Abstract
What you see is what you get—imaging techniques have long been essential for visualization and understanding of tissue development, homeostasis, and regeneration, which are driven by stem cell self-renewal and differentiation. Advances in molecular and tissue modeling techniques in the last decade are providing new imaging modalities to explore tissue heterogeneity and plasticity. Herein, we describe current state-of-the-art imaging modalities for tissue research at multiple scales, with a focus on explaining key tradeoffs such as spatial resolution, penetration depth, capture time/frequency, and moieties. We explore emerging tissue modeling and molecular tools which improve resolution, specificity, and throughput.
Introduction
A picture is worth a thousand words—a movie is worth more. As stem cells drive tissue development, homeostasis, and regeneration, understanding these complex, dynamic processes with histology alone is often insufficient. To address important aspects of stem cell biology, many imaging modalities have been leveraged to enable tissue visualization faster, over longer periods of time, at greater depths, across larger scales, or at higher resolutions to address application specific needs. When paired with powerful molecular and computational tools, many elusive features of stem cell biology can now be explored. This article will provide a comprehensive review of the state-of-the art imaging techniques available and their tradeoffs, their applications, and future directions which will further empower scientific discovery.
The sheer number of tools and rapidly developing adaptations can make selecting an appropriate tool challenging; this article aims to provide a guide to selection of technologies and techniques to explore any given hypothesis based on the specific needs. One of the first considerations when selecting an imaging paradigm is the scale of the phenomena being studied. Figure 1 summarizes many of the most powerful and widely-adapted imaging tools and adjunct technologies organized based on the field of view necessary – whole body, organs, tissues, or sub-cellular compartments. Each of the imaging techniques has fundamental limits in penetration depth and resolution, and the longer capture times needed to capture larger target areas reduce temporal sensitivity or spatial resolution. The adjunct technologies described include critically important tools enabling moiety, reducing detection burden, and supporting application specific needs.
Figure 1: Imaging methods and adjunct technologies depending on the scale.
Abbreviations used: MRI (magnetic resonance imaging), CT (computed tomography), NIRS (near infrared spectroscopy), uULM (ultrafast ultrasound localization microscopy), OCT (optical coherence tomography), MIMS (multi isotope mass spectroscopy), PAI/PAT (photoacoustic imaging/photoacoustic tomography), TAI (thermoacoustic imaging), LSCM/MPSC (laser scanning confocal microscopy/multipoint scanning confocal microscopy), AFM/ARFI (atomic force microscopy/acoustic radiation force impulse imaging), FISH (fluorescence in situ hybridization), FRET (Forester resonance energy transfer), DREADDs (Designer Receptor Exclusively Activated by Designer Drugs).
Imaging technologies are constantly under development to meet the diverse needs of researchers and clinicians. This development is bounded by the fundamental detection limits of each imaging technology. One familiar example in optics is the Abbe limit, which indicates that the resolution limit is the illuminated radius of an Airy disk (rAiry), determined by the wavelength of light used (λ) and the objective’s numerical aperture (NA),
Similarly, spatial and temporal resolution of all imaging technologies is limited by the physical characteristics of acoustic or electromagnetic waves used for detection and fundamental physics principles such as uncertainty. Research imaging methods are situated at different locations along the trade-off curves of spatial resolution, penetration depth, and temporal resolution, as graphically summarized in Figure 2. The key for choosing the best imaging modality is to understand the biological question so that the optimal combination of imaging tradeoffs and tissue models can be chosen for the intended application. For example, penetration depth continues to be a major challenge in imaging solid organs and thick or heterogeneous tissues (Zhang and Yao, 2018; Zhuang et al., 2019). Therefore, investigators may opt for smaller, less complex model systems to achieve higher spatiotemporal resolution, or select technologies to robustly image humans and large mammals, which remains important for translational research (Martinez-Milla et al., 2020; Rodrigues Simoes et al., 2020; Wolfram et al., 2019), as pharmacologic testing and medical device development often rely on large animals as the gold standard for pre-clinical studies.
Figure 2: Resolution and Penetration Depth.
Imaging modalities vary in imaging depth, spatial resolution, temporal resolution, and availability of molecular probes.
Novel methods like structured illumination, specialized fluorophores, and sophisticated post-processing algorithms are pushing the theoretical limits of spatial resolution, at the cost of reduced imaging speed and more complicated operation (Verdaasdonk et al., 2014). Implantable imaging technologies have some potential to address concerns about penetration depth, but given the ready absorption of visible light by biological tissues, advancements in technologies which are not reliant on light in the visible spectrum have the most promise (Sun et al., 2017; Umezawa et al., 2020). The development of specific and safe labeling methods for cells, proteins, and genes for clinical nuclear and acoustic imaging technologies is thus of significant interest (Errico et al., 2015; Miyaoka and Lehnert, 2020; Wolfram et al., 2019).
In this review, we discuss considerations for useful applications and translational promise of current imaging technologies. We will also address emerging trends in combinatorial approaches including multiple imaging modalities, auxiliary technologies with unique value for imaging biological phenomena, and the development of post-processing algorithms and pipelines. In addition, we will summarize the current state of the art for imaging and modeling stem cell biology to interrogate physiology from the whole-body scale to subcellular resolution.
Whole-Body Imaging
Whole-body imaging can detect systemic processes, like immune and metabolic responses. Imaging model systems on a whole-body scale requires careful consideration of tissue penetration, acquisition time, and detection sensitivity. Radiographic imaging techniques, including magnetic resonance imaging (MRI) (De Feyter et al., 2018; Johnson et al., 2002; Zubkov et al., 2018), computed tomography (CT) (Buck et al., 2008; Cassol et al., 2019; Martinez-Milla et al., 2020), positron emission tomography (PET) and single photon emission computed tomography (SPECT) (Benjamin L Franc; Golestani et al., 2010), have emerged as the mainstays in whole-body imaging. In vivo imaging systems (IVIS) are the most optimized light microscopy tool for whole-body applications (Luker et al., 2015; Pogue et al., 2010; Rothe et al., 2020). While advances in detection and signal processing have improved spatial resolution at the cost of reduced temporal resolution and greater computational demands (Ciobanu and Pennington, 2004; Maronpot et al., 2004; Miyaoka and Lehnert, 2020; Pandit et al., 2013; Slator et al., 2018), and new markers for tissue-specific radiographic targeting (De Feyter et al., 2018; Hare et al., 2016) have broadened the applications for these tools, whole-body imaging methods are largely not specific enough for most stem cell research applications. Thus, for the purposes of this review, we will focus on higher resolution and higher throughput technologies.
Organ Imaging
Imaging Techniques
To visualize organ development or function, a different set of imaging tools are employed to study intricate cellular processes like cell fate determination and stem cell dynamics (Chatzeli and Simons, 2020). The ability to repeatedly image a single living organism longitudinally provides unique opportunities for studying development, disease progression, wound healing, and numerous other applications. Figure 3a highlights the most widely used techniques discussed in this section, featuring representative images captured by each modality. Technologies are sorted into two broad categories related to their functional application – those with moiety guided by user-selected fluorescent probes, and those whose contrast is dependent on the mechanical properties of the sample.
Figure 3: Imaging technologies for organ visualization.
(A) Fluorescence and organ architecture can be imaged using different technical modalities in various tissues. Representative images shown here include multiphoton microscopy of the developing optic cup (Nakano et al., 2012), lightsheet microscopy of an optically cleared embryo (unpublished image courtesy of Dr. Qiang Huang and Dr. Xiling Shen at Duke University), ultrasound imaging of the kidney, and photoacoustic imaging of vasculature (unpublished images courtesy of Dr. Junjie Yao at Duke University). (B) Imaging windows placed in the cranial, dorsal, abdominal, and thoracic regions enable intravital microscopy. (Clockwise from the top) Representative imaging windows (IWs) and intravital microscopy (IM) images are from the following sources: brain IW and IM (Luckner et al., 2018); bone marrow IW (unpublished image courtesy of Dr. Gabi Bixel from Max-Planck-Institute for Molecular Biomedicine); bone marrow IM (Bixel et al., 2017); skin IW and IM (unpublished images courtesy of Haohua Tu and Stephen Boppart at the University of Illinois Biophotonics Imaging Laboratory); embryonic IW and IM, colonic IW and IM, and intestinal IW (unpublished images courtesy of Dr. Qiang Huang and Dr. Xiling Shen at Duke University); intestinal IM (Nobis et al., 2017); ovary IW and IM (Bochner et al., 2015); liver IW (unpublished image courtesy of Dr. Max Nobis and Dr. Paul Timpson at the Garvan Institute of Medical Research); liver IM (Nobis et al., 2017); kidney IW and IM (van den Berg et al., 2018b); pancreas IW (unpublished image courtesy of Dr. Qiang Huang and Dr. Xiling Shen at Duke University); pancreas IM (Nobis et al., 2017); breast IW (unpublished image courtesy of Dr. Max Nobis and Dr. Paul Timpson at the Garvan Institute of Medical Research); breast IM (Nobis et al., 2017); lung IW and IM (Lelkes et al., 2014).
Ultrasound (US) uses the mechanical contrast generated by acoustic backscattering to evaluate biomimetic properties of engineered tissues in vitro and provided insights regarding various in vivo physiological phenomena including vascular flow patterns (Errico et al., 2015; Zhang and Yao, 2018). US can safely detect tissue features tens of centimeters deep chronically with a kHz capture rate (Zhang and Yao, 2018). Clinical ultrasound machines have a spatial resolution of roughly 0.3 mm, but ultrafast ultrasound localization microscopy (uULM) has resolutions in the tens of microns (Errico et al., 2015; Zhang and Yao, 2018).
Several technologies have potential for subcellular imaging in vivo, but their limited molecular specificity has limited their relevance for stem cell research. Photoacoustic imaging (PAI), photoacoustic tomography (PAT) and thermoacoustic imaging (TAI) use short pulses of light or microwaves to cause thermoelastic expansion at the imaging target, resulting in an ultrasound emission (Zhu et al., 2020). While PAI spatial resolution is typically 100 μm, specialized systems can achieve 10 μm resolutions superficially and penetration depths up to 10 cm with reduced resolution (Martynowych et al., 2020; Weiss et al., 2020). Near infrared spectroscopy (NIRS) relies on optical backscattering to non-invasively sample topography (Scholkmann et al., 2014; Zhang and Yao, 2018). NIRS can penetrate up to 20 mm at the cost of reduced spatial resolution and signal-to-noise ratio, the best resolution limit being on the order of 10 μm (Scholkmann et al., 2014; Umezawa et al., 2020). Acoustic radiation force impulse (ARFI) imaging was developed as a non-invasive alternative to atomic force microscopy (AFM), which measures tissue deflection under short harmonic steady state radiation forces, mapping sample topography and stiffness tens of millimeters deep with spatial resolutions of nanometers (Nightingale, 2011; Rodrigues Simoes et al., 2020).
Multi-photon microscopy (MPM) is the most common imaging modality for in vivo imaging at single-cell resolution. MPM relies on simultaneous excitation of fluorophores using multiple photons of longer wavelengths than widefield or confocal microscopy; this limits excitation to a single focal plane, allows for deeper penetration, and reduces the risk of photobleaching or phototoxicity (Wichmann; Zhang and Yao, 2018). Two-photon microscopy is the most frequently utilized, but three- and four-photon microscopy are under development, which utilize longer wavelengths and have greater tissue penetration, higher resolution, and reduced autofluorescence (Qin et al., 2020; Wang et al., 2018; Zhuang et al., 2019). Typically, the XY resolution for multiphoton microscopy is 500 nm, while the Z resolution is 700 nm (Combs and Shroff, 2017; Sanderson et al., 2014). Capture rates are typically 1–30 FPS, but multi-frame femtosecond stroboscopic imaging (MFSI) techniques allow for femtosecond capture rates (Combs and Shroff, 2017; Martynowych et al., 2020). Sustained imaging for hours at a time is possible with appropriate environmental support due to the reduced laser power required. The up to 1mm penetration depth enables in vivo imaging with the support of intravital windows (Benninger and Piston, 2013; Coste et al., 2020; Gupta et al., 2017; Nakano et al., 2012; Nobis et al., 2017; Schafer et al., 2014; Wichmann; Zhang and Yao, 2018). An emerging technique unique to MPM, simultaneous label-free autofluorescence-multiharmonic (SLAM) microscopy enables differentiation of multiple distinct cellular and extracellular components in absence of designated probes using second and third harmonic generation, enabling targeted imaging of wild-type tissues and animals (Boppart et al., 2019; You et al., 2018). When paired with adjunct tools such as intravital windows and transgenic models, intravital imaging with MPM offers perhaps the best balance of spatiotemporal parameters for in vivo imaging, with decent penetration depths, high spatial resolution, relatively fast acquisition speeds, and safe chronic imaging.
Lightsheet microscopy uses thin sheets of light swept through the sample orthogonal to the objectives, providing efficient excitation at subcellular resolution and the best speeds for 3D light microscopy imaging. While lightsheet microscopy can be used to image live samples, penetration depths for untreated samples are typically in the hundreds of μm, limiting its value. Additionally, lightsheet samples must be embedded in a matrix to enable mounting; for live cell imaging, the 3D extracellular architecture selected has profound impacts on differentiation, morphology, and cellular behavior. Advances in optical clearing of tissues have enabled lightsheet microscopes to be used for in toto imaging of relatively large samples such as drosophila, C. elegans, zebrafish, mouse embryos, and even whole human organs (Combs and Shroff, 2017; McDole et al., 2018; McKey et al., 2020; Yue et al., 2020; Zhao et al., 2020). The resolution is as low as 330 nm, and images take only milliseconds to capture although scanning through many layers means even small samples take seconds to scan, and large samples take hours (Combs and Shroff, 2017).
