation of isolation for SARS-CoV-2 from international and national public health organisations (World Health Organization, European Centre for Disease Prevention and Control, United States Centre for Disease Control websites) were searched using two search strategies: (i) terms for ‘SARS-CoV-2’ and ‘viral clearance/shedding,’ on 23 May 2020 and (ii) terms for ‘SARS-CoV-2’ and ‘viral isolation/culture,’ on 1 July 2020. The latter search was done with specific terms because there was a paucity of studies with data from viral isolation/cultures identified in the previous iteration of the search; moreover, in order to include more recently published studies, we examined other reviews, including a living evidence review, until 29 September 2020 and added relevant references to our results [1,2,5-7,9]. All databases were searched from inception and searches were limited to English. Detailed information on search strategies undertaken in each database can be found in the Supplement (Appendix A). Additional studies were identified by reviewing the references of select high-impact articles, reports from reputable sources, and existing reviews. Any studies identified through other sources or contacts were manually added.
Inclusion criteria
We included studies presenting primary empirical data on duration of possible infectivity of SARS-CoV-2 in human populations using respiratory samples and reported in English. Articles that did not report data on duration of potential infectiousness in text, figures or tables were excluded. Studies reporting solely on pre-symptomatic or on convalescent period durations, incubation periods, serial intervals or on results based on statistical modelling were excluded. Studies, including reviews, that used data from other investigations were excluded; where relevant studies were identified, their references were reviewed to add relevant primary studies. When duplicate study reports were identified (i.e. a pre-print and a peer-reviewed journal article), the most recent version was included. Inclusion criteria are outlined in more detail in the Supplement (Appendix B).
Screening process
Titles and abstracts were screened for relevance independently by two reviewers. Full-text review was conducted independently by two reviewers for articles where relevance was not readily determined from title and abstracts, and any conflicts were resolved through consensus by the two reviewers.
Data extraction process
A draft data extraction form was developed and trialled across multiple reviewers to develop the final version. Extracted data fields included study characteristics (first author, publication status, study type, sample size), study population characteristics (age, hospitalisation, disease severity), method of determining infectious period (viral shedding, viral isolation), type of respiratory specimen(s) collected, reported durations (minimum, mean, median, maximum), whether cases without symptoms (asymptomatic or pre-symptomatic) were reported, whether the study focused solely on duration of communicability during the convalescent phase, how measurement of duration start and end was defined, and study quality. All extracted data were reviewed by a second reviewer.
Definitions
Sample size was defined as the total number of participants for whom data on communicability period was assessed. Disease severity associated with SARS-CoV-2 infection was classified according to the following definitions: mild referred to study populations reporting no symptoms or non-serious symptoms that did not require healthcare intervention, moderate severity included participants who required acute care and/or intervention, and severe disease referred to cases that required admission to the intensive care unit, critical intervention and/or resulted in death. Studies that included participants with mild, moderate and severe cases were classified as ‘mixed’; otherwise, if they included cases that were mild/moderate or moderate/severe, they were categorised as the higher level of severity. Hospitalisation status was determined as described in the study. Notably, in some jurisdictions, admission to hospital appeared to be part of routine isolation policies and so this description alone was not taken as an indicator of disease severity. Studies with children were those that included participants aged 19 years or younger. Respiratory samples included those taken from the upper (naso/oro-pharyngeal, nasal, throat, or saliva swabs) or lower respiratory tract (from sputum or bronchial lavage specimens). For the purposes of this review, the start of measurement of duration is referred to as ‘symptom onset’ and measurement end as ‘viral clearance’. The end point for measuring duration of viral isolation was the last reported day on which virus could be isolated and cultured (captured under ‘other’).
Assessment of study quality
An adaptation of the Mixed Methods Appraisal Tool was used to assess study quality [19]. Questions were concerned with the following: (i) the study had clear research questions or objectives, (ii) the collected data allowed the study to address the stated research question, (iii) the research question was aimed at understanding duration of communicability, (iv) there was a complete follow-up period defined to measure duration of communicability and (v) there was clarity about when measurement of communicability period started, about sample types collected and frequency of sample collection, about how long patients were followed (until viral clearance, study end, hospital discharge, death) and about the method of assessment of communicability. Owing to the emerging nature of this topic, we did not exclude studies from our synthesis or analysis based on study quality.
Analysis
All data processing and analysis was conducted using the statistical programming language R (version 4.0.0) [20]. For studies where more than one value was reported for duration (i.e. when data for multiple sample types were reported or results were presented in a stratified manner), the values corresponding to the higher duration were included for analysis. As analyses were generally aimed at identifying maximum reported durations, these values were pulled from each study and summarised. Raw data are available in Supplementary Table S1.
Results
Results of literature search
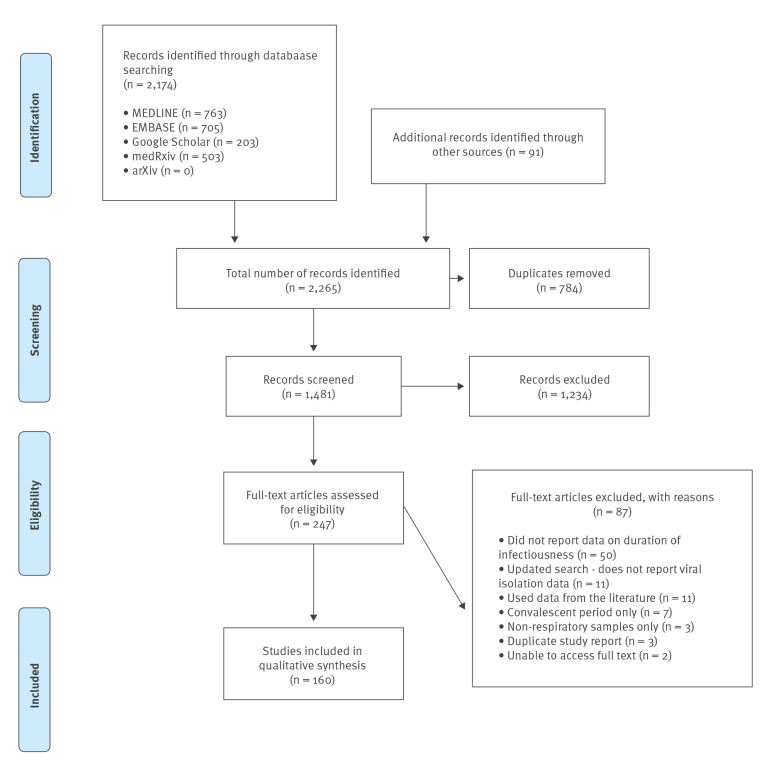
We retrieved, 2,174 records from database searches and 91 additional studies from reference chaining and other sources (2,265 total), of which 1,481 remained after removing duplicate records. Of these, 1,234 were excluded in screening and 87 in full-text review as they did not report data on duration of potential infectivity, used secondary data from the literature, reported only on the convalescent period, assessed non-respiratory samples only, the full text was not accessible, or they were duplicate reports of the same study. 160 studies were included in the final synthesis. The PRISMA flowchart in Figure 1 illustrates the study selection process.
Figure 1.
PRISMA flowchart diagram of included studies on the communicable period of SARS-CoV-2, March–September 2020 (n = 160)
SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
The PRISMA flowchart details the number of studies identified through our search and then included for analysis after screening for eligibility [18].
Study characteristics
Study populations
Information on study characteristics is presented in Table 1, overall and broken down by method of assessment. Of 160 total included studies, 17 studies assessed duration through both viral isolation and RT-PCR, while eight focused on viral isolation only and 135 included only results from RT-PCR. There was a wide range of study sample sizes (between one and 1,061), with a mean of 58.5 (standard deviation (SD): 118.9) and a median of 16.5 (interquartile range (IQR): 68.5). The majority of studies consisted of case series (n = 101; 63%), while 35 (22%) were case reports, cohort (n = 15; 9%), cross-sectional (n = 4; 3%), clinical trial (n = 3; 2%) and case control studies (n = 2; 1%) (Table 1). The results of the study quality assessment are included in Table 1.
