Abstract
Background
Hyperlipidemia plays an important role in the etiology of cardio-cerebrovascular disease. Over recent years, a number of studies have explored the impact of apolipoprotein genetic polymorphisms in hyperlipidemia, but considerable differences and uncertainty have been found in their association with different populations from different regions.
Results
A total of 59 articles were included, containing in total 13,843 hyperlipidemia patients in the case group and 15,398 healthy controls in the control group. Meta-analysis of the data indicated that APOA5–1131 T > C, APOA1 -75 bp, APOB XbaI, and APOE gene polymorphisms were significantly associated with hyperlipidemia, with OR values of 1.996, 1.228, 1.444, and 1.710, respectively. All P-values were less than 0.05.
Conclusions
Meta-analysis of the data indicated that the C allele of APOA5 1131 T > C, the A allele at APOA1-75 bp, the APOB XbaI T allele, and the ε2 and ε4 allele of APOE were each a risk factor for susceptibility for hyperlipidemia.
Keywords: Apolipoprotein, APO, Gene polymorphism, Hyperlipidemia, Meta-analysis
Background
Cardio-cerebrovascular disease is the leading cause of increased human mortality, globally [1]. Recently, studies have shown that the fatality rate from cardio-cerebrovascular disease accounts for approximately 30% of the total global death toll [2]. Hyperlipidemia is a chronic non-communicable disease caused by an imbalance in the structure of plasma lipids caused by a fat metabolism disorder [3]. It is the primary risk factor for atherosclerosis and the pathological basis for cardio-cerebrovascular disease [4]. In addition, a large number of manuscripts have demonstrated that hyperlipidemia is a pathogenic factor of digestive and urinary diseases such as diabetes, hepatopathy, and pancreatitis. Hyperlipidemia can be categorized as hypercholesteremia, hypertriglyceridemia, mixed hyperlipidemia, and low-density lipoproteinemia, etc. Medical research has established that the mechanism of hyperlipidemia is not only determined by environmental factors, such as long-term consumption of large quantities of saturated fatty acids, cholesterol, and sugar, it is also influenced by genetic factors at gene loci. There are multiple academic reports that apolipoprotein (APO) gene mutations are closely related to disorders of blood lipid metabolism [5]. APO is an important component of lipoprotein. So far, more than 20 forms of APO have been identified, including APOA, APOB, APOC, APOD, APOE, APOH, APOM, etc. [6]
Single nucleotide polymorphisms (SNPs) are changes to a single nucleic acid in a protein caused by the insertion, deletion, or substitution of a single nucleotide base in the gene sequence. Of the existing apolipoprotein candidate genes, researchers have correlated APOA1, APOA5, APOB, and APOE gene polymorphisms with hyperlipidemia. APOA1 and APOA5 genes are located in the long arm region of chromosome 11. APOA1 is located in the APOA1-C3-A4 gene cluster, the principal site controlling the expression of lipids and lipoproteins [7]. APOA5 is located downstream of APOA4, and its distance from the APOA1/C3/A4 gene cluster is approximately 30 kb. The APOA5 gene is most commonly altered at -1131 T > C, this polymorphism being closely associated with a number of diseases, such as hypertriglyceridemia and coronary heart disease [8]. The APOB gene is located in the short arm of chromosome 2 and contains 29 exons and 28 introns. The cleavage sites MspI and XbaI are located within exon 26 of the APOB gene. The EcoRI cleavage site is located within exon 29 [9]. A number of studies have clearly indicated that the APOB gene affects lipid metabolism to a certain extent. The APOE gene is located on chromosome 19 with a polymorphic gene structure. The isomers are encoded by the three alleles ε2, ε3, and ε4 [10], forming 6 genotypes E2/2, E3/3, E4/4, E2/3, E2/4, and E3/4, of which E3/3 is the most common within the population.
Over recent years, there have been multiple studies that have explored the correlation between genetic polymorphism and hyperlipidemia for the apolipoprotein gene loci described above, but there are great differences and uncertainties in different populations from different regions. Therefore, in the present review, we systematically searched the literature and reviewed case-control studies of hyperlipidemia. A meta-analysis was conducted to explore the relationship between APOA (A1-75bp, A1 + 83 bp, A5–1131T>C), APOB (MspI, XbaI, EcorI), and APOE with hyperlipidemia so that an evidence-base can be provided for the prevention and control of hyperlipidemia.
Results
Study characteristics
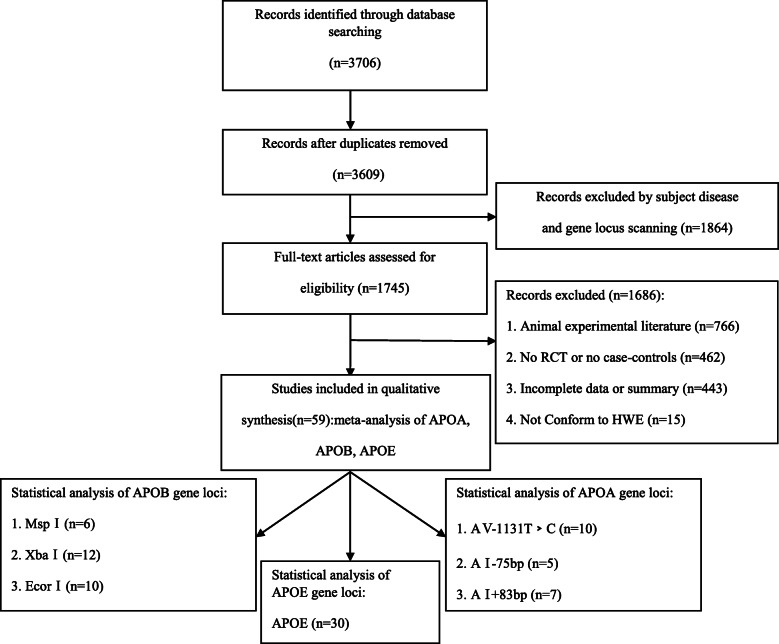
A total of 3706 articles were identified in the Chinese and English databases, of which 59 articles were finally selected, including 22 that analyzed APOA, 28 APOB, and 30 APOE. Three sites in the APOA gene were studied: A5–1131T > C was studied in 10 case-control studies that included 1211 cases and 1495 controls; A1-75bp was studied in 5 case-control studies that included 1284 cases and 1312 controls; and A1 + 83 bp was studied in 7 case-control studies that included 1452 cases and 1620 controls. The APOB gene was investigated at three sites: MspI was studied in 6 case-control studies that included a hyperlipidemia group, with 1155 cases and 1043 controls; XbaI was studied in 12 case-control studies that included 1900 cases and 1836 controls; and EcorI was studied in 10 case-control studies that included 1633 cases and 1686 controls. The APOE gene is co-coded by the three alleles, ε2, ε3, and ε4, for which 30 case control studies were studied that included 5208 cases in the hyperlipidemia group and 6406 cases in the control group. The NOS score of no study included in the review was less than 7. The comparison between case and control groups was highly credible. The specific process for literature retrieval is displayed in Fig. 1.
Fig. 1.
Flow diagram of the meta-analysis
Meta-analysis of APOA5–1131 T > C (rs662799)
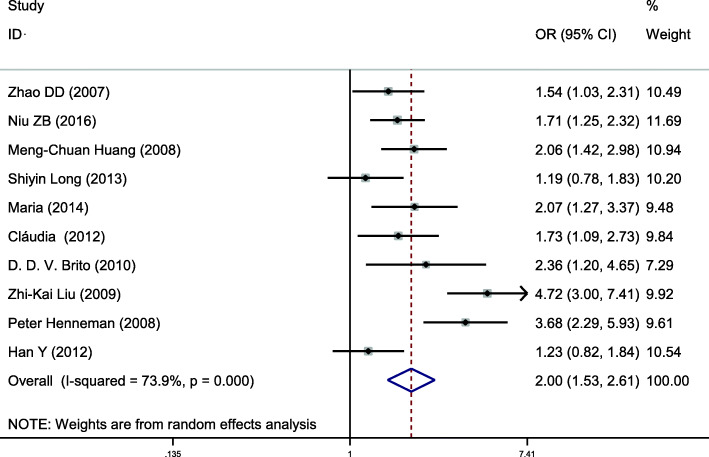
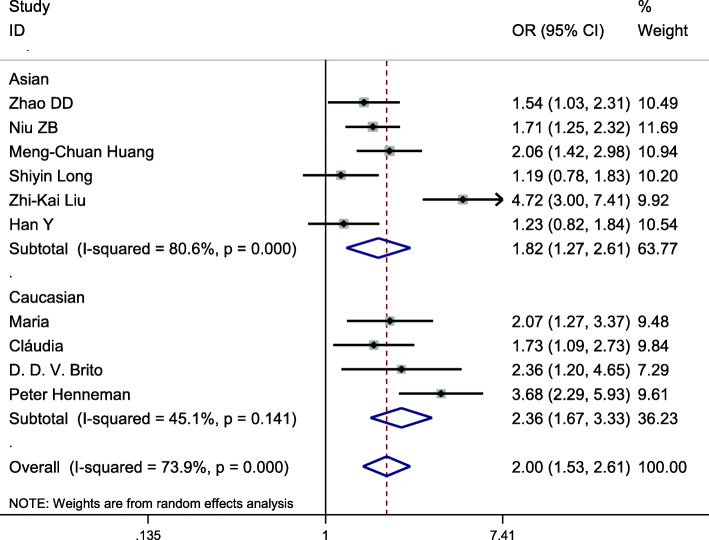
This gene locus was included in 10 case-control studies, involving a total of 2706 subjects, including 1211 in the hyperlipidemia group and 1496 in the control group. The baseline data and quality evaluation of each study are displayed in Table 1. Analysis of the relationship between C vs T alleles and hyperlipidemia (allele model) revealed substantial heterogeneity (I2 = 73.9%, P < 0.001), so a random-effects model was used to analyze the combined effects. Individuals with the C allele had a higher risk of hyperlipidemia than those with the T allele, a difference that was statistically significant (OR = 1.996, 95% CI = 1.529–2.606, P < 0.001) (Fig. 2). Other gene models at this site displayed consistent results (Table 2). Subgroup analysis by ethnicity demonstrated an increased risk of hyperlipidemia among Asians (OR = 1.818; 95% CI = 1.268–2.607, P = 0.001) and Caucasians (OR = 2.355; 95% CI = 1.665 ~ 3.331, P < 0.001) that had the C allele, using the allele model. Other gene models at this site displayed results that were consistent with this (Table 3, Fig. 3). Therefore, the single nucleotide polymorphism APOA5–1131 T > C was associated with hyperlipidemia, the C allele posing a risk factor for susceptibility to hyperlipidemia.
Table 1.