Molecular and Biochemical Tools
Optical clearing is a recently developed protocol which removes, replaces, or modifies optically opaque molecules to reduce the tissue’s refractive index (Bossolani et al., 2019; Chu et al., 2019; Fretaud et al., 2017; McKey et al., 2020). Several methods, including iDISCO, CUBIC, vDISCO, and SHANEL, have been developed for compatibility with various microscope systems and molecular tools, enabling impressive initiatives to image whole organs or organisms in toto. However, these processes cannot be carried out on living creatures, limiting the ability to perform longitudinal studies. In addition, the removal or modification of target epitopes during the clearing process makes this process impractical for some applications. When imaging reporter proteins, clearing quenches innate fluorescence, thus requiring staining with large volumes of antibodies or dyes. This limitation coupled with the poor tissue penetration of most probes pose significant challenges. Protocols for staining large samples and the use of nanobodies have improved outcomes (Bossolani et al., 2019; Chu et al., 2019; Fretaud et al., 2017). By combining optical clearing with lightsheet microscopy, structures which require high penetration depths or intact tissue architectures to image, like ovarian follicles or neural vasculature, have now been visualized at high resolutions.
Lineage tracing, a robust method for identifying all progeny of a single cell, is a critical tool in stem cell research to understand tissue development, homeostasis, and disease. Lineage tracing has encompassed several distinct methods over the years, including direct observation, labelling of cells with dyes or radiotracers, transfection, or genetic recombination. These latter two methods enable selective inducible silencing or expression of a genetic barcode for more controlled studies. By coupling intravital imaging with fluorescent reporters, tracking the lineage of stem cell in situ has elucidated both the structure of stem cell organization and contribution to functions like repair, niche repopulation, transplantation, and stress (Scaramozza et al., 2019). Cellular barcoding is a highly related technique; a custom barcode can be inserted into the genetic material of the cell using methods such as CRISPR or transposon tagging, enabling identification of the cell and its offspring (Sun et al., 2014; Wu et al., 2019). However, barcoding in this way also enables fate mapping, high throughput perturbation screening, sub-cellular anatomical tracing by functionalized barcoding, synaptic mapping, and molecular recording (Guernet et al., 2016; Kebschull and Zador, 2018).
More sophisticated fluorescent reporters provide additional capability to track cellular behaviors. Multiplexed reporters such as Brainbow (Cai et al., 2013) and Confetti (Snippert et al., 2010) have been used to generate transgenic models with inducible stochastic labeling of target cells. Specifically, as multiple spectrally separated fluorescent protein (so-called “XFPs”) cassettes undergo Cre-mediated recombination, the final genomic integration results in one combination of fluorophores to be expressed. Thus, each cell expresses one of over 100 distinct colors, allowing neighboring cells to be differentiated (Cai et al., 2013). Specific promoters and transient Cre-induction allow cell type-specific and lineage-specific tracing respectively, which allows daughter cells to be traced to study neural connectivity, clonality, and cell migration in the case of Brainbow (Brockway et al., 2019; Cai et al., 2013). Confetti has been used to preform lineage tracing from individual intestinal stem cells, both as short-term clonal tracing and long-term assessment of clonal evolution and coarsening under neutral drift (Han et al., 2019; Klein and Simons, 2011; Snippert et al., 2010). Florescence ubiquitin cell cycle indicator (FUCCI) enables visualization of the G1 to S phase transition by a color shift from green to red, which has been used to study cell cycle progression in vivo in a variety of contexts (e.g., stem cells and tumors) and animal models (Sakaue-Sawano et al., 2008; Zielke and Edgar, 2015). Genetically encoded calcium indicators such as GCaMP are popular tools that allow visual detection of neuronal firing based on fluorescent emission during physiologic calcium transients (Dana et al., 2019; Nakai et al., 2001).
Beyond reporting cellular activities, optogenetics enables researchers to control cellular behavior via light with high spatial and temporal precision (Deisseroth, 2011; Stone et al., 2019). Original applications focused on controlling neuronal activities using light-gated ion channel proteins such as channel rhodopsin and halorhodopsin. Since then, synthetic biology has greatly enriched the toolbox for optogenetic control of cellular pathways and functions with various modified photoreceptor proteins and regulatory elements (Dal Maschio et al., 2017; Gunaydin et al., 2010; Hermann et al., 2015; Kinjo et al., 2019; Pathak et al., 2013; Redchuk et al., 2018; Ronzitti et al., 2017; Stamatakis et al., 2018; Taslimi et al., 2014; Toettcher et al., 2013; Zhang et al., 2010).
Applications in Developmental and Regenerative Biology
Intravital imaging using implanted windows in combination with MPM and molecular tools described above have yielded valuable insights into processes involved in development and differentiation. Applying the various window types and placements highlighted in Figure 3b, several recent studies have revealed important features of how cranial, dermal, thoracic, and abdominal architecture evolve during development and regeneration.
While cranial windows were the earliest optical windows described in the literature, until recent advances in molecular tools and microscopy, characterizing the behavior of neural stem cells (NSCs) during development had remained elusive. Recent studies combining these technologies, however, have broadened our understanding of various aspects of neural differentiation, both in the context of neurotransmission in the developing brain as visualized using an embryonic window, and in the adult brain. In fact, in the young adult hippocampus, in vivo two-photon laser-scanning microscopy (TPLSM) has revealed that radial-like NSCs undergo limited rounds of symmetric and asymmetric divisions before being lost, while non-radial-like NSCs continue to undergo the asymmetric division required to serve as a major source of clonal expansion (Pilz et al., 2018). In addition, by tracing labelled stem cells, the precise number of the non-stem cell progeny from a single stem cell have been visualized (Obernier et al., 2018). When applied to neural stem cell populations like those of the subventricular zone (SVZ) and the subgranular zone (SGZ), novel technologies like lineage tracing and optogenetic modulation of neural circuits have elucidated key biological features which underlie murine brain function (Dal Maschio et al., 2017; Kinjo et al., 2019; Stamatakis et al., 2018). Other model systems have proven to be better suited for studies of early neural development, with longitudinal intravital imaging of the semi-transparent zebrafish embryo revealing non-random cell death occurring throughout the developing zebrafish brain and subcellular imaging enabling studies of myelinating oligodendrocytes and whole-brain calcium dynamics (Brockway et al., 2019; Early et al., 2018; Kim et al., 2017a). A single-cell lineage tracing system was also developed by labelling single cells in the brain, muscle, and hematopoietic system in zebrafish embryos (He et al., 2020). New technologies like CUBIC allow for optimal optical penetration for a whole-brain reconstruction of the larval zebrafish with co-registered functional reference atlases, which will support characterization of physiological neuronal processes, with insights for personalized medicine and drug screening (Fretaud et al., 2017; Hildebrand et al., 2017).
Within the cranial anatomy, the mouse calvarium presents another valuable intravital imaging target, enabling visualization of hematopoietic stem/progenitor cells (HSPCs) residing within the bone marrow (BM). While the microvasculature of the calvarium is particularly well suited for intravital imaging because of its thin and transparent quality, hematopoiesis within the BM niche, regardless of location, remains one of the most robust models of mammalian stem cell niches and behaviors (Lo Celso et al., 2009; Morrison and Scadden, 2014). Using live imaging, the function of individual HSPCs and their relationship to blood vessels, osteoblasts and endosteal surfaces under transplantation conditions has been visualized (Lo Celso et al., 2009; Pinho and Frenette, 2019). In tracking individual HSPCs engrafted into mice with impaired hematopoiesis, primitive cell localization was found to be influenced by both cell autonomous and non-autonomous factors, and spatiotemporal localization of HSPC populations were found to be driven by stage of differentiation (Koechlein et al., 2016; Lo Celso et al., 2009). Labelled dividing and non-dividing hematopoietic stem cells (HSCs) were observed mainly in perisinusoidal inches after irradiation (Acar et al., 2015). In addition, both non-hematopoietic (e.g. perivascular mesenchymal stems, endothelial cells, and osteoblasts) and HSC-derived (e.g. megakaryocytes, macrophages, and regulatory T cells) cells were crucial regulators in the niche and related with the positioning of HSPCs (Lo Celso et al., 2009; Pinho and Frenette, 2019). Another recent study using intravital imaging of the calvarial bone marrow found that long-term HSC (LT-HSCs) and multipotent progenitor cells (MPPs) resided in different locations, with time lapse studies revealing that LT-HSCs remained at steady state in vivo and, depending on spatial location, became either highly mobile or highly replicative following activation (Christodoulou et al., 2020).
While transplanted HPSCs home to microvasculature in the BM niche, circulating cells in the microvasculature are visible through another intravital imaging window—namely the dorsal skin fold chamber. In the context of stem cell biology, dorsal skin folds are valuable as a tractable model to image ectodermal differentiation into epidermis and hair follicles, including symmetrical stem cell self-renewal and long-term bone-derived skin dynamics (Dubois et al., 2018; Graf et al., 2013; Li et al., 2012). Intravital imaging has revealed that stem cells coordinate with neighboring differentiation events to respond to injury and maintain tissue homeostasis (Marsh et al., 2018; Mesa et al., 2018; Park et al., 2017). In hair follicles, a stem cell pool undergoing cycles of growth and regression exit from their quiescent state to divide and organize spatially after stimulation and distinct cells repopulated the stem cell compartment to sustain hair regeneration (Deschene et al., 2014; Mesa et al., 2015; Rompolas et al., 2012; Rompolas et al., 2013).
Within the peritoneal cavity, different organs including the small intestine, colon, liver, kidney, spleen, and pancreas have been visualized at single-cell resolutions using various abdominal windows (Bernier-Latmani and Petrova, 2016). Cell vitality and apoptosis, fluid transport, receptor-mediated endocytosis, blood flow, and recruitment of immune cells have been characterized using these intravital approaches (Kitamura et al., 2017; Nirmalraj et al., 2018; Sedin et al., 2019; Small et al., 2016; Zhang et al., 2014). The dynamics of the intestinal stem cell niche has been visualized using Lgr5-confetti mice with Lgr5+ stem cell labeling (Ritsma et al., 2014). Novel approaches to scaffolding coupled with intravital imaging have allowed studies to more faithfully recapitulate physiology by preventing peristaltic obstruction or preventing interference during stimulation experiments (Rakhilin et al., 2016; Rakhilin et al., 2019). This approach revealed that despite the absence of regenerative neural crest cells, enteric neurons had much higher turnover rates than expected (Kulkarni et al., 2017).
In the thorax, a minimally invasive, permanently implantable window, combined with microcartography, enabled tracking of cells longitudinally over the course of weeks (Entenberg et al., 2018; Lelkes et al., 2014). Labeled transplanted lung stem/progenitor cells were imaged in real-time as they differentiated into type I and II pneumocytes, underwent self-renewal, and preferentially localized to the terminal bronchioles (Wu et al., 2013). These studies have broadened our understanding of the developmental and regenerative mechanisms of adult physiology.
The final frontier in intravital imaging for development has remained the visualization of embryogenesis, which has at this point been accomplished in several mammalian and non-mammalian models. Between zygote and early organogenesis, a mouse embryo can be removed from the uterus and imaged using in toto imaging systems like optical coherence microscopy or light sheet fluorescence microscopy (McDole et al., 2018; Munoz and Trainor, 2019; Pantazis and Supatto, 2014; Takahashi et al., 2008). To observe the dramatic changes during mouse embryo development from gastrulation to early organogenesis, a self-adapting lightsheet microscope has also been developed. To reconstruct long-term cell tracks, a computational frame was developed to create a dynamic atlas of post-implantation mouse embryos with dynamic fate maps and tissue morphogenesis across the entire embryo (McDole et al., 2018). Our group developed an implantable window to observe organogenesis in the embryo from E9.5 onwards (Huang et al., 2020) This technique allowed visualization of the developing brain, autophagy in the retina, viral gene delivery, placental drug transfer, and LGR5+ stem cells in the developing gut.
Applications to Study Pathophysiological Processes
In addition to the previously described applications in studying developmental biology, intravital windows have also provided a powerful approach to understanding disease and disease response. For example, imaging the brain through cranial windows has been used to study neocortical plasticity and pathological processes like ischemic stroke, injury, and cancer (Dal Maschio et al., 2017; Kinjo et al., 2019; Luckner et al., 2018; Pilz et al., 2018; Stamatakis et al., 2018). Intravital imaging of the spinal cord in mice via an implanted chamber has been used to visualize axonal regeneration following lesioning. While axonal regeneration was observed within 6–24 hours, regrowth ultimately failed as the fledgling axons failed to navigate in the proper direction, leaving the lesioned site with massive microglia infiltration and a heterogenous dieback of axon stumps (Kerschensteiner et al., 2005).
In a similar response to hepatic injury, hepatocytes specifically were shown to dynamically undergo polyploidization, ploidy reversal, and aneuploidy—the so-called “ploidy conveyor”—to modulate regenerative capability (Matsumoto et al., 2020). The changes in resident hepatic cells including hepatocytes, Kupffer cells, stellate cells, and sinusoidal endothelial cells as well as their metabolic state during regeneration has been characterized (Alieva et al., 2014; Bonnardel et al., 2019; De Feyter et al., 2018). Following ischemia-reperfusion injuries in the heart, real time monitoring of heart cellular dynamics using two-photon imaging revealed neutrophil recruitment, cell permeability, and platelet microthrombus formation (Matsuura et al., 2018). Intravital imaging of the spleen, a complex organ which removes erythrocytes from circulation and functions as an immune organ, has been used to observe host-parasite interactions like malaria-infected models (De Niz et al., 2019; Stolp and Melican, 2016).
A particular area of interest in using intravital imaging in the recent years has been in imaging cancer dynamics in different contexts, whether during tumor growth itself or metastatic spread. Various windows have been used for this, including mammary intravital imaging which has been used to visualize the plasticity of cancer stem cells (CSCs) and how CSCs survive and recruit during tumor growth (Zomer et al., 2013). This cancer cell hierarchy was recapitulated in patient-derived organoids as well, with CSCs at the apex and a following of individual cells (Feng et al., 2018; Grange et al., 2014; Rios and Clevers, 2018; Shimokawa et al., 2017; Slator et al., 2018; van den Berg et al., 2018b). Similarly, in pancreatic tumors, certain transcriptional pathways and cellular adaptations have been associated with tumor lineages and hierarchies (Fox et al., 2016). (Park et al., 2020; Reissaus et al., 2019)Imaging of pancreatic neuroendocrine tumors showed processes such as angiogenesis and cell migration (Balan et al., 2019).
