Abstract
Prostate cancer (PCa) is characterized by a unique dependence on optimal androgen receptor (AR) activity where physiological androgen concentrations induce proliferation but castrate and supraphysiological levels suppress growth. This feature has been exploited in bipolar androgen therapy (BAT) for castrate resistant malignancies. Here, we investigate the role of the tumor suppressor protein p14ARF in maintaining optimal AR activity and the function of the AR itself in regulating p14ARF levels. We used a tumor tissue array of differing stages and grades to define the relationships between these components and identified a strong positive correlation between p14ARF and AR expression. Mechanistic studies utilizing CWR22 xenograft and cell culture models revealed that a decrease in AR reduced p14ARF expression and deregulated E2F factors, which are linked to p14ARF and AR regulation. Chromatin immunoprecipitation studies identified AR binding sites upstream of p14ARF. p14ARF depletion enhanced AR-dependent PSA and TMPRSS2 transcription, hence p14ARF constrains AR activity. However, p14ARF depletion ultimately results in apoptosis. In PCa cells, AR co-ops p14ARF as part of a feedback mechanism to ensure optimal AR activity for maximal prostate cancer cell survival and proliferation.
Keywords: p14ARF, prostate cancer, androgen receptor, apoptosis, E2F
1. Introduction
Prostate cancer (PCa) remains the most common non-cutaneous cancer of American men resulting in an estimated 29,430 deaths in 2018 [1].One distinguishing feature of PCa is its reliance on the androgen receptor (AR), a hormone-dependent transcription factor that controls cellular proliferation, apoptosis and expression of the PCa marker prostate specific antigen (PSA) [2–4]. Several studies reported that prostate cells are reliant on optimal AR activity [5, 6], which drive proliferation and survival, but suboptimal or supraphysiological levels result in growth arrest or apoptosis [6–8]. Given the vital role of AR in driving this malignancy, prostate cancer cells have developed mechanisms that ensure optimal AR activity.
The CDKN2A locus, encodes two tumor suppressor proteins, p14ARF and p16INK4A [9]. While the two share common exons, the first exon of each is unique and the two transcripts are regulated by different promoters. The protein products have no sequence similarity and have distinct functional properties, but expression of both is altered in multiple neoplasms [9]. The best studied function of p14ARF is its interaction with and inhibition of two ubiquitin ligases, MDM2 and ARF-BP1/Mule, which target p53 for degradation [10, 11] thus potentiating p53 activity [12]. p14ARF can interact with other proteins including B23, p63, p32, Miz1, p16INK4a, E2F and TBP-1 [13]. The p14ARF interaction with E2F results in decreased protein stability due to an increase in ubiquitin mediated degradation [14, 15]. E2Fs and p14ARF have reciprocal regulatory relationships, since in normal cells E2F1, E2F2 and E2F3a promote p14ARF transcription, while E2F3b silences p14ARF expression [16].
The 8-member E2F gene family encodes 12 E2F proteins [17]. Activator E2Fs 1–3a are mostly associated with proliferation and reported to be overexpressed in various malignancies. E2F3b and the remaining E2F proteins are more associated with transcription repression [17]. Activator E2Fs can interact with the RB tumor suppressor, and binding of the E2F/RB complex to E2F binding sites of multiple target genes promotes the assembly of repressor complexes that inhibit transcription [18]. Several studies reported elevated expression of E2F1 and E2F3 in prostate cancers [19], while cell culture studies demonstrated that E2F1 overexpression or RB depletion promoted castration resistance [20, 21].
Unlike in other malignancies where p14ARF is decreased or absent due to gene deletion or silencing, it is not decreased in prostate tumors [22]. Moreover, studies in a mouse model of prostate tumorigenesis have demonstrated that p14ARF ablation serves to retard rather than accelerate prostate tumorigenesis [23]. p14ARF can directly interact with AR to repress its activity [24] possibly by binding and degrading X-linked primate-specific melanoma antigen-A11. Thus, in prostate tumorigenesis, p14ARF’s role is likely to constrain AR activity in response to supraphysiological androgen levels. AR is also known to regulate its own expression, but the mechanism by which it does so remains unknown and the complex interactions and reciprocal regulation of p14ARF, p53, AR and E2F intersecting pathways are not well understood.
In the current study, we conducted IHC studies using a prostate tumor tissue array and identified a strong positive correlation between the nuclear expression of AR and nuclear p14ARF expression. Further analysis using a patient derived xenograft (PDX) and cell culture models confirmed a coordinated expression of AR and p14ARF, and AR-mediated regulation of E2F1–3. In addition, we identified AR binding to sequences upstream of p14ARF coding region and this region conferred androgen sensitivity. siRNA mediated reduction of p14ARF was accompanied by an increase in PSA and TMPRSS2 expression but a decrease in cell viability and induction of apoptosis. The results argue that AR, p14ARF and E2F1 serve as part of a feedback loop that maintains optimal AR activity to drive proliferation.
2. Materials & methods
2.1. Characteristics of prostate tumors
Clinical data of 78 patients who underwent radical retropubic prostatectomy at VA Northern California Health Care System (VANCHCS) between 1999 and 2004 were extracted from the VANCHCS archives. As of Jan 2018, 37 patients experienced biochemical recurrence (BCR), indicated by two consecutive increases in PSA >0.2 ng/ml as defined by the American Urological Association Prostate Guideline Update Panel [25], and 39 patients had died from the disease.
2.2. Tissue array Construction
This study utilized formalin fixed paraffin-embedded primary prostate tumor and surrounding non-tumor tissues of 78 patients who underwent radical retropubic prostatectomy at VANCHCS between 1996 and 2002. Patient characteristics are described in Table I. 60 um core samples were extracted from the specific areas of the donor blocks and arranged in triplicate in a tissue microarray (TMA) using a Beecher Instruments Manual Tissue Arrayer. Hematoxylin-eosin staining was used as a reference for interpreting the additional sections of the TMA.
Table I.
Patients and tumor characteristics.
| Total | Recurrent | Non-recurrent | p-value | |
|---|---|---|---|---|
| N | 78 | 37 | 41 | |
| Race | ||||
| Caucasian (%) | 51 | 23 (45.1%) | 28 (54.9 %) | 0.0826 |
| African American (%) | 19 | 11 (57.9 %) | 8 (42.1%) | |
| Other (%) | 8 | 3 (37.5%) | 5 (62.5%) | |
| Median Age of Surgery (Range) | 62 (41–74) | 64 (41–74) | 60 (50–72) | 0.142 |
| Median Pre-op PSA (Range) | 6.6 (1.0–53.6) | 7.3 (2.6–15.3) | 6.0 (1.0–53.5) | 0.223 |
| Stage | ||||
| T2 (%) | 59 | 22 (37.3%) | 37 (62.7) | 0.0111 |
| T3 (%) | 19 | 15 (78.9%) | 4 (21.1%) | |
| Median Gleason Grade (Range) | 6 (4–9) | 7 (5–9) | 6 (4–7) | 0.0009 |
| 10-year Survival | 69.84% | 64.86% | 73.17% | 0.415 |
2.3. Immunohistochemistry
Immunohistochemical studies were conducted as previously described [26]. Antihuman polyclonal rabbit IgG served as a negative control. The antibodies used were Ki67 (MIB-1,DAKO; dilution 1:100), p53 (sc-126, Santa Cruz Biotechnology, dilution 1:200), MDM2 (18–2403, Clone IF2, Invitrogen; dilution 1:25), MDM4 (MDMX Abcam 76362; dilution 1:2000), p14ARF (clone 4C6/4 MAB 3782; Chemicon International; dilution 1:250), AR (sc-816, Santa Cruz Biotechnology, dilution 1:500). The degree of staining was evaluated blindly in a semiquantitative fashion by a pathologist (F.U.M. and A.D.B.) noting both the intensity of staining as well as the percentage of cells exhibiting that intensity in the nucleus and the cytoplasm. The intensity of the staining was scored as no staining (0), weak (1), intermediate (2), and strong (3) staining. The composite score was the number of cells staining multiplied by the intensity of staining.