Main characteristics of the studies of APOA included in the review
| SNP | First author | Year | Area | Sample size | Age (y) | Source of control | Genotyping method | Cases | Controls | NOS | HWE | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | TT/GG/CC | CT/GA/CT | CC/AA/TT | TT/GG/CC | CT/GA/CT | CC/AA/TT | χ2 | P | |||||||
|
APOA5–1131 T>C |
Zhao DD [11] | 2007 | Beijing, China | 172 | 80 | NR | NR | HB | PCR-RFLP | 63 | 86 | 23 | 39 | 36 | 5 | 7 | 0.77 | 0.37 |
| Niu ZB [12] | 2016 | Shanghai, China | 156 | 262 | NR | NR | PB | MALDI-TOF | 68 | 68 | 20 | 153 | 94 | 15 | 9 | 0.01 | 0.91 | |
| Huang M [13] | 2008 | Taiwan, China | 76 | 240 | 59.57 ± 10.2 | 60.98 ± 13.58 | PB | PCR-RFLP | 15 | 41 | 20 | 99 | 111 | 30 | 8 | 0.02 | 0.9 | |
| Long SY [14] | 2013 | Hunan, China | 95 | 102 | 61 ± 12 | 62 ± 12 | HB | PCR-RFLP | 46 | 36 | 13 | 50 | 45 | 7 | 7 | 0.54 | 0.46 | |
| Maria [15] | 2014 | Napoli, Italian | 165 | 142 | 47.5 ± 12.2 | 43.9 ± 9.6 | HB | TaqMan | 111 | 49 | 5 | 117 | 23 | 2 | 7 | 0.49 | 0.48 | |
| Cláudia [16] | 2012 | Minas Gerais, Brazil | 108 | 107 | 48.4 ± 6.8 | 46.7 ± 6.6 | PB | PCR-RFLP | 52 | 52 | 4 | 71 | 33 | 3 | 7 | 0.13 | 72 | |
| Brito [17] | 2010 | Belo Horizonte, Brazil | 53 | 77 | 10.4 ± 2.7 | 11.2 ± 3.4 | HB | PCR-RFLP | 34 | 14 | 5 | 62 | 13 | 2 | 6 | 1.52 | 0.22 | |
| ZK Liu [18] | 2009 | Hongkong, China | 56 | 176 | 49.6 ± 12.3 | 50.1 ± 9.4 | HB | PCR | 9 | 27 | 20 | 101 | 61 | 11 | 7 | 0.19 | 0.66 | |
| Peter H [19] | 2008 | Netherlands | 254 | 240 | NR | NR | HB | PCR | 142 | 72 | 7 | 172 | 22 | 1 | 6 | 0.11 | 0.75 | |
| Han Y [8] | 2012 |
Hunan, China |
109 | 117 | 60.3 ± 12.1 | 62.9 ± 12.0 | HB | PCR-RFLP | 52 | 43 | 14 | 59 | 50 | 8 | 7 | 0.36 | 0.55 | |
| APOA1-75 bp | Huang G [20] | 2011 |
Xinjiang, China |
275 | 252 | 47.7 ± 7.9 | 48.23 ± 7.6 | HB | PCR-RFLP | 135 | 102 | 38 | 136 | 95 | 21 | 8 | 0.57 | 0.49 |
| Feng DW [7] | 2016 |
Xinjiang, China |
365 | 370 | 46.8 ± 15.9 | 45.21 ± 16.4 | PB | PCR | 248 | 104 | 13 | 280 | 83 | 7 | 9 | 0.09 | 0.77 | |
| Feng DW [7] | 2016 |
Xinjiang, China |
345 | 391 | 43.9 ± 14.3 | 41.5 ± 13.3 | PB | PCR | 250 | 87 | 7 | 299 | 86 | 5 | 9 | 0.18 | 0.67 | |
| Chi YH [21] | 2012 | Xinjiang,China | 200 | 200 | 58.5 ± 11.8 | 58.3 ± 11.5 | PB | PCR-RFLP | 116 | 82 | 2 | 124 | 73 | 5 | 7 | 2.31 | 1.29 | |
| Bora K [2] | 2017 | Assam, India | 100 | 100 | 43.1 ± 11.6 | 43.0 ± 11.6 | PB | PCR-RFLP | 62 | 35 | 3 | 60 | 33 | 7 | 8 | 0.68 | 0.41 | |
| APOA1+83 bp | Xie YJ [22] | 2011 |
Xinjiang, China |
150 | 150 | 56.8 ± 10.8 | 58.1 ± 10.5 | HB | PCR-RFLP | 126 | 24 | 0 | 130 | 20 | 0 | 7 | 0.77 | 0.38 |
| Ou HJ [5] | 2015 |
Xinjiang, China |
241 | 246 | 49.1 ± 0.7 | 48.3 ± 0.8 | HB | MALDI-TOF | 160 | 80 | 1 | 171 | 73 | 2 | 7 | 3.78 | 0.05 | |
| Feng DW [7] | 2016 |
Xinjiang, China |
365 | 370 | 46.8 ± 15.9 | 45.2 ± 16.4 | PB | PCR | 317 | 48 | 0 | 304 | 63 | 3 | 9 | 0.02 | 0.89 | |
| Feng DW [7] | 2016 | Xinjiang,China | 345 | 391 | 43.91 ± 14.27 | 41.51 ± 13.28 | PB | PCR | 299 | 44 | 1 | 330 | 57 | 3 | 9 | 0.1 | 0.76 | |
| Zhu H [23] | 2001 |
Sichuan, China |
134 | 255 | 54.7 ± 12.6 | 51.7 ± 10.9 | PB | PCR | 123 | 11 | 0 | 238 | 17 | 0 | 7 | 0.3 | 0.58 | |
| Jia LQ [24] | 2005 |
Sichuan, China |
118 | 109 | 58.1 ± 8.9 | 54.5 ± 9.6 | NR | PCR | 105 | 13 | 0 | 99 | 10 | 0 | 6 | 0.25 | 0.62 | |
| Bora K [2] | 2017 | Assam, India | 100 | 100 | 43.12 ± 11.64 | 42.95 ± 11.60 | PB | PCR-RFLP | 89 | 11 | 0 | 87 | 13 | 0 | 8 | 0.48 | 0.49 | |
SNP single nucleotide polymorphism, PB population-based; HB: hospital-based, HWE Hardy-Weinberg equilibrium, NR not reported
Fig. 2.
Pooled calculated OR for the association between the APOA5–1131 T > C allele and hyperlipidemia
Table 2.
Summary of the meta-analysis of the association of APOA gene polymorphisms with hyperlipidemia
| SNP | Analysis model | Genotype model | Heterogeneity(I2/P) | OR (95%CI) | P | Publication bias P |
|---|---|---|---|---|---|---|
| APOA5–1131 T>C | A | C vs T | 73.9%/ < 0.001 | 1.996(1.529 ~ 2.606) | < 0.001 | 0.353 |
| D | TC + CC vs TT | 71.2%/ < 0.001 | 2.179(1.565 ~ 3.035) | < 0.001 | 0.258 | |
| R | CC vs TC + TT | 5.5%/ 0.390 | 2.790(2.055 ~ 3.789) | < 0.001 | 0.991 | |
| C | CC vs TT | 45.7%/ 0.056 | 3.604(2.589 ~ 5.017) | < 0.001 | 0.899 | |
| TC vs TT | 67.2%/ 0.001 | 1.932(1.395 ~ 2.674) | < 0.001 | 0.465 | ||
| APOA1-75 bp | A | A vs G | 1.2%/ 0.400 | 1.228(1.067 ~ 1.413) | 0.004 | 0.086 |
| D | AA+GA vs GG | 0.0%/ 0.704 | 1.246(1.056 ~ 1.471) | 0.009 | 0.067 | |
| R | AA vs GA + GG | 15.9%/ 0.313 | 1.458(0.976 ~ 2.180) | 0.066 | 0.086 | |
| C | AA vs GG | 17.4%/ 0.304 | 1.520(1.008 ~ 2.291) | 0.046 | 0.086 | |
| GA vs GG | 0.0%/ 0.828 | 1.212(1.020 ~ 1.439) | 0.029 | 0.221 | ||
| APOA1 + 83 bp | A | T vs C | 0.0%/ 0.472 | 0.928(0.771 ~ 1.116) | 0.425 | 0.440 |
| D | TT + TC vs CC | 0.0%/ 0.478 | 0.950(0.780 ~ 1.157) | 0.607 | 0.371 | |
| R | TT vs TC + CC | 0.0%/ 0.799 | 0.310(0.076 ~ 1.271) | 0.104 | 0.315 | |
| C | TT vs CC | 0.0%/ 0.775 | 0.308(0.075 ~ 1.259) | 0.101 | 0.346 | |
| TC vs CC | 0.0%/ 0.607 | 0.967(0.793 ~ 1.180) | 0.740 | 0.466 |
A allelic model; D dominant model; R recessive model; C codominant model; Publication bias P: using Begg’s or Egger’s tests
Table 3.
Subgroup analysis by ethnicity of the APOA5–1131 T>C polymorphism on susceptibility to hyperlipidemia
| Ethnicity | Analysis model | Genotype model | OR (95%CI) | P |
|---|---|---|---|---|
| Asian | A | C vs T | 1.818(1.268 ~ 2.607) | 0.001 |
| D | TC + CC vs TT | 1.943(1.211 ~ 3.117) | 0.006 | |
| R | CC vs TC + TT | 2.794(2.011 ~ 3.883) | < 0.001 | |
| C | CC vs TT | 3.785(1.997 ~ 7.173) | < 0.001 | |
| TC vs TT | 1.622(1.060 ~ 2.482) | 0.026 | ||
| Caucasian | A | C vs T | 2.355(1.665 ~ 3.331) | < 0.001 |
| D | TC + CC vs TT | 1.943(1.918 ~ 3.749) | < 0.001 | |
| R | CC vs TC + TT | 2.790(2.055 ~ 3.789) | 0.016 | |
| C | CC vs TT | 3.282(1.392 ~ 7.739) | 0.007 | |
| TC vs TT | 2.600(1.873 ~ 3.609) | < 0.001 |
A allelic model; D dominant model; R recessive model; C codominant model
Fig. 3.
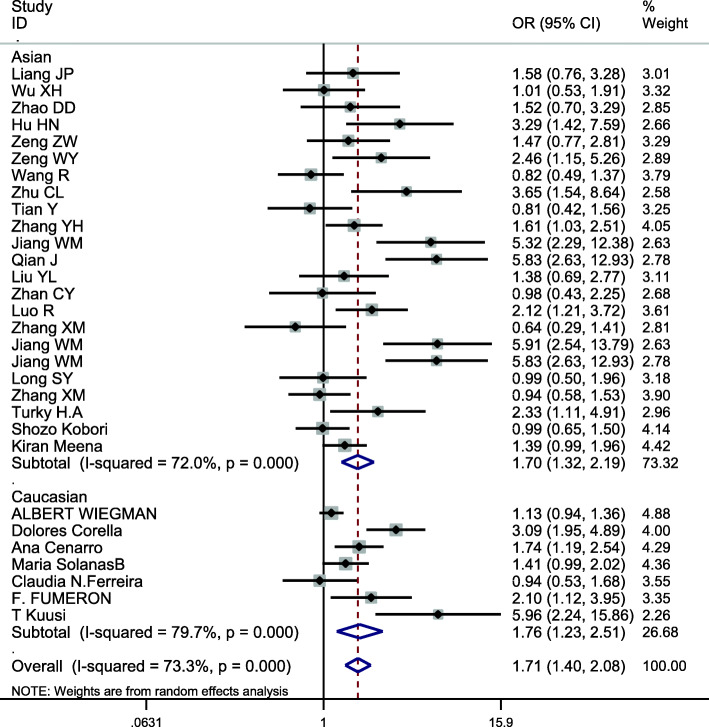
Subgroup analysis by ethnicity for the association between the APOA5–1131 T > C allele and the risk of hyperlipidemia
Meta-analysis of APOA1-75 bp (rs670)
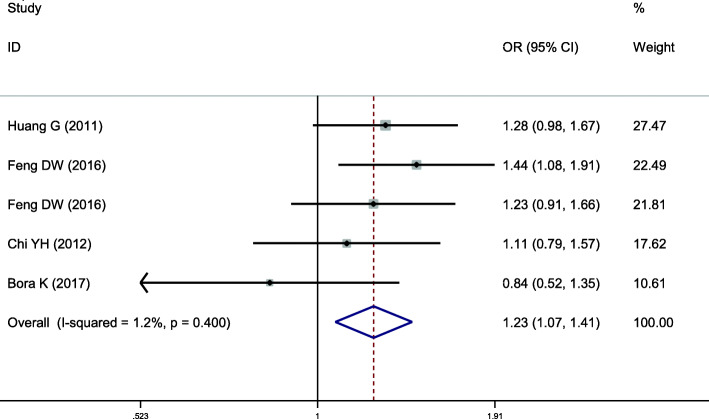
This location on APOA was included in 5 case-control studies, involving a total of 2596 subjects, of which 1284 were in the hyperlipidemia group and 1312 in the control group. Baseline data and quality evaluation are displayed in Table 1. There was no significant heterogeneity in the relationship between A vs G alleles and hyperlipidemia (allele model) (I2 = 1.2%, P = 0.400), and so a fixed-effects model was used to combine the effects. Individuals with the A allele had a higher risk of hyperlipidemia than those with the G allele, a difference that was statistically significant (OR = 1.228, 95% CI = 1.067–1.413, P = 0.004) (Fig. 4). The recessive model of this locus indicated that the difference was not statistically significant (P = 0.066). Other gene models at this site were consistent with this result, suggesting that the single nucleotide polymorphism APOA1-75 bp is associated with hyperlipidemia, the A allele being a risk factor for susceptibility to hyperlipidemia (Table 2).
Fig. 4.
Pooled calculated OR for the association between the APOA1-75 bp allele and hyperlipidemia
Meta-analysis of APOA1 + 83 bp (rs5069)
This site was included in 7 case-control studies, involving a total of 3072 subjects, including 1452 in the hyperlipidemia group and 1620 in the control group. The baseline data and quality evaluation of each study are shown in Table 1. Analysis of the relationship between A vs G alleles and hyperlipidemia (allele model) indicated that there was no significant heterogeneity (I2 = 0.0%, P = 0.472). Therefore, a fixed-effects model was selected to analyze the pooled effect. There was no significant difference in risk in individuals that carried the T allele compared with C (OR = 0.928, 95% CI = 0.771–1.116, P = 0.425). The P-values of other gene models at this locus were all higher than 0.05, suggesting that there was no significant difference. Thus, an association between APOA1 + 83 bp gene polymorphism and susceptibility to hyperlipidemia can be considered not to exist (Table 2).