The interaction of metastatic cells with new microenvironments has also been an area of significant interest, with implanted dorsal skin used to image circulating tumor cells and a recent study using a mounted intravital imaging apparatus used to visualize metastatic breast cancer cells in the vasculature in an unrestrained, awake murine model (Sasportas and Gambhir, 2014). Imaging of “pre-micrometastasis” in the liver yielded valuable insights into how metastasizing tumor cells seed in target tissues (Fumagalli et al., 2020; Ritsma et al., 2012).Tumor cell interactions with peripheral cells of the lung during invasion have also been imaged using multiphoton microscopy (Entenberg et al., 2018; Nakasone et al., 2012). In the context of cancer, the application of combined intravital imaging methods and fluorescent reporters have provided insights into how tumor cell interact with peripheral lung cells. Such studies have also elucidated novel biological functions like the crucial role of neutrophil extracellular traps (NETs) in triggering the metastatic growth of dormant tumor cells, a phenomenon which may pose an opportunity for therapeutic targeting (Albrengues et al., 2018).
Tissue Modeling and Imaging
In Vitro Tissue Models
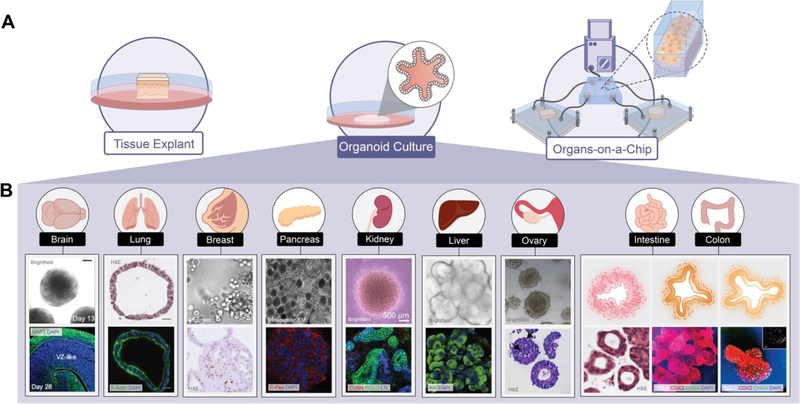
In vitro models provide an alternative to cost-prohibitive and technically challenging in vivo experiments; however, the aim of faithfully reproducing organ microenvironments has not yet been fully realized. The major model formats used to this end are depicted in Figure 4a. Supporting architecture and growth media conditions are both variables which can be strictly defined when rationally designing a biomimetic cellular microenvironment. To capture the complexity of in vivo systems in culture various 3D scaffolds, biodegradable structures which can be processed during cell seeding and growth, have been utilized. Physical properties like mechanical stress and elasticity of these synthetic extracellular environments have also been shown to influence cell fate determination (Engler et al., 2006; Fernandez-Sanchez et al., 2015).
Figure 4: Imaging of in vitro tissue models.
(A) Tissue explant, organoid cultures, and organs-on-a-chip provide in vitro tissue models. (B) Organoids recapitulate the morphology of the tissue from which cells are derived. Representative brightfield (BF), H&E, and/or fluorescent microscopy (FM) images from the following sources: brain BF and FM (Xiang et al., 2017); lung H&E and FM (McCauley et al., 2017); breast BF and H&E (Sachs et al., 2018); pancreatic TEM and FM (Seino et al., 2018); kidney BF and FM (van den Berg et al., 2018a); liver BF and FM (Hu et al., 2018); ovary BF and H&E (Maenhoudt et al., 2020); gastric, intestinal, and colonic illustration (Bruens and Snippert, 2017); gastric H&E (Yan et al., 2018); intestinal and colonic FM (Múnera et al., 2017).
Since the development of reprogrammed inducible pluripotent stem cells (iPSCs) from mouse embryonic or adult fibroblasts by defined factors (Takahashi and Yamanaka, 2006), iPSCs have been widely applied. In developmental biology, iPSCs provided a tractable model for physiological stem cells, with the capability to generate all type of cells in the body and self-organize into three-dimensional mini-organs (Booij et al., 2019). Human iPSCs, especially patient-derived iPSCs, have enormous potential to provide an autologous resource for human biology and disease research to study gene function, developmental pathways, disease mechanisms, cell therapy, and drug screening (Bruens and Snippert, 2017; Meijer et al., 2019; Zunder et al., 2015). Live high-resolution imaging provided the cellular dynamics of the reprogramming process and concluded that certain parts of the reprogramming process follow defined, not stochastic, steps (Smith et al., 2010). Visualization of subcellular structures enabled quantitative evaluation of the degrees of somatic cell reprogramming and iPSC differentiation (Nishimura et al., 2019). Tracking labelled iPSC-derived cells in explants models could longitudinally monitor their localization, survival, proliferation, integration, and differentiation and provide a platform for systematic interrogation of normal and disease states of these cells (Tian et al., 2019). In addition, iPSCs have been used to generate diverse organoids, including gut (Spence et al., 2011), liver (Ng et al., 2018), lung (Miller et al., 2019), and kidney (Takasato et al., 2016), which have provided great opportunity for developmental and disease modeling.
Organoids are models of organ development and disease in vitro, and an intermediate between 2D cell lines and living animals (Rios and Clevers, 2018). Several human pluripotent stem cell (hPSC)-derived organoids have been transplanted into mouse models and generated normal functional units in vivo (Rakotoson et al., 2019; van den Berg et al., 2018b). Ex vivo live imaging of organoids provides high resolution information on the cellular composition, cell morphology, cell-fate decisions, cell–cell interactions of intact biological samples, architecture, cell lineage decision, and differentiation (Pasca, 2019; Takebe and Wells, 2019). In addition to 3D scaffolding and architecture, organoid cultures require unique compositions of growth factors, with some general shared transcription factors needed to maintain stem-ness. These culture media requirements have been well described for most tissue types (Lee et al., 2007; Taguchi et al., 2014). However, the ECM parameters required for formation of organoids from tissues other than intestinal stem cells (Gjorevski et al., 2016) remain to be fully characterized. Organoid models of several key organs are pictured in Figure 4b, demonstrating the diversity of organoid cultures dependent on the organ which they seek to recapitulate.
The ability of organoids to reliably recapitulate many biological phenomena has spurred development of a few critical areas. Because these organoids more closely approximate physiology, patient-derived 3D organoids have been developed as clinical tools for evaluating patient heterogeneity and driving personalized therapies in a high throughput manner (Brazovskaja et al., 2019; Johnson et al., 2020). Organoids are also tractable models for most imaging methods discussed for tissue and cellular imaging, allowing deeper probing of cellular mechanisms which underlie organ physiology and disease. A simple clearing method makes it much more accessible to acquire 3D imaging on a cell-by-cell basis (Dekkers et al., 2019). The subcellular dynamics of DNA movement and chromosome segregation have also been imaged in organoids, making it possible to study these events in cell homeostasis and tissue remodeling (Matano et al., 2015; McKinley et al., 2018). Organoids remain most valuable as models for the cell-cell interactions and tissue organization (Dekkers et al., 2020; Dutta et al., 2017; Ganesh, 2019; Gjorevski et al., 2016; Hu et al., 2018; Lindeboom et al., 2018; McCauley et al., 2017; Múnera et al., 2017; Xiang et al., 2017). Biomimetic matrices and defined biochemical environments allow organoid cultures to recapitulate in vivo determinants of cellular behavior in a tightly controlled and reproducible experimental system. Computational tools may provide more high throughput pipelines for identifying these regulatory factors in the future.
Organoid culture techniques have provided an unprecedented opportunity to study cancer (Drost and Clevers, 2018; Yang et al., 2019). While the development of physiologically normal tissue organoids was already well underway prior to the first report describing intestinal cancer organoids in 2011 (Sato et al., 2011), it was followed by a rapid flurry of publications describing prostate (Gao et al., 2014; Karthaus et al., 2014), pancreatic (Boj et al., 2015; Seino et al., 2018), stomach (Yan et al., 2018), colorectal (Matano et al., 2015), liver (Broutier et al., 2017), breast (Sachs et al., 2018), bladder (Lee et al., 2018), ovarian (Maenhoudt et al., 2020), and esophageal (Drost et al., 2015; Vlachogiannis et al., 2018) cancer organoids. In coupling the high-efficiency generation of these organoids with cryopreservation techniques which maintained genomic integrity over time, biobanks of patient-derived tumor organoids were soon established, which have allowed studies into broader patterns of cancer development and progression (Calandrini et al., 2020; Jacob et al., 2020; Sachs et al., 2018). However, on a more individual basis, tumor organoids present a unique opportunity for personalized medicine, with the 3D structures grown from the patient tumor used for high throughput drug screening to guide clinical decision-making regarding treatments (Ganesh et al., 2019; Kim et al., 2019; Tiriac et al., 2018; Vlachogiannis et al., 2018). Indeed, the predictive value of patient-derived cancer organoids in reproducing patient therapeutic response was demonstrated in early studies in pancreatic cancer and other cancers since (de Witte et al., 2020; Huang et al., 2015; Ooft et al., 2019; Yao et al., 2020). Various studies have paved the way for more sophisticated manipulations of tumor organoids and their microenvironments which have yielded insights regarding tumor-pathogen interactions (McCracken et al., 2014; Scanu et al., 2015), mutational cascades driving tumorigenesis (Drost et al., 2015), and avenues for developing immunotherapies (Nozaki et al., 2016). Recent advances in microfluidic technologies to reproduce tumor-tissue interactions (Sontheimer-Phelps et al., 2019), bioengineered scaffold matrices (Gjorevski et al., 2016), and 3D bioprinted cellular architecture (Reid et al., 2019) will likely even further accelerate the acceptance of organoids as a well-established preclinical model for cancer biology. These tumor organoid applications highlight the need for robust, high-throughput imaging systems for genetic and drug screening platforms.
Organ-on-a-chip technology uses a microfabricated cell culture device to model functional units of an organ to study underlying molecular-and cellular-scale activities of different organs, to mimic whole-body physiology and disease states, and to predict human pharmacokinetic and pharmacodynamic (PK/PD) responses to novel therapeutics (Haddrick and Simpson, 2019; Yu and Choudhury, 2019). Using long-term high-resolution imaging of organ-on-a-chip models, the effects of changes in different biological conditions like microcirculation, perfusion, shear stress, nutrient supply, drug dosing, waste removal, and cell polarity between normal physical compartments (Rothbauer et al., 2018). Immune responses remain difficult to model, however more complex organoids-in-a-chip models are being developed to mimic tissue-tissue or multiorgan interactions in vitro (Park et al., 2019).
Tissue can also be explanted and embedded in an organotypic culture system with an imaging chamber. Long-term imaging of tissue explants has been described to track the dynamic behaviors of cells in tissue homeostasis and disease (Gericke et al., 2015; Germain et al., 2012). Due to the inaccessibility of many tissues during embryogenesis in vivo, tissue explants were widely used to acquire stem cell lineage analysis, the drivers of morphogenesis, the role of extraembryonic tissues, and the effect of maternal factors (Bedzhov and Zernicka-Goetz, 2014; Deglincerti et al., 2016; Kim et al., 2017b; Langenberg et al., 2003; Schauer et al., 2020; Shahbazi et al., 2016; Sonavane et al., 2017). Adult tissue explants have been used primarily as disease models to study cell homeostasis and drug screening (Ribeiro et al., 2017; Wang and Schwarz, 2009; Weaver and Hummon, 2013).
Imaging Techniques
Techniques for imaging tissues vary broadly in field of view, resolution, capture rate, and moieties. There is also a clear distinction between those which are suitable for use on live and fixed tissues. Many of the technologies discussed in the previous sections can also be used for imaging on the cellular or tissue level, but Figure 5 highlights some of the technologies discussed in this section which are generally not applicable to larger samples.
Figure 5: Imaging techniques used to interrogate tissues.
Tissue imaging methods focus on architecture, fluorescence, subcellular dynamics, and isotope incorporation. (Clockwise from the left): Optical coherence tomography (OCT) is particularly well suited for imaging superficial tissues including the retina (unpublished image courtesy of Dr. Junjie Yao at Duke University). Confocal microscopy can allow specific imaging of fluorescent markers (unpublished image courtesy of Aliesha O’Raw and Dr. Xiling Shen at Duke University). Multi-isotope mass spectrometry imaging (MIMS) of the murine intestinal crypt reveal sulfur-rich granules (arrows) surrounded by proliferating stem cells (Zhang et al., 2020). At the cellular level, spatial visualization of RNA transcripts is accomplished by MERFISH (Barbash et al., 2019) and cellular status indicators like Sytox can be imaged via high-throughput imaging methods like IncuCyte (Gong et al., 2017).
Tools for Live Cells
Optical coherence tomography (OCT) relies on optical backscattering to detect topography of the scanned tissue non-invasively based on the principles of low-coherence interferometry (Podoleanu, 2012; Zhang and Yao, 2018). OCT boasts an XY resolution of 10 μm and an axial resolution of 3 μm (Podoleanu, 2012; Zhang and Yao, 2018). Temporal resolutions vary from tens of milliseconds to seconds, with the fastest scanners having lower resolution and no axial scanning (Podoleanu, 2012). OCT penetration depths are limited to 2 mm, limiting their utility to explanted tissue, cell cultures, and superficial tissues such as the retina, as shown in Figure 5 (Chhablani et al., 2015; Dubois et al., 2018; Kim et al., 2016; Podoleanu, 2012).
Confocal microscopy is a form of light microscopy wherein the excitation and emission light are filtered through a pinhole (Combs and Shroff, 2017). As a result, the resolution in XY and Z planes is better than 700 nm and there is little out of focus light. Laser scanning confocal microscopes (LSCM) typically achieve 1–30 FPS, but multi-point/slit confocals (MPSC) can achieve frames in mere milliseconds although frame averaging is frequently used to improve signal fidelity (Combs and Shroff, 2017). Multiple wavelengths of spectrally separated light can be used simultaneously to detect multiple targets, and motorized programmable stages have made improvements in throughput, but speeds remain limited by exposure time and field of view. Standard confocal microscopes have penetration depths of under 100 μm, but near infrared microscopy (NIRM) can achieve depths of up to 500 μm (Combs and Shroff, 2017; Sun et al., 2017), sufficient for imaging sectioned tissues, cultured cells, thin tissues, and hollow organs in toto (Benninger and Piston, 2013; Combs and Shroff, 2017; Falsafi et al., 2020; Sun et al., 2017; Zhang and Yao, 2018). Confocal microscopes are widely available and are perhaps the most versatile, but the focused high-powered illumination typical of confocal microscopy has a higher risk of inducing photobleaching or phototoxicity than widefield microscopes, although MPSC is less severe than LSCM in this aspect (Combs and Shroff, 2017).