Table 1. Characteristics of included studies, overall and broken down by method of assessment of duration of communicability of SARS-CoV-2, March–September 2020 (n = 160).
| All studies (n = 160) |
Viral isolation only (n = 8) [38-44,46] |
RT-PCR only (n = 135) [12-16,48-177] |
Viral isolation and RT-PCR (n = 17) [21-37] |
|||||
|---|---|---|---|---|---|---|---|---|
| Sample size | ||||||||
| Minimum | 1 | 5 | 1 | 1 | ||||
| Maximum | 1,061 | 518 | 584 | 1,061 | ||||
| Mean (SD) | 58.5 (118.9) | 142.5 (167.8) | 49.3 (83.9) | 92 (252.3) | ||||
| Median (IQR) | 16.5 (68.5) | 97 (153.3) | 14 (64.5) | 12 (75.0) | ||||
| Study design | n | % | n | % | N | % | n | % |
| Case control | 2 | 1.3 | 0 | 0.0 | 2 | 1.5 | 0 | 0.0 |
| Case report | 35 | 21.9 | 0 | 0.0 | 30 | 22.2 | 5 | 29.4 |
| Case series | 101 | 63.1 | 7 | 87.5 | 84 | 62.2 | 10 | 58.8 |
| Clinical trial | 3 | 1.9 | 0 | 0.0 | 3 | 2.2 | 0 | 0.0 |
| Cohort | 15 | 9.4 | 0 | 0.0 | 15 | 11.1 | 0 | 0.0 |
| Cross-sectional | 4 | 2.5 | 1 | 12.5 | 1 | 0.7 | 2 | 11.8 |
| Measurement start | n | % | n | % | n | % | n | % |
| Hospital admission | 36 | 22.5 | 0 | 0.0 | 33 | 24.4 | 3 | 17.7 |
| Date of first positive test | 12 | 7.5 | 1 | 12.5 | 11 | 8.1 | 0 | 0.0 |
| Symptom onset | 91 | 56.9 | 7 | 87.5 | 72 | 53.3 | 12 | 70.6 |
| Other | 21 | 13.1 | 0 | 0.0 | 19 | 14.1 | 2 | 11.8 |
| Measurement end | n | % | n | % | n | % | n | % |
| Negative test | 114 | 71.3 | 3 | 37.5 | 105 | 77.8 | 7 | 41.2 |
| Discharge/death | 8 | 5.0 | 0 | 0.0 | 7 | 5.2 | 1 | 5.9 |
| Last positive test | 7 | 4.4 | 2 | 25.0 | 3 | 2.2 | 2 | 11.8 |
| Other | 31 | 19.4 | 3 | 37.5 | 20 | 14.8 | 7 | 41.2 |
| Region | n | % | n | % | n | % | n | % |
| Asia | 127 | 79.4 | 1 | 12.5 | 119 | 88.1 | 7 | 41.2 |
| Australia | 2 | 1.3 | 1 | 12.5 | 1 | 0.7 | 0 | 0.0 |
| Europe | 19 | 11.9 | 5 | 62.5 | 8 | 5.9 | 6 | 35.3 |
| Middle-East | 2 | 1.3 | 0 | 0.0 | 2 | 1.5 | 0 | 0.0 |
| North America | 10 | 6.3 | 1 | 12.5 | 5 | 3.7 | 4 | 23.5 |
| Age group | n | % | n | % | n | % | n | % |
| Children | 18 | 11.3 | 2 | 25.0 | 16 | 11.9 | 0 | 0.0 |
| Adults | 114 | 71.3 | 4 | 50.0 | 97 | 71.1 | 14 | 82.4 |
| Mixed | 28 | 17.5 | 2 | 25.0 | 23 | 17.0 | 3 | 17.6 |
| Hospitalisation status | n | % | n | % | n | % | n | % |
| Hospitalised | 136 | 85.0 | 3 | 37.5 | 119 | 88.1 | 14 | 82.4 |
| Not hospitalised | 4 | 2.5 | 0 | 0.0 | 4 | 3.0 | 0 | 0.0 |
| Mixed | 17 | 10.6 | 5 | 62.5 | 10 | 7.4 | 2 | 11.8 |
| Unclear | 3 | 1.9 | 0 | 0.0 | 2 | 1.5 | 1 | 5.9 |
| Disease severity | n | % | n | % | n | % | n | % |
| Mild | 36 | 22.5 | 0 | 0.0 | 33 | 24.4 | 3 | 17.7 |
| Moderate | 32 | 20.0 | 0 | 0.0 | 29 | 21.5 | 3 | 17.7 |
| Severe | 12 | 7.5 | 1 | 12.5 | 10 | 7.4 | 1 | 5.9 |
| Mixed | 51 | 31.9 | 6 | 75.0 | 38 | 28.1 | 7 | 41.2 |
| Unclear | 29 | 18.1 | 1 | 12.5 | 25 | 18.5 | 3 | 17.7 |
| Includes asymptomatic or pre-symptomatic cases | n | % | n | % | n | % | n | % |
| Yes | 69 | 43.1 | 2 | 25.0 | 62 | 45.9 | 5 | 29.4 |
| No | 91 | 56.9 | 6 | 75.0 | 73 | 54.1 | 12 | 70.6 |
| Publication status | n | % | n | % | n | % | n | % |
| Peer-reviewed journal | 134 | 83.8 | 5 | 62.5 | 112 | 83.0 | 17 | 100.0 |
| Preprint database | 26 | 16.3 | 3 | 37.5 | 23 | 17.0 | 0 | 0.0 |
| Study quality | n | % | n | % | n | % | n | % |
| 0 quality concerns | 31 | 19.4 | 3 | 37.5 | 26 | 19.3 | 2 | 11.8 |
| 1 quality concern | 43 | 26.9 | 1 | 12.5 | 36 | 26.7 | 6 | 35.3 |
| 2 quality concerns | 53 | 33.1 | 4 | 50.0 | 44 | 32.6 | 5 | 29.4 |
| 3 quality concerns | 27 | 16.9 | 0 | 0.0 | 23 | 17.0 | 4 | 23.5 |
| 4 quality concerns | 4 | 2.5 | 0 | 0.0 | 4 | 3.0 | 0 | 0.0 |
| 5 quality concerns | 2 | 1.3 | 0 | 0.0 | 2 | 1.5 | 0 | 0.0 |
IQR: interquartile range; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; SD: standard deviation.
The majority of studies were conducted in Asia (n = 127, 79%), with 19 studies conducted in Europe (12%), 10 in North America (6%), two in the Middle East (1%) and two in Australia (1%) (Table 1). Study populations had mixed disease severity (i.e. included mild, moderate and severe disease) in 51 studies (32%), and 29 studies (18%) did not provide information on disease severity. Thirty-six studies (23%) had participants with only mild symptoms, while 32 studies (20%) studied moderate symptoms and 12 (8%) severe disease. The majority of studies included only hospitalised patients (n = 136; 85%). Most of the studies included only adults (n = 114; 71%), while 18 studies (11%) focused solely on paediatric populations, and 28 studies (18%) had a mixed population of children and adults. Sixty-nine studies (43%) included asymptomatic or pre-symptomatic positive cases (Table 1).
Measurement of duration start and end
The starting point for measuring duration was primarily symptom onset (n = 91; 57%), followed by hospital admission (n = 36; 23%), when the patient first tested positive for SARS-CoV-2 (n = 12; 8%) or other (n = 21; 13%). The end point of duration measurement for most studies was the date of a single or consecutive negative RT-PCR test (n = 114; 71%), followed by discharge/death (n = 8; 5%), last positive test (n = 7; 4%) or other (n = 31; 19%).
Study quality
At the time of writing, 26 (16%) of the studies included in this review were pre-prints and thus had not undergone peer-review. Many of the remaining studies published in peer-review journals were letters to the editor or other short communications that do not fully report their methods. There was variation in study quality in both preprints and studies published in peer-reviewed journals; roughly half of the included studies (n = 86; 54%) had two or more study quality concerns (Table 1).
Overview of studies that measured durations using both RT-PCR tests and viral isolation experiments
There were 17 studies that investigated the duration of time until viral clearance through RT-PCR tests as well as viral isolation [21-37]. Eight other studies investigated viral isolation durations only [36,38-44]. Supplementary Table S2 displays detailed information on each individual study.
Table 2 provides an overview of the 12 studies that included measurements of the maximum time to viral clearance from both RT-PCR and viral isolation experiments. The studies included mixed clinical populations and determined the duration of viral viability either by taking a cross-section of diagnostic samples collected at different times from symptom onset (n = 2; 17%) or by serially collecting samples from the same individuals over time (n = 10; 83%). While not all studies reported the RT-PCR cycle threshold (Ct) values of samples that achieved viral isolation, of those that did, the range of Ct values with a successful isolation was 12.3–37.9.
Table 2. Overview of studies that included measurements of duration to SARS-CoV-2 clearance from both RT-PCR and viral isolation experiments, March–September 2020 (n = 12) .