Meta-analysis of APOB MspI (rs1801701)
This gene locus was included in 6 case-control studies, involving a total of 2198 subjects, including 1155 in the hyperlipidemia group and 1043 in the control group. Baseline data and quality evaluation are shown in Table 4. Analysis of the association between M- vs M+ alleles and hyperlipidemia (allele model) indicated no heterogeneity (I2 = 0.0%, P = 0.731), and do a fixed-effects model was selected to analyze the pooled effects. No significant difference in risk was found in individuals carrying the M- compared with the M+ allele (OR = 0.892, 95% CI = 0.756–1.053, P = 0.178). The P-values of other gene models at this site were also greater than 0.05, indicating that there was no significant difference. Thus, no association between genetic polymorphism of APOB MspI and risk of hyperlipidemia was found (Table 5).
Table 4.
Principal characteristics of the studies of APOB included in the review
| SNP | First author | Year | Area | Sample size | Age (y) | Source of control | Genotyping method | Cases | Controls | NOS | HWE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | M-M−/TT/ AA | M + M−/CT/ AG | M + M+ /CC/ GG | M-M−/TT/ AA | M + M−/CT/ AG | M + M+ /CC/ GG | χ2 | P | ||||||||
| APOB Msp | Cao WJ [25] | 2009 | Xinjiang, China | 100 | 90 | 46 ± 11 | 44 ± 11 | HB | PCR-RFLP | 0 | 4 | 95 | 0 | 3 | 87 | 6 | 0.03 | 0.87 | |
| Chi YH [26] | 2012 | Xinjiang, China | 247 | 221 | 48.7 ± 7.7 | 47.3 ± 6.2 | HB | PCR-RFLP | 9 | 70 | 168 | 6 | 67 | 148 | 7 | 0.24 | 0.63 | ||
| Huang G [20] | 2011 | Xinjiang, China | 275 | 252 | 47.7 ± 7.9 | 48.2 ± 7.6 | HB | PCR-RFLP | 25 | 68 | 182 | 22 | 69 | 161 | 8 | 3.43 | 0.06 | ||
| Jin YN [27] | 2015 | Chongqing,China | 157 | 180 | 48.1 ± 3.8 | 49.1 ± 4.2 | HB | DNA chips | 0 | 26 | 131 | 0 | 35 | 145 | 7 | 2.09 | 0.15 | ||
| Chi YH [21] | 2012 | Xinjiang, China | 200 | 200 | 58.5 ± 11.8 | 58.3 ± 11.5 | PB | PCR-RFLP | 6 | 66 | 128 | 12 | 64 | 124 | 7 | 0.91 | 0.34 | ||
| Selma [28] | 2000 | Sao Paulo, Brazil | 177 | 100 | 58 | 44 | HB | PCR | 2 | 25 | 150 | 1 | 24 | 75 | 6 | 0.37 | 0.54 | ||
| APOB XbaI | Qian J [29] | 2010 | Yunnan, China | 91 | 76 | 46.9 ± 11.4 | 47.5 ± 8.1 | HB | DNA chips | 0 | 7 | 84 | 1 | 11 | 64 | 7 | 0.42 | 0.51 | |
| Feng JS [30] | 1997 | Guangdong, China | 108 | 128 | 40–70 | HB | DNA probe | 0 | 8 | 100 | 0 | 11 | 117 | 6 | 0.26 | 0.61 | |||
| Ma ZZ [31] | 2012 | Guangdong, China | 250 | 250 | 45.50 ± 13.20 | PB | PCR-RFLP | 0 | 52 | 198 | 0 | 28 | 222 | 8 | 0.88 | 0.35 | |||
| Chi YH [26] | 2012 | Xinjiang, China | 247 | 221 | 48.7 ± 7.7 | 47.3 ± 6.2 | HB | PCR-RFLP | 4 | 54 | 189 | 3 | 41 | 177 | 7 | 0.13 | 0.72 | ||
| Xie YJ [22] | 2011 | Xinjiang, China | 150 | 150 | 56.8 ± 10.8 | 58.1 ± 10.5 | HB | PCR-RFLP | 2 | 29 | 119 | 0 | 12 | 138 | 7 | 0.26 | 0.61 | ||
| Jin YN [27] | 2015 | Chongqing,China | 157 | 180 | 48.1 ± 3.8 | 49.1 ± 4.2 | HB | DNA chips | 0 | 28 | 129 | 0 | 35 | 145 | 7 | 2.09 | 0.15 | ||
| Zhang PZ [32] | 2015 |
Beijing, China |
100 | 100 | 60.0 ± 5.0 | HB | PCR | 0 | 20 | 80 | 0 | 5 | 95 | 8 | 0.07 | 0.8 | |||
| Ou HJ [5] | 2015 | Xinjiang, China | 241 | 246 | 49.1 ± 0.7 | 48.3 ± 0.8 | HB | MALDI-TOF | 0 | 19 | 222 | 0 | 32 | 214 | 7 | 1.19 | 0.28 | ||
| Selma [28] | 2000 | Sao Paulo, Brazil | 177 | 100 | 58 | 44 | HB | PCR | 30 | 94 | 53 | 13 | 55 | 32 | 6 | 1.99 | 0.16 | ||
| Philippa [33] | 1987 | London, U.K. | 133 | 62 | NR | HB | PCR-RFLP | 43 | 59 | 31 | 12 | 38 | 12 | 6 | 3.16 | 0.08 | |||
| Gong LG [34] | 2003 | Liaoning, China | 115 | 150 | 54.2 ± 11.7 | 52.5 ± 13.1 | HB | PCR-RFLP | 1 | 29 | 85 | 0 | 12 | 138 | 6 | 0.26 | 0.61 | ||
| CHOONG [35] | 1999 | Singapore | 131 | 173 | NR | HB | PCR-RFLP | 0 | 25 | 106 | 0 | 21 | 152 | 6 | 0.72 | 0.4 | |||
| APOB EcorI | Qian J [29] | 2010 | Yunnan, China | 91 | 76 | 46.9 ± 11.4 | 47.5 ± 8.06 | HB | DNA chips | 0 | 13 | 78 | 0 | 3 | 73 | 7 | 0.03 | 0.86 | |
| Ma ZZ [31] | 2012 | Guangdong, China | 250 | 250 | 45.5 ± 13.2 | PB | PCR-RFLP | 0 | 41 | 209 | 0 | 28 | 222 | 8 | 0.88 | 0.35 | |||
| Huang G [20] | 2011 | Xinjiang, China | 275 | 252 | 47.7 ± 7.9 | 48.2 ± 7.6 | HB | PCR-RFLP | 12 | 73 | 190 | 10 | 77 | 165 | 8 | 0.07 | 0.79 | ||
| Xie YJ [22] | 2011 | Xinjiang, China | 150 | 150 | 56.8 ± 10.8 | 58.1 ± 10.5 | HB | PCR-RFLP | 1 | 55 | 94 | 0 | 19 | 131 | 7 | 0.69 | 0.41 | ||
| Jin YN [27] | 2015 | Chongqing,China | 157 | 180 | 48.1 ± 3.8 | 49.11 ± 4.2 | HB | DNA chips | 0 | 12 | 145 | 0 | 20 | 160 | 7 | 0.62 | 0.43 | ||
| Zhang PZ [32] | 2015 |
Beijing, China |
100 | 120 | 60.0 ± 5.0 | HB | PCR | 1 | 19 | 80 | 1 | 11 | 108 | 8 | 1.33 | 0.25 | |||
| Ou HJ [5] | 2015 | Xinjiang, China | 241 | 246 | 49.1 ± 0.7 | 48.3 ± 0.8 | HB | MALDI-TOF | 1 | 29 | 211 | 0 | 22 | 224 | 7 | 0.54 | 0.46 | ||
| Chi YH [21] | 2012 | Xinjiang, China | 200 | 200 | 58.5 ± 11.8 | 58.3 ± 11.5 | PB | PCR-RFLP | 6 | 52 | 142 | 6 | 56 | 138 | 7 | 0.01 | 0.91 | ||
| CHOONG [35] | 1999 | Singapore | 131 | 173 | NR | HB | PCR-RFLP | 0 | 9 | 122 | 0 | 16 | 157 | 6 | 0.41 | 0.52 | |||
| Timirci O [36] | 2010 | Capa-Istanbul, Turkey | 38 | 39 | 11.5 ± 3.6 | 11.4 ± 3.2 | HB | PCR | 0 | 4 | 34 | 0 | 4 | 35 | 7 | 0.11 | 0.74 | ||
SNP single nucleotide polymorphism, PB population-based; HB: hospital-based, HWE Hardy-Weinberg equilibrium, NR not reported
Table 5.
Summary of the results of the meta-analysis of the association of APOB gene polymorphisms and hyperlipidemia
| SNP | Analysis model | Genotype model | Heterogeneity(I2/P) | OR(95%CI) | P | Publication bias P |
|---|---|---|---|---|---|---|
| APOB MspI | A | M- vs M+ | 0.0%/ 0.731 | 0.892(0.756 ~ 1.053) | 0.178 | 0.452 |
| D | M-M−/M + M- Vs M + M+ | 0.0%/0.716 | 0.868(0.716 ~ 1.053) | 0.152 | 0.707 | |
| R | M-M-vs M + M−/M + M+ | 0.0%/ 0.513 | 0.932(0.596 ~ 1.456) | 0.757 | 0.908 | |
| C | M-M- vs M + M+ | 0.0%/ 0.555 | 0.903(0.574 ~ 1.421) | 0.660 | 0.883 | |
| M + M- vs M + M+ | 0.0%/ 0.654 | 0.864(0.705 ~ 1.057) | 0.156 | 0.746 | ||
| APOB XbaI | A | T vs C | 72.4%/ < 0.001 | 1.444(1.061 ~ 1.966) | 0.020 | 0.732 |
| D | TT + CT vs CC | 73.5%/ < 0.001 | 1.360(0.943 ~ 1.962) | 0.100 | 0.945 | |
| R | TT vs CT + CC | 0.0%/ 0.747 | 1.613(1.022 ~ 2.545) | 0.040 | 0.707 | |
| C | TT vs CC | 0.0%/ 0.774 | 1.432(0.851 ~ 2.411) | 0.017 | 0.724 | |
| CT vs CC | 73.5%/ < 0.001 | 1.322(0.912 ~ 1.917) | 0.140 | 0.837 | ||
| APOB EcorI | A | A vs G | 70.0%/ < 0.001 | 1.333(0.942 ~ 1.885) | 0.104 | 0.474 |
| D | AA+AG Vs GG | 72.9%/ < 0.001 | 1.366(0.924 ~ 2.020) | 0.118 | 0.283 | |
| R | AA vs AG + GG | 0.0%/ 0.942 | 1.183(0.628 ~ 2.229) | 0.603 | 0.221 | |
| C | AA vs GG | 0.0%/ 0.886 | 1.166(0.617 ~ 2.202) | 0.637 | 0.086 | |
| AG vs GG | 72.6%/ < 0.001 | 1.356(0.913 ~ 2.015) | 0.131 | 0.371 |
A allelic model; D dominant model; R recessive model; C codominant model; Publication bias P: using Begg’s or Egger’s tests
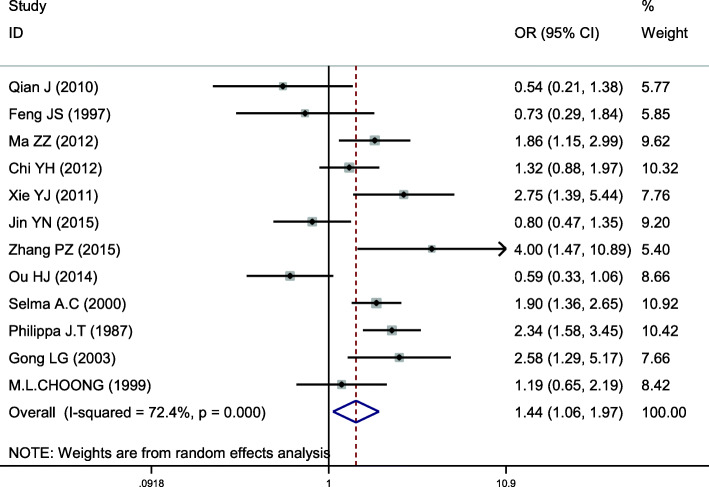
Meta-analysis of APOB XbaI (rs693)
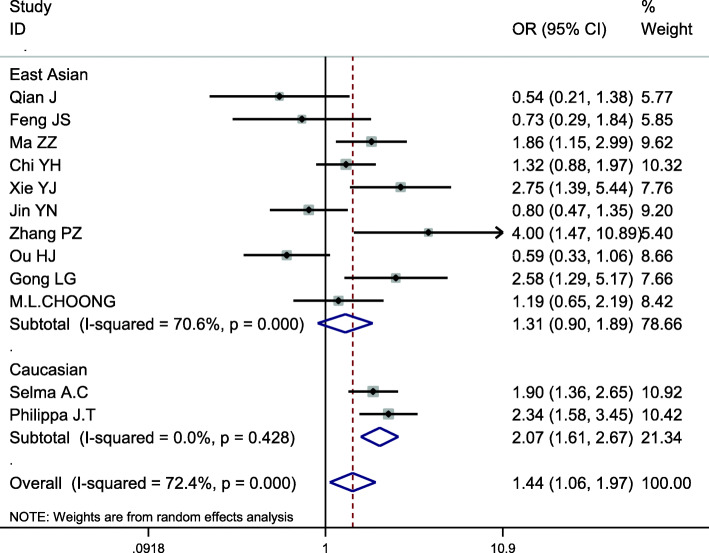
This site was included in 12 case-control studies, involving a total of 3736 subjects, including 1900 in the hyperlipidemia group and 1836 in the control group. Baseline data and quality evaluation are shown in Table 4. Analysis of the association between T vs C alleles and hyperlipidemia (allele model) indicated substantial heterogeneity (I2 = 72.4%,P < 0.001) and so a random-effects model was used to analyze the pooled effects. The risk of hyperlipidemia in the T allele population was higher than that with the C allele, the difference of which was statistically significant (OR = 1.444, 95% CI = 1.061–1.966, P = 0.020) (Fig. 5). There was no significant difference between the dominant and codominant models of this locus, with P-values of 0.100 and 0.140, respectively. The results of other gene models were consistent with those of the allele model (Table 5). Subgroup analysis by ethnicity displayed an increased risk of hyperlipidemia among Caucasians that carried the T allele when analyzed with the allele model, a difference that was statistically significant (OR = 2.074; 95% CI = 1.611–2.672, P < 0.001). However, no significant association was found in other gene models. We found that there was no significant association with risk of hyperlipidemia risk in Asians carrying the T allele using the allele model (OR = 1.305; 95% CI = 0.902–1.888, P = 0.158), other gene models displaying results consistent with those of the allele model (Table 6, Fig. 6). Therefore, an association between APOB XbaI gene single nucleotide polymorphism and hyperlipidemia in Asians was not considered to exist. However, the T allele at this locus could be considered a risk factor for hyperlipidemia in Caucasians.
