Abstract
Background
Mitogen-activated protein kinase (MAPK) cascades are involved with signal transduction in almost every aspect of plant growth and development, as well as biotic and abiotic stress responses. The evolutionary analysis of MAPKs and MKKs in individual or entire plant species has been reported, but the evolutionary patterns in the diverse inbred lines of Brachypodium distachyon are still unclear.
Results
We conducted the systematical molecular evolutionary analysis of B. distachyon. A total of 799 MAPKs and 618 MKKs were identified from 53 B. distachyon inbred lines. Remarkably, only three inbred lines had 16 MPKs and most of those inbred lines lacked MPK7-2 members, whereas 12 MKKs existed in almost all B. distachyon inbred lines. Phylogenetic analysis indicated that MAPKs and MKKs were divided into four groups as previously reported, grouping them in the same branch as corresponding members. MPK21-2 was the exception and fell into two groups, which may be due to their exon-intron patterns, especially the untranslated regions (UTRs). We also found that differential evolution patterns of MKK10 paralogues from ancient tandem duplicates may have undergone functional divergence. Expression analyses suggested that MAPKs and MKKs likely played different roles in different genetic contexts within various tissues and with abiotic stresses.
Conclusion
Our study revealed that UTRs affected the structure and evolution of MPK21-2 genes and the differential evolution of MKK10 paralogues with ancient tandem duplication might have functional divergences. Our findings provide new insights into the functional evolution of genes in closely inbred lines.
Keywords: Brachypodium distachyon, MAPK, MKK, Inbred lines, Tandem duplication, Gene expression, Evolution
Introduction
Mitogen-activated protein kinase (MAPK or MPK) signaling cascades play vital roles in the stress response, cell division, and developmental regulation. They are divided into three highly-conserved subfamilies that continuously act in a sequential manner in evolution and fundamental signaling transduction pathways (Rodriguez, Petersen & Mundy, 2010; Xu & Zhang, 2015; Jagodzik et al., 2018). The MAPK kinase kinases (MKKKs or MEKKs) are activated by extracellular cues and subsequently phosphorylate and activate the S/T-X3–5-S/T motif of downstream MAPK kinases (MAPKKs or MKKs), which, in turn, phosphorylate and activate MAPKs at their TXY activation loop (Rodriguez, Petersen & Mundy, 2010; Singh et al., 2012). Activated MPKs regulate downstream cellular targets, including regulatory and metabolic enzymes and transcription regulators (Joo et al., 2008, Guan et al., 2014).
Brachypodium distachyon (2n = 10) is an annual temperate grass with a close phylogenetic relationship to other temperate cereals and an intermediate position within the Pooideae subfamily (Soreng et al., 2015; Catalan et al., 2016). B. distachyon is desirable for its small physical stature, rapid life cycle, ability to self-fertilize, and small diploid genome size (Draper et al., 2001; Garvin et al., 2008). Highly efficient Agrobacterium-mediated transformation methods in Brachypodium have also been established (Vain et al., 2008; Vogel & Hill, 2008). Therefore, B. distachyon is widely used as a model plant for studying problems unique to cereals and grasses (Vogel et al., 2010; Brkljacic et al., 2011; Mur et al., 2011; Catalan et al., 2014). The morphological, molecular, and cytological analyses of diverse B. distachyon inbred lines have been conducted (Filiz et al., 2009; Vogel et al., 2009) and their nuclear and plastid genomes have been deep sequenced and annotated (Gordon et al., 2014; Gordon et al., 2017; Sancho et al., 2018). Further analysis showed that the inbred lines of B. distachyon are divided into three different genomic groups, including a mostly Extremely Delayed Flowering (EDF+) clade, a mostly Spanish (S+) clade, and a Turkish (T+) clade, based on their flowering phenotype and geographical substructure (Sancho et al., 2018).
To date, the evolutionary mechanisms of MAPK cascades in plants have indicated a diverse domain organization and novel activation loop variants (Mohanta et al., 2015) and/or distinct expansion mechanism (Jiang & Chu, 2018). A variety of single-gene duplication types emerge continuously and have involved in the plant’s adaptation to dramatically changing environments (Wang et al., 2009; Cuevas et al., 2016). However, whole-genome duplications (WGDs) are considered to be a major force in the evolution of morphological and physiological diversity (Soltis et al., 2009; Paterson et al., 2010). The ancient tandem duplication event occurred at the adjacent genes in the same chromosome, which are usually expanded or retained by an unequal crossing (Freeling, 2009). Tandem duplication often displays less expression difference and functional divergence than distant duplication (Makino & McLysaght, 2012; Ghanbarian & Hurst, 2015). However, there is limited information on the gene expansion mechanism and functional evolution of the MAPK cascades in diverse B. distachyon inbred lines, including for Bd21 (Chen et al., 2012; Jiang et al., 2015). We studied the evolutionary patterns of MAPKs and MKKs from different B. distachyon inbred lines. The phylogenetic relationships and the identification of MAPKs and MKKs were determined for 53 B. distachyon inbred lines. We investigated gene and domain construction patterns of the individual members with a certain divergence and focused on the evolutionary history of MKK10 paralogues in different B. distachyon inbred lines. This revealed various conservative and divergent tandem gene clusters. The expression patterns of these genes were analyzed in Bd21, BdTR8i, and Bd30-1 from three genetic groups in various tissues and abiotic stresses, and their potential functions were also investigated.
Methods and materials
Identification of MAPK and MKK gene family members
We downloaded gene information for MAPK and MKK from B. distachyon Bd21 from the PLAZA platform (https://bioinformatics.psb.ugent.be/plaza/) (Van Bel et al., 2018). BLASTP (Altschul et al., 1997) searches were conducted with a threshold of 90% identity; searches were performed with orthologous protein sequences using BdMAPKs and BdMKKs as queries in BrachyPan (https://brachypan.jgi.doe.gov/) (Gordon et al., 2017) to identify these genes in the 53 diverse B. distachyon inbred lines. Collected sequences were only accepted for scanning using InterPro software (Mitchell et al., 2019) if they harbored MAPK or MKK consensus sequences, including the activation loop TXY motif for MPKs, the active site motif D(L/I/V)K, and the phosphorylation target site S/T-X5-S/T within the activation loop for MKKs. The gene identifier information of these sequences was collected and is listed in Tables S1 and S2.
Gene structure and sequence alignments
The exon/intron structure of identified MAPKs and MKKs was performed using Gene Structure Display Server 2.0 (GSDS 2.0) software (http://gsds.gao-lab.org/). All of the full-length amino acid sequences were initially aligned using Clustal Omega with default parameters (http://www.ebi.ac.uk/Tools/msa/clustalo/). The domains and motifs of MAPKs and MKKs were scanned using InterProScan software (http://www.ebi.ac.uk/interpro/) (Jones et al., 2014). The structural schematic of all members of MAPK and MKK were executed according to InterProScan analysis results. The alignment logos of the protein conserved domain were generated using the WebLogo3 application (http://weblogo.threeplusone.com/).
Synteny and phylogenetic analyses
The phylogenetic relationships of all 53 B. distachyon inbred lines were generated in the BrachyPan project and visualized with the CorelDRAW X3 program. Phylogenetic trees were created based on the alignment of all MAPKs or MKKs using the maximum likelihood (ML) method with the Jones–Taylor–Thornton (JTT) model, 2,000 bootstrap values, and partial deletion by the MEGA 6.0 software, respectively (Tamura et al., 2013). The Neighbor Joining (NJ) Trees of the MAPKs or MKKs were also reconstructed with the same parameters using MEGA 6.0. We obtained the synteny information of duplicate genes and the tandem (TD) data from the PlantDGD database (http://pdgd.njau.edu.cn:8080/) (Qiao et al., 2019).
Plant sample preparation
We sowed BdTR8i, Bd21, and Bd30-1 seeds in ½ MS medium in the dark for 4 d at 25 °C and then transferred them to a soil mix. Plants were grown in a greenhouse under 14 h light (21 °C)/10 h dark (18 °C) photoperiods. We harvested the root, stem, leaf blade, and leaf sheath at the eight-to-nine leaf stage. Spikelet samples from B. distachyon were collected at the early flowering stages according to their different flowering times (Fig. S1). For the abiotic stress treatment, 2-week-old B. distachyon seedlings were dipped in ½ MS liquid medium containing 20% PEG 6000 and 200 mM NaCl, and then plants were collected after treatment for 3 h and 6 h, respectively. Moreover, seedlings were transferred to a growth chamber and heat-treated at 40 °C for 3 h and 6 h. All samples were flash frozen in liquid nitrogen and stored at −80 °C for RNA extraction.
Expression analysis
Total RNA was extracted from samples using Trizol reagent and 1–2 μg was reverse-transcribed into cDNA using PrimeScript RT Master Mix Perfect Real Time (TaKaRa, Beijing, China) according to the manufacturer’s instructions. The quality of total RNA was detected using Nanodrop1000 and its integrity was estimated by electrophoresis in 1.5% (w/v) agarose gel. The real-time quantitative polymerase chain reaction (RT-qPCR) was carried out in 10 µl reactions with 5–50 ng of first-stand cDNA products (four µl), five pmol of each primer (0.4 µl), five μl SYBR green master mix (2X), 0.2 µl ROX as a passive reference standard to normalize the SYBR fluorescent signal. The conditions for RT-qPCR were: initial activation at 95 °C for 5 min followed by 45 cycles of 95 °C for 30 s, and 60 °C for 30 s. Subsequently, the specificity of PCR products was monitored using a melting curve analysis (61–95 °C with fluorescence read every 0.5 °C). The B. distachyon actin (gene locus: Bradi2g24070) gene was used as an internal control for all RT-qPCR analyses; specific primers for MAPK and MKK were listed in Table S3. Three independent biological replicates were conducted for each experiment. The relative expression of MAPK and MKK genes was calculated using the 2−ΔΔCt method.
