Abstract
Summary
The PICKLE 3.0 upgrade refers to the enrichment of this human protein–protein interaction (PPI) meta-database with the mouse protein interactome. Experimental PPI data between mouse genetic entities are rather limited; however, they are substantially complemented by PPIs between mouse and human genetic entities. The relational scheme of PICKLE 3.0 has been amended to exploit the Mouse Genome Informatics mouse–human ortholog gene pair collection, enabling (i) the extension through orthology of the mouse interactome with potentially valid PPIs between mouse entities based on the experimental PPIs between mouse and human entities and (ii) the comparison between mouse and human PPI networks. Interestingly, 43.5% of the experimental mouse PPIs lacks a corresponding by orthology PPI in human, an inconsistency in need of further investigation. Overall, as primary mouse PPI datasets show a considerably limited overlap, PICKLE 3.0 provides a unique comprehensive representation of the mouse protein interactome.
Availability and implementation
PICKLE can be queried and downloaded at http://www.pickle.gr.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Protein InteraCtion KnowLegdE (PICKLE) meta-database has been developed to consistently integrate primary human protein–protein interaction (PPI) datasets (Gioutlakis et al., 2017; Klapa et al., 2013). Unique advantageous characteristics of PICKLE are (i) the use of the UniProtKB/Swiss-Prot (www.uniprot.org) reviewed human complete proteome (RHCP) as the reference protein set over which PPI datasets are integrated and (ii) the PPI dataset integration via the RHCP-based genetic information ontology network. Thus, primary PPIs along with the accompanying experimental evidence information are stored at the genetic information level of each source PPI database without a priori normalization to a particular preference level. Hence, the current, PICKLE 2.0, meta-database enables (i) the consistent reconstruction of the human PPI network at both the protein (UniProt) and gene levels and (ii) the experimental PPI evidence cross-checking between the primary datasets.
The objective of the PICKLE 3.0 upgrade is to enrich PICKLE with the mouse PPI network. Mus musculus, the laboratory mouse, is an important animal model for human normal and aberrant physiology, and for comparative functional genomic studies. However, a particularity of the mouse experimental PPI network over the human is that, in addition to interactions between mouse entities [], it comprises a large number of interactions between mouse and human genetic entities []. To accommodate for this feature, the PICKLE relational scheme was amended to include the subnetwork and mouse–human ortholog pair links, establishing correspondence through orthology between interactions in the and subnetworks and/or the human PICKLE network. Thus, the mouse experimental PPI network is extendable by orthology and comparisons between the mouse and human PPI networks can be made; these two features make PICKLE unique over existing mouse PPI meta-databases. The web interface of PICKLE 3.0 has been accordingly modified.
2 Implementation
PICKLE 3.0 builds on the structure of human PICKLE 2.0 (Gioutlakis et al., 2017), using the reviewed mouse complete proteome (RMCP) defined by UniProtKB/Swiss-Prot as the mouse reference protein set (Supplementary File S1A), and mining PPI information from IntAct (Orchard et al., 2014), BioGRID (Oughtred et al., 2019) and DIP (Salwinski et al., 2004) source databases on a semi-annual update rate. To accommodate for the large number of retrieved , the relational scheme of PICKLE 3.0 also includes the subnetwork, connected to both mouse and human genetic information ontology networks. Mouse–human ortholog gene pairs collected from the Mouse Genome Informatics database (Bult et al., 2019; Supplementary File S1B) link the two ontological networks and assist in establishing correspondence through orthology between interactions in the and subnetworks and/or the human PPI network of PICKLE. Through this correspondence, experimentally determined interactions of the subnetwork are extended into potentially valid PPIs between mouse genetic entities (. Moreover, corresponding through orthology interactions of the and/or subnetworks are merged into one interaction between mouse genetic entities. The resulted network is called the mouse ‘extended by orthology’, abbreviated as EbO, PPI network (Fig. 1 and Supplementary File S1C).
Fig. 1.
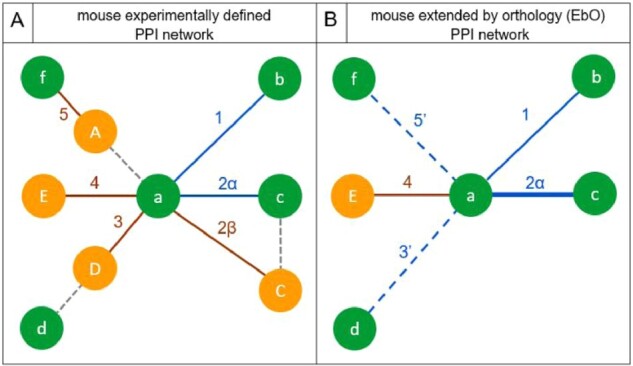
Schematic of the mouse EbO PPI network reconstruction process. The types of interactors for a mouse protein ‘a’ in the experimentally determined PPI network (A) and the EbO PPI network (B) are shown. Green and orange circles depict mouse and human genetic entities, respectively; the blue and brown straight-line edges depict experimentally determined PPI(m–m) and PPI(m–h), respectively; gray dashed edges in (A) link mouse–human ortholog pairs; thick blue straight edges in (B) indicate experimentally determined PPI(m–m), also supported by corresponding via orthology PPI(m–h); dashed blue edges depict potentially valid .
The PICKLE 3.0 web interface has been accordingly updated and the network visualization window in Cytoscape.js has been substantially enhanced over the previous version (Supplementary File S1D). In mouse PICKLE, the user can also visualize the EbO PPI network of the queried mouse entities and be informed whether mouse PPIs have corresponding through orthology PPIs in the human PICKLE.
