Abstract
Motivation
RNA molecules become attractive small molecule drug targets to treat disease in recent years. Computer-aided drug design can be facilitated by detecting the RNA sites that bind small molecules. However, very limited progress has been reported for the prediction of small molecule–RNA binding sites.
Results
We developed a novel method RNAsite to predict small molecule–RNA binding sites using sequence profile- and structure-based descriptors. RNAsite was shown to be competitive with the state-of-the-art methods on the experimental structures of two independent test sets. When predicted structure models were used, RNAsite outperforms other methods by a large margin. The possibility of improving RNAsite by geometry-based binding pocket detection was investigated. The influence of RNA structure’s flexibility and the conformational changes caused by ligand binding on RNAsite were also discussed. RNAsite is anticipated to be a useful tool for the design of RNA-targeting small molecule drugs.
Availability and implementation
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
In recent years, many experiments suggest that RNA becomes an attractive small molecule drug target to treat disease (Disney, 2019). A significant amount of efforts were made to study the interactions between RNA and small molecules, such as small molecule design (Ursu et al., 2019) and detection of small molecule–RNA binding motifs (Childs-Disney et al., 2018). In these studies, the preknowledge about the small molecule–RNA binding sites is essential. However, due to the high cost with wet-lab experiments, the structural details for small molecule–RNA interactions are unknown for most targets. This has motivated the development of computational algorithms to predict small molecule–RNA binding sites.
The existing prediction methods can be classified into two groups: structure-based and sequence-based. Structure-based approaches include MetalionRNA (Philips et al., 2012), LigandRNA (Philips et al., 2013), Rsite (Zeng et al., 2015) and RBind (Wang et al., 2018). MetalionRNA is a statistical potential-based method but can only predict the binding sites for metal ions (magnesium, sodium and potassium). LigandRNA is a knowledge-based potential to score the interactions between RNA and small molecules. Its input includes both the RNA structure and the small molecule’s binding poses, which are ranked according to the returned potential score. Rsite is a distance-based predictor to identify the functional sites of noncoding RNAs. RBind is the latest structure-based approach by constructing a structure-based nucleotide interaction network. As revealed in Wang et al. (2018), RBind has a high precision at the expense of a low recall. In contrast, there are very few sequence-based methods to predict small molecule–RNA binding sites. To the best of our knowledge, Rsite2 is the only sequence-based algorithm, which works based on predicted secondary structure (SS; Zeng and Cui, 2016).
In this work, we proposed a novel algorithm named RNAsite to predict small molecule–RNA binding sites. We first developed a structure-based method (RNAsite_str) by designing a group of novel structure-based features that effectively captured the preference toward small molecule–RNA binding sites. Second, a sequence profile-based predictor (RNAsite_seq) was designed to deal with the cases where no structure is available. These two methods are combined, yielding to the final approach RNAsite. Experiments on two independent test sets show that RNAsite is competitive compared with the state-of-the-art methods.
2 Materials and methods
2.1 Benchmark datasets
Two datasets were used to assess and compare the proposed method with existing methods. The first one contains 19 RNAs from the work of RBind (Wang et al., 2018) (denoted by RB19). This dataset was constructed based on the 251 structures in the work of LigandRNA (Philips et al., 2013) after filtering structures with simple helix topology, remaining 22 structures. In addition, three RNAs with more than one chain or pseudoknot interactions were further removed to enable the structure prediction by the RNAComposer program (Biesiada et al., 2016).
The second was constructed in this work from the Protein Data Bank (PDB) (Burley et al., 2019) according to the following procedure. First, in order to exclude the effects of other molecules (such as DNA, protein and so on), we downloaded the complex structures containing only RNAs with one or more small molecules (water was not considered). In total, we obtained 1673 RNA chains. Second, the following RNAs were removed: chain length is <20 or >1500; all small molecules in the structure are crystallization additives [a comprehensive list of crystallization additives was obtained from BioliP (Yang et al., 2012)]; no small molecules interact with the RNA structure. A nucleotide is defined as interacting with a small molecule if one of the atomic distances between the nucleotide and the small molecule is smaller than a specified distance cutoff (4 Å), as adopted in the work of RBind. Such nucleotides are defined as positive samples and others are defined as negative samples. 712 RNA chains remained after the above filtering.
Redundancy was removed for the above 712 RNA chains based on a combination of structure and sequence similarity clustering. First, the pairwise structure similarity TM-scoreRNA was calculated with the program RNA-align (Gong et al., 2019). If the TM-scoreRNA between two RNA structures is higher than 0.3, the one with the higher ratio of positive samples is kept (defined as the total number of positive samples in an RNA divided by the sequence length). For example, 22 structures sharing >0.3 structure similarity belong to the thiamin pyrophosphate riboswitches and only one of them is kept to remove redundancy. A total of 78 RNA chains was retained after this structure-similarity based filtering (denote by RB78). About 3/4 of the 78 RNA chains are used for training and the remaining 1/4 are for test. However, to make sure there is no redundancy between the training and the test set, a sequence-based clustering was performed. The 78 RNA sequences were then clustered into 57 clusters at 30% sequence similarity with cd-hit-est (Li and Godzik, 2006) and BLASTclust (Altschul et al., 1990) program. The split of these RNAs was based on the cluster information: 42 clusters containing 60 RNAs were used as training set (denoted by TR60) and 15 clusters containing 18 RNAs were used as independent test set (denoted by TE18). Due to the different procedure used above and the PDB update, 65 new structures are included in RB78 compared with RB19. In addition, 9 (resp. 4) structures from TR60 (resp. TE18) are the same as the structures in RB19. The detailed information about the RNA and the ligands in each RNA structure is available in Tables S1, S2 and S3.