| Reference | Study population description | Sample size | Sample types taken for isolation and sampling method | Longest time to viral clearance (RT-PCR) (in days) |
Longest time to viral clearance (isolation) (in days) |
Ct values |
|---|---|---|---|---|---|---|
| Arons et al. [22] | Patients in a skilled nursing facility with mixed disease severity; mean age: 78.6 years; 98% had a comorbidity. | 27 | NP and OP samples, collected at two time points, 1 week apart | 13 | 9 | RT-PCR Ct values ranged from 13.7 to 37.9 in positive samples |
| Bullard et al. [25] | All suspected COVID-19 cases had SARS-CoV-2 RT-PCR performed on samples at Cadham Provincial Laboratory. Median age of the patients sampled was 45 years (range: 30–59). | 90 (26 with positive viral isolation) | NP and endotracheal samples, from diagnostic samples of individuals who tested positive by RT-PCR from day 0 to 21 post symptom onset | 21 | 8 | Positive viral culture samples had lower Ct values than negative cultures (17 (IQR: 16–19) vs 27 (IQR: 22–33)). For every increase in unit in Ct value, the odds of a positive culture decreased by 32%. No growth in samples with Ct > 24. |
| Decker et al. [21] | 62-year-old male heart transplant recipient who was hospitalised with mild disease severity | 1 | Throat samples, collected serially at 10 time points until day 35 of illness | > 35 (patient still testing positive at study end) | 21 | Viral culture not successful in samples with RT-PCR Ct > 25 |
| Gautret et al. [29] | Hospitalised patients with age range of 18 to 88 years, 57.5% had at least one comorbidity. Three patients were transferred to ICU, one patient died. | 80 (53 with positive viral isolation) | NP samples, collected daily beginning at treatment | 12 | 9 | Not reported |
| Haveri et al. [30] | First COVID-19 case in Finland; hospitalised woman in her 30s from Wuhan with mild disease severity | 1 | NP samples, collected serially, on days 3, 4, 9, 10, 20 and 23; unclear when viral isolation was attempted | 8 | 4 | Ct values on day 4 for different RT-PCR targets: E (29.59), RdRp (30.87), N (31.78) |
| COVID-19 Investigation Team [26] | Convenience sample of the first 12 US patients confirmed to have COVID-19; five patients had underlying conditions. Median age was 53 years (range: 21–68); mild to moderate illness; seven patients hospitalised but none requiring mechanical ventilation and all showing recovery. | 12 (9 with positive viral isolation) | NP and OP samples, taken on days 1–9 from symptom onset; not attempted in later specimens | 29 | 9 | Positive viral isolation from samples with RT-PCR Ct values of 12.3–35.7 |
| Lescure et al. [27] | Patients were three men (aged 31 years, 48 years, and 80 years) and two women (aged 30 years and 46 years). | 5 | NP samples, taken from patients once only at days 2, 2, 6, 7, 9 since symptom onset. | 24 (until patient death) | 2 | Positive viral isolation in samples with RdRp Ct values of 23.6 and 24.4, E gene Ct of 22.8 and 20.0, RdRp IP Ct of 23.0 and 19.3, GAPDH (housekeeping gene) Ct of 26.5 and 25.6 |
| Liu et al. [31] | 50-year-old hospitalised woman with mild disease and no comorbidities | 1 | Throat and sputum samples, collected daily | 63 | 18 | Not reported |
| Million et al. [23] | Hospitalised patients with a mean age of 47.9 (SD 17.5). 973 patients (91.7%) had good clinical outcome; 38 had severe outcomes including death. | 1,061 (915 attempted, 204 positive viral isolations, 11 individuals with daily samples) | NP samples, collected daily for 11 participants | > 15 (patient still testing positive at study end) |
9 | Not reported |
| Perera et al. [32] | Hospitalised patients positive for COVID-19 with mixed disease severity and an age range of 17–75 years | 35 | NP, throat, sputum and saliva samples, not collected at predefined intervals; isolation attempted in all positive samples (n = 68) | > 30 | 8 | Not reported |
| Wölfel et al. [33] | Hospitalised, young-to-middle-aged patients with minimal pre-existing disease and mild symptoms; patients identified based on close contact with an index case and not based on symptoms | 9 | OP, NP, and sputum samples, collected daily beginning from 2 to 8 days from symptom onset; isolation attempted on multiple occasions from positive samples. | 28 | 8 | Not reported |
| Young et al. [34] | Patients hospitalised with COVID-19 with mixed disease severity, with an age range of 35–56 years. 38% had any comorbidity | 100 | NP serial samples taken on days 1, 3, 7, 14, 21 and 28 after enrolment; viral culture attempted from samples from 74 patients, but unclear when | 48 | 14 | Viral isolation not positive when Ct value was > 30 |
CPE: cytopathic effect; COVID-19: coronavirus disease; Ct: cycle threshold; E: envelope protein gene; GAPDH: glyceraldehyde-3-phosphate dehydrogenase gene (reference housekeeping gene); IgG: immunoglobulin G; IgM: immunoglobulin M; IQR: interquartile range; N: nucleocapsid protein gene; NP: nasopharyngeal; OP: oropharyngeal; RdRp: RNA-dependent RNA polymerase gene; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
Studies are presented in alphabetical order by first author.
Comparison of maximum duration of positive RT-PCR tests and successful viral isolation reported from studies that measured both
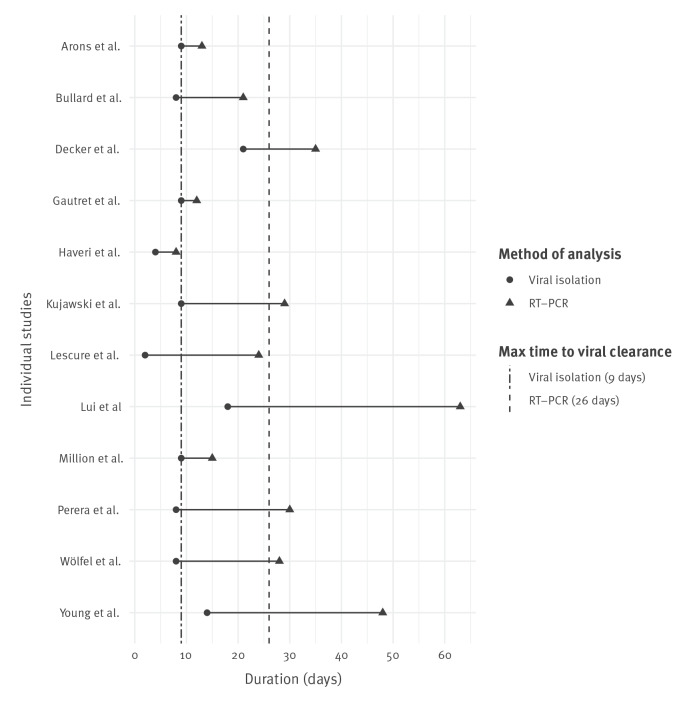
All studies that measured the longest time to viral clearance in both RT-PCR tests and viral isolation experiments reported positive RT-PCR test results after virus was no longer able to be isolated and cultured. The median duration after symptom onset that virus was successfully isolated was 9 days (IQR: 2.25; range: 2–21), while the corresponding median value for longest duration until viral clearance by RT-PCR was 26 days (IQR: 16.8; range: 8–63) (Figure 2). Three studies reported successful viral isolation beyond 10 days (Figure 2).
Figure 2.
Maximum reported durations of SARS-CoV-2 communicability from studies with data from RT-PCR tests and viral isolation experiments, March–September 2020 (n = 12)
SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
Each horizontal line represents data from the same study, with points indicating the longest time to viral clearance reported from each study for viral isolation (circles) and RT-PCR (triangles). Vertical lines represent median reported values across all studies on longest time to viral clearance from viral isolation experiments (dashed and dotted line) and RT-PCR tests (dashed line).
Maximum reported durations of positive RT-PCR tests and successful viral isolation across all studies
We conducted a final analysis on all included studies that had measurements of maximum duration to viral clearance (n = 142). In 20 studies that successfully achieved viral isolations, viable virus could be isolated from a case across a range of 2–32 days after symptom onset; the median and mean durations of these values across studies were 10.5 days (IQR: 10) and 12.8 days (SD: 7.3), respectively. From the 134 studies with data on maximum duration of positive RT-PCR test results, the shortest reported time until viral clearance was 5 days from symptom onset, while the maximum was 95 days, with respective median and mean values of 25 days (IQR: 19) and 28.8 days (SD: 15.8) (Table 3).
Table 3. Data on maximum duration of SARS-CoV-2 communicability from all studies reporting on isolation of live virus and detection of viral nucleic acid by RT-PCR from respiratory samples, March–September 2020 (n = 142) .
| Method of measurement | Number of included studies | Shortest reported duration | Longest reported duration | Longest reported duration mean (SD) |
Longest reported duration median (IQR) |
|---|---|---|---|---|---|
| Viral isolation | 20 | 2 | 32 | 12.8 (7.3) | 10.5 (10) |
| RT-PCR | 134 | 5 | 95 | 28.9 (15.8) | 25 (19) |
IQR: interquartile range; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; SD: standard deviation.
Note that some studies provided data for both viral isolation and viral shedding.
Discussion
This review contributes to the evidence on the period of potential infectivity after infection with SARS-CoV-2, showing that positive detection of viral nucleic acid by RT-PCR can far exceed the duration after which viral transmission may occur. Results from this review suggest that positive RT-PCR results after day 10 following symptom onset of COVID-19 are unlikely to indicate infectiousness. The conclusions of this review are also in line with recently updated guidelines that support relying on symptoms rather than RT-PCR test results for ending isolation precautions in non-healthcare settings [45].
Overall, results from this review concur with previous studies that also support a general 10-day isolation period. However, in reviewing the data from all studies reporting viral isolation, we identified important exceptions. The longest period that virus was isolated after symptom onset was 32 days from a hospitalised patient with severe disease [46]. A duration of 21 days was identified from a report of a single individual with mild symptoms but with significant underlying conditions including a recent heart transplant [21]. A separate case report of an individual without any significant comorbidities and also with mild disease symptoms reported viable virus until 18 days [11]. One case series of 129 hospitalised patients with moderate to severe disease, and mixed degrees of comorbidities, reported a maximum duration of viable virus of 20 days, with a median time to viral clearance in this sample of eight days [38].
Although these exceptions may not occur frequently, they highlight the need to be cautious when COVID-19 cases are being released from isolation into high-risk settings. Previous studies have suggested that there may be differences in duration of viral viability by disease severity with cases of more severe disease potentially having longer infectious viral shedding [7,39]. Despite such trends, the select cases highlighted herein suggest symptom presentation and disease severity do not always follow a pattern with respect to duration of viral viability. Of the studies assessing viral isolation, some also isolated and cultured virus from patients who were asymptomatic or during their convalescent disease period [21-24].