Fig. 5.
Pooled calculated OR for the association between the APOB XbaI allele and hyperlipidemia
Table 6.
Subgroup analysis by ethnicity of the APOB XbaI polymorphism on susceptibility to hyperlipidemia
| Ethnicity | Analysis model | Genotype model | OR(95%CI) | P |
|---|---|---|---|---|
| Asian | A | T vs C | 1.305(0.902 ~ 1.888) | 0.158 |
| D | TT + CT vs CC | 1.470(0.953 ~ 2.267) | 0.081 | |
| R | TT vs CT + CC | 1.476(0.507 ~ 4.300) | 0.475 | |
| C | TT vs CC | 1.569(0.542 ~ 4.541) | 0.406 | |
| CT vs CC | 1.466(0.960 ~ 2.238) | 0.077 | ||
| Caucasian | A | T vs C | 2.075(1.611 ~ 2.672) | < 0.001 |
| D | TT + CT vs CC | 0.985(0.640 ~ 1.518) | 0.947 | |
| R | TT vs CT + CC | 1.644(0.993 ~ 2.723) | 0.053 | |
| C | TT vs CC | 1.391(0.765 ~ 2.530) | 0.280 | |
| CT vs CC | 0.848(0.509 ~ 1.412) | 0.526 |
A allelic model; D dominant model; R recessive model; C codominant model
Fig. 6.
Subgroup analysis by ethnicity for the association between the APOB XbaI allele and the risk of hyperlipidemia
Meta-analysis of APOB EcorI (rs1042031)
This site was included in 10 case-control studies, involving a total of 3319 subjects, including 1633 in the hyperlipidemia group and 1686 in the control group. Baseline data and quality evaluation are shown in Table 4. Analysis of the association between A vs G alleles and hyperlipidemia (allele model) indicated heterogeneity (I2 = 70.0%, P < 0.001), so the pooled effects were analyzed using a random-effects model. There was no significant difference in risk in individuals carrying the A or G alleles (OR = 1.333, 95% CI = 0.942–1.885, P = 0.104). The results of other gene models at this site were consistent with this conclusion, and so no association between the genetic polymorphism of APOB Ecor I and susceptibility to hyperlipidemia (Table 5) can be considered to exist.
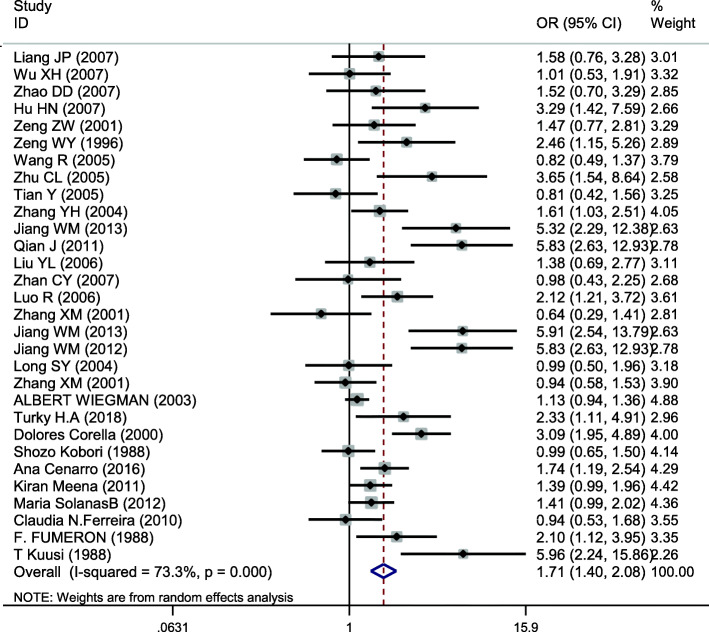
Meta-analysis of APOE
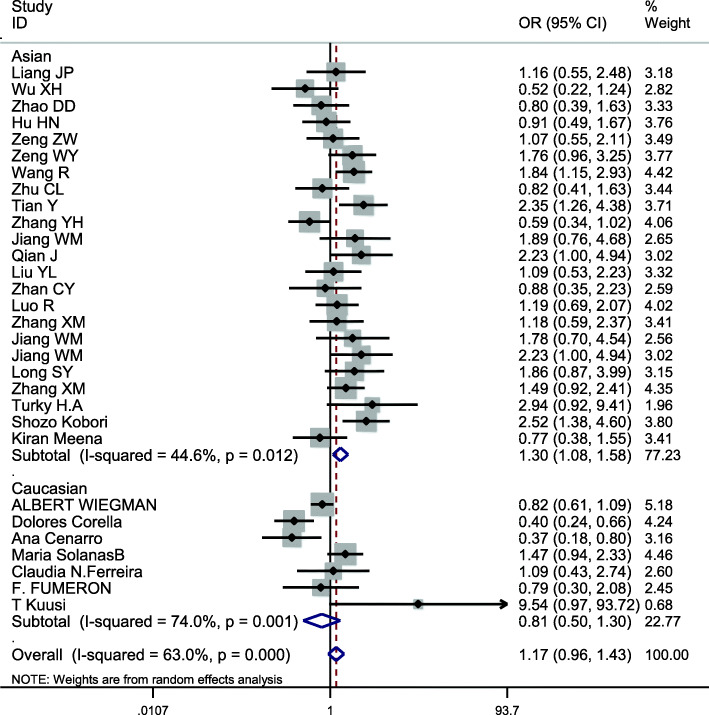
This site was included in 30 case-control studies, involving a total of 11,614 subjects, including 5208 in the hyperlipidemia group and 6406 in the control group. The baseline data and quality evaluation of the various studies are displayed in Table 7. The APOE ε3 allele was used as a reference to analyze the relationship between alleles and hyperlipidemia. Analysis of the data for ε2 (I2 = 63.0%, P < 0.001) and ε4 (I2 = 73.3%, P < 0.001) indicate that heterogeneity was present and so the pooled effects were analyzed using a random-effects model. The difference in risk between individuals with the ε2 and ε3 allele was not statistically significant (OR = 1.167, 95% CI = 0.955–1.426, P = 0.131). The risk of hyperlipidemia in individuals with the ε4 allele was higher than in those with the ε3 allele, a difference that was statistically significant (OR = 1.710, 95% CI = 1.405–2.083, P < 0.001) (Fig. 7). Because of heterogeneity, subgroup analysis by ethnicity was conducted, the results using the allele model demonstrating a risk of hyperlipidemia was different for Asians (OR = 1.304; 95% CI = 1.075–1.582, P = 0.007) for those with ε2 compared with the ε3 allele, but the association was not significant for Caucasians (OR = 0.807; 95% CI = 0.502–1.297, P = 0.376) (Fig. 8). There were significant differences in risk of hyperlipidemia, which was higher in both Asians (OR = 1.704; 95% CI = 1.325–2.192, P < 0.001) and Caucasians (OR = 1.759; 95% CI = 1.231–2.513, P = 0.002) with the ε4 allele than those carrying the ε3 allele (Fig. 9).
Table 7.
