Abstract
Fomitopsis pinicola is a common brown-rot fungal species found in northern hemisphere. It grows on many different gymnosperm and angiosperm trees. Recent studies show that it is a species complex; three species from North America and one species from Europe have been recognized in this complex. In the current study, six new species in the Fomitopsis pinicola complex were discovered from East Asia, based on morphological characters and phylogenetic analyses inferred from the sequence data of the internal transcribed spacer (ITS) regions, the second subunit of RNA polymerase II (RPB2), and the translation elongation factor 1-α gene (TEF). Detailed descriptions of the six new species are provided. Our results also indicates that species of the F. pinicola complex from East Asia usually have limited distribution areas and host specialization.
Keywords: brown-rot fungi, host specialization, multi-gene phylogeny, polypore, species complex
Introduction
Fomitopsis P. Karst. was established by Karsten and typified by F. pinicola (Sw.) P. Karst. (Karsten, 1881). It is characterized by a perennial or annual growth habit, is sessile to effused-reflexed, has tough to woody hard basidiocarps, has a white to tan or pinkish-colored pore surface with mostly small and regular pores, has a dimitic to trimitic hyphal system with clamped generative hyphae, and has a hyaline, thin-walled, smooth, and ellipsoid to subglobose basidiospores which are negative in Melzer’s reagent; it causes a brown rot (Ryvarden and Johansen, 1980; Gilbertson and Ryvarden, 1986; Ryvarden and Gilbertson, 1993; Núñez and Ryvarden, 2001; Han et al., 2016).
Fomitopsis pinicola has been intensively studied because it has the function of dispelling wind-evil and dampness, and has anti-tumor (Dai et al., 2009; Sun et al., 2016), antifungal, antioxidant, immunomodulation, and neuroprotective activities (Guler et al., 2009; Bao et al., 2015; Sun et al., 2016; Guo and Wolf, 2018). Högberg et al. (1999) showed that all European populations of F. pinicola belong to one intersterility group. Ryvarden and Stokland (2008) described Fomitopsis ochracea Ryvarden & Stokland from Alberta on Populus tremuloides that was distinguished from F. pinicola by substrate preference, basidiospore morphology, and match flame test to the lacquered pilei surface. Subsequently, this species was proven to belong to the F. pinicola complex (Haight et al., 2019). Haight et al. (2016) suggested that F. pinicola is a species complex comprised of at least four well-supported phylogenetic species, three in North America (F. ochracea and two previously undescribed species) and one in Europe (F. pinicola). Haight et al. (2019) described two new species: F. mounceae Haight & Nakasone and F. schrenkii Haight & Nakasone from North America in the F. pinicola complex. Until now, four species have been recognized in the F. pinicola complex; F. mounceae, F. ochracea, and F. schrenkii from North America and F. pinicola from Europe.
In some cases, fungi species boundaries based on morphology misrepresents the number of existing species (Leavitt et al., 2011). Due to geographic isolation, lack of migration, and genetic drift there may be variation among populations, although this genetic variation is not always obvious (Haight et al., 2016). Cryptic species of species complexes are proving to be extremely common in higher fungi, particularly those with wide geographic distributions or host ranges (Haight et al., 2016); they share similar morphological characteristics and phylogenetic relationships to known species. More cryptic species could be discovered by combining evidence of morphological characters, molecular data, host trees, and distribution areas in species complexes (Liu et al., 2021).
In recent years, taxonomic and phylogenetic studies of Fomitopsis have been carried out in China and several new species have been described (Li et al., 2013; Han et al., 2014, 2016; Han and Cui, 2015; Liu et al., 2019), but none have been focused on the F. pinicola complex. Samples collected from China were still identified as F. pinicola complex in these studies. With more and more specimens collected from different areas of China, Vietnam, and of East Asia, six new species of the F. pinicola complex have been discovered based on morphological characters and phylogenetic analysis of ITS + RPB2 + TEF gene regions.
Materials and Methods
Taxa Sampling and Morphological Study
The examined specimens were deposited in the herbarium of the Institute of Microbiology, Beijing Forestry University (BJFC, Beijing, P. R. China). Morphological descriptions and abbreviations used in this study follow Han et al. (2016) and Liu et al. (2019).
DNA Extraction and Molecular Analyses
The procedures for DNA extraction and polymerase chain reaction (PCR) used in this study were the same as described by Chen et al. (2017) and Song and Cui (2017). The primer pairs ITS5 and ITS4 for ITS regions, fRPB2-f5F and bRPB2-7.1R for the RPB2 gene, and EF1-983 F and EF1-1567R for the TEF gene used in this study are the same as in previous studies (White et al., 1990; Rehner, 2001; Matheny, 2005).
The PCR cycling schedules for different DNA sequences of ITS, RPB2, and TEF genes used in this study followed those used in Zhu et al. (2019) and Sun et al. (2020) with some modifications. The PCR procedure for ITS was the initial denaturation at 95°C for 3 min, followed by 35 cycles of denaturation at 94°C for 40 s, annealing at 54°C for 45 s, and extension at 72°C for 1 min, and a final extension at 72°C for 10 min. The PCR procedure for RPB2 was the initial denaturation at 94°C for 2 min, followed by 37 cycles of denaturation at 94°C for 45 s, annealing at 56°C for 90 s, and extension at 72°C for 2 min, and a final extension at 72°C for 10 min. The PCR procedure for TEF was the initial denaturation at 95°C for 3 min, followed by 35 cycles of denaturation at 94°C for 40 s, annealing at 54–57°C for 45 s and extension at 72°C for 1 min, and a final extension at 72°C for 10 min. The PCR products were purified and sequenced at the Beijing Genomics Institute (BGI), China, with the same primers. All newly generated sequences were deposited in GenBank (Table 1). Additional sequences for phylogenetic analyses were downloaded from GenBank (Table 1). All sequences were aligned in MAFFT 7 (Katoh and Standley, 2013)1 and manually adjusted in BioEdit (Hall, 1999). Alignments were spliced in Mesquite (Maddison and Maddison, 2017). The missing sequences were coded as “N,” ambiguous nucleotides were coded as “N” followed Chen et al. (2017). The final concatenated sequence alignment was deposited in TreeBase2 (submission ID: 27439).
TABLE 1.
A list of species, specimens, and GenBank accession numbers of sequences used in this study.
| Species name | Sample no. | Locality | GenBank accessions |
||
| ITS | RPB2 | TEF | |||
| Antrodia tanakae | Cui 9743 | China | KR605814 | KR610833 | KR610743 |
| A. tanakae | Yuan 1106 | China | KP715313 | KR610835 | KP715343 |
| Daedalea quercina | Dai 12152 | Czechia | KP171207 | KR610809 | KR610717 |
| D. quercina | Dai 2260 | Sweden | KR605792 | KR610808 | KR610718 |
| Fomitopsis abieticola | Cui 10532 | China | MN148230 | MN158174 | MN161745 |
| F. abieticola | Cui 10521 | China | MN148231 | — | MN161746 |
| F. betulina | Cui 10756 | China | KR605797 | KR610815 | KR610725 |
| F. betulina | Dai 11449 | China | KR605798 | KR610816 | KR610726 |
| F. cana | Cui 6239 | China | JX435777 | KR610761 | KR610661 |
| F. cana | Dai 9611 | China | JX435776 | KR610762 | KR610660 |
| F. durescens | Overholts 4215 | United States | KF937293 | — | — |
| F. durescens | O 10796 | Venezuela | KF937292 | KR610766 | KR610669 |
| F. hengduanensis | Cui 16259 | China | MN148232 | MN158175 | MN161747 |
| F. hengduanensis | Cui 17056 | China | MN148233 | MN158176 | MN161748 |
| F. kesiyae | Cui 16437 | Vietnam | MN148234 | MN158177 | MN161749 |
| F. kesiyae | Cui 16466 | Vietnam | MN148235 | MN158178 | MN161750 |
| F. massoniana | Cui 2848 | China | MN148236 | — | MN161751 |
| F. massoniana | Cui 9058 | China | MN148237 | — | MN161752 |
| F. massoniana | Cui 11288 | China | MN148238 | MN158179 | MN161753 |
| F. massoniana | Cui 11304 | China | MN148239 | — | MN161754 |
| F. meliae | Dai 10035 | China | KR605774 | — | KR610683 |
| F. meliae | Ryvarden 16893 | Unknown | KR605776 | KR610775 | KR610681 |
| F. mounceae | AFTOL ID 770 | United States | AY786056 | AY864874 | AY705967 |
| F. mounceae | DR 366 | United States | KF169624 | KF169693 | KF178349 |
| F. mounceae | JAG 08 19 | United States | KF169626 | KF169695 | KF178351 |
| F. mounceae | JEH 78 | Canada | KF169629 | KF169698 | KF178354 |
| F. mounceae | Tuomo Niemelä 2530 | Canada | MN148240 | — | MN161755 |
| F. mounceae | Teuvo Ahti 60351 | Canada | MN148241 | — | MN161756 |
| F. mounceae | OM 18782 | United States | MN148242 | — | MN161757 |
| F. mounceae | Spirin 8367 | United States | MN148243 | — | MN161758 |
| F. ochracea | HHB 19692 | United States | KF169594 | KF169663 | KF178319 |
| F. ochracea | HHB 19670 | United States | KF169593 | KF169662 | KF178318 |
| F. ochracea | JEH 38 | United States | KF169603 | KF169672 | KF178328 |
| F. ochracea | LT 18 | United States | KF169616 | KF169685 | KF178341 |
| F. ochracea | OM 18568 | United States | MN148244 | — | MN161759 |
| F. ochracea | OM 18673 | United States | MN148245 | — | MN161760 |
| F. ochracea | Spirin 8165 | United States | MN148246 | — | MN161761 |
| F. pinicola | FCUG 2056 | Sweden | KF169654 | KF169723 | KF178379 |
| F. pinicola | HK 19330 | Russia | KF169655 | KF169724 | KF178380 |
| F. pinicola | LT 323 | Estonia | KF169651 | KF169720 | KF178376 |
| F. pinicola | LT 319 | Estonia | KF169652 | KF169721 | KF178377 |
| F. pinicola | AT Fp 1 | Sweden | MK208852 | MK236362 | MK236359 |
| F. pinicola | AT Fp 2 | Sweden | MK208853 | MK236363 | MK236360 |
| F. schrenkii | FP 105881 R | United States | KF169641 | KF169710 | KF178366 |
| F. schrenkii | JEH 144 | United States | KF169621 | MK208857 | MK236355 |
| F. schrenkii | JEH 150 | United States | KF169622 | MK208858 | MK236356 |
| F. schrenkii | JV 1209/61 J | United States | MN148247 | MN158180 | MN161762 |
| F. schrenkii | Inkeri Ahonen 58 | United States | MN148248 | — | MN161763 |
| F. subpinicola | Cui 9836 | China | MN148249 | MN158181 | MN161764 |
| F. subpinicola | Cui 9819 | China | MN148250 | — | MN161765 |
| F. subpinicola | Dai 11101 | China | MN148251 | MN158182 | MN161766 |
| F. subpinicola | Dai 11206 | China | MN148252 | MN158183 | MN161767 |
| F. subpinicola | Dai 13480 | China | MN148253 | MN158184 | MN161768 |
| F. subpinicola | Yuan 4912 | China | MN148254 | — | MN161769 |
| F. subtropica | Cui 10578 | China | KR605787 | KR610791 | KR610698 |
| F. subtropica | Cui 10140 | China | JQ067651 | KR610789 | KR610699 |
| F. tianshanensis | Wei 1568 | China | MN148255 | — | MN161770 |
| F. tianshanensis | Wei 1473a | China | MN148256 | — | MN161771 |
| F. tianshanensis | Wei 1462a | China | MN148257 | — | MN161772 |
| F. tianshanensis | Cui 16821 | China | MN148258 | — | MN161773 |
| F. tianshanensis | Cui 16823 | China | MN148259 | — | MN161774 |
| F. tianshanensis | Cui 16825 | China | MN148260 | — | MN161775 |
| F. tianshanensis | Cui 16828 | China | MN148261 | — | MN161776 |
| F. tianshanensis | Cui 16830 | China | MN148262 | — | MN161777 |
| Laetiporus zonatus | Dai 13633 | China | KX354481 | KX354676 | KX354635 |
| L. zonatus | Cui 10404 | China | KF951283 | KT894797 | KX354639 |
| Niveoporofomes spraguei | JV 0509/62 | United States | KR605786 | KR610788 | KR610697 |
| N. spraguei | 4638 | France | KR605784 | KR610786 | KR610696 |
| Rhodofomes roseus | Cui 10520 | China | KC507162 | KR610783 | KR610692 |
| R. roseus | Cui 10633 | China | KR605782 | KR610784 | KR610693 |
| Rhodofomitopsis feei | LR 14115 | Costa Rica | KF999923 | — | — |
| R. feei | JV 0610/K9-Kout | Mexico | KF999922 | — | KR610673 |
| Rubellofomes cystidiatus | Cui 5481 | China | KF937288 | KR610765 | KR610667 |
| R. cystidiatus | Yuan 6304 | China | KR605769 | — | KR610668 |
New sequences are shown in bold.