Results
Identification and annotation of MPKs and MKKs in 53 diverse B. distachyon inbred lines
We identified the two gene families by searching homologous Bd21 sequences in the public BrachyPan database (Chen et al., 2012) to determine the conservation and divergence of MPKs and MKKs in 53 diverse B. distachyon inbred lines. All predicted MPKs and MKKs were named based on the similarity of their orthologous protein to that of A. thaliana and B. distachyon (Ichimura et al., 2002; Chen et al., 2012). Ultimately, a total of 799 MPKs and 618 MKKs were retrieved (Table 1; Tables S4 and S5). We found that most B. distachyon inbred lines had 14 or 15 MPKs apart from Bd21, BdTR3c, and Bd18-1, which had 16 members (Table 1). Further analysis showed that only seven inbred lines had the MPK7-2 gene, including Bd21, Bd2-3, BdTR3c, Bd18-1, S8iiC, Mur1, and Foz1 (Table S1). This may be the result of an incomplete annotation of the genome sequence or the long sequence of the MPK7-2 protein, which usually consists of 1,708 amino acid (aa) residues. Most B. distachyon inbred lines harbored 12 MKK members except Tek-4 (11), Bd3-1 (7), Adi-10 (10), Gaz-8 (5), ABR5 (11), Foz1 (11), and Jer1 (11) (Table 1; Table S5). The incomplete assembly of Tek-4 (77.82%), Bd3-1 (89.52%), Adi-10 (89.52%), and Gaz-8 (88.31%) may be the reason that relatively few MKK members have been identified (Gordon et al., 2017). Further analysis showed that the B. distachyon inbred lines lacked a particular MKK member; for example, MKK10-5 of Tek-4, MKK10-4 of ABR5, MKK4 of Foz1, MKK5 of Jer1, MKK10-3 and -4 of Adi-10 (Table S2). We also incorporated the available genomic detailed information from MPKs and MKKs (Tables S1 and S2).
Table 1. Number of B. distachyon inbred lines MPKs and MKKs identified genes from the BrachyPan database and their associated information.
| Genetic groups | Inbred line | Latitude (Gordon et al., 2017) | Longitude | Elevation (m) | Ploidy | MPKs | MKKs |
|---|---|---|---|---|---|---|---|
| EDF+ | Arn1 | 42° 15′ 23.44″ N | 0° 43′ 47.46″ E | 681 | – | 15 | 12 |
| Mon3 | 41° 39′ 4.75″ N | 0° 12′ 37.51″ W | 515 | diploid | 15 | 12 | |
| Bd1-1 | 39° 11′ 27.44″ N | 27° 36′ 28.59″ E | 141 | diploid | 14 | 12 | |
| ABR9 | – | – | – | – | 15 | 12 | |
| Bd29-1 | 44° 30′ 55″ N | 33° 33′ 23″ E | 260 | diploid | 15 | 12 | |
| Tek-4 | 41° 0′ 40.1″ N | 27° 31′ 8.8″ E | 20 | diploid | 14 | 11 | |
| BdTR7a | 39° 44′ 53.45″ N | 34° 39′ 1.15″ E | 1,035 | diploid | 15 | 12 | |
| Tek-2 | 41° 0′ 40.1″ N | 27° 31′ 8.8″ E | 20 | diploid | 15 | 12 | |
| BdTR8i | 37° 6′ 31.87″ N | 34° 4′ 17.06″ E | 2,385 | diploid | 15 | 12 | |
| T+ | Bd21 | 33° 45′ 39.18″ N | 44° 24′ 11.07″ E | 42 | diploid | 16 | 12 |
| Bd21-3 | 33° 45′ 39.19″ N | 44° 24′ 11.08″ E | 43 | diploid | 14 | 12 | |
| Bd3-1 | 33° 45′ 39.19″ N | 44° 24′ 11.08″ E | 43 | diploid | 14 | 7 | |
| Bd2-3 | 33° 45′ 39.18″ N | 44° 24′ 11.07″ E | 42 | diploid | 14 | 12 | |
| Adi-10 | 37° 46′ 14.5″ N | 38° 21′ 8.2″ E | 510 | diploid | 15 | 10 | |
| BdTR12c | 39° 44′ 53.45″ N | 34° 39′ 1.15″ E | 1,035 | diploid | 15 | 12 | |
| Adi-2 | 37° 46′ 14.5″ N | 38° 21′ 8.2″ E | 510 | diploid | 15 | 12 | |
| Adi-12 | 37° 46′ 14.5″ N | 38° 21′ 8.2″ E | 510 | diploid | 15 | 12 | |
| BdTR9k | 39° 45′ 10.62″ N | 30° 47′ 19.07″ E | 932 | diploid | 15 | 12 | |
| Kah-1 | 37° 44′ 2.3″ N | 38° 32′ 0.2″ E | 665 | diploid | 14 | 12 | |
| Kah-5 | 37° 44′ 2.3″ N | 38° 32′ 0.2″ E | 665 | diploid | 15 | 12 | |
| BdTR5i | 40° 23′ 37.13″ N | 32° 59′ 7.32″ E | 1,596 | diploid | 15 | 12 | |
| BdTR10c | 37° 46′ 41.64″ N | 31° 53′ 5.68″ E | 1,288 | diploid | 15 | 12 | |
| BdTR11a | 38° 25′ 0.42″ N | 28° 1′ 52.75″ E | 986 | diploid | 14 | 12 | |
| BdTR11i | 39° 44′ 17.39″ N | 28° 2′ 24.71″ E | 363 | diploid | 15 | 12 | |
| BdTR11g | 41° 25′ 17.86″ N | 27° 28′ 36.81″ E | 124 | diploid | 15 | 12 | |
| BdTR13c | 39° 24′ 46.28″ N | 32° 59′ 17.24″ E | 1,192 | diploid | 15 | 12 | |
| BdTR13a | 39° 45′ 23.35″ N | 32° 25′ 56.46″ E | 787 | diploid | 15 | 12 | |
| Bis-1 | 37° 52′ 35.6″ N | 41° 0′ 54.3″ E | 529 | diploid | 15 | 12 | |
| Koz-3 | 38° 9′ 8.2.6″ N | 41° 36′ 34.8″ E | 853 | diploid | 14 | 12 | |
| Koz-1 | 38° 9′ 8.2.6″ N | 41° 36′ 34.8″ E | 853 | diploid | 15 | 12 | |
| BdTR3c | 36° 46′ 58.92″ N | 32° 57′ 46.71″ E | 1,957 | diploid | 16 | 12 | |
| Gaz-8 | 37° 7′ 39.8″ N | 37° 23′ 26.9″ E | 891 | diploid | 15 | 5 | |
| BdTR1i | 38° 5′ 35.03″ N | 28° 34′ 59.02″ E | 841 | diploid | 15 | 12 | |
| BdTR2b | 40° 4′ 55.55″ N | 31° 19′ 52.01″ E | 667 | diploid | 15 | 12 | |
| BdTR2g | 40° 23′ 37.13″ N | 32° 59′ 7.32″ E | 1,596 | diploid | 15 | 12 | |
| Bd18-1 | 39° 22′ 4.25″ N | 33° 43′ 48.91″ E | 1,101 | diploid | 16 | 12 | |
| S+ | Bd30-1 | 36° 59′ 25.76″ N | 3° 33′ 31.44″ W | 1,220 | diploid | 15 | 12 |
| ABR5 | 42° 34′ 23.45″ N | 0° 33′ 49.39″ W | 828 | diploid | 15 | 11 | |
| Mig3 | 42° 8′ 52.76″ N | 0° 11′ 41.89″ W | 572 | diploid | 15 | 12 | |
| Uni2 | 42° 7′ 3.98″ N | 0° 26′ 42.81″ W | 480 | diploid | 15 | 12 | |
| Mur1 | 42° 06′ 18″ N | 0° 51′ 23″ E | 487 | diploid | 14 | 12 | |
| Foz1 | 42° 38′ 11.44″ N | 1° 18′ 17.42″ W | 434 | diploid | 15 | 11 | |
| ABR2 | 43° 36′ 15.343″ N | 3° 15′ 46.580″ E | 371 | diploid | 14 | 12 | |
| ABR3 | 42° 10′ 49.8″ N | 0° 4’ 23.2″ W | 1,928 | diploid | 14 | 12 | |
| ABR4 | 42° 15′ 45.54″ N | 0° 43′ 0.48″ E | 480 | – | 15 | 12 | |
| ABR6 | 42° 34′ 27.48″ N | 2° 11′ 5.39″ W | 484 | – | 15 | 12 | |
| ABR7 | 41° 35′ 23.86″ N | 4° 45′ 24.26″ W | 725 | – | 14 | 12 | |
| S8iiC | 41° 36′ 19.3″ N | 0° 08′ 38.4″ E | 144 | – | 15 | 12 | |
| Jer1 | 42° 3′ 16.56″ N | 0° 0′ 44.57″ W | 418 | – | 14 | 11 | |
| Per1 | 42° 44′ 13.34″ N | 1° 44′ 58.6″ W | 742 | – | 14 | 12 | |
| Luc1 | 42° 36′ 36.18″ N | 0° 53′ 35.48″ W | 597 | – | 15 | 12 | |
| RON2 | 42° 46′ 50″ N | 0° 57′ 48″ W | 594 | – | 15 | 12 | |
| Sig2 | 42° 36′ 46.55″ N | 1° 0′ 52.38″ W | 524 | – | 14 | 12 |
Phylogenetic classification of B. distachyon inbred lines MAPKs and MKKs
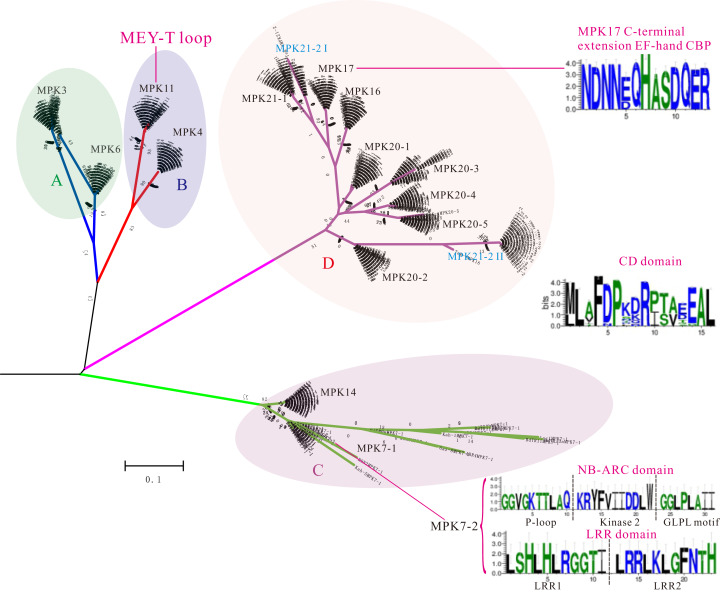
To investigate the phylogenetic relationship of MPK proteins in diverse B. distachyon inbred lines, the phylogeny of all identified 799 MPK protein sequences were performed using ML and NJ methods, respectively. As expected, all homologues for each of the 16 Bd21 MPKs (BdMPKs) were divided into four groups (A, B, C, and D) and clustered on the corresponding branch except Tek-4MPK16 (Fig. 1; Figs. S2 and S3). Tek-4MPK16 consisted of only 183 aa, while the other MPK16 members had 544 aa (Table S1). MPK21-2 had two branches, designated as type I and II (Table S6), indicating that it may have a certain functional divergence. In addition, MPK7-1 and MPK7-2 were located on same branch with a large discrepancy in their lengths (Fig. 1), suggesting functional divergence, which is supported by previous functional studies (Jiang et al., 2015).