High throughput and automated imaging systems for culture systems are becoming more popular. Commercial technologies such as IncuCyte automated cell imaging systems meet the need for high throughput screening assays with the support of newly released computational aids like the new Organoid QC module for automated imaging and quantification of organoid cultures. Recursion’s RxRx features open-source datasets from automated cell imaging accompanied by a deep learning platform to enable machine learning tools for biologic investigation. One new microscope aims to combine the 3D imaging speed of lightsheet microscopes with the resolution and penetration of MPM, resulting in faster imaging, less photobleaching, and better penetration depths than a confocal microscope with similar spatial resolution (Rakotoson et al., 2019). Another technology, THUNDER, achieves robust Z plane resolution and complete 3D imaging faster than confocal microscopes using computational clearing to remove out of focus light (Criscuolo et al., 2020). These high-throughput technologies are especially important in in vitro imaging, as these models are increasingly being adopted for both personalized medicine and drug development applications.
Multi-isotope mass spectrometry (MIMS) utilizes principles of secondary ion mass spectrometry and ion optics to detect metabolic activity. It is limited to superficial depths and therefore is largely used on cultures, biopsies, or fluid samples taken longitudinally. The ability to assess multiple targets of interest in a single test increases throughput, but run times per sample can be over ten minutes, and extensive post-processing is required to interpret the data. As shown in Figure 5, MIMS can serve as a powerful tool to understand the heterogeneity in metabolic uptake throughout cells in a specific structure, like the stem cells which surround the Paneth cells of the murine intestinal crypt. In addition, MIMS has been used to image in vivo lipid turnover in Drosophila and a series of small, conjugatable, orthogonal, and tunable fluorophores were used to image different small metabolites (e.g., lipids and sugars) in live cells and in zebrafish as well as metabolic heterogeneity in tumors (Benson et al., 2019; Steinhauser et al., 2012; Zhang et al., 2020).
Molecular and Biochemical Tools
While cellular complexity is challenging to characterize, advances in profiling biological characteristics of a cell—the genome, epigenome, transcriptome, metabolome, etc.—coupled with spatial information are powerful tools to reconstruct subcellular architecture. As such novel tools are currently in development to characterize this underlying architecture and its purpose in driving cell behavior on a scale of subcellular resolution. Many of these concepts have been reviewed elsewhere, but broadly, key advances in this area have been based around improving the resolution of molecular tools to both study and control cellular behaviors.
In modeling cellular dynamics, the advent of tools like CRISPR-Cas genome editing systems have transformed the accuracy and spatiotemporal control over DNA and RNA profiles. The most recent advances in CRISPR-Cas methodologies include using RNA-targeting Cas proteins to perturb both the epigenome and transcriptome, perturbations which can be coupled with super resolution microscopy to study subcellular dynamics (Pickar-Oliver and Gersbach, 2019). Through CRISPR based library knockdowns, in situ imaging provided the distribution of specific genetic perturbations to certain phenotypes to determine the genetic control of cell cycle and better characterize pancreatic adenocarcinomas (Camsund et al., 2020; Lytle et al., 2019).
Coupling such single cell methodologies with spatial measures has been a growing area of interest. Using ATAC-see, cell-type-specific spatial organization of the accessible genome and the coordinated process of neutrophil chromatin extrusion (NETosis) has been visualized (Chen et al., 2016). Similar to MERFISH, which allows spatial visualization of RNA transcripts as shown in Figure 5, FISSEQ provides a method for single cell sequencing using a confocal microscope, enabling spatial investigation of transcriptomes (Lee et al., 2015). By targeting RNA-binding proteins, the regulators of nuclear RNA localization were uncovered in subcellular compartments (Camsund et al., 2020; Wang et al., 2019a). CODEX, a method using oligonucleotide conjugated probes and custom computational pipeline, enabled delineation of rare cell populations in healthy spleens versus those of lupus model mice using standard widefield fluorescent microscopy. Multiplexed imaging with Oligopaint DNA–fluorescence in situ hybridization (FISH), RNA–FISH, and protein fluorescence and a three-dimensional assay for transposase-accessible chromatin-photoactivated localization microscopy (3D ATAC-PALM) were developed to image the accessible genome. To study the rapid morphological changes of organelles during embryogenesis, multiple organelles were marked with different fluorescence colors and simultaneously imaged in vivo. The preferential location, alignment, and distribution of different organelles during mitosis were tracked (Liu et al., 2018). High-resolution imaging was also used to analyze microtubule bundles, growing filaments, and chromosome movement in dividing cells (Xie et al., 2020). Recently, another nanoscale precision pipeline was developed to observe whole frozen cells by three-dimensional super-resolution and block-face electron microscopy to observe how specific subcellular components vary across the cellular volume (Hoffman et al., 2020).
Improvements in classic tools including FRET sensors and biosensor calibration are currently ongoing as well (Midde et al., 2018; Ombrato et al., 2019; Zhou et al., 2020), with advances in ultrasensitive signal amplification only recently allowing fine-tuning of the biosensor substrate detection ranges (Landry et al., 2018; Wan et al., 2019). The co-expression of these biosensors with agents which can modify cellular function (i.e. optogenetic channelrhodopsins, thermal or magnetic responsive proteins, etc.) are particularly powerful tools. As a representative example, chemogenetic control of oxidative stress via expression of a yeast D-amino acid oxidase (DAAO) allows highly specific intracellular H2O2 production upon administration of otherwise inert D-amino acid—an approach which has provided novel insights into the role of oxidative stress in dilated cardiomyopathy (Steinhorn et al., 2018). Similar “thermogenetics”, “magnetogenetics”, or “radiogenetics” tools remain a yet unexplored frontier in synthetic biology (Lee et al., 2014; Stanley et al., 2015; Urban and Roth, 2015; Wheeler et al., 2016). The further development of and combination of these tools pose significant potential to dissect cellular signaling and dynamics.
Future Directions
Numerous exciting developments are occurring rapidly in the field of imaging, both in terms of imaging technologies which push the boundaries of spatial and temporal resolution and in terms of supplemental techniques which allow the specific visualization of smaller and more complex cellular features and dynamics.
As our understanding of the complex dynamics involved in normal cellular function evolves, there continues to be a demand for imaging techniques which allow more comprehensive in vivo imaging of specific tissues, organs, and applications. Great strides have been made in the field of intravital imaging, due to the approach of customizing techniques to the specific organ of application. However, physical location, imaging depth, and physiologic motion continue to challenge investigators, especially those working on deep tissues and solid organs (Alieva et al., 2014; Bochner et al., 2015; Entenberg et al., 2018; Kitamura et al., 2017; Park et al., 2020; Rakhilin et al., 2019; Ritsma et al., 2012). Development of solutions allowing optical access to these hard to access tissues remains relevant, as do improvements to existing window technologies to enable intermittent manipulation, reduced foreign body response, improved safety and longevity of function, and ease of use. Improvements to organoid cultures and other more complex culture systems may also provide insight while reducing the burden on animal testing. These custom, application-focused techniques will continue to drive discovery in stem cell biology.
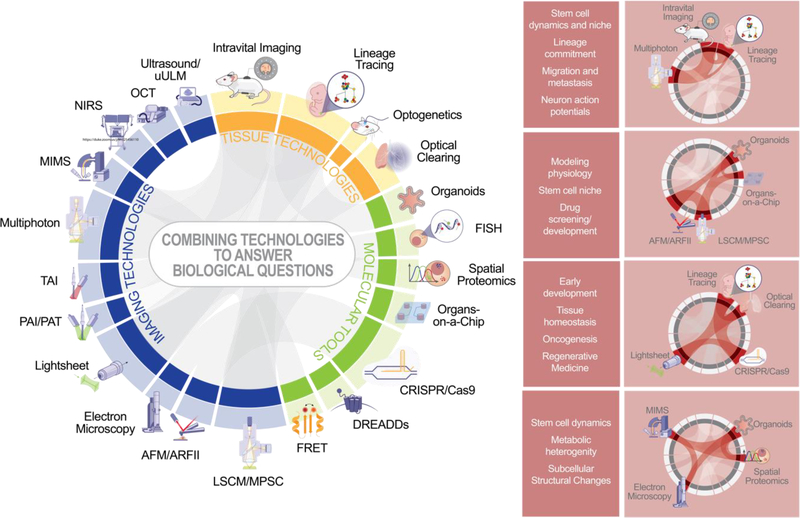
Regardless of the precise technology used to overcome challenges in acquiring relevant biological data, combinatorial approaches using imaging, molecular, and computational tools will be necessary to fully realize questions of interest. It is increasingly obvious that multimodal imaging and the integration of multiple forms of structural and functional data are necessary to fully understand both physiologic function and dysfunctional disease states (Celi et al., 2017; Falsafi et al., 2020; Martinez-Milla et al., 2020; Miyaoka and Lehnert, 2020; Ribeiro et al., 2017; Tian et al., 2019). Genomics, metabolomics, and other omics fields have provided dramatic insights into how and why we function as we do, but traditional omics methods lack spatial information and are not simultaneously monitored in conjunction with traditional imaging data and biometrics (Benson et al., 2019; Brazovskaja et al., 2019; Camsund et al., 2020; Madabhushi and Lee, 2016; Xie et al., 2020). Epigenetic modification, dynamic metabolic processes, and other omics features are not static nor homogeneous across cells and tissues within an organism. Several compelling tools such as ATAC-see, MERFISH, and RNAscope as well as new commercial technologies such as Nanostring’s GeoMx and 10X’s Visium hold great promise for identifying the spatial context for heterogeneity as identified by various omics tools (Barbash et al., 2019; Chen et al., 2016; Wang et al., 2019a). These tools provide both a means to understand functionality based on underlying organization, as well as allowing targeted tissue isolation and study, which is critically important to understand and capture rare cells, early oncogenic transformation, and other populations or phenomena which might otherwise be lost in bulk datasets. However, there are many data flows which still lack an integrated solution to explore spatial context. Furthermore, exploration of omics changes longitudinally remains a challenge.
Combining technologies and techniques which assess structural and functional data simultaneously, especially when used in conjunction with physiologic readouts, like those showcased in Figure 6, enables the best opportunity to robustly capture and describe biological phenomena. However, the resultant datasets will rely on improvement and expansion of analysis pipelines to make use of the combinatorial information. Machine learning and AI will likely play significant roles in addressing this important need, as the diversity of data becomes increasingly complex and diverse (de Haan et al., 2020; Madabhushi and Lee, 2016; Shi et al., 2020; Wang et al., 2019b). Exciting technologies in this space, such as Cell Painting, combine labeling protocols, automated imaging, and machine learning pipelines to provide high-throughput solutions to investigate biological questions (Bray et al., 2016). Others focus on utilizing clinical data to predict disease or treatment outcomes or expand our understanding of population biology (Byron et al., 2016; Madabhushi and Lee, 2016; Shi et al., 2020). Hybrid techniques like the use of decellularized organs to support organoids allow for investigation of harmonized data taken at the molecular, cellular, or tissue level, and implanting organoid derived tissues can allow for these to be further integrated with systemic outputs (Baptista et al., 2011; Giobbe et al., 2019; Tajima et al., 2019). Taken together, these new tools provide great potential for insight into stem cell biology. While these methods will require greater resources for data storage and processing, interdisciplinary cooperation between biologists, statisticians, and computer scientists will allow these powerful emerging tools to answer elusive biological questions and yield deeper insights into developmental and regenerative science.
Figure 6: Combinatorial approaches used to answer biological questions.
Through using different technologies in consort, it is possible to interrogate different biological functions at an unprecedented resolution.