Phylogenetic analyses approaches used in this study followed Han et al. (2016) and Cui et al. (2019). The congruences of the 3-gene (ITS, RPB2 and TEF) were evaluated with the incongruence length difference (ILD) test (Farris et al., 1994) implemented in PAUP∗ 4.0b10 (Swofford, 2002), under heuristic search and 1000 homogeneity replicates. The sequences of Daedalea quercina (L.) Pers obtained from GenBank were used as outgroups for the phylogeny of the Fomitopsis pinicola complex, and sequences of Laetiporus zonatus B.K. Cui & J. Song were used as outgroups for the phylogeny of the Fomitopsis pinicola complex and related taxa. A maximum parsimony (MP) analysis was performed in PAUP∗ version 4.0b10 (Swofford, 2002). A maximum likelihood (ML) analysis was performed in RAxmL v.7.2.8 with a GTR + G + I model (Stamatakis, 2006). Bayesian inference (BI) was calculated by MrBayes 3.1.2 (Ronquist and Huelsenbeck, 2003) with a general time reversible (GTR) model of DNA substitution and a gamma distribution rate variation across sites determined by MrModeltest 2.3 (Posada and Crandall, 1998; Nylander, 2004). Clade robustness was assessed using a bootstrap (BT) analysis with 1000 replicates (Felsenstein, 1985). The branch support was evaluated with a bootstrapping method of 1000 replicates (Hillis and Bull, 1993). Branches that received bootstrap supports for the MP and ML, greater than or equal to 75% and Bayesian posterior probabilities (BPP) greater than or equal to 0.95, were considered as significantly supported (Shen et al., 2019; Sun et al., 2020). The phylogenetic tree was visualized using FigTree v1.4.23.
Results
Molecular Phylogeny
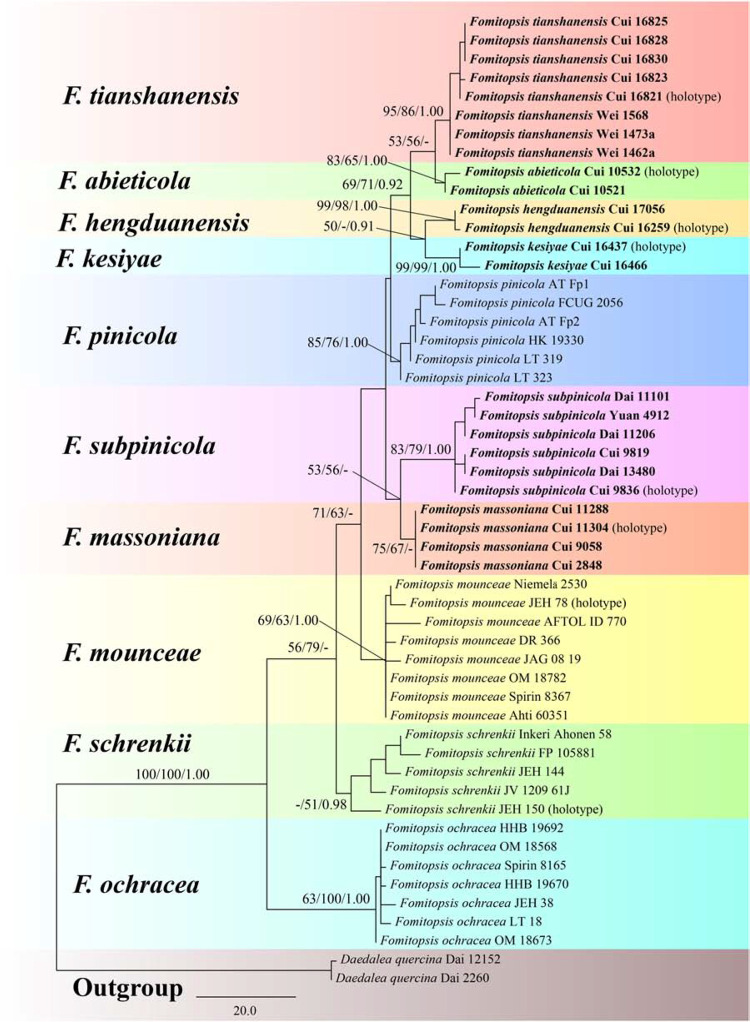
The combined 3-gene (ITS, RPB2, TEF) dataset to infer the phylogeny of species in the Fomitopsis pinicola complex included sequences from 52 fungal samples representing 11 taxa. The dataset had an aligned length of 1750 characters including gaps (553 characters for ITS, 641 characters for RPB2, 556 characters for TEF), of which 1403 characters were constant, 32 were variable and parsimony-uninformative, and 315 were parsimony-informative. Maximum parsimony analysis yielded 12 equally parsimonious trees (TL = 470, CI = 0.785, RI = 0.872, RC = 0.684, HI = 0.215). The best model for the combined ITS + RPB2 + TEF sequences dataset estimated and applied in the Bayesian analysis was GTR + I + G with equal frequency of nucleotides. Bayesian analysis and ML analysis resulted in a similar topology as the MP analysis, and only the MP tree inferred from the combined three-gene dataset is shown in Figure 1.
FIGURE 1.
Maximum parsimony tree illustrating the phylogeny of the Fomitopsis pinicola complex based on the combined sequences dataset of ITS + RPB2 + TEF. Daedalea quercina served as the outgroup. Branches are labeled with maximum likelihood bootstrap higher than 50%, maximum parsimony bootstrap proportions higher than 50% and Bayesian posterior probabilities more than 0.95. Bold names = New species.
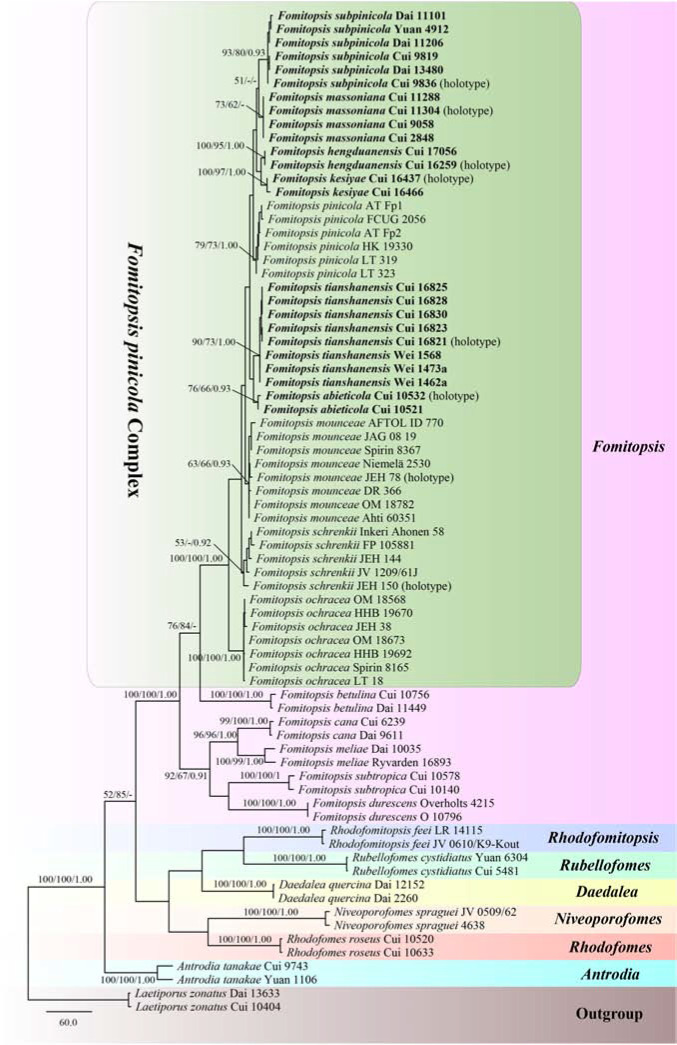
The combined three-gene (ITS, RPB2, TEF) dataset infer the phylogeny of species in the Fomitopsis pinicola complex and the related group included sequences from 74 fungal samples representing 21 taxa. The dataset had an aligned length of 1848 characters including gaps (641 characters for ITS, 641 characters for RPB2, 566 characters for TEF), of which 1063 characters were constant, 41 were variable and parsimony-uninformative, and 744 were parsimony-informative. MP analysis yielded 10 equally parsimonious trees (TL = 2428, CI = 0.521, RI = 0.762, RC = 0.397, HI = 0.479). The best model for the concatenate sequence dataset estimated and applied in the Bayesian inference was GTR + I + G with an equal frequency of nucleotides. Bayesian analysis and ML analysis resulted in a similar topology as the MP analysis, and only the MP tree inferred from the combined three-gene sequences dataset is shown in Figure 2.
FIGURE 2.
Maximum parsimony tree illustrating the phylogeny of the Fomitopsis pinicola complex and related group based on the combined sequences dataset of ITS + RPB2 + TEF. Laetiporus zonatus served as the outgroup. Branches are labeled with maximum likelihood bootstrap higher than 50%, maximum parsimony bootstrap proportions higher than 50% and Bayesian posterior probabilities more than 0.95. Bold names = New species.