Given the prolonged detection of viral nucleic acid by RT-PCR testing, there have been suggestions to incorporate additional testing metrics when results are being used to inform release from isolation [9,40], such as Ct values. This would be an important and relevant qualification to current protocol especially when considering that positive tests from RT-PCR may persist for several weeks [12-16,41,47]. While the available data on RT-PCR Ct values and viral viability were inconsistent, a number of studies have reported an upper Ct limit for successful viral isolation [9,22,25-28,40,41,46] which may help inform future threshold values.
This review highlights significant heterogeneity in the content and quality of the underlying literature which limits the ability to draw robust inferences to inform isolation protocols from the available data. There was a general paucity of high-quality evidence. We identified few studies that investigated viral isolation and many of the included studies had small sample sizes, making it difficult to draw robust findings from the single subject studies. The included studies suffer from selection bias and lack of generalisability, as case reports and case series often focus on highly specific clinical populations that do not represent the majority of COVID-19 cases. Given that data were often drawn from applied clinical settings, several of the included studies did not follow participants until viral clearance or collect samples at consistent intervals throughout the communicable period. A lack of reporting standards also resulted in incomplete information on valuable metrics such as viral load and Ct values. Finally, although this was not a focus of this review, studies that are able to demonstrate clinical confirmation of transmission are needed to better understand infectivity [47].
This study builds off of previous research by directly comparing results from all studies that measured both the maximum duration of positive RT-PCR test results and longest time to successful viral isolation, thereby adding robustness to interpreting overall findings from the literature. We have made the raw data extracted from all studies in this review available in Supplementary Table S1 for the benefit of other research or public health groups. This research identifies improvements for future research and reporting that should enable more robust syntheses of newly emerging evidence to better inform infection control policies for SARS-CoV-2.
Acknowledgements
We would like to thank Kelsey Furk and John Harding for their contributions to the development and design of this study.
Supplementary Data
Supplementary Data
Supplementary Data
Conflict of interest: MD is Chief Scientific officer at GenXys Health Care Systems; GenXys was not involved at any stage of this study. All other authors declare no conflicts of interest.
Authors’ contributions: MP designed the study, conducted analyses, and wrote the first draft of manuscript. CP contributed to study design, data cleaning and analysis, and implemented the search strategy. MP, CP, TN, AG, LDC, NF and MD contributed to screening of articles, data extraction, and critical review and approval of the final manuscript.
References
- 1. Joynt GM, Wu WK. Understanding COVID-19: what does viral RNA load really mean? Lancet Infect Dis. 2020;20(6):635-6. 10.1016/S1473-3099(20)30237-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Byrne AW, McEvoy D, Collins AB, Hunt K, Casey M, Barber A, et al. Inferred duration of infectious period of SARS-CoV-2: rapid scoping review and analysis of available evidence for asymptomatic and symptomatic COVID-19 cases. BMJ Open. 2020;10(8):e039856. 10.1136/bmjopen-2020-039856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fontana LM, Villamagna AH, Sikka MK, McGregor JC. Understanding viral shedding of severe acute respiratory coronavirus virus 2 (SARS-CoV-2): Review of current literature. Infect Control Hosp Epidemiol. 2020;1-10. 10.1017/ice.2020.1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jefferson T, Spencer EA, Brassey J, Heneghan C. Viral cultures for COVID-19 infectious potential assessment – a systematic review. Clin Infect Dis. 2020;ciaa1764. [DOI] [PMC free article] [PubMed]
- 5. Morone G, Palomba A, Iosa M, Caporaso T, De Angelis D, Venturiero V, et al. Incidence and persistence of viral shedding in COVID-19 post-acute patients with negativized pharyngeal swab: a systematic review. Front Med (Lausanne). 2020;7:562. 10.3389/fmed.2020.00562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weiss A, Jellingsø M, Sommer MOA. Spatial and temporal dynamics of SARS-CoV-2 in COVID-19 patients: A systematic review and meta-analysis. EBioMedicine. 2020;58:102916. 10.1016/j.ebiom.2020.102916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Walsh KA, Spillane S, Comber L, Cardwell K, Harrington P, Connell J, et al. The duration of infectiousness of individuals infected with SARS-CoV-2. J Infect. 2020;81(6):847-56. 10.1016/j.jinf.2020.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization (WHO). Criteria for releasing COVID-19 patients from isolation. Geneva: WHO; 2020 Jun. Available from: https://www.who.int/publications-detail-redirect/criteria-for-releasing-covid-19-patients-from-isolation
- 9.Jefferson T, Spencer E, Brassey J, Heneghan C. Viral cultures for COVID-19 infectivity assessment. Systematic review. medRxiv. 2020 Sep 29;2020.08.04.20167932.
- 10.Lu J, Peng J, Xiong Q, Liu Z, Lin H, Tan X, et al. Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR. medRxiv. 2020 Jun 17;2020.06.15.20131748. [DOI] [PMC free article] [PubMed]
- 11. Liu W-D, Chang S-Y, Wang J-T, Tsai M-J, Hung C-C, Hsu C-L, et al. Prolonged virus shedding even after seroconversion in a patient with COVID-19. J Infect. 2020;81(2):318-56. 10.1016/j.jinf.2020.03.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cao H, Ruan L, Liu J, Liao W. The clinical characteristic of eight patients of COVID-19 with positive RT-PCR test after discharge. J Med Virol. 2020;92(10):2159-64. 10.1002/jmv.26017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Xu X, Hu J, Chen Q, Xu F, Liang H, et al. Clinical Course and Risk Factors for Recurrence of Positive SARS-CoV-2 RNA: A Retrospective Cohort Study from Wuhan, China. medRxiv. 2020 May 12;2020.05.08.20095018. [DOI] [PMC free article] [PubMed]
- 14. Li J, Zhang L, Liu B, Song D. Case report: viral shedding for 60 days in a woman with COVID-19. Am J Trop Med Hyg. 2020;102(6):1210-3. 10.4269/ajtmh.20-0275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang JR, Deng DT, Wu N, Yang B, Li HJ, Pan XB. Persistent viral RNA positivity during the recovery period of a patient with SARS-CoV-2 infection. J Med Virol. 2020;92(9):1681-3. 10.1002/jmv.25940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Man Z, Jing Z, Huibo S, Bin L, Fanjun Z. Viral shedding prolongation in a kidney transplant patient with COVID-19 pneumonia. Am J Transplant. 2020;20(9):2626-7. 10.1111/ajt.15996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobbins M. Rapid review guidebook. Hamilton, ON: National Collaborating Centre for Methods and Tools. Hamilton: National Collaborating Centre for Methods and Tools; 2017. Available from: https://www.nccmt.ca/capacity-development/rapid-review-guidebook
- 18. Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group . Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hong QN, Fàbregues S, Bartlett G, Boardman F, Cargo M, Dagenais P, et al. The Mixed Methods Appraisal Tool (MMAT) version 2018 for information professionals and researchers. Educ Inf. 2018;34(4):285-91. 10.3233/EFI-180221 [DOI] [Google Scholar]
- 20.R Core Team. R: A language and environment for statistical computing. Vienna, Austria; 2020. Available from: http://www.R-project.org/
- 21. Decker A, Welzel M, Laubner K, Grundmann S, Kochs G, Panning M, et al. Prolonged SARS-CoV-2 shedding and mild course of COVID-19 in a patient after recent heart transplantation. Am J Transplant. 2020;20(11):3239-45. 10.1111/ajt.16133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, Jacobs JR, et al. Presymptomatic SARS-CoV-2 Infections and Transmission in a Skilled Nursing Facility. N Engl J Med. 2020;382(22):2081-90. 10.1056/NEJMoa2008457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Million M, Lagier JC, Gautret P, Colson P, Fournier PE, Amrane S, et al. Early treatment of COVID-19 patients with hydroxychloroquine and azithromycin: A retrospective analysis of 1061 cases in Marseille, France. Travel Med Infect Dis. 2020;35:101738. 10.1016/j.tmaid.2020.101738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kumar DS, O’Neill SB, Johnston JC, Grant JM, Sweet DD. SARS-CoV-2 infection in a 76-year-old man with initially negative results for nasopharyngeal swabs. CMAJ. 2020;192(20):E546-9. 10.1503/cmaj.200641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis. 2020;71(10):2663-6. 10.1093/cid/ciaa638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. COVID-19 Investigation Team . Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med. 2020;26(6):861-8. 10.1038/s41591-020-0877-5 [DOI] [PubMed] [Google Scholar]
- 27. Lescure FX, Bouadma L, Nguyen D, Parisey M, Wicky PH, Behillil S, et al. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. 2020;20(6):697-706. 10.1016/S1473-3099(20)30200-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim JY, Ko JH, Kim Y, Kim YJ, Kim JM, Chung YS, et al. Viral load kinetics of SARS-CoV-2 infection in first two patients in Korea. J Korean Med Sci. 2020;35(7):e86. 10.3346/jkms.2020.35.e86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, Sevestre J, et al. Clinical and microbiological effect of a combination of hydroxychloroquine and azithromycin in 80 COVID-19 patients with at least a six-day follow up: A pilot observational study. Travel Med Infect Dis. 2020;34:101663. 10.1016/j.tmaid.2020.101663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haveri A, Smura T, Kuivanen S, Österlund P, Hepojoki J, Ikonen N, et al. Serological and molecular findings during SARS-CoV-2 infection: the first case study in Finland, January to February 2020. Euro Surveill. 2020;25(11):2000266. 10.2807/1560-7917.ES.2020.25.11.2000266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu WD, Chang SY, Wang JT, Tsai MJ, Hung CC, Hsu CL, et al. Prolonged virus shedding even after seroconversion in a patient with COVID-19. J Infect. 2020;81(2):318-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perera RAPM, Tso E, Tsang OTY, Tsang DNC, Fung K, Leung YWY, et al. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis. 2020;26(11):2701-4. 10.3201/eid2611.203219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465-9. 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- 34. Young BE, Ong SWX, Ng LFP, Anderson DE, Chia WN, Chia PY, et al. Viral dynamics and immune correlates of COVID-19 disease severity. Clin Infect Dis. 2020;ciaa1280. 10.1093/cid/ciaa1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang D, Zhao P, Zhang D, Dong JH, Xu Z, Yang G, et al. Persistent viral presence determines the clinical course of the disease in COVID-19. J Allergy Clin Immunol Pract. 2020;8(8):2585-2591.e1. 10.1016/j.jaip.2020.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun J, Zhu A, Li H, Zheng K, Zhuang Z, Chen Z, et al. Isolation of infectious SARS-CoV-2 from urine of a COVID-19 patient. Emerg Microbes Infect. 2020;9(1):991-3. 10.1080/22221751.2020.1760144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim JM, Kim HM, Lee EJ, Jo HJ, Yoon Y, Lee NJ, et al. Detection and isolation of SARS-CoV-2 in serum, urine, and stool specimens of COVID-19 patients from the Republic of Korea. Osong Public Health Res Perspect. 2020;11(3):112-7. 10.24171/j.phrp.2020.11.3.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Kampen JJA, van de Vijver DAMC, Fraaij PLA, Haagmans BL, Lamers MM, Okba N, et al. Shedding of infectious virus in hospitalized patients with coronavirus disease-2019 (COVID-19): duration and key determinants. medRxiv. 2020 Jun 9;2020.06.08.20125310. [DOI] [PMC free article] [PubMed]
- 39. Singanayagam A, Patel M, Charlett A, Lopez Bernal J, Saliba V, Ellis J, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25(32):2001483. 10.2807/1560-7917.ES.2020.25.32.2001483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gniazdowski V, Morris CP, Wohl S, Mehoke T, Ramakrishnan S, Thielen P, et al. Repeat COVID-19 molecular testing: correlation with recovery of infectious virus, molecular assay cycle thresholds, and analytical sensitivity. medRxiv. 2020 Aug 6;2020.08.05.20168963.