Figure 1. Phylogenetic distribution of 799 MPKs among diverse B. distachyon inbred lines.
MPKs are divided into four clades (A–D). Unique to B. distachyon MPK17s, an EF-hand CBP domain is found in their C-terminal extensions. The group A, B and C carry TEY T-loops and group D MPKs carry TDY motifs, with the exception of MPK11s which carry an MEY motif. The CD domain is conserved in the MAPK family. The negatively charged amino acids (D and E) are expected to be exposed to the surface of the molecules. MPK7-2 also contained NB-ARC domain and LRR domain. NB: nucleotide-binding; LRR: leucine-rich repeat. ARC: APAF-1, R gene and CED-4.
We analyzed a total of 618 MKKs for their phylogenetic relationship with corresponding protein sequences using ML and NJ methods, respectively. Almost all of the orthologous genes for each of the 12 Bd21 MKKs (BdMKKs) had similar clustering patterns with corresponding branches and fell into four groups: A, B, C, and D (Fig. 2; Figs. S4 and S5). However, BdTR10cMKK10-3, Jer1MKK10-4, Mur1MKK10-5, and BdTR13cMKK10-5 were branched out from the other members, suggesting that these gene members may have diverged (Fig. 2). It is noteworthy that Mur1MKK10-5 (188 aa) and BdTR13cMKK10-5 (208 aa) had shorter amino acids than other members, which usually contained 332 aa (Table S2). BdTR10cMKK10-3 (163 aa) was also shorter relative to the normal MKK10-3 (344 aa). In contrast, Jer1MKK10-4 (424 aa) was longer when compared with the typical MKK10-4 (341 aa) (Table S2). These situations may affect their evolutionary relationship with MKKs from other B. distachyon inbred lines.
Figure 2. Phylogenetic distribution of 618 MKKs among diverse B. distachyon inbred lines.
MKKs are divided into four clades (A–D). Sequence features show in the form of web logos representing the NTF-2 domain conserved in the C-terminal extensions of MKK3s. Web logos analysis shows amino acid distribution of conserved S/T-X5-S/T motif in MKKs (groups A–C) and the docking site conserved in N-terminal extension of MKKs.
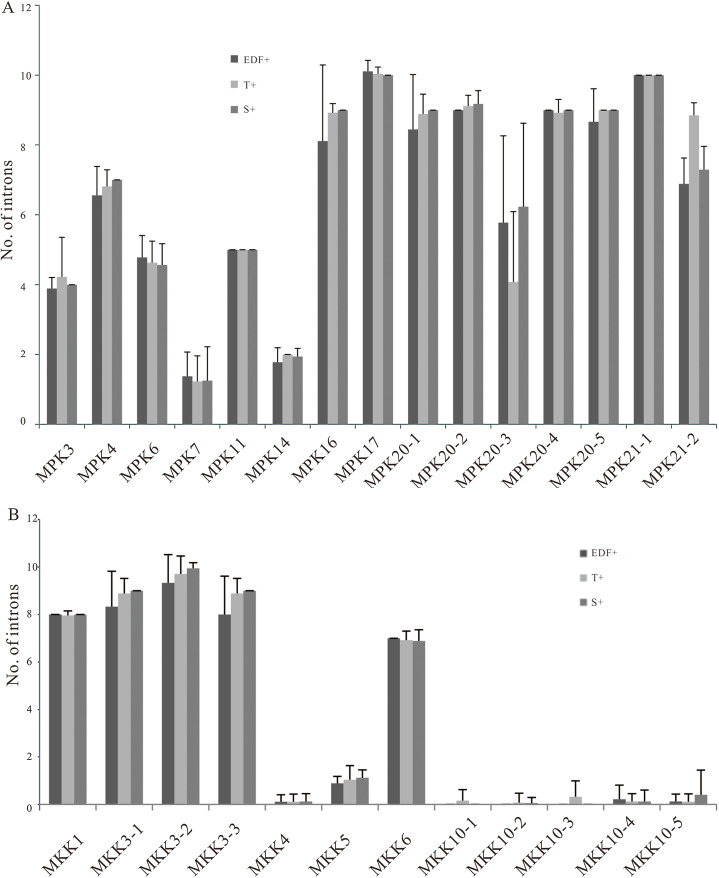
Exon-intron compositions and length variations of MPKs and MKKs in B. distachyon inbred lines
The abundance of non-protein-coding DNA within a genome, such as an intron, increased consistently with the genome complexity (Taft, Pheasant & Mattick, 2007). Intron pattern analyses can enhance our understanding of the structure and evolution of genes (Zhang et al., 2014). We also surveyed the exon-intron architecture of different MPKs and MKKs using GSDS software to elucidate the relationship or divergence among paralogues and orthologues. Most members showed similar exon-intron patterns with the intron number, exon length, and intron phase. The intron number was found to be relatively constant in three genetic groups, with the exceptions of MPK20-3, MPK21-2, and MKK3-3 (Fig. 3). Remarkably, the number of introns from MPK11, MPK21-1, and MKK1 in all B. distachyon inbred lines consistently contained 5, 10, and 8 introns, respectively (Fig. 3). Almost all MPK20-3 had three introns in group T+ apart from BdMPK20-3, Bd21-3MPK20-3, BdTR13aMPK20-3, BdTR3cMPK20-3, and Bd18-1MPK20-3 which had eight introns, while other two genetic groups of MPK20-3 displayed three or eight introns (Fig. 3A; Fig. S6). Moreover, the number of introns of MKK3-3 was highly consistent in group S+, and T+ usually contained nine introns. In contrast, the intron numbers in group EDF+ were highly variable; for example, ABR9MKK3-3, Bd29-1MKK3-3, Tek-4MKK3-3 had eight, six, and four introns, respectively (Fig. 3B; Fig. S7).
Figure 3. Intron number polymorphisms of MPKs (A) and MKKs (B) from 53 diverse B. distachyon inbred lines.
Different colors represent different genomic groups.
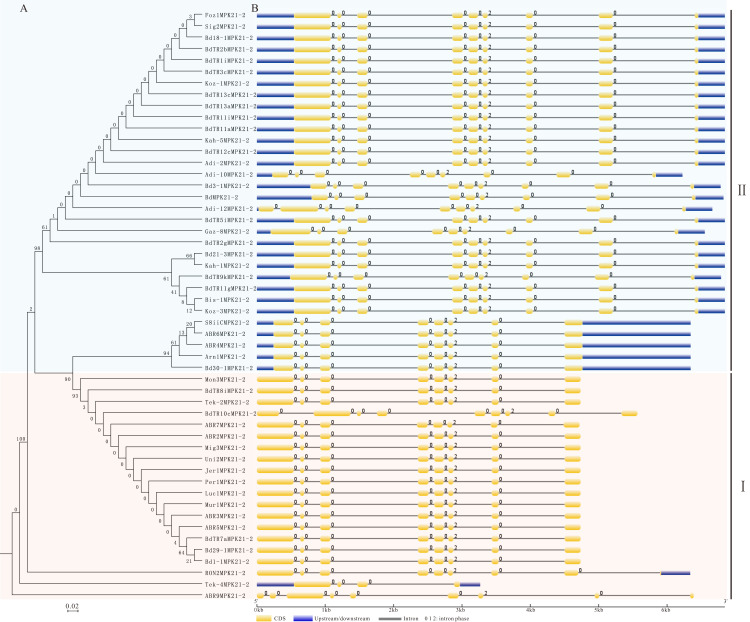
The exon-intron patterns of MPK21-2 fell into two groups, which coincided with their phylogenetic relationship mentioned above (Figs. 1 and 4). However, the phylogenetic relationship was not completely consistent between the reconstructed full length coding sequence (CDS) and their exon-intron patterns (Fig. 4). Further analysis showed that all type II MPK21-2s harbored UTRs, while type I MPK21-2s had no UTR except for RON2MPK21-2 and Tek-4MPK21-2 (Fig. 4). Most MPK21-2s had seven introns in group EDF+ and S+ or nine introns in group T+ except for BdMPK21-2, Bd3-1MPK21-2, and Adi-10MPK21-2 (Fig. 3A). These results suggest that the structure and evolution of these genes were influenced by intron patterns and may be affected by UTRs.