2.2 Overview of our prediction model
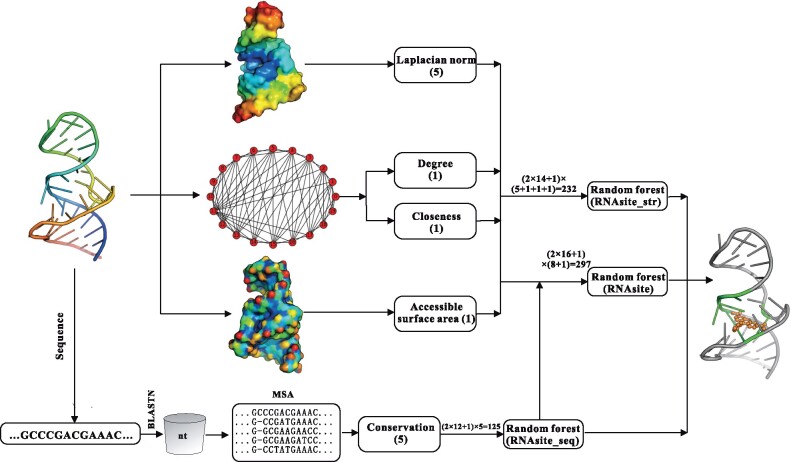
As shown in Figure 1, the proposed method, RNAsite, is composed of two independent components: a sequence-based module RNAsite_seq and a structure-based module RNAsite_str. When no structure is available, only RNAsite_seq is used to predict the binding nucleotides. When structure is available, additional prediction is done by RNAsite_str. In addition, the prediction from RNAsite_seq is added into the structure-based features and then fed into the random forest (RF) algorithm to make a consensus prediction in RNAsite, which shows improved performance over both RNAsite_str and RNAsite_seq.
Fig. 1.
The flowchart of RNAsite for small molecule–RNA binding sites prediction. RNAsite consists of two components, RNAsite_str and RNAsite_seq, which are structure- and sequence-based predictors, respectively. In RNAsite_str, based on the topological features and the LN of the structure, three groups of descriptors were obtained. In RNAsite_seq, the query sequence is searched against a sequence database (nt) with the BLASTN program to construct an MSA, from which conservation-based features are then designed. The prediction by RNAsite_seq is used as a feature, together with the structure-based features set in RNAsite. The classifiers for small molecule-binding sites prediction are trained with the RF algorithm
2.3 Sequence-based method RNAsite_seq
The RNA sequence is searched by BLASTN (Altschul et al., 1990) with E-value <0.001 against the NCBI’s nonredundant nucleotide sequence database (nt) to construct a multiple sequence alignment (MSA). The evolutionary conservation of each position in the RNA sequence is calculated from the MSA as follows. First, a weight is assigned to each of the sequence in the MSA based on a similar idea of the Henikoff–Henikoff scheme (Henikoff and Henikoff, 1994). For the jth position in the ith sequence of the MSA, a score wij is calculated as follows:
(1)
where fj is an integer indicating the number of occurring types of nucleotides at this position (≤4); Nij represents the type of nucleotide at the jth position of the ith sequence; f(Nij) is the number of the nucleotide Nij in the jth column of the MSA; second, the weight wi for each sequence in the MSA is calculated according to Eq. (2) as follows:
(2)
For each position in the RNA sequence, the weighted count for each nucleotide (gap included) is used as an evolutionary conservation score. As the nucleotides in an RNA are not independent with each other, to encode the ith nucleotide, its neighbors inside a window (size w) on each side are also considered. Thus, the total number of features for representing a nucleotide in RNAsite_seq is (2 × w+1) × 5. These features are fed into the RF algorithm for training and test.
2.4 Structure-based method RNAsite_str
When the structure (either experimentally solved or modeled) of an RNA is available, three different sets of structural descriptors are extracted, based on the topological features and the Laplacian norm (LN) of the structure. To our knowledge, the LN is applied to small molecule–RNA binding sites prediction for the first time. The traditional solvent accessibility (SA)-based features are also used here. Let L denote the number of nucleotides in an RNA.