- 41. L’Huillier AG, Torriani G, Pigny F, Kaiser L, Eckerle I. Culture-competent SARS-CoV-2 in nasopharynx of symptomatic neonates, children, and adolescents. Emerg Infect Dis. 2020;26(10):2494-7. 10.3201/eid2610.202403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basile K, McPhie K, Carter I, Alderson S, Rahman H, Donovan L, et al. Cell-based culture of SARS-CoV-2 informs infectivity and safe de-isolation assessments during COVID-19. Clin Infect Dis. 2020;ciaa1579. [DOI] [PMC free article] [PubMed]
- 43. Jeong HW, Kim SM, Kim HS, Kim YI, Kim JH, Cho JY, et al. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin Microbiol Infect. 2020;26(11):1520-4. 10.1016/j.cmi.2020.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ladhani SN, Chow JY, Janarthanan R, Fok J, Crawley-Boevey E, Vusirikala A, et al. Investigation of SARS-CoV-2 outbreaks in six care homes in London, April 2020. EClinicalMedicine. 2020;26:100533. 10.1016/j.eclinm.2020.100533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Centers for Disease Control and Prevention (CDC). Interim guidance on duration of isolation and precautions for adults with COVID-19. Atlanta: CDC. [Accessed: 17 Mar 2021]. Available from: https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html
- 46.Folgueira MD, Luczkowiak J, Lasala F, Perez-Rivilla A, Delgado R. Persistent SARS-CoV-2 replication in severe COVID-19. medRxiv. 2020 Jun 12;2020.06.10.20127837. [DOI] [PMC free article] [PubMed]
- 47. Cheng HY, Jian SW, Liu DP, Ng TC, Huang WT, Lin HH, et al. Contact tracing assessment of COVID-19 transmission dynamics in Taiwan and risk at different exposure periods before and after symptom onset. JAMA Intern Med. 2020;180(9):1156-63. 10.1001/jamainternmed.2020.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tu Y, Wei Y, Zhang D, Chen C, Hu X, Fei G. Analysis of factors affected the SARS-CoV-2 viral shedding time of COVID-19 patients in Anhui, China: a retrospective study. Research Square. 2020 Apr 6; 10.21203/rs.3.rs-20954/v1. [DOI]
- 49. Iwasaki S, Fujisawa S, Nakakubo S, Kamada K, Yamashita Y, Fukumoto T, et al. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J Infect. 2020;81(2):e145-7. 10.1016/j.jinf.2020.05.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng L, Liu J, Xu W, Luo Q, Deng K, Lin B, Gao Z. 2019 Novel Coronavirus can be detected in urine, blood, anal swabs and oropharyngeal swabs samples. medRxiv. 2020 Feb 25;2020.02.21.20026179
- 51. Lee NY, Li CW, Tsai HP, Chen PL, Syue LS, Li MC, et al. A case of COVID-19 and pneumonia returning from Macau in Taiwan: Clinical course and anti-SARS-CoV-2 IgG dynamic. J Microbiol Immunol Infect. 2020;53(3):485-7. 10.1016/j.jmii.2020.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, Liu C, Meng Q, Gui S, Wu Y, Cheng P, et al. A case report of moderate COVID-19 with an extremely long-term viral shedding period in China. Research Square. 2020 Aug 19; 10.21203/rs.3.rs-59700/v1 [DOI]
- 53. Jiehao C, Jin X, Daojiong L, Zhi Y, Lei X, Zhenghai Q, et al. A case series of children with 2019 Novel Coronavirus infection: clinical and epidemiological features. Clin Infect Dis. 2020;71(6):1547-51. 10.1093/cid/ciaa198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu P, Liang L, Chen C, Nie S. A child confirmed COVID-19 with only symptoms of conjunctivitis and eyelid dermatitis. Graefes Arch Clin Exp Ophthalmol. 2020;258(7):1565-6. 10.1007/s00417-020-04708-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mao LJ, Xu J, Xu ZH, Xia XP, Li B, He JG, et al. A child with household transmitted COVID-19. BMC Infect Dis. 2020;20(1):329. 10.1186/s12879-020-05056-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sanville B, Corbett R, Pidcock W, Hardin K, Sebat C, Nguyen MV, et al. A community-transmitted case of severe acute respiratory distress syndrome (SARS) due to SARS-CoV-2 in the United States. Clin Infect Dis. 2020;71(16):2222-6. 10.1093/cid/ciaa347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xu H, Liu E, Xie J, Smyth RL, Zhou Q, Zhao R, et al. A follow-up study of children infected with SARS-CoV-2 from western China. Ann Transl Med. 2020;8(10):623. 10.21037/atm-20-3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zheng Z, Dong M, Hu K. A preliminary evaluation on the efficacy of ozone therapy in the treatment of COVID-19. J Med Virol. 2020;92(11):2348-50. 10.1002/jmv.26040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hu Y, Shen L, Yao Y, Xu Z, Zhou J, Zhou H. A report of three COVID-19 cases with prolonged viral RNA detection in anal swabs. Clin Microbiol Infect. 2020;26(6):786-7. 10.1016/j.cmi.2020.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu Y, Chen X, Zou X, Luo H. A severe-type COVID-19 case with prolonged virus shedding. J Formos Med Assoc. 2020;119(10):1555-7. 10.1016/j.jfma.2020.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan L, Kang X, Zhang B, Zheng S, Liu B, Tu T, et al. A special case of COVID-19 with long duration of viral shedding for 49 days. medRxiv. 2020 Mar 27;2020.03.22.20040071
- 62. Qiu L, Jiao R, Zhang A, Chen X, Ning Q, Fang F, et al. A case of critically ill infant of coronavirus disease 2019 with persistent reduction of T Lymphocytes. Pediatr Infect Dis J. 2020;39(7):e87-90. 10.1097/INF.0000000000002720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kam KQ, Yung CF, Cui L, Tzer Pin Lin R, Mak TM, Maiwald M, et al. A well infant with coronavirus disease 2019 with high viral load. Clin Infect Dis. 2020;71(15):847-9. 10.1093/cid/ciaa201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fan Q, Pan Y, Wu Q, Liu S, Song X, Xie Z, et al. Anal swab findings in an infant with COVID-19. Pediatr Investig. 2020;4(1):48-50. 10.1002/ped4.12186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Seah IYJ, Anderson DE, Kang AEZ, Wang L, Rao P, Young BE, et al. Assessing viral shedding and infectivity of tears in Coronavirus Disease 2019 (COVID-19) patients. Ophthalmology. 2020;127(7):977-9. 10.1016/j.ophtha.2020.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen X, Zhang Y, Zhu B, Zeng J, Hong W, He X, et al. Associations of clinical characteristics and antiviral drugs with viral RNA clearance in patients with COVID-19 in Guangzhou, China: a retrospective cohort study. medRxiv. 2020 Apr 14;2020.04.09.20058941
- 67. Li C, Ji F, Wang L, Wang L, Hao J, Dai M, et al. Asymptomatic and human-to-human transmission of SARS-CoV-2 in a 2-family cluster, Xuzhou, China. Emerg Infect Dis. 2020;26(7):1626-8. 10.3201/eid2607.200718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jiang X, Luo M, Zou Z, Wang X, Chen C, Qiu J. Asymptomatic SARS-CoV-2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days. J Med Virol. 2020;92(10):1807-9. 10.1002/jmv.25941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Thevarajan I, Nguyen THO, Koutsakos M, Druce J, Caly L, van de Sandt CE, et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med. 2020;26(4):453-5. 10.1038/s41591-020-0819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lim J, Jeon S, Shin HY, Kim MJ, Seong YM, Lee WJ, et al. Case of the index patient who caused tertiary transmission of COVID-19 infection in Korea: the application of Lopinavir/Ritonavir for the treatment of COVID-19 infected pneumonia monitored by quantitative RT-PCR. J Korean Med Sci. 2020;35(6):e79. 10.3346/jkms.2020.35.e79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lombardi A, Consonni D, Carugno M, Bozzi G, Mangioni D, Muscatello A, et al. Characteristics of 1573 healthcare workers who underwent nasopharyngeal swab testing for SARS-CoV-2 in Milan, Lombardy, Italy. Clin Microbiol Infect. 2020;26(10):1413.e9-13. 10.1016/j.cmi.2020.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xu Y, Li X, Zhu B, Liang H, Fang C, Gong Y, et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26(4):502-5. 10.1038/s41591-020-0817-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Qiu H, Wu J, Hong L, Luo Y, Song Q, Chen D. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect Dis. 2020;20(6):689-96. 10.1016/S1473-3099(20)30198-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wolf GK, Glueck T, Huebner J, Muenchhoff M, Hoffmann D, French LE, et al. Clinical and epidemiological features of a family cluster of symptomatic and asymptomatic Severe Acute Respiratory Syndrome Coronavirus 2 infection. J Pediatric Infect Dis Soc. 2020;9(3):362-5. 10.1093/jpids/piaa060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Song R, Han B, Song M, Wang L, Conlon CP, Dong T, et al. Clinical and epidemiological features of COVID-19 family clusters in Beijing, China. J Infect. 2020;81(2):e26-30. 10.1016/j.jinf.2020.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen M, Fan P, Liu Z, Li J, Huang S, Wu, et al. Clinical characteristics of 10 children with COVID-19 outside of Wuhan in Hubei Province. Research Square. 2020 Nov 21; 10.21203/rs.3.rs-18255/v1 [DOI]