Figure 4. Gene structure of MPK21-2 genes.
(A) Maximum Likelihood phylogenetic trees of the full CDS sequences of genes encoding MPK21-2 from diverse B. distachyon inbred lines. (B) The exon/intron structure of each MPK21-2 gene was displayed. Yellow boxes represent exons, gray lines represent introns and blue boxes represent UTRs. The exons are drawn to scale.
Common conserved domain analysis of B. distachyon MPKs and MKKs
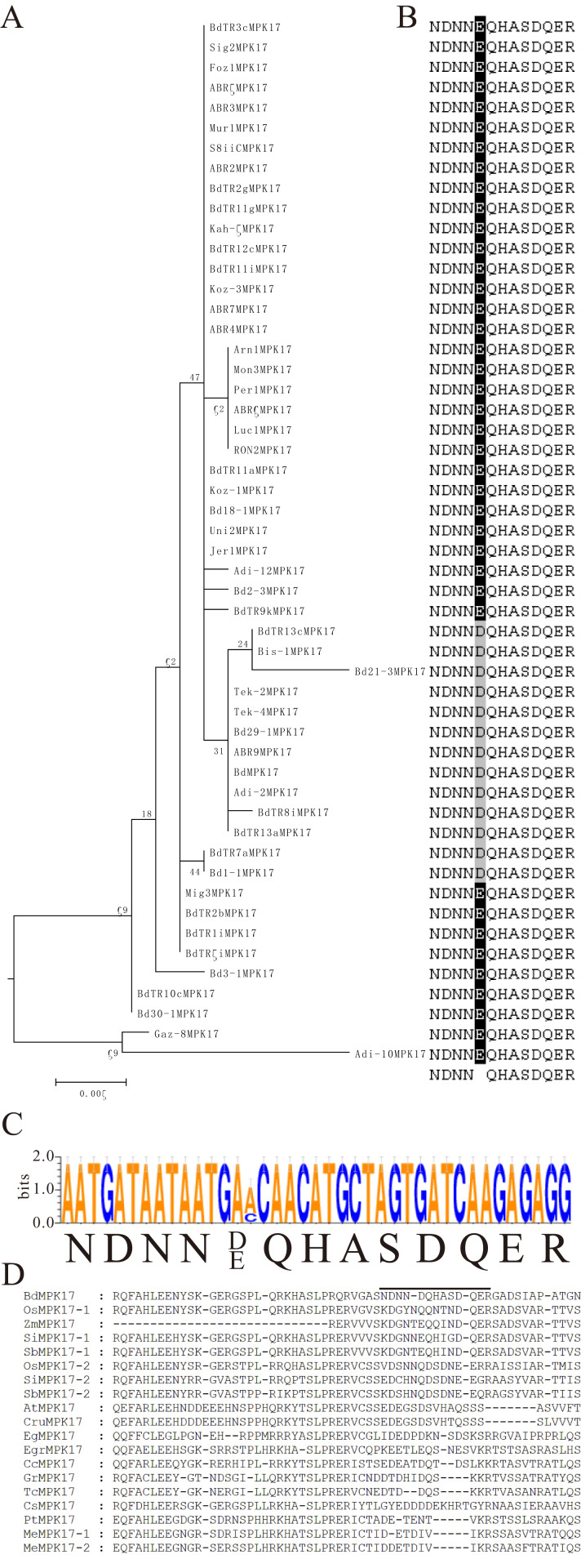
Previous research has reported that MAPKs had several conserved domains or signature sequences with vital structural or functional roles, including the GxGxxG motif in the nucleotide binding (NB) domain (Mohanta et al., 2015), the TXY motif in the activation loop (Xu & Zhang, 2015), D(I/L/V)K motif in the active site (Goyal et al., 2018), and the common docking (CD) domain in the C-terminal extension region outside the catalytic domain (Tanoue et al., 2000). Specifically, the threonine or tyrosine in the TXY motif as the activation loop plays pivotal roles in the signal transduction pathway. Remarkably, the average abundance of threonine or tyrosine (TXY), the most important enzymes in the B. distachyon inbred lines, were 4.67 and 3.91, respectively, and 4.62 and 3.81 in Bd21, respectively (Table S7), suggesting that these amino acids were relatively constant. We also found that the conserved domains of eleven MPKs reported in comprehensive plant species were highly conserved in individual MPKs of B. distachyon inbred lines (Figs. S8 and S9). These analyses revealed that the activation loop TEY in groups A, B, and C, and TDY in group D, were consistent with results from previous studies. Most notably, the activation loop TEY motif in all B. distachyon MPK11s was replaced by the MEY motif (Fig. 1). MAPKs harbored a CD domain featuring a cluster of negatively-charged amino acids with consensus sequences M/L-L-A/V-F-D-P-X2-R-P/I-T/S-A/V-X-E-A-L (Fig. 1) that bind the basic residues at the N-terminus of the docking site in MAPK-interaction proteins (Jiang et al., 2018). MPK7-2s belonged to leucine-rich repeat receptor kinases (LRR-RKs) that had LRR domains and an NB-ARC domain, only appearing in seven kinds of B. distachyon inbred lines (Fig. 1). Moreover, the DLK active site within the MPK7-1s signature was conserved; however, Luc1MPK7-1 and ABR6MPK7-1 were replaced with a DLN motif. Foz1MPK7-2 and Mur1MPK7-2 with DLN motifs were treated similarly (Fig. S8). Specifically, all MPK11s had a DLR motif instead of a DLK active site (Fig. S8). Furthermore, an elongation factor hand (EF-hand) calcium binding protein (CBP) with the consensus sequences “NDNNEQHASDQER” was observed in all B. distachyon MPK17s at their C-terminal end (Fis. 1, 5A and 5B). Further analysis indicated that 13 MPK17 members (group T+: BdTR13cMPK17, Bis-1MPK17, Bd21-3MPK17, BdMPK17, Adi-2MPK17 and BdTR13aMPK17; group EDF+: Tek-2MPK17, Tek-4MPK17, Bd29-1MPK17, ABR9MPK17, BdTR8iMPK17, BdTR7aMPK17, and Bd1-1MPK17) had a mutation in which E changed to D, which only resulted from a single nucleotide substitution of A changed to C (Fig. 5C). This mutation may be have no effect on the function due to the canonical EF-hand (for example the calcineurin B-like (CBL) protein), that are characterized by a conserved Asp (D) and Glu (E) residue with completely constant spacing (Kolukisaoglu et al., 2004; Jiang et al., 2020). Remarkably, the MPK17s exhibited the conserved domain specific to B. distachyon members compared to other plant species, especially eudicots (Fig. 5D).
Figure 5. Conservation and consensus pattern of the EF-hand motif of B. distachyon inbred lines MPK17s.
(A) Maximum Likelihood phylogenetic trees of the full sequences of genes encoding MPK17s from B. distachyon inbred lines. (B) ClustalW multiple-sequence alignment of the region containing the EF-hand motif within MPK17s. Conserved residues are shown in dark colors. (C) Consensus pattern and sequence logo of the EF-hand motif generated using the Weblogo3 application (http://weblogo.threeplusone.com/). The overall height of each stack indicates the sequence conservation at that position (measured in bits), whereas the height of symbols within the stack reflects the relative frequency of the corresponding base at that position. (D) ClustalW multiple-sequence alignment of the region containing the EF-hand motif within MPK17 in different plant species. The specific domain was marked by thick line. At: Arabidopsis thaliana; Bd: Brachypodium distachyon; Cru: Capsella rubella; Cc: Citrus clememtina; Cs: Cucumis sativus; Eg: Erythranthe guttata; Egr: Eucalyptus grandis; Gr: Gossypium raimondii; Me: Manihot esculenta; Os: Oryza sativa; Pt: Populus trichocarpa; Sb: Sorghum bicolor; Si: Setaria italica; Tc: Theobroma cacao; Zm: Zea mays.
