Abstract
Summary
Analysis of epitope-specific antibody repertoires has provided novel insights into the pathogenesis of inflammatory disorders, especially allergies. A novel multiplex immunoassay, termed Bead-Based Epitope Assay (BBEA), was developed to quantify levels of epitope-specific immunoglobulins, including IgE, IgG, IgA and IgD isotypes. bbeaR is an open-source R package, developed for the BBEA, provides a framework to import, process and normalize .csv data files exported from the Luminex reader, evaluate various quality control metrics, analyze differential epitope-binding antibodies with linear modeling, visualize results and map epitopes’ amino acid sequences to their respective primary protein structures. bbeaR enables streamlined and reproducible analysis of epitope-specific antibody profiles.
Availability and implementation
bbeaR is open-source and freely available from GitHub as an R package: https://github.com/msuprun/bbeaR; vignettes included.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Antibodies are part of the adaptive humoral immunity and offer essential protection against a variety of pathogens. When antibodies recognize a specific site on an antigenic protein, i.e. an epitope, an immune complex is formed, allowing the immune system to fight the pathogen (Lu et al., 2018; Sela-Culang et al., 2013). The variable domain of an antibody recognizes a wide range of molecules, while the constant domain defines its isotype (i.e. IgM, IgD, IgG, IgA and IgE) and effector functions (Lu et al., 2018). In the case of viral or bacterial infection, IgG and IgM antibodies could neutralize or help clear the pathogen (Forthal, 2014). In allergy, where an innocuous protein is recognized as an antigen, cross-linked IgE activates mast cells and basophils, which readily release inflammatory molecules such as histamine, heparin and tryptase, leading to allergic reactions (Burton and Oettgen, 2011). Quantifying epitope-specific antibodies has many applications, including (i) diagnostics, where the amount of IgE specific to different epitopes can indicate clinical reactivity and severity of allergy (Sackesen et al., 2019; Santos et al., 2020), Iii) prognostics, where isotypes induced during immunotherapy can identify responders (Suarez-Farinas et al., 2019; Sugimoto et al., 2016), (iii) insights into the natural disease evolution and pathogenesis (Suprun et al., 2020).
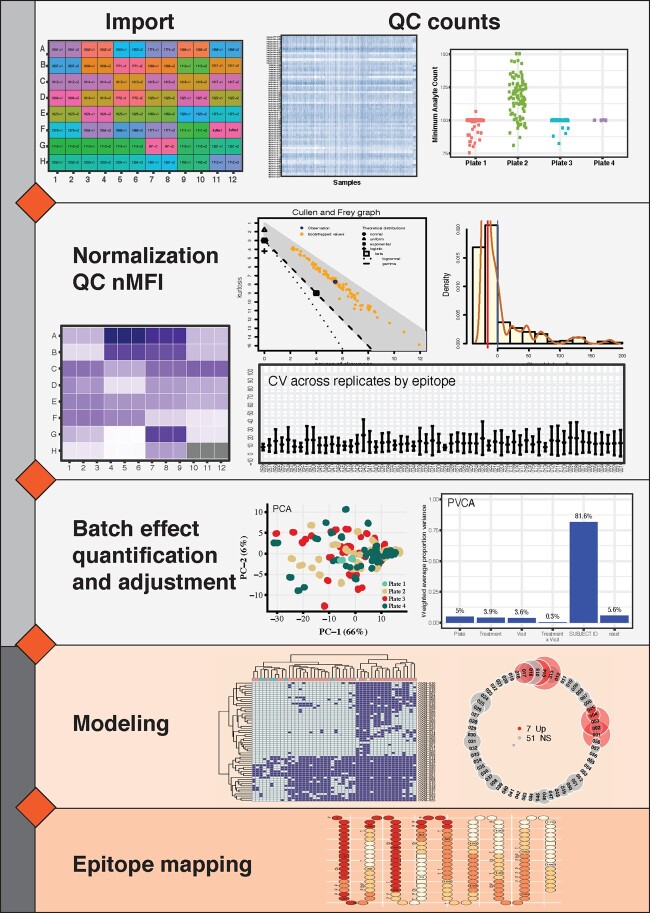
Bead-Based Epitope Assay (BBEA) is a Luminex-based high-throughput assay developed to measure the levels of many epitope-specific antibodies simultaneously and outperforms peptide microarrays (Suprun et al., 2019). Here, we present an open-source R package, bbeaR, which facilitates the management, preprocessing and quality control of BBEA data (Fig. 1). It creates a data structure that eases the differential antibodies analysis using limma or linear modeling and contains custom visualizations. bbeaR comes with a vignette with a detailed walkthrough of two examples, showcasing the package utility. While the framework is described using an example of food allergies, it can be applied to any disease or experimental condition where antibody responses are of interest. To the authors’ knowledge, bbeaR is the only R package currently available providing comprehensive pipeline for import, quality control and analytics.