The phylogenetic trees (Figures 1, 2) generated by Maximum parsimony, Maximum likelihood and Bayesian analyses showed that the six new species, Fomitopsis abieticola, F. hengduanensis, F. kesiyae, F. massoniana, F. subpinicola, and F. tianshanensis grouped together with F. mounceae, F. ochracea, F. pinicola, and F. schrenkii, thus, the species number of the F. pinicola complex increased to 10 around the world.
Taxonomy
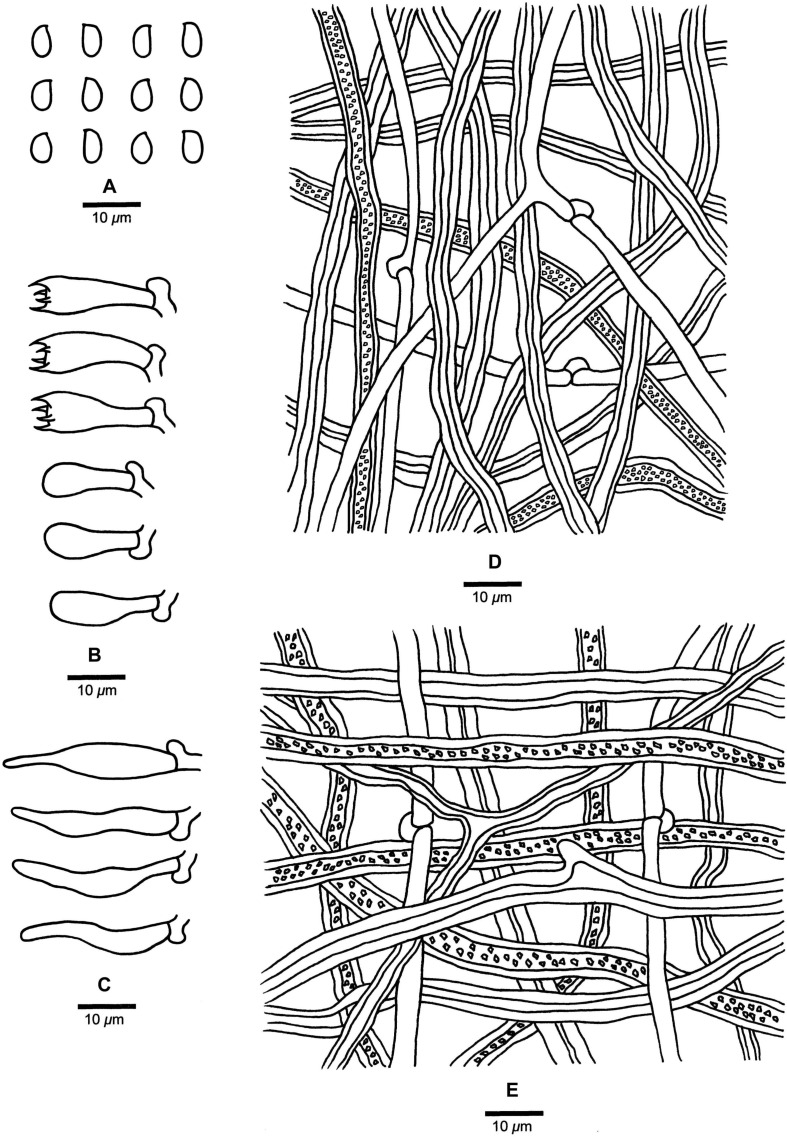
Fomitopsis abieticola B.K. Cui, M.L. Han & Shun Liu, sp. nov. (Figures 3A,B, 4).
FIGURE 3.
Basidiocarps of the Fomitopsis pinicola complex species. (A,B) F. abieticola; (C,D) F. hengduanensis; (E,F) F. kesiyae; (G,H) F. massoniana; (I,J) F. subpinicola; (K,L) F. tianshanensis. Bars: A,B,D,E,F = 2 cm; G,H = 1 cm; C,I,J = 3 cm; K,L = 5 cm.
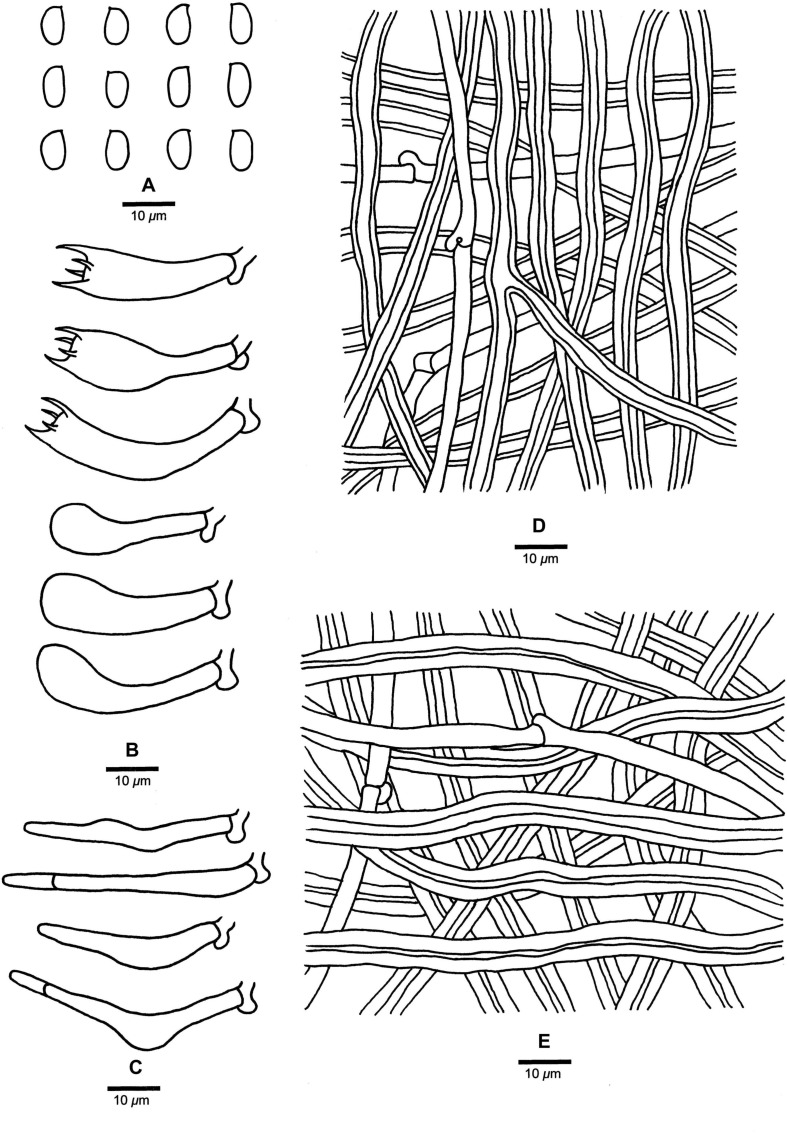
FIGURE 4.
Microscopic structures of Fomitopsis abieticola (drawn from the holotype). (A) Basidiospores; (B) Basidia and basidioles; (C) Cystidioles; (D) Hyphae from trama; (E) Hyphae from context. Bars: A–E = 10 μm.
MycoBank: MB 838908
Fomitopsis abieticola is characterized by its large pores (2–4 per mm), long cystidioles (17.5–50.2 × 4.3–9.5 μm), long basidia (20.8–40.5 × 5.5–11.5 μm) and big basidiospores (7–9 × 4–5 μm), and grows on Abies.
Type. — CHINA. Yunnan Province, Shangri-La County, Pudacuo National Park, on stump of Abies, 24 September 2011, Cui 10532 (Holotype, BJFC 011427).
Etymology. —Abieticola (Lat.), refers to the host tree genus Abies.
Basidiocarps. —Annual to perennial, pileate, sessile, solitary, hard corky, without odor or taste when fresh, woody hard and light in weight upon drying. Pilei semicircular to ungulate, projecting up to 6.5 cm long, 8.5 cm wide, 2.5 cm thick at base. Pileal surface cream to pinkish buff when fresh, becoming honey-yellow to grayish brown when dry, glabrous, small nodules appear near the base, rough, azonate; margin cream, slightly paler than pileal surface, obtuse. Pore surface cream to pinkish buff when fresh, becoming buff to curry-yellow when dry; sterile margin distinct, white to cream when fresh, becoming olivaceous buff to clay-buff when dry, up to 10 mm wide; pores round to angular, 2–4 per mm; dissepiments slightly thick to thick, entire. Context cream to straw-yellow, woody hard, up to 1.5 cm thick. Tubes concolorous with pore surface, woody hard, up to 1 cm long.
Hyphal structure. —Hyphal system dimitic; generative hyphae bearing clamp connections; skeletal hyphae IKI–, CB–; tissues unchanged in KOH.
Context. —Generative hyphae infrequent, hyaline, thin-walled, rarely branched, 2.5–5 μm in diam; skeletal hyphae dominant, yellowish brown to cinnamon brown, thick-walled with a narrow lumen to subsolid, rarely branched, straight, interwoven, 2.3–8.2 μm in diam.
Tubes. —Generative hyphae infrequent, hyaline, thin-walled, rarely branched, 1.9–3.2 μm in diam; skeletal hyphae dominant, hyaline, thick-walled with a wide lumen, occasionally branched, more or less straight, interwoven, 2.2–7.2 μm in diam. Cystidia absent, but fusoid cystidioles occasionally present, hyaline, thin-walled, 17.5–50.2 × 4.3–9.5 μm. Basidia clavate, bearing four sterigmata and a basal clamp connection, 20.8–40.5 × 5.5–11.5 μm; basidioles dominant, in shape similar to basidia, but smaller.
Spores. —Basidiospores oblong-ellipsoid to ellipsoid, hyaline, thin-walled, smooth, IKI–, CB–, 7–9(–9.2) × (3.2–)4–5 μm, L = 7.85 μm, W = 4.26 μm, Q = 1.83–1.89 (n = 60/2).
Type of rot. —Brown rot.
Additional specimen (paratype) examined: CHINA. Yunnan Province, Shangri-La County, Pudacuo National Park, on stump of Abies, 24 September 2011, Cui 10521 (BJFC 011416).