2.4.1 Laplacian norm
The LN has been applied to protein structure analysis (Bonnel and Marteau, 2012; Li et al., 2014) and function prediction (Liu and Liu, 2020; Sun et al., 2016). Here, we apply the LN to characterize RNA structures. The LN of each nucleotide is defined as the distance between a target nucleotide and the weighted center of its surrounding nucleotides. For convenience, the ‘C3’ atom is used as the representative of each nucleotide for all standard nucleotides. While for nonstandard nucleotides, an arbitrary heavy atom is used. In order to compute LN, a discrete Laplace operator is calculated using a Gaussian kernel defined as follows:
(3)
where pi and pj are the coordinate vectors for nucleotides i and j, respectively; ‖ ‖ is the L2 norm. The parameter σ in the Gaussian kernel is a scale factor. Under a given scale factor σ, the closer the two nucleotides are, the higher the Laplace operator is. And the Laplace operator decreases rapidly as the distance increases. Therefore, to highlight the distribution pattern of sequentially distant residues, the adjacent nucleotides are omitted when defining the discrete Laplace operator. By varying σ, the topology of a nucleotide can be described at various scales. A low σ measures deformation of each nucleotide locally and implies that only spatially close nucleotides will be considered in the computation of the Laplace operator. On the other hand, a high σ implies that more nucleotides in the RNA will be included. Here, the Laplace operators can be considered as the weights of surrounding nucleotides. Hence, the LN of each nucleotide at a given σ is defined as follows:
(4)
The value of LN can reflect the geometrical features of surface convexity/concavity. A high value of LN means the nucleotide position is convex in the RNA structure, while a low LN implies a concave position. The distances at 0, 1/4, 1/2, 3/4 and 1 quantile positions of this distribution are chosen as the scale factors. The complete distributions of LN are shown in Supplementary Figure S1. Thus, each nucleotide is encoded into a five-dimensional vector.
2.4.2 Topological features
For each RNA structure, we transform it into a nucleotide interaction network in which a node denotes a single nucleotide and an edge represents the existence of noncovalent interaction. In our definition, two nonconsecutive nucleotides in a sequence are connected in the network when they contain a pair of heavy atoms, one from each nucleotide, within the distance of 8 Å (Alipanahi et al., 2015; Wang et al., 2018). For each nucleotide, two network parameters are calculated: closeness (CL) assessing its global importance in the network and degree (DG) representing its local connectivity. CL is computed as the inverse of the average of the shortest distance to other nodes, while DG is the number of edges associated with the node. Note that similar features have been used in the work of RBind (Wang et al., 2018) and more information can be found there for the calculation of these two features.
2.4.3 Solvent accessibility
The last group of structural features we used is the SA. Each chain structure was submitted to the POPS package (Cavallo et al., 2003) with the probe diameter of 1.5 Å to calculate all nucleotide-specific accessible surface areas (ASAs).
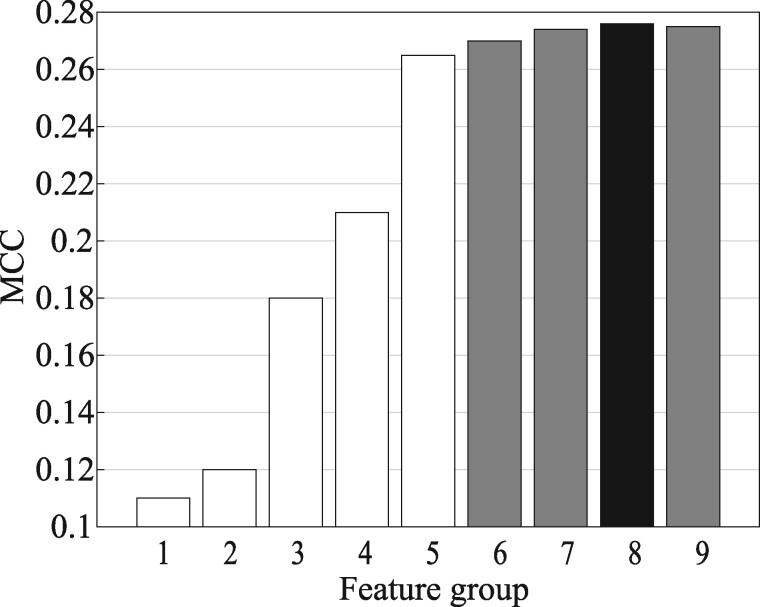
In addition to the above structural features, we also tested the effect of using SS including base-stacking, base-pairing and bulges computed by Lu et al. (2015). However, the prediction performance did not improve by adding the SS feature (seen in Figure 3). Thus, SS was not used in our method. More discussion about this is available in Section 3.2.
Fig. 3.
Predictive performance of RNAsite_str on TR60 from different feature groups. The features are divided into nine categories: five individual groups, (1) SS, (2) SA, (3) DG, (4) CL, (5) LN; four combined feature groups: (6) LN + CL, (7) LN + CL + DG, (8) LN + CL + DG + SA and (9) ALL (i.e. LN + CL + DG + SA + SS)
To summarize, a total of eight structural features were obtained. These features were linearly scaled to the range of [−1, 1] to make them comparable between different RNAs. Similar to RNAsite_seq, a sliding widow was used to incorporate the effects of neighboring nucleotides, resulting in a total of (2 × w + 1) × 8 features, which were fed into RF for training and test.