Main characteristics of the studies of APOE included in the review
| First author | Year | Area | Sample size | Age (y) | Source of control | Genotyping method | Cases | Controls | NOS | HWE | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | E2/E2 | E2/E3 | E2/E4 | E3/E3 | E3/E4 | E4/E4 | E2/E2 | E2/E3 | E2/E4 | E3/E3 | E3/E4 | E4/E4 | χ2 | P | |||||||
| Liang JP [37] | 2008 | Beijing,China | 210 | 94 | 58.48 | NR | HB | PCR-RFLP | 2 | 19 | 2 | 155 | 32 | 0 | 0 | 9 | 1 | 75 | 9 | 0 | 6 | 0.94 | 0.63 | |
| Wu XH [38] | 2007 | Xinjiang,China | 100 | 91 | 48.7 ± 10.5 | 43.1 ± 10.8 | HB | PCR-RFLP | 0 | 9 | 0 | 69 | 21 | 1 | 0 | 13 | 2 | 60 | 14 | 2 | 6 | 1.79 | 0.41 | |
| Zhao DD [11] | 2007 | Beijing,China | 172 | 80 | NR | HB | PCR-RFLP | 1 | 18 | 2 | 124 | 27 | 0 | 0 | 13 | 0 | 58 | 9 | 0 | 7 | 2.03 | 0.36 | ||
| Hu HN [39] | 2007 | Hubei,China | 165 | 108 | 60.5 ± 8.3 | 63.8 ± 6.2 | HB | ARMS-PCR | 0 | 26 | 0 | 109 | 27 | 3 | 0 | 20 | 0 | 81 | 7 | 0 | 7 | 2.2 | 0.33 | |
| Zeng ZW [40] | 2001 | Guangdong,China | 163 | 87 | 56.4 ± 3.2 | 58.0 ± 2.4 | HB | PCR-RFLP | 0 | 22 | 5 | 104 | 32 | 0 | 0 | 12 | 2 | 61 | 12 | 0 | 6 | 1.82 | 0.4 | |
| Zeng WY [41] | 1996 | Beijing,China | 133 | 122 | 41–60 | PB | PCR | 5 | 17 | 4 | 88 | 18 | 1 | 1 | 14 | 2 | 97 | 8 | 0 | 7 | 2.87 | 0.24 | ||
| Wang R [42] | 2005 | Sichuan,China | 206 | 250 | 52 | 51 | HB | PCR-RFLP | 0 | 46 | 2 | 135 | 22 | 1 | 2 | 28 | 1 | 182 | 35 | 2 | 7 | 1.9 | 0.39 | |
| Zhu CL [43] | 2005 | Hubei,China | 113 | 108 | 62.5 ± 7.2 | 63.8 ± 6.2 | HB | ARMS-PCR | 0 | 16 | 0 | 74 | 21 | 2 | 0 | 20 | 0 | 81 | 7 | 0 | 7 | 2.2 | 0.33 | |
| Tian Y [44] | 2005 | Sichuan,China | 103 | 146 | 56.9 ± 8.5 | 56.3 ± 9.8 | PB | PCR-RFLP | 2 | 23 | 1 | 64 | 12 | 1 | 1 | 15 | 1 | 102 | 27 | 0 | 8 | 2.53 | 0.28 | |
| Zhang YH [45] | 2004 | Beijing,China | 160 | 328 | 47.3 ± 13.8 | 40.1 ± 13.5 | PB | PCR-RFLP | 0 | 13 | 5 | 114 | 22 | 6 | 0 | 55 | 8 | 225 | 38 | 2 | 7 | 5.59 | 0.06 | |
| Jiang WM [46] | 2013 | Jiangsu,China | 102 | 100 | 48.4 ± 9.7 | 50.2 ± 15.1 | HB | DNA sequencing | 1 | 9 | 2 | 64 | 22 | 4 | 0 | 7 | 1 | 86 | 6 | 0 | 7 | 2.19 | 0.33 | |
| Qian J [47] | 2011 | Jiangsu,China | 212 | 100 | 54.6 ± 11.9 | 50.2 ± 15.1 | HB | DNA sequencing | 2 | 21 | 6 | 127 | 47 | 9 | 0 | 7 | 1 | 86 | 6 | 0 | 7 | 2.19 | 0.33 | |
| Liu YL [48] | 2006 | Shanxi,China | 72 | 95 | NR | HB | ARMS-PCR | 2 | 8 | 3 | 45 | 13 | 1 | 0 | 16 | 3 | 61 | 15 | 0 | 7 | 2.66 | 0.26 | ||
| Zhan CY [49] | 2007 | Beijing,China | 96 | 95 | 60.0 ± 8.3 | NR | HB | PCR | 0 | 9 | 0 | 75 | 12 | 0 | 0 | 9 | 1 | 75 | 9 | 1 | 7 | 1.75 | 0.42 | |
| Luo R [50] | 2006 | Hubei,China | 164 | 156 | 58.3 ± 7.1 | 53.1 ± 4.7 | HB | PCR-RFLP | 1 | 27 | 1 | 101 | 28 | 6 | 1 | 21 | 3 | 116 | 13 | 2 | 6 | 5.04 | 0.08 | |
| Zhang XM [51] | 2001 | Sichuan,China | 74 | 230 | 56.8 ± 12.4 | 51.3 ± 10.3 | PB | PCR-RFLP | 0 | 10 | 2 | 56 | 6 | 0 | 2 | 26 | 1 | 165 | 35 | 1 | 7 | 2.27 | 0.32 | |
| Jiang WM [52] | 2013 | Jiangsu,China | 93 | 100 | 56.0 ± 11.85 | 50.2 ± 15.1 | HB | DNA sequencing | 1 | 7 | 2 | 57 | 22 | 4 | 0 | 7 | 1 | 86 | 6 | 0 | 6 | 2.19 | 0.33 | |
| Jiang WM [53] | 2012 | Jiangsu,China | 212 | 100 | 54.6 ± 11.85 | 50.2 ± 15.1 | HB | DNA sequencing | 2 | 21 | 6 | 127 | 47 | 9 | 0 | 7 | 1 | 86 | 6 | 0 | 6 | 2.19 | 0.33 | |
| Long SY [54] | 2004 | Sichuan,China | 112 | 73 | 58.2 ± 7.9 | 55.1 ± 9.7 | PB | PCR-RFLP | 1 | 21 | 4 | 68 | 17 | 1 | 1 | 8 | 0 | 48 | 16 | 0 | 7 | 3.89 | 0.14 | |
| Zhang XM [55] | 2001 | Sichuan,China | 225 | 230 | 53.0 ± 15.5 | 51.3 ± 10.3 | PB | PCR-RFLP | 1 | 37 | 5 | 156 | 23 | 3 | 2 | 26 | 1 | 165 | 35 | 1 | 7 | 2.27 | 0.32 | |
| ALBERT [56] | 2003 | Amsterdam, Netherlands | 450 | 2018 | 10.8 | NR | HB | PCR | 0 | 50 | 10 | 243 | 135 | 12 | 13 | 261 | 45 | 1128 | 512 | 59 | 7 | 2.83 | 0.24 | |
| Turky H.A [57] | 2018 | Riyadh, Saudi Arabia | 104 | 100 | 57.8 ± 9.9 | 44.0 ± 6.3 | HB | TaqMan | 1 | 7 | 2 | 74 | 18 | 2 | 0 | 4 | 0 | 85 | 11 | 0 | 8 | 0.66 | 0.72 | |
| Corella [58] | 2000 | Valencia, Spain | 330 | 330 | 38.8 ± 9.1 | 37.6 ± 8.4 | PB | PCR | 0 | 17 | 5 | 237 | 69 | 2 | 3 | 50 | 1 | 252 | 23 | 1 | 7 | 1.28 | 0.53 | |
| Kobori [59] | 1988 | Kumamoto, Japan | 447 | 188 | 30–69 | HB | SRID | 9 | 49 | 7 | 323 | 47 | 12 | 0 | 12 | 1 | 143 | 30 | 2 | 7 | 0.39 | 0.82 | ||
| Cenarro [60] | 2016 | Zaragoza, Spain | 288 | 220 | 47.9 ± 11.5 | 44.8 ± 16.0 | HB | RT-PCR | 0 | 9 | 1 | 186 | 72 | 11 | 0 | 19 | 3 | 160 | 34 | 4 | 8 | 2.53 | 0.28 | |
| Kiran [61] | 2011 | New Delhi, India | 219 | 352 | 42.0 ± 7.9 | 35.2 ± 9.6 | HB | PCR-RFLP | 0 | 8 | 4 | 143 | 62 | 2 | 2 | 19 | 3 | 251 | 73 | 4 | 7 | 5.48 | 0.06 | |
| SolanasB [62] | 2012 | Zaragoza, Spain | 312 | 264 | 48.4 ± 9.7 | 43.5 ± 16.9 | HB | PCR | 11 | 25 | 5 | 189 | 65 | 8 | 1 | 27 | 4 | 183 | 45 | 4 | 8 | 0.46 | 0.79 | |
| N.Ferreira [63] | 2010 | Minas Gerais, Brasil | 109 | 107 | 48.4 ± 6.8 | 46.7 ± 6.6 | HB | PCR-RFLP | 0 | 10 | 0 | 77 | 18 | 4 | 0 | 9 | 0 | 72 | 25 | 1 | 7 | 2.26 | 0.32 | |
| FUMERON [64] | 1988 | Paris, France | 59 | 113 | NR | HB | PCR | 0 | 5 | 1 | 35 | 14 | 4 | 1 | 13 | 1 | 79 | 16 | 3 | 6 | 3.96 | 0.14 | ||
| T Kuusi [65] | 1988 | Helsinki, Finland | 21 | 21 | 45.2 ± 0.8 | 46.7 ± 1.5 | HB | PCR | 0 | 1 | 3 | 2 | 8 | 7 | 0 | 1 | 0 | 11 | 8 | 1 | 6 | 0.44 | 0.8 | |
SNP single nucleotide polymorphism, PB population-based, HB hospital-based, HWE Hardy-Weinberg equilibrium, NR not reported, SRID single radial immunodiffusion
Fig. 7.
Pooled calculated OR for the association between the APOE allele and hyperlipidemia
Fig. 8.
Subgroup analysis by ethnicity for the association between the APOE ε2 and ε3 alleles and the risk of hyperlipidemia
Fig. 9.
Subgroup analysis by ethnicity for the association between the APOE ε3 and ε4 alleles and the risk of hyperlipidemia
Correlations in the APOE genotype (E2/E2, E2/E3, E2/E4, E3/E4, E4/E4) and hyperlipidemia were analyzed using the wild type E3/E3 genotype as a reference. The heterogeneity, and OR and 95% CI values of these data are displayed in Table 8. The significance level was adjusted to α′ = α/(k-1) = 0.01. There was a significant difference in risk of hyperlipidemia between carriers of the E2/E4, E3/E4, and E4/E4 genotypes with carriers of the E3/E3 genotype, the P-values of which were < 0.01 in each case. To identify the source of significant heterogeneity, we conducted subgroup analysis based on ethnicity. The results demonstrated that there was a significant difference in risk of hyperlipidemia in carriers of all genotypes (E2/E2, E2/E3, E2/E4, E3/E4, E4/E4) compared with carriers of the E3/E3 genotype in Asians, while Caucasians carrying the E3/E4, E4/E4 genotypes were statistically different from those carrying E3/E3 (Table 9). Therefore, APOE gene polymorphisms can be considered to be closely associated with hyperlipidemia. For Asians, either the ε2 or ε4 allele was a risk factor for hyperlipidemia, while for Caucasians, only the ε4 allele was a risk factor.
Table 8.
Summary of the meta-analysis of the association of APOE gene polymorphisms with hyperlipidemia
| Genotype model | Heterogeneity(I2/P) | OR(95%CI) | P | publication bias P |
|---|---|---|---|---|
| E2/E2 | 0.0%/0.634 | 1.746(1.081 ~ 2.819) | 0.023 | 0.131 |
| E2/E3 | 50.3%/0.001 | 1.076(0.883 ~ 1.311) | 0.467 | 0.400 |
| E2/E4 | 0.0%/0.790 | 1.693(1.227 ~ 2.336) | 0.001 | 0.054 |
| E3/E4 | 67.8%/< 0.001 | 1.578(1.276 ~ 1.951) | < 0.001 | 0.073 |
| E4/E4 | 2.7%/ 0.424 | 2.346(1.723 ~ 3.195) | < 0.001 | 0.851 |
Publication bias P: using Begg’s or Egger’s tests
Table 9.
Subgroup analysis by ethnicity of APOE gene polymorphisms on susceptibility to hyperlipidemia
| Ethnicity | Genotype model | OR(95%CI) | P |
|---|---|---|---|
| Asian | E2/E2 | 2.062(1.131 ~ 3.761) | 0.003 |
| E2/E3 | 1.229(1.006 ~ 1.502) | 0.009 | |
| E2/E4 | 1.958(1.283 ~ 2.986) | 0.002 | |
| E3/E4 | 1.579(1.201 ~ 2.077) | 0.001 | |
| E4/E4 | 3.312(2.041 ~ 5.374) | < 0.001 | |
| Caucasian | E2/E2 | 1.248(0.549 ~ 2.841) | 0.597 |
| E2/E3 | 0.703(0.479 ~ 1.034) | 0.073 | |
| E2/E4 | 1.342(0.805 ~ 2.237) | 0.260 | |
| E3/E4 | 1.612(1.121 ~ 2.317) | 0.002 | |
| E4/E4 | 1.712(1.129 ~ 2.596) | 0.002 |
Publication bias and sensitivity analysis
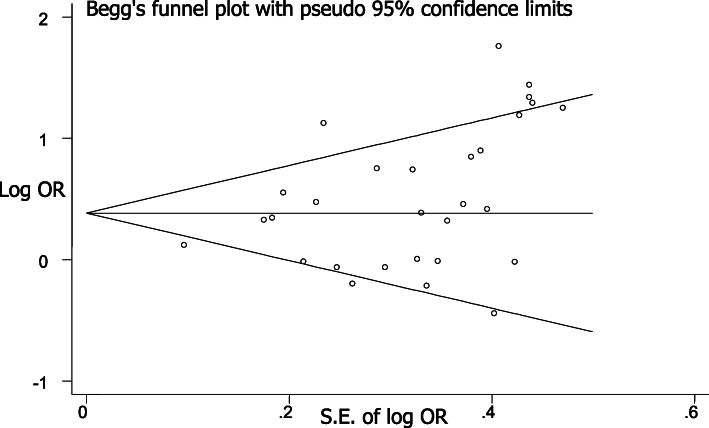
There was no apparent asymmetry in each Begg’s funnel plot (Fig. 10), indicating that publication bias was slight. In addition, statistical analysis of the symmetry of Begg’s funnel plots using an Egger’s test demonstrated that publication bias for each gene locus displayed P-values all > 0.05, indicating that publication bias was apparently not present.
Fig. 10.
Begg’s funnel plot for the APOE ε4 allele
For groups that deviated substantially in the analysis, meta-analysis was performed again after exclusion of the associated manuscripts, and OR and P-values re-calculated. Exclusion of the study [18] for APOA5–1131 T > C with the most deviating OR value using the allele model resulted in conclusions similar and consistent with those of the original data (OR = 1.800, 95% CI = 1.454–2.229, P < 0.001). The results indicated stability in the APOA1-75 bp and APOA1 + 83 bp allele models, with no literature having excessive deviation.