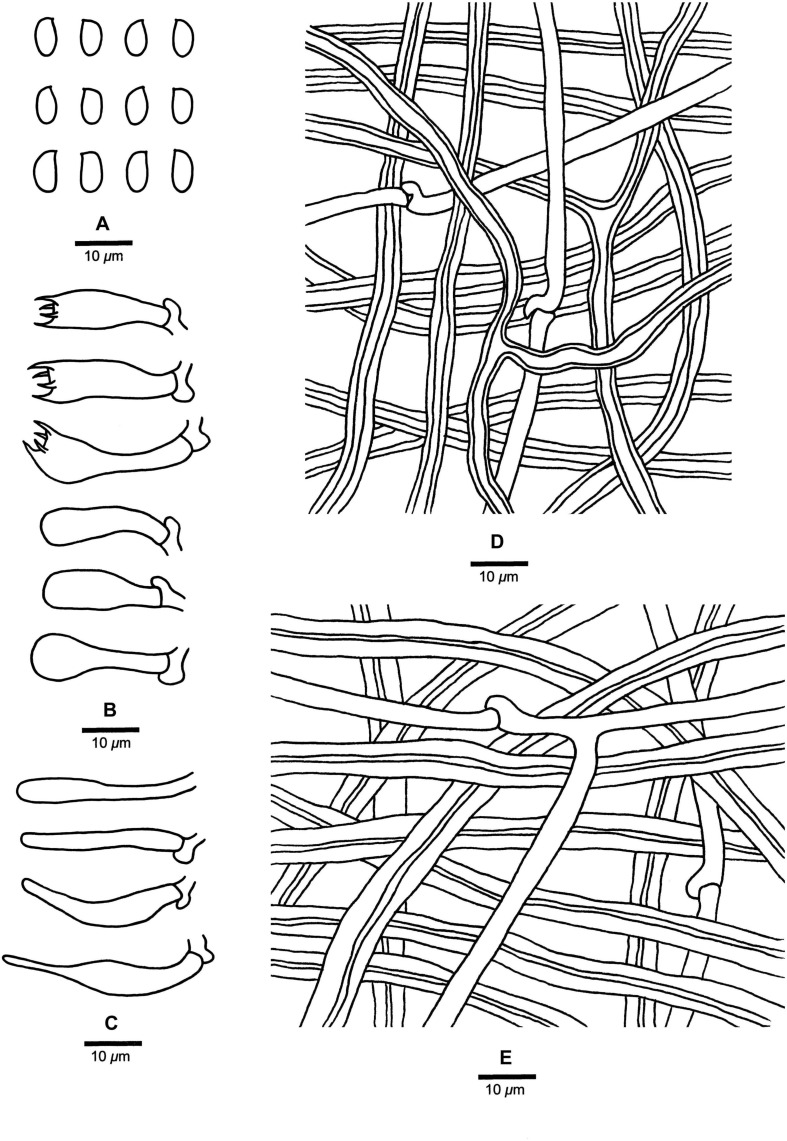
Fomitopsis hengduanensis B.K. Cui & Shun Liu, sp. nov. (Figures 3C,D, 5).
FIGURE 5.
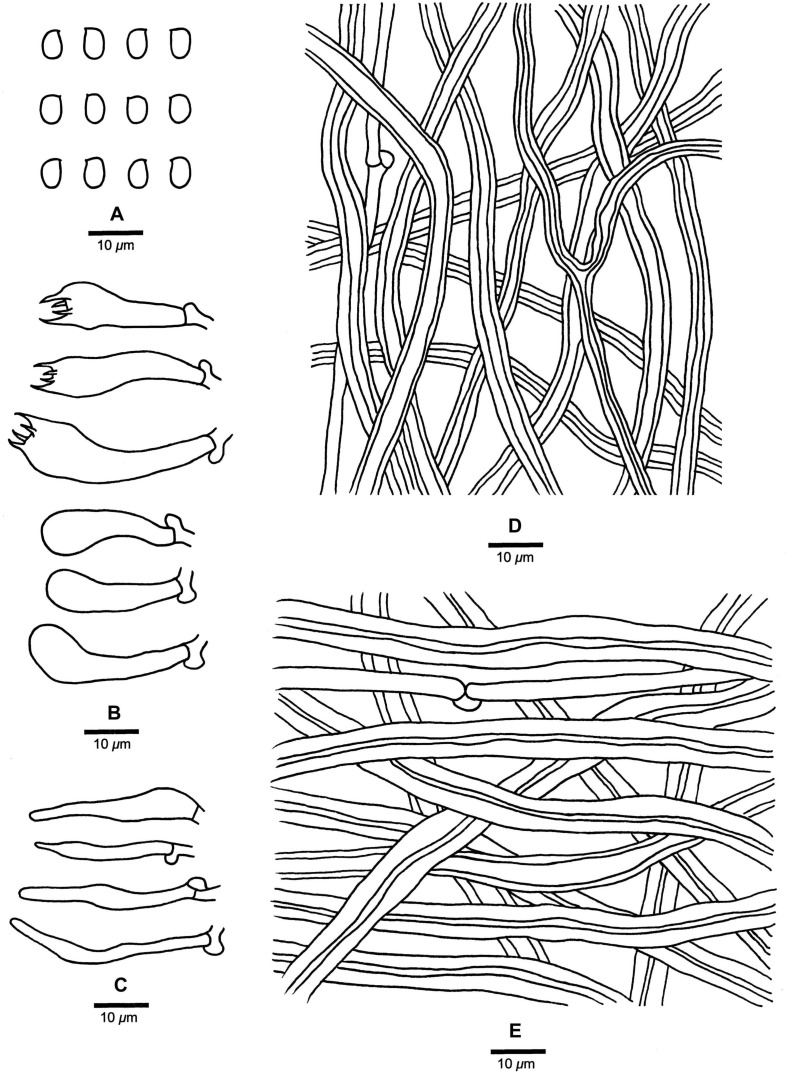
Microscopic structures of Fomitopsis hengduanensis (drawn from the holotype). (A) Basidiospores; (B) Basidia and basidioles; (C) Cystidioles; (D) Hyphae from trama; (E) Hyphae from context. Bars: A–E = 10 μm.
MycoBank: MB 838909
Fomitopsis hengduanensis is characterized by laccate pileus with pale dark gray to reddish brown surface at base and cream to flesh-pink toward the margin when fresh, oblong-ellipsoid to ellipsoid basidiospores (5.2–6 × 3.2–3.6 μm) and is distributed in high altitude areas of the Hengduan Mountains.
Type. —CHINA, Yunnan Province, Lanping County, Tongdian, Laojunshan of Hengduan Mountains, Luoguqing, on dead tree of Picea, 18 September 2017, Cui 16259 (Holotype, BJFC 029558).
Etymology. —Hengduanensis (Lat.), refers to the species distributed in the area of Hengduan Mountains.
Basidiocarps. —Annual to perennial, pileate, sessile, solitary, hard corky, without odor or taste when fresh, woody hard and light in weight upon drying. Pilei applanate, semicircular to ungulate, projecting up to 7.5 cm long, 9 cm wide, 3 cm thick at base. Pileal surface laccate, colors varied but usually pale dark gray to reddish brown at base and cream to flesh-pink toward the margin when fresh, curry-yellow, mouse-gray to reddish brown at base and buff to clay-buff toward the margin when dry, glabrous, sulcate, concentrically zonate; margin acute to obtuse. Pore surface white to cream when fresh, becoming buff to straw-yellow when dry; sterile margin distinct, cream, up to 4 mm wide; pores round to angular, 6–8 per mm; dissepiments thick, entire. Context cream to straw-yellow, woody hard, up to 1.4 cm thick. Tubes concolorous with pore surface, woody hard, up to 0.5 cm long.
Hyphal structure. —Hyphal system dimitic; generative hyphae bearing clamp connections; skeletal hyphae IKI–, CB–; tissues unchanged in KOH.
Context. —Generative hyphae infrequent, hyaline, thin- to slightly thick-walled, rarely branched, 1.9–4.3 μm in diam; skeletal hyphae dominant, hyaline to pale yellowish, thick-walled with a narrow lumen to subsolid, rarely branched, straight, interwoven, 2–8.5 μm in diam.
Tubes. —Generative hyphae infrequent, hyaline, thin-walled, rarely branched, 1.3–3.5 μm in diam; skeletal hyphae dominant, hyaline, thick-walled with a wide lumen, occasionally branched, more or less straight, interwoven, 1.7–7.5 μm in diam. Cystidia absent, but fusoid cystidioles occasionally present, hyaline, thin-walled, 13.2–36.5 × 2.5–5.4 μm. Basidia clavate, bearing four sterigmata and a basal clamp connection, 16.6–34.5 × 5.4–10.2 μm; basidioles dominant, in shape similar to basidia, but smaller.
Spores. —Basidiospores oblong-ellipsoid to ellipsoid, hyaline, thin-walled, smooth, IKI–, CB–, (5–)5.2–6(–6.2) × (3–)3.2–3.6(–4) μm, L = 5.44 μm, W = 3.41 μm, Q = 1.57–1.63 (n = 60/2).
Type of rot. —Brown rot.
Additional specimen (paratype) examined: CHINA. Yunnan Province, Lijiang, Yulong xueshan of Hengduan Mountains, on fallen trunk of Picea, 16 September 2018, Cui 17056 (BJFC 030355).
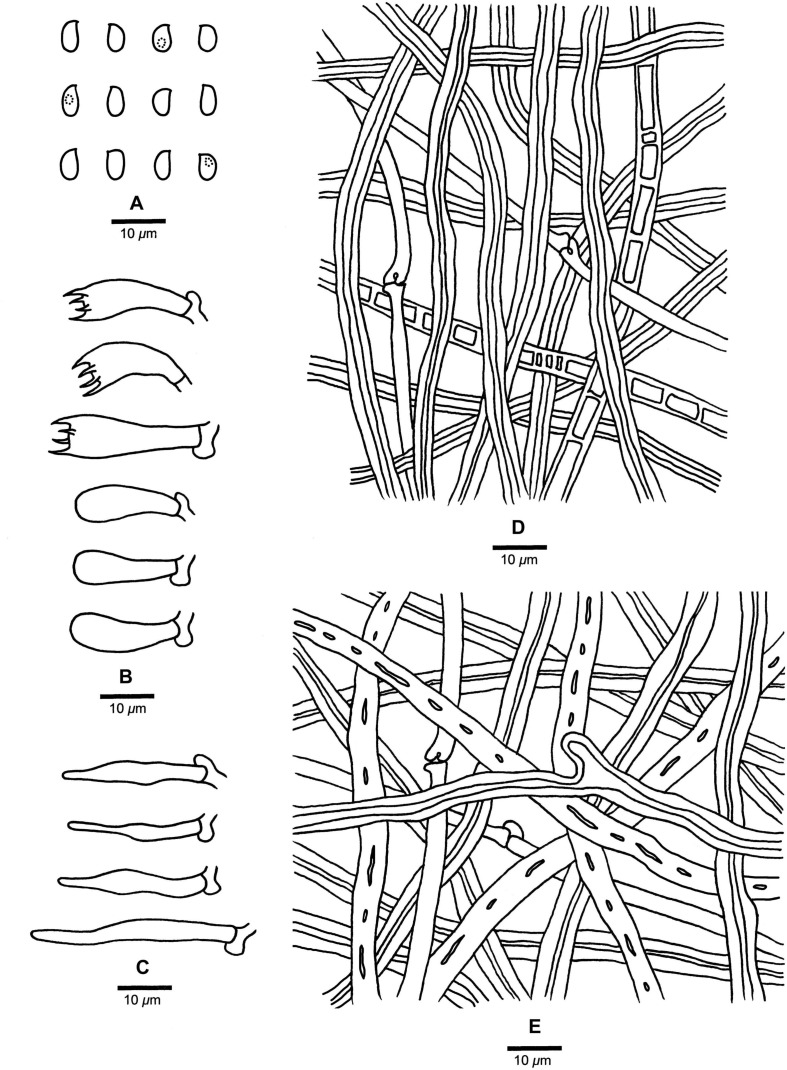
Fomitopsis kesiyae B.K. Cui & Shun Liu, sp. nov. (Figures 3E,F, 6).