As same as the MAPKs, MKKs also contained some important domains or motifs including the activation or T-loop S/T-X5-S/T motif (Asai et al., 2002), the docking site (K/R2–3X1–5L/IXL/I) in the N-terminal domain (Bardwell & Shah, 2006; Jiang & Chu, 2018), the GxGxxGxV motif in the NB domain and HK-X6-ALK motif in the ATP binding site (Hadiarto et al., 2006), and the active site D(I/L/V)K motif (Goyal et al., 2018). A detailed analysis of the conserved sequences of MKKs was displayed in alignment of individual MKKs (Figs. S10 and S11). Groups A, B, and C MKKs had the typical T-loop S/T-X5-S/T motif, while group D MKKs (MKK10s) had a part mutation in the phosphorylation site which coincided with a wide range of plant species (Fig. 2; Fig. S10) (Jiang & Chu, 2018). Interestingly, the average abundance of serine or threonine (S/T-X5-S/T), which were the most crucial amino acids in B. distachyon inbred lines, were 7.2 and 3.32, respectively, while the same results were 7.2 and 3.32 in Bd21 (Table S7), respectively. This suggests that these amino acids remained constant. We speculated that the MKKs may have experienced fundamental functional conservation. Further analysis showed that variations of some the conserved MKKs were also present. For instance, in addition to BdTR13CMKK4 being replaced by a DIL motif, the D(I/L/V)K active site within the signature of MKKs was conserved despite occasional variations (Fig. S10). We found the HRPTGRCYALK motif in the ATP binding site of MKK5 members, however, BdTR3cMKK5 was replaced by the HRPPGRCYALK motif (Fig. S10). Furthermore, our data showed that the nuclear transport factor 2 (NTF2) domains existed in all MKK3s from B. distachyon inbred lines (Fig. 2).
The differential evolution of MKK10 paralogs with tandem duplications
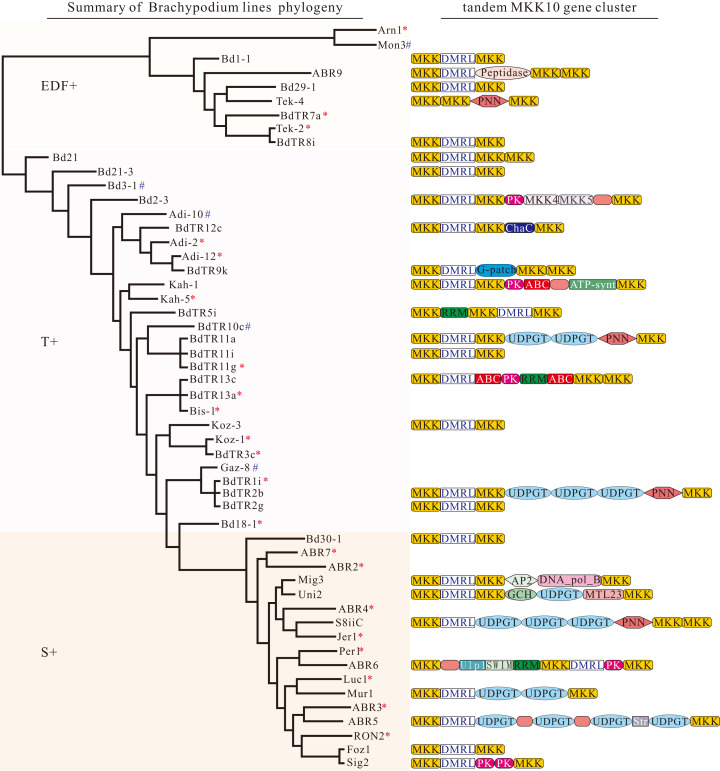
Gene duplication was the necessary material source for evolutionary novelty, leading to the gene responsible for the gene families (Lynch & Conery, 2000). In addition, some tandem duplication was observed in monocot MKK10 paralogues, such as in B. distachyon (Jiang & Chu, 2018). A 6,7-dimethyl-8-ribityllumazine (DMRL) synthase gene was observed between two MKK members (Jiang & Chu, 2018). We surveyed the duplicated genes in different B. distachyon inbred lines genome to further comprehend the duplication and evolutionary events of the B. distachyon MKK10 paralogues. As expected, most of MKK10 paralogues with the exception of five kinds of B. distachyon inbred lines (Mon3, Bd3-1, Adi-10, BdTR10c, and Gaz-8) presented tandem duplication in the canonical form of the MKK-DMRL-MKK model with occasional variations (Fig. 6). In addition, Tek-4 had the PNN (pinin) gene instead of the DMRL gene between two MKK gene members (Fig. 6). Twenty B. distachyon inbred lines had the same canonical model with Bd21 in the form of the MKK-DMRL-MKK-MKK model. Eight B. distachyon inbred lines possessed the tandem duplication in the form of the MKK-DMRL-MKK model (Fig. 6). Moreover, some small variations were also found in other B. distachyon inbred lines. For example, we also found the tandem gene clusters with the MKK-DMRL-UDPGT-UDPGT-MKK (UDPGT: UDP-glucoronosyl and UDP-glucosyl transferase) model in Mur1, MKK-DMRL-MKK-ChaC-MKK (ChaC: ChaC-like protein) model in BdTR12c, and MKK-DMRL-PK-PK-MKK (PK: protein tyrosine kinase) model in Sig2 (Fig. 6). These results indicated that the tandem MKK10 gene clusters in B. distachyon inbred lines originated in the common ancestral genomic contexts and a certain variation developed in order to adapt to the environment differences including light, temperature, or elevation.
Figure 6. The diversity and evolution of the fate of an ancestral locus having B. distachyon inbred lines MKK10 genes in tandem position.
Phylogenetic relationships among 53 diverse B. distachyon inbred lines were investigated. The phylogenetic tree is modified from BrachyPan (https://brachypan.jgi.doe.gov/). The variants of ancestral tandem MKK10 gene clusters in B. distachyon inbred lines are shown on the right. The red asterisk indicates the gene cluster models of these inbred lines are same as Bd21, while blue pound represents no tandem duplication events. Gene or protein names: MKK (MAPK kinase 10); DMRL ( DMRL synthase ); Peptidase (Peptidase_C48); PNN (pinin); ChaC (ChaC-like protein); G-patch (glycine rich nucleic binding domain); ATP-synt (ATP synthase subunit C); RRM (RNA recognition motif protein); UDPGT (UDP-glucoronosyl and UDP-glucosyl transferase); ABC (ABC transporter); PK (Protein tyrosine kinase); AP2 (AP2/EREBP transcription factor); DNA_pol_B (DNA polymerase family B); GCH (Predicted glycine cleavage system H protein); MTL23 (Methyltransferase-like protein 23); Ulp1 (Ulp1 protease); SWIM (SWIM zinc finger); Str (Strictosidine synthase).
Expression variation in the MPK and MKK gene family in three selected genomes
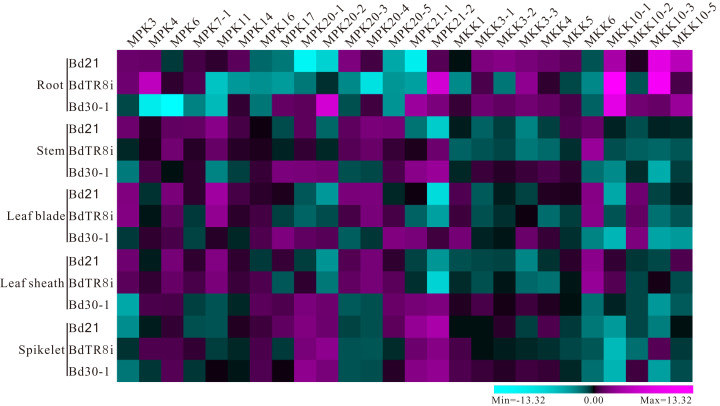
The expression profiles for five different tissues (root, stem, leaf blade, leaf sheath, and spikelet) and abiotic stresses (salt, drought, and heat) of MPK and MKK genes were performed (Table S8) to explore the expression variation in Bd21, BdTR8i, and Bd30-1, which belong to three different genetic groups. We observed the different expression levels of 15 MPK and 11 MKK genes apart from the MKK10-4 gene (Figs. 7 and 8), which may be involved with the barely detectable low expression level of the MKK10-4 gene observed in previous research (Jiang & Chu, 2018). Among these MPK and MKK genes, most genes showed distinct quantitative expression patterns in their different genetic backgrounds. For instance, the expression level of MPK4s at root was higher in BdTR8i and moderate in Bd21 compared with in Bd30-1 (Fig. 7). MPK3s had the same reaction after 6 h of drought treatment (Fig. 8). MKK10-3s had higher expression in spikelet’s in BdTR8i compared with in Bd21 and Bd30-1 (Fig. 7). Moreover, these genes also had similar expression patterns in different genetic backgrounds, such as MPK16s, MPK20-4s, MKK3-2s, and MKK10-1s in spikelet, MKK5s in leaf sheath, MPK6s and MKK10-2s in leaf blade, MPK16s and MKK10-5s in stem, MPK16s, and MPK20-5s and MKK10-1s in root (Fig. 7). MKK4s had a higher expression under drought conditions in BdTR8i compared with in Bd21 and Bd30-1 (Fig. 8). MPK3s had a relatively low expression under heat treatment in three backgrounds, while the expressions of MPK14s and MPK20-1s were opposite (Fig. 8). Expression variations were also observed in the different tissues and/or abiotic stresses. For example, MKK10-1s were more highly expressed in three genetic backgrounds in roots than in other tissues (Fig. 7). Almost all genes had higher expression levels after 6 h of exposure to three abiotic stresses in BdTR8i (Fig. 8). We further found that certain genes had unique expression profiles in specific tissues of a particular genetic background. For example, BdTR8iMPK4 was highly expressed in only the root (Fig. 7) and BdTR8iMKK1 was highly expressed in heat, salt, and drought stresses (Fig. 8). The striking variations of MPK and MKK gene family members expressed in different genetic contexts increases the diversity of the potential biological functions of these genes.
Figure 7. Quantitative expression variation of three diverse B. distachyon inbred lines MPK and MKK genes in five different tissues.