- 77. Hu Z, Song C, Xu C, Jin G, Chen Y, Xu X, et al. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci. 2020;63(5):706-11. 10.1007/s11427-020-1661-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pongpirul WA, Mott JA, Woodring JV, Uyeki TM, MacArthur JR, Vachiraphan A, et al. Clinical characteristics of patients hospitalized with Coronavirus Disease, Thailand. Emerg Infect Dis. 2020;26(7):1580-5. 10.3201/eid2607.200598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.An J, Liao X, Xiao T, Qian S, Yuan J, Ye H, et al. Clinical characteristics of the recovered COVID-19 patients with re-detectable positive RNA test. medRxiv. 2020 Mar 30;2020.03.26.20044222 [DOI] [PMC free article] [PubMed]
- 80. Kim ES, Chin BS, Kang CK, Kim NJ, Kang YM, Choi JP, et al. Clinical Course and Outcomes of Patients with Severe Acute Respiratory Syndrome Coronavirus 2 Infection: a Preliminary Report of the First 28 Patients from the Korean Cohort Study on COVID-19. J Korean Med Sci. 2020;35(13):e142. 10.3346/jkms.2020.35.e142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054-62. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Xu T, Chen C, Zhu Z, Cui M, Chen C, Dai H, et al. Clinical features and dynamics of viral load in imported and non-imported patients with COVID-19. Int J Infect Dis. 2020;94:68-71. 10.1016/j.ijid.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chen J, Qi T, Liu L, Ling Y, Qian Z, Li T, et al. Clinical progression of patients with COVID-19 in Shanghai, China. J Infect. 2020;80(5):e1-6. 10.1016/j.jinf.2020.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Danis K, Epaulard O, Bénet T, Gaymard A, Campoy S, Botelho-Nevers E, et al. Cluster of Coronavirus Disease 2019 (COVID-19) in the French Alps, February 2020. Clin Infect Dis. 2020;71(15):825-32. 10.1093/cid/ciaa424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Fang Z, Zhang Y, Hang C, Ai J, Li S, Zhang W. Comparisons of viral shedding time of SARS-CoV-2 of different samples in ICU and non-ICU patients. J Infect. 2020;81(1):147-78. 10.1016/j.jinf.2020.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. To KK, Tsang OT, Yip CC, Chan KH, Wu TC, Chan JM, et al. Consistent Detection of 2019 Novel Coronavirus in Saliva. Clin Infect Dis. 2020;71(15):841-3. 10.1093/cid/ciaa149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mandić-Rajčević S, Masci F, Crespi E, Franchetti S, Longo A, Bollina I, et al. Contact tracing and isolation of asymptomatic spreaders to successfully control the COVID-19 epidemic among healthcare workers in Milan (Italy). medRxiv. 2020 May 08;2020.05.03.20082818
- 88. Nicastri E, D’Abramo A, Faggioni G, De Santis R, Mariano A, Lepore L, et al. Coronavirus disease (COVID-19) in a paucisymptomatic patient: epidemiological and clinical challenge in settings with limited community transmission, Italy, February 2020. Euro Surveill. 2020;25(11):2000230. 10.2807/1560-7917.ES.2020.25.11.2000230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Qiancheng X, Jian S, Lingling P, Lei H, Xiaogan J, Weihua L, et al. Coronavirus disease 2019 in pregnancy. Int J Infect Dis. 2020;95:376-83. 10.1016/j.ijid.2020.04.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhu L, Gong N, Liu B, Lu X, Chen D, Chen S, et al. Coronavirus Disease 2019 pneumonia in immunosuppressed renal transplant recipients: a summary of 10 confirmed cases in Wuhan, China. Eur Urol. 2020;77(6):748-54. 10.1016/j.eururo.2020.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Liu Y, Liao W, Wan L, Xiang T, Zhang W. Correlation between relative nasopharyngeal virus RNA Load and lymphocyte count disease severity in patients with COVID-19. Viral Immunol. 2020;vim.2020.0062. 10.1089/vim.2020.0062 [DOI] [PubMed] [Google Scholar]
- 92. Zha L, Li S, Pan L, Tefsen B, Li Y, French N, et al. Corticosteroid treatment of patients with coronavirus disease 2019 (COVID-19). Med J Aust. 2020;212(9):416-20. 10.5694/mja2.50577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Arashiro T, Furukawa K, Nakamura A. COVID-19 in 2 persons with mild upper respiratory tract symptoms on a cruise ship, Japan. Emerg Infect Dis. 2020;26(6):1345-8. 10.3201/eid2606.200452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cai Q, Huang D, Ou P, Yu H, Zhu Z, Xia Z, et al. COVID-19 in a designated infectious diseases hospital outside Hubei Province, China. Allergy. 2020;75(7):1742-52. 10.1111/all.14309 [DOI] [PubMed] [Google Scholar]
- 95.Shastri A, Wheat J, Agrawal S, Chaterjee N, Pradhan K,Goldfinger M, et al. Delayed clearance of SARS-CoV2 in male compared to female patients: High ACE2 expression in testes suggests possible existence of gender-specific viral reservoirs. medRxiv. 2020 Apr 17;2020.04.16.20060566
- 96. Marchand-Senécal X, Kozak R, Mubareka S, Salt N, Gubbay JB, Eshaghi A, et al. Diagnosis and management of first case of COVID-19 in Canada: lessons applied from SARS-CoV-1. Clin Infect Dis. 2020;71(16):2207-10. 10.1093/cid/ciaa227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Han C, Duan C, Zhang S, Spiegel B, Shi H, Wang W, et al. Digestive symptoms in COVID-19 patients with mild disease severity: clinical presentation, stool viral RNA resting, and outcomes. Am J Gastroenterol. 2020;115(6):916-23. 10.14309/ajg.0000000000000664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wu F, Zhang W, Zhang L, Wang D, Wan Y. Discontinuation of antiviral drugs may be the reason for recovered COVID-19 patients testing positive again. Br J Hosp Med (Lond). 2020;81(4):1-2. 10.12968/hmed.2020.0156 [DOI] [PubMed] [Google Scholar]
- 99. Ma X, Su L, Zhang Y, Zhang X, Gai Z, Zhang Z. Do children need a longer time to shed SARS-CoV-2 in stool than adults? J Microbiol Immunol Infect. 2020;53(3):373-6. 10.1016/j.jmii.2020.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Qian GQ, Chen XQ, Lv DF, Ma AHY, Wang LP, Yang NB, et al. Duration of SARS-CoV-2 viral shedding during COVID-19 infection. Infect Dis (Lond). 2020;52(7):511-2. 10.1080/23744235.2020.1748705 [DOI] [PubMed] [Google Scholar]
- 101.Tan LV, Ngoc NM, That BTT, Uyen LTT, Hong NTT, Dung NTP, et al. Duration of viral detection in throat and rectum of a patient with COVID-19. medRxiv. 2020 Mar 16;2020.03.07.20032052
- 102. Lv DF, Ying QM, Weng YS, Shen CB, Chu JG, Kong JP, et al. Dynamic change process of target genes by RT-PCR testing of SARS-Cov-2 during the course of a Coronavirus Disease 2019 patient. Clin Chim Acta. 2020;506:172-5. 10.1016/j.cca.2020.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Xiao AT, Tong YX, Gao C, Zhu L, Zhang YJ, Zhang S. Dynamic profile of RT-PCR findings from 301 COVID-19 patients in Wuhan, China: A descriptive study. J Clin Virol. 2020;127:104346. 10.1016/j.jcv.2020.104346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lin A, He ZB, Zhang S, Zhang JG, Zhang X, Yan WH. Early risk factors for the duration of Severe Acute Respiratory Syndrome Coronavirus 2 viral positivity in patients with Coronavirus Disease 2019. Clin Infect Dis. 2020;71(16):2061-5. 10.1093/cid/ciaa490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xiao T, Wang Y, Yuan J, Ye H, Wei L, Liao X, et al. Early viral clearance and antibody kinetics of COVID-19 among asymptomatic carriers. meRxiv. 2020 May 2;2020.04.28.20083139 [DOI] [PMC free article] [PubMed]