For the APOB Xba I locus using the allele model, exclusion of the manuscript [32] with the largest deviation in OR value resulted in conclusions of the meta-analysis consistent with the original conclusions (OR = 1.365, 95% CI = 1.001–1.862, P = 0.049). Exclusion of the biased literature [36] that studied APOB Ecor I in Caucasians resulted in differences in the meta-analysis that were not statistically significant and consistent with the original conclusions (OR = 1.351, 95% CI = 0.940–1.941, P = 0.104). Sensitivity analysis of the allele model of APOB Msp I was performed, the results of which were consistent with the original conclusions (OR = 0.926, 95% CI = 0.779–1.102, P = 0.387).
Exclusion of the manuscript [65] with the greatest deviation in data for the ε2 allele of APOE resulted in conclusions for the meta-analysis that the ε2 allele was not associated with hyperlipidemia (OR = 1.150, 95% CI = 0.943–1.402, P = 0.167). Correspondingly, exclusion of the literature [65] with the largest deviation for the APOE ε4 allele resulted in conclusions consistent with those originally recorded, following recalculation, and so carrying the ε4 allele can be considered a risk factor for hyperlipidemia (OR = 1.657, 95% CI = 1.365–2.012, P < 0.001). To summarize, we conclude that there was no apparent inconsistency in the literature that would contradict our original conclusions, with good reliability.
Discussion
The present study found that allele C at APOA5–1131 T > C was a risk factor for hyperlipidemia, the A allele at AI-75 bp conferred susceptibility to hyperlipidemia, the T allele at APOB Xba I represents a preliminary pathogenic factor for hyperlipidemia in Caucasians, allele ε4 of the APOE gene is a risk factor for hyperlipidemia, and allele ε2 is a risk factor for hyperlipidemia in Asians.
The APOE gene, located on chromosome 19, contains 4 exons and 3 introns, with 3 isomers, and the functions by of regulating plasma total cholesterol (TC) and lipoprotein metabolism. APOE3 is the most common phenotype. A principal function is to bind low-density lipoprotein receptor (LDL-R) and APOE receptor as the ligand [66]. Compared with APOE3, the ability of APOE4 to bind to its receptor is relatively strong, resulting in the metabolism of chylomicrons (CMs) and very low-density lipoprotein (VLDL) residues to be accelerated and the conversion of VLDL to LDL to increase. Additionally, the rate of liver internalization and catabolism of CM and VLDL residues becomes accelerated, resulting in increased free cholesterol in hepatocytes with feedback that caused a down-regulation of LDL-R on their surface, resulting in a decrease in the metabolic rate of LDL [67]. Furthermore, the low intestinal cholesterol absorption capacity of ε4 carriers also increases, resulting in higher plasma levels of TC and LDL. This is consistent with the conclusion that the ε4 allele is a risk factor for hyperlipidemia in the present review. The study also found that the ε2 allele is harmful for blood lipid levels in the Asian population, but failed to establish the effects on blood lipid levels in the Caucasian population. This may be related to the imbalance of internal composition and the small sample size for Caucasians. Of course, we cannot rule out the possibility of a corresponding biological mechanism to explain why this locus has no harmful effects on Caucasians.
APOB is the principal protein component of LDL and plays a role in transportation of endogenous cholesterol to maintain its balance within the body. The APOB gene is located in region 23–24 of the short arm of human chromosome 2. The APOB gene plays a key role in the production, transport, and removal of LDL and VLDL from plasma and regulates the concentration of plasma cholesterol [68]. The polymorphism of the APOB XbaI restriction site is due to a mutation of nucleotide C → T at position 7673 of the APOB gene cDNA, which changes the codon sequence at position 2488 (ACC → ACT), thus producing an XbaI endonuclease recognition site. The T allele may be related to a reduction in LDL degradation rate mediated by the receptor [9]. A number of studies have also speculated that a single nucleotide polymorphism at this locus is a genetic marker and has linkage disequilibrium with other nearby DNA sequence variants that affect cholesterol levels [69]. Such a molecular mechanism could explain why the T allele is a risk factor for hyperlipidemia in Caucasians. Other studies further confirm our conclusions that this polymorphism of the APOB XbaI gene might increase the risk of cerebral infarction, and that the T allele is such a risk factor [70]. The T allele was associated with lower levels of HDL-C, which may be associated with the incidence of coronary heart disease [71].
The APOA1 gene is located in the terminal region of the long arm of chromosome 11 and consists of 3 introns and 4 exons. APOA1 is the main apolipoprotein to create high-density lipoprotein (HDL), maintaining the stability and integrity of the HDL structure, and promoting the esterification of cholesterol (TC) [72]. The APOA1-75 bp polymorphism not only destroys the endonuclease recognition site but also changes the GGGCCGG sequence which activates transcription. A change in the sequence may also affect the synthesis of APOA1 [73]. This mechanism is consistent with the conclusion that there is an association between the A1-75bp gene single nucleotide polymorphisms and hyperlipidemia. The APOA5 gene, located in 23 regions of the long arm of chromosome 11, has 1889 bps and consists of 4 exons, 2 introns, and 4 silencing molecules. APOA5 can reduce triglyceride (TG) and increase HDL, representing a protective factor for coronary heart disease [74]. Some of the manuscripts also clearly stated that the mutation APOA5–1131 T > C is closely related to increased triglyceride levels [75] and that the CC genotype of this locus was positively correlated with serum TG levels and negatively correlated with APOA5 levels [76].
A meta-analysis can effectively compensate for the lack of statistical efficacy and other problems within a single study. However, although the present review developed a scientifically-based and comprehensive search strategy with strict unified screening criteria, limitations still remain [77]: (1) There were few relevant Chinese and English manuscripts on the acquisition of particular gene loci, such as APOAI and APOB MspI, so the number of case-control studies included in the analysis was small, possibly reducing the effectiveness of the Egger’s and Begg’s tests, in addition to sensitivity analysis; (2) The data included in the review did not involve additional races, which led to heterogeneity. Although ethnic subgroup analysis can identify heterogeneity to some extent, we found that there was a small sample size in Caucasians for APOB XbaI, possibly the reason why the results of the genetic model were not consistent at this locus. (3) It is unknown whether there were statistical differences in sex and age among individuals included in the study; (4) The effects of gene-environmental interactions and genetic linkage disequilibrium were not considered. In the future, we shall include more reliable data in this respect and update the meta-analysis, thereby providing a more reliable evidence base for the prevention and control of hyperlipidemia from the perspective of the apolipoprotein gene.
Conclusions
In summary, the results of the present meta-analysis revealed that the C allele of APOA5 1131 T > C, the A allele at APOA1-75 bp, the APOB XbaI T allele, and the ε2 and ε4 alleles of APOE may represent genetic risk factors for susceptibility for hyperlipidemia. In addition, we found it is consistent with the present study on the pathological mechanisms of hyperlipidemia. However, there is a need for further large-scale studies, including larger case-control studies and analysis of other loci of the APO genes, to confirm our conclusions and elucidate the influence of gene-environment interactions.
Methods
Literature search strategy
The Pubmed, Web of Science, ScienceDirect, the Chinese National Knowledge Infrastructure database, the Chinese Wanfang database, and Database of Chinese science and technology periodicals were searched to identify studies that evaluated the association of APO gene polymorphisms with the risk of hyperlipidemia, where publication date was prior to June 9, 2020. The keywords “apolipoprotein”, “APO”, “hyperlipidemia”, “dyslipidemias”, “hypercholesteremia”, “hypertriglyceridemia”, “mixed hyperlipidemia”, “low density lipoproteinemia”, “APOA”, “APOB”, “APOC”, “APOD”, “APOE”, “APOA5–1131 T > C”, “rs662799”, “APOA1-75 bp”, “rs670”, “APOA1 + 83 bp”, “rs5069”, “APOB MspI”, “rs1801701”, “APOB XbaI”, “rs693”, “APOB EcorI”, “rs1042031”, “gene”, “polymorphism”, and “genetic polymorphism” were searched. The references of all eligible studies were also searched manually in order to find other studies missed during the initial search activity. The analysis followed the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) statement [78].
Identification of studies for inclusion
The inclusion criteria for the present meta-analysis were as follows: (1) studies that evaluated the association between APO and risk of hyperlipidemia; (2) studies with an appropriate statistical design and selection methods; (3) case-control and RCT studies; (4) diagnostic criteria for dyslipidemia that were clear and uniform [79]; (5) distribution of APO genotypes in controls group were consistent with the Hardy-Weinberg equilibrium (HWE); (6) allele typing methods were accurate; (7) data included in the studies were complete, without omissions. Duplicated data, reviews, abstracts, case reports, animal studies, and studies that did not meet the inclusion criteria were excluded.
Data extraction
Two reviewers (XNZ and QS) independently conducted literature screening and evaluation. The following information was extracted from each study for inclusion in the review: first author, year of publication, area, age, source of control, sample size of controls and cases, genotyping method, Hardy-Weinberg equilibrium (HWE), and the distribution of genotypes and frequencies of alleles in cases and controls. Any disputes were resolved by discussion with a third investigator.
Quality evaluation
The quality of the selected case-control studies was evaluated according to the Newcastle-Ottawa Quality Assessment Scale (NOS) [80], of which data with scores 0–3, 4–6 or 7–9 were low, moderate or high-quality, respectively [81].
Statistical analyses
The included hyperlipidemia data were analyzed by meta-analysis using Stata 11 software. The correlation between apolipoprotein gene polymorphism and hyperlipidemia was expressed by odds ratio (OR) and 95% confidence intervals (CIs). In order to better evaluate the presence of heterogeneity between the studies, an I2 test was also used. Where homogeneity (I2 < 50%) was identified in the meta-analysis, a fixed-effects model was adopted; otherwise, a random-effects model was used to integrate the incorporated data. The data were assessed using Egger’s and Begg’s tests to evaluate publication bias. Sensitivity analysis was conducted by deleting, in turn, the data from individual studies that had large deviations as identified in the results, then recalculating the OR value. All P-values were two-sided, with a significance threshold set at α = 0.05.
To explore the source of significant heterogeneity, subgroup analysis of race was performed. A total of 7 sites were included, of which 3 sites (APOA5–1131 T > C,APOB XbaI, and APOE) were evaluated by subgroup analysis of ethnicity, 2 sites (APOB MspI, and APOB EcorI) were analyzed by sensitivity analysis, as there was only one published study of different races in the literature that was not suitable for subgroup analysis. Race was not evaluated in 2 sites (APOA1-75 bp, APOA1 + 83 bp) by subgroup analysis due to the fact that the populations studied were the same race, and had no significant heterogeneity.
Acknowledgments
We would like to acknowledge all individuals who participated in this study. We thank all staff of the School of Public Health and the School of Health of Guizhou Medical University and the School of Public Health of Hebei Medical University for their collaboration.
Abbreviations
- APO
Apolipoprotein
- SNPs
Single nucleotide polymorphisms
- HWE
Hardy-Weinberg Equilibrium
- NOS
Newcastle-Ottawa Quality Assessment Scale
- TC
Total cholesterol
- LDL-R
Low-density lipoprotein receptor
- CM
Chylomicron
- VLDL
Very low-density lipoprotein
- HDL
High-density lipoprotein
Authors’ contributions
Writing-Original draft preparation: XNZ, QS; Methodology and data curation: QS, XNZ; Writing-review and editing: YQC, XR, and XNZ; Supervision: YC, QS. All authors have read and approved the final manuscript.
Funding
This work was supported by the First-Class Discipline Construction Project in Guizhou Province - Public Health and Preventive Medicine (no. 2017[85]), and by the 15th Provincial Capital Construction Project of Guizhou Development and Reform Commission in 2018 (no. [2018]1571); Soft Science Project of Yunyan District (no. [2016] 2). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data analysed in this study can be derived from publicly available databases.
Declarations
Ethics approval and consent to participate
This work has been approved by the Ethics Committee of Guizhou Medical University.
Consent for publication
Not applicable.
Competing interests
We declare that none of the work contained in this manuscript is published in any language or currently under consideration at any other journal, and there are no conflicts of interest to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiao-Ning Zhao and Quan Sun contributed equally to this work.