2.5 Statistical analysis of structural features
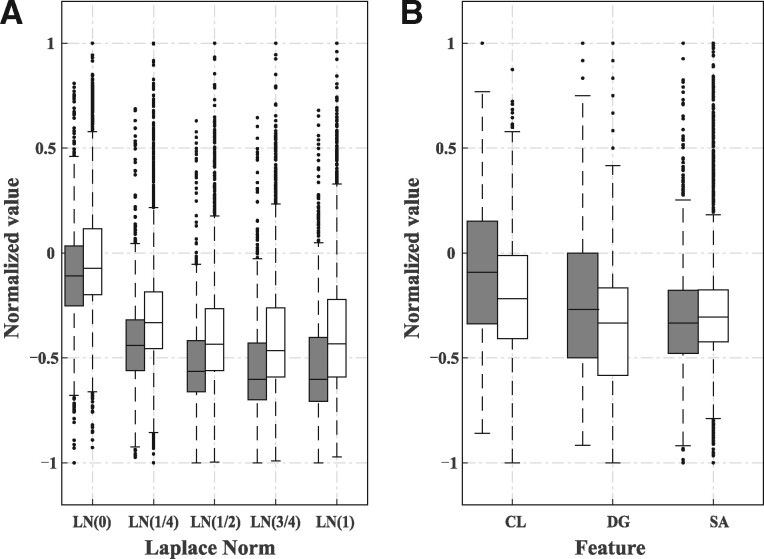
We performed a systematic comparison of the positive (the native small molecule–RNA binding sites) and negative samples of all structural features derived from structures on the dataset RB78. Figure 2A shows that the LNs of positive samples are consistently lower than those of negative samples from local to global scales, indicating that small molecule-binding nucleotides prefer relatively concave locations in RNA structures. In Figure 2B, the positive samples possess clearly higher CL values than the negative samples. A similar tendency holds for DG. This result indicates that small molecule-binding nucleotides mostly locate in the center of the distance-based nucleotides interaction networks and they physically interact with more nucleotides. The ASAs of the positive nucleotides are lower than the negative sites, probably because lower ASA values correspond to concave positions. Overall, the proposed features show discriminative characteristics between positive and negative samples.
Fig. 2.
Comparison of the distributions of the proposed structural features for small molecule-binding (gray bars) and nonbinding (white bars) nucleotides. (A) LN, (B) other features: CL, DG and SA. Each box plot consists of three parts: center of the data sample (bar), margin values (the ordinate values of the two short horizontal lines) and outliers (discrete points outside the margin values). Each bar contains 50% of the data in the middle of the sample and it consists three key values: median (short line in the middle of the bar), the upper quartile (short line at the top of the bar) and the lower quartile (short line at the bottom of the bar) of the sample data
2.6 Model construction and performance evaluation
In this work, we conducted five-fold cross validation (CV) on the training set and independent tests to evaluate our method. Due to random effects in RF, the CV was repeated 100 times and the average was reported. The Precision, Recall, Mathews correlation coefficient (MCC) and the area under the receiver operating characteristic curve (AUC) were used to assess the performance.
3 Results and discussions
3.1 Parameters optimization
All parameters including the sliding window size w and the number of trees n in RF were tuned to maximize the MCC on the training set TR60 based on five-fold CV. For the training set TR60, the samples were randomly divided into five subgroups. Four of them were used for training and the remaining one was used for test. The five-fold CV was repeated 100 times and the average was reported. To speed up the optimization, the number of trees was first fixed to 100 when optimizing the window size w. After the optimal window size was determined, a coarse-grid search was performed to optimize n.
The influence of n in RF to the performance of RNAsite_str is shown in Supplementary Figure S2. From the figure, we can see that the MCC is the lowest (0.2), when n is set to the default value 10. When n increases to 100, the MCC improves to 0.276 and becomes stable thereafter. Thus, the optimal value for n in RF was finally set to 100.
Supplementary Figure S3 shows the influence of the window size to the performance of RNAsite_str. The lowest MCC of 0.225 is obtained when the window size is zero, i.e. no neighboring nucleotides are used. When the window size is enlarged to 14, the MCC reaches the highest value (0.276). Hence, the optimal window size of RNAsite_str is set to 14. Similarly, the optimal window size of RNAsite_seq is set to 12 (Supplementary Figure S4). When the structure information is available, the final features used in RNAsite include the structural features and the prediction from RNAsite_seq. Therefore, the optimal window size of RNAsite is reoptimized. Supplementary Figure S5 shows the best window size for RNAsite is 16. For the sake of generality, these values are also used for the two independent test datasets without further optimization.
3.2 Analysis of feature contribution
The contribution of structural features to the method RNAsite_str was investigated based on five-fold CV on the training set TR60. The predictive quality measured by the average MCC is summarized in Figure 3. It shows that the novel structural descriptors rank at the top of all individual features. LN feature achieves the best performance, yielding MCC of 0.265. To avoid overtraining and remove redundant descriptors, we adopted the greedy algorithm, i.e. sequential forward selection (SFS), to produce the optimal feature groups. The SFS started with the feature, LN, showing the highest discriminatory capability between the positive and negative samples, as revealed by MCC. We iteratively selected a new feature which has the best performance from the remaining ones. This feature is retained if its combination with the kept ones results in higher MCC. This process is halted when the MCC value does not increase. Finally, the highest MCC (0.276) is achieved when all the features except SS are used together. As shown in Figure 3, the SS feature is reasonable with 0.11 MCC. However, the MCC value (0.276) does not increase when combining it with other features. This may be caused by the redundancy of the structural information between SS and other features. Statistical tests were performed to judge if the MCC improvements by the combined feature groups are significant or not, similar to the procedure used in Meng et al. (2018) and Su et al. (2019). The P-values for the tests are shown in Table S7. It indicates that the improvements by combining more feature groups except SS are significant at P-value <0.05. These data support the conclusion that these feature groups are in general complementary to each other. Therefore, the RNAsite_str was modeled with all structural features except SS.