Root, stem, leaf blade and leaf sheath samples were collected at the 8–9 leaf stage of BdTR8i, Bd21 and Bd30-1 seedling, respectively. While spikelet samples were harvested at B. distachyon flowering one to two weeks according to their different flower times.
Figure 8. Expression patterns of MPK and MKK genes in Bd21, BdTR8i and Bd30-1 seedlings under different abiotic stresses.
Discussion
Exon-intron compositions with conservative and divergent patterns
The conservation of exon length was associated with constraints of the gene function of organisms (Davila-Velderrain, Servin-Marquez & Alvarez-Buylla, 2014). The non-coding regions, such as the intron, may affect gene functions by a gradual deletion, which may be the result of recombination throughout the evolution of the intron (Hu, 2006). Therefore, we investigated the exon-intron composition of the corresponding MPKs and MKKs. Our results showed that the exon-intron architecture, including lengths and numbers of intron, intron phase, and lengths of UTR, was generally conserved in corresponding orthologs (Figs. S6 and S7). However, some variability was also found. For example, type II MPK21-2s harbored 5′-UTR and 3′-UTR which were absent in type I MPK21-2s (Fig. 4), although they were in agreement with their phylogenetic relationships (Fig. 1), indicating that they may have a difference in expression and functional divergence. The UTR length-dependent functional specificity significantly increases the coding capacity of the genome that regulates multiple plant process, including nutrient homeostasis, stress responses, and plant growth and development (Srivastava et al., 2018). In addition, there was a large difference in the intron lengths and numbers among B. distachyon MPK7-1s and MPK20-3s (Fig. S6). A detailed analysis showed that the fourth intron of BdTR8iMPK20-3 was shorter than BdMPK20-3 and Bd30-1MPK20-3 (Fig. S6). Moreover, BdTR8iMPK20-3 had a lower expression level in roots and spikelets compared to the other corresponding members as described in a previous study that intron lengths were correlated with gene expression (Rose et al., 2016). Our findings indicated that the exon-intron composition affect the evolutionary patterns and expression efficiency of MPK and MKK orthologs.
Tandem duplications contributed to MKK10s gene expansion
Our analysis suggested that MKK10 paralogs undergo an ancient tandem duplication event with differential evolution. Further examination of tandem MKK10 gene clusters revealed that a DMRL gene often occurred (Fig. 6). These results are supported by previous studies (Jiang & Chu, 2018), indicating that they were derived from common ancestral genomic contexts. However, some variations have also been found among B. distachyon inbred lines such as an insertion of RRM (RNA recognition motif protein) instead of the DMRL gene (Fig. 6). This may result in a difference of gene expression. Indeed, tandem duplicates generally show more similar expression patterns than remote duplicates (Dai, Xiong & Dai, 2014; Lan & Pritchard, 2016) and preferentially retain the cis-PPIs (protein–protein interactions) after WGD (Makino & McLysaght, 2008, 2012). Therefore, ancient tandem duplications of MKK10s may have contributed to gene expansion and function conservation and/or divergence during the evolution process of monocots.
Expression divergence of MAPKs and MKKs within three B. distachyon genetic groups
Tissue-specific expression patterns of MAPK and MKK genes have been characterized with corresponding functions in plant growth and development. For instance, the expression levels of AtMKK10 are high in pollen but do not appear in shoot apices, mesophyll cells, or mature leaves (Yoo et al., 2008), indicating a potential role in flower tissues (Jiang & Chu, 2018). CaMPK19-2 genes are highly expressed in roots and stems in pepper, while CaMPK1 is highly expressed in in leaves (Liu et al., 2015), which indicates that these genes are expressed preferentially in different tissues and developmental stages (Wei et al., 2014). We investigated the tissue-specific expression profiles in different B. distachyon inbred lines. The result indicated that most MPK and MKK genes had quantitative distinct expression patterns among the three different genetic contexts in different tissues and various abiotic stresses. For example, MPK17 had higher expression levels in the root, stem, leaf blade, and salt treatment in Bd30-1 compared with Bd21 and BdTR8i (Figs. 7 and 8). These results are indicative of the distinct function of MPK17s, which may result from the nonsynonymous substitutions at some pivotal amino acid sites in EF-hand CBP motif in their C-terminal extensions (Fig. 5) as described previously (Yang et al., 2019). Moreover, MKK10-3 and MKK10-5 had similar expression patterns in the leaf blade in Bd30-1 and BdTR8i and distinct profiles in Bd21 (Fig. 7). These results coincide with the tandem gene cluster model (Fig. 6) and are supported by previous reports that physically linked genes (tandem duplicates) usually had less expression differences than distant genes (Ghanbarian & Hurst, 2015; Lan & Pritchard, 2016). Furthermore, the MKK3-2 gene had similar patterns under heat and salt condition (Fig. 8). Taken together, these results suggest that MAPKs and MKKs had an expression divergence which was correlated with the differential evolution in B. distachyon inbred lines.
Conclusion
A total of 799 MPK and 618 MKK genes were retrieved from 53 kinds of B. distachyon inbred lines based on their conserved TXY or S/T-X5-S/T domain, respectively, using bioinformatics approaches. Phylogenetic analyses showed that most MAPKs and MKKs clustered into same branch, with the exception of MPK21-2s, which was divided into two groups, designated as type I and II. Further analysis found that the divergence of MPK21-2 may be involved with the presence of UTRs. MKK10s expanded during the evolutionary process by ancient tandem duplications with a differential model. This may have resulted in expression differences and functional divergence. We discovered that the expression of the MPK and MKK gene members varied in different tissues and across abiotic stresses in three different genetic contexts, suggesting that these genes may have diverse biological functions. Taken together, our results revealed a more comprehensive understanding of the function and evolutionary patterns of MAPKs and MKKs in diverse B. distachyon inbred lines.
Supplemental Information
Five MPKs including Gaz-8MPK4, Kah-1MPK20-4, Mon3MPK7-1, Tek-4MPK16, Tek-4MPK20-1, were excluded from entire sequences for reconstructing NJ tree.
Nine MKKs including Adi-10MKK5, Bd3-1MKK4, BdTR10cMKK10-5, Bd29-1MKK4, BdTR12cMKK3-1, BdTR5iMKK5, BdTR9kMKK10-5, Tek-4MKK3-1, Tek-4MKK3-3, were excluded from entire sequences for reconstructing NJ tree.
Funding Statement
This work was supported by the Shanghai Sailing Project (19YF1414800) chaired by Min Jiang. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Min Jiang conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Peng Li performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Wei Wang analyzed the data, prepared figures and/or tables, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The data of RT-qPCR experiments are available in the Supplementary File.
References
- Altschul et al. (1997).Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai et al. (2002).Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415(6875):977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- Bardwell & Shah (2006).Bardwell L, Shah K. Analysis of mitogen-activated protein kinase activation and interactions with regulators and substrates. Methods. 2006;40(3):213–223. doi: 10.1016/j.ymeth.2006.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brkljacic et al. (2011).Brkljacic J, Grotewold E, Scholl R, Mockler T, Garvin DF, Vain P, Brutnell T, Sibout R, Bevan M, Budak H, Caicedo AL, Gao CX, Gu Y, Hazen SP, Holt BF, Hong SY, Jordan M, Manzaneda AJ, Mitchell-Olds T, Mochida K, Mur LAJ, Park CM, Sedbrook J, Watt M, Zheng SJ, Vogel JP. Brachypodium as a model for the grasses: today and the future. Plant Physiology. 2011;157(1):3–13. doi: 10.1104/pp.111.179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalan et al. (2014).Catalan P, Chalhoub B, Chochois V, Garvin DF, Hasterok R, Manzaneda AJ, Mur LAJ, Pecchioni N, Rasmussen SK, Vogel JP, Voxeur A. Update on the genomics and basic biology of Brachypodium International Brachypodium Initiative (IBI) Trends in Plant Science. 2014;19(7):414–418. doi: 10.1016/j.tplants.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Catalan et al. (2016).Catalan P, Lopez-Alvarez D, Bellosta C, Villar L. Updated taxonomic descriptions, iconography, and habitat preferences of Brachypodium distachyon, B-stacei, and B-hybridum (Poaceae) Anales Del Jardin Botanico De Madrid. 2016;73(1):e028. doi: 10.3989/ajbm.2428. [DOI] [Google Scholar]