Acknowledgments
We thank Dr. Michael Hancock for his review and comments. We thank Dr. Gabriele Bixel at the Max Planck Institute for Molecular Biomedicine (Münster, Germany), Dr. Max Nobis and Dr. Paul Timpson at the Garvan Institute of Medical Research (Darlinghurst, Australia), Haohua Tu and Stephen Boppart at the University of Illinois Biophotonics Imaging Laboratory (Urbana, IL), Al Johnson at the Duke University Center for In Vivo Microscopy (Durham, NC), and Junjie Yao at Duke University (Durham, NC) for providing previously unpublished images for our figures. The work was supported by NIH R35GM122465 (X.S.), DK119795 (X.S), NSFC81670468 (Q.H), 2017KJXX-43(Q.H), and 2018SF-208 (Q.H).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acar M, Kocherlakota KS, Murphy MM, Peyer JG, Oguro H, Inra CN, Jaiyeola C, Zhao Z, Luby-Phelps K, and Morrison SJ (2015). Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 526, 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrengues J, Shields MA, Ng D, Park CG, Ambrico A, Poindexter ME, Upadhyay P, Uyeminami DL, Pommier A, Küttner V, et al. (2018). Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science 361, eaao4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alieva M, Ritsma L, Giedt RJ, Weissleder R, and van Rheenen J (2014). Imaging windows for long-term intravital imaging: General overview and technical insights. Intravital 3, e29917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balan M, Trusohamn M, Ning FC, Jacob S, Pietras K, Eriksson U, Berggren PO, and Nyqvist D (2019). Noninvasive intravital high-resolution imaging of pancreatic neuroendocrine tumours. Sci Rep 9, 14636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista PM, Siddiqui MM, Lozier G, Rodriguez SR, Atala A, and Soker S (2011). The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology (Baltimore, Md) 53, 604–617. [DOI] [PubMed] [Google Scholar]
- Barbash S, Persson T, Lorenzen E, Kazmi MA, Huber T, and Sakmar TP (2019). Detection of concordance between transcriptional levels of GPCRs and receptor-activity-modifying proteins. iScience 11, 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedzhov I, and Zernicka-Goetz M (2014). Self-Organizing Properties of Mouse Pluripotent Cells Initiate Morphogenesis upon Implantation. Cell 156, 1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin L Franc, P.D.A., Carina Mari, Bruce H Hasegawa Small-Animal SPECT and SPECT/CT: Important Tools for Preclinical Investigation. [DOI] [PubMed]
- Benninger RK, and Piston DW (2013). Two-photon excitation microscopy for the study of living cells and tissues. Curr Protoc Cell Biol Chapter 4, Unit 4 11 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson S, Fernandez A, Barth ND, de Moliner F, Horrocks MH, Herrington CS, Abad JL, Delgado A, Kelly L, Chang Z, et al. (2019). SCOTfluors: Small, Conjugatable, Orthogonal, and Tunable Fluorophores for In Vivo Imaging of Cell Metabolism. Angew Chem Int Ed Engl 58, 6911–6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier-Latmani J, and Petrova TV (2016). High-resolution 3D analysis of mouse small-intestinal stroma. Nat Protoc 11, 1617–1629. [DOI] [PubMed] [Google Scholar]
- Bixel MG, Kusumbe AP, Ramasamy SK, Sivaraj KK, Butz S, Vestweber D, and Adams RH (2017). Flow dynamics and HSPC homing in bone marrow microvessels. Cell reports 18, 1804–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner F, Fellus-Alyagor L, Kalchenko V, Shinar S, and Neeman M (2015). A Novel Intravital Imaging Window for Longitudinal Microscopy of the Mouse Ovary. Sci Rep 5, 12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boj SF, Hwang C-I, Baker LA, Chio IIC, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, and Spector MS (2015). Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnardel J, T’Jonck W, Gaublomme D, Browaeys R, Scott CL, Martens L, Vanneste B, De Prijck S, Nedospasov SA, Kremer A, et al. (2019). Stellate Cells, Hepatocytes, and Endothelial Cells Imprint the Kupffer Cell Identity on Monocytes Colonizing the Liver Macrophage Niche. Immunity 51, 638–654 e639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booij TH, Price LS, and Danen EHJ (2019). 3D Cell-Based Assays for Drug Screens: Challenges in Imaging, Image Analysis, and High-Content Analysis. SLAS Discov 24, 615–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boppart SA, You S, Li L, Chen J, and Tu H (2019). Simultaneous label-free autofluorescence-multiharmonic microscopy and beyond. APL Photonics 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossolani GDP, Pintelon I, Detrez JD, Buckinx R, Thys S, Zanoni JN, De Vos WH, and Timmermans JP (2019). Comparative analysis reveals Ce3D as optimal clearing method for in toto imaging of the mouse intestine. Neurogastroenterol Motil 31, e13560. [DOI] [PubMed] [Google Scholar]
- Bray M-A, Singh S, Han H, Davis CT, Borgeson B, Hartland C, Kost-Alimova M, Gustafsdottir SM, Gibson CC, and Carpenter AE (2016). Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nature Protocols 11, 1757–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazovskaja A, Treutlein B, and Camp JG (2019). High-throughput single-cell transcriptomics on organoids. Curr Opin Biotechnol 55, 167–171. [DOI] [PubMed] [Google Scholar]
- Brockway NL, Cook ZT, O’Gallagher MJ, Tobias ZJC, Gedi M, Carey KM, Unni VK, Pan YA, Metz MR, and Weissman TA (2019). Multicolor lineage tracing using in vivo time-lapse imaging reveals coordinated death of clonally related cells in the developing vertebrate brain. Dev Biol 453, 130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarró LM, Bradshaw CR, Allen GE, Arnes-Benito R, Sidorova O, and Gaspersz MP (2017). Human primary liver cancer–derived organoid cultures for disease modeling and drug screening. Nature medicine 23, 1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruens L, and Snippert HJ (2017). Expanding the Tissue Toolbox: Deriving Colon Tissue from Human Pluripotent Stem Cells. Cell Stem Cell 21, 3–5. [DOI] [PubMed] [Google Scholar]
- Buck AK, Nekolla S, Ziegler S, Beer A, Krause BJ, Herrmann K, Scheidhauer K, Wester HJ, Rummeny EJ, Schwaiger M, et al. (2008). SPECT/CT. Journal of nuclear medicine : official publication, Society of Nuclear Medicine 49, 1305–1319. [DOI] [PubMed] [Google Scholar]
- Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, and Craig DW (2016). Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genet 17, 257–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Cohen KB, Luo T, Lichtman JW, and Sanes JR (2013). Improved tools for the Brainbow toolbox. Nature Methods 10, 540–547. [PubMed] [Google Scholar]
- Calandrini C, Schutgens F, Oka R, Margaritis T, Candelli T, Mathijsen L, Ammerlaan C, van Ineveld RL, Derakhshan S, and de Haan S (2020). An organoid biobank for childhood kidney cancers that captures disease and tissue heterogeneity. Nature Communications 11, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camsund D, Lawson MJ, Larsson J, Jones D, Zikrin S, Fange D, and Elf J (2020). Time-resolved imaging-based CRISPRi screening. Nat Methods 17, 86–92. [DOI] [PubMed] [Google Scholar]
- Cassol F, Portal L, Richelme S, Dupont M, Boursier Y, Arechederra M, Auphan-Anezin N, Chasson L, Laprie C, Fernandez S, et al. (2019). Tracking Dynamics of Spontaneous Tumors in Mice Using Photon-Counting Computed Tomography. iScience 21, 68–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celi S, Martini N, Pastormerlo LE, Positano V, and Berti S (2017). Multimodality Imaging for Interventional Cardiology. Curr Pharm Des 23, 3285–3300. [DOI] [PubMed] [Google Scholar]
- Chatzeli L, and Simons BD (2020). Tracing the Dynamics of Stem Cell Fate. Cold Spring Harb Perspect Biol 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Shen Y, Draper W, Buenrostro JD, Litzenburger U, Cho SW, Satpathy AT, Carter AC, Ghosh RP, East-Seletsky A, et al. (2016). ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat Methods 13, 1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhablani J, Sharma A, Goud A, Peguda HK, Rao HL, Begum VU, and Barteselli G (2015). Neurodegeneration in Type 2 Diabetes: Evidence From Spectral-Domain Optical Coherence Tomography. Invest Ophthalmol Vis Sci 56, 6333–6338. [DOI] [PubMed] [Google Scholar]
- Christodoulou C, Spencer JA, Yeh SA, Turcotte R, Kokkaliaris KD, Panero R, Ramos A, Guo G, Seyedhassantehrani N, Esipova TV, et al. (2020). Live-animal imaging of native haematopoietic stem and progenitor cells. Nature 578, 278–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu LA, Lu CH, Yang SM, Liu YT, Feng KL, Tsai YC, Chang WK, Wang WC, Chang SW, Chen P, et al. (2019). Rapid single-wavelength lightsheet localization microscopy for clarified tissue. Nat Commun 10, 4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciobanu L, and Pennington CH (2004). 3D micron-scale MRI of single biological cells. Solid State Nucl Magn Reson 25, 138–141. [DOI] [PubMed] [Google Scholar]
- Combs CA, and Shroff H (2017). Fluorescence Microscopy: A Concise Guide to Current Imaging Methods. Curr Protoc Neurosci 79, 2 1 1–2 1 25. [DOI] [PubMed] [Google Scholar]
- Coste A, Oktay MH, Condeelis JS, and Entenberg D (2020). Intravital Imaging Techniques for Biomedical and Clinical Research. Cytometry Part A : the journal of the International Society for Analytical Cytology 97, 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo D, Avolio R, Calice G, Laezza C, Paladino S, Navarra G, Maddalena F, Crispo F, Pagano C, Bifulco M, et al. (2020). Cholesterol Homeostasis Modulates Platinum Sensitivity in Human Ovarian Cancer. Cells 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Maschio M, Donovan JC, Helmbrecht TO, and Baier H (2017). Linking Neurons to Network Function and Behavior by Two-Photon Holographic Optogenetics and Volumetric Imaging. Neuron 94, 774–789 e775. [DOI] [PubMed] [Google Scholar]
- Dana H, Sun Y, Mohar B, Hulse BK, Kerlin AM, Hasseman JP, Tsegaye G, Tsang A, Wong A, Patel R, et al. (2019). High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nature methods 16, 649–657. [DOI] [PubMed] [Google Scholar]
- De Feyter HM, Behar KL, Corbin ZA, Fulbright RK, Brown PB, McIntyre S, Nixon TW, Rothman DL, and de Graaf RA (2018). Deuterium metabolic imaging (DMI) for MRI-based 3D mapping of metabolism in vivo. Sci Adv 4, eaat7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan K, Rivenson Y, Wu Y, and Ozcan A (2020). Deep-Learning-Based Image Reconstruction and Enhancement in Optical Microscopy. Proceedings of the IEEE 108, 30–50. [Google Scholar]
- De Niz M, Meehan GR, and Tavares J (2019). Intravital microscopy: Imaging host-parasite interactions in lymphoid organs. Cell Microbiol 21, e13117. [DOI] [PubMed] [Google Scholar]
- de Witte CJ, Valle-Inclan JE, Hami N, Lõhmussaar K, Kopper O, Vreuls CPH, Jonges GN, van Diest P, Nguyen L, and Clevers H (2020). Patient-derived ovarian cancer organoids mimic clinical response and exhibit heterogeneous inter-and intrapatient drug responses. Cell reports 31, 107762. [DOI] [PubMed] [Google Scholar]
- Deglincerti A, Croft GF, Pietila LN, Zernicka-Goetz M, Siggia ED, and Brivanlou AH (2016). Self-organization of the in vitro attached human embryo. Nature 533, 251–254. [DOI] [PubMed] [Google Scholar]
- Deisseroth K (2011). Optogenetics. Nat Methods 8, 26–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers JF, Alieva M, Wellens LM, Ariese HCR, Jamieson PR, Vonk AM, Amatngalim GD, Hu H, Oost KC, Snippert HJG, et al. (2019). High-resolution 3D imaging of fixed and cleared organoids. Nat Protoc 14, 1756–1771. [DOI] [PubMed] [Google Scholar]
- Dekkers JF, Whittle JR, Vaillant F, Chen H-R, Dawson C, Liu K, Geurts MH, Herold MJ, Clevers H, and Lindeman GJ (2020). Modeling Breast Cancer Using CRISPR-Cas9–Mediated Engineering of Human Breast Organoids. JNCI: Journal of the National Cancer Institute 112, 540–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschene ER, Myung P, Rompolas P, Zito G, Sun TY, Taketo MM, Saotome I, and Greco V (2014). β-Catenin activation regulates tissue growth non-cell autonomously in the hair stem cell niche. Science 343, 1353–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, and Clevers H (2018). Organoids in cancer research. Nature Reviews Cancer 18, 407–418. [DOI] [PubMed] [Google Scholar]
- Drost J, Van Jaarsveld RH, Ponsioen B, Zimberlin C, Van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, and Begthel H (2015). Sequential cancer mutations in cultured human intestinal stem cells. Nature 521, 43–47. [DOI] [PubMed] [Google Scholar]
- Dubois A, Levecq O, Azimani H, Siret D, Barut A, Suppa M, Del Marmol V, Malvehy J, Cinotti E, Rubegni P, et al. (2018). Line-field confocal optical coherence tomography for high-resolution noninvasive imaging of skin tumors. J Biomed Opt 23, 1–9. [DOI] [PubMed] [Google Scholar]
- Dutta D, Heo I, and Clevers H (2017). Disease modeling in stem cell-derived 3D organoid systems. Trends in molecular medicine 23, 393–410. [DOI] [PubMed] [Google Scholar]
- Early JJ, Cole KL, Williamson JM, Swire M, Kamadurai H, Muskavitch M, and Lyons DA (2018). An automated high-resolution in vivo screen in zebrafish to identify chemical regulators of myelination. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, and Discher DE (2006). Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689. [DOI] [PubMed] [Google Scholar]
- Entenberg D, Voiculescu S, Guo P, Borriello L, Wang Y, Karagiannis GS, Jones J, Baccay F, Oktay M, and Condeelis J (2018). A permanent window for the murine lung enables high-resolution imaging of cancer metastasis. Nat Methods 15, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico C, Pierre J, Pezet S, Desailly Y, Lenkei Z, Couture O, and Tanter M (2015). Ultrafast ultrasound localization microscopy for deep super-resolution vascular imaging. Nature 527, 499–502. [DOI] [PubMed] [Google Scholar]
- Falsafi SR, Rostamabadi H, Assadpour E, and Jafari SM (2020). Morphology and microstructural analysis of bioactive-loaded micro/nanocarriers via microscopy techniques; CLSM/SEM/TEM/AFM. Adv Colloid Interface Sci 280, 102166. [DOI] [PubMed] [Google Scholar]
- Feng Y, Tamadon A, and Hsueh AJW (2018). Imaging the ovary. Reprod Biomed Online 36, 584–593. [DOI] [PubMed] [Google Scholar]
- Fernandez-Sanchez ME, Barbier S, Whitehead J, Béalle G, Michel A, Latorre-Ossa H, Rey C, Fouassier L, Claperon A, and Brullé L (2015). Mechanical induction of the tumorigenic β-catenin pathway by tumour growth pressure. Nature 523, 92–95. [DOI] [PubMed] [Google Scholar]