3.3 Comparison with existing methods
RBind and Rsite are two structure-based methods for small molecule-binding nucleotides prediction (Wang et al., 2018; Zeng et al., 2015). Rsite2 is an SS-based computational prediction to identify the potential functional sites in RNA molecules (Zeng and Cui, 2016). To demonstrate the effectiveness and robustness of the proposed method, we assessed the performance of our method with existing publicly available methods on two independent datasets, TE18 and RB19. We used the standalone programs RBind (http://zhaolab.com.cn/RBind), Rsite and Rsite2 (http://www.cuilab.cn/rsite) to collect the corresponding predictions of two test sets. RNAsite obtained a very high accuracy when directly applying the model trained on TR60 to make prediction on RB19. This is likely because 12 RNAs in the training set TR60 share >80% sequence identity with the RNAs in RB19. To deal with this issue, the leave-one-out test was applied on RB19 to evaluate the accuracy.
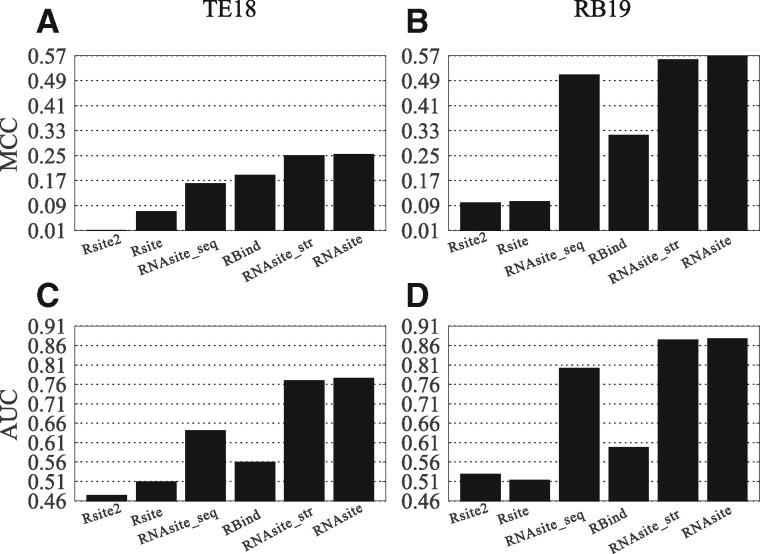
When experimental structures are used, Figure 4 presents the MCC and AUC, the two most objective metrics, on both datasets. The results for other metrics are listed in Tables S4 and S5. The figure shows that the sequence-based predictor RNAsite_seq has slightly lower MCC values than structure-based predictor RNAsite_str on both datasets, suggesting the importance of structure-based descriptors. The combination of both methods in RNAsite yields improved MCC to 0.253 and 0.567 on TE18 and RB19, respectively, both higher than other methods. Similar observations can be obtained based on the AUC data in Figure 4. We note that the AUC values for RBind are even lower than our sequence-based predictor RNAsite_seq on both datasets. This is maybe because the prediction by RBind is in binary rather than probability format, which is necessary for AUC calculation. In addition, Tables S4 and S5 show RNAsite has comparable precision with RBind but much higher recall on both datasets.
Fig. 4.
The MCC and AUC of all compared methods with the input of experimental structure. (A) and (C) the respective MCC and AUC for the dataset TE18, while (B) and (D) are for the dataset RB19
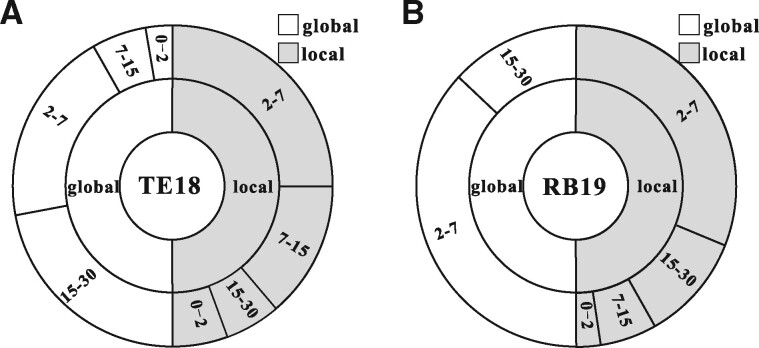
As many RNAs do not have experimentally solved structures, we tested our methods on predicted structure models. The RNAComposer server (Biesiada et al., 2016) was used to predict the structure for each RNA in the test datasets TE18 and RB19. First, we calculated the accuracy of the predicted structures, measured by the full-chain root-mean-square deviation (RMSD) (called global RMSD) and RMSD for the small molecule-binding residues (called local RMSD). Figure 5 and Supplementary Figure S6 show that the predicted structure models have reasonable accuracy. For the dataset TE18, the average global and local RMSDs are 11.732 and 7.469 Å, respectively. There are 11 models with local RMSD <7 Å. The predicted models for the RB19 dataset have higher accuracy with global average global and local RMSDs are 9.042 and 7.014 Å, respectively. As shown in Table 1, the accuracies for all methods decrease when the modeled structures are used. In spite of this, our methods outperform other methods when the same set of predicted models are used. Statistical tests indicate that the improvement of all our methods over existing methods is significant at the level of 0.05, with detailed data in Tables S8 and S9.