2.4. Cell culture and transfection of siRNA
LNCaP and 22Rv1 cells were obtained from American Type Culture Collection and used under passage 20. LNCaP AI cells were generated in our laboratory by continuous culture of LNCaP cells in androgen-free media [27]. Cells were propagated in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) or charcoal stripped fetal bovine serum (CCS), 2 mmol/L L-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin (Invitrogen) at 37°C and 5% CO2. 3 ×105 cells were plated in 60 mm dishes and transfected 24h later using with Lipofectamine 2000 or RNAi Max (Invitrogen) and 100 nM siRNA. The siRNA sequences are listed in Supplementary Materials and Methods. The transfection mix was removed 24h later and replaced with normal growth medium. Cells were harvested 72h post-transfection.
2.5. Western immunoblot analysis
Following washes with cold PBS, cells were directly placed in a radioimmunoprecipitation assay lysis buffer that contained a protease inhibitor cocktail (Sigma). Thirty micrograms of protein were separated on 8%, 10%, or 12% SDS–PAGE gels and transferred to 0.22 mm GE Nitrocellulose supported membrane (BioExpress). The membrane was blocked with 5% nonfat dry milk in PBS and 0.1% Tween-20. The following antibodies were used: from Santa Cruz Biotechnology AR (sc-816), p14ARF (sc-8340), E2F1 (sc-251), E2F3 (sc-878), GAPDH (sc-32233), from Cell Signaling: tubulin (2125), Novus Biologicals: E2F2 (NB100–92380). Proteins were detected using enhanced chemiluminescence (GE Healthcare).
2.6. RNA preparation and realtime PCR
Total cellular RNA was prepared utilizing RNeasy mini kit (Qiagen, Inc. CA) based on the manufacturer’s protocol. cDNA was synthesized from RNA using the QuantaBio qScript XLT cDNA SuperMix (Qiagen). Realtime PCR was conducted using the QuantaBio PerfeCTa SYBR Green FastMix Low Rox kit. HPRT1 was used as the endogenous expression standard. Primer sequences are in Supplementary Materials and Methods. Data was collected with ViiA 7 (Applied Biosystems) and analyzed using the efficiency corrected relative standard curve method.
2.7. Chromatin Immunoprecipitation
Chip studies were conducted using the Magna ChIP™ HiSens Chromatin Immunoprecipitation Kit (Millipore) with the following modifications. 3 ug of normal rabbit IgG (Santa Cruz Biotechnology) was added to the lysate for 1.5 hr at 4°C and precleared with 40 ul of protein A/G magnetic beads cells at room temperature. The precleared chromatin was split into aliquots. 1.5 ug of E2F3, E2F1, ChIPAb+ Androgen Receptor- ChIP Validated Antibody (Millipore) and normal rabbit IgG (Santa Cruz Biotechnology) were added to separate aliquots. Following cleanup, the DNA was fractionated on a 1% low melt agarose gel and the region between 200 to 500 bp was excised and purified using the QIAquick gel extraction kit (Qiagen, Inc.). Quantification of the recovered DNA products was determined by qPCR using primer sets (Supplementary Materials and Methods) using PerfeCTa SYBR Green FastMix Low ROX according to manufacturer’s recommendations (Quanta Biosciences, Gaithersburg, MD). Recovered products after ChIP were normalized to the respective negative control (IgG) using the formula ΔCt = Cttarget product or input – CtIgG, and further calculated as percent of input.
2.8. Cloning and transfection
Oligonucleotides containing the putative AR binding sequences or mutated sequences which included HindIII and BamH1 overhanging ends were cloned into the HindIII and BamH1 sites upstream of TK minimal promoter-luciferase plasmid (a generous gift from Dr. Hongwu Chen, University of California, Davis). Cloned sequences were p14ARF-2wt: tcatgttctgat; p14ARF2-mut: tcatattatgat; p14ARF-4wt: tcttctaccagctgtctaggggctaaaacatttgttca; p14ARF-4mut: tattataccagctgtctaggggctaaaacatttattaa. The luciferase constructs or parent plasmid along with B-gal expression plasmid were transfected into LNCaP and cultured in steroid depleted media or following addition of 1nM R1881 for 24h (Sigma-Aldrich). 48h following transfection cells were harvested and subject to luciferase and B-gal assays. Each sample’s luciferase value was normalized to its respective β-galactosidase control. Relative luciferase units (RLU) were then calculated for cells in DHT treated condition relative to vehicle treatment. RLU for vector control in steroid depleted conditions was then set to “1” and the RLU of the plotted relative to this parameter. (*p<0.05, Student’s t-test).
2.9. Flow Cytometry
LNCaP cells were transfected with either a control oligonucleotide or p14ARF targeting siRNA in triplicate. Cells were harvested at 3 or 6 days post transfection, fixed in 70% ethanol, and prepared for flow cytometry using phosphate buffered saline, DNase-free RNase A, and propidium iodide (PI) stained. Flow cytometry was carried out with BD CellQuest Pro data acquisition software. Cell cycle data was analyzed using Flowing Software (version 2.5.1 Turku Bioimaging). Data is displayed as the mean +/− the standard deviation.
2.10. Caspase 3/7 assay
Cells were plated at 100,000 cells per well in 6 well plates, 3 wells per treatment. Cells were transfected using Lipofectamine 2000 and the manufacturers associated protocol with siRNA concentrations of 150 nM. Cells were incubated for 4 hours before media was added to dilute the transfection media. CellEvent Caspase 3/7 reagent (Invitrogen) was applied, per manufacturers protocol, post transfection and imaged using an EVOS FL 2 Auto Imaging System.
2.11. Animal Studies
4–5-week old nu/nu athymic male mice were obtained from Harlan Sprague Dawley, Inc. (Indianapolis, IN) and implanted subcutaneously with sustained release testosterone pellets (12.5 mg, 90-day release; Innovative Research of America). Suspensions of CWR22 cells were made in a 1:1 solution of RPMI 1640 (GIBCO) and Matrigel solubilized basement membrane (BD Biosciences). Xenografts were established by subcutaneous injections of 2.5 × 106 cells/injection into the right flank. When palpable tumors were observed, animals were (i) left intact (sham operated) or (ii) castrated by bilateral orchiectomy and removal of the testosterone pellet. The animals were followed for approximately four weeks, after which the mice were euthanized, and the tumors were resected. The study was conducted under an approved IACUC protocol.