FIGURE 6.
Microscopic structures of Fomitopsis kesiyae (drawn from the holotype). (A) Basidiospores; (B) Basidia and basidioles; (C) Cystidioles; (D) Hyphae from trama; (E) Hyphae from context. Bars: A–E = 10 μm.
MycoBank: MB 838910
Fomitopsis kesiyae is characterized by its buff yellow to orange-yellow buff pileal surface when fresh, reddish brown to yellowish brown when dry, and grows on Pinus kesiya and is distributed in tropical areas of Vietnam.
Type. — VIETNAM. Dam Dong Province, Da Lat, Bidoup Nui Ba National Park, on living tree of Pinus kesiya, 15 October 2017, Cui 16437 (Holotype, BJFC 029736).
Etymology. — Kesiyae (Lat.), refers to the host tree species Pinus kesiya.
Basidiocarps. — Annual, pileate, sessile, hard corky, without odor or taste when fresh, woody hard and light in weight upon drying. Pilei applanate, semicircular to sectorial, projecting up to 4.7 cm long, 6.5 cm wide, 4 cm thick at base. Pileal surface laccate, buff yellow to orange-yellow buff when fresh, becoming reddish brown to yellowish brown when dry, glabrous, sulcate, azonate; margin cream, distinctly paler than the pileal surface, obtuse. Pore surface white to cream when fresh, olivaceous buff to cinnamon-buff when dry; sterile margin distinct, buff to honey-yellow, up to 3 mm wide; pores round to angular, 6–8 per mm, dissepiments thick, entire. Context cream to straw-yellow, corky, up to 1.2 cm thick. Tubes concolorous with pore surface, hard corky, up to 1 cm long.
Hyphal structure. — Hyphal system dimitic; generative hyphae bearing clamp connections; skeletal hyphae IKI–, CB–; tissues unchanged in KOH. Small polyhedric or irregular crystals present among context and tubes.
Context. — Generative hyphae infrequent, hyaline, thin- to slightly thick-walled, rarely branched, 1.9–4.2 μm in diam; skeletal hyphae dominant, yellowish brown to cinnamon brown, thick-walled with a wide to narrow lumen, occasionally branched, straight to flexuous, interwoven, 2.2–9.2 μm in diam.
Tubes. — Generative hyphae infrequent, hyaline, thin-walled, occasionally branched, 1.9–3 μm in diam; skeletal hyphae dominant, hyaline to pale yellowish, thick-walled with a wide to narrow lumen, rarely branched, more or less straight, interwoven, 1.7–6.2 μm in diam. Cystidia absent, but fusoid cystidioles occasionally present, hyaline, thin-walled, 11.5–30.4 × 2.6–6 μm. Basidia clavate, bearing four sterigmata and a basal clamp connection, 16–20.3 × 4.8–7.2 μm; basidioles dominant, in shape similar to basidia, but smaller.
Spores. — Basidiospores oblong-ellipsoid to ellipsoid, hyaline, thin-walled, smooth, IKI–, CB–, (4.5–)4.8–5.3(–6) × (2.8–)3–3.5(–4) μm, L = 5.04 μm, W = 3.11 μm, Q = 1.60–1.65 (n = 60/2).
Type of rot. — Brown rot.
Additional specimen (paratype) examined: VIETNAM. Dam Dong Province, Da Lat, Bidoup Nui Ba National Park, on fallen trunk of Pinus kesiya, 16 October 2017, Cui 16466 (BJFC 029765).
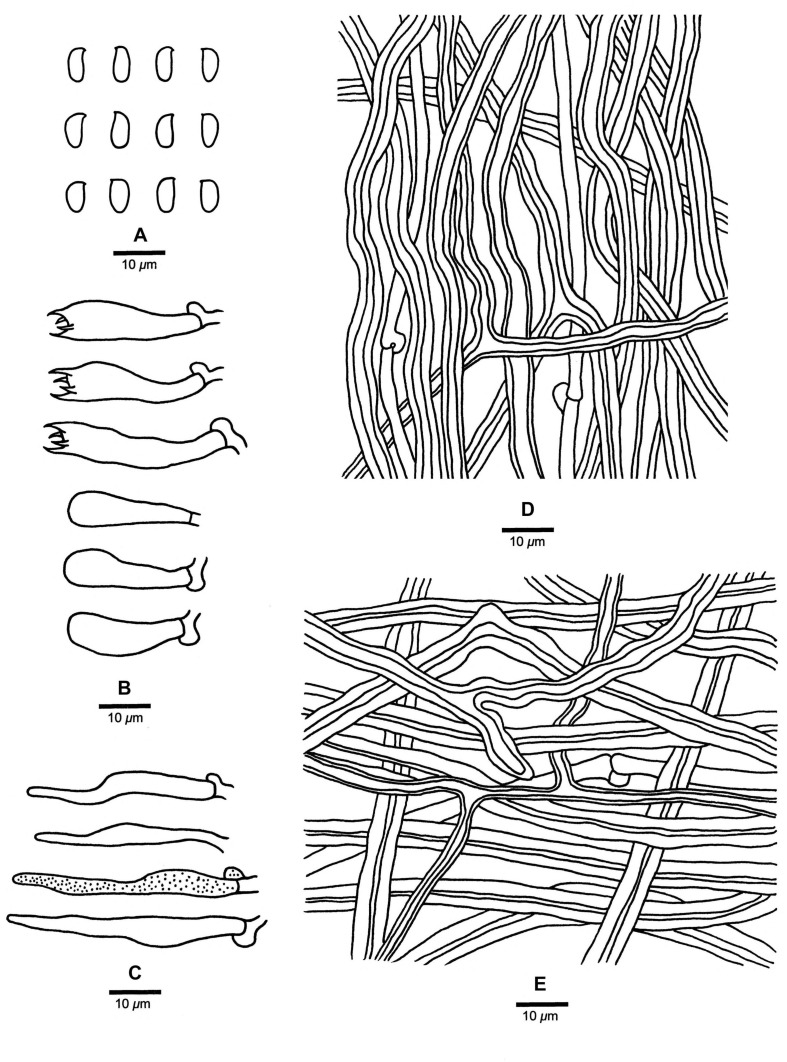
Fomitopsis massoniana B.K. Cui, M.L. Han & Shun Liu, sp. nov. (Figures 3G,H, 7).
FIGURE 7.
Microscopic structures of Fomitopsis massoniana (drawn from the holotype). (A) Basidiospores; (B) Basidia and basidioles; (C) Cystidioles; (D) Hyphae from trama; (E) Hyphae from context. Bars: A–E = 10 μm.
MycoBank: MB 838911
Fomitopsis massoniana is characterized by its effused-reflexed to pileate basidiocarps, applanate to triquetrous or irregular pilei with buff-yellow to apricot-orange pileal surface when fresh, buff to grayish brown when dry, a white to cream pore surface when fresh, cream to buff when dry, and grows on Pinus massoniana.
Type. — CHINA. Fujian Province, Wuping County, Liangyeshan Nature Reserve, on dead tree of Pinus massoniana, 25 October 2013, Cui 11304 (Holotype, BJFC 015420).
Etymology. — Massoniana (Lat.), refers to the host tree species Pinus massoniana.
Basidiocarps. — Annual, effused-reflexed to pileate, sessile, hard corky, without odor or taste when fresh, woody hard and light in weight upon drying. Pilei applanate to triquetrous or irregular, projecting up to 4 cm long, 4.2 cm wide, 1.5 cm thick at base. Pileal surface laccate, buff-yellow to apricot-orange when fresh, becoming buff to grayish brown when dry, glabrous, sulcate, azonate; margin white to cream, obtuse. Pore surface white to cream when fresh, turning cream to buff when dry; sterile margin distinct, cream, up to 4 mm wide; pores round, 5–7 per mm, dissepiments thick, entire. Context cream to straw-yellow, woody hard, up to 0.8 cm thick. Tubes concolorous with pore surface, woody hard, up to 0.4 cm long.
Hyphal structure. — Hyphal system dimitic; generative hyphae bearing clamp connections; skeletal hyphae IKI–, CB–; tissues unchanged in KOH.
Context. — Generative hyphae infrequent, hyaline, thin- to slightly thick-walled, occasionally branched, 2–4.5 μm in diam; skeletal hyphae dominant, hyaline, thick-walled with a narrow lumen to subsolid, rarely branched, straight, interwoven, 2.2–8.2 μm in diam.
Tubes. — Generative hyphae infrequent, hyaline, thin-walled, rarely branched, 1.8–4 μm in diam; skeletal hyphae dominant, hyaline, thick-walled with a narrow lumen to narrow lumen, occasionally branched, more or less straight, interwoven, 2–7.2 μm in diam. Cystidia absent, but fusoid cystidioles occasionally present, hyaline, thin-walled, 14.8–36 × 3.8–6 μm. Basidia clavate, bearing four sterigmata and a basal clamp connection, 17–26.5 × 5.5–7.9 μm; basidioles dominant, in shape similar to basidia, but smaller.
Spores. — Basidiospores oblong-ellipsoid, hyaline, thin-walled, smooth, IKI–, CB–, (5.8–)6.2–7.3(–7.6) × (3–)3.3–4 μm, L = 6.91 μm, W = 3.53 μm, Q = 1.93–1.99 (n = 90/3).
Type of rot. — Brown rot.
Additional specimens (paratypes) examined: CHINA. Fujian Province, Wuping County, Liangyeshan Nature Reserve, on dead tree of Pinus massoniana, 25 October 2013, Cui 11288 (BJFC 015404); Wuyishan County, Longchuan Valley, on dead tree of Pinus massoniana, 16 October 2005, Cui 2848 (BJFC 000719); Guangdong Province, Fengkai County, Heishiding Nature Reserve, on fallen trunk of Pinus massoniana, 2 July 2010, Cui 9058 (BJFC 007996).
Fomitopsis subpinicola B.K. Cui, M.L. Han & Shun Liu, sp. nov. (Figures 3I,J, 8).
FIGURE 8.
Microscopic structures of Fomitopsis subpinicola (drawn from the holotype). (A) Basidiospores; (B) Basidia and basidioles; (C) Cystidioles; (D) Hyphae from trama; (E) Hyphae from context. Bars: A–E = 10 μm.
MycoBank: MB 838912
Fomitopsis subpinicola is characterized by its apricot-orange, scarlet to fuscous pileal surface when fresh, reddish brown to dark brown when dry, occasionally septated skeletal hyphae and is distributed in Northeast China.
Type. — CHINA. Heilongjiang Province, Yichun, Fenglin Nature Reserve, on fallen trunk of Pinus koraiensis, 2 August 2011, Cui 9836 (Holotype, BJFC 010729).
Etymology. — Subpinicola (Lat.), refers to the new species resembling F. pinicola in morphology.