Fig. 5.
RMSD distributions of the predicted structure models on TE18 (A) and RB19 (B). Each pie chart is divided into two equal parts, to present the global and local RMSD distributions, respectively. The RMSD values are divided into four intervals: 0–2, 2–7, 7–15 and 15–30 Å
Table 1.
MCC and AUC of our methods and other existing methods with input of predicted structures
| Methods | TE18 |
RB19 |
||
|---|---|---|---|---|
| MCC | AUC | MCC | AUC | |
| Rsite2 | 0.01 | 0.474 | 0.099 | 0.529 |
| Rsite | 0.055 | 0.496 | 0.051 | 0.496 |
| RBind | 0.141 | 0.540 | 0.187 | 0.558 |
| RNAsite_seq | 0.16 | 0.641 | 0.508 | 0.801 |
| RNAsite_str | 0.185 | 0.695 | 0.445 | 0.806 |
| RNAsite | 0.186 | 0.703 | 0.526 | 0.834 |
In addition, we compared our method with two other methods, which are not designed for small molecule-binding site prediction but are related with RNA-small molecule binding. The first one is Weinreb’s method (denoted by DCA) (Weinreb et al., 2016), which predicts the intern-ucleotide interactions based on direct coupling analysis (DCA). This comparison is based on the fact that nucleotides contributing to an interface with other molecules would be under selective pressure. We installed and ran the method DCA locally with MSA input for each RNA in our test set TE18. The top predicted nucleotide pairs are considered as binding sites. However, it only returned predictions for four RNAs. On these four RNAs, DCA has a higher recall at the expense of lower precision, resulting to very low MCC and AUC (Table S10).
The second one is InfoRNA (Disney et al., 2016), which is for RNA motif mining by comparing the target RNA sequence against a database of known RNA motif small molecule-binding partners. The nucleotides in the returned RNA motif are regarded as the small molecule-binding sites. InfoRNA only returned results for seven RNAs. On these RNAs, InfoRNA does not perform well compared with our method (Table S11). This is probably because the main purpose of InfoRNA is for the design of small molecule targeting RNA rather than binding site prediction.
3.4 Why is the accuracy on RB19 higher than that on TE18?
We note that all metrics on the RB19 dataset are significantly higher than the TE18 dataset for all methods. To explain this data, we visually checked the small molecule–RNA complexes (the experimental structures) on the two test sets. In addition, the prediction by RBind rather than RNAsite was used here to make this analysis unbiased. We find that the location of small molecules in an RNA structure has a significant impact on the predictive accuracy for predicted binding sites, illustrated by two examples in Supplementary Figure S7. As shown in Supplementary Figure S7A (PDB ID: 1Q8N), when the small molecule is embedded in the RNA structure, the binding nucleotides in this RNA are easier to be identified, as revealed by the relatively high Precision and Recall values (1 and 0.43, respectively). On the contrary, when the small molecule is on the surface of the RNA structure, binding nucleotides are more difficult to be recognized. For example, the Precision and Recall are all 0 for the example shown in Supplementary Figure S7B (PDB ID: 5BJO). Based on this observation, we divided the RNAs into two groups. A target is called a hard target when the small molecule appears on the surface of the RNA structure; and an easy target is defined when the small molecule locates in the concave region of the RNA structure. The small molecule–RNA complexes in TE18 were divided into 10 easy and 8 hard targets. For the easy targets, the MCC, Precision, Recall and AUC of RBind are 0.296, 0.867, 0.227 and 0.596, respectively, which are close to the ones on the dataset RB19. This result indicates that most of the RNAs on RB19 dataset are easy targets. In addition, the lower RMSD of the structure models for RB19 is consistent with this conclusion as well.
3.5 Performance on metal ions and non-metal ion small molecules
The information about the small molecules is summarized in Tables S1 and S2. Because the metal ions may have different properties with other non-metal ion small molecules, we divided the small molecules into two types: metal ions and non-metal ions. Based on such division, the RNA structures in the test set TE18 are divided into three subsets: metal ions only (4 structures), no metal ions (12 structures) and mixed (2 structures). RNAsite_str’s performance on these subsets is presented in Table S12. It suggests that the metal ions-only structures have lower accuracy than other structures. This is probably because many metal ions are often on the surface of the RNA structure and do not form well-shaped binding pockets, making it more difficult to predict. It may be also because the placement for metal ions in the RNA structure was made through computational predictions and thus could contain noises. We tried to calculate the average B-factor for the metal ions in the RNA structure but this B-factor did not seem to correlate the prediction accuracy of binding site prediction. Another possible reason is because the training structures are dominated by non-metal ion small molecules. In future, it should be worth of developing ion-specific methods, such as MetalionRNA for metal ions-binding site prediction (Philips et al., 2012).