2.12. Statistical Analysis
Clinical Information was compared between patients with biochemical recurrent disease (BCR) and no-BCR using Wald Chi-square statistics. Median staining levels were compared between tumor and non-tumor areas from the same subject using Mann-Whitney tests. The correlations between staining levels and demographic characteristics were estimated using Spearmans’ Rank Order Correlation. Survival analyses used Weibull models and was conducted using SAS version 9.3. Mouse tumor data were analyzed by normalization of all measurements to pre-operation (sham or castration) measurements for each individual mouse, then mean and standard errors calculated for the aggregate group. Graphs were generated using Excel or SigmaPlot 12.0. All RT PCR analyses of mRNA levels were conducted in triplicate. Student’s t-test was used to determine statistical significance (P≤ 0.05).
3. Results
3.1. p14ARF levels correlated significantly with AR
An analysis of prostate cancer patient data found no significant difference between patients who experienced BCR vs those who did not, with respect to race, age or median pre-operative PSA (Table I). However, pathological stage and biopsy Gleason grade, known predictors of BCR, significantly correlated with subsequent recurrence (Table I). There was no significant difference in 10-year survival rates between patients with BCR vs no BCR (Table I) (p=0.415). Overall survival (OS) correlated with age of patient (p=0.009) but no other clinical parameters.
We examined whether there was correlation between AR expression and expression of p53 pathway components. TMA sections were stained with antibodies directed against p53, MDM2, MDM4 and p14ARF, known regulators of p53 that are deregulated in multiple malignancies [28]. Sections were also stained with an AR antibody and the Ki67 proliferation marker antibody (Figure 1A). As expected, the tumor cells were highly proliferative, exhibiting high Ki67 staining in comparison to non-cancer areas (p<0.001) (Figure 1). Further, Ki67 scores correlated with clinical stage (Pearson Product Moment = 0.239, p=0.0407), but not with OS, BCR, or pathological stage. There was no overall difference between the levels of p53, MDM4 and p14ARF or nuclear MDM2 between PCa and non-tumor prostate, but levels of cytoplasmic MDM2 were significantly up-regulated in tumor vs non-tumor tissue (p=0.009) (Table 2, supplementary Table I). Comparison of nuclear protein expression in the tumors revealed weak correlations between all components of the p53 pathway with AR and Ki67. However, there was a strong significant correlation between p14ARF and AR (Pearson Product Moment R = 0.521, p=1.25 × 105 between nuclear AR and nuclear p14ARF in cancer tissue) (Table 2, Supplementary Table I).
FIGURE 1. Immunohistochemical staining of benign, Interim grade and high-grade prostate cancer.

Hemotoxylin and eosine (H&E) staining was used to define tumor morphology. Brown staining is indicative of antibody positive staining, while blue staining demonstrates hemotoxylin counterstain to demonstrate localization of cytoplasmic and nuclear material. Ki67 nuclear in the nucleus demonstrates Ki67 expression. There was a lack of Ki67 staining in non-tumor tissue. For immunohistochemical staining for AR in tumor vs non-tumor areas of prostatectomy specimens, AR staining in the luminal epithelia, but not the stroma, was scored. Since AR was observed in both the nucleus (N) and the cytoplasm (C), these areas were scored separately. The staining was mostly nuclear in nature but there was no significant difference in overall AR staining between tumor and non-tumor areas. Immunohistochemical staining for p14ARF in tumor and non-tumor revealed expression in both. p14ARF levels correlated with AR. The association was significant (p=0.0005).
Table 2.
A. Spearman Rank Order Correlation of nuclear expression in tumors of p53 pathway components, AR and Ki67 in all patients. The pair(s) of variables with positive correlation coefficients and P values below 0.050 tends to increase together. For the pairs with negative correlation coefficients and P values below 0.050, one variable tends to decrease while the other increases. For pairs with P values greater than 0.050, there is no significant relationship between the two variables.
| MDM2 | MDM4 | AR | Ki67 | 14ARF | ||
|---|---|---|---|---|---|---|
| p53 | correlation | 0.236 | 0.315 | 0.32 | 0.284 | 0.399 |
| P Value | 0.0562 | 0.012 | 0.00757 | 0.0191 | 0.00106 | |
| N | 66 | 63 | 69 | 68 | 56 | |
| MDM2 | correlation | 0.274 | 0.272 | 0.211 | 0.464 | |
| P Value | 0.0396 | 0.0284 | 0.718 | 0.0000987 | ||
| N | 57 | 65 | 67 | 66 | ||
| MDM4 | correlation | 0.117 | 0.359 | 0.368 | ||
| P Value | 0.369 | 0.00539 | 0.00543 | |||
| N | 61 | 59 | 56 | |||
| AR | correlation | 0.241 | 0.521 | |||
| P Value | 0.0514 | 0.0000125 | ||||
| N | 66 | 64 | ||||
| Ki67 | correlation | 0.343 | ||||
| P Value | 0.493 | |||||
| N | 66 |
3.2. AR transcriptionally regulated p14ARF expression in animal and cell models of prostate cancer
Previous reports showed that p14ARF regulates the expression of AR [24, 29], however, the influence of AR on p14ARF is unknown. We examined the effect of AR inhibition on p14ARF expression in the CWR22 patient-derived xenograft (PDX) model. CWR22 is an established model that is known to be androgen dependent [30, 31]. Immediately following castration of CWR22 tumor-bearing mice, the AR levels fall drastically, and tumor growth is inhibited; however, eventually tumors relapse [26, 32], thus recapitulating the disease course documented in PCa patients [30]. 4–5-week-old athymic male nu/nu mice were subcutaneously transplanted with CWR22 cells. When the tumors were palpable, the mice underwent castration (n=5) or sham operation (n=6). 29 days following the surgery, the mice were euthanized, and the tumors were harvested. Tumors in castrated mice were significantly smaller than those in intact mice. Immunohistochemical (IHC) studies confirmed that castration resulted in a reduction of AR levels. Moreover, AR localization was mostly cytoplasmic in the castrated mice; in contrast, in intact mice, AR levels were much high and were mostly nuclear with low levels in the cytoplasm (Figure 2A). The proliferation index (nuclear Ki67) precipitously declined following castration. p14ARF expression was mostly nuclear and decreased following castration. These results were confirmed by western blotting (Figure 2B) which showed a decrease of AR protein in the tumors from castrated animals. Importantly, RT-PCR analysis showed that p14ARF mRNA levels were reduced in the tumors from castrated animals (Figure 2B) indicating the decrease was at the transcriptional level.
FIGURE 2. Expression of AR and p14ARF in CWR22 tumor in intact and castrated mice.
(A) Immunohistochemical staining for AR in CWR22 xenograph tumors from intact and castrated mice. Expression (brown) was scored as low or high in the nuclear or cytoplasmic compartments. The slides were counterstained with Hematoxylin (blue/purple) to visualize the cells. The white arrows note the predominantly nuclear staining in tumors from intact animals and the cytoplasmic staining in tumors from castrated animals. The study utilized a total of 11 tumors. N= nuclear staining, C= cytoplasmic staining. (B) Western immunoblot analysis of CWR22 tumors from intact and castrated animals (top panel). Tubulin served as a loading control. The levels of p14ARF in the tumors were assessed by realtime PCR and standardized to the first control tumor (lower panel). (C) LNCaP cells cultured in androgen containing (FBS) express higher levels of AR and p14ARF protein than cells cultured in steroid depleted media (CSS). Actin served as a loading control. Decreased AR expression resulted in a decrease of p14ARF. (D) p14ARF mRNA levels are reduced in cells cultured in CSS media. Addition of DHT results in increased levels of p14ARF mRNA. (E) p14ARF mRNA levels are lower in castrate resistant LNCaP AI cells following siRNA mediated AR ablation. * P≤ 0.05. Protein expression was quantified using Image J and standardized to the loading control.