Basidiocarps. — Annual, pileate, sessile, hard corky, without odor or taste when fresh, woody hard and light in weight upon drying. Pilei applanate, circular to sectorial, projecting up to 7.5 cm long, 8.5 cm wide, 4.5 cm thick at base. Pileal surface laccate, apricot-orange, scarlet to fuscous when fresh, becoming reddish brown to dark brown upon drying, glabrous, sulcate, azonate; margin white to cream, distinctly paler than the pileal surface, obtuse. Pore surface white to cream when fresh, turning buff yellow to buff when dry; sterile margin distinct, white to cream, up to 6 mm wide; pores round, 6–8 per mm, dissepiments thick, entire. Context cream to straw-yellow, woody hard, up to 1.2 cm thick. Tubes concolorous with pore surface, woody hard, up to 0.5 cm long.
Hyphal structure. — Hyphal system dimitic; generative hyphae bearing clamp connections; skeletal hyphae IKI–, CB–; tissues unchanged in KOH.
Context. — Generative hyphae infrequent, hyaline, thin- to slightly thick-walled, rarely branched, 2–3.2 μm in diam; skeletal hyphae dominant, yellowish brown to cinnamon brown, thick-walled with a narrow lumen to subsolid, occasionally branched, straight, interwoven, 2.2–6.8 μm in diam.
Tubes. — Generative hyphae infrequent, hyaline, thin-walled, rarely branched, 1.8–3 μm in diam; skeletal hyphae dominant, yellowish brown to cinnamon brown, thick-walled with a wide to narrow lumen, occasionally septate, without clamps, rarely branched, straight, interwoven, 1.9–6.2 μm in diam. Cystidia absent, but fusoid cystidioles occasionally present, hyaline, thin-walled, 14.5–34.6 × 3.2–7.2 μm. Basidia clavate, bearing four sterigmata and a basal clamp connection, 16–24.5 × 4.5–9 μm; basidioles dominant, in shape similar to basidia, but smaller.
Spores. — Basidiospores oblong-ellipsoid to ellipsoid, hyaline, thin-walled, smooth, IKI–, CB–, (4–)4.3–5.5(–5.9) × (2.5–)2.7–3.3(–3.5) μm, L = 4.94 μm, W = 2.97 μm, Q = 1.65–1.69 (n = 90/3).
Type of rot. — Brown rot.
Additional specimens (paratypes) examined: CHINA. Heilongjiang Province, Yichun, Fenglin Nature Reserve, on fallen trunk of Pinus koraiensis, 1 August 2011, Cui 9819 (BJFC 010712); Tangyuan County, Daliangzihe Forest Park, on living tree of Pinus koraiensis, 26 August 2008, Yuan 4912 (BJFC 015654); Inner Mongolia, Genhe, Greater Khingan Mountains Nature Reserve, on Larix, 28 August 2009, Dai 11101 (BJFC 015660), Dai 11206 (BJFC 015661); Jilin Province, Antu County, Changbaishan Nature Reserve, on fallen trunk of Betula, 7 September 2013, Dai 13480 (BJFC 014941).
Fomitopsis tianshanensis B.K. Cui & Shun Liu, sp. nov. (Figures 3K,L, 9).
FIGURE 9.
Microscopic structures of Fomitopsis tianshanensis (drawn from the holotype). (A) Basidiospores; (B) Basidia and basidioles; (C) Cystidioles; (D) Hyphae from trama; (E) Hyphae from context. Bars: A–E = 10 μm.
MycoBank: MB 838913
Fomitopsis tianshanensis is characterized by its effused-reflexed to pileate basidiocarps with soft corky texture when fresh, large pores (1–3 per mm) and long tubes (up to 2.5 cm), grows on Picea and is distributed in Tianshan Mountains Xinjiang, China.
Type. — CHINA. Xinjiang Autonomous Region, Fukang County, Tianshan Tianchi Nature Reserve, on fallen trunk of Picea schrenkiana, 4 July 2018, Cui 16821 (Holotype, BJFC 030120).
Etymology. — Tianshanensis (Lat.), refers to the species located at the Tianshan regions.
Basidiocarps. — Annual to perennial, effused-reflexed to pileate, sessile, soft corky, without odor or taste when fresh, hard corky and light in weight upon drying. Pilei applanate, semicircular to ungulate, projecting up to 11 cm long, 20 cm wide, 7 cm thick at base. Pileal surface dark bluish gray to yellowish brown when fresh, becoming fawn to deep olive when dry, slightly velutinate, small nodules appear near the base, rough, azonate; margin cream to cinnamon, obtuse to acute. Pore surface cream to pinkish buff when fresh, becoming faint yellow to light pink when dry; sterile margin distinct, cream to buff, up to 3 mm wide; pores mostly round, occasionally angular, 1–3 per mm, dissepiments thick, entire. Context cream to buff, corky, up to 3.5 cm thick. Tubes concolorous with pore surface, hard corky, up to 2.5 cm long.
Hyphal structure. — Hyphal system dimitic; generative hyphae bearing clamp connections; skeletal hyphae IKI–, CB–; tissues unchanged in KOH.
Context. — Generative hyphae infrequent, hyaline, thin- to slightly thick-walled, rarely branched, 2–4 μm in diam; skeletal hyphae dominant, yellowish brown to cinnamon brown, thick-walled with a narrow lumen to subsolid, occasionally branched, straight to flexuous, interwoven, 2.2–7.2 μm in diam.
Tubes. — Generative hyphae infrequent, hyaline, thin-walled, rarely branched, 1.9–3.2 μm in diam; skeletal hyphae dominant, hyaline to pale yellowish, thick-walled with a wide lumen, occasionally branched, straight to flexuous, 2–6.9 μm in diam, interwoven. Cystidia absent, sometimes skeletal hyphae penetrated into the hymenium, but not forming typical catahymenium; cystidioles present, fusoid, hyaline, thin-walled, 15.5–44 × 3.3–6.5 μm. Basidia clavate, bearing four sterigmata and a basal clamp connection, 17–32.5 × 4.2–9.5 μm; basidioles dominant, in shape similar to basidia, but smaller.
Spores. — Basidiospores oblong-ellipsoid, sometimes tapering at apiculus, hyaline, thin-walled, smooth, IKI–, CB–, (6–)6.3–7(–7.2) × (3–)3.2–3.8(–4) μm, L = 6.62 μm, W = 3.52 μm, Q = 1.85–1.93 (n = 90/3).
Type of rot. — Brown rot.
Additional specimens (paratypes) examined: CHINA. Xinjiang Autonomous Region, Urumqi, Nanshan Park, on fallen trunk of Picea schrenkiana, 5 July 2018, Cui 16823 (BJFC 030122), Cui 16825 (BJFC 030124), Cui 16828 (BJFC 030127); Shawan County, Lujiaowan Park, on stump of Picea schrenkiana, 6 July 2018, Cui 16830 (BJFC 030129).
Other Specimens of the Fomitopsis pinicola Complex Examined
Fomitopsis mounceae. CANADA. 3 August 2000, Teuvo Ahti 60351 (H); on Picea glauca, 25 July 1982, Tuomo Niemelä 2530 (H). UNITED STATES. On Betula, 15 September 2014, OM 18782 (H); on Tsuga heterophylla, 11 September 2014, Spirin 8367 (H).
Fomitopsis ochracea. UNITED STATES. OM 18568 (H), OM 18673 (H); on Picea, Spirin 8165 (H).
Fomitopsis schrenkii. UNITED STATES. 7 September 1992, Inkeri Ahonen 58 (BJFC 013921); Turkey Creek, Chricahua Mountain, Arizona, on Douglas fir, September 2012, Josef Vlasák 1209/61-J (BJFC 015604).
Fomitopsis pinicola. FINLAND. Helsinki, Vantaa, Tamisto Nature Reserve, on fallen trunk of Picea, 16 August 2012, Dai 12869 (BJFC 013149), Dai 12870 (BJFC 013150). ITALY. Roma, Trentino Altoadie, Trento, Molveno, on Picea, 28 April 2005, Dai 6553 (IFP). POLAND. On dead tree of Pinus, 3 October 2014, Dai 14841 (BJFC 017955); Lesser Poland Voivodeship, Gorce National Park, on Picea abies, 9 July 1985, Pekka Nuorteva (H).
Discussion
Based on the phylogenetic analyses, 10 species of the Fomitopsis pinicola complex grouped together (Figures 1, 2), including six new species from East Asia: F. abieticola, F. hengduanensis, F. kesiyae, F. massoniana, F. subpinicola, and F. tianshanensis. The main morphological characters of species in the F. pinicola complex are provided in Table 2.
TABLE 2.
A comparison of species in the Fomitopsis pinicola complex.
| Species | Distribution | Basidio- | Pileal surface | Host | Pores (per mm) | Cystidia (μm) | Basidiospores | References | |
| carps | when fresh | L × W (μm) | Q = L/W | ||||||
| F. abieticola | Southwestern China | Annual to perennial; pileate | Cream to pinkish buff | Abies | 2–4 | 17.5–50.2 × 4.3–9.5 | 7–9 × 4–5 | 1.83–1.89 | This study |
| F. hengduanensis | High altitude areas of the Hengduan Mountains of southwestern China | Annual to perennial; pileate | Pale dark gray to reddish brown at base and cream to flesh-pink toward the margin | Picea, Pinus | 6–8 | 13.2–36.5 × 2.5–5.4 | 5.2–6 × 3.2–3.6 | 1.57–1.63 | This study |
| F. kesiyae | Tropical areas of Vietnam | Annual; pileate | Buff yellow to orange-yellow buff | Pinus kesiya | 6–8 | 11.5–30.4 × 2.6–6 | 4.8–5.3 × 3–3.5 | 1.60–1.65 | This study |
| F. massoniana | Southeastern China | Annual; effused-reflexed to pileate | Buff-yellow to apricot-orange | Pinus massoniana | 5–7 | 14.8–36 × 3.8–6 | 6.2–7.3 × 3.3–4 | 1.85–1.9 | This study |
| F. mounceae | Canada, northern United States | Perennial; pileate | Brownish orange to black at base and pale orange to grayish orange toward the margin | Abies, Betula, Larix, Picea, Populus, Pseudotsuga | 3–5 | 16–35 × 3–6.5 | 5.8–6.6 × 3.4–4 | 1.7–1.9 | Haight et al., 2019 |
| F. ochracea | Canada, northern United States | Perennial; pileate | Brownish gray to grayish brown at base and orange White to pale orange toward the margin | Abies, Picea, Populus | 4–5 | 20–40 × 4–6.5 | 5.1–5.9 × 3.6–4 | 1.3–1.4 | Haight et al., 2019 |
| F. pinicola | Europe | Perennial; pileate | Brownish orange to black at base and buff-yellow to cinnamon toward the margin | Mostly on Picea and Pinus, occasionally on other different gymnosperm or angiosperm wood | 4–6 | 18–60 × 3–9 | 6–9 × 3–4.5 | 1.8–2.2 | Ryvarden and Melo, 2014; Haight et al., 2016, 2019 |
| F. schrenkii | Western and southwestern United States | Perennial; effused-reflexed to pileate | Grayish orange to olive brown at base and grayish orange to grayish yellow toward the margin | Abies, Picea, Pinus, Pseudotsuga | 3–4 | 16–30 × 3–8 | 5.7–6.7 × 3.7–4.2 | 1.5–1.6 | Haight et al., 2019 |
| F. subpinicola | Northeastern China | Annual; pileate | Apricot-orange, scarlet to fuscous | Mostly on Pinus koraiensis, occasionally on Larix, Betula | 6–8 | 14.5–34.6 × 3.2–7.2 | 4.3–5.5 × 2.7–3.3 | 1.65–1.69 | This study |
| F. tianshanensis | Tianshan Mountains of northwestern China | Annual to perennial, effused-reflexed to pileate | Dark bluish gray to yellowish brown | Picea | 1–3 | 15.5–44 × 3.3–6.5 | 6.3–7 × 3.2–3.8 | 1.9–1.96 | This study |
In the phylogenetic trees (Figures 1, 2), Fomitopsis abieticola is closely related to F. tianshanensis. Morphologically, both F. abieticola and F. tianshanensis have an annual to perennial growth habit, cream to pinkish pore surface when fresh, and large pores, but F. tianshanensis differs in its soft corky basidiocarps when fresh, and usually grows on Picea. Fomitopsis schrenkii has similar pores (3–4 per mm), but it has smaller basidia (12–22 × 6–8 μm), and slightly wider basidiospores (5.7–6.7 × 3.7–4.2 μm). Fomitopsis hengduanensis was also discovered in the Yunnan Province, but F. hengduanensis differs with smaller pores (6–8 per mm) and smaller basidiospores (5.2–6 × 3.2–3.6 μm).