- 106. Zeng QL, Yu ZJ, Gou JJ, Li GM, Ma SH, Zhang GF, et al. Effect of convalescent plasma therapy on viral shedding and survival in patients with Coronavirus Disease 2019. J Infect Dis. 2020;222(1):38-43. 10.1093/infdis/jiaa228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Young BE, Ong SWX, Kalimuddin S, Low JG, Tan SY, Loh J, et al. Epidemiologic features and clinical course of patients infected With SARS-CoV-2 in Singapore. JAMA. 2020;323(15):1488-94. 10.1001/jama.2020.3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Pan Y, Yu X, Du X, Li Q, Li X, Qin T, et al. Epidemiological and clinical characteristics of 26 asymptomatic Severe Acute Respiratory Syndrome Coronavirus 2 carriers. J Infect Dis. 2020;221(12):1940-7. 10.1093/infdis/jiaa205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Lo IL, Lio CF, Cheong HH, Lei CI, Cheong TH, Zhong X, et al. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int J Biol Sci. 2020;16(10):1698-707. 10.7150/ijbs.45357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831-1833.e3. 10.1053/j.gastro.2020.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Cai Q, Yang M, Liu D, Chen J, Shu D, Xia J, et al. Experimental treatment with Favipiravir for COVID-19: an open-label control study. Engineering (Beijing). 2020;6(10):1192-8. 10.1016/j.eng.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Qi L, Yang Y, Jiang D, Tu C, Wan L, Chen X, et al. Factors associated with the duration of viral shedding in adults with COVID-19 outside of Wuhan, China: a retrospective cohort study. Int J Infect Dis. 2020;96:531-7. 10.1016/j.ijid.2020.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Hu X, Xing Y, Jia J, Ni W, Liang J, Zhao D, et al. Factors associated with negative conversion of viral RNA in patients hospitalized with COVID-19. Sci Total Environ. 2020;728:138812. 10.1016/j.scitotenv.2020.138812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Xu K, Chen Y, Yuan J, Yi P, Ding C, Wu W, et al. Factors associated with prolonged viral RNA shedding in patients with Coronavirus Disease 2019 (COVID-19). Clin Infect Dis. 2020;71(15):799-806. 10.1093/cid/ciaa351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Yan D, Liu XY, Zhu YN, Huang L, Dan BT, Zhang GJ, et al. Factors associated with prolonged viral shedding and impact of lopinavir/ritonavir treatment in hospitalised non-critically ill patients with SARS-CoV-2 infection. Eur Respir J. 2020;56(1):2000799. 10.1183/13993003.00799-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zhang J, Wang S, Xue Y. Fecal specimen diagnosis 2019 novel coronavirus-infected pneumonia. J Med Virol. 2020;92(6):680-2. 10.1002/jmv.25742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Mansour A, Atoui R, Kanso K, Mohsen R, Fares Y, Fares J. First Case of an Infant with COVID-19 in the Middle East. Cureus. 2020;12(4):e7520. 10.7759/cureus.7520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Cheng SC, Chang YC, Fan Chiang YL, Chien YC, Cheng M, Yang CH, et al. First case of Coronavirus Disease 2019 (COVID-19) pneumonia in Taiwan. J Formos Med Assoc. 2020;119(3):747-51. 10.1016/j.jfma.2020.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Ghinai I, McPherson TD, Hunter JC, Kirking HL, Christiansen D, Joshi K, et al. First known person-to-person transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the USA. Lancet. 2020;395(10230):1137-44. 10.1016/S0140-6736(20)30607-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Scott SE, Zabel K, Collins J, Hobbs KC, Kretschmer MJ, Lach M, et al. First mildly ill, nonhospitalized case of Coronavirus Disease 2019 (COVID-19) without viral transmission in the United States-Maricopa County, Arizona, 2020. Clin Infect Dis. 2020;71(15):807-12. 10.1093/cid/ciaa374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wajnberg A, Mansour M, Leven E, Bouvier NM, Patel G, Firpo A, et al. Humoral immune response and prolonged PCR positivity in a cohort of 1343 SARS-CoV 2 patients in the New York City region. medRxiv. 2020 May 5;2020.04.30.20085613
- 122. Gautret P, Lagier J-C, Parola P, Hoang VT, Meddeb L, Mailhe M, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;56(1):105949. 10.1016/j.ijantimicag.2020.105949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Tang W, Cao Z, Han M, Wang Z, Chen J, Sun W, et al. Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: open label, randomised controlled trial. BMJ. 2020;369:m1849. 10.1136/bmj.m1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mallat J, Hamed F, Balkis N, Mohamed MA, Mooty, M, Malik A. Hydroxychloroquine is associated with slower viral clearance in clinical COVID-19 patients with mild to moderate disease: A retrospective study. medRxiv. 2020 May 2;2020.04.27.20082180 [DOI] [PMC free article] [PubMed]
- 125.Chen X, Hu W, Ling J, Mo P, Zhang Y, Jiang Q, et al. Hypertension and diabetes delay the viral clearance in COVID-19 patients. medRxiv, 2020 Mar 24;2020.03.22.20040774
- 126.Fu W, Chen Q, Wang T. Letter to the Editor: Three cases of redetectable positive SARS-CoV-2 RNA in recovered COVID-19 patients with antibodies. J Med Virol. 2020;92(11):2298-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huang J, Mao T, Li S, Wu L, Xu Z, Li H, et al. Long period dynamics of viral load and antibodies for SARS-CoV-2 infection: an observational cohort study. medRxiv. 2020 Apr 27;2020.04.22.20071258
- 128.Lan X, Shao C, Zeng X, Wu Z, Xu Y. Lopinavir-ritonavir alone or combined with arbidol in the treatment of 73 hospitalized patients with COVID-19: a pilot retrospective study. medRxiv. 2020 Apr 29;2020.04.25.20079079 [DOI] [PubMed]
- 129. Fang X, Mei Q, Yang T, Li L, Wang Y, Tong F, et al. Low-dose corticosteroid therapy does not delay viral clearance in patients with COVID-19. J Infect. 2020;81(1):147-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shabrawishi MH, Naser AY, Alwafi H, Aldobyany AM, Touman AA. Negative nasopharyngeal SARS-CoV-2 PCR conversion in Response to different therapeutic interventions. medRxiv. 2020 May 11;2020.05.08.20095679 [DOI] [PMC free article] [PubMed]
- 131. Wang X, Zhou Q, He Y, Liu L, Ma X, Wei X, et al. Nosocomial outbreak of COVID-19 pneumonia in Wuhan, China. Eur Respir J. 2020;55(6):2000544. 10.1183/13993003.00544-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Chen L, Liu M, Zhang Z, Qiao K, Huang T, Chen M, et al. Ocular manifestations of a hospitalised patient with confirmed 2019 novel coronavirus disease. Br J Ophthalmol. 2020;104(6):748-51. 10.1136/bjophthalmol-2020-316304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Liu F, Xu A, Zhang Y, Xuan W, Yan T, Pan K, et al. Patients of COVID-19 may benefit from sustained Lopinavir-combined regimen and the increase of Eosinophil may predict the outcome of COVID-19 progression. Int J Infect Dis. 2020;95:183-91. 10.1016/j.ijid.2020.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Yang R, Gui X, Xiong Y. Patients with respiratory symptoms are at greater risk of COVID-19 transmission. Respir Med. 2020;165:105935. 10.1016/j.rmed.2020.105935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Ling Y, Xu SB, Lin YX, Tian D, Zhu ZQ, Dai FH, et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J (Engl). 2020;133(9):1039-43. 10.1097/CM9.0000000000000774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Zhang B, Liu S, Dong Y, Zhang L, Zhong Q, Zou Y, et al. Positive rectal swabs in young patients recovered from coronavirus disease 2019 (COVID-19). J Infect. 2020;81(2):e49-52. 10.1016/j.jinf.2020.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Qu YM, Kang EM, Cong HY. Positive result of Sars-Cov-2 in sputum from a cured patient with COVID-19. Travel Med Infect Dis. 2020;34:101619. 10.1016/j.tmaid.2020.101619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Luo A. Positive SARS-Cov-2 test in a woman with COVID-19 at 22 days after hospital discharge: A case report. Journal of Traditional Chinese Medical Sciences. 2020;7(4):413-7. 10.1016/j.jtcms.2020.04.001 [DOI] [Google Scholar]
- 139. Xing Y, Mo P, Xiao Y, Zhao O, Zhang Y, Wang F. Post-discharge surveillance and positive virus detection in two medical staff recovered from coronavirus disease 2019 (COVID-19), China, January to February 2020. Euro Surveill. 2020;25(10):2000191. 10.2807/1560-7917.ES.2020.25.10.2000191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Xiao AT, Tong YX, Zhang S. Profile of RT-PCR for SARS-CoV-2: a preliminary study from 56 COVID-19 patients. Clin Infect Dis. 2020;71(16):2249-51. 10.1093/cid/ciaa460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong X, et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5(5):434-5. 10.1016/S2468-1253(20)30083-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zhang WY, Yu LQ, Huang JA, Zeng DX. Prolonged Viral RNA Shedding Duration in COVID-19. Am J Ther. 2020;Publish Ahead of Print. 10.1097/MJT.0000000000001200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Xing YH, Ni W, Wu Q, Li WJ, Li GJ, Wang WD, et al. Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. J Microbiol Immunol Infect. 2020;53(3):473-80. 10.1016/j.jmii.2020.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Chen D, Xu W, Lei Z, Huang Z, Liu J, Gao Z, et al. Recurrence of positive SARS-CoV-2 RNA in COVID-19: A case report. Int J Infect Dis. 2020;93:297-9. 10.1016/j.ijid.2020.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Wang H, Li Y, Wang F, Du H, Lu X. Rehospitalization of a Recovered Coronavirus Disease 19 (COVID-19) Child With Positive Nucleic Acid Detection. Pediatr Infect Dis J. 2020;39(6):e69-70. 10.1097/INF.0000000000002690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen X, Ling J, Mo P, Zhang Y, Jiang Q, Ma Z, et al. Restoration of leukomonocyte counts is associated with viral clearance in COVID-19 hospitalized patients. medRxiv. 2020 Mar 6;2020.03.03.20030437.