Previous studies established that culture in steroid depleted CSS as compared to FBS significantly reduces AR levels in castrate sensitive LNCaP cells [27, 33]. Therefore, we investigated whether the same treatment would affect p14ARF levels. LNCaP cells cultured in CSS expressed lower levels of AR and p14ARF protein (Figure 2C) and exhibited decreased p14ARF mRNA expression (Figure 2D). However, the transcription of p16, the other protein encoded by this locus was elevated on steroid depletion, indicating that the two transcripts are differentially regulated (Supplementary Figure 2). To further confirm that the decrease in p14ARF was due to depletion of androgen, not other steroids, cells cultured in steroid depleted media were supplemented with 1nM DHT. Cells supplemented with DHT expressed higher levels of p14ARF compared to those in CSS alone, confirming that this gene was subject to androgen-dependent regulation (Figure 2D). LNCaP AI cells are an AR dependent but castration resistant LNCaP derivative developed in our lab [27]. AR levels were depleted using an AR specific siRNA. AR depletion significantly reduced the levels of p14ARF mRNA (Figure 2E), establishing that p14ARF transcription was dependent on the AR. These results support previous studies showing that supraphysiological androgen levels result in p14ARF mRNA elevation in LNCaP cells [34].
3.3. The effect of AR on p14ARF is independent of its effect on the E2F family
Several studies have shown that p14ARF is transcriptionally controlled by members of the E2F family [35–37]. Hence, we investigated whether the E2F family of transcriptional factors mediate the transcriptional effects of AR on p14ARF. Since our data indicate that the AR regulates p14ARF, we also assessed the expression of E2F1, −2, −3a, −3b in the presence or absence of AR activation. Analysis utilizing CWR22 xenograft tumors from sham-operated and castrated mice showed that the mRNA expression of E2F1, −2, −3a significantly decreased (P˂ 0.001) while E2F3b levels significantly increased P˂ 0.001) in tumors from castrated mice (Figure 3A, left panel). Moreover, AR and E2F1–3a protein levels were decreased in xenografts from castrated mice, while E2F3b levels were elevated, compared to intact mice, demonstrating a coordination between mRNA and protein expression (Figure 3A, right panel). To determine whether this deregulation of E2Fs is observed in other cell models, the same analysis was conducted in LNCaP cells cultured in FBS and CSS media. The results recapitulate those obtained in the CWR22 study, where in the absence of steroids E2F1, 2 and 3a transcript and protein expression decreased, while E2F3b transcript and protein levels increased (Figure 3B, left panel). Thus, the expression of E2F1–3b is androgen-regulated in hormone-sensitive PCa cells.
FIGURE 3. Expression of E2F1, 2 3a and 3b in LNCaP cells.
(A) mRNA expression was analyzed by realtime PCR in CWR22 xenografts from intact and castrated mice (left panel). The data reflect triplicate analysis (mean +/− standard deviation. The differences in expression between the xenograft from sham and castrated mice was calculated using unpaired Student t test, *P ˂ 0.001. Western immunoblot analysis of the same xenograft samples (right panel). The arrows denote E2F3a and b. (B) mRNA expression of E2F1, 2, 3a and 3b in LNCaP cells cultured in the presence (FBS) and absence (CSS) of steroids. Data reflect triplicate analysis of at least 2–3 independent biological replicates (mean +/− standard deviation) (C) Realtime PCR analysis of E2F1, 3a, 3b and p14ARF mRNA expression following siRNA-mediated decrease of E2F3b, E2F1, E2F3a. Studies of siRNA-mediated decrease of E2F3b was conducted in LNCaP cells cultured in steroid depleted media, while the E2F31 and E2F3a studies were conducted in LNCaP cells cultured in steroid contacting media. (D) AR mRNA and protein levels are decreased in LNCaP cells following siRNA mediated decrease of E2F1. Protein expression was quantified using Image J and standardized to the loading control.
Next, we investigated the effects of E2Fs on p14ARF expression. Since E2F3b has previously been shown to specifically repress p19ARF (mouse ortholog of human p14ARF) during mouse development [36], and E2F3b expression was elevated in the absence of androgen signaling, we assessed the effect of E2F3b depletion on p14ARF transcription. The study used LNCaP cells cultured in steroid depleted media where E2F3b levels were highest while p14ARF levels were low. Unexpectedly E2F3b depletion did not relieve p14ARF repression (Figure 3C left) but resulted in a decrease of p14ARF transcripts suggesting that in this cellular context, E2F3b is not repressing p14ARF transcription, but may contribute to p14ARF activation. A decrease of E2F3a had a small negative effect (Figure 3C, right). siRNA-mediated depletion of both forms also reduced p14ARF expression (Supplementary Figure 1A). E2F1 has been shown to positively regulate p14ARF expression in non-prostate cells [38]. Therefore, the effect of E2F1 on p14ARF transcription was analyzed in LNCaP cells proliferating in the presence of androgens. E2F1 depletion effectively reduced p14ARF transcription (Figure 3D right), however, E2F1 depletion also resulted in reduction of AR (Figure 3D). Given this result it was unclear if AR or E2F1 was regulating p14ARF.
3.4. AR binds upstream of p14ARF encoding CDKN2A gene
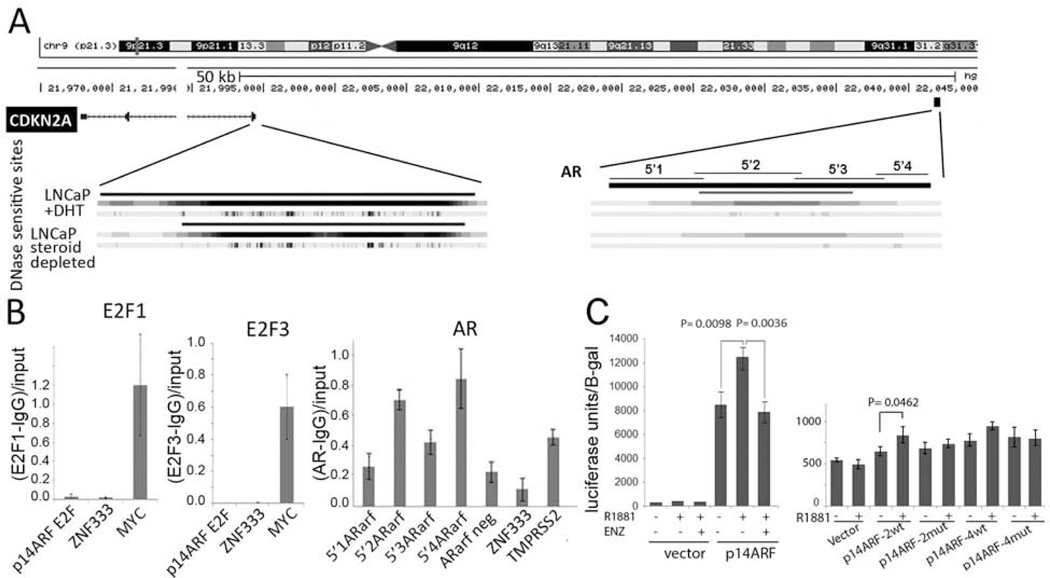
An examination of the p14ARF promoter and immediate surrounding regions did not identify a canonical AR response element (ARE). However, AR binding at great distances can affect gene transcription [39, 40]. Therefore, we used published studies to determine if an AR binding site had been detected near the p16INK4A/p14ARF locus. AR ChIP-seq studies conducted in LNCaP cells [41] identified an AR binding site ~45KB upstream of the p14ARF promoter (Figure 4A). An inspection of this sequence did not identify canonical AR binding sites, but several AR half-sites were present (Supplementary Material and Methods). Further analysis of LNCaP ENCODE data revealed that the AR binding site was coincident with a DNase sensitive region that was detected in LNCaP cells and was enhanced following DHT treatment. This region was not DNase sensitive in non-prostate tumor lines. Additionally, this region contains binding sites for the FOXA1 and GATA transcription factors, two factors that have been shown to augment AR-dependent gene expression [39].