Phylogenetically, Fomitopsis hengduanensis grouped together with F. kesiyae (Figures 1, 2). Morphologically, F. hengduanensis and F. kesiyae share a white to cream pore surface when fresh and have similar pores, but F. kesiyae differs in having a buff yellow to orange-yellow buff pileal surface when fresh, reddish brown to yellowish brown when dry, and grows on the Pinus kesiya tree. Fomitopsis pinicola, F. schrenkii and F. mounceae all have similar pilei, but they have larger pores (4–6 per mm in F. pinicola, 3–4 per mm in F. schrenkii, 3–5 per mm in F. mounceae; Table 2). Fomitopsis subpinicola has similar sized pores, but it has smaller basidia (16.1–24.5 × 4.5–9 μm) and basidiospores (4.3–5.5 × 2.7–3.3 μm) and is distributed in south northeast China.
Fomitopsis kesiyae was described from Vietnam on the tree of Pinus kesiya. Phylogenetically, two sampled specimens of F. kesiyae formed a high supported lineage (99% ML, 99% MP, 1.00 BPP) and are closely related to F. hengduanensis (Figures 1, 2). However, F. hengduanensis differs in having larger basidia (16.6–34.5 × 5.4–10.2 μm) and basidiospores (5.2–6 × 3.2–3.6 μm). Fomitopsis subpinicola has similar sized pores (6–8 per mm) and basidiospores (4.3–5.5 × 2.7–3.3 μm), but it has an apricot-orange, scarlet to fuscous pileal surface when fresh, and is reddish brown to dark brown when dry. Fomitopsis massoniana has a similar colored pileal surface when fresh and similar sized pores (5–7 per mm), but it has larger basidiospores (6.2–7.3 × 3.3–4 μm) and grows on Pinus massoniana rather than Pinus kesiya.
Morphologically, Fomitopsis massoniana is similar to F. kesiyae; both species have an annual growth habit and a similar colored pileal surface when fresh. However, F. kesiyae differs in having smaller basidiospores (4.8–5.3 × 3–3.5 μm), is distributed in Vietnam, and grows on Pinus kesiya. Fomitopsis hengduanensis has similar sized pores (6–8 per mm), but compared to F. massoniana, F. hengduanensis has larger sized basidiocarps, a laccate pileal surface with pale dark gray to reddish brown at base and cream to flesh-pink toward the margin when fresh, and smaller sized basidiospores (5.2–6 × 3.2–3.6 μm). Phylogenetically, these two species are distinct from each other (Figures 1, 2). Fomitopsis massoniana is closely related to F. subpinicola, they share a cream to buff pore surface and have similar sized pores, but F. subpinicola has smaller basidiospores (4.3–5.5 × 2.7–3.3 μm).
Fomitopsis subpinicola can be easily separated from F. pinicola by its apricot-orange, scarlet to fuscous pileal surface when fresh, reddish brown to dark brown when dry, smaller basidiospores (4.3–5.5 × 2.7–3.3 μm) and is located in the Northeast of China. Phylogenetically, F. subpinicola is closely related to F. massoniana. Morphologically, F. subpinicola is similar to F. massonian, which has an annual growth habit and white to cream pore surface when fresh. But F. massoniana differs by its effused-reflexed to pileate basidiocarps, lager basidiospores (6.2–7.3 × 3.3–4 μm) and grows on the Pinus massoniana tree. Fomitopsis hengduanensis and F. kesiyae have similar sized pores (6–8 per mm), but F. hengduanensis has larger sized basidia (16.6–34.5 × 5.4–10.2 μm), F. kesiyae has a buff yellow to orange-yellow buff pileal surface when fresh, reddish brown to yellowish brown when dry, and grows on the Pinus kesiya tree.
Phylogenetically, specimens of Fomitopsis tianshanensis formed a well-supported lineage (Figures 1, 2) and are grouped with F. abieticola. But F. abieticola differs from F. tianshanensis in having large sized cystidioles (17.5–50.2 × 4.3–9.5 μm), basidia (20.8–40.5 × 5.5–11.5 μm), and basidiospores (7–9 × 4–5 μm). Fomitopsis pinicola also grows mainly on Picea, but it has smaller pores (4–6 per mm), a brownish orange to black pileal surface at base and buff-yellow to cinnamon toward the margin when fresh, and is distributed in Europe.
Previously, Fomitopsis pinicola was used as a broad species concept, specimens from Europe, North America, and East Asia were all identified as F. pinicola based on morphological characters (Gilbertson and Ryvarden, 1986; Ryvarden and Gilbertson, 1993; Núñez and Ryvarden, 2001; Dai, 2012; Ryvarden and Melo, 2014). Recent phylogenetic analyses indicated that F. pinicola is a species complex and represent several different species. Haight et al. (2016) proposed four well-supported clades: one lineage F. pinicola sensu stricto from Europe and three lineages from North America. Subsequently, Haight et al. (2019) identified these three lineages as F. mounceae, F. ochracea, and F. schrenkii from North America, and F. pinicola is restricted to Eurasia and does not occur in North America. In the current study, samples previously identified as F. pinicola from China and Vietnam in East Asia represent six distinct species. Furthermore, our results indicated that species of the F. pinicola complex usually have limited distribution areas and host specialization. In East Asia, F. abieticola is distributed in southwestern China and grows on Abies; F. hengduanensis is distributed in high altitude areas of the Hengduan Mountains of southwestern China, and it mostly grows on Picea, occasionally on other gymnosperm wood; F. kesiyae is distributed in tropical areas of Vietnam and grows only on Pinus kesiya; F. massoniana is distributed in southeastern China and grows only on Pinus massoniana; F. subpinicola was found in northeastern China and grows mainly on Pinus koraiensis, occasionally on other gymnosperm or angiosperm wood; F. tianshanensis is distributed in Tianshan Mountains of northwestern China and only grows on Picea schrenkiana. In Europe, F. pinicola is widespread, and it mostly grows on Picea and Pinus, occasionally on other different gymnosperm or angiosperm wood (Ryvarden and Melo, 2014). In North America, F. mounceae and F. ochracea is distributed in Canada and the northern United States, and they grow on different gymnosperm or angiosperm wood (Ryvarden and Stokland, 2008; Haight et al., 2019); F. schrenkii is distributed in western and southwestern regions of the United States and mostly grows on different gymnosperm wood, rarely on angiosperm wood (Haight et al., 2019). Among the wood-rotting fungi, some other polypore genera also have limited distribution areas and host specializations, such as Bondarzewia Singer (Song et al., 2016), Heterobasidion Bref. (Chen et al., 2015; Yuan et al., 2020), Laetiporus Murrill (Song and Cui, 2017), and Sanghuangporus Sheng H. Wu, L.W. Zhou & Y.C. Dai (Zhu et al., 2019).
Key to Accepted Species of Fomitopsis pinicola Complex
| 1. | Distribution in East Asia | 2 |
| 1. | Distribution in North America or Europe | 7 |
| 2. | Pores < 5 per mm | 3 |
| 2. | Pores 5–8 per mm | 4 |
| 3. | Basidiospores oblong-ellipsoid to ellipsoid, 7–9 × 4–5 μm | F. abieticola |
| 3. | Basidiospores oblong-ellipsoid, 6.3–7 × 3.2–3.8 μm | F. tianshanensis |
| 4. | Distribution in tropical areas | F. kesiyae |
| 4. | Distribution in temperate areas | 5 |
| 5. | Basidiocarps annual to perennial; pileal surface cream to flesh-pink toward the margin when fresh | F. hengduanensis |
| 5. | Basidiocarps annual; pileal surface white to cream toward the margin when fresh | 6 |
| 6. | Basidiospores oblong-ellipsoid, 6.2–7.3 × 3.3–4 μm | F. massoniana |
| 6. | Basidiospores oblong-ellipsoid to ellipsoid, 4.3–5.5 × 2.7–3.3 μm | F. subpinicola |
| 7. | Pilei never with reddish brown band | F. ochracea |
| 7. | Pilei with reddish brown band | 8 |
| 8. | Distribution in Europe; basidia < 20 μm long | F. pinicola |
| 8. | Distribution in North America; basidia > 20 μm long | 9 |
| 9. | Basidiospores ellipsoid to cylindrical, Q = 1.6–1.9 | F. mounceae |
| 9. | Basidiospores ellipsoid to broadly cylindrical, Q = 1.5–1.6 | F. schrenkii |
Data Availability Statement
The datasets presented in this study can be found in an online repository. The name of the repository and accession number can be found below: https://treebase.org/treebase-web/search/study/summary.html?id=27994&x-access-code=ab2495717aaf081f0557e6680c381710&agreement=ok, submission ID: 27994.
Author Contributions
B-KC and SL designed the experiment. SL, M-LH, D-MW, and B-KC prepared the samples. SL, T-MX, and YW conducted the molecular experiments and analyzed the data. SL, D-MW, and B-KC drafted the manuscript. All the authors approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We express our gratitude to Drs. Yu-Cheng Dai (China), Josef Vlasák (Czech Republic), and the curators of herbaria of H and IFP for loaning the specimens. Drs. De-Wei Li (United States), Jie Song (China), and Ms. Xing Ji (China) are thanked for companionship during field collections.