- Fox RG, Lytle NK, Jaquish DV, Park FD, Ito T, Bajaj J, Koechlein CS, Zimdahl B, Yano M, Kopp J, et al. (2016). Image-based detection and targeting of therapy resistance in pancreatic adenocarcinoma. Nature 534, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fretaud M, Riviere L, Job E, Gay S, Lareyre JJ, Joly JS, Affaticati P, and Thermes V (2017). High-resolution 3D imaging of whole organ after clearing: taking a new look at the zebrafish testis. Sci Rep 7, 43012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli A, Oost KC, Kester L, Morgner J, Bornes L, Bruens L, Spaargaren L, Azkanaz M, Schelfhorst T, Beerling E, et al. (2020). Plasticity of Lgr5-Negative Cancer Cells Drives Metastasis in Colorectal Cancer. Cell Stem Cell 26, 569–578.e567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh K, Wu C, O’Rourke KP, Szeglin BC, Zheng Y, Sauvé C-EG, Adileh M, Wasserman I, Marco MR, and Kim AS (2019). A rectal cancer organoid platform to study individual responses to chemoradiation. Nature Medicine 25, 1607–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh K, Wu C, O’Rourke KP, Szeglin BC, Zheng Y, Sauvé CEG, Adileh M, Wasserman I, Marco MR, Kim AS and Shady M (2019). A rectal cancer organoid platform to study individual responses to chemoradiation. Nature medicine 25, 1607–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, and Arora VK (2014). Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gericke M, Weyer U, Braune J, Bechmann I, and Eilers J (2015). A method for long-term live imaging of tissue macrophages in adipose tissue explants. Am J Physiol Endocrinol Metab 308, E1023–1033. [DOI] [PubMed] [Google Scholar]
- Germain RN, Robey EA, and Cahalan MD (2012). A decade of imaging cellular motility and interaction dynamics in the immune system. Science 336, 1676–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giobbe GG, Crowley C, Luni C, Campinoti S, Khedr M, Kretzschmar K, De Santis MM, Zambaiti E, Michielin F, Meran L, et al. (2019). Extracellular matrix hydrogel derived from decellularized tissues enables endodermal organoid culture. Nature Communications 10, 5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, Clevers H, and Lutolf MP (2016). Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564. [DOI] [PubMed] [Google Scholar]
- Golestani R, Wu C, Tio RA, Zeebregts CJ, Petrov AD, Beekman FJ, Dierckx RA, Boersma HH, and Slart RH (2010). Small-animal SPECT and SPECT/CT: application in cardiovascular research. Eur J Nucl Med Mol Imaging 37, 1766–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y-N, Guy C, Olauson H, Becker JU, Yang M, Fitzgerald P, Linkermann A, and Green DR (2017). ESCRT-III acts downstream of MLKL to regulate necroptotic cell death and its consequences. Cell 169, 286–300. e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf BW, Chaney EJ, Marjanovic M, Adie SG, De Lisio M, Valero MC, Boppart MD, and Boppart SA (2013). Long-term time-lapse multimodal intravital imaging of regeneration and bone-marrow-derived cell dynamics in skin. Technology 1, 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange C, Tapparo M, Bruno S, Chatterjee D, Quesenberry PJ, Tetta C, and Camussi G (2014). Biodistribution of mesenchymal stem cell-derived extracellular vesicles in a model of acute kidney injury monitored by optical imaging. Int J Mol Med 33, 1055–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guernet A, Mungamuri SK, Cartier D, Sachidanandam R, Jayaprakash A, Adriouch S, Vezain M, Charbonnier F, Rohkin G, Coutant S, et al. (2016). CRISPR-Barcoding for Intratumor Genetic Heterogeneity Modeling and Functional Analysis of Oncogenic Driver Mutations. Mol Cell 63, 526–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin LA, Yizhar O, Berndt A, Sohal VS, Deisseroth K, and Hegemann P (2010). Ultrafast optogenetic control. Nat Neurosci 13, 387–392. [DOI] [PubMed] [Google Scholar]
- Gupta K, Li Q, Fan JJ, Fong ELS, Song Z, Mo S, Tang H, Ng IC, Ng CW, and Pawijit P (2017). Actomyosin contractility drives bile regurgitation as an early response during obstructive cholestasis. Journal of Hepatology 66, 1231–1240. [DOI] [PubMed] [Google Scholar]
- Haddrick M, and Simpson PB (2019). Organ-on-a-chip technology: turning its potential for clinical benefit into reality. Drug Discov Today 24, 1217–1223. [DOI] [PubMed] [Google Scholar]
- Han S, Fink J, Jörg DJ, Lee E, Yum MK, Chatzeli L, Merker SR, Josserand M, Trendafilova T, Andersson-Rolf A, et al. (2019). Defining the Identity and Dynamics of Adult Gastric Isthmus Stem Cells. Cell Stem Cell 25, 342–356.e347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare DJ, Jones MW, Wimmer VC, Jenkins NL, de Jonge MD, Bush AI, and McColl G (2016). High-resolution complementary chemical imaging of bio-elements in Caenorhabditis elegans. Metallomics 8, 156–160. [DOI] [PubMed] [Google Scholar]
- He S, Tian Y, Feng S, Wu Y, Shen X, Chen K, He Y, Sun Q, Li X, Xu J, et al. (2020). In vivo single-cell lineage tracing in zebrafish using high-resolution infrared laser-mediated gene induction microscopy. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann A, Liewald JF, and Gottschalk A (2015). A photosensitive degron enables acute light-induced protein degradation in the nervous system. Curr Biol 25, R749–750. [DOI] [PubMed] [Google Scholar]
- Hildebrand DGC, Cicconet M, Torres RM, Choi W, Quan TM, Moon J, Wetzel AW, Scott Champion A, Graham BJ, Randlett O, et al. (2017). Whole-brain serial-section electron microscopy in larval zebrafish. Nature 545, 345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DP, Shtengel G, Xu CS, Campbell KR, Freeman M, Wang L, Milkie DE, Pasolli HA, Iyer N, Bogovic JA, et al. (2020). Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Gehart H, Artegiani B, LÖpez-Iglesias C, Dekkers F, Basak O, van Es J, de Sousa Lopes SMC, Begthel H, and Korving J (2018). Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 175, 1591–1606. e1519. [DOI] [PubMed] [Google Scholar]
- Huang L, Holtzinger A, Jagan I, BeGora M, Lohse I, Ngai N, Nostro C, Wang R, Muthuswamy LB, and Crawford HC (2015). Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell–and patient-derived tumor organoids. Nature medicine 21, 1364–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Cohen MA, Alsina FC, Devlin G, Garrett A, McKey J, Havlik P, Rakhilin N, Wang E, Xiang K, et al. (2020). Intravital imaging of mouse embryos. Science 368, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Salinas RD, Zhang DY, Nguyen PT, Schnoll JG, Wong SZH, Thokala R, Sheikh S, Saxena D, and Prokop S (2020). A patient-derived glioblastoma organoid model and biobank recapitulates inter-and intra-tumoral heterogeneity. Cell 180, 188–204. e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GA, Cofer GP, Gewalt SL, and Hedlund LW (2002). Morphologic phenotyping with MR microscopy: the visible mouse. Radiology 222, 789–793. [DOI] [PubMed] [Google Scholar]
- Johnson J, Sharick JT, Skala MC, and Li L (2020). Sample preparation strategies for high-throughput mass spectrometry imaging of primary tumor organoids. J Mass Spectrom 55, e4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, Van Boxtel R, Wongvipat J, Dowling CM, Gao D, Begthel H, and Sachs N (2014). Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159, 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebschull JM, and Zador AM (2018). Cellular barcoding: lineage tracing, screening and beyond. Nat Methods 15, 871–879. [DOI] [PubMed] [Google Scholar]
- Kerschensteiner M, Schwab ME, Lichtman JW, and Misgeld T (2005). In vivo imaging of axonal degeneration and regeneration in the injured spinal cord. Nat Med 11, 572–577. [DOI] [PubMed] [Google Scholar]
- Kim DH, Kim J, Marques JC, Grama A, Hildebrand DGC, Gu W, Li JM, and Robson DN (2017a). Pan-neuronal calcium imaging with cellular resolution in freely swimming zebrafish. Nat Methods 14, 1107–1114. [DOI] [PubMed] [Google Scholar]
- Kim K, Yu SY, Kwak HW, and Kim ES (2016). Retinal Neurodegeneration Associated With Peripheral Nerve Conduction and Autonomic Nerve Function in Diabetic Patients. Am J Ophthalmol 170, 15–24. [DOI] [PubMed] [Google Scholar]
- Kim M, Mun H, Sung CO, Cho EJ, Jeon H-J, Chun S-M, Shin TH, Jeong GS, Kim DK, and Choi EK (2019). Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nature communications 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Prochazka J, and Bush JO (2017b). Live Imaging of Mouse Secondary Palate Fusion. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo T, Terai K, Horita S, Nomura N, Sumiyama K, Togashi K, Iwata S, and Matsuda M (2019). FRET-assisted photoactivation of flavoproteins for in vivo two-photon optogenetics. Nat Methods 16, 1029–1036. [DOI] [PubMed] [Google Scholar]
- Kitamura T, Pollard JW, and Vendrell M (2017). Optical Windows for Imaging the Metastatic Tumour Microenvironment in vivo. Trends Biotechnol 35, 5–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, and Simons BD (2011). Universal patterns of stem cell fate in cycling adult tissues. Development 138, 3103–3111. [DOI] [PubMed] [Google Scholar]
- Koechlein CS, Harris JR, Lee TK, Weeks J, Fox RG, Zimdahl B, Ito T, Blevins A, Jung S-H, Chute JP, et al. (2016). High-resolution imaging and computational analysis of haematopoietic cell dynamics in vivo. Nature Communications 7, 12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S, Micci MA, Leser J, Shin C, Tang SC, Fu YY, Liu L, Li Q, Saha M, Li C, et al. (2017). Adult enteric nervous system in health is maintained by a dynamic balance between neuronal apoptosis and neurogenesis. Proc Natl Acad Sci U S A 114, E3709–E3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry BP, Palanki R, Dyulgyarov N, Hartsough LA, and Tabor JJ (2018). Phosphatase activity tunes two-component system sensor detection threshold. Nature communications 9, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenberg T, Brand M, and Cooper MS (2003). Imaging brain development and organogenesis in zebrafish using immobilized embryonic explants. Dev Dyn 228, 464–474. [DOI] [PubMed] [Google Scholar]
- Lee GY, Kenny PA, Lee EH, and Bissell MJ (2007). Three-dimensional culture models of normal and malignant breast epithelial cells. Nature methods 4, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HM, Giguere PM, and Roth BL (2014). DREADDs: novel tools for drug discovery and development. Drug Discov Today 19, 469–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R, Turczyk BM, Yang JL, Lee HS, Aach J, et al. (2015). Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nature Protocols 10, 442–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Hu W, Matulay JT, Silva MV, Owczarek TB, Kim K, Chua CW, Barlow LJ, Kandoth C, and Williams AB (2018). Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell 173, 515–528. e517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelkes E, Headley MB, Thornton EE, Looney MR, and Krummel MF (2014). The spatiotemporal cellular dynamics of lung immunity. Trends in immunology 35, 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JL, Goh CC, Keeble JL, Qin JS, Roediger B, Jain R, Wang Y, Chew WK, Weninger W, and Ng LG (2012). Intravital multiphoton imaging of immune responses in the mouse ear skin. Nat Protoc 7, 221–234. [DOI] [PubMed] [Google Scholar]
- Lindeboom RG, van Voorthuijsen L, Oost KC, Rodríguez-Colman MJ, Luna-Velez MV, Furlan C, Baraille F, Jansen PW, Ribeiro A, and Burgering BM (2018). Integrative multi-omics analysis of intestinal organoid differentiation. Molecular systems biology 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TL, Upadhyayula S, Milkie DE, Singh V, Wang K, Swinburne IA, Mosaliganti KR, Collins ZM, Hiscock TW, Shea J, et al. (2018). Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Celso C, Fleming HE, Wu JW, Zhao CX, Miake-Lye S, Fujisaki J, Côté D, Rowe DW, Lin CP, and Scadden DT (2009). Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature 457, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckner M, Burgold S, Filser S, Scheungrab M, Niyaz Y, Hummel E, Wanner G, and Herms J (2018). Label-free 3D-CLEM using endogenous tissue landmarks. Iscience 6, 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker KE, Pata P, Shemiakina II, Pereverzeva A, Stacer AC, Shcherbo DS, Pletnev VZ, Skolnaja M, Lukyanov KA, Luker GD, et al. (2015). Comparative study reveals better far-red fluorescent protein for whole body imaging. Sci Rep 5, 10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle NK, Ferguson LP, Rajbhandari N, Gilroy K, Fox RG, Deshpande A, Schürch CM, Hamilton M, Robertson N, and Lin W (2019). A multiscale map of the stem cell state in pancreatic adenocarcinoma. Cell 177, 572–586. e522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madabhushi A, and Lee G (2016). Image analysis and machine learning in digital pathology: Challenges and opportunities. Med Image Anal 33, 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenhoudt N, Defraye C, Boretto M, Jan Z, Heremans R, Boeckx B, Hermans F, Arijs I, Cox B, and Van Nieuwenhuysen E (2020). Developing Organoids from Ovarian Cancer as Experimental and Preclinical Models. Stem cell reports. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maronpot RR, Sills RC, and Johnson GA (2004). Applications of magnetic resonance microscopy. Toxicol Pathol 32 Suppl 2, 42–48. [DOI] [PubMed] [Google Scholar]
- Marsh E, Gonzalez DG, Lathrop EA, Boucher J, and Greco V (2018). Positional Stability and Membrane Occupancy Define Skin Fibroblast Homeostasis In Vivo. Cell 175, 1620–1633 e1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Milla J, Galan-Arriola C, Carnero M, Cobiella J, Perez-Camargo D, Bautista-Hernandez V, Rigol M, Solanes N, Villena-Gutierrez R, Lobo M, et al. (2020). Translational large animal model of hibernating myocardium: characterization by serial multimodal imaging. Basic Res Cardiol 115, 33. [DOI] [PubMed] [Google Scholar]
- Martynowych D, Veysset D, Maznev AA, Sun Y, Kooi SE, and Nelson KA (2020). Multi-frame interferometric imaging with a femtosecond stroboscopic pulse train for observing irreversible phenomena. Rev Sci Instrum 91, 033711. [DOI] [PubMed] [Google Scholar]
- Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, and Sato T (2015). Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med 21, 256–262. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Wakefield L, Tarlow BD, and Grompe M (2020). In Vivo Lineage Tracing of Polyploid Hepatocytes Reveals Extensive Proliferation during Liver Regeneration. Cell Stem Cell 26, 34–47 e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura R, Miyagawa S, Fukushima S, Goto T, Harada A, Shimozaki Y, Yamaki K, Sanami S, Kikuta J, Ishii M, et al. (2018). Intravital imaging with two-photon microscopy reveals cellular dynamics in the ischeamia-reperfused rat heart. Sci Rep 8, 15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley KB, Hawkins F, Serra M, Thomas DC, Jacob A, and Kotton DN (2017). Efficient derivation of functional human airway epithelium from pluripotent stem cells via temporal regulation of Wnt signaling. Cell stem cell 20, 844–857. e846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken KW, Catá EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai Y-H, Mayhew CN, Spence JR, and Zavros Y (2014). Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDole K, Guignard L, Amat F, Berger A, Malandain G, Royer LA, Turaga SC, Branson K, and Keller PJ (2018). In Toto Imaging and Reconstruction of Post-Implantation Mouse Development at the Single-Cell Level. Cell 175, 859–876 e833. [DOI] [PubMed] [Google Scholar]
- McKey J, Cameron LA, Lewis D, Batchvarov IS, and Capel B (2020). Combined iDISCO and CUBIC tissue clearing and lightsheet microscopy for in toto analysis of the adult mouse ovarydagger. Biol Reprod 102, 1080–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Stuurman N, Royer LA, Schartner C, Castillo-Azofeifa D, Delling M, Klein OD, and Vale RD (2018). Cellular aspect ratio and cell division mechanics underlie the patterning of cell progeny in diverse mammalian epithelia. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer M, Rehbach K, Brunner JW, Classen JA, Lammertse HCA, van Linge LA, Schut D, Krutenko T, Hebisch M, Cornelisse LN, et al. (2019). A Single-Cell Model for Synaptic Transmission and Plasticity in Human iPSC-Derived Neurons. Cell Rep 27, 2199–2211 e2196. [DOI] [PubMed] [Google Scholar]
- Mesa KR, Kawaguchi K, Cockburn K, Gonzalez D, Boucher J, Xin T, Klein AM, and Greco V (2018). Homeostatic Epidermal Stem Cell Self-Renewal Is Driven by Local Differentiation. Cell Stem Cell 23, 677–686.e674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa KR, Rompolas P, Zito G, Myung P, Sun TY, Brown S, Gonzalez DG, Blagoev KB, Haberman AM, and Greco V (2015). Niche-induced cell death and epithelial phagocytosis regulate hair follicle stem cell pool. Nature 522, 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midde K, Sun N, Rohena C, Joosen L, Dhillon H, and Ghosh P (2018). Single-cell imaging of metastatic potential of cancer cells. iScience 10, 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, Dye BR, Ferrer-Torres D, Hill DR, Overeem AW, Shea LD, and Spence JR (2019). Generation of lung organoids from human pluripotent stem cells in vitro. Nat Protoc 14, 518–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka RS, and Lehnert A (2020). Small animal PET: a review of what we have done and where we are going. Phys Med Biol. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, and Scadden DT (2014). The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Múnera JO, Sundaram N, Rankin SA, Hill D, Watson C, Mahe M, Vallance JE, Shroyer NF, Sinagoga KL, and Zarzoso-Lacoste A (2017). Differentiation of human pluripotent stem cells into colonic organoids via transient activation of BMP signaling. Cell stem cell 21, 51–64. e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz WA, and Trainor PA (2019). Mouse Embryo Culture for the Study of Neural Crest Cells. Methods Mol Biol 1976, 107–119. [DOI] [PubMed] [Google Scholar]
- Nakai J, Ohkura M, and Imoto K (2001). A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nature biotechnology 19, 137–141. [DOI] [PubMed] [Google Scholar]
- Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, Saito K, Yonemura S, Eiraku M, and Sasai Y (2012). Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell stem cell 10, 771–785. [DOI] [PubMed] [Google Scholar]
- Nakasone ES, Askautrud HA, Kees T, Park J-H, Plaks V, Ewald AJ, Fein M, Rasch MG, Tan Y-X, and Qiu J (2012). Imaging tumor-stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer cell 21, 488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SS, Saeb-Parsy K, Blackford SJI, Segal JM, Serra MP, Horcas-Lopez M, No DY, Mastoridis S, Jassem W, Frank CW, et al. (2018). Human iPS derived progenitors bioengineered into liver organoids using an inverted colloidal crystal poly (ethylene glycol) scaffold. Biomaterials 182, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale K (2011). Acoustic Radiation Force Impulse (ARFI) Imaging: a Review. Curr Med Imaging Rev 7, 328–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirmalraj P, Lehner R, Thompson D, Rothen-Rutishauser B, and Mayer M (2018). Subcellular Imaging of Liquid Silicone Coated-Intestinal Epithelial Cells. Sci Rep 8, 10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Ishiwata H, Sakuragi Y, Hayashi Y, Fukuda A, and Hisatake K (2019). Live-cell imaging of subcellular structures for quantitative evaluation of pluripotent stem cells. Sci Rep 9, 1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobis M, Herrmann D, Warren SC, Kadir S, Leung W, Killen M, Magenau A, Stevenson D, Lucas MC, and Reischmann N (2017). A RhoA-FRET biosensor mouse for intravital imaging in normal tissue homeostasis and disease contexts. Cell reports 21, 274–288. [DOI] [PubMed] [Google Scholar]
- Nozaki K, Mochizuki W, Matsumoto Y, Matsumoto T, Fukuda M, Mizutani T, Watanabe M, and Nakamura T (2016). Co-culture with intestinal epithelial organoids allows efficient expansion and motility analysis of intraepithelial lymphocytes. Journal of gastroenterology 51, 206–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernier K, Cebrian-Silla A, Thomson M, Parraguez JI, Anderson R, Guinto C, Rodas Rodriguez J, Garcia-Verdugo JM, and Alvarez-Buylla A (2018). Adult Neurogenesis Is Sustained by Symmetric Self-Renewal and Differentiation. Cell Stem Cell 22, 221–234.e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ombrato L, Nolan E, Kurelac I, Mavousian A, Bridgeman VL, Heinze I, Chakravarty P, Horswell S, Gonzalez-Gualda E, and Matacchione G (2019). Metastatic-niche labelling reveals parenchymal cells with stem features. Nature 572, 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooft SN, Weeber F, Dijkstra KK, McLean CM, Kaing S, van Werkhoven E, Schipper L, Hoes L, Vis DJ, and van de Haar J (2019). Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Science Translational Medicine 11, eaay2574. [DOI] [PubMed] [Google Scholar]
- Pandit P, Johnston SM, Qi Y, Story J, Nelson R, and Johnson GA (2013). The utility of micro-CT and MRI in the assessment of longitudinal growth of liver metastases in a preclinical model of colon carcinoma. Academic radiology 20, 430–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazis P, and Supatto W (2014). Advances in whole-embryo imaging: a quantitative transition is underway. Nat Rev Mol Cell Biol 15, 327–339. [DOI] [PubMed] [Google Scholar]
- Park I, Hong S, Hwang Y, and Kim P (2020). A Novel Pancreatic Imaging Window for Stabilized Longitudinal In Vivo Observation of Pancreatic Islets in Murine Model. Diabetes Metab J 44, 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Gonzalez DG, Guirao B, Boucher JD, Cockburn K, Marsh ED, Mesa KR, Brown S, Rompolas P, Haberman AM, et al. (2017). Tissue-scale coordination of cellular behaviour promotes epidermal wound repair in live mice. Nat Cell Biol 19, 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SE, Georgescu A, and Huh D (2019). Organoids-on-a-chip. Science 364, 960–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca SP (2019). Assembling human brain organoids. Science 363, 126–127. [DOI] [PubMed] [Google Scholar]
- Pathak GP, Vrana JD, and Tucker CL (2013). Optogenetic control of cell function using engineered photoreceptors. Biol Cell 105, 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickar-Oliver A, and Gersbach CA (2019). The next generation of CRISPR–Cas technologies and applications. Nature Reviews Molecular Cell Biology 20, 490–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilz GA, Bottes S, Betizeau M, Jorg DJ, Carta S, Simons BD, Helmchen F, and Jessberger S (2018). Live imaging of neurogenesis in the adult mouse hippocampus. Science 359, 658–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho S, and Frenette PS (2019). Haematopoietic stem cell activity and interactions with the niche. Nat Rev Mol Cell Biol 20, 303–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podoleanu AG (2012). Optical coherence tomography. J Microsc 247, 209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue BW, Leblond F, Krishnaswamy V, and Paulsen KD (2010). Radiologic and near-infrared/optical spectroscopic imaging: where is the synergy? AJR Am J Roentgenol 195, 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Alifu N, Lam JWY, Cui Y, Su H, Liang G, Qian J, and Tang BZ (2020). Facile Synthesis of Efficient Luminogens with AIE Features for Three-Photon Fluorescence Imaging of the Brain through the Intact Skull. Advanced Materials. [DOI] [PubMed] [Google Scholar]
- Rakhilin N, Barth B, Choi J, Munoz NL, Kulkarni S, Jones JS, Small DM, Cheng YT, Cao Y, LaVinka C, et al. (2016). Simultaneous optical and electrical in vivo analysis of the enteric nervous system. Nat Commun 7, 11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhilin N, Garrett A, Eom CY, Chavez KR, Small DM, Daniel AR, Kaelberer MM, Mejooli MA, Huang Q, Ding S, et al. (2019). An intravital window to image the colon in real time. Nat Commun 10, 5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakotoson I, Delhomme B, Djian P, Deeg A, Brunstein M, Seebacher C, Uhl R, Ricard C, and Oheim M (2019). Fast 3-D Imaging of Brain Organoids With a New Single-Objective Planar-Illumination Two-Photon Microscope. Front Neuroanat 13, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redchuk TA, Kaberniuk AA, and Verkhusha VV (2018). Near-infrared light-controlled systems for gene transcription regulation, protein targeting and spectral multiplexing. Nature protocols 13, 1121–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JA, Palmer X-L, Mollica PA, Northam N, Sachs PC, and Bruno RD (2019). A 3D bioprinter platform for mechanistic analysis of tumoroids and chimeric mammary organoids. Scientific reports 9, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissaus CA, Pineros AR, Twigg AN, Orr KS, Conteh AM, Martinez MM, Kamocka MM, Day RN, Tersey SA, Mirmira RG, et al. (2019). A Versatile, Portable Intravital Microscopy Platform for Studying Beta-cell Biology In Vivo. Sci Rep 9, 8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro AJS, Schwab O, Mandegar MA, Ang YS, Conklin BR, Srivastava D, and Pruitt BL (2017). Multi-Imaging Method to Assay the Contractile Mechanical Output of Micropatterned Human iPSC-Derived Cardiac Myocytes. Circ Res 120, 1572–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios AC, and Clevers H (2018). Imaging organoids: a bright future ahead. Nat Methods 15, 24–26. [DOI] [PubMed] [Google Scholar]
- Ritsma L, Ellenbroek SIJ, Zomer A, Snippert HJ, de Sauvage FJ, Simons BD, Clevers H, and van Rheenen J (2014). Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature 507, 362–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritsma L, Steller EJ, Beerling E, Loomans CJ, Zomer A, Gerlach C, Vrisekoop N, Seinstra D, van Gurp L, Schafer R, et al. (2012). Intravital microscopy through an abdominal imaging window reveals a pre-micrometastasis stage during liver metastasis. Sci Transl Med 4, 158ra145. [DOI] [PubMed] [Google Scholar]
- Rodrigues Simoes AP, Cristina Maronezi M, Andres Ramirez Uscategui R, Garcia Kako Rodrigues M, Sitta Gomes Mariano R, Tavares de Almeida V, Jose Correia Santos V, Del Aguila da Silva P, Ricardo Russiano Vicente W, and Antonio Rossi Feliciano M (2020). Placental ARFI elastography and biometry evaluation in bitches. Anim Reprod Sci 214, 106289. [DOI] [PubMed] [Google Scholar]
- Rompolas P, Deschene ER, Zito G, Gonzalez DG, Saotome I, Haberman AM, and Greco V (2012). Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature 487, 496–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Mesa KR, and Greco V (2013). Spatial organization within a niche as a determinant of stem-cell fate. Nature 502, 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronzitti E, Conti R, Zampini V, Tanese D, Foust AJ, Klapoetke N, Boyden ES, Papagiakoumou E, and Emiliani V (2017). Submillisecond Optogenetic Control of Neuronal Firing with Two-Photon Holographic Photoactivation of Chronos. The Journal of Neuroscience 37, 10679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer M, Zirath H, and Ertl P (2018). Recent advances in microfluidic technologies for cell-to-cell interaction studies. Lab Chip 18, 249–270. [DOI] [PubMed] [Google Scholar]
- Rothe K, Babaian A, Nakamichi N, Chen M, Chafe SC, Watanabe A, Forrest DL, Mager DL, Eaves CJ, and Dedhar S (2020). Integrin-Linked Kinase Mediates Therapeutic Resistance of Quiescent CML Stem Cells to Tyrosine Kinase Inhibitors. Cell Stem Cell. [DOI] [PubMed] [Google Scholar]
- Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, Balgobind AV, Wind K, Gracanin A, and Begthel H (2018). A living biobank of breast cancer organoids captures disease heterogeneity. Cell 172, 373–386. e310. [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. (2008). Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ, Smith I, Parker I, and Bootman MD (2014). Fluorescence microscopy. Cold Spring Harb Protoc 2014, pdb top071795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasportas LS, and Gambhir SS (2014). Imaging circulating tumor cells in freely moving awake small animals using a miniaturized intravital microscope. PLoS One 9, e86759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van Den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, and Siersema PD (2011). Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772. [DOI] [PubMed] [Google Scholar]
- Scanu T, Spaapen RM, Bakker JM, Pratap CB, Wu L. e., Hofland I, Broeks A, Shukla VK, Kumar M, and Janssen H (2015). Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell host & microbe 17, 763–774. [DOI] [PubMed] [Google Scholar]
- Scaramozza A, Park D, Kollu S, Beerman I, Sun X, Rossi DJ, Lin CP, Scadden DT, Crist C, and Brack AS (2019). Lineage Tracing Reveals a Subset of Reserve Muscle Stem Cells Capable of Clonal Expansion under Stress. Cell Stem Cell 24, 944–957 e945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer R, Leung HM, and Gmitro AF (2014). Multi-modality imaging of a murine mammary window chamber for breast cancer research. BioTechniques 57, 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer A, Pinheiro D, Hauschild R, and Heisenberg CP (2020). Zebrafish embryonic explants undergo genetically encoded self-assembly. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedin J, Giraud A, Steiner SE, Ahl D, Persson AEG, Melican K, Richter-Dahlfors A, and Phillipson M (2019). High Resolution Intravital Imaging of the Renal Immune Response to Injury and Infection in Mice. Front Immunol 10, 2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seino T, Kawasaki S, Shimokawa M, Tamagawa H, Toshimitsu K, Fujii M, Ohta Y, Matano M, Nanki K, and Kawasaki K (2018). Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell 22, 454–467. e456. [DOI] [PubMed] [Google Scholar]