FIGURE 4. ChIP analysis of AR binding to sequences upstream of the p14ARF gene.
(A) A schematic of the p14ARF (CDKN2A) gene. The heavy black block notes the location of an identified AR binding sites. An analysis of DNase sensitive sites from ENCODE revealed that in LNCaP cells regions proximal to the promoter regions (left) and AR binding region were DNase sensitive. (B) ChIP analysis of E2F1 and E2F3 binding to the p14ARF promoter, and AR binding at sequences spanning the identified AR binding region. IgG antibodies served as a negative control. An E2F site in the myc promoter served as a positive control for E2F1 and E2F3, and the TMPRSS2 gene AR binding site served as a positive control for AR. ZNF333 served as a negative control. A sequence upstream of the p14ARF promoter, but outside the identified AR binding sites (AR arf neg) served as an additional negative control. Following quantification of the products by RT PCR the values were normalized to the IgG negative control and calculated as percent input. C. Luciferase assay of the control minimal TK promoter-luciferase vector and the vector harboring the AR binding region. The plasmid harboring the AR binding region exhibited elevated luciferase activity which was sensitive to androgens. Plasmid were transfected into LNCaP in triplicate. B-gal co-transfected plasmid served as a transfection control.
To determine if the putative AR binding site truly bound AR, we performed AR ChIP in LNCaP cells. The ZNF333 site served as a negative control, while the TMPRSS2 AR binding site was a positive control. A series of primer sets (p5’1–5’4 ARarf) spanning this 428 bp sequence (Supplementary Material and Methods) were used in chromatin immunoprecipitation studies (ChIP) and confirmed that AR binds to this region in LNCaP cells (Figure 4B). Significant AR binding was detected in the 5’2 and 5’4 regions, indicating the presence of at least one, and possibly two binding sites. ChIP analysis was also used to detect E2F1 and E2F3a binding to the promoter region that has been shown to contain an E2F binding site [42]. While E2F1 and E2F3a binding to E2F sites in Myc promoter [43] was readily detected, there was no binding to the previously identified E2F site on the p14ARF promoter, suggesting that in this cellular context, E2F does not predominantly drive p14ARF transcription.
The putative AR binding region was cloned into a minimal TK promoter-luciferase vector and transfected into LNCaP cells (Material and Methods). After transfection, the cells were placed into steroid depleted medium and supplemented with the synthetic androgen 1nM R1881 or with vehicle (Figure 4C, left panel). The region contained positive regulatory sequences since promoter activity was elevated. The addition of R1881 further enhanced transcription from this plasmid but did not affect transcription from the control vector. The putative AR binding sites in region 2 and region 4, p14ARF-2 and p14ARF-4, respectively were cloned into the minimal promoter vector. These sites did not alter promoter activity in the absence of androgens, but p14ARF-2 exhibited a sensitivity to androgen (Figure 4C, right panel). Androgen activation of P14ARF-4 promoter activity did not reach statistical significance. Taken together, these results suggest that p14ARF transcription is regulated by the AR.
3.5. p14ARF depletion reduces cell viability but increases PSA and TMPRSS2 expression
siRNA-mediated depletion of mRNA and protein (Figure 5A) in LNCaP cells did not alter p53 or AR protein levels (Figure 5A), but p27 protein levels were significantly elevated. E2F1 and E2F2 mRNA and protein were decreased indicating that p14ARF did not cause the degradation of E2F1 and 2. Unexpectedly, both E2F3a and E2F3b mRNA and proteins were elevated. The latter result is opposite from what was observed following decreased AR expression (Figure 5A).
FIGURE 5. siRNA-mediated decrease of p14ARF in LNCaP cells reduces cell viability.
(A) p14ARF targeting siRNA was used to reduce p14ARF expression. Western immunoblots were used to detect the protein expression. qPCR was used to quantitate the fold change of mRNA. A non-targeting siRNA served as a control, and GAPDH served as a gel loading control. (B) siRNA mediated-decrease of p14ARF reduced LNCaP cell viability 4 and 6 days after transfection. (C) Flow cytometry conducted 6 days after transfection assessed cell cycle alterations. (D) top panel- images of LNCaP cells 5 days after transfection with p14ARF targeting siRNA. Bottom panel- caspase 3/7 assay 5 days after p14ARF depletion. E. PSA and TMPRSS2 transcript levels were analyzed by RT PCR 3 days after p14ARF depletion. F. Summary of the effects of low, optimal, supraphysiological androgen levels and p14ARF depletion on prostate cancer cells. n/a (not available). Protein expression was quantified using Image J and standardized to the loading control.
p14ARF depletion reduced cell viability (Figure 5B), a result that is consistent with a reduction of E2F1 and 2 and increase in p27. This effect was evident 4 days post transfection but was greater 6 days post transfection. Flow cytometry did not detect significant cell cycle changes at the 3 day or 6-day timepoint post transfection (Figure 5C), but we noted that 4 days following transfection the cells had a slightly altered morphology and by day 5 this change was clearly apparent and suggested an apoptotic response (Figure 5D). Caspase 3/7 assays confirmed that p14ARF depletion results in a significant apoptosis (Figure 5D). As previously reported p14ARF depletion results in a time dependent increased PSA and TMPRSS2 expression (Figure 5E), indicative of elevated AR activity. These results argue that p14ARF serves to constrain AR activity. In the absence of p14ARF, AR activity is elevated, but cell viability is decreased due to an induction of apoptosis.