3.6 Potential improvement by combining with binding pocket detection
When structure information is available, it is evident to detect the binding pocket, which usually maps to the concave region in the structure. There are many binding pocket detection programs for protein structure but very few for RNA structure. To the best of our knowledge, there is only one publically available package for RNA pocket detection, i.e. Fpocket (Le Guilloux et al., 2009). Fpocket detects pockets based on Voronoi tessellation and alpha spheres and it works for both protein and RNA structures.
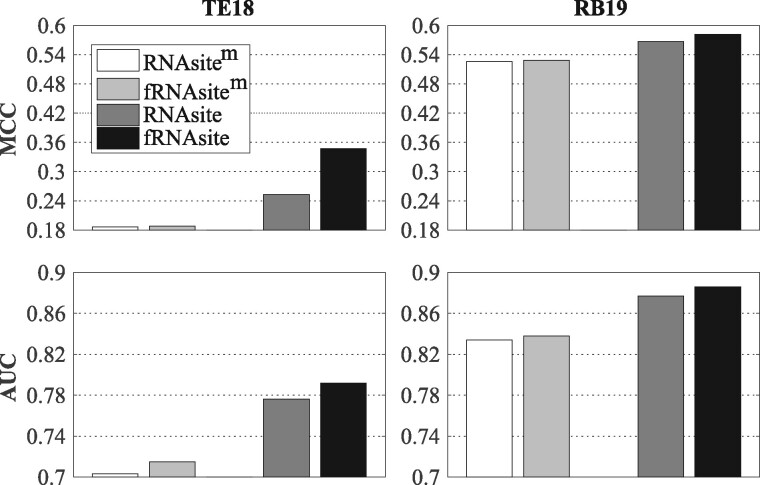
We combine the pocket detection into RNAsite to see if further improvement could be achieved. First, Fpocket returns a few binding pockets for each RNA structure, which are sorted based on the number of nucleotides. Second, the nucleotides of the target sequence are encoded by 1 if they are within the first pocket; and 0 otherwise. After combing this feature with previous ones of RNAsite, we retrained an RF-based predictor and tested it on the previous datasets. For the sake of convenience, this new predictor is named as fRNAsite. As shown in Figure 6, when the native structure is used, MCC and AUC on both test sets are significantly improved in fRNAsite. This is expected as the native structures are in holo form with bound small molecule and geometry-based pocket detection can easily recognize the small molecule-binding regions from the holo structures. However, when the input structures are computational models, the pocket detection seems to be useless to improve the accuracy. This is probably because the modeled structures are in apo form without bound small molecule. In addition, some of the modeled structures have low resolution. In reality, this is the case for most RNAs. Thus, we did not combine pocket detection in RNAsite.
Fig. 6.
Comparison between fRNAsite and RNAsite. RNAsitem and fRANsitem represent the respective RNAsite and fRANsite with input of predicted structure model
3.7 The impact of the dynamic feature of RNA structure
As RNA molecular is usually more flexible, which was not considered in the design of our method. We tested if the consideration of the dynamic feature of RNA structure could improve our method or not here. The GROMACS package (Abraham et al., 2015) was applied to perform molecular dynamics simulations to generate five alternative structural configurations for each RNA structure in the dataset RB78. The two network-based features, CL and DG, were extracted from each conformation. Hence, 10 additional structural features were added for each target. We retrained the RF models on TR60 and tested on the TE18. With these new features, we got slightly higher Recall on TE18, but lower values for other metrics (Table S6). Other new features may be designed to make full use of the dynamic nature of RNA structure in future.
3.8 The impact of holo and apo structures
When RNA bind to small molecules, there may be some conformational changes. By comparing the RNAs in the test dataset TE18 with other RNAs in PDB, we detected four RNAs that have both holo and apo structures. We evaluated the performance of RNAsite_str on these four paired structures.
Table 2 shows that two RNAs (on the 2nd and the 3rd rows) have larger conformational changes, as reflected by the high RMSD values (5.05 and 1.71 Å, respectively). For these two RNAs, the MCC values decrease significantly. For the remaining two RNAs (on the 4th and the 5th rows), the conformational changes are relatively low (RMSD <1 Å). For the third RNA, its MCC values are very low for both the holo (379 dB) and the apo (1mmeB) structures. For the last RNA, the MCC for the holo structure (2misA) is 1, which decreases for the apo structure but is still reasonable (0.752). To summarize, the accuracy for binding residues prediction for holo structures is usually higher than the apo structures. This is probably because our prediction model was trained on holo structures only. Extended training on both holo and apo structures should be helpful for improving the binding residues prediction accuracy on apo structures, which will be investigated in future work.
Table 2.
MCC of RNAsite_str for four RNAs from the test set TE18 that have apo and holo structures
| PDB ID | RMSD (Å) | Conformation |
|
|---|---|---|---|
| holo | apo | ||
| 430dA (1sclA) | 5.05 | 1 | 0.033 |
| 2jukA (1pjyA) | 1.71 | 0.436 | 0.27 |
| 379dB (1mmeB) | 0.91 | 0.031 | 0.011 |
| 2misA (2l5zA) | 0.64 | 1 | 0.752 |
Note: The PDB IDs outside/inside the brackets are for the holo/apo structures.