Fig. 1.
Functionality schematic of bbeaR for data import, normalization, quality control (QC), analysis and visualizations
2 Materials and methods
BBEA is run on 96-well microplates, allowing the measurement of many plasma/serum epitope-specific antibodies on many patients simultaneously (Breen, 2017). We recommend that experiments are randomized using PlateDesigner (Suprun and Suárez-Fariñas, 2019), to avoid confounding by technical factors, such as plate or well position. PlateDesigner’s experimental metadata can be uploaded directly to the Luminex reader, avoiding data entry errors. The assay’s readout, exported from Luminex as a comma-separated values (.csv) file for each plate, is a starting point of the bbeaR pipeline.
Csv files from each plate are imported simultaneously, extracting counts and Median Fluorescence Intensity (MFI) tables, along with metadata about the plate run. The MFI is provided for each Luminex bead, i.e. an analyte, which uniquely corresponds to an individual epitope, and annotations are mapped. The counts undergo a first round of quality controls, where samples or antibodies can be removed if they do not reach sufficient reads, e.g. at least 25 counts per analyte (Bjornstal et al., 2011). Then, for each epitope-specific antibody j and sample i, the MFI signal is normalized as
where BG are background wells. The distribution of nMFI is evaluated using Cullen-Frey plots and histograms, and agreement between technical replicates assessed by plate-layout plots, coefficients of variation (CV) and intra-class correlation coefficients (ICC). If an experiment consists of several microplate runs, plate effect can be identified using principal component analysis (PCA) and the nMFI can be adjusted for plate and well effects using a limma model (Silver et al., 2009; Smyth, 2004; Suprun et al., 2019) or other approaches like SVA/ComBat (Leek et al., 2012).
The differential epitope-specific antibodies can be identified by comparing patient groups, using either classical linear models or limma (Silver et al., 2009; Smyth, 2004). We have implemented custom net circle visualization plots that present fold-changes and P-values across antibodies ordered by the epitopes’ position on the protein. Additionally, amino acid sequences of the epitopes are mapped to the protein using the topology plot and can be quite useful when a 3D crystal structure is not available. This plot extracts protein characteristics from the UniProt database, and displays additional information, i.e. sites of disulfide bridges, glycosylation and enzymatic cleavage.
3 Usage example
The features of bbeaR are illustrated in two detailed examples, provided as vignettes: (i) comparison of egg epitope-specific IgE antibodies in egg-allergic children and controls (Suprun et al., 2020) and (ii) immunomodulation of milk epitope-specific IgEs in patients undergoing oral immunotherapy (Suarez-Farinas et al., 2019) (Supplementary Files S1 and S2). While the egg experiment was performed on one microplate, the milk experiment was performed on four plates and required a plate effect adjustment. A brief walkthrough of the milk allergy project is outlined below.
Levels of IgE specific to 66 allergenic milk epitopes were measured before and after treatment in 47 milk-allergic patients undergoing two years of immunotherapy with either omalizumab or placebo adjuvant (Suarez-Farinas et al., 2019). First, to make sure the MFI values are valid, it is important to have sufficient numbers of analyte (that represent epitopes) counts. In all of the plates, minimum counts were above 75, which is considered reliable (Bjornstal et al., 2011). nMFI values are stored in ExpressionSet object along with epitope annotation and patients’ phenotypic or clinical information, allowing for a seamless use of commonly used -omic methods. PCA visualization indicates the presence of a plate effect, which can be quantified by principal variance component analysis (Bushel, 2019; Li et al., 2009) to account for ∼5% of the total variability. Since the samples were randomly allocated to plates, the plate effect was adjusted using linear models. The distribution of the nMFIs is investigated through the Cullen–Frey plot (Delignette-Muller and Dutang, 2015), which is shown to be close to log-normal. As such, parametric models using the nMFI can be used in downstream analyses. Reliability across technical replicates are measured by CV and ICC. After the reliability is ascertained, the replicates are averaged. Changes from baseline to the end of treatment in placebo and omalizumab groups are modeled with limma and show decreased levels of more epitope-specific IgEs in the placebo arm, visualized by the net circle plots.
4 Conclusion
bbeaR provides a flexible functionality for analyzing epitope-specific antibodies quantified with the BBEA or other Luminex-based platforms. After data loading and pre-processing, the workflow is similar to that of microarray and RNA-seq pipelines, allowing users to implement other tools developed for these high-throughput technologies. bbeaR allows seamless and reproducible profiling of epitope-specific antibodies in R.