4. Discussion
In this study we identified a strong correlation between AR and p14ARF expression. The well-defined p14ARF effect on p53 protein stability [10, 11] was not detected in the current study, suggesting an alternative role for p14ARF in PCa. In the mouse PTEN deletion model of prostate tumorigenesis, p19ARF (mouse p14ARF homolog) loss does not accelerate but inhibits PCa development [23]. These observations suggest that in PCa, the role of p14ARF differs from that seen in other contexts and the strong correlation we observed between p14ARF and AR in primary prostate tumors suggests a link between these proteins. Analyses in the CWR22 xenograft and LNCaP models confirmed that diminishing AR activity reduces p14ARF expression. This complements previous studies showing that supraphysiological androgen levels elevated p14ARF mRNA in LNCaP cells [34]. Taken together, the studies show that AR activates p14ARF transcription.
p14ARF transcription has been shown to be controlled by multiple factors that include AML/Runx1 [44], TBX3 [45], STAT3 [46], Pit [47], as well as E2F/RB [36]. The RB/E2F network figures prominently in prostate tumorigenesis. An increase of E2F1 or depletion of RB elevates AR expression and promotes the acquisition of castration resistance [20, 48]. Higher E2F3a levels are predictive of a poor clinical outcome [19]. An E2F binding site upstream of the AR gene, provides a mechanism by which deregulated E2F/RB enhances AR expression [48]. The current study uncovered a reciprocal relationship where restricting AR activity results in a precipitous decline in the expression of the activator E2F1 and 2, a more modest decrease of E2F3a but a significant increase in E2F3b. Depletion of E2F1 or E2F3 retards cell cycle progression (Supplementary Figure 1B) The effects of decreased AR signaling were obtained in several contexts and are consistent with a cell cycle arrest. While it has not been established how AR regulates E2F1,2 and 3a transcription, one possible mechanism may involve myc. Myc transcription is positively regulated by AR [49] and myc in turn can drive transcription of activator E2Fs [50, 51]. In PCa cells, AR and activator E2Fs may be part of a positive feed-forward regulatory loop. While E2F3b, a transcription factor more associated with transcriptional repression rather than activation, was a suspected link between AR and p14ARF, depletion of E2F3b did not lead to de-repression of p14ARF. Therefore, in this context E2F3b is not an intermediary between AR and p14ARF regulation.
p14ARF can bind to growth promoting transcription factors E2F1, E2F2 and E2F3 resulting in their degradation and sequestration in the nucleolus [15, 52]. Hence, p14ARF depletion should result in increased levels of these proteins. However, we found that in the context of PCa cells, p14ARF depletion does not result in an elevation of E2F1 and 2, but rather a decline in both protein and mRNA levels. Interestingly, E2F3a and b protein and mRNA levels are elevated, arguing that E2F proteins are differentially regulated by p14ARF.
Given the importance of maintaining optimal AR activity in PCa cells, AR employs p14ARF as a component of a negative feedback mechanism to limit its own activity and ensure maximal cell viability. This is analogous to the p53/MDM2 feedback loop where high p53 levels promote MDM2 transcription, which in turn limits p53 levels [53]. Therefore, in the context of PCa, a major role of p14ARF is to maintain optimal AR activity since supraphysiological androgen levels are as detrimental to PCa cell proliferation as castrate levels.
The biphasic effect of androgens on prostate tumor cell proliferation has been reported in multiple studies [54–56]. Castrate androgen levels reduce cell proliferation and decrease expression of prostate specific transcripts such as PSA and TMPRSS2. Supraphysiological androgen levels also reduce proliferation, an effect that is consistent with the decrease in E2F1 and 2 expression and increase in cyclin dependent kinase inhibitor p27 protein. However, supraphysiological androgen levels enhance PSA and TMPRESS transcription due to the enhanced activity of the AR (Figure 5F). This AR duality where androgen depletion and super elevated levels both inhibit prostate cancer cell viability is exploited in the clinical setting by the use of bipolar androgen therapy where androgen levels are cycled from castration to the supraphysiological range [57, 58].
Our studies show that p14ARF depletion and supraphysiological androgen levels share several features. Both result in decreased cell viability, an increase of p27, a decrease of E2F1 and 2, and an increase of E2F3b. Studies of supraphysiological testosterone treatment of castrate resistant patient derived xenografts found that responding malignancies exhibited E2F1 and 2 decrease and p27 increases [8]. We found that PSA and TMPRSS2 expression is increased following p14ARF depletion, a result that is consistent with a previous report that these proteins are elevated at supraphysiological androgen level [59]. However, an important difference between supraphysiological androgen levels and p14ARF depletion is that p14ARF depletion promotes apoptosis not growth arrest and senescence [8, 34, 59]. Since elevated AR activity is linked to a growth arrest, increased apoptosis following p14ARF depletion is likely due to an alteration in activity of other p14ARF targets. Using pharmacological means to repress p14ARF or modulate relevant p14ARF targets, in conjunction with an elevation of androgen levels, may redirect PCa cells to undergo apoptosis rather than growth arrest and senescence.
Supplementary Material
Highlights.
There is a correlation between AR and p14ARF expression in prostate cancers
The AR directly regulates p14ARF transcription to constrain its own activity
P14ARF deletion ultimately results in apoptosis of prostate cancer cells
Acknowledgments
Grant support
Funding: VA MERIT awards BX00004000 and BX003458 to PMG and MM, respectively and National Cancer Institute CA133209 (PMG).
Footnotes
Conflicts of interest
The authors report no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2018, CA Cancer J Clin, 68 (2018) 7–30. [DOI] [PubMed] [Google Scholar]
- [2].Taplin ME, Balk SP, Androgen receptor: a key molecule in the progression of prostate cancer to hormone independence, Journal of cellular biochemistry, 91 (2004) 483–490. [DOI] [PubMed] [Google Scholar]
- [3].Sadi MV, Walsh PC, Barrack ER, Immunohistochemical study of androgen receptors in metastatic prostate cancer. Comparison of receptor content and response to hormonal therapy, Cancer, 67 (1991) 3057–3064. [DOI] [PubMed] [Google Scholar]
- [4].Coffey RN, Watson RW, O’Neill AJ, Mc Eleny K, Fitzpatrick JM, Androgen-mediated resistance to apoptosis, The Prostate, 53 (2002) 300–309. [DOI] [PubMed] [Google Scholar]
- [5].Sonnenschein C, Olea N, Pasanen ME, Soto AM, Negative controls of cell proliferation: human prostate cancer cells and androgens, Cancer Res, 49 (1989) 3474–3481. [PubMed] [Google Scholar]