References
- 1.Ye H, Zhou A, Hong Q, Tang L, Xu X, Xin Y, et al. Positive association between APOA5 rs662799 polymorphism and coronary heart disease: a case-control study and meta-analysis [J]. PLoS One. 2015;10(8):e135683. 10.1371/journal.pone.0135683. [DOI] [PMC free article] [PubMed]
- 2.Bora K, Pathak MS, Borah P, Hussain MI, Das D. Association of the Apolipoprotein A-I gene polymorphisms with cardiovascular disease risk factors and Atherogenic indices in patients from Assam, Northeast India. Balkan J Med Genet. 2017;20(1):59–70. doi: 10.1515/bjmg-2017-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Li J, Wang Y, et al. Acupuncture and related therapies for hyperlipidemia [J] Medicine. 2020;99(49):e23548. doi: 10.1097/MD.0000000000023548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miao J, Zang X, Cui X, et al. Autophagy, hyperlipidemia, and atherosclerosis [J] Adv Exp Med Biol. 2020;1207:237–264. doi: 10.1007/978-981-15-4272-5-18. [DOI] [PubMed] [Google Scholar]
- 5.Ou HJ, Huamg G, Liu W, et al. Relationship of the APOA5/A4/C3/A1 gene cluster and APOB gene polymorphisms with dyslipidemia [J] Genet Mol Res. 2015;14(3):9277–9290. doi: 10.4238/2015.August.10.8. [DOI] [PubMed] [Google Scholar]
- 6.Meng Q, Zhang XH, Zhang XW. Meta-anaylsis on association of ApoE gene polymorphism with hyperlipidemia. Chinese Preventive Medicine. 2015;16:304–307. doi: 10.16506/j.1009-6639.2015.04.017. [DOI] [Google Scholar]
- 7.Feng DW. Association between Polymorphisms of APOA1 Gene and Susceptibility for Uyghur and Kazak’s Dyslipidemia [D] Shihezi city: Shihezi University; 2016. [Google Scholar]
- 8.Han Y. Association between the Subclasses of HDL and APOA5 Gene Polymorphism in Hypertriglyceridemia [D] Hengyang city: University of South China; 2012. [Google Scholar]
- 9.Zhang PZ, Tian Y. Relationship of Apolipoprotein B and E gene polymorphisms to dyslipidemia and the influence of exercise training. China Sport Science. 2006;26:65–69. doi: 10.16469/j.css.2006.10.010. [DOI] [Google Scholar]
- 10.Zhang Y, Zeng T, Xu J, Liu L. Apolipoprotein gene polymorphism in coronary heart disease. Adv Cardiovasc Dis. 2019;40:1294–1297. doi: 10.16806/j.cnki.issn.1004-3934.2019.09.028. [DOI] [Google Scholar]
- 11.Zhao DD. Relationship between apolipoprotein A5, C3 and E gene polymorphisms and phlegm and blood stasis syndrome and therapeutic effect in patients with hyperlipidemia [D] Beijing: China Academy of Chinese Medical Sciences; 2007. [Google Scholar]
- 12.Niu ZB. Association study between lipid metabolism-related genepolymorphisms and polymorphisms and hyperlipidemia in aged patients withlong-term aerobic exercise [D] Shanghai: Shanghai University of Sport; 2016. [Google Scholar]
- 13.Huang MC, Wang TN, Wang HS, Sung YC, Ko YC, Chiang HC. The -1131T>C polymorphism in the apolipoprotein A5 gene is related to hypertriglyceridemia in Taiwanese aborigines. Kaohsiung J Med Sci. 2008;24(4):171–179. doi: 10.1016/S1607-551X(08)70114-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long SY, Chen ZJ, Han Y, Christopher DM, Zhang CP, Yang Y, et al. Relationship between the distribution of plasma HDL subclasses and the polymorphisms of APOA5 in hypertriglyceridemia. Clin Biochem. 2013;46(9):733–9. 10.1016/j.clinbiochem.2013.03.003. [DOI] [PubMed]
- 15.Di Taranto MD, Staiano A, D'Agostino MN, D'Angelo A, Bloise E, Morgante A, et al. Association of USF1 and APOA5 polymorphisms with familial combined hyperlipidemia in an Italian population. Mol Cell Probes. 2015;29(1):19–24. doi: 10.1016/j.mcp.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira CN, Carvalho MG, Fernandes AP, Santos IR, Rodrigues KF, Lana AMQ, et al. The polymorphism -1131T>C in apolipoprotein A5 gene is associated with dyslipidemia in Brazilian subjects. Gene. 2013;516(1):171–5. 10.1016/j.gene.2012.12.016. [DOI] [PubMed]
- 17.Brito DD, Fernandes AP, Gomes KB, Coelho FF, Cruz NG, Sabino AP, et al. Apolipoprotein A5-1131T>C polymorphism, but not APOE genotypes, increases susceptibility for dyslipidemia in children and adolescents. Mol Biol Rep. 2011;38(7):4381–8. 10.1007/s11033-010-0565-5. [DOI] [PubMed]
- 18.Liu ZK, Hu M, Baum L, Thomas GN, Tomlinson B. Associations of polymorphisms in the apolipoprotein A1/C3/A4/A5 gene cluster with familial combined hyperlipidaemia in Hong Kong Chinese. Atherosclerosis. 2010;208(2):427–432. doi: 10.1016/j.atherosclerosis.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 19.Henneman P, van der Sman-de Beer F, Moghaddam PH, Huijts P, Stalenhoef AFH, Kastelein JJP, et al. The expression of type III hyperlipoproteinemia: involvement of lipolysis genes. Eur J Hum Genet. 2009;17(5):620–8. 10.1038/ejhg.2008.202. [DOI] [PMC free article] [PubMed]
- 20.Huang G, Zhong H, Re HM, Mao HM, Chi YH. Association of polymorphisms of apoB genes EcoRI, XbaI, and MspI and apoAI gene - 75 bp and + 83 bp with dyslipidemia in Kazaks. The Journal of Practical Medicine. 2011;27:3518–3522. doi: 10.3969/j.issn.1006-5725.2011.19.026. [DOI] [Google Scholar]
- 21.Chi YH. Relationship between ApoB gene EcoRI, XbaI, MspI and apoAI gene-75bp, + 83bp polymorphisms and blood lipids [D]. Shihezi city: Shihezi University; 2012. (in Chinese) doi: 10.7666/d.D284991.
- 22.Xie YJ. The Association of APoB and APoAI Gene Polymorphism With DysliPidemia in Han Chinese of Xinjiang Shihezi [D] Shihezi city: Shihezi University; 2011. [Google Scholar]
- 23.Zhu H, Liu Y, Bai H, Liu BW. Apolipoprotein AI gene MspI polymorphism in relation to endogenous hypertriglyceridemia in Chinese population. Chin J Arterioscler. 2001;9:332–336. doi: 10.3969/j.issn.1007-3949.2001.04.017. [DOI] [Google Scholar]
- 24.Jia LQ, Bai H, Fu MD, Xu YH, Gou LT. Relationship of subclasses of serum HDL and Apo A-I gene polymorphism in hyperlipidemia. Chinese J Pathophysiol. 2006;04:796–800. doi: 10.3321/j.issn:1000-4718.2006.04.039. [DOI] [Google Scholar]
- 25.Cao WJ, Sheng L, Yang J, Zhou D, Cheng J. Relationship between MspI polymorphism of apolipoprotein B gene and blood-fat of Hazakh inhabitant. J Pract Med Techniques. 2009;16:770–772. doi: 10.3969/j.issn.1671-5098.2009.10.002. [DOI] [Google Scholar]
- 26.Chi YH, Huang G, Xie YJ, Guo ZL. Study on relationship between joint action of EcoR I, Xba I and Msp I polymorphisms of apoB gene and dyslipidemia. Journal of Clinical and Experimental Medicine. 2012;11:481–483. doi: 10.3969/j.issn.1671-4695.2012.07.001. [DOI] [Google Scholar]
- 27.Jin YN, Zhou L, Tang M, Zhang MJ, Tang XJ. Relationship between ApoB gene Msp I/Xba I/EcoR I polymorphisms and serum lipid level in male Han population in Chongqing,China. Acad J Second Mil Univ. 2015;36(9):966–71. (in Chinese). 10.3724/SP.J.1008.2015.00966, Relationship betweenApoBgeneMspI/XbaI/EcoRI polymorphisms and serum lipid level in maleHanpopulation in Chongqing, China.
- 28.Cavalli SA, Hirata MH, Salazar LA, Diament J, Forti N, Giannini SD, et al. Apolipoprotein B gene polymorphisms: prevalence and impact on serum lipid concentrations in hypercholesterolemic individuals from Brazil. Clin Chim Acta. 2000;302(1-2):189–203. 10.1016/s0009-8981(00)00367-3. [DOI] [PubMed]
- 29.Qian J, Hu DC, Zhao XL, Shao JC. Study on relationship between apolipoprotein B gene polymorphisms frequencies distributionand and essential hyperlipidemia of an nationality in Kunming area. Int J Lab Med. 2010;31:1262–1264. doi: 10.3969/j.issn.1673-4130.2010.11.026. [DOI] [Google Scholar]
- 30.Feng JS, Xie XQ, Lin CL. Apolipoprotein B Gene Polymorphisms in Patients with Hyperlipidemia or Coronary Heart Disease. J Jinan Univ (Natural Science & Medicine Edition) 1997;18:14–18. doi: 10.1007/BF02951625. [DOI] [Google Scholar]
- 31.Ma ZZ, Huang WB, He FP, Zhang SB. Relationship between apolipoprotein B gene polymorphisms and lipid levels in Yao population of Yuebei area. J Mol Diagn Ther. 2012;4:333–335. doi: 10.3969/j.issn.1674-6929.2012.05.013. [DOI] [Google Scholar]
- 32.Zhang PZ, Tian Y. Influence of Apolipoprotein B gene polymorphisms over effect of exercise on blood lipid. China Sport Science. 2015;35:38–47. doi: 10.16469/j.css.201505005. [DOI] [Google Scholar]
- 33.Talmud PJ, Barni N, Kessling AM, Carlsson P, Darnfors C, Bjursell G, et al. Apolipoprotein B gene variants are involved in the determination of serum cholesterol levels: a study in normo- and hypelipidaemic individuals. Atherosclerosis. 1987;67(1):81–9. 10.1016/0021-9150(87)90267-x. [DOI] [PubMed]
- 34.Gong LG, Liu XR, Qiu GB, Li HF, Cui XW. Analysis of XbaI polymorphism in the ApoB gene to hypertriglyceridemics in Chinese population. Chin J Lab Diagn. 2003;7:306–308. doi: 10.3969/j.issn.1007-4287.2003.04.012. [DOI] [Google Scholar]
- 35.Choong ML, Sethi SK, Koay ES. Effects of intragenic variability at 3 polymorphic sites of the apolipoprotein B gene on serum lipids and lipoproteins in a multiethnic Asian population. Hum Biol. 1999;71:381–397. [PubMed] [Google Scholar]
- 36.Timirci O, Darendeliler F, Bas F, Arzu EH, Umit Z, Isbir T. Comparison of lipid profiles in relation to APOB EcoRI polymorphism in obese children with hyperlipidemia. In Vivo. 2010;24:65–69. [PubMed] [Google Scholar]
- 37.Liang JP, Yang HM, Sheng T, Han LB, Yuan YJ, Niu XH, et al. Study on the relationship between ApoE gene polymorphism and plasma lipid levels of phlegm-blood-stasis syndrome of hyperlipemia. China J Trad Chin Med Pharm. 2008;23:633–635. [Google Scholar]
- 38.Wu XH, Cheng J, Zhou ZZ, Qin JM, He L. Relationship between apolipoprotein E gene polymorphism and blood lipids in Kazakh population in Xinjiang. Chinese J Clin Laboratory Sci. 2007;25:447–449. doi: 10.13602/j.cnki.jcls.2007.06.037. [DOI] [Google Scholar]
- 39.Hu HN, Chen W, Yang G, Lv M. Association of polymorphisms of apoprotein E and lipid levels with hyperlipidemia. Chin J Tissue Engineering Res. 2007;11:1453–1456. doi: 10.3321/j.issn:1673-8225.2007.08.027. [DOI] [Google Scholar]
- 40.Zeng ZW, Peng S, Peng J, Gong WX. Relationship between apolipoprotein E gene polymorphism and hyperlipidemia. Guangdong Med J. 2001;22:120–121. doi: 10.13820/j.cnki.gdyx.2001.02.019. [DOI] [Google Scholar]
- 41.Zeng WW, Lv XY, Chen BS. The study on the association between PolymorPhism of APoliPoProtein E gene and HyPerliPidemia. Chin J Arterioscler. 1996;4:185–189. [Google Scholar]
- 42.Wang R, Xie RL, Huang WF, Yang MQ. Apolipoprotein E gene polymorphism and its relationship with type IV hyperlipidemia. Sichuan Med J. 2005;26:400–401. doi: 10.16252/j.cnki.issn1004-0501-2005.04.027. [DOI] [Google Scholar]