- Chen et al. (2012).Chen LH, Hu W, Tan SL, Wang M, Ma ZB, Zhou SY, Deng XM, Zhang Y, Huang C, Yang GX, He GY. Genome-wide identification and analysis of MAPK and MAPKK gene families in Brachypodium distachyon. PLOS ONE. 2012;7(10):e46744. doi: 10.1371/journal.pone.0046744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas et al. (2016).Cuevas HE, Zhou CB, Tang HB, Khadke PP, Das S, Lin YR, Ge ZX, Clemente T, Upadhyaya HD, Hash CT, Paterson AH. The evolution of photoperiod-insensitive flowering in sorghum, a genomic model for panicoid grasses. Molecular Biology and Evolution. 2016;33(9):2417–2428. doi: 10.1093/molbev/msw120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, Xiong & Dai (2014).Dai ZM, Xiong YY, Dai XH. Neighboring genes show interchromosomal colocalization after their separation. Molecular Biology and Evolution. 2014;31(5):1166–1172. doi: 10.1093/molbev/msu065. [DOI] [PubMed] [Google Scholar]
- Davila-Velderrain, Servin-Marquez & Alvarez-Buylla (2014).Davila-Velderrain J, Servin-Marquez A, Alvarez-Buylla ER. Molecular evolution constraints in the floral organ specification gene regulatory network module across 18 angiosperm genomes. Molecular Biology and Evolution. 2014;31(3):560–573. doi: 10.1093/molbev/mst223. [DOI] [PubMed] [Google Scholar]
- Draper et al. (2001).Draper J, Mur LAJ, Jenkins G, Ghosh-Biswas GC, Bablak P, Hasterok R, Routledge APM. Brachypodium distachyon: a new model system for functional genomics in grasses. Plant Physiology. 2001;127(4):1539–1555. doi: 10.1104/pp.010196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiz et al. (2009).Filiz E, Ozdemir BS, Budak F, Vogel JP, Tuna M, Budak H. Molecular, morphological, and cytological analysis of diverse Brachypodium distachyon inbred lines. Genome. 2009;52(10):876–890. doi: 10.1139/G09-062. [DOI] [PubMed] [Google Scholar]
- Freeling (2009).Freeling M. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annual Review of Plant Biology. 2009;60(1):433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- Garvin et al. (2008).Garvin DF, Gu YQ, Hasterok R, Hazen SP, Jenkins G, Mockler TC, Mur LAJ, Vogel JP. Development of genetic and genomic research resources for Brachypodium distachyon, a new model system for grass crop research. Crop Science. 2008;48(S1):S69–S84. doi: 10.2135/cropsci2007.06.0332tpg. [DOI] [Google Scholar]
- Ghanbarian & Hurst (2015).Ghanbarian AT, Hurst LD. Neighboring genes show correlated evolution in gene expression. Molecular Biology and Evolution. 2015;32(7):1748–1766. doi: 10.1093/molbev/msv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon et al. (2017).Gordon SP, Contreras-Moreira B, Woods DP, Marais DLD, Burgess D, Shu SQ, Stritt C, Roulin AC, Schackwitz W, Tyler L, Martin J, Lipzen A, Dochy N, Phillips J, Barry K, Geuten K, Budak H, Juenger TE, Amasino R, Caicedo AL, Goodstein D, Davidson P, Mur LAJ, Figueroa M, Freeling M, Catalan P, Vogel JP. Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nature Communications. 2017;8(1):2184. doi: 10.1038/s41467-017-02292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon et al. (2014).Gordon SP, Priest H, Marais DLD, Schackwitz W, Figueroa M, Martin J, Bragg JN, Tyler L, Lee CR, Bryant D, Wang WQ, Messing J, Manzaneda AJ, Barry K, Garvin DF, Budak H, Tuna M, Mitchell-Olds T, Pfender WF, Juenger TE, Mockler TC, Vogel JP. Genome diversity in Brachypodium distachyon: deep sequencing of highly diverse inbred lines. Plant Journal. 2014;79(3):361–374. doi: 10.1111/tpj.12569. [DOI] [PubMed] [Google Scholar]
- Goyal et al. (2018).Goyal RK, Tulpan D, Chomistek N, Fundora DGP, West C, Ellis BE, Frick M, Laroche A, Foroud NA. Analysis of MAPK and MAPKK gene families in wheat and related Triticeae species. BMC Genomics. 2018;19(1):178. doi: 10.1186/s12864-018-4545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan et al. (2014).Guan YF, Meng XZ, Khanna R, LaMontagne E, Liu YD, Zhang SQ. Phosphorylation of a WRKY Transcription Factor by MAPKs is required for pollen development and function in arabidopsis. PLoS Genetics. 2014;10(5):e1004384. doi: 10.1371/journal.pgen.1004384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadiarto et al. (2006).Hadiarto T, Nanmori T, Matsuoka D, Iwasaki T, Sato K, Fukami Y, Azuma T, Yasuda T. Activation of arabidopsis MAPK kinase kinase (AtMEKK1) and induction of AtMEKK1-AtMEK1 pathway by wounding. Planta. 2006;223(4):708–713. doi: 10.1007/s00425-005-0126-7. [DOI] [PubMed] [Google Scholar]
- Hu (2006).Hu KJ. Intron exclusion and the mystery of intron loss. FEBS Letters. 2006;580(27):6361–6365. doi: 10.1016/j.febslet.2006.10.048. [DOI] [PubMed] [Google Scholar]
- Ichimura et al. (2002).Ichimura K, Shinozaki K, Tena G, Sheen J, Henry Y, Champion A, Kreis M, Zhang SQ, Hirt H, Wilson C, Heberle-Bors E, Ellis BE, Morris PC, Innes RW, Ecker JR, Scheel D, Klessig DF, Machida Y, Mundy J, Ohashi Y, Walker JC, Grp M. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends in Plant Science. 2002;7(7):301–308. doi: 10.1016/S1360-1385(02)02302-6. [DOI] [PubMed] [Google Scholar]
- Jagodzik et al. (2018).Jagodzik P, Tajdel-Zielinska M, Ciesla A, Marczak M, Ludwikow A. Mitogen-activated protein kinase cascades in plant hormone signaling. Frontiers in Plant Science. 2018;9:1387. doi: 10.3389/fpls.2018.01387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang & Chu (2018).Jiang M, Chu ZQ. Comparative analysis of plant MKK gene family reveals novel expansion mechanism of the members and sheds new light on functional conservation. BMC Genomics. 2018;19(1):407. doi: 10.1186/s12864-018-4793-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang et al. (2018).Jiang M, Niu C, Cao JM, Ni DA, Chu ZQ. In silico-prediction of protein-protein interactions network about MAPKs and PP2Cs reveals a novel docking site variants in Brachypodium distachyon. Scientific Reports. 2018;8(1):15083. doi: 10.1038/s41598-018-33428-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang et al. (2015).Jiang M, Wen F, Cao JM, Li P, She J, Chu ZQ. Genome-wide exploration of the molecular evolution and regulatory network of mitogen-activated protein kinase cascades upon multiple stresses in Brachypodium distachyon. BMC Genomics. 2015;16(1):228. doi: 10.1186/s12864-015-1452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang et al. (2020).Jiang M, Zhao CL, Zhao MF, Li YZ, Wen GS. Phylogeny and evolution of Calcineurin B-Like (CBL) gene family in grass and functional analyses of rice CBLs. Journal of Plant Biology. 2020;63(2):117–130. doi: 10.1007/s12374-020-09240-y. [DOI] [Google Scholar]
- Jones et al. (2014).Jones P, Binns D, Chang HY, Fraser M, Li WZ, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo et al. (2008).Joo S, Liu Y, Lueth A, Zhang SQ. MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant Journal. 2008;54(1):129–140. doi: 10.1111/j.1365-313X.2008.03404.x. [DOI] [PubMed] [Google Scholar]
- Kolukisaoglu et al. (2004).Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J. Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiology. 2004;134(1):43–58. doi: 10.1104/pp.103.033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan & Pritchard (2016).Lan X, Pritchard JK. Coregulation of tandem duplicate genes slows evolution of subfunctionalization in mammals. Science. 2016;352(6288):1009–1013. doi: 10.1126/science.aad8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2015).Liu ZQ, Shi LP, Liu YY, Tang Q, Shen L, Yang S, Cai JS, Yu HX, Wang RZ, Wen JY, Lin YQ, Hu J, Liu CL, Zhang YW, Mou SL, He SL. Genome-wide identification and transcriptional expression analysis of mitogen-activated protein kinase and mitogen-activated protein kinase kinase genes in Capsicum annuum. Frontiers in Plant Science. 2015;6:780. doi: 10.3389/fpls.2015.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch & Conery (2000).Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Makino & McLysaght (2008).Makino T, McLysaght A. Interacting gene clusters and the evolution of the vertebrate immune system. Molecular Biology and Evolution. 2008;25(9):1855–1862. doi: 10.1093/molbev/msn137. [DOI] [PubMed] [Google Scholar]
- Makino & McLysaght (2012).Makino T, McLysaght A. Positionally biased gene loss after whole genome duplication: evidence from human, yeast, and plant. Genome Research. 2012;22(12):2427–2435. doi: 10.1101/gr.131953.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell et al. (2019).Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang HY, El-Gebali S, Fraser MI, Gough J, Haft DR, Huang HZ, Letunic I, Lopez R, Luciani A, Madeira F, Marchler-Bauer A, Mi HY, Natale DA, Necci M, Nuka G, Orengo C, Pandurangan AP, Paysan-Lafosse T, Pesseat S, Potter SC, Qureshi MA, Rawlings ND, Redaschi N, Richardson LJ, Rivoire C, Salazar GA, Sangrador-Vegas A, Sigrist CJA, Sillitoe I, Sutton GG, Thanki N, Thomas PD, Tosatto SCE, Yong SY, Finn RD. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Research. 2019;47(D1):D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanta et al. (2015).Mohanta TK, Arora PK, Mohanta N, Parida P, Bae H. Identification of new members of the MAPK gene family in plants shows diverse conserved domains and novel activation loop variants. BMC Genomics. 2015;16(1):58. doi: 10.1186/s12864-015-1244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur et al. (2011).Mur LAJ, Allainguillaume J, Catalan P, Hasterok R, Jenkins G, Lesniewska K, Thomas I, Vogel J. Exploiting the Brachypodium Tool Box in cereal and grass research. New Phytologist. 2011;191(2):334–347. doi: 10.1111/j.1469-8137.2011.03748.x. [DOI] [PubMed] [Google Scholar]