- [6].Kokontis JM, Hay N, Liao S, Progression of LNCaP prostate tumor cells during androgen deprivation: hormone-independent growth, repression of proliferation by androgen, and role for p27Kip1 in androgen-induced cell cycle arrest, Mol Endocrinol, 12 (1998) 941–953. [DOI] [PubMed] [Google Scholar]
- [7].Geck P, Maffini MV, Szelei J, Sonnenschein C, Soto AM, Androgen-induced proliferative quiescence in prostate cancer cells: the role of AS3 as its mediator, Proceedings of the National Academy of Sciences of the United States of America, 97 (2000) 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lam HM, Nguyen HM, Labrecque MP, Brown LG, Coleman IM, Gulati R, Lakely B, Sondheim D, Chatterjee P, Marck BT, Matsumoto AM, Mostaghel EA, Schweizer MT, Nelson PS, Corey E, Durable Response of Enzalutamide-resistant Prostate Cancer to Supraphysiological Testosterone Is Associated with a Multifaceted Growth Suppression and Impaired DNA Damage Response Transcriptomic Program in Patient-derived Xenografts, Eur Urol, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ozenne P, Eymin B, Brambilla E, Gazzeri S, The ARF tumor suppressor: structure, functions and status in cancer, International journal of cancer. Journal international du cancer, 127 (2010) 2239–2247. [DOI] [PubMed] [Google Scholar]
- [10].Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D, Nucleolar Arf sequesters Mdm2 and activates p53, Nature cell biology, 1 (1999) 20–26. [DOI] [PubMed] [Google Scholar]
- [11].Chen D, Kon N, Li M, Zhang W, Qin J, Gu W, ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor, Cell, 121 (2005) 1071–1083. [DOI] [PubMed] [Google Scholar]
- [12].Saporita AJ, Maggi LB Jr., Apicelli AJ, Weber JD, Therapeutic targets in the ARF tumor suppressor pathway, Current medicinal chemistry, 14 (2007) 1815–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fontana R, Ranieri M, La Mantia G, Vivo M, Dual Role of the Alternative Reading Frame ARF Protein in Cancer, Biomolecules, 9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Rizos H, Scurr LL, Irvine M, Alling NJ, Kefford RF, p14ARF regulates E2F-1 ubiquitination and degradation via a p53-dependent mechanism, Cell Cycle, 6 (2007) 1741–1747. [DOI] [PubMed] [Google Scholar]
- [15].Martelli F, Hamilton T, Silver DP, Sharpless NE, Bardeesy N, Rokas M, DePinho RA, Livingston DM, Grossman SR, p19ARF targets certain E2F species for degradation, Proceedings of the National Academy of Sciences of the United States of America, 98 (2001) 4455–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Iaquinta PJ, Aslanian A, Lees JA, Regulation of the Arf/p53 tumor surveillance network by E2F, Cold Spring Harbor symposia on quantitative biology, 70 (2005) 309–316. [DOI] [PubMed] [Google Scholar]
- [17].Kent LN, Leone G, The broken cycle: E2F dysfunction in cancer, Nat Rev Cancer, 19 (2019) 326–338. [DOI] [PubMed] [Google Scholar]
- [18].Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR, The E2F transcription factor is a cellular target for the RB protein, Cell, 65 (1991) 1053–1061. [DOI] [PubMed] [Google Scholar]
- [19].Foster CS, Falconer A, Dodson AR, Norman AR, Dennis N, Fletcher A, Southgate C, Dowe A, Dearnaley D, Jhavar S, Eeles R, Feber A, Cooper CS, Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome, Oncogene, 23 (2004) 5871–5879. [DOI] [PubMed] [Google Scholar]
- [20].Libertini SJ, Tepper CG, Guadalupe M, Lu Y, Asmuth DM, Mudryj M, E2F1 expression in LNCaP prostate cancer cells deregulates androgen dependent growth, suppresses differentiation, and enhances apoptosis, The Prostate, 66 (2006) 70–81. [DOI] [PubMed] [Google Scholar]
- [21].Sharma A, Comstock CE, Knudsen ES, Cao KH, Hess-Wilson JK, Morey LM, Barrera J, Knudsen KE, Retinoblastoma tumor suppressor status is a critical determinant of therapeutic response in prostate cancer cells, Cancer Res, 67 (2007) 6192–6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang Z, Rosen DG, Yao JL, Huang J, Liu J, Expression of p14ARF, p15INK4b, p16INK4a, and DCR2 increases during prostate cancer progression, Mod Pathol, 19 (2006) 1339–1343. [DOI] [PubMed] [Google Scholar]
- [23].Chen Z, Carracedo A, Lin HK, Koutcher JA, Behrendt N, Egia A, Alimonti A, Carver BS, Gerald W, Teruya-Feldstein J, Loda M, Pandolfi PP, Differential p53-independent outcomes of p19(Arf) loss in oncogenesis, Science signaling, 2 (2009) ra44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lu W, Xie Y, Ma Y, Matusik RJ, Chen Z, ARF represses androgen receptor transactivation in prostate cancer, Mol Endocrinol, 27 (2013) 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cookson MS, Aus G, Burnett AL, Canby-Hagino ED, D’Amico AV, Dmochowski RR, Eton DT, Forman JD, Goldenberg SL, Hernandez J, Higano CS, Kraus SR, Moul JW, Tangen C, Thrasher JB, Thompson I, Variation in the definition of biochemical recurrence in patients treated for localized prostate cancer: the American Urological Association Prostate Guidelines for Localized Prostate Cancer Update Panel report and recommendations for a standard in the reporting of surgical outcomes, J Urol, 177 (2007) 540–545. [DOI] [PubMed] [Google Scholar]
- [26].Mooso BA, Vinall RL, Tepper CG, Savoy RM, Cheung JP, Singh S, Siddiqui S, Wang Y, Bedolla RG, Martinez A, Mudryj M, Kung HJ, Devere White RW, Ghosh PM, Enhancing the effectiveness of androgen deprivation in prostate cancer by inducing Filamin A nuclear localization, Endocr Relat Cancer, 19 (2012) 759–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen L, Siddiqui S, Bose S, Mooso B, Asuncion A, Bedolla RG, Vinall R, Tepper CG, Gandour-Edwards R, Shi X, Lu XH, Siddiqui J, Chinnaiyan AM, Mehra R, Devere White RW, Carraway KL 3rd, Ghosh PM, Nrdp1-mediated regulation of ErbB3 expression by the androgen receptor in androgen-dependent but not castrate-resistant prostate cancer cells, Cancer Res, 70 (2010) 5994–6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wasylishen AR, Lozano G, Attenuating the p53 Pathway in Human Cancers: Many Means to the Same End, Cold Spring Harb Perspect Med, 6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Minges JT, Grossman G, Zhang P, Kafri T, Wilson EM, Post-translational Down-regulation of Melanoma Antigen-A11 (MAGE-A11) by Human p14-ARF Tumor Suppressor, The Journal of biological chemistry, 290 (2015) 25174–25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wainstein MA, He F, Robinson D, Kung HJ, Schwartz S, Giaconia JM, Edgehouse NL, Pretlow TP, Bodner DR, Kursh ED, et al. , CWR22: androgen-dependent xenograft model derived from a primary human prostatic carcinoma, Cancer Res, 54 (1994) 6049–6052. [PubMed] [Google Scholar]
- [31].Nagabhushan M, Miller CM, Pretlow TP, Giaconia JM, Edgehouse NL, Schwartz S, Kung HJ, de Vere White RW, Gumerlock PH, Resnick MI, Amini SB, Pretlow TG, CWR22: the first human prostate cancer xenograft with strongly androgen-dependent and relapsed strains both in vivo and in soft agar, Cancer Res, 56 (1996) 3042–3046. [PubMed] [Google Scholar]
- [32].Kim D, Gregory CW, French FS, Smith GJ, Mohler JL, Androgen receptor expression and cellular proliferation during transition from androgen-dependent to recurrent growth after castration in the CWR22 prostate cancer xenograft, Am J Pathol, 160 (2002) 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Berns EM, de Boer W, Mulder E, Androgen-dependent growth regulation of and release of specific protein(s) by the androgen receptor containing human prostate tumor cell line LNCaP, The Prostate, 9 (1986) 247–259. [DOI] [PubMed] [Google Scholar]