Abbreviations
- BI
Bayesian inference
- BJFC
Herbarium of the Institute of Microbiology, Beijing Forestry University
- BGI
Beijing Genomics Institute
- BPP
Bayesian posterior probabilities
- BT
Bootstrap
- CB
Cotton Blue
- CB–
acyanophilous
- GTR + I + G
General time reversible + proportion invariant + gamma
- IKI
Melzer’s reagent
- IKI–
neither amyloid nor dextrinoid
- ILD
incongruence length difference test
- ITS
internal transcribed spacer
- KOH
5% potassium hydroxide
- L
mean spore length (arithmetic average of all spores)
- ML
maximum likelihood
- MP
maximum parsimony
- MPT
most parsimonious tree
- n (a/b)
number of spores (a) measured from given number (b) of specimens
- Q
variation in the L/W ratios between the specimens studied
- RPB2
DNA-directed RNA polymerase II subunit 2
- TL
tree length
- W
mean spore width (arithmetic average of all spores)
- CI
consistency index
- RI
retention index
- RC
rescaled consistency index
- HI
homoplasy index
- TEF
translation elongation factor 1-α.
Funding. This research was supported by the National Natural Science Foundation of China (Nos. U2003211, 31750001), the Scientific and Technological Tackling Plan for the Key Fields of Xinjiang Production and Construction Corps (No. 2021AB004) Beijing Forestry University Outstanding Young Talent Cultivation Project (No. 2019JQ03016), and the Biodiversity Survey and Assessment Project of the Ministry of Ecology and Environment, China (No. 2019HJ2096001006).
References
- Bao H. Y., Sun Q., Huang W., Sun X., Bau T., Li Y. (2015). Immunological regulation of fermentation mycelia of Fomitopsis pinicola on mice. Mycosystema 34 287–292. [Google Scholar]
- Chen J. J., Cui B. K., Zhou L. W., Korhonen K., Dai Y. C. (2015). Phylogeny, divergence time estimation, and biogeography of the genus Heterobasidion (Basidiomycota, Russulales). Fungal Divers. 71 185–200. 10.1007/s13225-014-0317-2 [DOI] [Google Scholar]
- Chen Y. Y., Wu F., Wang M., Cui B. K. (2017). Species diversity and molecular systematics of Fibroporia (Polyporales. Basidiomycota) and its related genera. Mycol. Prog. 16 521–533. 10.1007/s11557-017-1285-1 [DOI] [Google Scholar]
- Cui B. K., Li H. J., Ji X., Zhou J. L., Song J., Si J., et al. (2019). Species diversity, taxonomy and phylogeny of Polyporaceae (Basidiomycota) in China. Fungal Divers. 97 137–392. 10.1007/s13225-019-00427-4 [DOI] [Google Scholar]
- Dai Y. C. (2012). Polypore diversity in China with an annotated checklist of Chinese polypores. Mycoscience 53 49–80. 10.1007/s10267-011-0134-3 [DOI] [Google Scholar]
- Dai Y. C., Yang Z. L., Cui B. K., Yu C. J., Zhou L. W. (2009). Species diversity and utilization of medicinal mushrooms and fungi in China (Review). Int. J. Med. Mushrooms 11 287–302. 10.1615/IntJMedMushr.v11.i3.80 31502759 [DOI] [Google Scholar]
- Farris J. S., Källersjö M., Kluge A. G., Kluge A. G., Bult C. (1994). Testing significance of incongruence. Cladistics 10 315–319. 10.1006/clad.1994.1021 [DOI] [Google Scholar]
- Felsenstein J. (1985). Confidence intervals on phylogenies: an approach using the bootstrap. Evolution 39 783–791. 10.1111/j.1558-5646.1985.tb00420.x [DOI] [PubMed] [Google Scholar]
- Gilbertson R. L., Ryvarden L. (1986). North American polypores, Vol. 1. Oslo: Fungiflora. [Google Scholar]
- Guler P., Akata I., Kutluer F. (2009). Antifungal activities of Fomitopsis pinicola (Sw.: Fr.) Karst. and Lactarius vellereus (Pers.). Fr. Afr. J. Biotechnol. 8 3811–3813. 10.5897/AJB09.719 [DOI] [Google Scholar]
- Guo S. S., Wolf D. R. (2018). Study on neuroprotective effects of water extract of Fomitopsis pinicola on dopaminergic neurons in vitro. Chin. J. Pharma. 15 582–586. [Google Scholar]
- Haight J. E., Laursen G. A., Glaeser J. A., Taylor D. L. (2016). Phylogeny of Fomitopsis pinicola: a species complex. Mycologia 108 925–938. 10.3852/14-225R1 [DOI] [PubMed] [Google Scholar]
- Haight J. E., Nakasone K. K., Laursen G. A., Redhead S. A., Taylor D. L., Glaeser J. A. (2019). Fomitopsis mounceae and F. schrenkii—two new species from North America in the F. pinicola complex. Mycologia 111 339–357. 10.1080/00275514.2018.1564449 [DOI] [PubMed] [Google Scholar]
- Hall T. A. (1999). Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. (Oxford) 41 95–98. 10.1021/bk-1999-0734.ch008 [DOI] [Google Scholar]
- Han M. L., Cui B. K. (2015). Morphological characters and molecular data reveal a new species of Fomitopsis (Polyporales) from southern China. Mycoscience 56 168–176. 10.1016/j.myc.2014.05.004 [DOI] [Google Scholar]
- Han M. L., Chen Y. Y., Shen L. L., Song J., Vlasák J., Dai Y. C., et al. (2016). Taxonomy and phylogeny of the brown-rot fungi: Fomitopsis and its related genera. Fungal Divers. 80 343–373. 10.1007/s13225-016-0364-y [DOI] [Google Scholar]
- Han M. L., Song J., Cui B. K. (2014). Morphology and molecular phylogeny for two new species of Fomitopsis (Basidiomycota) from South China. Mycol. Prog. 13 905–914. 10.1007/s11557-014-0976-0 [DOI] [Google Scholar]
- Hillis D. M., Bull J. J. (1993). An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biodivers. 42 182–192. 10.1093/sysbio/42.2.182 [DOI] [Google Scholar]
- Högberg N., Holdenreider O., Stenlid J. (1999). Population structure of the wood decay fungus Fomitopsis pinicola. Heredity 83 354–360. 10.1038/sj.hdy.6885970 [DOI] [PubMed] [Google Scholar]
- Karsten P. A. (1881). Symbolae ad mycologiam Fennicam. 8. Acta Soc. Fauna Flora Fenn. 6 7–13. [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt S. D., Fankhauser J. D., Leavitt D. H., Porter L. D., Johnson L. A., Clair L. L. S. (2011). Complex patterns of speciation in cosmopolitan “rock posy” lichens—discovering and delimiting cryptic fungal species in the lichen-forming Rhizoplaca melanophthalma species-complex (Lecanoraceae, Ascomycota). Mol. Phylogenet. Evol. 59 587–602. 10.1016/j.ympev.2011.03.020 [DOI] [PubMed] [Google Scholar]
- Li H. J., Han M. L., Cui B. K. (2013). Two new Fomitopsis species from southern China based on morphological and molecular characters. Mycol. Prog. 12 709–718. 10.1007/s11557-012-0882-2 [DOI] [Google Scholar]
- Liu S., Shen L. L., Wang Y., Xu T. M., Gates G., Cui B. K. (2021). Species diversity and molecular phylogeny of Cyanosporus (Polyporales, Basidiomycota). Front. Microbiol. 12:631166. 10.3389/fmicb.2021.631166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Song C. G., Cui B. K. (2019). Morphological characters and molecular data reveal three new species of Fomitopsis (Basidiomycota). Mycol. Prog. 18 1317–1327. 10.1007/s11557-019-01527-w [DOI] [Google Scholar]
- Maddison W. P., Maddison D. R. (2017). Mesquite: a modular system for evolutionary analysis, Version 3.2. http://mesquiteproject.org [Google Scholar]
- Matheny P. B. (2005). Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe. Agaricales). Mol. Phylogenet. Evol. 35 1–20. 10.1016/j.ympev.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Núñez M., Ryvarden L. (2001). East Asian polypores 2. Synop. Fungorum 14 170–522. [Google Scholar]
- Nylander J. A. A. (2004). MrModeltest v2. Program distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- Posada D., Crandall K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14 817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Rehner S. (2001). Primers for Elongation Factor 1-a (EF1-a). Available online at: http://ocid.nacse.org/research/deephyphae/EF1primer.pdf (accessed on 20 May 2020). [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Ryvarden L., Gilbertson R. L. (1993). European polypores 1. Abortiporus–Lindtneria. Synop. Fungorum 6 1–387. [Google Scholar]
- Ryvarden L., Johansen I. (1980). A preliminary polypora flora of East Africa. Oslo: Fungiflora. [Google Scholar]
- Ryvarden L., Melo I. (2014). Poroid fungi of Europe. Synop. Fungorum 31 1–455. [Google Scholar]
- Ryvarden L., Stokland J. (2008). Fomitopsis ochracea nova species. Synop. Fungorum 25 44–46. [Google Scholar]
- Shen L. L., Wang M., Zhou J. L., Xing J. H., Cui B. K., Dai Y. C. (2019). Taxonomy and phylogeny of Postia. Multi-gene phylogeny and taxonomy of the brown-rot fungi: Postia (Polyporales, Basidiomycota) and related genera. Persoonia 42 101–126. 10.3767/persoonia.2019.42.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Cui B. K. (2017). Phylogeny, divergence time and historical biogeography of Laetiporus (Basidiomycota, Polyporales. BMC Evol. Biol. 17:102. 10.1186/s12862-017-0948-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Chen J. J., Wang M., Chen Y. Y., Cui B. K. (2016). Phylogeny and biogeography of the remarkable genus Bondarzewia (Basidiomycota. Russulales). Sci. Rep. 6:34568. 10.1038/srep34568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics 22 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Sun Q., Huang W., Bao H. Y., Bau T., Li Y. (2016). Anti-tumor and antioxidation activities of solid fermentation products of Fomitopsis pinicola. Mycosystema 35 965–974. [Google Scholar]
- Sun Y. F., Costa-Rezende D. H., Xing J. H., Zhou J. L., Zhang B., Gibertoni T. B., et al. (2020). Multi-gene phylogeny and taxonomy of Amauroderma s.lat. (Ganodermataceae). Persoonia 44 206–239. 10.3767/persoonia.2020.44.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. L. (2002). PAUP∗. Phylogenetic analysis using parsimony (∗and other methods). Version 4.0b10. Sunderland, MA: Sinauer Associates, 10.1111/j.0014-3820.2002.tb00191.x [DOI] [Google Scholar]
- White T. J., Bruns T. D., Lee S., Taylor J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics in PCR Protocols, a guide to methods and applications. Cambridge, M: A: Academic Press. 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Yuan Y., Chen J. J., Korhonen K., Martin F. M., Dai Y. C. (2020). An updated global species diversity and phylogeny in the forest pathogenic genus Heterobasidion (Basidiomycota. Russulales). Front. Microbiol. 11:596393. 10.3389/fmicb.2020.596393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Song J., Zhou J. L., Si J., Cui B. K. (2019). Species diversity, phylogeny, divergence time and biogeography of the genus Sanghuangporus (Basidiomycota). Front. Microbiol. 10:812. 10.3389/fmicb.2019.00812 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in an online repository. The name of the repository and accession number can be found below: https://treebase.org/treebase-web/search/study/summary.html?id=27994&x-access-code=ab2495717aaf081f0557e6680c381710&agreement=ok, submission ID: 27994.