- Paterson et al. (2010).Paterson AH, Freeling M, Tang HB, Wang XY. Insights from the comparison of plant genome sequences. Annual Review of Plant Biology. 2010;61(1):349–372. doi: 10.1146/annurev-arplant-042809-112235. [DOI] [PubMed] [Google Scholar]
- Qiao et al. (2019).Qiao X, Li QH, Yin H, Qi KJ, Li LT, Wang RZ, Zhang SL, Paterson AH. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biology. 2019;20(1):38. doi: 10.1186/s13059-019-1650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, Petersen & Mundy (2010).Rodriguez MCS, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annual Review of Plant Biology. 2010;61(1):621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- Rose et al. (2016).Rose AB, Carter A, Korf I, Kojima N. Intron sequences that stimulate gene expression in Arabidopsis. Plant Molecular Biology. 2016;92(3):337–346. doi: 10.1007/s11103-016-0516-1. [DOI] [PubMed] [Google Scholar]
- Sancho et al. (2018).Sancho R, Cantalapiedra CP, Lopez-Alvarez D, Gordon SP, Vogel JP, Catalan P, Contreras-Moreira B. Comparative plastome genomics and phylogenomics of Brachypodium: flowering time signatures, introgression and recombination in recently diverged ecotypes. New Phytologist. 2018;218(4):1631–1644. doi: 10.1111/nph.14926. [DOI] [PubMed] [Google Scholar]
- Singh et al. (2012).Singh R, Lee MO, Lee JE, Choi J, Park JH, Kim EH, Yoo RH, Cho JI, Jeon JS, Rakwal R, Agrawal GK, Moon JS, Jwa NS. Rice mitogen-activated protein kinase interactome analysis using the yeast two-hybrid system. Plant Physiology. 2012;160(1):477–487. doi: 10.1104/pp.112.200071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis et al. (2009).Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng CF, Sankoff D, De Pamphilis CW, Wall PK, Soltis PS. Polyploidy and Angiosperm diversification. American Journal of Botany. 2009;96(1):336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- Soreng et al. (2015).Soreng RJ, Peterson PM, Romaschenko K, Davidse G, Zuloaga FO, Judziewicz EJ, Filgueiras TS, Davis JI, Morrone O. A worldwide phylogenetic classification of the Poaceae (Gramineae) Journal of Systematics and Evolution. 2015;53(2):117–137. doi: 10.1111/jse.12150. [DOI] [Google Scholar]
- Srivastava et al. (2018).Srivastava AK, Lu YM, Zinta G, Lang ZB, Zhu JK. UTR-dependent control of gene expression in plants. Trends in Plant Science. 2018;23(3):248–259. doi: 10.1016/j.tplants.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft, Pheasant & Mattick (2007).Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29(3):288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- Tamura et al. (2013).Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis Version 6.0. Molecular Biology and Evolution. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue et al. (2000).Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nature Cell Biology. 2000;2(2):110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- Vain et al. (2008).Vain P, Worland B, Thole V, McKenzie N, Opanowicz M, Fish LJ, Bevan MW, Snape JW. Agrobacterium-mediated transformation of the temperate grass Brachypodium distachyon (genotype Bd21) for T-DNA insertional mutagenesis. Plant Biotechnology Journal. 2008;6(3):236–245. doi: 10.1111/j.1467-7652.2007.00308.x. [DOI] [PubMed] [Google Scholar]
- Van Bel et al. (2018).Van Bel M, Diels T, Vancaester E, Kreft L, Botzki A, Van de Peer Y, Coppens F, Vandepoele K. PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Research. 2018;46(D1):D1190–D1196. doi: 10.1093/nar/gkx1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel & Hill (2008).Vogel J, Hill T. High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Reports. 2008;27(3):471–478. doi: 10.1007/s00299-007-0472-y. [DOI] [PubMed] [Google Scholar]
- Vogel et al. (2010).Vogel JP, Garvin DF, Mockler TC, Schmutz J, Rokhsar D, Bevan MW, Barry K, Lucas S, Harmon-Smith M, Lail K, Tice H, Grimwood J, McKenzie N, Huo NX, Gu YQ, Lazo GR, Anderson OD, You FM, Luo MC, Dvorak J, Wright J, Febrer M, Idziak D, Hasterok R, Lindquist E, Wang M, Fox SE, Priest HD, Filichkin SA, Givan SA, Bryant DW, Chang JH, Wu HY, Wu W, Hsia AP, Schnable PS, Kalyanaraman A, Barbazuk B, Michael TP, Hazen SP, Bragg JN, Laudencia-Chingcuanco D, Weng YQ, Haberer G, Spannagl M, Mayer K, Rattei T, Mitros T, Lee SJ, Rose JKC, Mueller LA, York TL, Wicker T, Buchmann JP, Tanskanen J, Schulman AH, Gundlach H, De Oliveira AC, Maia LD, Belknap W, Jiang N, Lai JS, Zhu LC, Ma JX, Sun C, Pritham E, Salse J, Murat F, Abrouk M, Bruggmann R, Messing J, Fahlgren N, Sullivan CM, Carrington JC, Chapman EJ, May GD, Zhai JX, Ganssmann M, Gurazada SGR, German M, Meyers BC, Green PJ, Tyler L, Wu JJ, Thomson J, Chen S, Scheller HV, Harholt J, Ulvskov P, Kimbrel JA, Bartley LE, Cao PJ, Jung KH, Sharma MK, Vega-Sanchez M, Ronald P, Dardick CD, De Bodt S, Verelst W, Inze D, Heese M, Schnittger A, Yang XH, Kalluri UC, Tuskan GA, Hua ZH, Vierstra RD, Cui Y, Ouyang SH, Sun QX, Liu ZY, Yilmaz A, Grotewold E, Sibout R, Hematy K, Mouille G, Hofte H, Pelloux J, O’Connor D, Schnable J, Rowe S, Harmon F, Cass CL, Sedbrook JC, Byrne ME, Walsh S, Higgins J, Li PH, Brutnell T, Unver T, Budak H, Belcram H, Charles M, Chalhoub B, Baxter I, Initiative IB. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463(7281):763–768. doi: 10.1038/463591e. [DOI] [PubMed] [Google Scholar]
- Vogel et al. (2009).Vogel JP, Tuna M, Budak H, Huo NX, Gu YQ, Steinwand MA. Development of SSR markers and analysis of diversity in Turkish populations of Brachypodium distachyon. BMC Plant Biology. 2009;9(1):88. doi: 10.1186/1471-2229-9-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2009).Wang XY, Gowik U, Tang HB, Bowers JE, Westhoff P, Paterson AH. Comparative genomic analysis of C4 photosynthetic pathway evolution in grasses. Genome Biology. 2009;10(6):R68. doi: 10.1186/gb-2009-10-6-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei et al. (2014).Wei CJ, Liu XQ, Long DP, Guo Q, Fang Y, Bian CK, Zhang DY, Zeng QW, Xiang ZH, Zhao AC. Molecular cloning and expression analysis of mulberry MAPK gene family. Plant Physiology and Biochemistry. 2014;77:108–116. doi: 10.1016/j.plaphy.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Xu & Zhang (2015).Xu J, Zhang SQ. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends in Plant Science. 2015;20(1):56–64. doi: 10.1016/j.tplants.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Yang et al. (2019).Yang Q, Han XM, Gu JK, Liu YJ, Yang MJ, Zeng QY. Functional and structural profiles of GST gene family from three Populus species reveal the sequence-function decoupling of orthologous genes. New Phytologist. 2019;221(2):1060–1073. doi: 10.1111/nph.15430. [DOI] [PubMed] [Google Scholar]
- Yoo et al. (2008).Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature. 2008;451(7180):789–U781. doi: 10.1038/nature06543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2014).Zhang GJ, Li C, Li QY, Li B, Larkin DM, Lee C, Storz JF, Antunes A, Greenwold MJ, Meredith RW, Odeen A, Cui J, Zhou Q, Xu LH, Pan HL, Wang ZJ, Jin LJ, Zhang P, Hu HF, Yang W, Hu J, Xiao J, Yang ZK, Liu Y, Xie QL, Yu H, Lian JM, Wen P, Zhang F, Li H, Zeng YL, Xiong ZJ, Liu SP, Zhou L, Huang ZY, An N, Wang J, Zheng QM, Xiong YQ, Wang GB, Wang B, Wang JJ, Fan Y, Da Fonseca RR, Alfaro-Nunez A, Schubert M, Orlando L, Mourier T, Howard JT, Ganapathy G, Pfenning A, Whitney O, Rivas MV, Hara E, Smith J, Farre M, Narayan J, Slavov G, Romanov MN, Borges R, Machado JP, Khan I, Springer MS, Gatesy J, Hoffmann FG, Opazo JC, Hastad O, Sawyer RH, Kim H, Kim KW, Kim HJ, Cho S, Li N, Huang YH, Bruford MW, Zhan XJ, Dixon A, Bertelsen MF, Derryberry E, Warren W, O’Brien SJ, Griffin D, Johnson WE, Haussler D, Ryder OA, Willerslev E, Graves GR, Alstrom P, Fjeldsa J, Mindell DP, Edwards SV, Braun EL, Rahbek C, Burt DW, Houde P, Zhang Y, Yang HM, Wang J, Jarvis ED, Gilbert MTP, Wang J, Avian Genome Consortium Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014;346:1311–1320. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Five MPKs including Gaz-8MPK4, Kah-1MPK20-4, Mon3MPK7-1, Tek-4MPK16, Tek-4MPK20-1, were excluded from entire sequences for reconstructing NJ tree.
Nine MKKs including Adi-10MKK5, Bd3-1MKK4, BdTR10cMKK10-5, Bd29-1MKK4, BdTR12cMKK3-1, BdTR5iMKK5, BdTR9kMKK10-5, Tek-4MKK3-1, Tek-4MKK3-3, were excluded from entire sequences for reconstructing NJ tree.
Data Availability Statement
The following information was supplied regarding data availability:
The data of RT-qPCR experiments are available in the Supplementary File.