Supplementary Material
Acknowledgements
The authors thank Robert Getts, Helena L. Chang, Rohit Raghunathan and Ronaldo Silva for the technical input on the development of the package.
Funding
M.S. was funded by the Integrated Pharmacological Sciences Training grant from the National Institute of General Medical Sciences (NIGMS) [T32GM062754]. H.A.S. was funded in part by a grant from National Institute of Allergy and Infectious Diseases (NIAID/NIH) [UM1 AI109565]. M.S-.F. was partially supported by an institutional grant from AllerGenis LLC.
Conflict of Interest: none declared.
Data availability
The data underlying this article are available as part of the bbeaR package.
Contributor Information
Maria Suprun, Department of Pediatrics, Allergy and Immunology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.
Randall J Ellis, Departments of Neuroscience and Psychiatry, Addiction Institute of Mount Sinai, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.
Hugh A Sampson, Department of Pediatrics, Allergy and Immunology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.
Mayte Suárez-Fariñas, Department of Population Health Science and Policy, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA; Department of Genetics and Genomic Sciences, Icahn Institute for Data Science and Genomic Technology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.
References
- Bjornstal O. et al. (2011) The impact of decreased bead count to determine concentrations of amyloid beta1-42, total-tau, and phosphorylated-tau181 in human cerebrospinal fluid using xMAP technology. J. Pharm. Sci., 100, 4655–4663. [DOI] [PubMed] [Google Scholar]
- Breen E.J. (2017) Protein multiplexed immunoassay analysis with R. Methods Mol. Biol., 1619, 495–537. [DOI] [PubMed] [Google Scholar]
- Burton O.T., Oettgen H.C. (2011) Beyond immediate hypersensitivity: evolving roles for IgE antibodies in immune homeostasis and allergic diseases. Immunol. Rev., 242, 128–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushel P. (2019) pvca: Principal Variance Component Analysis (PVCA). R package version 1.26.0. https://bioconductor.org/packages/release/bioc/html/pvca.html (10 May 2020, date last accessed).
- Delignette-Muller M.L., Dutang C. (2015) fitdistrplus: an R package for fitting distributions. 64, 34. [Google Scholar]
- Forthal D.N. (2014) Functions of antibodies. Microbiol. Spectr., 2, 1–17. [PMC free article] [PubMed] [Google Scholar]
- Leek J.T. et al. (2012) The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics, 28, 882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. (2009) Principal variance components analysis: estimating batch effects in microarray gene expression data. In: Scherer, A. (ed.) Batch Effects and Noise in Microarray Experiments: Sources and Solutions. John Wiley & Sons, Ltd. pp. 141–154. [Google Scholar]
- Lu L.L. et al. (2018) Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol., 18, 46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackesen C. et al. (2019) A new Luminex-based peptide assay to identify reactivity to baked, fermented, and whole milk. Allergy, 74, 327–336. [DOI] [PubMed] [Google Scholar]
- Santos A.F. et al. (2020) IgE to epitopes of Ara h 2 enhance the diagnostic accuracy of Ara h 2-specific IgE. Allergy, 75, 2309–2318. [DOI] [PubMed] [Google Scholar]
- Sela-Culang I. et al. (2013) The structural basis of antibody–antigen recognition. Front. Immunol., 4, 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J.D. et al. (2009) Microarray background correction: maximum likelihood estimation for the normal-exponential convolution. Biostatistics, 10, 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol., 3, Article3. [DOI] [PubMed] [Google Scholar]
- Suarez-Farinas M. et al. (2019) Predicting development of sustained unresponsiveness to milk oral immunotherapy using epitope-specific antibody binding profiles. J. Allergy Clin. Immunol., 143, 1038–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M. et al. (2016) Differential response in allergen-specific IgE, IgGs, and IgA levels for predicting outcome of oral immunotherapy. Pediatr. Allergy Immunol., 27, 276–282. [DOI] [PubMed] [Google Scholar]
- Suprun M. et al. (2020) Ovomucoid epitope-specific repertoire of IgE, IgG4, IgG1, IgA1 and IgD antibodies in egg allergic children. Allergy, 75, 2633–2643. [DOI] [PMC free article] [PubMed]
- Suprun M. et al. (2019) Novel Bead-Based Epitope Assay is a sensitive and reliable tool for profiling epitope-specific antibody repertoire in food allergy. Sci. Rep., 9, 18425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suprun M., Suárez-Fariñas M. (2019) PlateDesigner: a web-based application for the design of microplate experiments. Bioinformatics, 35, 1605–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available as part of the bbeaR package.