4 Conclusions
We presented a new method RNAsite for small molecule–RNA binding sites prediction by combining sequence and structure information. The sequence-based features are obtained based on sequence profile, reflecting position-specific evolutionary conservation of nucleotides. The structure-based features include LN, nucleotides interaction network-based topological features and SA. RNAsite was shown to be competitive with the state-of-the-art methods on two independent test sets. When predicted structure models were used, RNAsite outperforms other methods by a large margin, probably due to its combination of sequence- and structure-based descriptors. We explored the possibility of improving RNAsite by geometry-based binding pocket detection. It suggests that RNAsite can be enhanced by the inclusion of pocket detection for experimental structures. However, for predicted structure models, pocket detection does not contribute to RNAsite. In addition, we also considered and discussed the influence of conformational flexibility and conformational changes caused by ligand binding on RNAsite.
Funding
This work has been supported by the National Natural Science Foundation of China (NSFC 11871290 and 61873185), Fok Ying-Tong Education Foundation (161003) and KLMDASR.
Conflict of Interest: none declared.
Supplementary Material
Contributor Information
Hong Su, School of Mathematical Sciences, Nankai University, Tianjin 300071, China.
Zhenling Peng, Center for Applied Mathematics, Tianjin University, Tianjin 300072, China.
Jianyi Yang, School of Mathematical Sciences, Nankai University, Tianjin 300071, China.
References
- Abraham M.J. et al. (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1–2, 19–25. [Google Scholar]
- Alipanahi B. et al. (2015) Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol., 33, 831–838. [DOI] [PubMed] [Google Scholar]
- Altschul S.F. et al. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Biesiada M. et al. (2016) Automated RNA 3D structure prediction with RNAComposer. Methods Mol. Biol., 1490, 199–215. [DOI] [PubMed] [Google Scholar]
- Bonnel N., Marteau P.F. (2012) LNA: fast protein structural comparison using a Laplacian characterization of tertiary structure. IEEE/ACM Trans. Comput. Biol. Bioinform., 9, 1451–1458. [DOI] [PubMed] [Google Scholar]
- Burley S.K. et al. (2019) RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res., 47, D464–D474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallo L. et al. (2003) POPS: a fast algorithm for solvent accessible surface areas at atomic and residue level. Nucleic Acids Res., 31, 3364–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs-Disney J.L. et al. (2018) A massively parallel selection of small molecule-RNA motif binding partners informs design of an antiviral from sequence. Chem, 4, 2384–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disney M.D. (2019) Targeting RNA with small molecules to capture opportunities at the intersection of chemistry, biology, and medicine. J. Am. Chem. Soc., 141, 6776–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disney M.D. et al. (2016) Inforna 2.0: a platform for the sequence-based design of small molecules targeting structured RNAs. ACS Chem. Biol., 11, 1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S. et al. (2019) RNA-align: quick and accurate alignment of RNA 3D structures based on size-independent TM-scoreRNA. Bioinformatics, 35, 4459–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J.G. (1994) Position-based sequence weights. J. Mol. Biol., 243, 574–578. [DOI] [PubMed] [Google Scholar]
- Le Guilloux V. et al. (2009) Fpocket: an open source platform for ligand pocket detection. BMC Bioinform., 10, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. et al. (2014) Quantifying sequence and structural features of protein-RNA interactions. Nucleic Acids Res., 42, 10086–10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics, 22, 1658–1659. [DOI] [PubMed] [Google Scholar]
- Liu H.F., Liu R. (2020) Structure-based prediction of post-translational modification cross-talk within proteins using complementary residue- and residue pair-based features. Brief. Bioinform., 21, 609–620. [DOI] [PubMed] [Google Scholar]
- Lu X.J. et al. (2015) DSSR: an integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Res., 43, e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q. et al. (2018) CoABind: a novel algorithm for coenzyme A (CoA)- and CoA derivatives-binding residues prediction. Bioinformatics, 34, 2598–2604. [DOI] [PubMed] [Google Scholar]
- Philips A. et al. (2012) MetalionRNA: computational predictor of metal-binding sites in RNA structures. Bioinformatics, 28, 198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips A. et al. (2013) LigandRNA: computational predictor of RNA-ligand interactions. RNA, 19, 1605–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H. et al. (2019) Improving the prediction of protein-nucleic acids binding residues via multiple sequence profiles and the consensus of complementary methods. Bioinformatics, 35, 930–936. [DOI] [PubMed] [Google Scholar]
- Sun J. et al. (2016) CRHunter: integrating multifaceted information to predict catalytic residues in enzymes. Sci. Rep., 6, 34044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursu A. et al. (2019) Methods to identify and optimize small molecules interacting with RNA (SMIRNAs). Drug Disc. Today, 24, 2002–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. et al. (2018) RBind: computational network method to predict RNA binding sites. Bioinformatics, 34, 3131–3136. [DOI] [PubMed] [Google Scholar]
- Weinreb C. et al. (2016) 3D RNA and functional interactions from evolutionary couplings. Cell, 165, 963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. et al. (2012) BioLiP: a semi-manually curated database for biologically relevant ligand-protein interactions. Nucleic Acids Res., 41, D1096–D1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng P., Cui Q. (2016) Rsite2: an efficient computational method to predict the functional sites of noncoding RNAs. Sci. Rep., 6, 19016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng P. et al. (2015) Rsite: a computational method to identify the functional sites of noncoding RNAs. Sci. Rep., 5, 9179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.