- 147. Fu Y, Han P, Zhu R, Bai T, Yi J, Zhao X, et al. Risk factors for viral RNA shedding in COVID-19 patients. Eur Respir J. 2020;56(1):2001190. 10.1183/13993003.01190-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Colavita F, Lapa D, Carletti F, Lalle E, Bordi L, Marsella P, et al. SARS-CoV-2 isolation from ocular secretions of a patient with COVID-19 in Italy with prolonged viral RNA detection. Ann Intern Med. 2020;173(3):242-3. 10.7326/M20-1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Zhang JF, Yan K, Ye HH, Lin J, Zheng JJ, Cai T. SARS-CoV-2 turned positive in a discharged patient with COVID-19 arouses concern regarding the present standards for discharge. Int J Infect Dis. 2020;97:212-4. 10.1016/j.ijid.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Huang Y, Chen S, Yang Z, Guan W, Liu D, Lin Z, et al. SARS-CoV-2 viral load in clinical samples from critically ill patients. Am J Respir Crit Care Med. 2020;201(11):1435-8. 10.1164/rccm.202003-0572LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177-9. 10.1056/NEJMc2001737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Han MS, Seong MW, Heo EY, Park JH, Kim N, Shin S, et al. Sequential analysis of viral load in a neonate and her mother infected with Severe Acute Respiratory Syndrome Coronavirus 2. Clin Infect Dis. 2020;71(16):2236-9. 10.1093/cid/ciaa447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Peng J, Wang M, Zhang G, Lu E. Seven discharged patients turning positive again for SARS-CoV-2 on quantitative RT-PCR. Am J Infect Control. 2020;48(6):725-6. 10.1016/j.ajic.2020.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Le TQM, Takemura T, Moi ML, Nabeshima T, Nguyen LKH, Hoang VMP, et al. Severe Acute Respiratory Syndrome Coronavirus 2 Shedding by Travelers, Vietnam, 2020. Emerg Infect Dis. 2020;26(7):1624-6. 10.3201/eid2607.200591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Alshami AA, Alattas RA, Anan HF, Qahtani HSA, Mulhim MAA, Alahilmi AA, et al. Silent disease and loss of taste and smell are common manifestations of SARS-COV-2 infection in a quarantine facility: first report from Saudi Arabia. medRxiv. 2020 May 19;2020.05.13.20100222.
- 156. Lu Y, Li Y, Deng W, Liu M, He Y, Huang L, et al. Symptomatic infection is associated with prolonged duration of viral shedding in mild Coronavirus Disease 2019: a retrospective study of 110 children in Wuhan. Pediatr Infect Dis J. 2020;39(7):e95-9. 10.1097/INF.0000000000002729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565-74. 10.1016/S1473-3099(20)30196-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Yuan J, Zou R, Zeng L, Kou S, Lan J, Li X, et al. The correlation between viral clearance and biochemical outcomes of 94 COVID-19 infected discharged patients. Inflamm Res. 2020;69(6):599-606. 10.1007/s00011-020-01342-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Zhou B, She J, Wang Y, Ma X. Duration of viral shedding of discharged patients with severe COVID-19. Clin Infect Dis. 2020;71(16):2240-2. 10.1093/cid/ciaa451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Le HT, Nguyen LV, Tran DM, Do HT, Tran HT, Le YT, et al. The first infant case of COVID-19 acquired from a secondary transmission in Vietnam. Lancet Child Adolesc Health. 2020;4(5):405-6. 10.1016/S2352-4642(20)30091-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Hill KJ, Russell CD, Clifford S, Templeton K, Mackintosh CL, Koch O, et al. The index case of SARS-CoV-2 in Scotland. J Infect. 2020;81(1):147-78. 10.1016/j.jinf.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Van Vinh Chau N, Lam VT, Dung NT, Yen LM, Minh NNQ, Hung LM, et al. The natural history and transmission potential of asymptomatic Severe Acute Respiratory Syndrome Coronavirus 2 Infection. Clin Infect Dis. 2020;71(10):2679-87. 10.1093/cid/ciaa711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Chen Y, Chen L, Deng Q, Zhang G, Wu K, Ni L, et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J Med Virol. 2020;92(7):833-40. 10.1002/jmv.25825 [DOI] [PubMed] [Google Scholar]
- 164. Zhao F, Yang Y, Wang Z, Li L, Liu L, Liu Y. The time sequences of respiratory and rectal viral shedding in patients with Coronavirus Disease 2019. Gastroenterology. 2020;159(3):1158-1160.e2. 10.1053/j.gastro.2020.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Chang D, Mo G, Yuan X, Tao Y, Peng X, Wang FS, et al. Time kinetics of viral clearance and resolution of symptoms in Novel Coronavirus infection. Am J Respir Crit Care Med. 2020;201(9):1150-2. 10.1164/rccm.202003-0524LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Su JW, Wu WR, Lang GJ, Zhao H, Sheng JF. Transmission risk of patients with COVID-19 meeting discharge criteria should be interpreted with caution. J Zhejiang Univ Sci B. 2020;21(5):408-10. 10.1631/jzus.B2000117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Shen C, Wang Z, Zhao F, Yang Y, Li J, Yuan J, et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323(16):1582-9. 10.1001/jama.2020.4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kim MS, Jang S-W, Park Y-K, Kim B, Hwang T-H, Kang SH, et al. Treatment response to Hydroxychloroquine, Lopinavir/Ritonavir, and antibiotics for moderate COVID 19: a first report on the pharmacological outcomes from South Korea. medRxiv. 2020 May 18;2020.05.13.20094193.
- 169. Hung IF, Lung KC, Tso EY, Liu R, Chung TW, Chu MY, et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395(10238):1695-704. 10.1016/S0140-6736(20)31042-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Zhou R, Li F, Chen F, Liu H, Zheng J, Lei C, et al. Viral dynamics in asymptomatic patients with COVID-19. Int J Infect Dis. 2020;96:288-90. 10.1016/j.ijid.2020.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Liu Y, Yan LM, Wan L, Xiang TX, Le A, Liu JM, et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020;20(6):656-7. 10.1016/S1473-3099(20)30232-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Lui G, Ling L, Lai CK, Tso EY, Fung KS, Chan V, et al. Viral dynamics of SARS-CoV-2 across a spectrum of disease severity in COVID-19. J Infect. 2020;81(2):318-56. 10.1016/j.jinf.2020.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tan W, Lu Y, Zhang J, Wang J, Dan Y, Tan Z, et al. Viral kinetics and antibody responses in patients with COVID-19. medRxiv. 2020 Mar 26;2020.03.24.20042382.
- 174. Zheng S, Fan J, Yu F, Feng B, Lou B, Zou Q, et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. 10.1136/bmj.m1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20(4):411-2. 10.1016/S1473-3099(20)30113-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Yuan C, Zhu H, Yang Y, Cai X, Xiang F, Wu H, et al. Viral loads in throat and anal swabs in children infected with SARS-CoV-2. Emerg Microbes Infect. 2020;9(1):1233-7. 10.1080/22221751.2020.1771219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang N, Gong Y, Meng F, Bi Y, Yang P, Wang F. Virus shedding patterns in nasopharyngeal and fecal specimens of COVID-19 patients. medRxiv. 2020 Mar 30;2020.03.28.20043059. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.