- [34].Roediger J, Hessenkemper W, Bartsch S, Manvelyan M, Huettner SS, Liehr T, Esmaeili M, Foller S, Petersen I, Grimm MO, Baniahmad A, Supraphysiological androgen levels induce cellular senescence in human prostate cancer cells through the Src-Akt pathway, Mol Cancer, 13 (2014) 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].del Arroyo AG, El Messaoudi S, Clark PA, James M, Stott F, Bracken A, Helin K, Peters G, E2F-dependent induction of p14ARF during cell cycle re-entry in human T cells, Cell cycle (Georgetown, Tex, 6 (2007) 2697–2705. [DOI] [PubMed] [Google Scholar]
- [36].Aslanian A, Iaquinta PJ, Verona R, Lees JA, Repression of the Arf tumor suppressor by E2F3 is required for normal cell cycle kinetics, Genes & development, 18 (2004) 1413–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Komori H, Enomoto M, Nakamura M, Iwanaga R, Ohtani K, Distinct E2F-mediated transcriptional program regulates p14ARF gene expression, EMBO J, 24 (2005) 3724–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Elliott MJ, Dong YB, Yang H, McMasters KM, E2F-1 Up-Regulates c-Myc and p14ARF and Induces Apoptosis in Colon Cancer Cells, Clinical Cancer Research, 7 (2001) 3590–3597. [PubMed] [Google Scholar]
- [39].Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M, A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth, Mol Cell, 27 (2007) 380–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Decker KF, Zheng D, He Y, Bowman T, Edwards JR, Jia L, Persistent androgen receptor-mediated transcription in castration-resistant prostate cancer under androgen-deprived conditions, Nucleic acids research, 40 (2012) 10765–10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Massie CE, Lynch A, Ramos-Montoya A, Boren J, Stark R, Fazli L, Warren A, Scott H, Madhu B, Sharma N, Bon H, Zecchini V, Smith DM, Denicola GM, Mathews N, Osborne M, Hadfield J, Macarthur S, Adryan B, Lyons SK, Brindle KM, Griffiths J, Gleave ME, Rennie PS, Neal DE, Mills IG, The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis, EMBO J, 30 (2011) 2719–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Komori H, Enomoto M, Nakamura M, Iwanaga R, Ohtani K, Distinct E2F-mediated transcriptional program regulates p14ARF gene expression, EMBO J, 24 (2005) 3724–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rabinovich A, Jin VX, Rabinovich R, Xu X, Farnham PJ, E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites, Genome research, 18 (2008) 1763–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nishimoto N, Arai S, Ichikawa M, Nakagawa M, Goyama S, Kumano K, Takahashi T, Kamikubo Y, Imai Y, Kurokawa M, Loss of AML1/Runx1 accelerates the development of MLL-ENL leukemia through down-regulation of p19ARF, Blood, 118 (2011) 2541–2550. [DOI] [PubMed] [Google Scholar]
- [45].Platonova N, Scotti M, Babich P, Bertoli G, Mento E, Meneghini V, Egeo A, Zucchi I, Merlo GR, TBX3, the gene mutated in ulnar-mammary syndrome, promotes growth of mammary epithelial cells via repression of p19ARF, independently of p53, Cell and tissue research, 328 (2007) 301–316. [DOI] [PubMed] [Google Scholar]
- [46].Pencik J, Schlederer M, Gruber W, Unger C, Walker SM, Chalaris A, Marie IJ, Hassler MR, Javaheri T, Aksoy O, Blayney JK, Prutsch N, Skucha A, Herac M, Kramer OH, Mazal P, Grebien F, Egger G, Poli V, Mikulits W, Eferl R, Esterbauer H, Kennedy R, Fend F, Scharpf M, Braun M, Perner S, Levy DE, Malcolm T, Turner SD, Haitel A, Susani M, Moazzami A, Rose-John S, Aberger F, Merkel O, Moriggl R, Culig Z, Dolznig H, Kenner L, STAT3 regulated ARF expression suppresses prostate cancer metastasis, Nature communications, 6 (2015) 7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Diaz-Rodriguez E, Garcia-Lavandeira M, Perez-Romero S, Senra A, Canibano C, Palmero I, Borrello MG, Dieguez C, Alvarez CV, Direct promoter induction of p19Arf by Pit-1 explains the dependence receptor RET/Pit-1/p53-induced apoptosis in the pituitary somatotroph cells, Oncogene, 31 (2012) 2824–2835. [DOI] [PubMed] [Google Scholar]
- [48].Sharma A, Yeow WS, Ertel A, Coleman I, Clegg N, Thangavel C, Morrissey C, Zhang X, Comstock CE, Witkiewicz AK, Gomella L, Knudsen ES, Nelson PS, Knudsen KE, The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression, J Clin Invest, 120 (2010) 4478–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bernard D, Pourtier-Manzanedo A, Gil J, Beach DH, Myc confers androgen-independent prostate cancer cell growth, J Clin Invest, 112 (2003) 1724–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Leone G, Sears R, Huang E, Rempel R, Nuckolls F, Park CH, Giangrande P, Wu L, Saavedra HI, Field SJ, Thompson MA, Yang H, Fujiwara Y, Greenberg ME, Orkin S, Smith C, Nevins JR, Myc requires distinct E2F activities to induce S phase and apoptosis, Mol Cell, 8 (2001) 105–113. [DOI] [PubMed] [Google Scholar]
- [51].Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B, Genomic targets of the human c-Myc protein, Genes Dev, 17 (2003) 1115–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sherr CJ, Divorcing ARF and p53: an unsettled case, Nature reviews. Cancer, 6 (2006) 663–673. [DOI] [PubMed] [Google Scholar]
- [53].Zhou X, Liao JM, Liao WJ, Lu H, Scission of the p53-MDM2 Loop by Ribosomal Proteins, Genes & cancer, 3 (2012) 298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Guillemette C, Hum DW, Belanger A, Evidence for a role of glucuronosyltransferase in the regulation of androgen action in the human prostatic cancer cell line LNCaP, The Journal of steroid biochemistry and molecular biology, 57 (1996) 225–231. [DOI] [PubMed] [Google Scholar]
- [55].Esquenet M, Swinnen JV, Heyns W, Verhoeven G, Control of LNCaP proliferation and differentiation: actions and interactions of androgens, 1alpha,25-dihydroxycholecalciferol, all-trans retinoic acid, 9-cis retinoic acid, and phenylacetate, The Prostate, 28 (1996) 182–194. [DOI] [PubMed] [Google Scholar]
- [56].Henttu P, Liao SS, Vihko P, Androgens up-regulate the human prostate-specific antigen messenger ribonucleic acid (mRNA), but down-regulate the prostatic acid phosphatase mRNA in the LNCaP cell line, Endocrinology, 130 (1992) 766–772. [DOI] [PubMed] [Google Scholar]
- [57].Schweizer MT, Wang H, Luber B, Nadal R, Spitz A, Rosen DM, Cao H, Antonarakis ES, Eisenberger MA, Carducci MA, Paller C, Denmeade SR, Bipolar androgen therapy for men with androgen ablation naive prostate cancer: Results from the phase II BATMAN study, The Prostate, (2016). [DOI] [PubMed] [Google Scholar]
- [58].Mohammad OS, Nyquist MD, Schweizer MT, Balk SP, Corey E, Plymate S, Nelson PS, Mostaghel EA, Supraphysiologic Testosterone Therapy in the Treatment of Prostate Cancer: Models, Mechanisms and Questions, Cancers (Basel), 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Tsihlias J, Zhang W, Bhattacharya N, Flanagan M, Klotz L, Slingerland J, Involvement of p27Kip1 in G1 arrest by high dose 5 alpha-dihydrotestosterone in LNCaP human prostate cancer cells, Oncogene, 19 (2000) 670–679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.