3 Results
In PICKLE 3.0 (release 1) (Table 1 and Supplementary File S2A), with 17 021 UniProt IDs in the RMCP, the default mouse PPI network at protein (UniProt) level comprises 11 026 direct interactions for 5087 mouse UniProt IDs in the subnetwork, and 5396 direct interactions between 2209 mouse and 2255 human UniProt IDs in the subnetwork. A set of 1709 mouse UniProt IDs have interactions in both subnetworks and 500 have interactions only with human proteins. These data indicate that, currently, only ∼33% of the RMCP has known experimental direct PPIs supported by 6616 references, compared to 81% of the RHCP with 191 113 direct PPIs supported by 42 121 references in the human PICKLE 3.0 (release 1) dataset. Notably, 82% of the 6616 supporting references are provided uniquely by one of the three source databases [IntAct: 2265 (36.7%), BioGRID: 3128 (50.7%), DIP: 119 (1.9%)]. Hence, the overlap between the primary PPI datasets and particularly between the two major ones (IntAct and BioGRID) is substantially limited (Supplementary File S2B). Remarkably, mouse PICKLE comprises more than double interactions than IntAct and 61% more than BioGRID in the PPI(m–m) subnetwork, and 78% more interactions than IntAct and more than double than BioGRID in the PPI(m–h) subnetwork.
Table 1.
Mouse experimental and EbO PPI network at protein (UniProt) level
| UniProt IDs |
PPIs | Refs | ||
|---|---|---|---|---|
| Mouse | Human | |||
| PPI(m–m) subnetwork | ||||
| Experimental | 5087 | — | 11 026 | 4946 |
| EbO | 6008 | — | 14 734 | 6532 |
| PPI(m–h) subnetworka | ||||
| Experimental | 2209 (1709) | 2255 | 5396 | 2552 (882) |
| EbO | 373 (318) | 177 | 490 | 232 (148) |
The number of common mouse UniProt IDs and references with the respective set in the subnetwork is shown in parenthesis.
Through the extension by orthology, the mouse EbO PPI network has 3708 potentially valid in addition to the 11 026 experimental , while1166 are represented by their corresponding experimental (Table 1 and Supplementary File S2C). Both the experimentally determined and EbO networks follow the scale-free structure, with EbO displaying better power-law fit and increased connectivity of the nodes in the largest component (Supplementary File S2C). The maximum degree for a protein is 212 in the EbO compared to 174 for the experimentally determined. Indeed, the extension through orthology of the experimental PPI(m–m) network expands certain neighborhoods by combining the information from both the PPI(m–m) and the PPI(m–h) (see examples in Supplementary File S2C). Notably, comparison through orthology indicated 4614 and 1840 having no corresponding PPI in the human PICKLE (Supplementary File S2D). These represent 43.5% of the 14 843 mouse PPIs with both interactors having human orthologs. PICKLE 3.0 provides this unique information to the user, which can motivate the design of targeted experiments to further investigate these differences.
Funding
This work was supported mainly by ELIXIR-GR (MIS 5002780); partly by EATRIS-GR (MIS 5028091); and INSPIRED (MIS 5002550) projects in the Action ‘Reinforcement of the Research and Innovation Infrastructure’; and partly by ΒΙΤΑΔ-ΔΕ (MIS 5002469), project in the ‘Action for the Strategic Development on the Research and Technological Sector’; of the Greek NSRF 2014-2020 Operational Program ‘Competitiveness, Entrepreneurship and Innovation’, co-financed by Greece and EU (European Regional Development Fund).
Conflict of Interest: none declared.
Data availability
PICKLE data are available for download at http://www.pickle.gr.
Supplementary Material
Contributor Information
Georgios N Dimitrakopoulos, Laboratory of General Biology, School of Medicine, University of Patras, Patras, Greece; Metabolic Engineering and Systems Biology Laboratory, Institute of Chemical Engineering Sciences, Foundation for Research and Technology Hellas (FORTH/ICE-HT), Patras, Greece.
Maria I Klapa, Metabolic Engineering and Systems Biology Laboratory, Institute of Chemical Engineering Sciences, Foundation for Research and Technology Hellas (FORTH/ICE-HT), Patras, Greece.
Nicholas K Moschonas, Laboratory of General Biology, School of Medicine, University of Patras, Patras, Greece; Metabolic Engineering and Systems Biology Laboratory, Institute of Chemical Engineering Sciences, Foundation for Research and Technology Hellas (FORTH/ICE-HT), Patras, Greece.
References
- Bult C.J. et al. ; the Mouse Genome Database Group. (2019) Mouse genome database (MGD) 2019. Nucleic Acids Res., 47, D801–D806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioutlakis A. et al. (2017) PICKLE 2.0: a human protein-protein interaction meta-database employing data integration via genetic information ontology. PLoS One, 12, e0186039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapa M.I. et al. (2013) Reconstruction of the experimentally supported human protein interactome: what can we learn? BMC Syst. Biol., 7, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard S. et al. (2014) The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res., 42, D358–D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oughtred R. et al. (2019) The BioGRID interaction database: 2019 update. Nucleic Acids Res., 47, D529–D541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salwinski L. et al. (2004) The database of interacting proteins: 2004 update. Nucleic Acids Res., 32, D449–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PICKLE data are available for download at http://www.pickle.gr.