- 43.Zhu CL, Zhou X, Liu F, Hu HL. The relationship between polymorphisms of Apolipoprotein E gene and Serum lipid. Chinese J Arteriosclerosis. 2005;13:203–206. doi: 10.3969/j.issn.1007-3949.2005.02.021. [DOI] [Google Scholar]
- 44.Tian Y, Long SY, Xu YH, Fu MD, Zhang XM, Liu BW. Study on apoE gene polymorphism and subclasses of serum high density lipoprotein in type IV hyperlipidemia. Chin J Med Genet. 2005;22:100–102. doi: 10.3760/j.issn:1003-9406.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 45.Zhang YH, Dou XF, Shang W, Dou XF, Wang YF, Liu YQ, et al. Association between familial combined hyperlipidemia (FCHL) and Apolipoprotein E polymorphism. Chinese J Hypertension. 2004;22:29–32. doi: 10.16439/j.cnki.1673-7245.2004.02.008. [DOI] [Google Scholar]
- 46.Jiang WM, Fang ZY, Zhu CL, Tang SH. Effects of ApoE gene polymorphism on anti-inflammatory action of Xuezhikang capsule. Chinese J Integrated Traditional Western Med. 2013;33(1):35–39. doi: 10.7661/CJIM.2013.1.35. [DOI] [PubMed] [Google Scholar]
- 47.Qian J, Jiang WM, Chen XH, Zhu CL, Xie L. Study on the relation between ApoE gene polymorphisms and the plasma lipids in Hyperlipemia patients. Chinese General Pract. 2011;14:840–842. doi: 10.3969/j.issn.1007-9572.2011.08.010. [DOI] [Google Scholar]
- 48.Liu YL, Li JK, Yan ZQ, Chen YJ. Correlation between apolipoprotein E gene polymorphism and plasma lipid level. J Fourth Mil Med Univ. 2006;27:460–461. doi: 10.3321/j.issn:1000-2790.2006.05.022. [DOI] [Google Scholar]
- 49.Zhan CY. Study on the relationship between Apolipoprotein E gene polymorphism and blood lipid level and lipid-regulating effect in hyperlipidemia with phlegm and blood stasis syndrome [D] Beijing: Beijing University of Chinese Medicine; 2006. [Google Scholar]
- 50.Luo R, Chen WY. Relationship between apolipoprotein E gene polymorphism and hyperlipidemia. Chinese Journal of Coal Industry Medicine. 2006;9:246–247. doi: 10.3969/j.issn.1007-9564.2006.03.036. [DOI] [Google Scholar]
- 51.Zhang XM, Bai H, Liu BW, Fan P, Zhang RJ, Xu YH, et al. Study on apoE Gene Polymorphism in Chinese Type IIb Hyperlipidemia. J Sichuan Univ (Medical Science Edition) 2001;32:179–182. doi: 10.3969/j.issn.1672-173X.2001.02.006. [DOI] [PubMed] [Google Scholar]
- 52.Jiang WM, Chen XH, Tang SH, Zhu CL, Xie L. Relationship between TCM syndrome differentiation and ApoE exon 4 polymorphism and dyslipidemia in patients with hyperlipidemia complicated with hypertension. The third National Youth Forum on Cardiovascular Diseases with Integrated traditional Chinese and Western Medicine and the second Symposium of Cardiovascular Committee of Xinjiang Society of Integrated traditional Chinese and Western Medicine; 2013 Sep 20; Urumqi, Xinjiang, China. Xinjiang (China): The third National Youth Forum on Cardiovascular Diseases with Integrated traditional Chinese and Western Medicine and the second Symposium of Cardiovascular Committee of Xinjiang Society of Integrated traditional Chinese and Western Medicine; 2004.p. 172–177. (in Chinese).
- 53.Jiang WM, Zhu CL, Liu J, Tang SH. Correlation analysis of TCM syndrome characteristics with ApoE gene polymorphisms in 212 hyperlipemia patients. China Journal of Traditional Chinese Medicine and Pharmacy. 2012;27:1458–1460. [Google Scholar]
- 54.Long SY, Zhang XM, Fu MD, Xu YH, Liu BW. Relationship of APOE gene polymorphism with subclasses of serum high density lipoprotein in hyperlipidemia. Chin J Med Genet. 2004;21:83–86. doi: 10.3760/j.issn:1003-9406.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 55.Zhang XM, Liu BW, Bai H, Fan P. Study on apoE gene polymorphism in Chinese endogenous hypertriglycerdemia. Chin J Med Genet. 2001;18:21–25. [PubMed] [Google Scholar]
- 56.Wiegman A, Sijbrands EJG, Rodenburg J, Defesche JC, de Jongh S, Bakker HD, et al. The apolipoprotein epsilon4 allele confers additional risk in children with familial hypercholesterolemia. Pediatr Res. 2003;53(6):1008–12. 10.1203/01.PDR.0000064580.23308.CB. [DOI] [PubMed]
- 57.Alharbi TH, Batais MA, Hasanato RM, Alharbi FK, Khan IA, Alharbi KK. Role of Apolipoprotein E gene polymorphism in the risk o f familial hypercholesterolemia: a case-control study. Acta Biochim Pol. 2018;65:415–420. doi: 10.18388/abp.2017-2344. [DOI] [PubMed] [Google Scholar]
- 58.Corella D, Guillén M, Portolés O, Sabater A, Cortina S, Folch J, et al. Apolipoprotein E gene polymorphism and risk of hypercholesterolemia: a case control study in a working population of Valencia. Med Clin. 2000;115(5):170–5. 10.1016/s0025-7753(00)71498-9. [DOI] [PubMed]
- 59.Kobori S, Nakamura N, Uzawa H, Shichiri M. Influence of apolipoprotein E polymorphism on plasma lipid and apolipoprotein levels, and clinical characteristics of type III hyperlipoproteinemia due to apolipoprotein E phenotype E2/2 in Japan. Atherosclerosis. 1988;69(1):81–88. doi: 10.1016/0021-9150(88)90291-2. [DOI] [PubMed] [Google Scholar]
- 60.Cenarro A, Etxebarria A, de Castro-Orós I, Stef M, Bea AM, Palacios L, et al. The p.Leu167del mutation in APOE gene causes autosomal dominant hypercholesterolemia by Down-regulation of LDL receptor expression in hepatocytes. J Clin Endocrinol Metab. 2016;101(5):2113–21. 10.1210/jc.2015-3874. [DOI] [PubMed]
- 61.Meena K, Misra A, Vikram N, Ali S, Pandey RM, Luthra K. Cholesterol ester transfer protein and apolipoprotein E gene polymorphisms in hyperlipidemic Asian Indians in North India. Mol Cell Biochem. 2011;352(1-2):189–196. doi: 10.1007/s11010-011-0753-1. [DOI] [PubMed] [Google Scholar]
- 62.Solanas-Barca M, de Castro-Orós I, Mateo-Gallego R, Cofán M, Plana N, Puzo J, et al. Apolipoprotein E gene mutations in subjects with mixed hyperlipidemia and a clinical diagnosis of familial combined hyperlipidemia. Atherosclerosis. 2012;222(2):449–55. 10.1016/j.atherosclerosis.2012.03.011. [DOI] [PubMed]
- 63.Ferreira CN, Carvalho MG, Fernandes APSM, Lima LM, Loures-Valle AA, Dantas J, et al. Comparative study of apolipoprotein-E polymorphism and plasma lipid levels in dyslipidemic and asymptomatic subjects, and their implication in cardio/cerebro-vascular disorders. Neurochem Int. 2010;56(1):177–82. 10.1016/j.neuint.2009.09.016. [DOI] [PubMed]
- 64.Fumeron F, Rigaud D, Bertiere MC, Bardon S, Dely C, Apfelbaum M. Association of apolipoprotein epsilon 4 allele with hypertriglyceridemia in obesity. Clin Genet. 1988;34(4):258–264. doi: 10.1111/j.1399-0004.1988.tb02873.x. [DOI] [PubMed] [Google Scholar]
- 65.Kuusi T, Taskinen MR, Solakivi T, Kauppinen-Mäkelin R. Role of apolipoproteins E and C in type V hyperlipoproteinemia. J Lipid Res. 1988;29(3):293–298. doi: 10.1016/S0022-2275(20)38536-9. [DOI] [PubMed] [Google Scholar]
- 66.Mahley RW. Apolipoprotein E: from cardiovascular disease to neurodegenerative disorders [J] J Mol Med. 2016;94(7):739–746. doi: 10.1007/s00109-016-1427-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mahley RW. Central nervous system lipoproteins [J] Arterioscler Thromb Vasc Biol. 2016;36(7):1305–1315. doi: 10.1161/ATVBAHA.116.307023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pencina MJ, D'Agostino RB, Zdrojewski T, et al. Apolipoprotein B improves risk assessment of future coronary heart disease in the Framingham heart study beyond LDL-C and non-HDL-C [J] Eur J Prev Cardiol. 2015;22(10):1321–1327. doi: 10.1177/2047487315569411. [DOI] [PubMed] [Google Scholar]
- 69.Benn M, Nordestgaard BG, Jensen JS, Grande P, Sillesen H, Tybjaerg-Hansen A. Polymorphism in APOB associated with increased low-density lipoprotein levels in both genders in the general population. J Clin Endocrinol Metab. 2005;90(10):5797–5803. doi: 10.1210/jc.2005-0974. [DOI] [PubMed] [Google Scholar]
- 70.Niu C, Luo Z, Yu L, et al. Associations of the APOB rs693 and rs17240441 polymorphisms with plasma APOB and lipid levels: a meta-analysis.[J] Lipids Health dis. 2017;16(1):166. doi: 10.1186/s12944-017-0558-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Renges HH, Wile DB, McKeigue PM, Marmot MG, Humphries SE. Apolipoprotein B gene polymorphisms are associated with lipid levels in men of south Asian descent. Atherosclerosis. 1991;91:267–275. doi: 10.1016/0021-9150(91)90174-2, 3. [DOI] [PubMed]
- 72.Toptas B, Gormus U, Ergen A, et al. Comparison of lipid profiles with APOA1 MspI polymorphism in obese children with hyperlipidemia [J] In Vivo. 2011;25(3):425–430. [PubMed] [Google Scholar]
- 73.Yin RX, Li YY, Lai CQ. Apolipoprotein A1/C3/A5 haplotypes and serum lipid levels [J] Lipids Health Dis. 2011;10(1):140. doi: 10.1186/1476-511X-10-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li YY, Wu XY, Xu J, Qian Y, Zhou CW, Wang B. Apo A5 -1131T/C, FgB -455G/A, −148C/T, and CETP TaqIB gene polymorphisms and coronary artery disease in the Chinese population: a meta-analysis of 15,055 subjects [J] Mol Biol Rep. 2013;40(2):1997–2014. doi: 10.1007/s11033-012-2257-9. [DOI] [PubMed] [Google Scholar]
- 75.Vrablik M, Hubacek JA, Dlouha D, Satny M, Adamkova V, Ceska R. Strong association between APOA5 gene polymorphisms and Hypertriglyceridaemic episodes [J] Folia Biol (Praha) 2019;65(4):188–194. doi: 10.14712/fb2019065040188. [DOI] [PubMed] [Google Scholar]
- 76.Kim M, Kim M, Yoo HY, Lee E, Chae JS, Lee SH, et al. A promoter variant of the APOA5 gene increases atherogenic LDL levels and arterial stiffness in hypertriglyceridemic patients. PLoS One. 2017;12(12):e186693. doi: 10.1371/journal.pone.0186693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhai GH, Ma JL, Li MF. Association between apolipoprotein A5-1131T/C polymorphism and type 2 diabetes mellitus in Chinese Han population: a meta-analysis. Int J Lab Med. 2019;40:1321–1324. (in Chinese) https://doi.org/10.3969/j.issn.1673-4130.2019.11.010.
- 78.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rhee E, Kim HC, Kim JH, et al. 2018 guidelines for the management of dyslipidemia [J] Korean J Intern Med. 2019;34(4):723–771. doi: 10.3904/kjim.2019.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wells GA, Shea B, O'Connell D, Peterson J, Welch V, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp
- 81.Lo CK, Mertz D, Loeb M. Newcastle-Ottawa scale: comparing reviewers' to authors' assessments [J] BMC Med Res Methodol. 2014;14(1):45. doi: 10.1186/1471-2288-14-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data analysed in this study can be derived from publicly available databases.