- Shahbazi MN, Jedrusik A, Vuoristo S, Recher G, Hupalowska A, Bolton V, Fogarty NME, Campbell A, Devito LG, Ilic D, et al. (2016). Self-organization of the human embryo in the absence of maternal tissues. Nature Cell Biology 18, 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Wang J, Shi J, Wu Z, Wang Q, Tang Z, He K, Shi Y, and Shen D (2020). Review of Artificial Intelligence Techniques in Imaging Data Acquisition, Segmentation and Diagnosis for COVID-19. IEEE Rev Biomed Eng. [DOI] [PubMed] [Google Scholar]
- Shimokawa M, Ohta Y, Nishikori S, Matano M, Takano A, Fujii M, Date S, Sugimoto S, Kanai T, and Sato T (2017). Visualization and targeting of LGR5(+) human colon cancer stem cells. Nature 545, 187–192. [DOI] [PubMed] [Google Scholar]
- Slator PJ, Hutter J, McCabe L, Gomes ADS, Price AN, Panagiotaki E, Rutherford MA, Hajnal JV, and Alexander DC (2018). Placenta microstructure and microcirculation imaging with diffusion MRI. Magn Reson Med 80, 756–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small DM, Sanchez WY, and Gobe GC (2016). Intravital Multiphoton Imaging of the Kidney: Tubular Structure and Metabolism. Methods Mol Biol 1397, 155–172. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Nachman I, Regev A, and Meissner A (2010). Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol 28, 521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. (2010). Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144. [DOI] [PubMed] [Google Scholar]
- Sonavane PR, Wang C, Dzamba B, Weber GF, Periasamy A, and DeSimone DW (2017). Mechanical and signaling roles for keratin intermediate filaments in the assembly and morphogenesis of Xenopus mesendoderm tissue at gastrulation. Development 144, 4363–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer-Phelps A, Hassell BA, and Ingber DE (2019). Modelling cancer in microfluidic human organs-on-chips. Nature Reviews Cancer 19, 65–81. [DOI] [PubMed] [Google Scholar]
- Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, et al. (2011). Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis AM, Schachter MJ, Gulati S, Zitelli KT, Malanowski S, Tajik A, Fritz C, Trulson M, and Otte SL (2018). Simultaneous Optogenetics and Cellular Resolution Calcium Imaging During Active Behavior Using a Miniaturized Microscope. Front Neurosci 12, 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Sauer J, Kane RS, Dordick JS, and Friedman JM (2015). Remote regulation of glucose homeostasis in mice using genetically encoded nanoparticles. Nat Med 21, 92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauser ML, Bailey AP, Senyo SE, Guillermier C, Perlstein TS, Gould AP, Lee RT, and Lechene CP (2012). Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature 481, 516–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhorn B, Sorrentino A, Badole S, Bogdanova Y, Belousov V, and Michel T (2018). Chemogenetic generation of hydrogen peroxide in the heart induces severe cardiac dysfunction. Nature Communications 9, 4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolp B, and Melican K (2016). Microbial pathogenesis revealed by intravital microscopy: pros, cons and cautions. FEBS Lett 590, 2014–2026. [DOI] [PubMed] [Google Scholar]
- Stone OJ, Pankow N, Liu B, Sharma VP, Eddy RJ, Wang H, Putz AT, Teets FD, Kuhlman B, Condeelis JS, et al. (2019). Optogenetic control of cofilin and αTAT in living cells using Z-lock. Nat Chem Biol 15, 1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Wang Y, Zhang H, and Qian J (2017). Near-infrared laser scanning confocal microscopy and its application in bioimaging. Optical and Quantum Electronics 50. [Google Scholar]
- Sun J, Ramos A, Chapman B, Johnnidis JB, Le L, Ho YJ, Klein A, Hofmann O, and Camargo FD (2014). Clonal dynamics of native haematopoiesis. Nature 514, 322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A, Kaku Y, Ohmori T, Sharmin S, Ogawa M, Sasaki H, and Nishinakamura R (2014). Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell stem cell 14, 53–67. [DOI] [PubMed] [Google Scholar]
- Tajima A, Pradhan I, Geng X, Trucco M, and Fan Y (2019). Construction of Thymus Organoids from Decellularized Thymus Scaffolds. Methods Mol Biol 1576, 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S (2006). Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Nomura T, and Osumi N (2008). Transferring genes into cultured mammalian embryos by electroporation. Dev Growth Differ 50, 485–497. [DOI] [PubMed] [Google Scholar]
- Takasato M, Er PX, Chiu HS, and Little MH (2016). Generation of kidney organoids from human pluripotent stem cells. Nat Protoc 11, 1681–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe T, and Wells JM (2019). Organoids by design. Science 364, 956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taslimi A, Vrana JD, Chen D, Borinskaya S, Mayer BJ, Kennedy MJ, and Tucker CL (2014). An optimized optogenetic clustering tool for probing protein interaction and function. Nat Commun 5, 4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian R, Gachechiladze MA, Ludwig CH, Laurie MT, Hong JY, Nathaniel D, Prabhu AV, Fernandopulle MS, Patel R, Abshari M, et al. (2019). CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron 104, 239–255 e212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiriac H, Belleau P, Engle DD, Plenker D, Deschênes A, Somerville TD, Froeling FE, Burkhart RA, Denroche RE, and Jang G-H (2018). Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer discovery 8, 1112–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher JE, Weiner OD, and Lim WA (2013). Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa M, Sera T, Yokota H, Takematsu M, Morita M, Yeroslavsky G, Kamimura M, and Soga K (2020). Computed Tomography for In Vivo Deep Over-1000 nm Near-Infrared Fluorescence Imaging. J Biophotonics, e202000071. [DOI] [PubMed] [Google Scholar]
- Urban DJ, and Roth BL (2015). DREADDs (designer receptors exclusively activated by designer drugs): chemogenetic tools with therapeutic utility. Annual review of pharmacology and toxicology 55, 399–417. [DOI] [PubMed] [Google Scholar]
- van den Berg CW, Ritsma L, Avramut MC, Wiersma LE, van den Berg BM, Leuning DG, Lievers E, Koning M, Vanslambrouck JM, and Koster AJ (2018a). Renal subcapsular transplantation of PSC-derived kidney organoids induces neo-vasculogenesis and significant glomerular and tubular maturation in vivo. Stem cell reports 10, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg CW, Ritsma L, Avramut MC, Wiersma LE, van den Berg BM, Leuning DG, Lievers E, Koning M, Vanslambrouck JM, Koster AJ, et al. (2018b). Renal Subcapsular Transplantation of PSC-Derived Kidney Organoids Induces Neo-vasculogenesis and Significant Glomerular and Tubular Maturation In Vivo. Stem Cell Reports 10, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaasdonk JS, Stephens AD, Haase J, and Bloom K (2014). Bending the rules: widefield microscopy and the Abbe limit of resolution. J Cell Physiol 229, 132–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, Lampis A, Eason K, Huntingford I, and Burke R (2018). Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359, 920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, Volpetti F, Petrova E, French C, Maerkl SJ, and Wang B (2019). Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals. Nature chemical biology 15, 540–548. [DOI] [PubMed] [Google Scholar]
- Wang C, Lu T, Emanuel G, Babcock HP, and Zhuang X (2019a). Imaging-based pooled CRISPR screening reveals regulators of lncRNA localization. Proc Natl Acad Sci U S A 116, 10842–10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Rivenson Y, Jin Y, Wei Z, Gao R, Gunaydin H, Bentolila LA, Kural C, and Ozcan A (2019b). Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat Methods 16, 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Ouzounov DG, Wu C, Horton NG, Zhang B, Wu CH, Zhang Y, Schnitzer MJ, and Xu C (2018). Three-photon imaging of mouse brain structure and function through the intact skull. Nat Methods 15, 789–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, and Schwarz TL (2009). Imaging axonal transport of mitochondria. Methods Enzymol 457, 319–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver EM, and Hummon AB (2013). Imaging mass spectrometry: from tissue sections to cell cultures. Adv Drug Deliv Rev 65, 1039–1055. [DOI] [PubMed] [Google Scholar]
- Weiss LE, Shalev Ezra Y, Goldberg S, Ferdman B, Adir O, Schroeder A, Alalouf O, and Shechtman Y (2020). Three-dimensional localization microscopy in live flowing cells. Nat Nanotechnol. [DOI] [PubMed] [Google Scholar]
- Wheeler MA, Smith CJ, Ottolini M, Barker BS, Purohit AM, Grippo RM, Gaykema RP, Spano AJ, Beenhakker MP, Kucenas S, et al. (2016). Genetically targeted magnetic control of the nervous system. Nat Neurosci 19, 756–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann S.W.H.a.J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. [DOI] [PubMed]
- Wolfram F, Gullmar D, Bottcher J, Schubert H, Bischoff S, Reichenbach JR, and Lesser TG (2019). Assessment of MR imaging during one-lung flooding in a large animal model. MAGMA 32, 581–590. [DOI] [PubMed] [Google Scholar]
- Wu SS, Lee JH, and Koo BK (2019). Lineage Tracing: Computational Reconstruction Goes Beyond the Limit of Imaging. Mol Cells 42, 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TJ, Tzeng YK, Chang WW, Cheng CA, Kuo Y, Chien CH, Chang HC, and Yu J (2013). Tracking the engraftment and regenerative capabilities of transplanted lung stem cells using fluorescent nanodiamonds. Nat Nanotechnol 8, 682–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tanaka Y, Patterson B, Kang Y-J, Govindaiah G, Roselaar N, Cakir B, Kim K-Y, Lombroso AP, and Hwang S-M (2017). Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell stem cell 21, 383–398. e387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Dong P, Chen X, Hsieh TS, Banala S, De Marzio M, English BP, Qi Y, Jung SK, Kieffer-Kwon KR, et al. (2020). 3D ATAC-PALM: super-resolution imaging of the accessible genome. Nat Methods 17, 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan HH, Siu HC, Law S, Ho SL, Yue SS, Tsui WY, Chan D, Chan AS, Ma S, and Lam KO (2018). A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell stem cell 23, 882–897. e811. [DOI] [PubMed] [Google Scholar]
- Yang L, Yang S, Li X, Li B, Li Y, Zhang X, Ma Y, Peng X, Jin H, and Fan Q (2019). Tumor organoids: From inception to future in cancer research. Cancer letters 454, 120–133. [DOI] [PubMed] [Google Scholar]
- Yao Y, Xu X, Yang L, Zhu J, Wan J, Shen L, Xia F, Fu G, Deng Y, and Pan M (2020). Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell stem cell 26, 17–26. e16. [DOI] [PubMed] [Google Scholar]
- You S, Tu H, Chaney EJ, Sun Y, Zhao Y, Bower AJ, Liu YZ, Marjanovic M, Sinha S, Pu Y, et al. (2018). Intravital imaging by simultaneous label-free autofluorescence-multiharmonic microscopy. Nat Commun 9, 2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, and Choudhury D (2019). Microfluidic bioprinting for organ-on-a-chip models. Drug Discov Today 24, 1248–1257. [DOI] [PubMed] [Google Scholar]
- Yue Y, Zong W, Li X, Li J, Zhang Y, Wu R, Liu Y, Cui J, Wang Q, Bian Y, et al. (2020). Long-term, in toto live imaging of cardiomyocyte behaviour during mouse ventricle chamber formation at single-cell resolution. Nat Cell Biol 22, 332–340. [DOI] [PubMed] [Google Scholar]
- Zhang F, Gradinaru V, Adamantidis AR, Durand R, Airan RD, de Lecea L, and Deisseroth K (2010). Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nat Protoc 5, 439–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Guillermier C, De Raedt T, Cox AG, Maertens O, Yimlamai D, Lun M, Whitney A, Maas RL, and Goessling W (2020). Imaging mass spectrometry reveals tumor metabolic heterogeneity. iScience, 101355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jeon M, Rich LJ, Hong H, Geng J, Zhang Y, Shi S, Barnhart TE, Alexandridis P, Huizinga JD, et al. (2014). Non-invasive multimodal functional imaging of the intestine with frozen micellar naphthalocyanines. Nat Nanotechnol 9, 631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YS, and Yao J (2018). Imaging Biomaterial-Tissue Interactions. Trends Biotechnol 36, 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Todorov MI, Cai R, Maskari RA, Steinke H, Kemter E, Mai H, Rong Z, Warmer M, Stanic K, et al. (2020). Cellular and Molecular Probing of Intact Human Organs. Cell 180, 796–812 e719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Nguyen L, Arnesano C, Ando Y, Raval M, Rodgers JT, Fraser S, Lu R, and Shen K (2020). Non-invasive optical biomarkers distinguish and track the metabolic status of single hematopoietic stem cells. iScience, 100831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Feng T, Cheng Q, Wang X, Du S, Sato N, Kuniyil Ajith Singh M, and Yuan J (2020). Towards Clinical Translation of LED-Based Photoacoustic Imaging: A Review. Sensors (Basel) 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z, He C, Du Y, Wen W, Zhang G, Zhao Y, Tao M, Hu Z, Wang K, and Qiu P (2019). Refractive index and pulse broadening characterization using oil immersion and its influence on three-photon microscopy excited at the 1700-nm window. J Biophotonics 12, e201800263. [DOI] [PubMed] [Google Scholar]
- Zielke N, and Edgar BA (2015). FUCCI sensors: powerful new tools for analysis of cell proliferation. WIREs Developmental Biology 4, 469–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer A, Ellenbroek SI, Ritsma L, Beerling E, Vrisekoop N, and Van Rheenen J (2013). Intravital imaging of cancer stem cell plasticity in mammary tumors. Stem Cells 31, 602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubkov M, Hurshkainen AA, Brui EA, Glybovski SB, Gulyaev MV, Anisimov NV, Volkov DV, Pirogov YA, and Melchakova IV (2018). Small-animal, whole-body imaging with metamaterial-inspired RF coil. NMR Biomed 31, e3952. [DOI] [PubMed] [Google Scholar]
- Zunder ER, Lujan E, Goltsev Y, Wernig M, and Nolan GP (2015). A continuous molecular roadmap to iPSC reprogramming through progression analysis of single-cell mass cytometry. Cell Stem Cell 16, 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]